Abstract
NYVAC has been engineered as a safe, attenuated vaccinia virus (VV) vector for use in vaccination against a broad spectrum of pathogens and tumors. Due to the interest in NYVAC-based vectors as vaccines and current phase I/II clinical trials with this vector, there is a need to analyze the human host response to NYVAC infection. Using high-density cDNA microarrays, we found 368 differentially regulated genes after NYVAC infection of HeLa cells. Clustering of the regulated genes identified six discrete gene clusters with altered expression patterns. Clusters 1 to 3 represented 47.5% of the regulated genes, with three patterns of gene activation kinetics, whereas clusters 4 to 6 showed distinct repression kinetics. Quantitative real-time reverse transcription-PCR analysis of selected genes validated the array data. Upregulated transcripts correlated with genes implicated in immune responses, including those encoding interleukin-1 receptor 2 (IL-1R2), IL-6, ISG-15, CD-80, and TNFSF7. NYVAC upregulated several intermediates of apoptotic cascades, including caspase-9, correlating with its ability to induce apoptosis. NYVAC infection also stimulated the expression of NF-κB1 and NF-κB2 as well as that of NF-κB target genes. Expression of the VV host range K1L gene during NYVAC infection prevented NF-κB activation, but not the induction of apoptosis. This study is the first overall analysis of the transcriptional response of human cells to NYVAC infection and provides a framework for future functional studies to evaluate this vector and its derivatives as human vaccines.
Attenuated strains of vaccinia virus (VV) were developed in response to the need for safer vaccines (37). NYVAC is a derivative of the VV Copenhagen strain in which 18 open reading frames were specifically deleted from the parental viral genome; genes involved in host range, virulence, and pathogenesis were thus lost (49). NYVAC-derived vectors are able to express antigens from a wide range of species (50). A number of examples have been reported, using NYVAC as a recombinant vaccine delivery system against pathogens and tumors (5, 10, 31, 46). Clinical trials using NYVAC-based vectors show a good safety profile, with induction of high levels of immunity against heterologous antigens (24, 34).
Since phase I/II clinical trials using this vector are currently under way, particularly for human immunodeficiency virus type 1 (HIV-1) (www.eurovacc.org), it is essential to obtain a comprehensive understanding of the effect of NYVAC infection on human host gene expression (34). DNA microarray technology allows monitoring of the expression of several thousand individual genes (22) and has been used to identify the differential expression of cellular genes in response to infection by several animal viruses, including VV strains WR (Western Reserve) and MVA (modified vaccinia Ankara) (15, 16, 29, 55).
Host genes that govern vital cell processes such as replication, transcription, and translation are downregulated during the course of WR infection. The expression of genes involved in apoptosis or the proteasome-ubiquitin degradation pathway is also repressed; few genes are upregulated after WR infection (15). A larger number of genes than that seen with WR is upregulated during strain MVA infection; most encode immune modulator proteins, some of which may be involved in host resistance and immune modulation during MVA infection (16). Wiskott-Aldrich syndrome protein (WASP) family members are upregulated after MVA or WR infection (15, 16). This family includes the gene that codes for the N-WASP protein, implicated in the actin-mediated motility of VV as a mechanism for intercellular viral spreading (8, 11). Additional studies show that WASP is required for VV pathogenesis (17). The results of microarray analysis have implicated cell signaling events and cell proteins as important regulators of VV infection.
For this study, we used cDNA microarray technology to analyze host gene expression changes in HeLa cell cultures following NYVAC infection. This is the first large-scale analysis of the transcriptional response of HeLa cells to NYVAC infection. It provides a better understanding of the mechanisms of vaccine protection against VV infection and will aid in the development of NYVAC recombinant-based vaccines against pathogens and tumors.
MATERIALS AND METHODS
Cells, viruses, and infection conditions.
HeLa cells (ATCC) were cultured in Dulbecco's medium supplemented with 10% newborn bovine serum and antibiotics. The VV WR strain was cultured in monkey BSC-40 cells, purified by sucrose gradient banding, and titrated on BSC-40 cells by a plaque assay. The NYVAC and MVA strains were cultured in BHK-21 cells, purified by sucrose gradient banding, and titrated on BHK-21 cells by immunostaining of fixed infected cultures with a polyclonal anti-VV protein antibody (12).
Microarray production.
The Research Genetics 40K sequence-verified human cDNA clone library (http://www.resgen.com/products/SVHcDNA.php3) was used to generate cDNA arrays as described previously (15). Slides contained 15,360 cDNAs, of which 13,295 correspond to known genes and 2,065 correspond to control genes. The cDNAs were printed on CMT-GAPS II slides (Corning) with a Microgrid II apparatus (BioRobotics).
Microarray hybridization.
Total RNA was isolated from purified NYVAC-infected (5 PFU/cell) or mock-infected HeLa cells cultured in 10-cm plates, using Ultraspect-II RNA (Biotecx). RNAs were then purified with Megaclear (Ambion), and their integrity was confirmed using an Agilent 2100 bioanalyzer (Agilent). Uninfected samples were isolated at each infection time point and processed in parallel with infected cells. Two independent replicates were processed for analysis. Total RNA (1.5 μg for each replicate) was amplified using an amino allyl MessageAmp aRNA kit (Ambion), and approximately 25 to 40 μg of amplified RNA (aRNA) was obtained. The mean aRNA size was 1,500 nucleotides. For each sample, 5 μg aRNA was labeled with Cy3 or Cy5 (CyDye postlabeling reactive dye pack; Amersham) and purified. We measured Cy3 and Cy5 incorporation using 1 μl of probe in a Nanodrop spectrophotometer (Nanodrop Technologies Inc.). For each hybridization, Cy3 and Cy5 probes (150 pmol each) were mixed, dried in a Speed-Vac machine, and resuspended in 9 μl of RNase-free water. Two dye-swapped hybridizations were performed; in one, the mock-infected sample was Cy3 labeled and the NYVAC-infected sample was Cy5 labeled; in the second, the labeling was reversed. Double labeling was used to abolish dye-specific labeling and hybridization differences. Slides were prehybridized and hybridized as described previously (15, 16), dried by centrifugation, and scanned on an Axon 4000B instrument (Axon). Images and raw data were obtained using GenePix 5.0 software (Axon) and were processed using SOLAR software (Bioalma, Spain). Briefly, the background was subtracted from the signal, log10 (signal) values were plotted versus log2 (ratio) values, and Lowess normalization was used to adjust most spots to a log ratio of 0. This was calculated for all four replicates, and a table was obtained with mean signals, degrees of change, log ratios, standard deviations of the log ratios, and z scores (40).
Gene expression analysis.
The original data set, containing 13,295 clones per slide, was prepared for clustering. Genes with an interreplicate standard deviation of >1 were removed from the analysis. The resulting data set was reduced to 8,722 transcripts that showed consistent expression values among the four replicates. The z score (a measure of the proximity of one value [log ratio] to other values with similar signals) was used to eliminate genes that did not show significant expression under at least one experimental condition (40). Only genes with a z score of >2 were thus selected for clustering. A new data set was created with the 368 transcripts that passed the filter. After data preprocessing, genes were clustered using Kohonen's self-organizing map (25). The resulting seven-by-five map was analyzed using the Engene software package (13) at http://www.engene.cnb.uam.es.
Quantitative real-time RT-PCR.
RNAs (1 μg) were reverse transcribed using the Superscript first-strand synthesis system for reverse transcription-PCR (RT-PCR) (Invitrogen). A 1:40 dilution of the RT reaction mixture was used for quantitative PCR. The primer and probe sets used to amplify the genes for PCNT2, WASL, NF-κB2, interleukin-7 (IL-7), IL-6, gamma interferon (IFN-γ), CASP-9, ATF-3, GADD34, and APEXL2 were purchased from Applied Biosystems. RT-PCRs were performed according to Assay-on-Demand, optimized for TaqMan Universal PCR master mix with no AmpErase UNG (15, 16). All samples were assayed in duplicate. Threshold cycle (CT) values were used to plot a standard curve in which the CT decreased in linear proportion to the log of the template copy number. Correlation values of standard curves were always >99%.
Gel retardation assay.
Nuclear extracts (3 μg) from HeLa cells grown in 6-cm-well plates, either uninfected or infected with WR, MVA, or NYVAC at 5 PFU/cell, were analyzed using the synthetic [α-32P]dCTP-labeled double-stranded wild-type HIV enhancer oligonucleotide 5′-AGCTTACAAGGGACTTTCCGCTGGGGACTTTCCAGGGA-3′, containing two κB consensus motifs, as described previously (3). For supershift assays, anti-p50 antibody (1 μl; Santa Cruz) was added 15 min before the labeled probe.
ELISA.
IL-6 secreted into the medium of NYVAC-, MVA-, or WR-infected HeLa cells (5 PFU/cell) in 12-well plates was measured with a quantitative human IL-6 kit (BD Biosciences). Supernatants (100 μl) from uninfected or infected HeLa cells at 2, 6, and 16 h postinfection (hpi) were used for enzyme-linked immunosorbent assays (ELISAs). Captured IL-6 was quantified at 450 nm in a spectrophotometer. Duplicate samples were measured in two independent experiments.
Apoptosis assay.
HeLa cells cultured in 12-well plates were infected (5 PFU/cell) with the indicated viruses and harvested at 24 hpi for determination of the absorbance (405 nm). Cell death was detected with an ELISA kit (Roche) which uses mouse anti-DNA and -histone monoclonal antibodies to estimate the amount of cytoplasmic histone-associated DNA.
Western blotting.
HeLa cells grown in six-well plates were infected (5 PFU/cell) with WR, MVA, or NYVAC and collected, and cell extracts were prepared at 2, 6, and 16 hpi by lysis in buffer (50 mM Tris-HCl, pH 8.0, 0.5 M NaCl, 10% NP-40, 1% sodium dodecyl sulfate [SDS]) on ice for 5 min. Protein lysates (100 μg) were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) on 14% or 8% gels, transferred to nitrocellulose membranes, and incubated with the following antibodies: anti-caspase-9 (Oncogene), -PARP (Cell Signaling), -actin (Santa Cruz), -E3L (a gift from B. L. Jacobs), -A14L (43), -NF-κB1, which recognizes p105 and p50 (Santa Cruz), -IκBα (Santa Cruz), -ATF-3 (Santa Cruz), -initiation factor 2α-P (IF-2α-P; Biosource), and -eIF-2α (Santa Cruz), followed by secondary antibodies (mouse and rabbit peroxidase conjugates). Protein expression was detected using ECL reagents (Amersham).
