Abstract
Single-stranded genomes of adeno-associated virus (AAV) are packaged into preformed capsids. It has been proposed that packaging is initiated by interaction of genome-bound Rep proteins to the capsid, thereby targeting the genome to the portal of encapsidation. Here we describe a panel of mutants with amino acid exchanges in the pores at the fivefold axes of symmetry on AAV2 capsids with reduced packaging and reduced Rep-capsid interaction. Mutation of two threonines at the rim of the fivefold pore nearly completely abolished Rep-capsid interaction and packaging. This suggests a Rep-binding site at the highly conserved amino acids at or close to the pores formed by the capsid protein pentamers. A different mutant (P. Wu, W. Xiao, T. Conlon, J. Hughes, M. Agbandje-McKenna, T. Ferkol, T. Flotte, and N. Muzyczka, J. Virol. 74:8635-8647, 2000) with an amino acid exchange at the interface of capsid protein pentamers led to a complete block of DNA encapsidation. Analysis of the capsid conformation of this mutant revealed that the pores at the fivefold axes were occupied by VP1/VP2 N termini, thereby preventing DNA introduction into the capsid. Nevertheless, the corresponding capsids had more Rep proteins bound than wild-type AAV, showing that correct Rep interaction with the capsid depends on a defined capsid conformation. Both mutant types together support the conclusion that the pores at the fivefold symmetry axes are involved in genome packaging and that capsid conformation-dependent Rep-capsid interactions play an essential role in the packaging process.
Viruses package their genomes into a protein shell either by association of structural proteins with the viral DNA or by translocation of viral genomes into preformed capsids. The insertion of viral genomes into preassembled capsids has been shown for viruses with double-stranded (ds) linear genomes such as bacteriophages or herpesviruses. These viruses first assemble an icosahedral procapsid which contains, in most cases, a dodecameric complex of virus-encoded portal protein, positioned asymmetrically at one of the 12 icosahedral vertices (16, 19, 29, 32, 41). This so-called portal or connector complex forms a channel through which the viral DNA enters and exits the capsid. Packaging enzymes bind to the portal vertex and mediate cleavage of monomeric units from concatemeric DNA as well as translocation of the genomes into the capsid in an ATP-dependent fashion (5, 11). There are a number of single-stranded DNA (ssDNA) and ssRNA viruses described, such as φX174, φ6, or picornaviruses, which package their genomes into preformed capsids as well, and this is also the postulated assembly pathway for parvoviruses (16, 25, 31).
The adeno-associated virus type 2 (AAV2) is a member of the parvovirus family, which requires coinfection with helper viruses, such as adenovirus (Ad) and herpesvirus, for efficient reproduction (30). AAV2 has a nonenveloped icosahedral capsid that encloses a 4.7-kilobase-long ssDNA genome containing two large open-reading frames, rep and cap (38). The Rep proteins encoded by the rep gene are required for DNA replication, DNA encapsidation, and control of gene expression (6, 25, 30). The viral capsid is formed by three structural proteins (VPs) designated VP1, VP2, and VP3, which are encoded by the cap gene. The VPs become expressed from the p40 promoter in an approximate molar ratio of 1:1:10 by use of alternative splicing and an uncommon start codon for VP2 (2, 4, 40). This ratio is also maintained in the assembled capsid (23). The VPs share a common central and C-terminal region with N-terminal extensions of 65 amino acids common for VP1 and VP2 and an additional 138 N-terminal amino acids forming the unique portion of VP1.
The AAV2 capsid structure has been resolved by electron cryomicroscopy and X-ray crystallography (27, 47). The capsids contain 60 copies of VPs, arranged with T=1 icosahedral symmetry. Striking structural features are a dimple-like depression at the twofold symmetry axes, three elongated spikes surrounding the threefold axes, and 12 narrow pores at the fivefold axes. Neither the AAV2 structure nor other parvovirus X-ray structures could resolve the precise localization of the N-terminal regions of VP1 and VP2 so far. However, there is evidence that N termini of AAV2 VPs—probably including the N-terminal extensions of VP1 and VP2—are organized in globular structures at the twofold axes inside native virions (26, 27). The VP1 and VP2 N termini can become exposed, most likely through the pores at the fivefold symmetry axes, thereby displaying a phospholipase A2 activity required for efficient AAV2 infection (3, 12, 26).
The assembly of AAV2 has been proposed to occur via introduction of ss genomes into preformed capsids (for a review, see reference 39). Myers and Carter (31) hypothesized that replicated, strand-displaced ssDNA is initially associated with preassembled capsids, which are then slowly converted into DNA-filled virions. Capsid assembly requires only the expression of VPs (43). It occurs already at early stages of a productive infection and assembled capsids first accumulate in nucleoli as detected by indirect immunofluorescence (43). At later time points of an infection, Rep proteins, capsids, and viral DNA are codistributed in larger compartments throughout the nucleoplasm (21, 43), most likely, the sites for encapsidation of viral genomes. The ssDNA is proposed to be targeted to the capsid by Rep proteins, which are not only able to bind specifically to the inverted terminal repeats of the genome but are also able to form complexes with capsids (9). Additionally, Rep-capsid complexes based on the covalent linkage of Rep78 to the 5′ end of encapsidated ssDNA have been observed (33, 34). Genome encapsidation is suggested to proceed in a preferred direction from the 3′ end, and the helicase activity of the small Rep proteins, present in encapsidation complexes, is proposed to drive translocation of full-length ssDNA into capsids (6, 25). The AAV2 Rep40 helicase is a member of the SF3 helicase family and is structurally related to the AAA+ family of motor proteins (ATPases associated with diverse cellular activities) (22). Structural features directly imply the requirement of Rep40 oligomerization for ATP hydrolysis and helicase activity. A hexameric ring-like model similar to that for other SF3 helicases could be predicted for the Rep40 helicase (22). Mutation of the DNA-binding amino acids of Rep40 results in a significant packaging defect (48). However, the portal through which AAV2 ss genomes are inserted into capsids remains unknown. We previously could show that mutation of amino acids surrounding the fivefold pore on AAV2 capsids affected genome packaging (3), indicating that a channel at the fivefold symmetry axes may be the portal for DNA encapsidation.
The aim of this study was to further characterize the packaging defect caused by mutations of amino acids surrounding the fivefold pores. These mutants showed a reduced amount of Rep proteins in complexes with capsids and a decreased but complete packaging of viral genomes. In particular, mutation of the pore-rim resulted in a drastic reduction in Rep-capsid interactions and in a strong packaging defect. This supports our interpretation that the packaging complex, including Rep proteins, binds to the pore at the fivefold axes and uses it as a portal for DNA translocation into the capsid. In contrast, the R432A mutant, a previously described packaging mutant (46) with an amino acid exchange at the interpentamer surface, is characterized by a complete block of packaging. Packaging of this mutant seems to be inhibited by occupation of the channels at the fivefold axes by VP1 and VP2 N termini already in empty capsids. Surprisingly, these capsids showed increased association with Rep proteins. In general, we provide evidence that a defined conformation of preassembled AAV2 capsids—involving correct interactions with Rep proteins—is necessary for successful DNA encapsidation.
MATERIALS AND METHODS
Plasmids and site-directed mutagenesis.
The pTAV2.0 construct (18) contains the entire AAV2 genome from pAV-2 (28), including both inverted terminal repeats cloned into the BamHI site of Bluescript II. Plasmid pTAV2.1 is based on pTAV2.0 but contains a mutated Rep52/40 translation start site to prevent expression of the small Rep proteins (25). The constructs N335A, del219/220, D219A, and S338A are based on pTAV2.0 with the designated mutations in the cap open reading frame (3). Plasmid pCMV-VP (43) expresses the AAV2 VPs under control of the cytomegalovirus promoter. Plasmid pDG expresses the AAV2 Rep and VP and all essential Ad helper proteins (15), and construct pDGΔVP is based on plasmid pDG but prevents expression of the VPs (9). In plasmid pDM, the rep-cap open reading frame of plasmid pDG has been replaced by the full-length AAV2 genome (26). As a template for site-directed mutagenesis reactions, plasmid pJ407 (24), containing the BamHI-NotI fragment of pTAV2.0 cloned into pUC131, was used. Mutagenesis was performed by using the QuickChange site-directed mutagenesis kit (Stratagene, Amsterdam, The Netherlands) according to the manufacturer's manual. For each mutant, two complementary PCR primers were designed containing the substitution, flanked by 10 to 15 homologous base pairs on each site of the mutation. Mutant plasmids were identified by DNA sequencing. The SwaI-BsiWI fragment (pore mutants) or the BsiWI-SnaBI fragment (R432A mutant) containing the mutation was then subcloned into the pTAV2.0 backbone. The complete fragments were sequenced to exclude additional mutations.
