Abstract
A reevaluation of the transcription profile of Aleutian mink disease parvovirus (AMDV)-infected CRFK cells at either 32°C or 37°C has determined that strain AMDV-G encodes six species of mRNAs produced by alternative splicing and alternative polyadenylation of a pre-mRNA generated by a single promoter at the left end of the genome. Three different splicing patterns are used, and each type is found polyadenylated at either the 3′ end of the genome (the distal site) or at a site in the center of the genome (the proximal site). All spliced species accumulate similarly over the course of infection, with the R2 RNA predominant throughout. The R2 RNA, which contains and can express the NS2 coding region, encodes the viral capsid proteins VP1 and VP2.
Aleutian mink disease parvovirus (AMDV) is an autonomous parvovirus (13) which causes a number of important clinical and pathological syndromes in mink, including abortion (1, 25), acute pneumonia in neonatal mink (1, 2, 5, 7), and chronic immune complex-mediated glomerulonephritis and arteritis in adult mink (1, 4, 18, 27, 29, 30). Strains of AMDV differ reproducibly in their pathogenicity in animals (18). In adult mink, the virulent AMDV Utah 1 strain, which replicates abortively in cell culture (13, 19), produces a persistent infection associated with severe dysfunction of the immune system (6, 12, 18). Conversely, the AMDV-G strain, a variant of the Utah 1 strain which was adapted to grow permissively in CRFK cells at 32°C, is nonpathogenic in adult mink (13). The genome of AMDV-G is approximately 97.5% identical to that of AMDV Utah 1 (9).
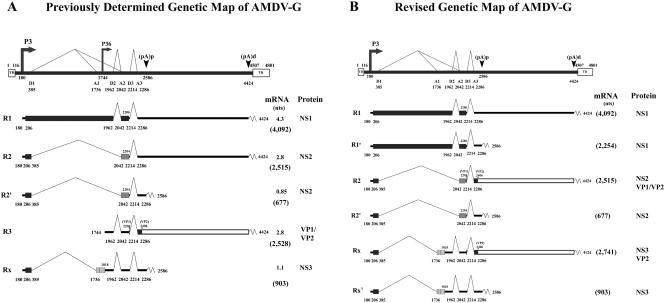
A detailed map of transcripts generated by AMDV-G infection of CRFK cells was generated a number of years ago (3). That analysis revealed that ADMV generates a more complex transcription profile than other parvoviruses of nonprimate mammals (34). Two promoters, one at map unit 3 and another at map unit 36, were identified, as were the use of a complex splicing pattern and internal polyadenylation site (see Fig. 3A). The left open reading frame (ORF) of AMDV has been shown to encode the viral nonstructural proteins, and the right ORF encodes the virion proteins (13, 14, 16).
FIG. 3.
Previously determined and revised transcription maps of AMDV-G. The genome of AMDV-G is shown to scale, with the major transcription units, including the inverted hairpin termini (TR), promoters, splice donors, and acceptors, as well as (pA)p and (pA)d. The AMDV proteins that are predicted to be encoded from each RNA are indicated. The sizes of all RNAs as determined by mobility in the Northern assays are shown (in kilobases), and the predicted sizes of the RNAs, minus polyadenylated tails, are given in parentheses (in nt). Within the nonstructural region, the black bar indicates NS1-encoding ORF1, the striped box indicates NS3-encoding ORF2, and the gray box indicates NS2-encoding ORF3. The large capsid protein-encoding ORF shared by VP1 and VP2 in the right-hand end of the genome is read in ORF3, indicated by an open box.
Unlike results for other parvoviruses with an internal capsid gene promoter, previous studies found that the AMDV P36 promoter was extremely weak compared to the corresponding P38 promoters of minute virus of mice and canine parvovirus, following transfection of CRFK cells, even when supplied with the potentially trans-activating AMDV NS1 protein (15, 36). It was suggested that the weakness of this putative promoter contributed to the persistence this virus displays during its normal life cycle (15, 36, 37); however, the abundance of the reported P36 transcripts also did not correlate with levels of VP1 and VP2 found during infection of CRFK cells in culture (37).
In this study, we have reevaluated the transcription profile of AMDV. In contrast to previous studies (3), we show that during productive infection of CRFK cells at both 37°C and 32°C only RNAs generated by the viral P3 promoter are detected by RNase protection assays. P3-generated pre-mRNAs are processed to generate six RNA products. These RNAs are alternatively spliced in three patterns, and each alternatively spliced type is found polyadenylated either at the 3′ end of the genome, at the distal polyadenylation site [(pA)d], or at a site in the center of the genome, at the proximal polyadenylation site [(pA)p]. Additionally, we demonstrate that the R2 RNA, the predominant RNA generated throughout infection, encodes the viral capsid proteins VP1 and VP2. Thus, the transcription profile of AMDV most closely resembles that of the erythroviruses B19 and simian parvovirus (SPV).
MATERIALS AND METHODS
Cells and virus.
The Crandall feline kidney cell line, CRFK (ATCC CCL-94), was obtained from Marshall Bloom (Rocky Mountain Laboratory, NIAID). Cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal calf serum at 37°C in 5% CO2. A molecularly cloned stock of AMDV-G, also obtained from M. Bloom, was propagated and assayed in CRFK cells as previously reported (8, 10, 21). CRFK cells were infected with AMDV-G at 1 fluorescence-forming unit/cell (13, 28).
Plasmids. (i) Construction of AMDV expression plasmids.