Immunofluorescence.
HeLa cells cultured on coverslips were infected with NYVAC or WR (5 PFU/cell). At 6 and 16 hpi, cells were washed with phosphate-buffered saline, fixed with 4% paraformaldehyde, and permeabilized with 0.1% Triton X-100 in phosphate-buffered saline (room temperature, 10 min). Cells were incubated with the primary antibody anti-ATF-3, -caspase-9 (Calbiochem), -T7-tag (Novagen), or -E3L, followed by fluorescein- or Texas red-conjugated isotype-specific secondary antibodies and the DNA staining reagent ToPro (Molecular Probes). Images were obtained using a Bio-Rad Radiance 2100 confocal laser microscope.
Transfection assay.
HeLa cells cultured on coverslips were infected (0.1 PFU/cell) with NYVAC, and after 1 h, were transfected with the expression plasmid pK1L (3 μg DNA) using JetPEI (Q-Biogene). At 40 h posttransfection, cells were fixed and analyzed by immunofluorescence. The pK1L expression plasmid was derived from pMJ601 by insertion of the VV (WR strain) K1L gene, containing the code for T7-Tag at the N terminus, under the transcriptional control of the VV early-late promoter p7.5 (kindly provided by A. Alcamí). The transfection efficiency was approximately 80%, with ≤5% interexperiment variation.
RESULTS
Cellular gene expression pattern after NYVAC infection.
cDNA microarray technology was used to analyze the overall changes in human cellular gene expression following NYVAC infection. Due to the interest in NYVAC as a live recombinant vaccine vector, it is important to define the effects of NYVAC infection on human host gene expression. We thus used human HeLa cells, as many fundamental studies in poxvirus biology are based on this cell line.
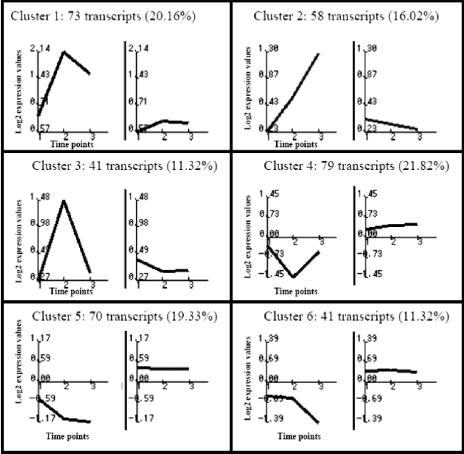
We used chips carrying cDNAs from 13,295 human genes to profile gene expression in HeLa cells which were mock infected or infected for 2, 6, and 16 h. We analyzed mRNA duplicates isolated at 2, 6, and 16 hpi in two independent experiments and identified 368 genes that were differentially expressed after NYVAC infection, which represent 2.8% of the genes in the array. Following detailed analysis of their profiles, we grouped these genes into six main clusters according to their behavior at three time points during NYVAC infection. The expression clusters of genes that were up- or downregulated after infection and that passed the filtering conditions are depicted in Supplementary Fig. 1 on our website (http://www.cnb.uam.es/∼sguerra/JVirol/supplementary.pdf). The clusters appear to represent specific regulation patterns; the average profile for each cluster is shown in Fig. 1.
FIG. 1.
Characteristic expression patterns represented in clusters 1 to 6. Mean values (left) and standard deviations (right) of the expression profiles of genes assigned to each cluster are shown. Experimental points on the x axis are indicated as follows: 1 for 2 h, 2 for 6 h, and 3 for 16 hpi. The y axis shows normalized expression values. Each value in parentheses shows the percentage of genes in each cluster with reference to the total of 368 differentially expressed genes.
Clusters 1 to 3 correspond to three patterns of gene activation kinetics, whereas clusters 4 to 6 show distinct repression kinetics. Cluster 1 contains 73 transcripts (20.16%) that are upregulated during infection; this is the only cluster that shows generalized induction from 2 to 16 hpi. Cluster 2 contains 58 transcripts (16.02%), including genes with induction maintained from 6 to 16 hpi that are unaffected at 2 hpi. Cluster 3 contains 41 transcripts (11.32%) that are upregulated at 6 hpi but return to basal levels by 16 hpi. Cluster 4 contains 79 transcripts (21.82%) that are downregulated at 6 hpi and are unaltered at the other times studied. Clusters 5 and 6 contain 111 transcripts (30.7%); both show downregulation throughout infection, with distinct profiles.
Representative human genes that are upregulated in NYVAC infection (clusters 1 to 3) are classified according to their biological functions in Table 1. Genes whose expression was repressed after NYVAC infection represented 52.5% of the 368 genes; examples of human genes downregulated by NYVAC in clusters 5 and 6 are detailed in supplementary Table 1 on our website (http://www.cnb.uam.es/∼sguerra/JVirol/supplementary.pdf).
TABLE 1.
Representative human genes in clusters 1 to 3 (upregulated by NYVAC)
| Cluster, function, and gene name | Accession no. | Gene symbola | Change (fold) at indicated time (hpi)
|
||
|---|---|---|---|---|---|
| 2.00 | 6.00 | 16.00 | |||
| Cluster 1 | |||||
| Cell cycle, apoptosis | |||||
| V-Jun avian sarcoma virus 17 oncogene homolog | W96155 | JUN | 2.14 | 13.75 | 8.01 |
| Activating transcription factor 3 | H21042 | ATF3 | 2.42 | 8.36 | 6.10 |
| Caspase-9, apoptosis-related cysteine protease | AA281152 | CASP9 | 2.59 | 2.37 | 1.98 |
| Dual-specificity phosphatase 2 | AA759046 | DUSP2 | 2.43 | 5.25 | 1.56 |
| JunB proto-oncogene | T99236 | JUNB | 1.96 | 2.64 | 1.65 |
| Cytochrome P450, subfamily I (dioxin-inducible) | N21019 | CYP1B1 | 1.89 | 2.73 | 2.15 |
| Cytoskeleton | |||||
| Connective tissue growth factor | AA598794 | CTGF | 4.24 | 11.45 | 5.14 |
| Pleckstrin homology-like domain, family A, member 1 | AA258396 | PHLDA1 | 4.19 | 8.24 | 5.28 |
| Pleckstrin homology-like domain, family A, member 1 | AA258396 | PHLDA1 | 3.85 | 5.22 | 4.11 |
| Calcineurin-binding protein calsarcin-1 | AA065090 | CS-1 | 1.70 | 2.30 | 2.55 |
| Kinesin family member 5A | AA984728 | KIF5A | 1.89 | 5.45 | 4.64 |
| Immune system | |||||
| Tumor necrosis factor (ligand) superfamily, member 7 | AI347622 | TNFSF7 | 1.79 | 3.08 | 3.41 |
| Coagulation factor III (thromboplastin, tissue factor) | AI313387 | F3 | 1.58 | 2.04 | 1.53 |
| Interleukin-6 (interferon beta 2) | N98591 | IL6 | 2.02 | 5.85 | 2.68 |
| Signaling | |||||
| Early growth response 1 | AA488533 | EGR1 | 4.74 | 10.38 | 6.83 |
| Nuclear factor kappa light polypeptide gene enhancer in B cells 2 (p49/p100) | AA952897 | NFKB2 | 1.53 | 2.02 | 1.79 |
| Serum-inducible kinase | AA460152 | SNK | 1.57 | 6.49 | 2.44 |
| Tissue factor pathway inhibitor 2 | AA399473 | TFPI2 | 1.62 | 1.77 | 2.60 |
| Brain-derived neurotrophic factor | AA262988 | BDNF | 1.78 | 4.35 | 2.12 |
| Small inducible cytokine subfamily A, member B | AI268937 | SCYA8 | 1.76 | 3.04 | 2.89 |
| Miscellaneous | |||||
| Nucleoporin, 88 kDa | AA488609 | NUP88 | 3.52 | 6.49 | 4.23 |
| DHHC1 protein | W80739 | LOC51304 | 1.96 | 8.09 | 4.32 |
| Ubiquitin-conjugating enzyme E2M | AA449119 | UBE2M | 1.52 | 3.06 | 2.91 |
| Prostagtandin-endoperoxide synthase 2 | AA644211 | PTGS2 | 1.80 | 2.90 | 2.68 |
| Small nuclear ribonucleoprotein polypeptides B and B1 | AA279662 | SNRPB | 1.66 | 2.85 | 2.87 |
| Hypothetical protein A-211C6.1 | N39229 | LOC57149 | 1.55 | 2.74 | 2.54 |
| Cluster 2 | |||||
| Adhesion and cytoskeleton | |||||
| Collagen, type VII, alpha 1 | AA598507 | COL7A1 | 1.46 | 2.82 | 2.26 |
| Tight junction protein 1 (zona occludens 1) | H50344 | TJP1 | 1.18 | 1.93 | 2.96 |
| Protocadherin alpha 9 | AA437139 | PCDHA9 | 1.21 | 1.31 | 2.04 |
| Cell cycle, apoptosis | |||||
| Myeloid cell leukemia sequence 1 (BCL2-related) | AA488674 | MCL1 | 1.31 | 3.61 | 2.18 |
| Growth arrest and DNA-damage-inducible 34 | AA460168 | GADD34 | 1.21 | 2.48 | 1.93 |
| SRC homology three (SH3) and cysteine-rich domain | AA031284 | STAC | 1.11 | 2.41 | 2.84 |