Transfection of 293T cells and preparation of virus supernatants.
Transfection was carried out by calcium phosphate precipitation as described previously (3). Briefly, cells were seeded at 1.5 × 106 cells per 10-cm dish. The next day, they were transfected with 12 μg DNA per dish by calcium phosphate precipitation at 37°C. Sixteen hours posttransfection, the medium was removed and replaced with fresh medium containing adenovirus type 5 (multiplicity of infection of 2) and incubated at 37°C for a further 2 days. Cells were harvested and lysed by three rounds of freeze-thawing (−80°C and 37°C) in TNEM buffer (0.15 M NaCl, 50 mM Tris, 1 mM EDTA, 5 mM MgCl, pH 8.0). For immunoprecipitations, cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, 0.15 M NaCl, 0.1% sodium dodecyl sulfate, 1% NP-40, 0.5% sodium deoxycholate). Cell debris was removed by centrifugation at 1,000 × g for 20 min.
Analyses of viral protein expression.
Western blot analysis was performed on equal portions of harvested cells according to standard methods (17). Monoclonal antibodies 303.9 and B1, specific for Rep and VP, respectively, were used as described previously (44).
Titration of AAV2 stocks.
Capsid titers were determined by an enzyme-linked immunosorbent assay (ELISA) (14). Genomic titers and infectious viral titers were determined as described previously (3).
Analyses of associated and packaged DNA.
Comparison of capsid-associated and encapsidated viral genomes was carried out as described previously (3).
Immunoprecipitation.
Precipitation was performed using protein A-Sepharose (2 mg per sample) (CL-4B; Amersham Pharmacia, Freiburg, Germany) in NET-N buffer (20 mM Tris, 100 mM NaCl, 1 mM EDTA, 0.5% NP-40, pH 7.5) containing protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany). Protein A-Sepharose was added to 600 μl of A20 or the non-AAV-related IVA7 hybridoma supernatant for capsid precipitation or to 600 μl αRep90 or the non-AAV-related αIP1918 serum (1:100 in NET-N) for Rep precipitation and incubated with gentle inversion for 1 h at 4°C. The protein A-Sepharose-antibody complex was washed twice with NET-N to remove unbound antibody and then added to virus supernatants in RIPA buffer containing equal amounts of capsids. The samples were incubated overnight at 4°C with gentle inversion, washed four times with NET-N and analyzed by Western blot analysis with antibody B1 or 303.9.
Continuous sucrose gradient.
Virus supernatants in TNEM buffer obtained from two 10-cm dishes (600 μl) were loaded onto a 11-ml 10 to 30% sucrose gradient (sucrose in TNEM buffer containing 10 mM EDTA and protease inhibitor cocktail). After centrifugation at 160,000 × g for 2 h at 4°C (SW41 rotor; Beckman) 500-μl fractions were collected. Fifteen microliters of each fraction was processed for Western blot analysis. Monoclonal antibodies 303.9 and B1 were used to detect Rep and VP, respectively.
Native dot blot assay.
60S or 110S fractions of continuous sucrose gradients (50 μl) were subjected to different temperatures for 5 min in a total volume of 200 μl phosphate-buffered saline containing 1 mM MgCl2 and 2.5 mM KCl. Samples were then incubated on ice for 5 min and transferred to Protran nitrocellulose membranes (Schleicher & Schuell, Dassel, Germany). Membranes were processed as described previously (3), by using monoclonal antibodies A20, A1, A69, and B1 (44, 45).
Graphics.
Pictures were made by using Rasmol software, version 2.7.2.1.1 (36). The structure of AAV2 was obtained from the Brookhaven Protein Data Bank, entry 1LP3 (47).
Statistical analysis.
Data were analyzed by an unpaired two-tailed Student's t test and are shown as means ± standard deviations. Data were considered significant at a P value of less than 0.05.
RESULTS
Characterization of packaging mutants.
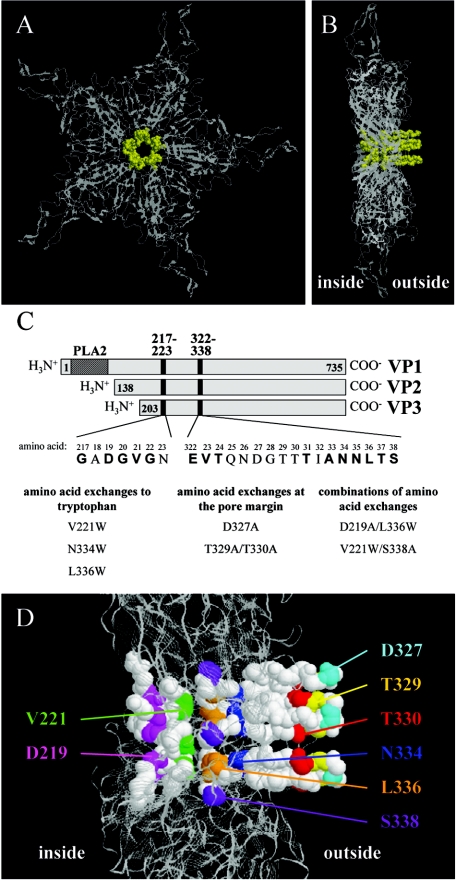
The structure of AAV2 virions revealed open pores of approximately 12 Å in diameter at the fivefold symmetry axes of the icosahedral capsid (Fig. 1A) (27, 47). These pores are encircled by a protrusion at the outside and a funnel-like structure at the inside of the capsid (Fig. 1B). These two structures are formed by amino acids 217 to 223 and 322 to 338 of the VP sequence, which show a high level of conservation between the AAV serotypes (Fig. 1C; see Fig. 8). We have recently shown that mutation of several conserved pore-forming amino acids led to reduced capsid assembly, genome packaging, and infectivity (3). These three phenotypes were assigned to different residues, but they overlapped in some positions. While the infectivity phenotype could be linked to an impaired ability to externalize VP1 N termini through the pores at the fivefold axis (see also reference 26), the precise mechanism(s) which led to reduced packaging remained unresolved. In order to elucidate the role of pore-forming amino acids in the packaging process, we generated new mutants of selected residues of the pore structure (Fig. 1C and D). We exchanged amino acids for tryptophan, a large and bulky residue, which may be able to block the pore and prevent packaging more efficiently. In addition, we chose amino acids at the pore margin which might interfere with binding of the packaging complex. And finally, we combined different mutations showing an effect on genome packaging.
FIG. 1.
Establishment of new AAV2 pore mutants. A backbone model of a VP pentamer is shown from the outside of the capsid (A) and in a side view (B); amino acids which form the pore at the fivefold symmetry axis are represented as space-filling models in yellow. (C) Schematic representation of the pore-forming amino acids (position 217 to 223 and 322 to 338 of the VP sequence) shared by the three capsid proteins. Bold lettering indicates amino acids which are conserved between the AAV serotypes 1 to 11. Three groups of mutations were established as indicated. (D) Localization of established mutations within the pore structure. A backbone model of a pentamer is shown as a side view and amino acids which form the pore are represented as space-filling models; residues subjected to site-directed mutagenesis are shown in different colors.
FIG. 8.
Alignment of pore-forming amino acids of the 11 AAV serotypes described so far. The pore-forming amino acids of AAV2 (positions 217 to 223 and positions 322 to 338 of the VP sequence) were aligned to the corresponding VP region of the other 10 AAV serotypes; bold lettering indicates conserved amino acids. Similarity significance value cutoffs (Consensus) were more than 60% (capital letters) and more than 35% (lowercase letters).