The cytomegalovirus (CMV)-driven AMDV nonreplicating plasmid CMVNSCap was constructed by linking the CMV promoter to nucleotides (nts) 180 to 4560 of the AMDV-G genome (taken from the infectious clone pXXI Q-3-15 [8]) (see Fig. 1C, top) in the pBluescript SK+ (Strategene) background. All nucleotide numbers for AMDV-G used in the manuscript refer to GenBank accession no. M20036 (8). CMVNSCapmP36 was constructed by silent mutation of the TATA box and initiator region, between nts 2383 and 2399 (see Fig. 5A for diagram). CMV1744Cap, CMV1761Cap, CMV1841Cap, and CMV2044Cap were constructed by deleting AMDV nts 180 to 1743, nts 180 to 1760, nts 180 to 1840, and nts 180 to 2043, respectively, from CMVNSCap. CMVNSCapmD1 was constructed by debilitating the large intron donor site at nt 384 from AAG GTT AGC to AGC ATG CCA. CMVΔD1A1Cap and CMVΔD1A2Cap (see Fig. 5) were made by exact deletion of intronic sequences D1-A1 (nts 385 to 1736) and D1-A2 (nts 385 to 2042), respectively, in the CMVNSCap backbone. CMVNSHACapmD1 and CMVΔD1A2CapNSHA (see Fig. 5) were made by insertion of hemagglutinin (HA) tags into the N termini of the NS1 ORF in CMVNSCapmD1 and CMVΔD1A2Cap, respectively. The transfection control plasmid, pC1GFPHA, was made by insertion of an HA tag at the C terminus of the green fluorescent protein (GFP) gene in pC1GFP (Clontech).
FIG. 1.
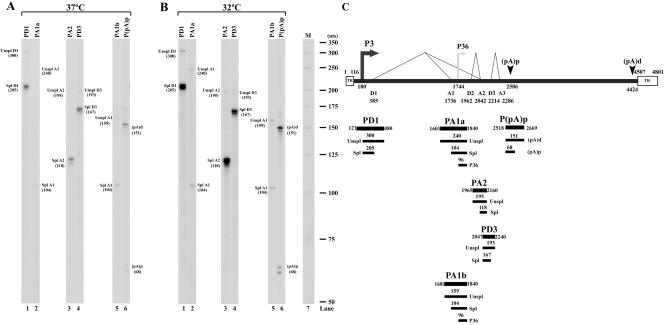
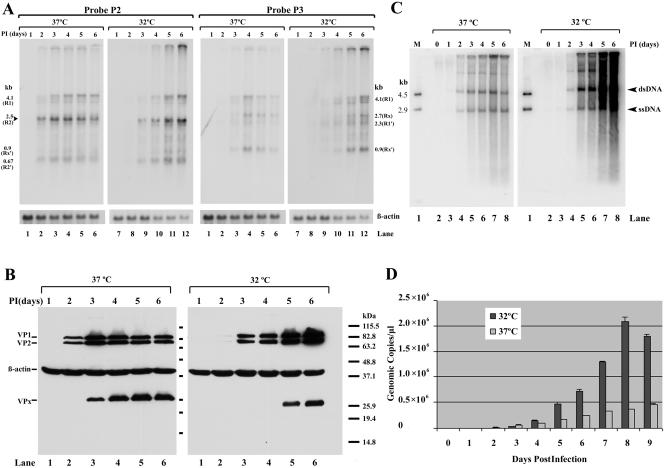
RNase protections of AMDV RNA. CRFK cells were infected with AMDV-G at 37°C (A) and 32°C (B). Total RNA isolated 6 days after infection was used for RNase protection assay. The probes are indicated at the top of each lane, and the identity of the RNA species protected by each band is indicated to either the right or the left. The probes are diagrammed in panel C, in relation to the previously published map of AMDV, with the predicted protected bands shown below each probe. The promoters are shown by arrows and the splice junctions by thin lines. Donor and acceptor sites are indicated, and the P36 promoter is depicted as a gray arrow to indicate that its presence is not confirmed by the experiments in the manuscript. A 32P-labeled RNA ladder (32) with the respective sizes indicated to the left is presented in panel B, lane 7. Unspl, unspliced; Spl, spliced; TR, inverted hairpin terminus.
FIG. 5.
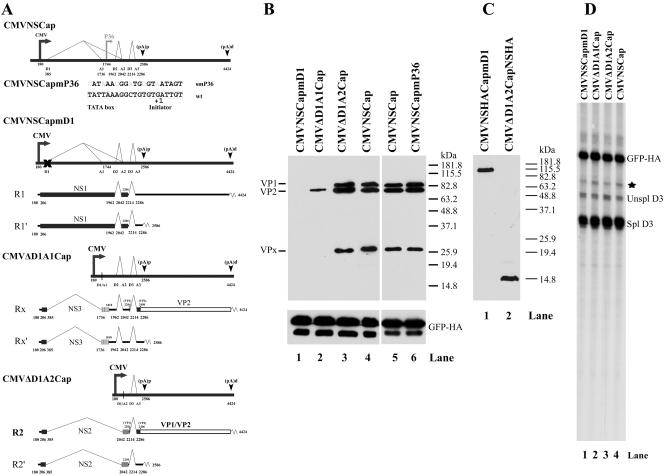
R2 RNA encodes VP1 and VP2. (A) CMV-driven AMDV-G capsid constructs transfected into CRFK cells (as described in Results). The predicted RNA products they generate, their relevant ORFs (black, frame 1; striped, frame 2; gray, frame 3), and their protein products are indicated. Within the nonstructural region, the black bar indicates NS1-encoding ORF1, the striped box indicates NS3-encoding ORF2, and the gray box indicates NS2-encoding ORF3. The large capsid protein-encoding ORF shared by VP1 and VP2 in the right-hand end of the genome is read in ORF3, indicated by an open box. (B) Two days later, total cell lysates were separated on SDS-10% PAGE gels and analyzed by Western blotting, and total RNA was analyzed by RNase protection assay using probe D3 (D). Expression of HA-tagged GFP following transfection of an expression plasmid (C1GFPHA) was used as an internal control and is shown at the bottom of panel B and as the GFP-HA band in panel D. (C) Proteins generated from HA-tagged NS1 and NS2 constructs were separated on SDS-12% PAGE gels and analyzed by Western blotting using a monoclonal antibody to the HA tag (Sigma). (D) RNase protection assay using probe PD3, which spans the D3 donor at nt 2214 and thus detects all RNAs generated by these constructs, showing that they all make equivalent amounts of RNA. The band marked by a star was likely the undigested probe. Unspl, unspliced; Spl, spliced.
(ii) Clones used to generate probes for RNase protection assay.
To map the transcription profile of AMDV-G by RNase protection assay, probes PD1, PA1a, PA1b, PA2, PD3, and P(pA)p (see Fig. 1C) were constructed by cloning the following regions of AMDV-G into BamHI-HindIII-digested pGEM4Z (Promega): nts 121 to 480 (PD1), nts 1601 to 1840 (PA1a), nts 1681 to 1840 (PA1b), nts 1965 to 2160 (PA2), nts 2047 to 2240 (PD3), and nts 2518 to 2669 [P(pA)p].