| Growth arrest and DNA-damage-inducible 135 | AA404666 | GADD135 | 1.32 | 2.98 | 2.06 |
| Mitochondrial translational release factor 1 | AI347695 | MTRF1 | 1.19 | 2.06 | 2.21 |
| DnaJ (Hsp40) homolog, subfamily B, member 2 | AA455298 | DNAJB2 | 1.22 | 2.69 | 2.39 |
| Ras homolog gene family, member E | W86282 | ARHE | 1.08 | 2.68 | 2.35 |
| GRO3 oncogene | AA935273 | GRO3 | 1.20 | 2.62 | 2.38 |
| v-Myc avian myelocytomatosis viral oncogene homolog | AA464600 | MYC | 1.05 | 2.93 | 1.65 |
| v-Myc avian myelocytomatosis viral oncogene homolog | W87741 | MYC | 1.08 | 2.93 | 1.59 |
| v-Akt murine thymoma viral oncogene homolog 2; | AA457097 | AKT2 | 1.22 | 2.15 | 1.57 |
| Murine leukemia viral (Bmi-1) oncogene homolog | AA478036 | BMI1 | 1.15 | 1.36 | 3.18 |
| Immune system | |||||
| CD80 antigen (CD28 antigen ligand 1, B7-1 antigen) | AA983817 | CD80 | 1.25 | 2.02 | 2.70 |
| Interleukin-1 receptor, type II | H78386 | IL1R2 | 1.29 | 1.58 | 4.02 |
| Interferon stimulated protein | |||||
| Interferon-stimulated protein, 15 kDa | AA406020 | ISG15 | 1.21 | 1.14 | 3.39 |
| Metabolism | |||||
| Fatty acid desaturase 3 | AI123992 | FADS3 | 1.45 | 2.94 | 2.41 |
| Carboxypeptidase A2 (pancreatic) | AA844831 | CPA2 | 1.48 | 2.83 | 3.45 |
| Miscellaneous | |||||
| Discs, large (Drosophila) homolog 1 | AA521155 | DLG1 | 1.24 | 2.16 | 2.80 |
| CGI-20 protein | R02578 | LOC51608 | 1.22 | 2.11 | 2.94 |
| Bromodomain, testis specific | AA454222 | BRDT | 1.37 | 1.56 | 2.17 |
| RNA binding motif protein 5 | AA456007 | RBM5 | 1.18 | 3.20 | 2.42 |
| Spermidine/spermine N1-acetyltransferase | AA598631 | SAT | 1.20 | 2.51 | 2.62 |
| Spermidine/spermine N1-acetyltransferase | AA011215 | SAT | 1.20 | 2.47 | 2.25 |
| Splicing factor, arginine/serine-rich 10 | AI583623 | SFRS10 | 1.19 | 4.41 | 3.21 |
| CBP/p300-interacting transactivator 2 | AA115078 | CITED2 | 1.19 | 4.86 | 1.87 |
| Solute carrier family 2, member 3 | AA406552 | SLC2A3 | 1.43 | 2.12 | 1.77 |
| RAE1 (RNA export 1, from Schizosaccharomyces pombe) homolog | AA504128 | RAE1 | 1.39 | 2.07 | 1.82 |
| Apg12 (autophagy 12, S. cerevisiae)-like | AA251737 | APG12L | 1.17 | 2.70 | 2.07 |
| Discs, large (Drosophila) homolog 1 | H24708 | DLG1 | 1.35 | 1.89 | 3.15 |
| Signaling | |||||
| Nuclear factor kappa light polypeptide gene enhancer | AA451716 | NFKB1 | 1.19 | 2.13 | 1.92 |
| A kinase (PRKA) anchor protein (gravin) 12 | AA478543 | AKAP12 | 1.43 | 4.55 | 2.86 |
| Phorbol-12-myristate-13-acetate-induced protein 1 | AA458838 | PMAIP1 | 1.10 | 2.46 | 2.93 |
| Glutamate receptor, ionotropic, N-methyl d-aspartate 2D | AI565972 | GRIN2D | 1.30 | 2.36 | 2.24 |
| Receptor tyrosine kinase-like orphan receptor 2 | N94921 | ROR2 | 1.26 | 1.68 | 2.46 |
| Immediate-early response 3 | AA457705 | IER3 | 1.44 | 1.38 | 2.99 |
| Prostaglandin E receptor 3 (subtype EP3) | AA406362 | PTGER3 | 1.29 | 1.22 | 4.32 |
| Solute carrier family 17 (sodium phosphate) | H60423 | SLC17A2 | 1.29 | 1.60 | 2.14 |
| Adaptor-related protein complex 3, delta 1 subunit | AA630776 | AP3D1 | 1.18 | 2.05 | 2.34 |
| Translation and transcription | |||||
| Splicing factor (CC1.3) | H47069 | CC1.3 | 1.24 | 2.36 | 2.40 |
| Splicing factor (CC1.3) | AA171948 | CC1.3 | 1.04 | 1.89 | 2.26 |
| Splicing factor, proline/glutamine-rich | AA425853 | SFPQ | 1.09 | 1.69 | 2.55 |
| Wiskott-Aldrich family | |||||
| Wiskott-Aldrich syndrome-like protein | AI261600 | WASL | 1.22 | 1.62 | 2.60 |
| Cluster 3 | |||||
| Adhesion and cytoskeleton | |||||
| Claudin 1 | AA194833 | CLDN1 | 1.48 | 2.29 | 1.30 |
| Catenin (cadherin-associated protein), alpha 1 (102 kDa) | AA676957 | CTNNA1 | 1.76 | 2.20 | 1.48 |
| Collagen, type III, alpha 1 | COL3A1 | COL3A1 | 1.19 | 2.03 | 1.09 |
| Cadherin 18, type 2 | AA865745 | CDH18 | 0.27 | 2.17 | 1.24 |
| Cell cycle, apoptosis | |||||
| BCL2-associated athanogene | AI017240 | BAG1 | 1.06 | 2.05 | 1.32 |
| FOS-like antigen 1 | T89996 | FOSL1 | 1.19 | 2.08 | −1.10 |
| CDC10 (cell division cycle 10, S. cerevisiae), homolog | AA633993 | CDC10 | 1.07 | 3.76 | 1.45 |
| Transforming growth factor beta-stimulated protein TSC-22 | AA664389 | TSC22 | 1.20 | 3.16 | 1.41 |
| Transformer-2 alpha (hTra-2 alpha) | AA701944 | HSU53209 | 1.25 | 2.05 | 1.46 |
| Signaling | |||||
| Delodinase, iodothyronine, type II | AA864322 | DIO2 | 1.17 | 3.86 | 1.40 |
| Mitogen-activated protein kinase kinase 3 | H08749 | MAP2K3 | 1.73 | 2.15 | 1.47 |
| Phosphatidylinositol-4-phosphate 5-kinase, type I, gamma | AA482251 | PIP5K1C | 1.25 | 2.17 | 1.36 |
| Phosphatidylinositol-4-phosphate 5-kinase, type I, alpha | AA255881 | PIP5K1A | 1.08 | 2.31 | 1.44 |
| Immune system | |||||
| Insulin-like growth factor binding protein 3 | AA598601 | IGFBP3 | 1.14 | 4.58 | 1.56 |
| Chemokine (C-X-C motif), receptor 4 (fusin) | T62636 | CXCR4 | 1.47 | 2.40 | 1.41 |
| Inhibitor of growth 1 family, member 1 | N33574 | ING1 | 1.49 | 2.24 | 1.76 |
| Coagulation factor III (thromboplastin, tissue factor) | AI313387 | F3 | 1.31 | 2.23 | 1.39 |
| Miscellaneous | |||||
| Serum/glucocorticoid-regulated kinase | AI375353 | SGK | 1.12 | 2.89 | 1.41 |
| Cofactor required for Sp1 transcriptional activation | N92735 | CRSP6 | 1.19 | 2.11 | 1.49 |
| Tryptophan 2,3-dioxygenase | T72398 | TDO2 | 1.26 | 2.12 | 1.44 |
| Transiation and transcription | |||||
| Slug (chicken homolog), zinc finger protein | H57310 | SLUG | 1.07 | 3.29 | 1.47 |
| Eukaryotic translation initiation factor 5A | H99842 | EIF5A | 1.10 | 2.78 | 1.14 |
| Zinc finger protein 274 | AI085519 | ZNF274 | 1.07 | 2.08 | 1.33 |
Representative human gene targets of NF-κB are underlined.
Confirmation of microarray data by quantitative real-time RT-PCR.
We used real-time RT-PCR to verify the transcriptional changes in selected genes, as detected by microarray analysis. We analyzed 10 genes, 6 of which were upregulated (WASL, NF-κB-2, CASP-9, ATF-3, GADD-34, and IL-6), 2 of which were downregulated (IL-7 and IFNγ), and 2 of which were unaltered (PCNT2 and APEXL); HPRT was used as an internal control (Table 2). The assay was performed with the same RNA samples from infected and uninfected HeLa cells used in microarray experiments. The expression patterns and relative mRNA abundances of the genes selected concurred with the microarray data in all cases, with only slight variations, validating the microarray results.
TABLE 2.
Confirmation of microarray data by real-time RT-PCR
| Gene product | Fold change at indicated time (hpi)
|
|||||
|---|---|---|---|---|---|---|
| Microarray
|
RT-PCR
|
|||||
| 2 | 6 | 16 | 2 | 6 | 16 | |
| IL-6 | 2.02 | 5.85 | 2.68 | 3.2 | 4.65 | 3.25 |
| WASL | 1.22 | 1.62 | 2.6 | 2.6 | 2.03 | 4.3 |
| NF-κB2 | 1.53 | 2.02 | 1.79 | 1.68 | 2.87 | 2.06 |
| IL-7 | 0.80 | 0.55 | 0.88 | 0.95 | 0.63 | 0.79 |
| IFN-γ | 1.08 | 0.95 | 0.74 | 1.20 | 0.96 | 0.85 |
| APEXL2 | 1.18 | 1.0 | 1.45 | 1.01 | 0.98 | 1.25 |
| PCNT2 | 1.01 | 1.30 | 1.07 | 1.10 | 1.28 | 1.20 |
| CASP-9 | 2.59 | 2.37 | 1.98 | 2.98 | 3.02 | 2.10 |
| GADD-34 | 1.21 | 2.48 | 1.93 | 1.90 | 2.82 | 1.80 |
| ATF-3 | 2.42 | 8.36 | 6.10 | 2.31 | 9.21 | 5.69 |
Target verification and activation of representative cell proteins after NYVAC infection.
Although changes in mRNA levels do not necessarily represent changes in protein expression, we used various approaches to analyze whether the gene expression changes detected by microarray analysis correlated with protein levels and the activities of selected gene products.
(i) Caspase-9 activation.