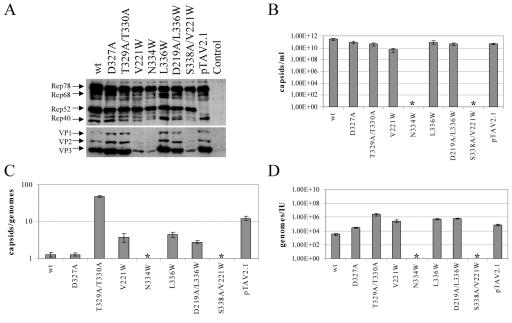
Protein expression levels of wild-type (wt) and mutant AAV2 were analyzed by transient transfection of the respective plasmids followed by Western blot analysis. Extracts of transfected cells showed no significant influence of the mutations on the expression of the four Rep proteins except a weak reduction in the levels of Rep68 and Rep40 in V221W, N334W, and S338A/V221W mutants (Fig. 2A). Four mutants (D327A, T329A/T330A, L336W, and D219A/L336W mutants) showed also no reduction in VP expression levels compared to those in the wt. The capsid titers of these mutants were also comparable to those of the wt (Fig. 2B; Table 1). A reduced VP expression (Fig. 2A) combined with impaired capsid assembly (Fig. 2B; Table 1) could be observed for three mutants (V221W, N334W, and S338A/V221W mutants). The packaging efficiency of the mutants was quantified by calculating the ratios of capsids to genomes and was compared to that of wt AAV2 (Fig. 2C; Table 1). Three mutants (V221W, L336W, and D219A/L336W mutants) showed a three- to fivefold reduction in genome packaging, whereas packaging of the T329A/T330A mutant was about 50-fold reduced. This value greatly exceeded the reduction seen for a small Rep-deficient mutant described previously (pTAV2.1) (25). Interestingly, the two threonine residues 329 and 330 are located at the pore margin where the interaction with the packaging complex is expected to occur. The D327A mutant, however, was not affected in packaging. Infectious virus titers, determined as replicative units in the presence of Ad5, were reduced in all mutants compared to that in wt AAV2 (data not shown). The ratios of genomes to infectious units were calculated to quantify the effect on infectivity, corrected for the reduction in packaging (Fig. 2D; Table 1). These results confirm and extend our previous observations (3) that mutations in pore-forming amino acids can affect three steps of the AAV2 life cycle: capsid assembly, genome packaging, and infectivity (Table 1).
FIG. 2.
Characterization of new AAV2 pore mutants. Viral supernatants were generated from 293T cells transfected with AAV2 wt or mutated genomic plasmids or a small Rep-deficient mutant (pTAV2.1) (25) or Bluescript II as controls. Supernatants were assayed for viral protein expression (Western blot analysis using monoclonal anti-Rep [303.9] or anti-VP [B1] antibodies) (A) or ELISA-based AAV2 capsid titer (B). Influence of the mutations on genome packaging and infectivity was quantified by calculating the ratios of capsids to genomes (C) and genomes to infectious units (IU) (D), respectively. Means ± standard deviations from at least four independent experiments are shown; an asterisk indicates mutants for which capsids could not be detected in the ELISA (detection limit, 5 × 108 capsids/ml).
TABLE 1.
Classification of new AAV2 pore mutants
| Mutation(s) | Mean value ± SDa
|
Classe | ||
|---|---|---|---|---|
| Assembly reduction with respect to wtb | Packaging reduction with respect to wtc | Infectivity reduction with respect to wtd | ||
| D327A | 4.4 ± 2.9f | 1.0 ± 0.2 | 11.3 ± 6.3 | I |
| T329A/T330A | 7.8 ± 4.1f | 47.1 ± 8.8f | 8.3E2 ± 2.7E2f | P+I |
| V221W | 40.8 ± 20.1f | 2.9 ± 0.4f | 1.1E2 ± 0.5E2f | A+P+I |
| N334W | UD | UD | UD | A |
| L336W | 4.9 ± 2.6f | 4.7 ± 0.6f | 2.0E2 ± 0.7E2f | P+I |
| D219A/L336W | 6.7 ± 4.4f | 2.6 ± 0.4f | 2.1E2 ± 0.3E2f | P+I |
| S338A/V221W | UD | UD | UD | A |
Values are means plus or minus standard deviations from at least four independent experiments. UD, undetectable.
Assembly is expressed as the number of assembled capsids, as determined by a capsid ELISA (14), and compared to the capsid titer of wt AAV2.
Packaging was measured as the ratio of capsids to viral genomes and compared to the ratio for wt AAV2. The number of assembled capsids was determined by a capsid ELISA (14); packaged DNA was quantified by a dot blot assay.
Infectivity is expressed as the ratio of genomes to infectious units and compared to the ratio for wt AAV2. Packaged DNA was measured by a dot blot assay; infectious AAV2 particles were quantified by a dot blot replication assay.
Mutants were classified according to their defects as follows: A, assembly (more than 10-fold reduction compared to that in the wt); I, infectivity (more than 10-fold reduction compared to that in the wt); P, packaging (more than 2-fold reduction compared to that in the wt).
In comparison with the wt, the P value was <0.05.
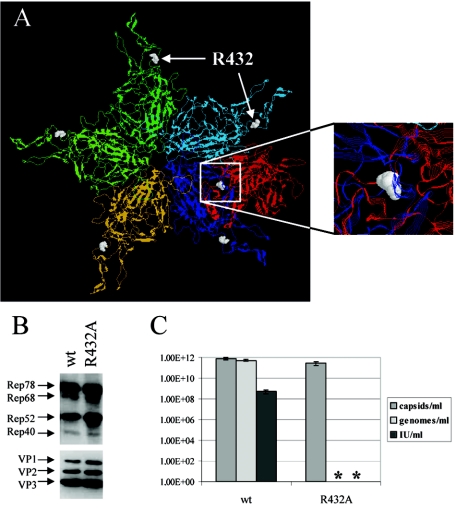
Several packaging mutants with alterations in amino acids apart from the pore at the fivefold axis of the capsid have been described (37, 46). We were interested in the exchange of arginine 432, which is located at a site where VP subunits of different pentamers interact (Fig. 3A), for alanine (46). This amino acid is neither accessible from the outside of the capsid nor accessible from the inside and therefore has probably an indirect effect on packaging, possibly by preventing conformational rearrangements needed for establishing a functional packaging complex. Rep and VP expression as well as the capsid titer of the mutant was comparable to that of the wt (Fig. 3B and C). However, neither genomes nor infectious units could be determined, indicating a strong packaging defect in accordance with the data from Wu et al. (46).
FIG. 3.
Characterization of the R432A packaging mutant. (A) A backbone model of a VP pentamer and a neighboring dimer is shown from the outside of the capsid; the subunits are shown in different colors (the pentamer is shown in cyan, blue, orange, and green; the dimer is shown in cyan and red) and amino acid R432 is represented as a space-filling model shown in white. The inset shows the R432 of the red subunit of the dimer interacting with the blue subunit of the pentamer. Viral supernatants were generated from 293T cells transfected with an AAV2 wt or mutated genomic plasmid. Freeze-thaw supernatants were assayed for viral protein expression (Western blot analysis using monoclonal anti-Rep [303.9] or anti-VP [B1] antibodies) (B) or ELISA-based AAV2 capsid titer, quantity of encapsidated AAV2 DNA, and infectious viral titer (C). Means ± standard deviations from at least four independent experiments are shown; an asterisk indicates that neither genomes nor infectious particles were detected for the R432A mutant. IU, infectious units.
Analysis of DNA replication and capsid-associated DNA versus encapsidated DNA.
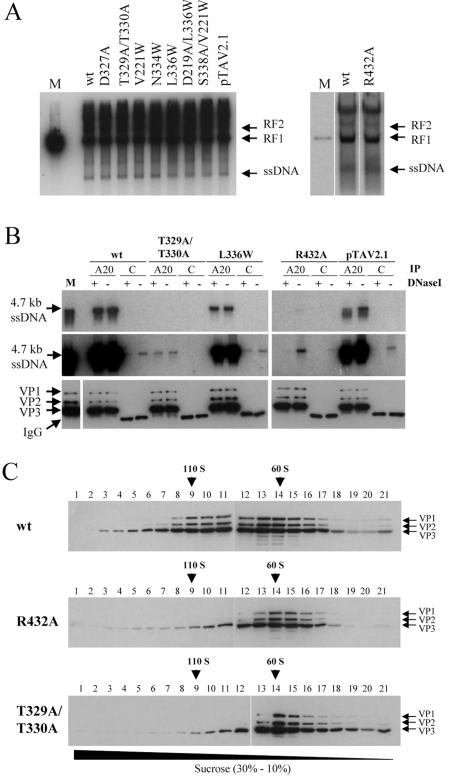
To show that the observed reduction in genome packaging was not due to inefficient DNA replication, Hirt extracts were prepared (20) from 293T cells transfected with wt or mutant AAV2 genomic plasmids and superinfected with Ad5. Southern blot analyses of DpnI-digested viral DNAs showed no differences in viral replication between the wt and any of the mutants (Fig. 4A). All mutants showed levels of ss and dsDNAs comparable to that of the wt.