All of the DNA constructs were sequenced at the DNA core facility of the University of Missouri—Columbia to ensure that they were as predicted.
RNA isolation and RNase protection assay.
Total RNA was isolated using guanidine isothiocyanate and purified by CsCl ultracentrifugation as previously reported (35). RNase protection assays were performed with 10 μg of total RNA as previously reported (23, 35). Probes were generated from linearized templates by in vitro transcription as previous reported (35). RNA hybridizations were done in substantial probe excess, and signals were quantified with a Molecular Imager FX system and Quantity One version 4.2.2 image software (Bio-Rad, Hercules, CA) or with Fuji FLA 3000 and Fuji Multi Gauge v2.3 software (FUJIFILM Medical Systems USA., Inc.). Relative molar ratios of individual species of RNAs were determined after adjustment for the number of 32P-labeled uridines in each protected fragment as previously described (35).
Northern blot analysis.
Northern analyses were done as previously described (26), using 5 μg of total RNA. The 32P-labeled DNA probes P1, P2, P3, and P6 spanned AMDV-G genome sequences nts 180 to 4506, nts 180 to 1600, nts 1121 to 1960, and nts 1121 to 1600, respectively (see Fig. 2 for diagram). The probe used for the internal β-actin control was purchased from Ambion, Inc. (Austin, TX).
FIG. 2.
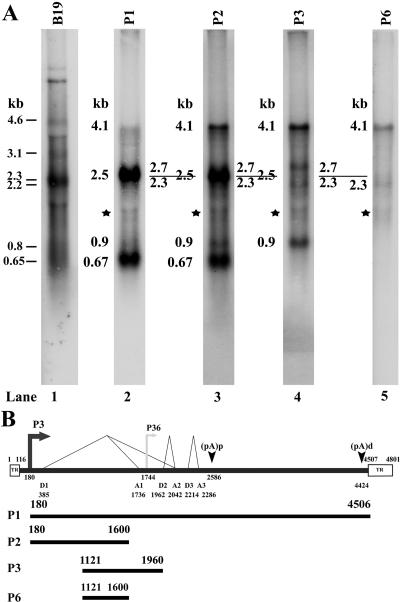
Northern analysis of RNA generated during AMDV-G infection of CRFK cells at 37°C. Total RNA isolated from AMDV-G-infected CRFK cells 6 days postinfection at 37°C was used for Northern analysis. RNA from uninfected CRFK cells was also assayed as a control and showed no hybridization (data not shown). (A) The sizes of the bands are shown to the left of each panel and are described in the text and in the legend for Fig. 3. RNA from B19-transfected COS cells, hybridized with a B19 genomic clone, was used for marker purposes (31) (lane 1). The probes used for hybridization are shown at the top of each lane. The bands indicated by the stars in lanes 2 to 5 run at the borders of the abundant unlabeled ribosomal RNAs and are likely to represent radioactivity excluded from this area of the gel. (B) The probes are diagrammed with respective nucleotide numbers and are shown in relation to the previously published map of AMDV, as described in the legend for Fig. 1. TR, inverted hairpin terminus.
Southern blot analysis.
AMDV-G DNA replicative forms produced during infection at either 32°C or 37°C were visualized by Southern analysis. Cells were collected and washed twice with phosphate-buffered saline (PBS). Then, cells were lysed in 2% sodium dodecyl sulfate and treated with proteinase K (0.5 mg/ml). Samples were run on a 1% agarose gel, and Southern blot analysis was performed as described previously (22, 38), using the probe P1 (see Fig. 2B). Blots were exposed to a Fuji phosphorimaging screen, and the DNA forms were quantified by using Fuji Multi Gauge v2.3 software where indicated.
Western blot analysis.
Western blotting was performed on cell extracts taken at various times postinfection with 1 fluorescence-forming unit/cell or, where indicated, 48 h posttransfection, using 1 μg of DNA/well of a six-well plate with Lipofectamine and Plus reagent (Gibco BRL) as previously described (33). Cells were harvested, washed with phosphate-buffered saline twice, and then lysed in 200 μl of 1× Laemmli protein loading buffer containing 1× proteinase inhibitor cocktail (Roche). A 36-μl aliquot of the total lysed protein was loaded onto a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and subjected to immunoblot analysis as previously described (20), using a monoclonal antibody to AMDV VP2 amino acids 428 to 446 (hybridoma cell line 282.20.1.4) (11). Monoclonal antibody mAbCam8224 (AbCam, Ltd.) was used to detect β-actin as an internal control, and monoclonal antibody HA-7 (Sigma, St. Louis, MO) was used to detect all HA-tagged proteins. In some experiments, the images were captured by charged-coupled-device camera (Fuji LAS-3000 imaging system) for quantification by Fuji Multi Gauge v2.3 software.
Real-time PCR quantification of AMDV-G virus production.
Approximately 1 × 106 AMDV-G-infected CRFK cells were collected, washed twice with PBS, and resuspended in 5 ml of PBS. Cell suspensions were frozen and thawed three times and centrifuged at 10,000 rpm at 4°C for 10 min. Benzonase (5 μl, 25 U/μl; Novagen) was added to 200 μl of the supernatant, and the solution was incubated at 37°C for 10 h. Viral DNA was purified by a QIAamp DNA blood mini kit (QIAGEN) and finally eluted in 200 μl of double-distilled H2O. The CMVNSCap plasmid was used as a control (1 genomic copy = 5 × 10−12 μg) to establish the standard curve for absolute quantification by using TaqMan technology with an Applied Biosystems 7500 system (Foster City, Calif.). The amplicon and the TaqMan minor groove binder probe were designed by Primer Express 2.0.0 software (Applied Biosystems). Their sequences are as follows: forward primer, 5′GGCTCAAATTGATGA3′ (nts 214 to 228); reverse primer, 5′CCTTCACCGTCGTTAATCTCCTT3′ (nts 288 to 266); and minor groove binder probe, 5′CAGAGGAGACTGCAGGAC3′ (nts 230 to 247). TaqMan universal PCR master mix (Applied Biosystems) was used for amplification with the standard protocol.