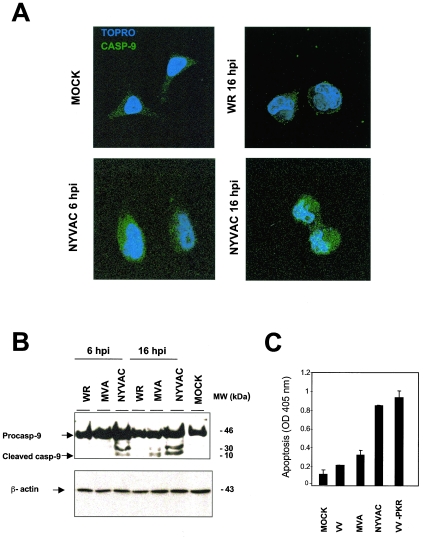
The caspase-9 gene was differentially expressed following NYVAC infection (Tables 1 and 2); thus, we analyzed the effect of the mRNA increase on the protein level. Immunofluorescence experiments indicated that transcriptional upregulation of caspase-9 after NYVAC infection corresponded with an increase in active caspase-9 protein levels, confirming the microarray data. Whereas no active caspase-9 signal was found in control or WR-infected HeLa cells, a distinct punctate pattern was observed in NYVAC-infected cells at 6 and 16 hpi (Fig. 2A). In addition, we used Western blotting to measure the levels of procaspase cleavage to activated caspase-9 in HeLa cells infected with NYVAC compared to those in cells infected with other VV strains. Caspase-9 activation was detected, with a 10-kDa cleavage product from the 46-kDa procaspase. We observed a clear increment in procaspase-9 and a notable increase in active caspase-9 after NYVAC infection. In MVA-infected cells, weakly active caspase-9 was observed at 16 hpi, whereas there was no evidence of active caspase-9 in WR-infected HeLa cells at any time postinfection (Fig. 2B).
FIG. 2.
NYVAC-induced apoptosis in HeLa cells. (A) Protein validation of microarray data by immunofluorescence analysis of the effect of NYVAC infection on caspase-9 distribution in HeLa cells. Mock-, WR-, or NYVAC-infected cells were incubated at the indicated times with an anti-caspase-9 antibody (Calbiochem) that recognizes the active form of caspase-9, followed by the appropriate conjugated secondary antibody and the ToPro reagent. The samples were analyzed by confocal immunofluorescence microscopy. (B) Activation of caspase-9 after WR, MVA, or NYVAC infection. Total proteins (100 μg) were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted at various times (6 and 16 hpi) with an anti-caspase-9 antibody (Oncogene) that recognizes the procaspase and the cleaved active form of caspase-9. Caspase-9 presented a proform (46 kDa) that it is cleaved into a heterodimer of 35-kDa and 10-kDa chains. The molecular sizes (in kilodaltons) of the proteins are indicated on the right. Actin levels revealed that the same amount of protein was loaded into each lane. (C) Apoptosis assays after WR, MVA, or NYVAC infection determined the amount of cytoplasmic histone-associated DNA by ELISA. HeLa cells grown in 12-well plates were infected (5 PFU/cell) with the viruses indicated and harvested at 24 hpi for determinations with an ELISA kit of the absorbance at 405 nm. As a positive control for apoptosis, we used VV-PKR-infected HeLa cells. Duplicate samples were measured in two independent experiments.
This apparent caspase activation in NYVAC-infected cells prompted us to compare apoptosis in HeLa cells infected with different VV strains. To measure apoptosis, we determined the amount of cytoplasmic histone-associated DNA in mock-, WR-, MVA-, or NYVAC-infected cells (Fig. 2C). As a positive control, we infected HeLa cells with an inducible VV recombinant expressing protein kinase R (PKR) in an isopropyl-β-d-thiogalactopyranoside (IPTG)-dependent manner, which produces apoptosis (27). Apoptosis levels in NYVAC-infected cells at 24 hpi were similar to those in VV-PKR-infected cells. Apoptosis was not detected in mock- or WR-infected cells, whereas low apoptosis levels were found after MVA infection. Altogether, these results indicated that NYVAC upregulates several intermediates of apoptotic cascades, in correlation with the ability of this virus to induce apoptosis.
(ii) Activation of effector caspases.
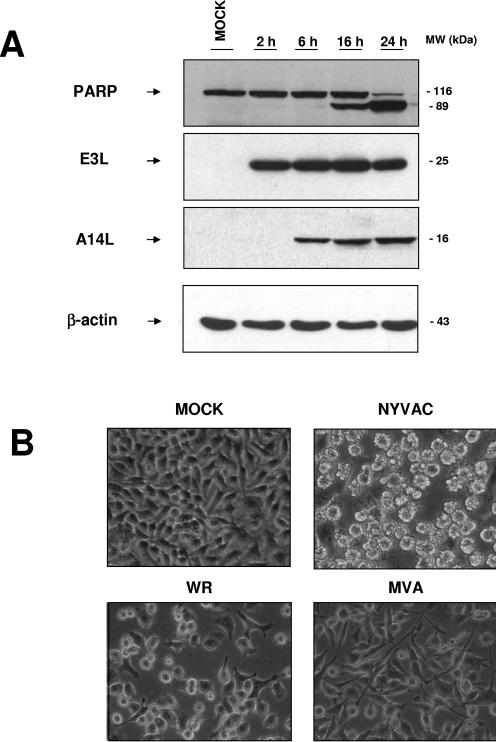
Apoptosis is mediated by the activation of caspase-8 or -9, leading to the activation of effector caspases-3 and -7, which cleave specific substrates, including PARP-1. This enzyme catalyzes the formation of poly(ADP-ribose) polymers on acceptor proteins involved in the maintenance of chromatin structure, which indicates activation of the apoptotic cascade (47). To study apoptosis induction after NYVAC infection, we measured the kinetics of apoptosis in Western blot analysis by the detection of PARP-1, using an antibody that recognizes both the full-length and the cleaved protein. As shown in Fig. 3A, the 89-kDa cleaved PARP-1 fragment became evident at 16 hpi. As a control of infection, we measured the virus gene expression patterns of representative early (E3L) and late (A14L) VV proteins (Fig. 3A).
FIG. 3.
Apoptosis induction kinetics during NYVAC infection. (A) Time course of PARP-1 cleavage during NYVAC infection. HeLa cells were infected with NYVAC, and at the indicated times, total proteins (100 μg) were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with anti-PARP-1, which recognizes both the full-length and the cleaved protein. An 89-kDa cleavage product of PARP-1 was observed at 16 hpi. As a measure of infection, we used immunoblotting to visualize the viral proteins E3L and A14L. The molecular sizes (in kilodaltons) of the proteins are indicated on the right. Actin levels showed that the same amount of protein was loaded in each lane. (B) Apoptotic phenotype of NYVAC-infected HeLa cells. Monolayer cultures of HeLa cells were left uninfected or infected (5 PFU/cell) with the different VV strains for 16 h, and the apoptotic phenotype was visualized by phase-contrast microscopy. NYVAC-infected cells showed characteristic apoptosis, defined by a rounded, floating, phase-bright, wrinkled shape.
By phase-contrast microscopy, we monitored apoptosis induction in WR-, MVA-, or NYVAC-infected HeLa cells (5 PFU/cell). A characteristic apoptotic phenotype was manifested at 16 hpi in NYVAC-infected cells, defined by the rounded, floating, phase-bright, and wrinkled shape of the cells (Fig. 3B). At 16 hpi, a nonapoptotic phenotype was observed in WR- and MVA-infected cells (Fig. 3B). WR produced a pronounced cytopathic effect, including a change in cell morphology from an elongated spindle-shaped form to a rounded form. Compared to WR, MVA produced a minor cytopathic effect, with a characteristic bipolar phenotype as described previously (12).
(iii) Enhanced NF-κB expression and activation.
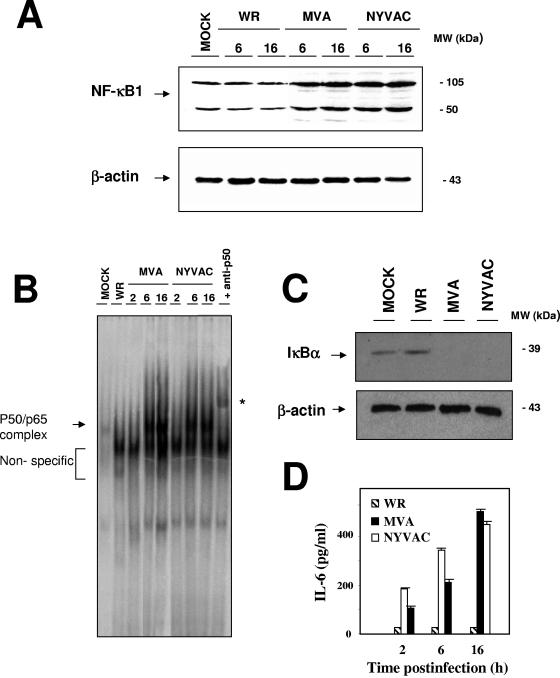
To further validate the microarray data, we measured NF-κB1 by Western blotting of NYVAC- compared to mock-, WR-, or MVA-infected HeLa cells, using an antibody that recognizes both the p105 and p50 subunits of NF-κB1 (Fig. 4A). In WR-infected cells, NF-κB1 protein levels (p105 and p50) were similar to those in mock-infected cells. In MVA- and NYVAC-infected cells, the NF-κB1 signal was higher than that in mock- or WR-infected cells due to enhanced NF-κB1 expression, as shown by microarray analysis (Table 1). To evaluate whether the NYVAC-induced increase in NF-κB component mRNA and protein expression corresponded to NF-κB activation, nuclear extracts from mock-, NYVAC-, MVA-, or WR-infected HeLa cells were incubated with radiolabeled oligonucleotides containing a consensus NF-κB binding site sequence and analyzed by electrophoretic mobility shift assays (EMSAs). The MVA and NYVAC strains activated NF-κB at 6 and 16 hpi, whereas NF-κB activation was not observed in WR-infected nuclear extracts at 6 and 16 hpi (Fig. 4B). NYVAC-infected HeLa cells at 16 hpi produced a specific complex that showed a supershift when a specific anti-NF-κB p50 antibody was added to nuclear extracts. NYVAC produced an increase in NF-κB protein levels and transcription factor activity (Fig. 4A and B). NF-κB translocation from the cytoplasm to the nucleus is governed by IκBα degradation; we thus evaluated IκBα levels by Western blotting of cytoplasmic extracts from mock-, WR-, MVA-, or NYVAC-infected cells at 4 hpi. IκBα was localized in the cytoplasm in mock- and WR-infected cells (Fig. 4C), whereas cytoplasmic IκBα was greatly reduced in MVA- and NYVAC-infected cells, concurring with NF-κB activation in the nucleus (Fig. 4B).