FIG. 4.
Analysis of DNA replication, capsid-associated versus encapsidated viral genomes, and empty capsid formation. (A) Low-molecular-weight replicative form (RF) and single-stranded DNAs were isolated from cells transfected with plasmids harboring an AAV2 wt genome, genomes with mutated VP sequences, or genomes of a small Rep-deficient mutant (pTAV2.1) by a modified Hirt extraction procedure; AAV2 DNAs were detected by Southern blotting using a rep-specific probe (XhoI-BamHI fragment of plasmid pBSΔTR18) (42); a 4.7-kb AAV2 fragment (dsDNA) was used as a marker (lane M). (B) Capsids were immunoprecipitated (IP) from freeze-thawed lysates obtained from 293T cells transfected with AAV2 wt or mutated genomic plasmids or a small Rep-deficient mutant (pTAV2.1) by protein A-Sepharose-bound antibody A20. Control (C) precipitations were performed with a non-AAV-related monoclonal antibody. Immunoprecipitates were not digested (−) or digested (+) with DNase I to discriminate between capsid-associated and encapsidated genomes. Coprecipitated DNAs were isolated and electrophoresed on neutral agarose gels. AAV2 genomes were detected by Southern blotting as described for panel A; wt AAV2 ssDNA was used as a marker (lane M). Shown are autoradiographs of a short exposure time (top panel) and a longer exposure time (middle panel). A portion of each immunoprecipitate was tested by Western blotting using antibody B1 to control the recovery of capsids; extract from Ad5- and AAV2-coinfected HeLa cells was used as a marker (lane M) (bottom panel). (C) Extracts from 293T cells transfected with AAV2 wt or mutated genomic plasmids were analyzed on continuous sucrose gradients (10 to 30% sucrose). VPs were detected by Western blot analysis using monoclonal antibody B1. The 110S and 60S positions were determined by using purified DNA-containing and empty AAV2 particles, respectively.
To distinguish between capsid-associated and encapsidated viral genomes, a Southern blot analysis was performed with viral DNAs which were recovered from immunoprecipitates of wt or selected examples of mutant virions (Fig. 4B). Prior to DNA extraction, the immunoprecipitates were not treated or treated with DNase I. For wt AAV2, full-length genomes were completely protected against DNase I treatment, indicating efficient packaging of the 4.7-kb ssDNA (Fig. 4B, top panel). For comparison, the small Rep-deficient construct (pTAV2.1) (see also reference 25) showed that part of the capsid-associated DNA was nuclease sensitive. For the L336W mutant, which showed a fivefold-reduced capsid-per-genome ratio (Fig. 2C; Table 1), the capsid-coprecipitated DNA was completely protected against DNase I treatment; however, the level of packaged DNA was reduced compared to wt AAV2 (Fig. 4B). This was also characteristic for the other pore mutants with a packaging defect described previously (3). The T329A/T330A mutant showed very small amounts of encapsidated full-length genomes, which were detectable only after prolonged exposure times (Fig. 4B, middle panel). Again, packaging was complete and not partial, although this was at a very low level. AAV2 DNA coprecipitated with the R432A mutant—which had a similar low level—was completely sensitive to DNase I treatment, demonstrating that viral genomes were associated with the mutant capsid but not encapsidated (Fig. 4B, upper and middle panel). Immunoprecipitations with a non-AAV-related monoclonal antibody (Fig. 4B) controlled the specificity of the precipitations and Western blot analysis of A20-precipitated samples by using antibody B1 verified that comparable amounts of capsids were analyzed (Fig. 4B, bottom panel). These data clearly demonstrate that pore mutants and the R432A mutant showed different characteristics with respect to capsid-associated and encapsidated genomes. While pore mutants packaged only reduced levels of genomes into capsids, the R432A mutant showed small amounts of capsid-bound DNA, which were not encapsidated.
Sedimentation analysis on continuous sucrose gradients verified that the two strongest packaging mutants formed stable, empty particles with sedimentation characteristics similar to empty wt AAV2 capsids (Fig. 4C).
Rep-capsid interactions.
It was previously shown that Rep proteins are associated with capsids in the presence of DNA packaging by either a direct or DNA-mediated interaction (33, 44). Dubielzig et al. (9) showed later a DNA-independent Rep-capsid interaction as well as Rep-Rep interactions, suggesting a role for such complexes in some steps of the packaging process, e.g., by mediating the docking of the genome to the capsid. This led us to hypothesize that capsid-associated Rep proteins play a role in AAV2 DNA packaging, which was experimentally confirmed for the small Rep proteins (25). We also previously hypothesized that the pores at the fivefold symmetry axes of AAV2 capsids would form the portal for DNA encapsidation (3). Consequently, docking of the packaging machinery, involving large and small Rep proteins in complex with replicated DNA, is expected to take place at or close to the pores at the fivefold symmetry axis. We were therefore interested if AAV2 pore mutants which exhibited a packaging defect showed altered association of Rep proteins with the packaging complex at the capsid.
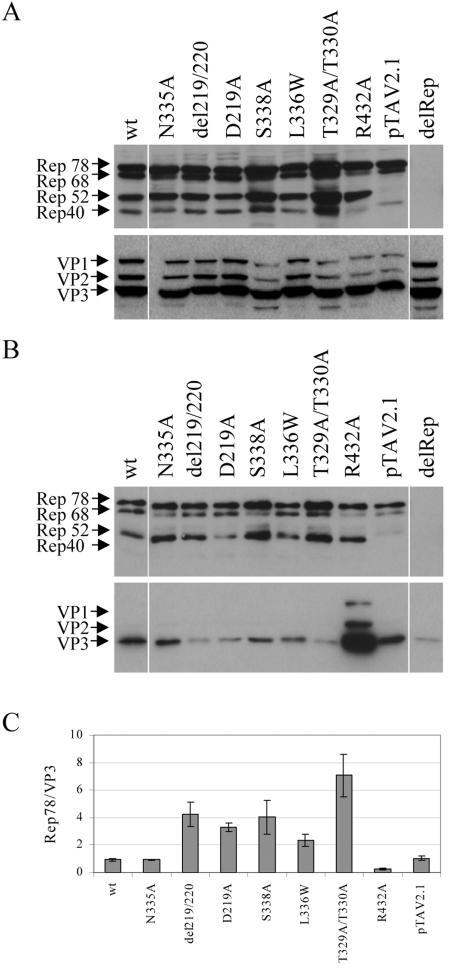
We analyzed the Rep interactions with the capsids by a coimmunoprecipitation assay using a polyclonal Rep antiserum to precipitate Rep proteins in complex with capsids involved in packaging from RIPA extracts of cells transfected with the respective mutant plasmids. In this experiment, we included mutants with strong packaging defects, as well as one mutant which was not impaired in packaging. Expression controls indicated the presence of comparable amounts of Rep and VP in the cell extracts (Fig. 5A). Precipitated Rep proteins and coprecipitated capsid proteins were detected by Western blot analysis using antibodies 303.9 and B1, respectively (Fig. 5B). Rep78 and Rep52 as well as smaller amounts of Rep68 were precipitated with the Rep-antiserum, correlating with the expression levels of these proteins. In accordance with previous observations (9), we could not detect precipitated Rep40 in these experiments, most likely due to the lower expression level of this protein. But the fact that capsids could be coprecipitated with Rep antisera when Rep40 was overexpressed together with VPs (9) suggests that also the smallest Rep protein could form complexes with capsids. Capsids of the N335A mutant, a pore mutant which showed no reduction in genome packaging (3), could be coprecipitated with the Rep antiserum as well as wt capsids (Fig. 5B). This is also the case for the small Rep-deficient mutant (pTAV2.1), indicating that the large Rep proteins are sufficient for coprecipitation of capsids. In contrast, capsids of all pore mutants with a packaging defect showed reduced coprecipitation of capsids with Rep proteins compared to wt, suggesting a reduced or weakened association of Rep proteins with the capsids. Surprisingly, the R432A mutant showed greatly increased amounts of coprecipitated capsids compared to wt, pointing to a different packaging defect. In the absence of Rep proteins, a weak background of precipitated capsids could be detected (Fig. 5B). To quantify the differences in Rep-capsid coprecipitation, the ratio of precipitated Rep78 to coprecipitated VP3 was calculated from three independent experiments by using the program ImageQuant (Fig. 5C). This analysis confirmed an approximate 2.5- to 7.5-fold reduction in VP3 coprecipitation of the packaging pore mutants compared to wt. Notably, the reduction in Rep-capsid coprecipitation correlated with the packaging defect of the respective mutants. The T329A/T330A mutant, which showed the strongest packaging reduction compared to wt, exhibited the most pronounced effect on Rep-capsid coprecipitation. For del219/220, D219A (3), and L336W mutants, with a fivefold-reduced packaging, and the S338A mutant (3), with a ninefold-reduced packaging, the reduction in coprecipitation of capsids with Rep proteins was not as strong. An approximate fivefold increase in coprecipitated VP3 was noted for the R432A mutant.