RESULTS
During productive infection of CRFK cells, all detectable AMDV-G RNAs were generated from the P3 promoter.
Reevaluation of the RNAs generated by AMDV-G during productive infection of CRFK cells by RNase protection and Northern analysis has suggested modifications to the published AMDV transcription map.
RNase protection assays.
To determine relative usage of known donors, acceptors, and the internal polyadenylation site, as well as to map the initiation sites of AMDV-generated RNAs, RNase protection assays were performed. The probes employed in this study and the predicted products of the RNase protection assays are shown in Fig. 1C, relative to the previously published AMDV map (3). Assays performed on RNA generated following infection at either 37°C (Fig. 1A) or 32°C (Fig. 1B) are presented. The RNA patterns generated at the two temperatures were indistinguishable from one another; however, more RNA was generated at 32°C, consistent with increased replication at that temperature, as discussed further below.
To determine levels of RNA spliced at the first donor (D1), probe PD1, which spans the P3 RNA initiation site and the previously identified large intron donor at nt 385, was used. PD1 protected bands of approximately 205 and 300 nts (Fig. 1A and B, lanes 1), which corresponded to spliced and unspliced RNAs, respectively, across this region. The sizes of these bands confirmed that the RNA initiation site was at approximately nt 180, and their relative abundances suggested that from this region, approximately 10-fold more spliced than unspliced RNA was generated during infection.
To determine usage of internal donors and acceptors, probes PA2 and PD3 were used. PA2, which spans the A2 acceptor region, protected bands of approximately 118 nts and 195 nts, which corresponded to RNA spliced using A2 or unspliced in this region, respectively (Fig. 1A and B, lanes 3). More than 95% of AMDV-generated RNA was spliced in this region. Probe PD3, which spans the D3 donor site and assays levels of excision of the third intron (D3-A3), protected primarily a single band of approximately 167 nts, demonstrating that more than 95% of AMDV RNA excised the third intron (Fig. 1A and B, lanes 4). Results using probes spanning the D2 donor and A3 acceptor were consistent with these conclusions (data not shown).
To determine the relative usage of the internal polyadenylation site [(pA)p], probe P(pA)p, which spans p(A)p, was used. P(pA)p protected bands of approximately 151 and 68 nts, which correspond to RNAs that either read through or are polyadenylated at the internal site, respectively (Fig. 1A and B, lanes 6). Quantification of the relative abundances of these bands demonstrated that approximately half of the RNAs generated by AMDV were polyadenylated at (pA)p. The size of the band protected by the internally polyadenylated RNA mapped the RNA cleavage site to approximately nt 2586.
Probes PA1a and PA1b were designed to map the start site of RNA generated by the previously identified P36 promoter. Surprisingly, PA1a only detected bands of approximately 240 and 104 nts, and PA1b only detected bands of approximately 159 and 104 nts in these assays, which represented unspliced RNA protecting the complete probe and RNA spliced using the A1 acceptor, respectively (Fig. 1A and B, lanes 2 and 5). RNAs generated from a P36 promoter previously mapped to a TATA box at nt 1744 (3) would protect fragments of probes PA1a and PA1b of 96 nts or less, and such bands are not detectable in our assay. These results suggested that if RNA was generated from a P36 promoter it was present only at only very low levels, likely incompatible with the levels of capsid protein-encoding RNAs expected with a productive infection. This opened the possibility that, contrary to previous suggestion (3), the abundant capsid proteins were encoded from another RNA molecule.
Northern blot analysis.
Four primary bands were detected using a full-length AMDV probe (P1): a band of approximately 4.1 kb, a broad band between approximately 2.7 and 2.3 kb, a band of approximately 0.9 kb, and a band of approximately 0.7 kb (Fig. 2A, lane 2). A similar profile was seen with previously published studies (3). To determine the origin of these bands, individual probes, diagrammed in Fig. 2B, were used for hybridization.
Probe P2, which spans nts 180 to 1600 and which should hybridize with all P3-generated RNAs but not to potential P36-generated RNAs, detected the same set of RNAs as the full-length probe, albeit at a slightly different ratio (Fig. 2A, lane 3).
P36-generated RNA, if present, would be predicted to be approximately 2.5 kb in length, essentially the same size as the R2 RNA (see Fig. 3B). Following hybridization with the genomic probe P1, these two RNAs would be expected to appear together within the broad band present in that area of the blot, and so we probed the same RNA with the P3 probe, which spans nts 1121 to 1960 (Fig. 2B). Probe P3 would hybridize to a P36-generated product but not to the R2 RNA because its 3′ terminus lies upstream of the A2 acceptor utilized by R2 (Fig. 2B). However, probe P3 did not detect a significant RNA migrating at 2.5 kb (the size of the potential R3 product) (Fig. 2A, lane 4), corroborating the RNase protection assay results described above, which failed to detect a P36-generated AMDV RNA. These results also indicate that the 2.5-kb band detected by probes P1 and P2 was the R2 RNA.
As expected, the P3 probe also hybridized to the 4.1-kb and 0.9-kb bands, which represent the previously identified R1 and RX RNAs, respectively (Fig. 2A, lane 4, and Fig. 3A). However, two additional bands not previously reported were also revealed. A band of 2.7 kb, whose size is consistent with an RNA extending to the 3′ polyadenylation site [(pA)d] from which all three introns have been removed, was detected (Fig. 2A, lane 4) (termed RX in Fig. 3B). Another minor band of 2.3 kb, whose size is consistent with an RNA in which the two small introns had been removed and which was polyadenylated internally at (pA)p, was also detected (Fig. 2A, lane 4) (termed R1′ in Fig. 3B). The existence of the 2.3-kb R1′ RNA was confirmed by hybridization with the P6 probe spanning nts 1121 to 1600 (Fig. 2B), which also demonstrated that the 2.3-kb R1′ RNA was present a very low level compared to the R1 RNA (Fig. 2A, lane 5).
Our RNase protection and Northern blot data are summarized in Fig. 3B, which presents a modified transcription profile for AMDV. The previously determined map is reproduced for comparison (Fig. 3A) (3). In summary, we have detected six RNAs, all generated from the P3 promoter, which fall into three classes based on splicing pattern. One species of each class was polyadenylated internally at (pA)p and one at the 3′ end at (pA)d. We also propose a slight alteration of the nomenclature for these RNAs, as shown in Fig. 3B.