FIG. 4.
NYVAC-induced NF-κB signaling. (A) Validation of microarray data by NF-κB protein levels and comparison between WR, MVA, and NYVAC infections. Total proteins (100 μg) were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with anti-NF-κB1 antibody at various times (6 and 16 hpi). Actin levels indicated that the same amount of protein was loaded into each lane. The molecular sizes (in kilodaltons) of the proteins are indicated on the right. (B) Autoradiogram of NF-κB activity, determined by EMSA performed with nuclear extracts of mock-, MVA-, and NYVAC-infected HeLa cells at the indicated times. WR, nuclear extract from WR-infected HeLa cells at 16 hpi. The arrow indicates the 32P-labeled NF-κB/DNA complex and the nonspecific signal, and the asterisk indicates the supershift of NF-κB after incubation of the nuclear extracts with the 32P-labeled probe and anti-p50 antibody. (C) IκΒα degradation kinetics of WR-, MVA-, and NYVAC-infected HeLa cells. Cytoplasmic extracts were prepared, as previously described (45), from uninfected and poxvirus-infected (5 PFU/cell) HeLa cells. IκΒα levels were detected by immunoblotting at 4 hpi. The molecular sizes (in kilodaltons) of the proteins are indicated on the right. Actin levels indicated that the same amount of protein was loaded into each lane of the gel. (D) Levels of IL-6 secreted from WR-, MVA-, and NYVAC-infected HeLa cells (5 PFU/cell), as determined by ELISA. Protein levels of IL-6 in supernatants of infected cells were measured at 2, 6, and 16 hpi. Duplicate samples were measured in two independent experiments.
NF-κB plays an important role in the expression of inflammatory cytokines, including IL-6 (21). To correlate NF-κB activation with IL-6 expression, we used ELISA to quantify the levels of secreted IL-6 in WR-, MVA-, or NYVAC-infected HeLa cells at 2, 6, and 16 hpi. We observed a strong IL-6 increase over time after MVA or NYVAC infection. The increase in IL-6 protein levels following NYVAC infection is in complete agreement with the upregulation of IL-6 mRNA levels detected by microarray analysis and real-time RT-PCR (Tables 1 and 2). IL-6 synthesis is augmented due to the increase in transcription, which results in higher levels of protein translation and secretion. When HeLa cells were infected with WR, we did not observe NF-κB activation, and IL-6 was not detected by ELISA in cell-free supernatants (Fig. 4B and D). NF-κB activation by NYVAC infection occurs by 2 hpi (observed by IL-6 secretion in Fig. 4D) and before apoptosis induction (observed at 16 hpi by PARP-1 cleavage in Fig. 3).
(iv) Enhanced expression of ATF-3 transcription factor.
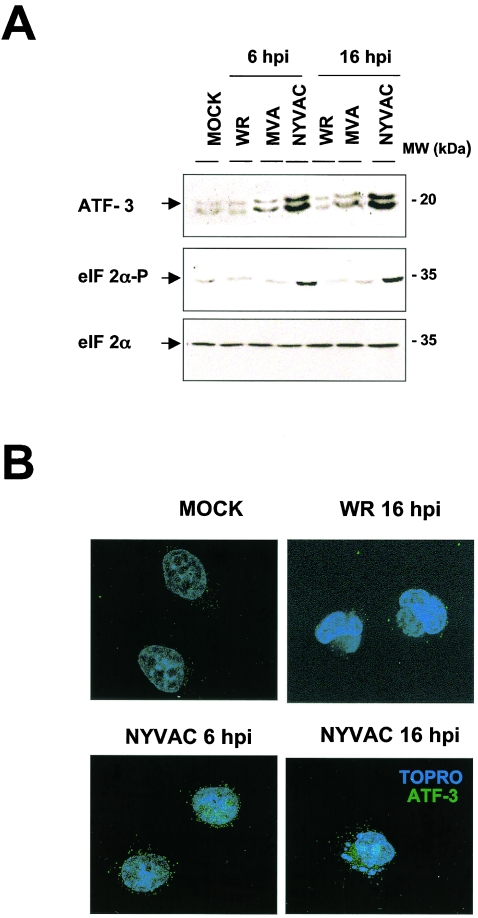
NF-κB pathway activation contributes to the transcription of NF-κB-dependent genes, such as ATF-3 (activating transcription factor 3), which is specifically upregulated after NYVAC or MVA infection and unaltered by WR infection. ATF-3 protein levels increased after NYVAC infection compared to those in mock-, WR-, or MVA-infected HeLa cells (Fig. 5A, upper panel). More-than-fourfold increases in ATF-3 protein levels were evident at 6 and 16 hpi in NYVAC-infected cells compared to controls, as determined by densitometric analyses (data not shown). These data concur with microarray and real-time RT-PCR results. In an immunofluorescence assay, whereas ATF-3 was nearly undetectable in uninfected and WR-infected cells, there was a clear increase in ATF-3 staining intensity after NYVAC infection, with a mainly nuclear and perinuclear localization (Fig. 5B).
FIG. 5.
ATF-3 protein expression is induced by NYVAC infection. (A) Levels of ATF-3, phosphorylated eIF2α at Ser-51, and total eIF2α were measured by immunoblot analysis of extracts of HeLa cells after WR, MVA, and NYVAC infection. Total proteins (100 μg) were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with anti-ATF-3, anti-phosphorylated-eIF2α, or anti-eIF2α antibody at various times (6 and 16 hpi). The molecular sizes (in kilodaltons) of the proteins are indicated on the right. The antibody against ATF-3 recognizes two or three bands, depending on the running conditions, perhaps as a result of posttranslational modifications of the transcription factor, which can be cell dependent. (B) Immunofluorescence analysis of the effect of NYVAC infection on ATF-3 distribution in HeLa cells. Mock-, WR-, and NYVAC-infected cells were labeled with anti-ATF-3 (Santacruz) at the indicated times, followed by the appropriate conjugated secondary antibody and ToPro reagent. The samples were analyzed by confocal immunofluorescence microscopy.
Phosphorylation of the α subunit of eukaryotic initiation factor 2 (eIF-2α) enhances the transcription of genes involved in stress-sensing pathways, such as the genes for ATF-4, GADD34, and GADD153 (57). NYVAC infection induced mRNA expression of the GADD34 (Tables 1 and 2) and GADD153 (Table 1) genes, suggesting that eIF-2α may be phosphorylated after NYVAC infection. To determine the effect of NYVAC infection on eIF-2α phosphorylation, we measured specific eIF-2α phosphorylation at Ser-51 by Western blotting of mock-, WR-, MVA-, or NYVAC-infected HeLa cells. Clear eIF-2α phosphorylation was observed at 6 and 16 hpi in NYVAC-infected cells but not in WR- or MVA-infected cells (Fig. 5A, lower panels). Thus, we found a correlation of enhanced ATF-3 expression at the mRNA and protein levels and enhanced GADD34 and GADD153 expression at the RNA level with eIF-2α phosphorylation, suggesting that these stress pathways could be involved in apoptosis induction by NYVAC infection.
VV K1L gene prevents NF-κB activation but not apoptosis induction after NYVAC infection.
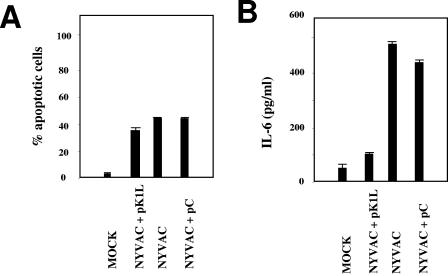
The VV K1L host range gene acts as an NF-κB inhibitor (45). Since NYVAC lacks the K1L gene, we analyzed whether apoptosis induction and NF-κB activation by NYVAC can be inhibited by the K1L gene. NYVAC-infected HeLa cells were transfected with a plasmid containing the K1L gene with a T7-Tag tag or with a control empty plasmid (pC), and apoptosis was measured by immunofluorescence. The K1L gene is necessary for VV replication in RK13 cells (38, 41), and after infection-transfection, we confirmed that the transfected K1L gene rescued the host range by producing plaques in RK13 cells (data not shown). Apoptotic cells were counted based on the characteristic increase in nucleus granularity and the appearance of apoptotic bodies. Cells were costained with E3L antibody to detect VV, ToPro for DNA detection, and anti-T7-Tag to detect transfected cells. The apoptotic phenotype of K1L-transfected NYVAC-infected cells was comparable to that found in the absence of K1L (not shown). We observed similar numbers of apoptotic cells after K1L expression as for pC-transfected cells (Fig. 6A), indicating that the K1L gene was not responsible for the apoptotic phenotype after NYVAC infection. To analyze the ability of K1L to repress NF-κB activation after NYVAC infection, we used ELISA to quantify secreted IL-6, an indicator of NF-κB activity, in supernatants from NYVAC-infected HeLa cells that were untransfected or transfected with pC or pK1L. We observed a marked decrease in secreted IL-6 in NYVAC-infected HeLa cells after pK1L transfection (Fig. 6B). This result indicated that K1L expression inhibits NF-κB activity in NYVAC-infected cells.
FIG. 6.
K1L is necessary for NF-κB inhibition but not for apoptotic induction in NYVAC infection. (A) Ectopic K1L expression in NYVAC-infected HeLa cells does not inhibit apoptosis during NYVAC infection. HeLa cells grown on coverslips were infected with NYVAC at 0.1 PFU/cell, and at 1 hpi, cells were transfected (3 μg) with an expression plasmid containing the K1L gene with T7-Tag (pK1L) or with an empty control plasmid (pC). At 40 h posttransfection, cells were fixed and processed to measure apoptosis by immunofluorescence, using antibodies directed against the E3L protein (red), T7-Tag (green), and ToPro for DNA staining (blue). The ratios of infected apoptotic cells to infected nonapoptotic cells were determined by counting (about 2,000 cells per experiment); each point represents the average of two independent experiments. (B) Ectopic K1L expression inhibits NF-κB activation in NYVAC infection. Levels of IL-6 secreted from cells infected and transfected as described above were quantified by ELISA. Duplicate samples were measured in two independent experiments.