FIG. 5.
Effect of different mutations on the formation of packaging complexes. 293T cells were transfected with AAV2 wt, newly mutated genomic plasmids (L336W, T329A/T330A, and R432A mutants) or previously described mutated genomic plasmids (N335A pore mutant without a packaging defect and del219/220, D219A, and S338A pore mutants with packaging defects [3]; pTAV2.1, a small Rep-deficient mutant [25]; delRep, plasmid pCMV-VP expressing only VPs [43]). (A) Cell extracts were prepared in RIPA buffer and analyzed for viral protein expression by Western blot analysis using monoclonal anti-Rep (303.9) or anti-VP (B1) antibodies. (B) Rep-capsid complexes were immunoprecipitated from RIPA extracts by using a polyclonal anti-Rep antiserum and analyzed by Western blotting to detect precipitated Rep proteins and coprecipitated capsid proteins. A control reaction was carried out using extracts of cells that had been transfected with a plasmid only expressing the three VPs (lane delRep). (C) The amounts of coprecipitated VPs were quantified by calculating the ratios of precipitated Rep78 to coprecipitated VP3 by using the program ImageQuant (Molecular Dynamics), and these were compared to the ratio of wt AAV2. Means ± standard deviations from at least three independent experiments are shown.
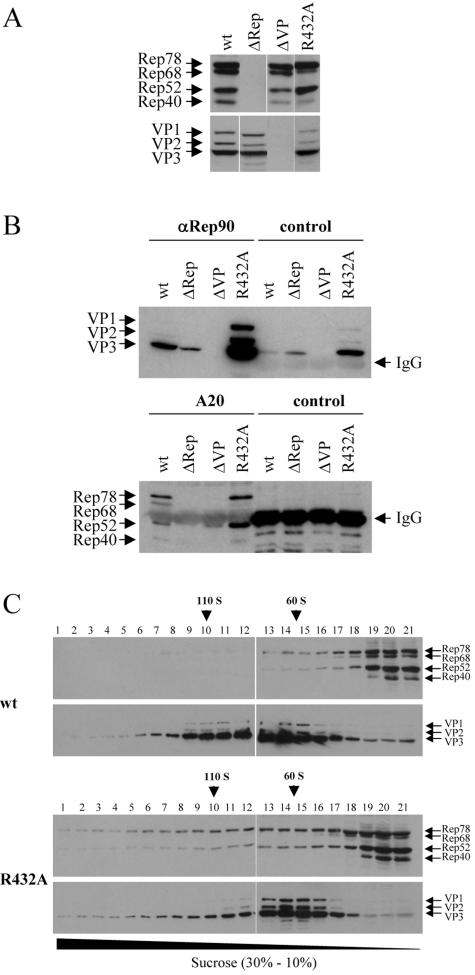
The increased coprecipitation of capsids with an antiserum against Rep proteins observed for the R432A mutant could be nonspecific due to the formation of capsid aggregates or to less soluble capsids rather than due to increased occupation of capsids by Rep proteins. To exclude these possibilities, we performed coimmunoprecipitations in both directions, i.e., precipitation with a Rep antiserum and detection of VPs as well as precipitation with an anticapsid antibody and detection of coprecipitated Rep proteins. Both precipitations were normalized for expression of capsid proteins or Rep proteins alone (Fig. 6A). Distinctly larger amounts of capsids could be coprecipitated with the Rep antiserum in the case of the R432A mutant compared to wt, as shown above (see also Fig. 5). Only small levels of all VPs are precipitated with the Rep antiserum when VPs were expressed in the absence of Rep (ΔRep). Control precipitations with a non-AAV-related polyclonal antiserum showed a background level of capsid precipitation which is higher for the R432A mutant than for wt and for wt in the ΔRep situation, indicating a higher background level of nonspecific immunoprecipitation (Fig. 6B, upper panel). With the capsid-specific antibody A20, Rep78 and Rep52 as well as smaller amounts of Rep68 and Rep40 could be coprecipitated from extracts of cells transfected with wt AAV2 plasmids (Fig. 6B, lower panel). However, from extracts of cells transfected with the R432A mutant, significantly higher levels of Rep78 and Rep52 were coprecipitated with the capsids (Fig. 6B). Controls with ΔRep and ΔVP extracts and precipitation with a control antibody demonstrated the specificity of the precipitations.
FIG. 6.
Increased capacity of the R432A mutant to form complexes between capsids and Rep proteins. 293T cells were transfected with AAV2 wt or R432A-mutated genomic plasmids. As controls, cells were transfected with plasmids expressing only VPs (pCMV-VP [ΔRep]) or only Rep (pDGΔVP [ΔVP]) proteins. (A) Cell extracts were prepared in RIPA buffer and analyzed for viral protein expression by Western blot analysis using monoclonal anti-Rep (303.9) or anti-VP (B1) antibodies. (B) Rep-capsid complexes were immunoprecipitated from RIPA extracts by using a polyclonal anti-Rep antiserum (αRep90) or a capsid-specific monoclonal antibody (A20) and analyzed by Western blotting to detect coprecipitated capsids (top panels) or Rep proteins (bottom panels). Control precipitations were also performed with a non-AAV-related polyclonal antiserum (control) (top panels) or a non-AAV-related monoclonal antibody (control) (bottom panels). (C) Extracts from 293T cells transfected with an AAV2 wt or R432A-mutated genomic plasmid were analyzed on continuous sucrose gradients (10 to 30% sucrose) in the absence of detergents. AAV2 proteins were detected by Western blot analysis using monoclonal anti-Rep (303.9) or anti-VP (B1) antibodies. The 110S and 60S positions were determined by using purified DNA-containing and empty AAV2 particles, respectively.
The possibly increased association of Rep proteins with capsids was also analyzed by cosedimentation of Rep proteins with capsids (Fig. 6C). Extracts of cells transfected with plasmids harboring either an AAV2 wt genome or a R432A mutant genome were fractionated by sucrose gradient centrifugation and analyzed by Western blotting using VP-specific (B1) and Rep protein-specific (303.9) antibodies. Figure 6C shows that significantly higher levels of Rep78 and Rep52 cosedimented with capsids of the R432A mutant compared to wt. This observation further supports the assumption that the R432A mutant shows altered, increased interactions between Rep proteins and capsids. The increased Rep-capsid association does not, however, provide a conclusive explanation for the packaging defect of this mutant.
Altered capsid-conformation of the R432A mutant indicates occupation of fivefold pores by VP N termini.
