Abstract
Foamy viruses (FVs) are nonpathogenic retroviruses infecting many species of mammals, notably primates, cattle, and cats. We have examined whether members of the apolipoprotein B-editing catalytic polypeptide-like subunit (APOBEC) family of antiviral cytidine deaminases restrict replication of simian FV. We show that human APOBEC3G is a potent inhibitor of FV infectivity in cell culture experiments. This antiviral activity is associated with cytidine editing of the viral genome. Both molecular FV clones and primary uncloned viruses were susceptible to APOBEC3G, and viral infectivity was also inhibited by murine and simian APOBEC3G homologues, as well as by human APOBEC3F. Wild-type and bet-deleted viruses were similarly sensitive to this antiviral activity, suggesting that Bet does not significantly counteract APOBEC proteins. Moreover, we did not detect FV sequences that may have been targeted by APOBEC in naturally infected macaques, but we observed a few G-to-A substitutions in humans that have been accidentally contaminated by simian FV. In infected hosts, the persistence strategy employed by FV might be based on low levels of replication, as well as avoidance of cells expressing large amounts of active cytidine deaminases.
Foamy viruses (FVs) or spumaretroviruses represent a large family of retroviruses that have been isolated in various mammals (for recent reviews, see references 11, 15, and 35). They are highly prevalent in nonhuman primates, and at least 11 different simian FV (SFV) viral subtypes have been described in monkey species (7, 15, 44). FVs are particularly well adapted to their natural hosts. SFVs may have cospeciated with Old World primates for >30 million years, making them the oldest known vertebrate RNA viruses (63). These viruses are innocuous in naturally or experimentally infected animals, in which they induce persistent, lifelong infections (35). SFVs are readily transmitted via saliva, and seroprevalence exceeds rates of 70% in some species (7, 15, 44). Several cases of simian-to-human transmissions have been reported, generally after bites or scratches (8, 22, 23, 69). Up to 2% of humans in contact with monkeys harbor anti-SFV antibodies or are positive by PCR analysis (22). As for monkeys, human infection is so far considered to be nonpathogenic. Moreover, there is no evidence for secondary transmission, suggesting that humans are dead-end hosts for these retroviruses.
In cell culture, FVs generally provoke characteristic foamlike cytopathic effects (CPEs) and large syncytia and display a wide tropism (24, 35). FVs establish persistent productive infection in human hematopoietic cell lines, as well as acute infection in primary human lymphocytes (43, 51, 75). The CPE varies according to the cell type. For instance, killing of infected cells was observed with primary CD4 lymphocytes but not with CD8 cells (43). In infected monkeys and humans, various hematopoietic cell types harbor viral sequences. It was initially reported that CD8+ T cells may represent a viral reservoir in monkeys (African green monkeys [AGM] and chimpanzees) and in some humans (66). This tropism for CD8+ cells was not observed in another study of a patient infected by AGM FV, which detected FV in monocytes and B cells and not in CD8 lymphocytes (8). The primary viral reservoir is thus not well characterized in vivo.
The replication strategy of FVs differs in some aspects from that of other retroviruses, presenting similarities with the life cycle of pararetrovirus (i.e., hepatitis B virus [HBV]), as well as of endogenous retroviruses (11, 36, 48, 73). For instance, reverse transcription occurs to a large extent in the producer cell, leading to the presence of double-stranded viral DNA in the extracellular virion (12, 48, 73). Other properties include the formation of a specific pol mRNA, the budding of virions into the endoplasmic reticulum rather at the cell surface, and the presence of an internal promoter, which drives the synthesis of the early regulatory proteins Bel1 (or Tas) and Bet. Tas is a nuclear transactivator essential for viral replication, which also acts as a suppressor of cellular antiviral microRNA (29). Bet plays an important role in the establishment and control of viral persistence in vitro and in vivo (8, 50). In chronically infected cells, a spliced Tas-defective viral genome may negatively interfere with the replication of parental virus, probably by the production of Bet (4, 50). Although Bet and an Env-Bet fusion protein are secreted by infected cells and can be taken up by neighboring cells, the role of this phenomenon is not fully understood (16, 30, 34). Besides infecting novel naive cells, FVs share with endogenous retroviruses the ability to retrotranspose within the producer cell (20, 21). Retrotransposition is a copy-and-paste process leading to the integration in the host cell genome of cDNA which had been reverse transcribed intracellularly (5). The physiological relevance of this “replicative shortcut” remains to be characterized (20).