DISCUSSION
Poxvirus vector technology has provided reagents for high-level expression of proteins for vaccine use. Due to their infection efficiencies, high gene product expression levels, and safety, attenuated poxvirus vectors are useful for gene delivery in immunotherapy (10, 35). NYVAC is a highly attenuated VV strain, generated by the specific deletion of 18 open reading frames from the viral genome, which affects host range, virulence, and pathogenesis genes (32, 49, 50). This vector has been applied as a recombinant vaccine delivery system in animal models and target species, including humans (34, 37). NYVAC-based vectors have been analyzed as vaccines for several tumors (23, 46) and are protective against pathogens such as retroviruses (5, 10, 31), Japanese encephalitis virus (24), and Plasmodium falciparum (34). The potential use of variola virus as a biological weapon requires the development of safer smallpox vaccines, using attenuated VV strains such as MVA and NYVAC (4, 9).
The use of these vectors as vaccines requires a better understanding of the impact of NYVAC infection in the human host, including the identification of differentially regulated genes and their functions. Using high-throughput cDNA microarray screening of over 15,000 genes in NYVAC-infected HeLa cells, we identified transcriptional alterations in a defined subset of human genes. When grouped into functional categories, a large proportion of altered transcripts were from genes involved in apoptosis, adhesion, cytoskeletal architecture, cell cycling, and immune modulation.
Apoptosis is regarded as an innate cellular response that limits virus propagation. The importance of apoptosis as an antiviral defense against poxvirus infection is suggested by the finding that a number of poxvirus genes encode proteins that interfere with apoptosis in specific cell types (30, 39). During NYVAC infection, we found an upregulation of the caspase-9 gene, an apoptosis-linked gene. Increased caspase-9 protein levels reflected an increase in caspase-9 activity and effector caspases, as determined by the appearance of a cleavage product from the target 89-kDa PARP-1 protein. Although we detected caspase-9 activation at 6 hpi, as shown by cleavage of the 46-kDa procaspase, apoptosis and PARP-1 cleavage were not evident until 16 hpi. These temporal differences in NYVAC-induced apoptosis may be due to distinct target specificities and the sequence of biochemical events. The role of apoptosis in poxvirus biology is not well understood; previous studies of K1L gene-defective viruses found no correlation between apoptosis suppression and virus growth (7), and similar results were obtained by inactivation of the cowpox virus crmA gene (42). CrmA/SPI-2 is the best-characterized apoptosis modulator encoded by poxvirus family members; it inhibits apoptosis by preventing caspase-8 activity (51, 56). An antiapoptotic function was previously described for the VV E3L gene product that blocks PKR activity (27, 29) and was recently shown for the viral protein F1L, which inhibits mitochondrial release of cytochrome c, which is essential for apoptosis induction (48, 52). B13R and B14R, encoding the interleukin-1β-converting enzyme inhibitor, are also involved in antiapoptotic functions (39, 49). Since the antiapoptotic genes F1L and E3L are present in the NYVAC genome, other genes with antiapoptotic functions, such as B13R and B14R or others with unknown functions that are absent in NYVAC, may be the cause for apoptotic induction of this virus. Our results indicate that the K1L gene does not act as an inhibitor of apoptosis during NYVAC infection, as we did not observe a blockade of apoptosis by ectopic expression of this gene (Fig. 6).
With regard to immune modulators, our microarray experiments showed NF-κB upregulation, which induces the transcription of several genes that encode cytokines or are essential for immune response activation (14, 21). NF-κB forms a heterodimer composed of 50- and 65-kDa subunits (p50 and p65) which is retained in the cytoplasm by its inhibitor, IκΒα. Specific stimuli cause proteolytic degradation of IκΒα in the cytoplasm and subsequent NF-κB protein translocation to the nucleus, where it transactivates certain genes (14, 21). Our microarray data indicated upregulated expression for both NF-κB1 and NF-κB2 messengers during NYVAC infection (Table 1). NF-κB2 upregulation was confirmed by real-time RT-PCR. The increase in NF-κB1 mRNA corresponded to increased levels of the p105 and p50 proteins (Fig. 4A). NYVAC infection also upregulated NF-κB transcriptional activity (Fig. 4B). NF-κB was activated early in infection, although EMSA did not detect activation at 2 hpi, when upregulation of IL-6, an indicator of NF-κB activation, was observed by real-time RT-PCR and microarray analysis. The NF-κB activation pathway contributes to the transcription of κB-dependent genes. Many transcripts upregulated by NYVAC are known NF-κB target genes (Table 1), although other κB-dependent genes such as TNF, A20, CCL2, CXCL1, and IL-8 were not upregulated during NYVAC infection. In view of the dual NF-κB-activating effect and apoptotic phenotype triggered by NYVACinfection, it is not surprising to find that not all of the NF-κB-dependent genes are upregulated by virus infection. One of the NF-κB-dependent genes that is upregulated is ATF-3, a stress-inducible gene that encodes an ATF/CREB transcription factor (18, 19, 28). Recent reports describe the effect of ATF-3 in apoptosis induction (20, 53); ATF-3, considered a proapoptotic factor, might be involved in apoptosis activation in NYVAC-infected cells, although its physiological relevance in this system remains to be established. NYVAC infection also induced mRNAs such as those encoding GADD34 and GADD153, whose activation is eIF-2α phosphorylation dependent; eIF-2α phosphorylation in NYVAC infection was confirmed by Western blotting (Fig. 5A).
MVA, another attenuated VV strain, also activates NF-κB (16, 36). NF-κB activation mediated by NYVAC and MVA may reflect the loss of a specific viral gene from the parental viral genome, deleted during the generation of MVA and NYVAC. Indeed, the K1L gene was deleted in NYVAC (49), and the viral K1L protein inhibits NF-κB activation by preventing IκΒα degradation (45). Since both the MVA and NYVAC genomes lack K1L, which is present in VV, we studied the effect of the absence of K1L on NF-κB activation by measuring IL-6 levels in NYVAC-infected HeLa cells transfected with an expression plasmid bearing the K1L gene. IL-6 levels were considerably lower in K1L-expressing cells than in control-transfected cells, indicating that K1L inhibits NF-κB activity during NYVAC infection (Fig. 6B). K1L inhibition of NF-κB activity was independent of NYVAC-induced apoptosis, as indicated by counting NYVAC-infected apoptotic cells expressing K1L (Fig. 6A).
Although NF-κB activation and apoptosis induction might be negative factors in the NYVAC cycle, the triggering of these mechanisms may favor a better immune response to a recombinant vector. NF-κB activation from a recombinant NYVAC infection should stimulate and amplify the immune response against recombinant products and facilitate clearance of the vector. The role of apoptosis in the immune response is still unclear, but antigens produced by apoptotic cells are reported to increase antigen immunogenicity (54). Dendritic cells that phagocytose apoptotic bodies from virus-infected cells can present viral antigens to cytotoxic T cells, inducing a cytotoxic response (1). It remains to be defined whether apoptosis induced by a NYVAC-based vector increases antigen immunogenicity in comparison with that induced by MVA vectors or NYVAC vectors unable to induce apoptosis by gene targeting.
We compared expression profiles of representative genes obtained in this study with those obtained from MVA infections (16) (Table 3). In infected HeLa cells, one group of genes is upregulated by both MVA and NYVAC, whereas others are upregulated selectively by MVA or NYVAC. Both viruses trigger cellular transcription factors, including NF-κB, Myc, c-Jun, ATF-3, DUSP2, and CTGF, and cell adhesion molecules such as the kinesin KIF5C. EGR1 (early gene response 1) was also upregulated by MVA and NYVAC, as also reported for MVA-ΔE3L-infected HeLa cells (29), and the MEK/ERK/RSK2/Elk-1/EGR-1 signaling pathway is necessary for VV multiplication (2). The immunomodulatory molecules IL-6, ISG15, and CD80 were upregulated in NYVAC- and MVA-infected cells, whereas IL-7, IL-1A, IL-8, and IL-15 were upregulated after MVA but not NYVAC infection. This indicates that MVA and NYVAC induce distinct proinflammatory cytokine profiles in vitro. In HeLa cells, WR infection also upregulates CD80 (15), but IL-6 and ISG15 upregulation is specific to MVA and NYVAC infections. ISG15 was first identified as an interferon-stimulated gene (6, 26) that affects IFN-α/β signal transduction; its expression increases markedly after viral infection (16, 33, 55). IFN-α/β-induced ISG15 activation suggests a role for ISG15 in the innate immune response to viral infection (44).
TABLE 3.
Representative gene expression profiling of MVA and NYVAC infections of human HeLa cellsa
| Expression profile and gene product name | Gene symbol | Fold change with MVA
|
Fold change with NYVAC
|
||||
|---|---|---|---|---|---|---|---|
| 2 hpi | 6 hpi | 16 hpi | 2 hpi | 6 hpi | 16 hpi | ||
| Increased in MVA and NYVAC infections | |||||||
| Early growth response 1 | EGR1 | 4.17 | 4.60 | 1.94 | 4.74 | 10.38 | 6.83 |
| Nuclear factor (erythroid-derived 2)-like 3 | NFE2L3 | 2.36 | 2.36 | 2.52 | 1.19 | 2.13 | 1.32 |
| Nuclear factor kappa light polypeptide 2 | NFKB2 | 2.03 | 2.22 | 2.34 | 1.53 | 2.02 | 1.79 |
| Interleukin-6 | IL6 | 2.65 | 2.14 | 1.56 | 2.02 | 5.85 | 2.68 |
| Interferon-stimulated protein, 15 kDa | ISG15 | 2.57 | 2.25 | 1.61 | 1.21 | 1.14 | 3.39 |
| CD80 antigen | CD80 | 1.75 | 2.58 | 2.06 | 1.25 | 2.02 | 2.7 |
| Dual-specificity phosphatase 5 | DUSP5 | 3.01 | 2.91 | 1.87 | 1.98 | 4.16 | 2.18 |
| JunB proto-oncogene | JUNB | 2.93 | 3.92 | 1.06 | 2.16 | 2.14 | 1.32 |
| v-Jun avian sarcoma virus 17 oncogene homolog | JUN | 2.17 | 2.02 | 1.31 | 2.14 | 13.75 | 8.01 |
| v-Myc avian myelocytomatosis viral oncogene homolog | MYC | 2.22 | 1.53 | 1.05 | 1.05 | 2.93 | 1.65 |
| Activating transcription factor 3 | ATF3 | 1.78 | 1.98 | 0.99 | 2.42 | 8.36 | 6.10 |
| Dual-specificity phosphatase 2 | DUSP2 | 3.93 | 3.59 | 1.85 | 2.43 | 5.25 | 1.56 |
| Connective tissue growth factor | CTGF | 5.77 | 4.51 | 1.12 | 4.24 | 11.45 | 5.14 |
| Kinesin family member 5A | KIF5A | 2.05 | 1.88 | 1.73 | 1.89 | 5.45 | 4.64 |
| Increased in MVA infection | |||||||
| Interleukin-7 | IL7 | 6.1 | 5.3 | 3.2 | 0.80 | 0.55 | 0.88 |
| B7 protein | B7 | 1.97 | 1.75 | 3.53 | 1.58 | 1.58 | 1.78 |
| CD47 antigen | CD47 | 5.35 | 4.08 | 3.16 | 1.02 | 0.76 | 1.03 |
| Mitogen-activated protein kinase kinase 5 | MAP2K5 | 2.37 | 2.74 | 1.69 | 1.02 | 0.76 | 1.03 |
| Nuclear factor kappa light polypeptide epsilon | NFKBIE | 2.41 | 1.61 | 1.12 | 1.18 | 1.27 | 1.42 |
| Nuclear factor of activated T cells, cytoplasmic 3 | NFATC3 | 3.46 | 2.97 | 1.30 | 1.01 | 1.34 | 1.34 |
| Increased in NYVAC infection | |||||||
| Caspase-9, apoptosis-related cysteine protease | CASP9 | 1.1 | 0.82 | 0.32 | 2.59 | 2.37 | 1.98 |
| Nucleoporin (88 kDa) | NUP88 | 0.87 | 0.93 | 0.92 | 3.52 | 6.49 | 4.23 |
| Pleckstrin homology-like domain, family A, member 1 | PHLDA1 | 1.01 | 0.95 | 0.99 | 4.19 | 8.24 | 5.28 |
MVA data are from a previous report (16).