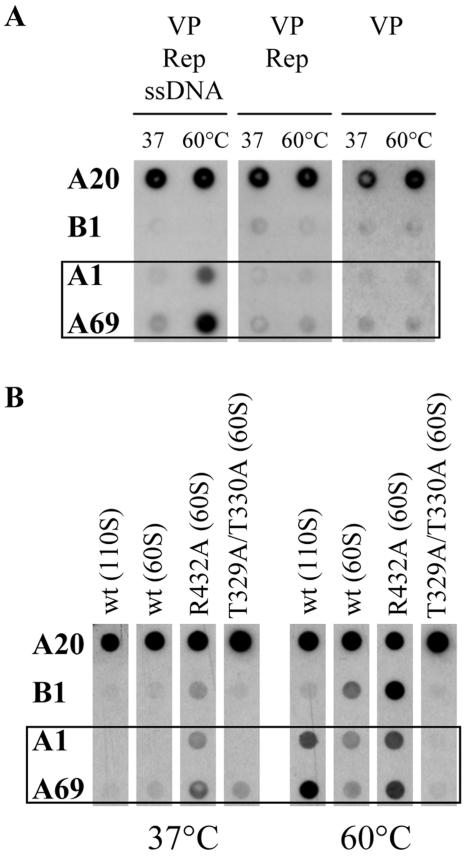
We could previously show that N termini of VP1 and VP2, probably located in globular structures at the twofold axes inside the capsid, become exposed through the fivefold pores of DNA containing capsids upon a short heat shock (26, 3). This is not the case for recombinant empty capsids (26) or capsids assembled in presence of all packaging components except DNA (Fig. 7A). Hence, the preencapsidated and encapsidated stages of the capsids are defined by different structural conformations. Based on this premise, we speculated that mutation of R432, which is located at the interaction surface of neighboring pentamers, may have an effect on these conformations. We therefore examined the R432A mutant in a native dot blot assay for its ability to undergo a conformational change upon a short heat treatment (Fig. 7B). For comparison, we included the other strong packaging mutant, the T329A/T330A mutant, in this analysis. As outlined previously (3), reactions of monoclonal antibodies with their defined epitopes (45) were indicative of the confomational changes occurring within intact capsids. While antibody A20 recognizes whole capsids, antibody A1 detects an epitope on the N terminus of VP1, antibody A69 reacts with an epitope common to VP1 and VP2 and antibody B1 recognizes the very C-terminal part of all three VPs. Extracts of cells transfected with plasmids either harboring an AAV2 wt genome or a genome with the respective packaging mutation were fractionated by sucrose gradient centrifugation. Aliquots of the 60S and 110S fractions, containing empty and DNA-containing particles, respectively, were incubated for 5 min at 37 or 60°C and then transferred to nitrocellulose membranes under nondenaturing conditions. As previously shown (3), N termini of VP1 and VP2 are not detectable at 37°C and become exposed on intact DNA-containing AAV2 wt capsids after treatment at 60°C (wt 110S fraction) (Fig. 7B). A slightly different behavior could be detected for AAV2 wt 60S fractions (Fig. 7B) comprising unpackaged, partially packaged, and defective interfering particles, which showed less accessible N termini and, additionally, a reaction with the C terminus-specific antibody B1 after 60°C incubation. The packaging-deficient T329A/T330A pore mutant was not able to expose the VP1 and VP2 N termini after 60°C treatment (T329A/T330A 60S) (Fig. 7B), which confirms our previously published data that most mutations of pore-forming amino acids impede the ability of AAV2 capsids to externalize their VP1 N termini (3). In sharp contrast to these mutants, capsids of the R432A packaging mutant showed a reaction with the N terminus-specific antibodies (A1 and A69) and the C terminus-specific antibody (B1) already at 37°C (R432A 60S) (Fig. 7B). This reaction was increased after incubation of mutant capsids at 60°C. This result suggests that the R432A mutant is not able to encapsidate genomes because the pores at the fivefold axes are occupied by VP1 and VP2 N termini, thereby blocking the portal for genome encapsidation. In addition, this altered capsid conformation may allow binding of Rep proteins to more than one site of the capsid.
FIG. 7.
Analysis of capsid conformations at the fivefold pores. Extracts from 293T cells transfected with an AAV2 wt or mutated genomic plasmid were fractionated by sucrose gradient centrifugation (10 to 30% sucrose). Aliquots of the 110S (DNA-containing particles) and 60S (empty or partially packaged particles) fractions were incubated at the designated temperatures for 5 min, applied to nitrocellulose membranes under nondenaturing conditions, and then reacted with antibodies A20, B1, A1, and A69. (A) Analyzed were the 110S fraction of lysates obtained from cells which were transfected with AAV2 wt genomic plasmids (pDM [VP Rep ssDNA]) as well as the 60S fractions obtained from lysates of cells transfected with a plasmid expressing AAV2 Rep and VPs (pDG [VP Rep]) or a plasmid expressing only VPs (pCMV-VP [VP]). (B) The 110S and 60S fractions of lysates obtained from cells which were transfected with AAV2 wt genomic plasmids (pTAV2.0 [wt]) were compared with the 60S fractions obtained from lysates of cells transfected with genomic plasmids of the packaging-deficient R432A and T329A/T330A mutants.
DISCUSSION
In this report, we have compared three types of AAV2 packaging mutants with respect to their patterns of capsid-associated and encapsidated DNA and to their Rep-capsid interactions in a productive infection. The small Rep-deficient mutant was used as a reference for normal Rep-capsid interactions but reduced and incomplete packaging of capsid-associated genomes (25). A panel of mutants with exchanges of amino acids forming the pores at the fivefold axes of symmetry all showed reduced but complete DNA encapsidation linked with reduced Rep-capsid interactions. Finally, a capsid packaging mutant (46) with an amino acid exchange located at the capsid protein interface (R432A) showed very small amounts of capsid-associated DNA and no detectable packaging. Surprisingly, this mutant was characterized by an increased association of Rep proteins with capsids. These results provoke the question if these seemingly contradicting results can be interpreted on the basis of the currently established genome packaging model for AAV2. The model postulates that (i) ssDNA becomes encapsidated into preformed capsids (31), (ii) the docking of the genomes to the capsids involves targeting of the AAV inverted terminal repeats (ITRs) to the capsid by binding of Rep proteins to both the capsid and the ITRs (9), (iii) the translocation into the capsid involves Rep helicase/ATPase activity (25, 48), (iv) translocation occurs in the 3′ to 5′ direction (25), and (v) one pore at the fivefold symmetry axis of the capsid may function as a portal for genome introduction (3). Supportive evidence for several of these aspects has also been reported for autonomous parvoviruses (7, 10, 49).
If a pore at the fivefold symmetry axis acts as the portal for genome encapsidation into preformed capsids (3), one would postulate that one should be able to plug the pore by introducing bulky amino acid residues into the channel. This should prevent packaging completely rather than reduce it five- to ninefold, as was observed previously (3). We intended to generate such mutants by placing tryptophan residues at the most narrow positions of the channel (V221W, N334W, and L336W) or by introducing a tryptophan at a position of the channel opposite to the amino acids exchanged in the strongest packaging pore mutants which we have described previously (D219A/L336W and V221W/S338A mutants) (3). Two of these mutants showed no detectable capsid assembly (N334W and S338A/V221W mutants) in correlation with reduced capsid protein steady-state levels. We already noticed previously that some mutations of pore-forming amino acids resulted in reduced capsid protein levels and reduced capsid assembly (3). It is not clear if reduced assembly is a consequence of low capsid protein levels due to increased protein instability of the mutant protein itself or if the low capsid protein level is a consequence of inhibited capsid assembly and greater accessibility of nonassembled VPs to protein degradation. If the latter is the case, these mutants would point to pentamer formation as a sensitive step in the capsid assembly process. It is less likely that the observation of reduced Rep68 and Rep40 levels in three assembly mutants (V221W, N334W, and S338A/V221W mutants) indicates reduced splicing and hence reduced capsid protein expression, because reduced Rep68 levels were not observed in three other assembly mutants (3).
The three other capsid pore mutants, the V221W, L336W, and D219/L336W mutants, showed reduced packaging but again by only three- to fivefold. Molecular modeling of the amino acid exchanges into the capsid structure suggests that passage of the DNA through the channel is not completely prevented. If one assumes that the pores have some flexibility in diameter, bulky amino acids could lead only to a hindered or slowed passage through the pore into the capsid. However, if packaging is more slowed down in mutants with a stronger packaging defect, one would expect an increased amount of genomes which are not completely packaged in these mutants. This was not observed for any of the packaging pore mutants. Alternatively, reduced but complete packaging could be explained by the assumption that the pore mutations affect docking of Rep-DNA complexes to the capsid. This interpretation is supported by combined mutation of amino acids T329 and T330, which are located close to the outer rim of the pore, to alanine. The T329A/T330A mutant showed the strongest reduction in encapsidated genomes and the strongest reduction in Rep-capsid interaction. However, as mentioned above, the small amount of capsid-coprecipitated DNA seemed to be completely encapsidated. Also, the other relatively strong packaging pore mutants analyzed for Rep-capsid interaction showed complete packaging but a reduced interaction in the Rep-capsid coimmunoprecipitation assay. Contrary, Rep-capsid coprecipitation was comparable to that of wt in extracts of cells transfected with a pore mutant which had no packaging phenotype as well as with the small Rep-deficient mutant or with small Rep mutants with a defect in the helicase motor activity (see also reference 48). This indicates that mutations in the pore-forming amino acids which interfered with packaging may lead—directly or indirectly—to a reduced number of successful docking events of Rep-bound genomes to the capsid. It further suggests that the interaction of Rep proteins with the capsids may rely upon the entire structure of the pore, since mutations of amino acids which are located deeper inside the cylinder also interfered with these interactions. If, however, a packaging initiation complex was formed, genome encapsidation seemed to occur more or less normally. This does not exclude that, additionally, passage through the packaging portal could be affected by the mutations. However, this hindrance was not strong enough that larger amounts of partially packaged genomes were detected.