Human immunodeficiency virus (HIV), HBV, and some endogenous retroviruses are restricted by members of the apolipoprotein B-editing catalytic polypeptide-like subunit (APOBEC) cytidine deaminase family (13, 14, 26, 39, 54, 58, 60, 65, 67). APOBEC3G and APOBEC3F are cytoplasmic proteins that are incorporated into HIV virions and which deaminate cytosine residues to uracil (C to U) in nascent DNA. Most of the uracil-containing viral DNAs are then degraded, likely by cellular enzymes, prior to integration. Some molecules escape degradation, and the resulting proviruses contain numerous guanosine-to-adenosine (G-to-A) substitutions in the plus strand (18, 31, 39, 41, 58, 62). Human APOBEC3G and APOBEC3F, as well as rat APOBEC1, can also induce C-to-T mutations in the HIV genome, albeit at a low frequency (2, 3, 17, 68, 72, 77). Furthermore, editing-independent anti-HIV effects of APOBEC3G have recently been demonstrated, indicating that multiple mechanisms mediate the restriction activity of this family of proteins (9, 45). Interestingly, the HIV Vif protein neutralizes the APOBEC3G-mediated antiviral effect by promoting proteasomal degradation of the enzyme (10, 42, 59, 76).
We have examined here whether APOBEC family members restrict replication of FV. We show that human APOBEC3G is a potent inhibitor of FV infectivity and induces a G-to-A editing of the viral genome. Very recently, two reports indicated that FV Bet proteins inhibit the antiviral activity of APOBEC3 (37, 49). Löchelt et al. observed that Bet-deficient and not wild-type (WT) feline FVs were susceptible to feline APOBEC3 editing. APOBEC was present in Bet-deficient and not in WT FV particles, and edited viral genomes were detected in released virions, indicating that cytidine deamination occurred in producing cells (37). Russell et al. reported that several APOBEC3 proteins inhibit the infectivity of a simian FV-based viral vector, with a partial rescue of infectivity when Bet was overexpressed (49). However, we show here that Bet does not efficiently counteract APOBEC activity against FV or HIV, suggesting that this regulatory protein is not a functional analogue of Vif. We also report that besides human APOBEC3G, human APOBEC3F and murine and simian APOBEC3G homologues restrict FV replication in cell culture experiments. In addition, by using a sensitive PCR method capable of selectively amplifying G-to-A hypermutated genomes (61), we did not detect any FV sequences that have been targeted by APOBEC3 in infected macaques. We analyzed peripheral blood mononuclear cells (PBMCs) from humans contaminated by simian FVs and observed a few G-to-A substitutions in viral genomes. Collectively, these results indicate that FVs are efficiently restricted by APOBEC cytidine deaminase, likely impacting the life style of this peculiar class of retroviruses in their animal and human hosts.
MATERIALS AND METHODS
Cells and plasmids.
Cell lines were grown in Dulbecco's modified essential medium containing GlutaMAX I and sodium pyruvate (Invitrogen) and supplemented with 10% fetal calf serum (Sigma) and antibiotics. The following cell lines were used: 293T cells, FAB cells (BHK21-derived indicator cells containing a β-galactosidase gene [β-gal gene] under the control of the FV long terminal repeat [LTR]) (74), HeLa cells, P4 cells (HeLa CD4 indicator cells containing a β-gal gene under the control of the HIV LTR) (55), and HeLa-hA3G cells (HeLa cells stably expressing human APOBEC3G, obtained through the NIH AIDS Research and Reference Reagent Program (27). The following APOBEC expression plasmids were used: hA3G (carrying a V5 tag), was a gift from Allan Hance (31); hA1, hA2, hA3B, hA3C, hA3F, and Agm3G were obtained through the NIH AIDS Research and Reference Reagent Program (41, 77); and mA1, mA2, mA3 were a gift from N. Landau (41). Chimpanzee APOBEC3G (CpzA3G) was obtained from the cDNA of Pan troglodytes PBMCs and cloned with a C-terminal V5 tag in pCDNA6 plasmid. The GenBank accession number for CpzA3G is DQ185513. As expected (41), CpzA3G restricted Δvif and not WT HIV (not shown). The cytomegalovirus (CMV)-Bet expression vector has been previously described (30). The following plasmids encoding for infectious FV clones were used: FV clone 13 (WT FV, containing an heterologous CMV promoter) (38), Δbet (an isogenic provirus carrying a frameshift mutation in the bet open reading frame leading to a C-terminal-truncated Bet protein) (50), and pHSRV2 (a FV provirus, containing its natural LTR promoter (38). WT HIV and Δvif proviruses (from the NL4-3 HIV-1 strain) were a kind gift of Allan Hance (31).