The R2 transcript increased significantly and proportionally to viral capsid production during productive AMDV infection.
If the previously reported P36-generated R3 RNA is present at undetectably low levels during productive AMDV infection, which RNA encoded the abundant capsid proteins? Only the three AMDV-G transcripts that are polyadenylated at (pA)d (R1, R2, and RX) contain the capsid-coding ORF. The AUG codon that initiates VP1 is at nt 2204, and the AUG codon that initiates VP2 is at nt 2406.
AMDV-G infection of CRFK cells is productive, featuring the amplification of substantial levels of new virus (13), which reaches higher levels in these cells at 32°C than at 37°C. During low-multiplicity AMDV-G infection of CRFK cells, as capsid proteins (Fig. 4B), replicative DNA forms (Fig. 4C), and full virion production (Fig. 4D) increased, the AMDV P3-generated RNAs increased coordinately (Fig. 4A, probe P2), while no RNAs generated from the putative P36 promoter could be detected (Fig. 4A, probe P3). As expected, the increase in viral macromolecular synthesis was greater and sustained for a longer period at 32°C than at 37°C (Fig. 4). While all P3-generated RNAs increased proportionally with the increases in capsid proteins, replicative DNA forms, and DNA-containing virions, the R2 RNAs were predominant throughout (Fig. 4A, probe P2), suggesting that these RNAs may be a suitable source of the capsid proteins.
FIG. 4.
Time course of viral macromolecular synthesis following AMDV-G infection of CRFK cells at 32°C and 37°C. CRFK cells were infected with AMDV-G at 37°C or 32°C as indicated, and total RNA, DNA, capsid proteins, and generated virus were isolated at daily intervals. (A) Northern blot with probe P2 or P3 as indicated. The identities of the transcripts are shown to the left or right of the panels. R2 increased coordinately with capsid protein production (probe P2), while no RNAs initiated at P36 were detected (probe P3). Accumulation of β-actin RNA is shown as an internal control. (B) Western blot analysis showing accumulation of capsid proteins VP1 and VP2 at the time points and temperatures indicated. The accumulation of β-actin is shown as an internal control. VPx is an uncharacterized band that is likely a breakdown product of the capsid proteins. Markers are shown to the right. (C) Accumulation of replicating AMDV DNA at the times and temperatures shown. The replicative intermediates (double-stranded DNA [dsDNA] and single-stranded DNA [ssDNA]) are indicated. Lane 1 contains DNA markers of 4.5 and 2.9 kb. (D) Accumulation of AMDV virus as determined by quantitative PCR at either 37°C or 32°C at the indicated times postinfection.
R2 RNA encodes VP1 and VP2.
To determine whether the R2 RNA was capable of encoding the AMDV capsid proteins, we constructed a CMV-driven plasmid from which the D1-A2 intron was precisely removed (CMVΔD1A2Cap) (Fig. 5). Following transfection of CRFK cells, this construct, which generated only the R2 and R2′ transcripts (data not shown), efficiently generated VP1 and VP2 to levels equivalent to those generated by the parent-CMV-driven NSCap construct (Fig. 5B, compare lanes 3 and 4). These results suggested that the R2 RNA was capable of encoding VP1 and VP2. When the NH2 terminus of the NS2 ORF of this construct was tagged with HA, a small protein the predicted size of NS2 could be detected (Fig. 5C, lane 2), suggesting that either R2 or R2′ encoded the NS2 protein.
A CMV-driven construct (CMVΔD1A1Cap) (Fig. 5A) from which the D1-A1 intron was removed and which generated only the RX and RX′ RNAs (data not shown) produced significantly lower levels of capsid protein production (Fig. 5B, compare lane 2 to lanes 3 and 4), although this construct generated as much total RNA as did CMVNSCap and CMVΔD1A2Cap (Fig. 5D, compare lanes 2, 3, and 4). Because the RX RNA is also present at low levels during viral infection, it is unlikely that these RNAs are a major source of VP1 and VP2. A similar CMV-driven construct (CMVNSCapmD1) (Fig. 5A), in which the D1 donor of parent CMVNSCap was destroyed by mutation, such that only R1 and R1′ were produced (data not shown), was unable to generate VP1 or VP2 (Fig. 5B, compare lane 1 to lanes 3 and 4). However, it too generated levels of total RNA similar to those of CMVNSCap (Fig. 5D, compare lanes 1 and 4). When the NH2-terminal NS1 ORF of CMVNSCapmD1 was tagged with HA, expression of NS1 was confirmed (Fig. 5C, lane 1).
These experiments strongly suggested that, contrary to previous suggestion (3), the AMDV R2 RNAs were capable of generating the viral capsid proteins. As they were the predominant RNA species produced during viral infection, this provided the likely explanation for capsid protein production in the absence of detectable P36-generated RNAs. Consistent with these results, a CMV-driven NSCap construct in which the putative P36 TATA box and initiator sequence was mutated (CMVNSCapmP36) (Fig. 5A) was fully capable of generating VP1 and VP2 (Fig. 5B, compare lanes 5 and 6).
RNA elements upstream of VP1 AUG modulate expression of AMDV capsid proteins.
In the previous section, we have shown that although P3-generated R1, R2, and RX all contain the capsid protein ORF and the VP1- and VP2-initiating AUGs (at nts 2204 and 2406, respectively), only the R2 RNA efficiently generated the capsid proteins. This suggested that there were limitations on capsid protein expression governed at the level of translation of these RNAs. A further example of the control of translation of the capsid protein-coding RNA was apparent in the following experiment.