Other interpretations are conceivable. Rep-capsid coprecipitation could indicate the result of reduced packaging and not the cause of reduced packaging due to reduced initiation of packaging. In other words, the Rep-cap coprecipitations could be the result of Rep proteins passively bound at the 5′ end of the DNA in the packaging process (33, 34). A similar observation has been made for NS1 associated with the capsid of minute virus of mice (8). If this is the case, the packaging machinery would have to be host cell-derived because the virus does not encode further energy-converting proteins apart from Rep proteins. In addition, the increased Rep protein coprecipitation of the R432A mutant contradicts this interpretation because in this mutant, no significant amounts of DNA could be coprecipitated with the capsids which could bring down DNA-bound Rep proteins. Also, the fact that Rep52, which does not stably associate with the ITRs, is coprecipitated with the capsid argues for capsid-associated rather than DNA- associated Rep proteins. Another alternative interpretation suggests that the DNA enters the capsid through a pore which is not visible on those capsids which have been structurally analyzed so far, which means on capsids after completion of packaging or on empty capsids before packaging. We observed upon heat treatment at 65°C a capsid species sedimenting slightly below 60S (3). This resembles the sedimentation characteristics of the proposed packaging intermediate “SP” observed by Myers and Carter (31). This capsid species did react with the C terminus-specific antibody B1 and had lost its DNA. However, we could not identify such capsids, under the conditions of a productive infection, in the region of the 60S fractions by using the antibody B1 in the native dot blot assay (Fig. 7A). The opening of a packaging portal at the threefold symmetry axis also cannot be excluded. In this case, all the mutations at the fivefold axis would have to have an indirect effect on packaging at the putative portal at the threefold axis, which is unlikely. Furthermore, there are several other viruses with icosahedral capsids—such as bacteriophages, herpesviruses, and picornaviruses—which encapsidate their genomes through a packaging portal at 1 of the 12 fivefold vertices (16, 29). Visualization of the packaging complex of AAV2 by structural analysis could definitively clarify the question where the packaging portal of this virus is located. However, the presented data strongly support our hypothesis that one of the channels at the fivefold axes acts as the genome entry site.
Further support for our packaging model implying Rep docking to the capsid and usage of the pore at the fivefold axis of symmetry as a packaging portal comes from the R432A packaging mutant, which had first been described by Wu et al. (46). As mentioned above, this mutant demonstrated increased Rep binding in the absence of DNA, supporting the Rep-capsid interaction concept. However, why then is packaging prevented? The most plausible interpretation is supplied by the observation that at 37°C the N termini of VP1 and VP2 are already accessible to antibodies A1 and A69 (Fig. 7B), indicating that the pores are occupied by these protein domains and may not permit introduction of the DNA genome from the opposite direction. The reaction of this mutant with the B1 antibody in addition indicates an altered capsid conformation, which may explain the increased ability of the mutant capsid to bind Rep proteins, possibly by interaction of Rep proteins with binding sites at more than 1 of the 12 fivefold vertices. This, however, does not explain why so little DNA is associated with capsid-bound Rep proteins. We assume that detection of capsid-associated and not encapsidated DNA is based on Rep-Rep interactions (9) which might not be very stable and might also depend on a defined composition of the Rep-capsid complexes to which Rep proteins in complex with the genome bind. In this context, it is worthwhile to mention that the amount of capsid-associated but not encapsidated full-length DNA in the small Rep-deficient mutant—which is believed to occur by protein-protein interactions—is also relatively low compared to the partially encapsidated DNA, which is more stably associated with the capsid.
Our analysis of AAV packaging mutants provides evidence that mutations in the fivefold pore and in particular at the rim of the pore leads to a reduced Rep-capsid interaction which in turn results in reduced packaging initiation. This observation suggests that Rep binding to the capsid occurs probably at or close to the pores at the fivefold axes. Most amino acids forming the pores are highly conserved among the 11 different AAV serotypes sequenced so far (Fig. 8). Surprisingly, T329 and T330 are not conserved, although they exhibited the strongest packaging defect when they were both mutated to alanine. However, these amino acids are never exchanged together or for alanine in different serotypes, and mutation of T329A/T330A might have an indirect effect on the conserved T331 of the pore structure. Therefore, if packaging occurs by binding of Rep-DNA complexes in this region, one could understand the high degree of cross-packaging of AAV2 vector genomes into capsids of different serotypes (1, 13, 35). In addition, the interaction domain of Rep proteins with the capsids was mapped to the ATPase/helicase motif (9), which is also conserved among different AAV serotypes.
To permit the use of the pores, both as a packaging portal during virus assembly and as a channel to expose infection-relevant N-terminal domains during infection (3), two restrictions of the pore structure have to be maintained: (i) empty capsids—with or without Rep proteins bound—should not allow exposure of the VP1/VP2 N termini (this has been observed in this study and in a previous study [26]), and (ii) binding of Rep to the capsid has to be restricted to one pore at the capsid. This could be achieved by introduction of a specific capsid conformation upon occupation of one pore by Rep proteins or by binding of so far unidentified factors that prevent packaging initiation at multiple sites. Our previous observation that Rep-capsid complex formation was observed only when Rep proteins and capsid proteins were present together during capsid assembly may be relevant to explain this restriction and support the first hypothesis. Rep-capsid complexes could not be formed by adsorption of Rep to preformed capsids (9), suggesting that Rep-capsid complex formation is a coassembly process in which binding of Rep proteins to one pore may prevent further binding to the other pores. It seems possible that in the R432A mutant, this conformation-dependent restriction is not maintained. In conclusion, the data presented here support the idea that initiation of packaging involves the interaction of Rep-bound genomes with Rep-capsid complexes and that this process greatly depends on a defined conformation of preassembled capsids.
Acknowledgments
We thank S. Kronenberg for generating the R432A mutant and K. Schmidt for technical assistance. We are grateful to W. Von der Lieth for modeling the channel diameter of selected mutants.