Virus production and infection.
FVs produced from plasmids were obtained by transfection of 293T cells. When stated, APOBEC expression plasmids were cotransfected at the indicated FV:APOBEC plasmid ratio. After overnight incubation, fresh medium was added. Media and cells were harvested 2 days later, frozen and thawed three times, clarified by a 10-min centrifugation at 6,000 rpm at 4°C, and filtered through a 0.45-μm-pore-size filter. Similar amounts of viruses, as judged by Western blot analysis with anti-Gag FV antibodies, were used to infect indicator FAB cells (74). Measurement of β-Gal activity was carried out 48 h later. HeLa and HeLa-hA3G cells were infected with two viral strains (T1FV and T2FV) isolated from two naturally infected macaques. High and low levels of inoculum were used for infections, corresponding, respectively, to pure and 1:5 dilutions of the viral preparations. Cells were then grown up to 16 days postinfection (p.i.). At the indicated days, cell samples were fixed with glutaraldehyde (0.5% in phosphate-buffered saline [PBS]) and stained with Giemsa (5% in PBS). Syncytia and viral foci were then scored under a binocular microscope. HIV production, titration, and infection of P4 indicator cells were performed as previously described (56).
Analysis of human specimens.
We identified four hunters from southern Cameroon infected by gorilla FV, likely through severe bites that occurred some 10 to 30 years ago (S. Calatini et al., manuscript in preparation). These four men were FV seropositive by Western blot analysis. High-molecular-weight DNA was extracted from buffy coats and analyzed by PCR for FV sequences. Informed oral consent to perform this analysis was obtained from each individual.
Animals.
A colony of 57 Celebes macaques (Macaca tonkeana) (25) housed in the Strasbourg Primatology Center were investigated for the presence of simian FV infection. This colony was established from three animals originating from the central part of Sulawesi (Indonesia) and brought to the center in 1972. Since then, the animals have been maintained for ethological studies and isolated from other monkey species. The animals have never been infected with any biological material. Plasma samples were investigated for FV antibodies by a Western blot assay that uses a chimpanzee FV strain as viral antigens. Criteria for Western blot seropositivity were the presence of a clear reactivity to both FV Gag p68 and p72.
Virus isolation from naturally infected animals.
Virus isolation was carried out with samples from animals showing a strong FV seropositivity. PBMCs were isolated; maintained for 2 days in RPMI medium containing 20% fetal calf serum, antibiotics, and phytohemagglutinin (3 μg/ml); and further stimulated with interleukin 2 (100 U/ml). After 4 days of growth, PBMCs were cocultivated with BHK-21 cells. Cultures were checked daily for syncytial CPE typical of FV infection. The presence of FV in BHK-21 cells was verified by immunofluorescence analysis. Supernatants from infected BHK-21 cells were filtered (0.45-μm-pore-size filters) and frozen until further use.
Western blot analysis.
At 24 h or 48 h after transfection, cells were lysed in PBS-1% NP-40 supplemented with protease inhibitors (Roche). Lysates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis using 4 to 12% NuPage gels (Invitrogen). The following antibodies were used: anti-FV rabbit serum and anti-Bet monoclonal antibodies (MAb) D11 (30), anti-HIV Gag p24 MAb (47), and anti-V5 MAb (Invitrogen) to detect V5-tagged hA3G.
Detection of FV sequences by PCR.