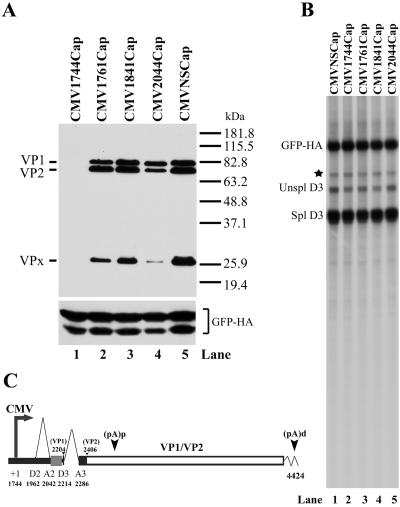
As shown in Fig. 5 and again in Fig. 6A, lane 1, a construct in which the CMV promoter replaced the P3 promoter (CMVNSCap) was fully capable of generating both VP1 and VP2 in CRFK cells. However, when the CMV promoter was fused to the capsid protein-coding transcription unit at nt 1744, at the putative P36 initiation site immediately downstream of acceptor A1 and upstream of the first small intron (CMV1744Cap) (Fig. 6), no capsid proteins were produced following CRFK cell transfection (Fig. 6A, lane 1), although the levels of spliced AMDV RNA were as high as for CMVNSCap (Fig. 6B, compare lanes 1 and 2). These results implied that a P36-initiated RNA would not be competent for capsid protein expression in CRFK cells, consistent with results described above. However, as the CMV promoter was moved closer to the initiating AUG of VP1, production of the capsid proteins was regained (Fig. 6A, lanes 2 to 4), from similar amounts of spliced RNA (Fig. 6B, lanes 3 to 5). This suggested that cis-acting elements upstream of the initiating AUG of VP1 in these constructs played a modulating role in the production of protein from these RNAs and perhaps explains previous results that demonstrated that the AMDV capsid gene from nts 2059 to 4762 expressed VP1 and VP2 when inserted into a vaccinia virus vector (16). This vaccinia virus construct was shown to generate more VP1 than VP2 (rather than more VP2 than VP1, as seen during AMDV infection). Interestingly, a construct in which we fused the CMV promoter to the capsid gene further downstream, at nt 2044 rather than nt 1744, also generated more VP1 than VP2 (CMV2044Cap) (Fig. 6A, lane 4), although there was no change in the ratio of spliced to unspliced RNAs generated by this construct (Fig. 6B, lane 5). This observation further implicates the importance of RNA sequences upstream of the VP1-initiating AUG in proper expression of the AMDV capsid proteins.
FIG. 6.
Capsid protein production is governed by sequences upstream of VP1 AUG. The constructs used (described in Results) are shown in panel C, as are the relevant ORFs used and potential products generated. The protein products generated by these constructs are shown in the Western blot in panel A. Panel B is an RNase protection assay using probe PD3, showing that all constructs generate similar amounts of RNA. Expression of HA-tagged GFP, following transfection of C1GFPHA, was used as an internal control and is shown at the bottom of panel A and as the GFP-HA band in panel B. The band marked by a star was likely the undigested probe. Unspl, unspliced; Spl, spliced.
DISCUSSION
In this study, we have reevaluated the transcription profile generated during AMDV-G infection of CRFK cells. Contrary to previous experiments, we have found evidence for use of only a single promoter, P3, at the left end of the genome. P3-generated pre-mRNAs were processed to generate six mRNAs. These pre-mRNAs were alternatively spliced in three patterns, all of which were present as species polyadenylated at (pA)d, the distal site at the right end of the genome (R1, R2, and RX) and at the proximal internal site, (pA)p (R1′, R2′, and RX′) (Fig. 3). Surprisingly, RNase protection assays could not detect RNAs initiated at the previously identified P36 promoter, following productive AMDV-G infections of CRFK cells at either 37°C or 32°C or following plasmid transfection. Thus, the expression profile of AMDV-G is similar to that of the erythroviruses B19 (24) and simian parvovirus (20, 40), which also generate genetic diversity by alternative splicing and polyadenylation of a single pre-mRNA generated from a promoter at the left end of the genome.
A P36 promoter was originally demonstrated for AMDV by primer extension assay, and its initiation site was mapped to nt 1744 (3); however, the P36 promoter appeared to be much weaker than the P3 promoter. In addition, P36 promoter activity was quite low when assayed in reporter constructs, and it showed only low levels of transactivation by the AMDV NS1 protein (15, 36). In our studies, we have found no evidence of P36 activity in CRFK cells even at late times during productive infection when capsid protein levels were high. In addition, artificial R3 transcripts expressed from a CMV promoter were not capable of expressing the capsid proteins (CMV1744Cap) (Fig. 6), further arguing against a role for the P36-generated RNAs during productive infection. A previously published report identified a band by RNase protection assay that was thought to be R3 (37). It appears that this band may have represented the RX and RX′ RNAs which utilize an acceptor (A1) located at nt 1736, only 8 nts upstream of the putative R3 start site.
We have not excluded the possibility that rare R3 transcripts might be detectable by sensitive reverse transcription PCR assays; however, it seems unlikely that RNAs present only at that level of detection could be a major source of capsid protein production during productive infection. On the other hand, the putative P36 promoter region does contain both a TATA box and an initiator signal (3, 15), and its location is similar to that of the P38 promoters of minute virus of mice and canine parvovirus (17, 36). Therefore, it remains possible that the P36 promoter is active either at a particular stage in the viral life cycle or in certain cell types within a host animal, where it may generate levels of R3 RNAs that are biologically important. It has been suggested that low levels of capsid accumulation from a weak P36 promoter could play a major role in establishment of the low-level persistent infection seen with Aleutian disease of mink (4, 15, 36, 37), but perhaps AMDV has evolved another mechanism to modulate expression of the capsid genes.
In most parvovirus infections, the level of capsid-encoding transcripts becomes predominant during late stages of infection. This is often accomplished by transactivation of the capsid gene promoter. In productive AMDV infection of CRFK cells, however, as is also the case for B19 (24) and SPV (20, 40), individual mRNAs are generated posttranscriptionally from a single pre-mRNA by a combination of alternative polyadenylation and alternative splicing. There is no evidence that processing of P3-generated pre-mRNAs changes during the course of infection (J. Qiu and D. Pintel, unpublished data), and thus all AMDV RNAs generated during infection increase coordinately with replicating DNA and in concert with increased virus and capsid protein production. However, because R2 is the predominant RNA generated at all times postinfection, it seemed most likely to encode the capsid proteins. Indeed, transfection of a plasmid engineered to generate only the R2 and R2′ mRNAs (CMVΔD1A2Cap) (Fig. 5) resulted in production of levels of the capsid proteins VP1 and VP2 equivalent to that produced following transfection of a genomic clone (CMVNSCap) (Fig. 5), and a genomic construct in which the putative P36 promoter was destroyed (CMVNSCapmP36) (Fig. 5) was similarly competent. We conclude that, contrary to previous suggestion (3), the virion proteins are encoded by the R2 RNA. Unlike B19 and SPV, however, for which the two capsid proteins VP1 and VP2 are encoded by differently spliced mRNAs, the AMDV-G VP1 and VP2 proteins are both encoded by the fully spliced R2 molecule. How expression of the R2 RNAs, which contain both initiating AUGs, is controlled to generate the appropriate relative levels of these two proteins is not yet known.