REFERENCES
- 1.Auricchio, A., G. Kobinger, V. Anand, M. Hildinger, E. O'Connor, A. M. Maguire, J. M. Wilson, and J. Bennett. 2001. Exchange of surface proteins impacts on viral vector cellular specificity and transduction characteristics: the retina as a model. Hum. Mol. Genet. 10:3075-3081. [DOI] [PubMed] [Google Scholar]
- 2.Becerra, S. P., F. Koczot, P. Fabisch, and J. A. Rose. 1988. Synthesis of adeno-associated virus structural proteins requires both alternative mRNA splicing and alternative initiations from a single transcript. J. Virol. 62:2745-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bleker, S., F. Sonntag, and J. A. Kleinschmidt. 2005. Mutational analysis of narrow pores at the fivefold symmetry axes of adeno-associated virus type 2 capsids reveals a dual role in genome packaging and activation of phospholipase A2 activity. J. Virol. 79:2528-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cassinotti, P., M. Weitz, and J. D. Tratschin. 1988. Organization of the adeno-associated virus (AAV) capsid gene: mapping of a minor spliced mRNA coding for virus capsid protein 1. Virology 167:176-184. [PubMed] [Google Scholar]
- 5.Catalano, C. E. 2000. The terminase enzyme from bacteriophage lambda: a DNA-packaging machine. Cell Mol. Life Sci. 57:128-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chejanovsky, N., and B. J. Carter. 1989. Mutagenesis of an AUG codon in the adeno-associated virus rep gene: effects on viral DNA replication. Virology 173:120-128. [DOI] [PubMed] [Google Scholar]
- 7.Cotmore, S. F., and P. Tattersall. 2005. Encapsidation of minute virus of mice DNA: aspects of the translocation mechanism revealed by the structure of partially packaged genomes. Virology 336:100-112. [DOI] [PubMed] [Google Scholar]
- 8.Cotmore, S. F., and P. Tattersall. 1989. A genome-linked copy of the NS-1 polypeptide is located on the outside of infectious parvovirus particles. J. Virol. 63:3902-3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubielzig, R., J. A. King, S. Weger, A. Kern, and J. A. Kleinschmidt. 1999. Adeno-associated virus type 2 protein interactions: formation of pre encapsidation complexes. J. Virol. 73:8989-8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farr, G. A., and P. Tattersall. 2004. A conserved leucine that constricts the pore through the capsid fivefold cylinder plays a central role in parvoviral infection. Virology 323:243-256. [DOI] [PubMed] [Google Scholar]
- 11.Fujisawa, H., and M. Morita. 1997. Phage DNA packaging. Genes Cells 2:537-545. [DOI] [PubMed] [Google Scholar]
- 12.Girod, A., C. E. Wobus, Z. Zadori, M. Ried, K. Leike, P. Tijssen, J. A. Kleinschmidt, and M. Hallek. 2002. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J. Gen. Virol. 83:973-978. [DOI] [PubMed] [Google Scholar]
- 13.Grimm, D. 2002. Production methods for gene transfer vectors based on adeno-associated virus serotypes. Methods 28:146-157. [DOI] [PubMed] [Google Scholar]
- 14.Grimm, D., A. Kern, M. Pawlita, F. Ferrari, R. Samulski, and J. Kleinschmidt. 1999. Titration of AAV-2 particles via a novel capsid ELISA: packaging of genomes can limit production of recombinant AAV-2. Gene Ther. 6:1322-1330. [DOI] [PubMed] [Google Scholar]
- 15.Grimm, D., A. Kern, K. Rittner, and J. A. Kleinschmidt. 1998. Novel tools for production and purification of recombinant adenoassociated virus vectors. Hum. Gene Ther. 9:2745-2760. [DOI] [PubMed] [Google Scholar]
- 16.Guo, P. 1994. Principles, perspectives and potential applications in viral assembly. Semin. Virol. 5:27-37. [Google Scholar]
- 17.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual, 1988 ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Heilbronn, R., A. Burkle, S. Stephan, and H. zur Hausen. 1990. The adeno-associated virus rep gene suppresses herpes simplex virus-induced DNA amplification. J. Virol. 64:3012-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hendrix, R. W. 1998. Bacteriophage DNA packaging: RNA gears in a DNA transport machine. Cell 94:147-150. [DOI] [PubMed] [Google Scholar]
- 20.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 21.Hunter, L. A., and R. J. Samulski. 1992. Colocalization of adeno-associated virus Rep and capsid proteins in the nuclei of infected cells. J. Virol. 66:317-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.James, J. A., C. R. Escalante, M. Yoon-Robarts, T. A. Edwards, R. M. Linden, and A. K. Aggarwal. 2003. Crystal structure of the SF3 helicase from adeno-associated virus type 2. Structure (Cambridge) 11:1025-1035. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, F. B., H. L. Ozer, and M. D. Hoggan. 1971. Structural proteins of adenovirus-associated virus type 3. J. Virol. 8:860-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kern, A., K. Schmidt, C. Leder, O. J. Muller, C. E. Wobus, K. Bettinger, C. W. Von der Lieth, J. A. King, and J. A. Kleinschmidt. 2003. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J. Virol. 77:11072-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.King, J. A., R. Dubielzig, D. Grimm, and J. A. Kleinschmidt. 2001. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J. 20:3282-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kronenberg, S., B. Bottcher, C. W. von der Lieth, S. Bleker, and J. A. Kleinschmidt. 2005. A conformational change in the adeno-associated virus type 2 capsid leads to the exposure of hidden VP1 N termini. J. Virol. 79:5296-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kronenberg, S., J. A. Kleinschmidt, and B. Bottcher. 2001. Electron cryo-microscopy and image reconstruction of adeno-associated virus type 2 empty capsids. EMBO Rep. 2:997-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laughlin, C. A., J. D. Tratschin, H. Coon, and B. J. Carter. 1983. Cloning of infectious adeno-associated virus genomes in bacterial plasmids. Gene 23:65-73. [DOI] [PubMed] [Google Scholar]
- 29.Moore, S. D., and P. E. Prevelige, Jr. 2002. DNA packaging: a new class of molecular motors. Curr. Biol 12:R96-R98. [DOI] [PubMed] [Google Scholar]
- 30.Muzyczka, N., and K. I. Berns. 2001. Parvoviridae: the viruses and their replication, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 31.Myers, M. W., and B. J. Carter. 1980. Assembly of adeno-associated virus. Virology 102:71-82. [DOI] [PubMed] [Google Scholar]
- 32.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prasad, K. M., and J. P. Trempe. 1995. The adeno-associated virus Rep78 protein is covalently linked to viral DNA in a preformed virion. Virology 214:360-370. [DOI] [PubMed] [Google Scholar]
- 34.Prasad, K. M., C. Zhou, and J. P. Trempe. 1997. Characterization of the Rep78/adeno-associated virus complex. Virology 229:183-192. [DOI] [PubMed] [Google Scholar]
- 35.Rabinowitz, J. E., F. Rolling, C. Li, H. Conrath, W. Xiao, X. Xiao, and R. J. Samulski. 2002. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J. Virol. 76:791-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sayle, R. A., and E. J. Milner-White. 1995. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 20:374. [DOI] [PubMed] [Google Scholar]
- 37.Shi, W., G. S. Arnold, and J. S. Bartlett. 2001. Insertional mutagenesis of the adeno-associated virus type 2 (AAV2) capsid gene and generation of AAV2 vectors targeted to alternative cell- surface receptors. Hum. Gene Ther. 12:1697-1711. [DOI] [PubMed] [Google Scholar]
- 38.Srivastava, A., E. W. Lusby, and K. I. Berns. 1983. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J. Virol. 45:555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Timpe, J., J. Bevington, J. Casper, J. D. Dignam, and J. P. Trempe. 2005. Mechanisms of adeno-associated virus genome encapsidation. Curr. Gene Ther. 5:273-284. [DOI] [PubMed] [Google Scholar]
- 40.Trempe, J. P., and B. J. Carter. 1988. Alternate mRNA splicing is required for synthesis of adeno-associated virus VP1 capsid protein. J. Virol. 62:3356-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valpuesta, J. M., and J. L. Carrascosa. 1994. Structure of viral connectors and their function in bacteriophage assembly and DNA packaging. Q. Rev. Biophys. 27:107-155. [DOI] [PubMed] [Google Scholar]
- 42.Weger, S., A. Wistuba, D. Grimm, and J. A. Kleinschmidt. 1997. Control of adeno-associated virus type 2 cap gene expression: relative influence of helper virus, terminal repeats, and Rep proteins. J. Virol. 71:8437-8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wistuba, A., A. Kern, S. Weger, D. Grimm, and J. A. Kleinschmidt. 1997. Subcellular compartmentalization of adeno-associated virus type 2 assembly. J. Virol. 71:1341-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wistuba, A., S. Weger, A. Kern, and J. A. Kleinschmidt. 1995. Intermediates of adeno-associated virus type 2 assembly: identification of soluble complexes containing Rep and Cap proteins. J. Virol. 69:5311-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wobus, C. E., B. Hugle-Dorr, A. Girod, G. Petersen, M. Hallek, and J. A. Kleinschmidt. 2000. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J. Virol. 74:9281-9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu, P., W. Xiao, T. Conlon, J. Hughes, M. Agbandje-McKenna, T. Ferkol, T. Flotte, and N. Muzyczka. 2000. Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J. Virol. 74:8635-8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie, Q., W. Bu, S. Bhatia, J. Hare, T. Somasundaram, A. Azzi, and M. S. Chapman. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. USA 99:10405-10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoon-Robarts, M., A. G. Blouin, S. Bleker, J. A. Kleinschmidt, A. K. Aggarwal, C. R. Escalante, and R. M. Linden. 2004. Residues within the B' motif are critical for DNA binding by the superfamily 3 helicase Rep40 of adeno-associated virus type 2. J. Biol. Chem. 279:50472-50481. [DOI] [PubMed] [Google Scholar]
- 49.Yuan, W., and C. R. Parrish. 2001. Canine parvovirus capsid assembly and differences in mammalian and insect cells. Virology 279:546-557. [DOI] [PubMed] [Google Scholar]