A fragment of the integrase gene of gorilla and macaque FV genomes was amplified by a nested procedure. To increase sensitivity and specificity, a hot-start PCR was performed for both amplifications. First-round primers for gorilla FV genome amplification were 5′ CCTAAACAATATACATATTATATRRAA and 5′ AAGAAYATGYAAATATYYRTTGAG; for the second round, primers were 5′ AAATAATTCCTYYYTCCTTARAA and 5′ ATGAAAAATTTATYAAAAGGYTTTYAA, where Y = T/C and R = A/G. First-round primers for macaque FV amplification were 5′ CGGAGTTGCTACAARRACATWATCC and 5′ YYTGTYATACYATCGACWACTA, while primers 5′ CGGTCTCTTCYAAGTATTRGTRGCCT and 5′ TYGACTACTACAAGGACATGTAAAT were used for the second round. Hypermutated genomes were identified by differential DNA denaturation PCR (3DPCR) in a two-round procedure (61). The first reaction involved standard amplification. The reaction parameters were 95°C for 5 min, followed by 35 cycles (each consisting of 95°C for 1 min, 45°C for 30 s, and 72°C for 30 s), and finally 10 min at 72°C for gorilla and macaque FV genomes. Differential amplification occurred in the second round by using the equivalent of 0.5 μl of the first-round reaction as input. Cycling condition were 95°C or 85°C (for FV gorilla samples) or 83°C (for FV macaque samples) for 5 min, followed by 35 cycles (each consisting of 95, 85, and 83°C for 1 min; 45°C for 30 s; and 72°C for 30 s), and 10 min at 72°C. The buffer conditions were 2.5 mM MgCl2, 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 200 μM each deoxynucleoside triphosphate, 100 μM each primer, and 2.5 U of Taq DNA polymerase (Cetus) in a final volume of 50 μl. Second-round PCR for gorilla and macaque FV amplifications yielded a 244-bp and 217-bp fragment, respectively. PCR products were purified from agarose gels (Qiaex II kit; QIAGEN, France) and ligated in the TOPO TA cloning vector (Invitrogen, France). After transformation of XL1-Blue cells, individual clones were picked and analyzed. Sequencing was performed using Thermosequenase (USB; Amersham) on an ABI373A sequencer. Analysis of FV genomes present in HeLa and HeLa-hA3G cells was done with internal primers, by performing a single-round PCR under normal conditions. About 200 sequences were analyzed for each point of infected HeLa-hA3G cells. For naturally occurring FV sequences present in macaques and in humans, about 20 sequences were analyzed per point.
RESULTS
hA3G inhibits SFV replication.
To study the effect of hA3G on FV infectivity, viral particles were produced by transfection of a FV provirus (clone 13) (32, 38) in 293T cells in the presence of increasing doses of an expression plasmid for hA3G. Viral infectivity was measured in a single-round assay of replication, based on the infection of BHK21-derived indicator cells containing a β-gal gene under the control of the FV LTR (FAB cells) (74). Western blot analysis showed that hA3G did not significantly affect the production of Gag proteins, which appeared as a characteristic doublet at 72 and 68 kDa (Fig. 1A). hA3G was detected with an expected apparent molecular mass of 46 kDa. A minor band of 23 kDa was also observed with anti-hA3G MAbs and likely corresponded to a natural degradation product of the deaminase (see below). Interestingly, hA3G markedly decreased FV infectivity (Fig. 1B). The virus was very sensitive to this inhibitory effect, which was observed at low molar ratios of hA3G:FV expression plasmids (Fig. 1B). We then examined whether the viral Bet protein interfered with hA3G effect. With this aim, we tested the susceptibility to hA3G of Δbet, an isogenic provirus carrying a premature stop codon in the bet open reading frame, leading to a C-terminal-truncated Bet protein (32, 50). Viral production was similar for FV and Δbet, as judged by Gag expression (not shown), whereas the 62-kDa Bet protein was only detected in cells infected with the WT virus (Fig. 1C). In the absence of hA3G, a small decrease of viral infectivity was detected with Δbet, compared to FV, as previously reported (Fig. 1C) (74). hA3G efficiently inhibited Δbet, and a similar antiviral activity was observed against both viruses in a dose-response analysis of hA3G expression level (Fig. 1C). Occasionally, when low levels of hA3G were present, we observed that WT FV was slightly more resistant to the cytidine deaminase than Δbet (Fig. 1C). However, these differences were not significant (not shown). In comparison, hA3G potently inhibited HIV Δvif, and not WT HIV, whatever the levels of hA3G expressed (see Fig. S1 in the supplemental material). This strongly suggests that Bet does not significantly counteract the activity of hA3G against the clone 13 FV strain.
FIG. 1.
Human APOBEC3G (hA3G) restricts FV infection. (A) hA3G does not affect the production of FV virions. hA3G and FV virions (clone 13 provirus) were produced in 293T cells by transfection of plasmids (at the ratio indicated). Expression of FV Gag and hA3G was assessed by Western blot analysis of cell lysates. FV proteins were detected with an anti-Gag serum and hA3G proteins were detected with an anti-V5 tag MAb. Apparent molecular masses (in kilodaltons) are indicated on the right. (B) hA3G inhibits infectivity of wild-type and Δbet FV. The infectivity of viral preparations described in the legend to panel A was tested on FAB indicator cells, which express β-Gal upon FV infection. β-Gal levels obtained with FV and Δbet produced without hA3G were set at 100%. (C) Comparison of the infectivity of FV and Δbet. Similar amounts of virions were used to infect indicator FAB cells, and infectivity of FV was set at 100%. Bet expression was detected in cells infected with FV and not with Δbet. A CMV-Bet expression plasmid was transfected as a control. (D) hA3G inhibits infectivity of the FV strain HSRV2. hA3G and FV virions (HSRV2 provirus) were produced, and viral infectivity was tested as described above. Data are means ± standard deviation (SD) of triplicates and are representative of two to five independent experiments.