In view of our findings, we propose that NYVAC infection activates a sequence of survival and apoptotic responses in the injured cell. NYVAC induces NF-κB activation early in infection and will trigger the expression of several gene classes, each of which might be required at different times of infection for cell survival. The NF-κB-activated early signals will not be able to overcome the cell death process caused by the virus infection, and cells will ultimately die by apoptosis. Some of the NF-κB-induced genes, such as ATF-3, together with other stress signals, such as GADD34 and GADD153 and other unknown signals, might promote apoptosis. Since apoptosis is a late event in NYVAC infection, most of the virus cycle will be completed, and hence the contribution of apoptosis to virus replication will be minimized. While the poxvirus genome encodes several genes that antagonize apoptosis and NF-κB activation, it is likely that these virus genes have a major impact on the evasion of the immune system by the virus rather than on virus growth in culture. We demonstrated that the K1L viral protein prevented NF-κB activation but not apoptosis induction in NYVAC infection, although the viral gene(s) involved in NYVAC-induced apoptosis remains unknown.
Overall, our results provide a basis for future functional evaluations of the NYVAC vector for use in human vaccines. Gene expression profiling permits detailed analyses of the impact of poxvirus vectors on immune system target cells after vaccination and the development of more effective poxvirus vaccines against a broad spectrum of pathogens and tumors.
Acknowledgments
We are indebted to R. Bablanian for critically reviewing the manuscript. We thank J. Tartaglia (Aventis-Pasteur) for the generous gift of NYVAC, and we are grateful to B. L. Jacobs for the anti-E3L antibody and A. Alcamí for the K1L expression plasmid. We thank S. Gutiérrez for confocal microscopy, V. Jiménez for expert technical assistance, and C. Mark and P. Martinez for editorial assistance.
S.G. was supported by a contract from the EU Project on Vaccinia Vectors. A.P.-M. was partially supported by the Spanish CAM grant GR/SAL/0653/2004 and the Ramón y Cajal Program. This work was supported by grants from the Spanish Ministry of Education and Science (BIO2004-03954), the Spanish Foundation for AIDS Research (FIPSE 36344/02), Fundación Botín, and the European Union (EuroVac QLRT-PL-1999-01321, Vaccinia Vectors QLK2-CT-2002-01867). The Department of Immunology and Oncology was founded and is supported by the Spanish Council for Scientific Research (CSIC) and Pfizer.
REFERENCES
- 1.Albert, M. L. 2004. Death-defying immunity: do apoptotic cells influence antigen processing and presentation? Nat. Rev. Immunol. 4:223-231. [DOI] [PubMed] [Google Scholar]
- 2.Andrade, A. A., P. N. G. Silva, A. C. T. C. Pereira, L. P. de Sousa, P. C. P. Ferreira, R. T. Gazzinelli, E. G. Kroon, C. Ropert, and C. A. Bonjardim. 2004. The vaccinia virus-stimulated mitogen-activated protein kinase (MAPK) pathway is required for virus multiplication. Biochem. J. 381:437-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arenzana-Seisdedos, F., J. Thompson, M. S. Rodriguez, F. Bachelerie, D. Thomas, and R. T. Hay. 1995. Inducible nuclear expression of newly synthesized I kappa B alpha negatively regulates DNA-binding and transcriptional activities of NF-kappa B. Mol. Cell. Biol. 15:2689-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belyakov, I. M., P. Earl, A. Dzutsev, V. A. Kuznetsov, M. Lemon, L. S. Wyatt, J. T. Snyder, J. D. Ahlers, G. Franchini, B. Moss, and J. A. Berzofsky. 2003. Shared modes of protection against poxvirus infection by attenuated and conventional smallpox vaccine viruses. Proc. Natl. Acad. Sci. USA 16:9458-9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson, J., C. Chougnet, M. Robert-Guroff, D. Montefiori, P. Markham, G. Shearer, R. C. Gallo, M. Cranage, E. Paoletti, K. Limbach, D. Venzon, J. Tartaglia, and G. Franchini. 1998. Recombinant vaccine-induced protection against the highly pathogenic simian immunodeficiency virus SIVmac251: dependence on route of challenge exposure. J. Virol. 72:4170-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blomstrom, D. C., D. Fahey, R. Kutny, B. D. Korant, and E. Knight. 1986. Molecular characterization of the interferon-induced 15-kDa protein. Molecular cloning and nucleotide and amino acid sequence. J. Biol. Chem. 261:8811-8816. [PubMed] [Google Scholar]
- 7.Chung, C. S., I. A. Vasilevskaya, S. C. Wang, C. H. Bair, and W. Chang. 1997. Apoptosis and host restriction of vaccinia virus in RK13 cells. Virus Res. 52:121-132. [DOI] [PubMed] [Google Scholar]
- 8.Cudmore, S., P. Cossart, G. Griffiths, and M. Way. 1995. Actin-based motility of vaccinia virus. Nature 378:636-638. [DOI] [PubMed] [Google Scholar]
- 9.Edghill-Smith, Y., D. Venzon, T. Karpova, J. McNally, J. Nacsa, W. P. Tsai, E. Tryniszewska, M. Moniuszko, J. Manischewitz, L. R. King, S. J. Snodgrass, J. Parrish, P. Markham, M. Sowers, D. Martin, M. G. Lewis, J. A. Berzofsky, I. M. Belyakov, B. Moss, J. Tartaglia, M. Bray, V. Hirsch, H. Golding, and G. Franchini. 2003. Modelling a safer smallpox vaccination regimen, for human immunodeficiency virus type 1-infected patients in immunocompromised macaques. J. Infect. Dis. 8:1181-1191. [DOI] [PubMed] [Google Scholar]
- 10.Franchini, G., S. Gurunathan, L. Baglyos, S. Plotkin, and J. Tartaglia. 2004. Poxvirus-based vaccine candidates for HIV: two decades of experience with special emphasis on canarypox vectors. Exp. Rev. Vaccines 3:75-88. [DOI] [PubMed] [Google Scholar]
- 11.Frischknecht, F., V. Moreau, S. Rottger, I. Reckmann, C. Superti-Furga, and M. Way. 1999. Actin based motility of vaccinia virus mimics receptor tyrosine kinase signalling. Nature 404:1007-1011. [DOI] [PubMed] [Google Scholar]
- 12.Gallego-Gómez, J. C., C. Risco, D. Rodriguez, P. Cabezas, S. Guerra, J. L. Carrascosa, and M. Esteban. 2003. Differences in virus-induced cell morphology and virus maturation between the WR and MVA strains of vaccinia virus. J. Virol. 77:10606-10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García de la Nava, J., D. Franco Santaella, J. Cuenca Alba, J. M. Carazo, O. Trelles, and A. Pascual-Montano. 2003. Engene: the processing and exploratory analysis of gene expression data. Bioinformatics 19:657-658. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh, S., and M. Karin. 2002. Missing pieces in the NF-kappaB puzzle. Cell 109:81-96. [DOI] [PubMed] [Google Scholar]
- 15.Guerra, S., L. A. López-Fernandez, A. Pascual-Montano, M. Muñoz, K. Harshman, and M. Esteban. 2003. Cellular gene expression survey of vaccinia virus infection of human HeLa cells. J. Virol. 77:6493-6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerra, S., L. A. López-Fernandez, R. Conde, A. Pascual-Montano, M. Muñoz, K. Harshman, and M. Esteban. 2004. Microarray analysis reveals characteristic changes of host cell gene expression in response to attenuated modified vaccinia virus Ankara (MVA) infection of human HeLa cells. J. Virol. 78:5820-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guerra, S., M. Aracil, R. Conde, A. Bernad, and M. Esteban. 2005. Wiskott-Aldrich syndrome protein is needed for vaccinia virus pathogenesis. J. Virol. 79:2133-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hai, T., C. D. Wolfgang, D. K. Marsee, A. E. Allen, and U. Sivaprasad. 1999. ATF3 and stress responses. Gene Exp. 7:321-335. [PMC free article] [PubMed] [Google Scholar]
- 19.Hai, T., and M. G. Hartman. 2001. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene 1:1-11. [DOI] [PubMed] [Google Scholar]
- 20.Hartman, M. G., D. Lu, M. L. Kim, G. J. Kociba, T. Shukri, J. Buteau, X. Wang, W. L. Frankel, D. Guttridge, M. Prentki, S. T. Grey, D. Ron, and T.Hai. 2004. Role for activating transcription factor 3 in stress-induced beta-cell apoptosis. Mol. Cell. Biol. 13:5721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayden, M. S., and S. Ghosh. 2004. Signaling to NF-kappaB. Genes Dev. 18:2195-2224. [DOI] [PubMed] [Google Scholar]
- 22.Hughes, T. R., M. J. Marton, A. R. Jones, C. J. Roberts, R. Stoughton, C. D. Armour, H. A. Bennett, E. Coffey, H. Dai, Y. D. He, M. J. Kidd, A. M. King, M. R. Meyer, D. Slade, P. Y. Lum, S. B. Stepaniants, D. D. Shoemaker, D.Gachotte, K. Chakraburtty, J. Simon, M. Bard, and S. H. Friend. 2000. Functional discovery via a compendium of expression profiles. Cell 102:109-126. [DOI] [PubMed] [Google Scholar]
- 23.Jourdier, T. M., C. Moste, M. C. Bonnet, F. Delisle, J. P. Tafani, P. Devauchelle, J. Tartaglia, and P. Moingeon. 2003. Local immunotherapy of spontaneous feline fibrosarcomas using recombinant poxviruses expressing interleukin 2 (IL-2). Gene Ther. 26:2126-2132. [DOI] [PubMed] [Google Scholar]
- 24.Kanesa-thasan, N., J. J. Smucny, C. H. Hoke, D. H. Marks, E. Konishi, I. Kurane, D. B. Tang, D. W. Vaughn, P. W. Mason, and R. E. Shope. 2000. Safety and immunogenicity of NYVAC-JEV and ALVAC-JEV attenuated recombinant Japanese encephalitis virus-poxvirus vaccines in vaccinia-nonimmune and vaccinia-immune humans. Vaccine 19:483-491. [DOI] [PubMed] [Google Scholar]
- 25.Kohonen, T. 1997. Self-organizing maps, 2nd ed. Springer-Verlag, Heidelberg, Germany.