Furthermore, the R2 RNAs have the potential to generate the nonstructural NS2 protein. Mutation of the internal polyadenylation site in CMVΔD1A2Cap resulted in a plasmid from which only R2 (and no longer R2′) was generated, and this plasmid produced levels of NS2 and both capsid proteins VP1 and VP2 similar to levels produced by the parent plasmid (Qiu and Pintel, unpublished). Thus, it appears that in addition to both capsid proteins, the R2 transcripts can express NS2. If this holds true during viral infection, it would likely add a potential additional constraint on the expression of the AMDV capsid proteins. Generally speaking, such a translation strategy would be expected to limit expression of the capsid proteins, and this is currently under investigation. As shown by others (13) and as mentioned above, the relative production of AMDV-G in CRFK cells is modest; approximately 1,000-fold less AMDV-G is produced in these infections than during productive infection by mink enteritis virus (37, 39) or minute virus of mice (Qiu and Pintel, unpublished). In addition, the chronic immune complex-mediated Aleutian disease of mink is characterized by slow viral replication, which has been proposed by others (4) to allow the development of the classical characteristics of the disease. Decreased production of capsid proteins would be expected to be a rate-limiting step for single-stranded DNA amplification and hence for virion production. Dual capsid protein expression from an R2 RNA further limited by the presence of the NS2 gene may play a determining role in the ability of AMDV to establish and maintain a latent infection.
Acknowledgments
We thank Gregory E. Tullis for critical reading of the manuscript. We are very grateful to Marshall E. Bloom and Sonja Best (Rocky Mountain Laboratory, NIH, NIAID, Hamilton, MT) for providing cells, virus, antisera, advice, and encouragement, which helped initiate this study.
This work was supported by PHS grants, RO1 AI56310 and RO1 AI21302 from NIAID to D.P.
REFERENCES
- 1.Alexandersen, S. 1986. Acute interstitial pneumonia in mink kits: experimental reproduction of the disease. Vet. Pathol. 23:579-588. [DOI] [PubMed] [Google Scholar]
- 2.Alexandersen, S., and M. E. Bloom. 1987. Studies on the sequential development of acute interstitial pneumonia caused by Aleutian disease virus in mink kits. J. Virol. 61:81-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexandersen, S., M. E. Bloom, and S. Perryman. 1988. Detailed transcription map of Aleutian mink disease parvovirus. J. Virol. 62:3684-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexandersen, S., M. E. Bloom, and J. Wolfinbarger. 1988. Evidence of restricted viral replication in adult mink infected with Aleutian disease of mink parvovirus. J. Virol. 62:1495-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexandersen, S., M. E. Bloom, J. Wolfinbarger, and R. E. Race. 1987. In situ molecular hybridization for detection of Aleutian mink disease parvovirus DNA by using strand-specific probes: identification of target cells for viral replication in cell cultures and in mink kits with virus-induced interstitial pneumonia. J. Virol. 61:2407-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexandersen, S., S. Larsen, A. Cohn, A. Uttenthal, R. E. Race, B. Aasted, M. Hansen, and M. E. Bloom. 1989. Passive transfer of antiviral antibodies restricts replication of Aleutian mink disease parvovirus in vivo. J. Virol. 63:9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alexandersen, S., A. Uttenthal-Jensen, and B. Aasted. 1986. Demonstration of non-degraded Aleutian disease virus (ADV) proteins in lung tissue from experimentally infected mink kits. Arch. Virol. 87:127-133. [DOI] [PubMed] [Google Scholar]
- 8.Bloom, M. E., S. Alexandersen, C. F. Garon, S. Mori, W. Wei, S. Perryman, and J. B. Wolfinbarger. 1990. Nucleotide sequence of the 5′-terminal palindrome of Aleutian mink disease parvovirus and construction of an infectious molecular clone. J. Virol. 64:3551-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bloom, M. E., S. Alexandersen, S. Perryman, D. Lechner, and J. B. Wolfinbarger. 1988. Nucleotide sequence and genomic organization of Aleutian mink disease parvovirus (ADV): sequence comparisons between a nonpathogenic and a pathogenic strain of ADV. J. Virol. 62:2903-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bloom, M. E., B. D. Berry, W. Wei, S. Perryman, and J. B. Wolfinbarger. 1993. Characterization of chimeric full-length molecular clones of Aleutian mink disease parvovirus (ADV): identification of a determinant governing replication of ADV in cell culture. J. Virol. 67:5976-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bloom, M. E., S. M. Best, S. F. Hayes, R. D. Wells, J. B. Wolfinbarger, R. McKenna, and M. Agbandje-McKenna. 2001. Identification of Aleutian mink disease parvovirus capsid sequences mediating antibody-dependent enhancement of infection, virus neutralization, and immune complex formation. J. Virol. 75:11116-11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bloom, M. E., R. E. Race, W. J. Hadlow, and B. Chesebro. 1975. Aleutian disease of mink: the antibody response of sapphire and pastel mink to Aleutian disease virus. J. Immunol. 115:1034-1037. [PubMed] [Google Scholar]
- 13.Bloom, M. E., R. E. Race, and J. B. Wolfinbarger. 1980. Characterization of Aleutian disease virus as a parvovirus. J. Virol. 35:836-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christensen, J., T. Storgaard, B. Bloch, S. Alexandersen, and B. Aasted. 1993. Expression of Aleutian mink disease parvovirus proteins in a baculovirus vector system. J. Virol. 67:229-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christensen, J., T. Storgaard, B. Viuff, B. Aasted, and S. Alexandersen. 1993. Comparison of promoter activity in Aleutian mink disease parvovirus, minute virus of mice, and canine parvovirus: possible role of weak promoters in the pathogenesis of Aleutian mink disease parvovirus infection. J. Virol. 67:1877-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clemens, D. L., J. B. Wolfinbarger, S. Mori, B. D. Berry, S. F. Hayes, and M. E. Bloom. 1992. Expression of Aleutian mink disease parvovirus capsid proteins by a recombinant vaccinia virus: self-assembly of capsid proteins into particles. J. Virol. 66:3077-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clemens, K. E., and D. J. Pintel. 1988. The two transcription units of the autonomous parvovirus minute virus of mice are transcribed in a temporal order. J. Virol. 62:1448-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hadlow, W. J., R. E. Race, and R. C. Kennedy. 1983. Comparative pathogenicity of four strains of Aleutian disease virus for pastel and sapphire mink. Infect. Immun. 41:1016-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hahn, E. C., L. Ramos, and A. J. Kenyon. 1977. Expression of Aleutian mink disease antigen in cell culture. Infect. Immun. 15:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, Z., J. Qiu, F. Cheng, Y. Chu, Y. Yoto, M. G. O'Sullivan, K. E. Brown, and D. J. Pintel. 2004. Comparison of the transcription profile of simian parvovirus with that of the human erythrovirus B19 reveals a number of unique features. J. Virol. 78:12929-12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayer, L. W., B. Aasted, C. F. Garon, and M. E. Bloom. 1983. Molecular cloning of the Aleutian disease virus genome: expression of Aleutian disease virus antigens by a recombinant plasmid. J. Virol. 48:573-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naeger, L. K., J. Cater, and D. J. Pintel. 1990. The small nonstructural protein (NS2) of the parvovirus minute virus of mice is required for efficient DNA replication and infectious virus production in a cell-type-specific manner. J. Virol. 64:6166-6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naeger, L. K., R. V. Schoborg, Q. Zhao, G. E. Tullis, and D. J. Pintel. 1992. Nonsense mutations inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes Dev. 6:1107-1119. [DOI] [PubMed] [Google Scholar]
- 24.Ozawa, K., J. Ayub, Y. S. Hao, G. Kurtzman, T. Shimada, and N. Young. 1987. Novel transcription map for the B19 (human) pathogenic parvovirus. J. Virol. 61:2395-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Padgett, G. A., J. R. Gorham, and J. B. Henson. 1967. Epizootiologic studies of Aleutian disease. I. Transplacental transmission of the virus. J. Infect. Dis. 117:35-38. [DOI] [PubMed] [Google Scholar]
- 26.Pintel, D., D. Dadachanji, C. R. Astell, and D. C. Ward. 1983. The genome of minute virus of mice, an autonomous parvovirus, encodes two overlapping transcription units. Nucleic Acids Res. 11:1019-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porter, D. D. 1986. Aleutian disease: a persistent parvovirus infection of mink with a maximal but ineffective host humoral immune response. Prog. Med. Virol. 33:42-60. [PubMed] [Google Scholar]
- 28.Porter, D. D., A. E. Larsen, N. A. Cox, H. G. Porter, and S. C. Suffin. 1977. Isolation of Aleutian disease virus of mink in cell culture. Intervirology 8:129-144. [DOI] [PubMed] [Google Scholar]
- 29.Porter, D. D., A. E. Larsen, and H. G. Porter. 1969. The pathogenesis of Aleutian disease of mink. I. In vivo viral replication and the host antibody response to viral antigen. J. Exp. Med. 130:575-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porter, D. D., A. E. Larsen, and H. G. Porter. 1973. The pathogenesis of Aleutian disease of mink. III. Immune complex arteritis. Am. J. Pathol. 71:331-344. [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu, J., L. Kakkola, F. Cheng, C. Ye, M. Söderlund-Venermo, K. Hedman, and D. J. Pintel. 2005. Human circovirus TT virus genotype 6 expresses six proteins following transfection of a full-length clone. J. Virol. 79:6505-6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu, J., R. Nayak, G. E. Tullis, and D. J. Pintel. 2002. Characterization of the transcription profile of adeno-associated virus type 5 reveals a number of unique features compared to previously characterized adeno-associated viruses. J. Virol. 76:12435-12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu, J., and D. J. Pintel. 2002. The adeno-associated virus type 2 Rep protein regulates RNA processing via interaction with the transcription template. Mol. Cell. Biol. 22:3639-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu, J., Y. Yoto, G. E. Tullis, and D. Pintel. Parvovirus RNA processing strategies. In M. E. Bloom (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom, in press.
- 35.Schoborg, R. V., and D. J. Pintel. 1991. Accumulation of MVM gene products is differentially regulated by transcription initiation, RNA processing and protein stability. Virology 181:22-34. [DOI] [PubMed] [Google Scholar]
- 36.Storgaard, T., J. Christensen, B. Aasted, and S. Alexandersen. 1993. cis-Acting sequences in the Aleutian mink disease parvovirus late promoter important for transcription: comparison to the canine parvovirus and minute virus of mice. J. Virol. 67:1887-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Storgaard, T., M. Oleksiewicz, M. E. Bloom, B. Ching, and S. Alexandersen. 1997. Two parvoviruses that cause different diseases in mink have different transcription patterns: transcription analysis of mink enteritis virus and Aleutian mink disease parvovirus in the same cell line. J. Virol. 71:4990-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tullis, G. E., L. R. Burger, and D. J. Pintel. 1993. The minor capsid protein VP1 of the autonomous parvovirus minute virus of mice is dispensable for encapsidation of progeny single-stranded DNA but is required for infectivity. J. Virol. 67:131-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uttenthal, A., S. Larsen, E. Lund, M. E. Bloom, T. Storgard, and S. Alexandersen. 1990. Analysis of experimental mink enteritis virus infection in mink: in situ hybridization, serology, and histopathology. J. Virol. 64:2768-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vashisht, K., K. S. Faaberg, A. L. Aber, K. E. Brown, and M. G. O'Sullivan. 2004. Splice junction map of simian parvovirus transcripts. J. Virol. 78:10911-10919. [DOI] [PMC free article] [PubMed] [Google Scholar]