- 26.Korant, B. D., D. C. Blomstrom, G. J. Jonak, and E. Knight. 1984. Interferon-induced proteins. Purification and characterization of a 15,000-dalton protein from human and bovine cells induced by interferon. J. Biol. Chem. 23:14835-14839. [PubMed] [Google Scholar]
- 27.Lee, S. B., and M. Esteban. 1994. The interferon-induced double-stranded RNA-activated protein kinase induces apoptosis. Virology 199:491-496. [DOI] [PubMed] [Google Scholar]
- 28.Liang, G., A. D. Wolfgang, B. P. C. Chen, T. H. Chen, and T. Hai. 1996. ATF3 gene: genome organization, promoter and regulation. J. Biol. Chem. 271:1695-1701. [DOI] [PubMed] [Google Scholar]
- 29.Ludwig, H., J. Mages, C. Staib, M. H. Lehmann, R. Lang, and G. Sutter. 2005. Role of viral factor E3L in modified vaccinia virus Ankara infection of human HeLa cells: regulation of the virus life cycle and identification of differentially expressed host genes. J. Virol. 79:2584-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McFadden, G. 2005. Poxvirus tropism. Nat. Rev. Microbiol. 3:201-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Myagkikh, M., S. Alipanah, P. D. Markham, J. Tartaglia, E. Paoletti, R. C. Gallo, G. Franchini, and M. Robert-Guroff. 1996. Multiple immunizations with attenuated poxvirus HIV type 2 recombinants and subunit boosts required for protection of rhesus macaques. AIDS Res. Hum. Retrovir. 12:985-992. [DOI] [PubMed] [Google Scholar]
- 32.Nájera, J. L., C. E. Gómez, M. M. Gherardi, E. Domingo-Gil, and M. Esteban. Unpublished data.
- 33.Nicholl, M. J., L. H. Robinson, and C. M. Preston. 2000. Activation of cellular interferon-responsive genes after infection of human cells with herpes simplex virus type 1. J. Gen. Virol. 9:2215-2218. [DOI] [PubMed] [Google Scholar]
- 34.Ockenhouse, C. F., P. F. Sun, D. E. Lanar, B. T. Wellde, B. T. Hall, K. Kester, J. A. Stoute, A. Magill, U. Krzych, L. Farley, R. A. Wirtz, J. C. Sadoff, D. C. Kaslow, S. Kumar, L. W. Church, J. M. Crutcher, B. Wizel, S. Hoffman, A. Lalvani, A. V. Hill, J. A. Tine, K. P. Guito, C. de Taisne, R. Anders, W. R. Ballou, et al. 1998. Phase I/IIa safety, immunogenicity, and efficacy trial of NYVAC-Pf7, a pox-vectored, multiantigen, multistage vaccine candidate for Plasmodium falciparum malaria. J. Infect. Dis. 6:1664-1673. [DOI] [PubMed] [Google Scholar]
- 35.Odin, L., M. Favrot, D. Poujol, J. P. Michot, P. Moingeon, J. Tartaglia, and I. Puisieux. 2001. Canarypox virus expressing wild type p53 for gene therapy in murine tumors mutated in p53. Cancer Gene Ther. 2:87-98. [DOI] [PubMed] [Google Scholar]
- 36.Oie, L. K., and D. Pickup. 2001. Cowpox and other members of the orthopoxvirus genus interfere with the regulation of NF-κB activation. Virology 288:175-187. [DOI] [PubMed] [Google Scholar]
- 37.Paoletti, E. 1996. Applications of poxvirus vectors to vaccination: an update. Proc. Natl. Acad. Sci. USA 21:11349-11353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perkus, M. E., S. J. Goebel, S. W. Davis, G. P. Johnson, K. Limbach, E. K. Norton, and E. Paoleti. 1990. Vaccinia virus host range genes. Virology 179:276-286. [DOI] [PubMed] [Google Scholar]
- 39.Pogo, B. G., S. M. Melana, and J. Blaho. 2004. Poxvirus infection and apoptosis. Int. Rev. Immunol. 23:61-74. [DOI] [PubMed] [Google Scholar]
- 40.Quackenbush, J. 2002. Microarray data normalization and transformation. Nat. Genet. 32:496-501. [DOI] [PubMed] [Google Scholar]
- 41.Ramsey-Ewing, A. L., and B. Moss. 1996. Complementation of a vaccinia virus host-range K1L gene deletion by the nonhomologous CP77 gene. Virology 222:75-86. [DOI] [PubMed] [Google Scholar]
- 42.Ray, C. A., and D. J. Pickup. 1996. The mode of death of pig kidney cells infected with cowpox virus is governed by the expression of the crmA gene. Virology 217:384-391. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez, J. R., C. Risco, J. L. Carrascosa, M. Esteban, and D. Rodríguez. 1998. Vaccinia virus 15-kilodalton (A14L) protein is essential for assembly and attachment of viral crescents to virosomes. J. Virol. 72:1287-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ritchie, K. J., C. S. Hahn, K. I. Kim, M. Yan, D. Rosario, L. Li, J. C. de la Torre, and D. E. Zhang. 2004. Role of ISG15 protease UBP43 (USP18) in innate immunity to viral infection. Nat. Med. 12:1374-1378. [DOI] [PubMed] [Google Scholar]
- 45.Shisler, J. L., and X. L. Jin. 2004. The vaccinia virus K1L gene product inhibits host NF-kappaB activation by preventing IkappaBalpha degradation. J. Virol. 78:3553-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sivanandham, M., P. Shaw, S. F. Bernik, E. Paoletti, and M. K. Wallack. 1998. Colon cancer cell vaccine prepared with replication-deficient vaccinia viruses encoding B7.1 and interleukin-2 induce antitumor response in syngeneic mice. Cancer Immunol. Immunother. 5:261-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soldani, C., and A. I. Scovassi. 2002. Poly(ADP-ribose) polymerase-1 cleavage during apoptosis: an update. Apoptosis 7:321-328. [DOI] [PubMed] [Google Scholar]
- 48.Stewart, T. L., S. T. Wasilenko, and M. Barry. 2005. Vaccinia virus F1L protein is a tail-anchored protein that functions at the mitochondria to inhibit apoptosis. J. Virol. 79:1084-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tartaglia, J., M. E. Perkus, J. Taylor, E. K. Norton, J. C. Audonnet, W. I. Cox, S. W. Davis, J. van der Hoeven, B. Meignier, and M. Riviere. 1992. NYVAC: a highly attenuated strain of vaccinia virus. Virology 188:217-232. [DOI] [PubMed] [Google Scholar]
- 50.Tartaglia, J., W. I. Cox, S. Pincus, and E. Paoletti. 1994. Safety and immunogenicity of recombinants based on the genetically-engineered vaccinia strain, NYVAC. Dev. Biol. Stand. 82:125-129. [PubMed] [Google Scholar]
- 51.Tewari, M., W. G. Telford, R. A. Miller, and V. M. Dixit. 1995. CrmA, a poxvirus-encoded serpin, inhibits cytotoxic T-lymphocyte-mediated apoptosis. J. Biol. Chem. 270:22705-22708. [DOI] [PubMed] [Google Scholar]
- 52.Wasilenko, S. T., T. L. Stewart, A. F. Meyers, and M. Barry. 2003. Vaccinia virus encodes a previously uncharacterized mitochondrial-associated inhibitor of apoptosis. Proc. Natl. Acad. Sci. USA 24:14345-14350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yan, C., M. S. Jamaluddin, B. Aggarwal, J. Myers, and D. D. Boyd. 2005. Gene expression profiling identifies activating transcription factor 3 as a novel contributor to the proapoptotic effect of curcumin. Mol. Cancer Ther. 2:233-241. [PubMed] [Google Scholar]
- 54.Ying, H., T. Z. Zaks, R. F. Wang, K. R. Irvine, U. S. Kammula, F. M. Marincola, W. W. Leitner, and N. P. Restifo. 1999. Cancer therapy using a self-replicating RNA vaccine. Nat. Med. 5:823-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu, H., J. P. Cong, G. Mantora, T. Gineras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu, W., A. Cowie, G. W. Wasfy, L. Z. Penn, B. Leber, and D. W. Andrews. 1996. Bcl-2 mutants with restricted subcellular location reveal spatially distinct pathways for apoptosis in different cell types. EMBO J. 15:4130-4141. [PMC free article] [PubMed] [Google Scholar]
- 57.Zinszner, H., M. Kuroda, X. Z. Wang, N. Batchvarova, R. T. Lightfoot, H. Remotti, J. L. Stevens, and D. Ron. 1998. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 12:982-995. [DOI] [PMC free article] [PubMed] [Google Scholar]