We extended this observation to other viral isolates. In clone 13 FV, a heterologous CMV promoter drives expression of viral proteins (32, 38). We first examined whether hA3G was active against the infectious molecular clone pHSRV2, which includes its natural FV LTR promoter (52). Again, transient expression of hA3G significantly inhibited HSRV2 infectivity (Fig. 1D). We also studied the susceptibility of primary FV strains, isolated from naturally infected macaques (7). To study the effect of hA3G, we compared viral spread in control HeLa cells and in HeLa cells stably expressing hA3G (HeLa-hA3G cells) (27). In the later cells, production of HIV is Vif dependent (27; data not shown). Two primary isolates (T1FV and T2FV) were isolated from two different macaques and analyzed. In control HeLa cells, as previously reported (46), replication of T1FV occurred slowly, and syncytia were observed at day 6 p.i. and increased progressively in size and number over time (Fig. 2A). At day 16 p.i., about 1,200 syncytia/well were visible. Of note, the number of syncytia correlated with the viral input (Fig. 2B). In HeLa-hA3G cells, the number of syncytia was greatly reduced. For instance, at day 16 p.i., according to the viral input, 50 to 200 syncytia/well were detected (Fig. 2B). Similar results were obtained with the viral strain T2FV (not shown). Altogether, these results indicate that hA3G efficiently restricts SFV replication. This antiviral effect was observed with various viral isolates, with either single-cycle or multiple-round replication analyses, and with different cell types in which the deaminase was either transiently or stably expressed.
FIG. 2.
Susceptibility of a primary FV strain to human APOBEC3G. (A) Viral replication and syncytium formation in HeLa and HeLa-hA3G cells. The indicated cells were infected at a high multiplicity of infection with T1FV, a primary viral strain isolated from a naturally infected macaque. At various days postinfection, cells were stained with Giemsa to visualize syncytium formation and virus-induced cytopathic effect. (B) Quantification of syncytium formation in HeLa and HeLa-hA3G cells. Cells were infected with T1FV (at either a high or low multiplicity of infection value) and cultured up to 16 days p.i. Numbers of syncytia per well (from six-well plates) were scored at the indicated time points. Data are representative of three independent experiments.
Absence of a Vif-like activity of Bet.
That hA3G alters replication of various FV strains, including primary isolates and WT and Δbet clone 13 proviruses, strongly suggests that Bet does not neutralize hA3G activity. To confirm this point, we examined whether Bet, when expressed in the absence of other FV proteins, exhibits a Vif-like function on HIV replication. A Bet expression vector was cotransfected with WT HIV or HIV Δvif proviruses in the absence or presence of hA3G. Gag p24, Bet, and hA3G expression in transfected cells was verified by Western blot analysis (Fig. 3A), while viral release was measured by a Gag p24 enzyme-linked immunosorbent assay (not shown). Gag p24 was similarly detected with WT HIV and HIV Δvif, with or without Bet. Interestingly, the steady-state levels of the hA3G 46-kDa species were greatly reduced in cells expressing WT HIV, compared to Δvif. This reduction was associated with an increase of the 23-kDa species (Fig. 3A). Therefore, Vif induces hA3G degradation and the accumulation of a 23-kDa degradative intermediate product. In contrast, the presence of Bet did not affect hA3G steady-state levels (Fig. 3A). Infectivity of the HIV virions was then assessed with P4 indicator cells. As expected, in the absence of Bet, the infectivity of Δvif, but not that of WT HIV, was inhibited by hA3G (Fig. 3B). Similar results were obtained when Bet was coexpressed (Fig. 3) and with various amounts of hA3G expression plasmid transfected (not shown). Therefore, Bet does not complement the Δvif defect. This observation, together with that of a similar antiviral activity of hA3G on WT and Δbet FV, indicates that Bet is not a functional analogue of Vif and does not efficiently counteract the antiviral activity of hA3G.
FIG. 3.
FV Bet does not rescue the HIV Δvif defect. (A) HIV Vif but not FV Bet decreases hA3G steady-state levels. 293T cells were transfected with HIV or Δvif proviruses and, when stated, with hA3G- and Bet-encoding plasmids. Expression of hA3G was assessed by Western blot analysis of cell lysates. As controls, levels of HIV Gag and FV Bet were also visualized. Apparent molecular masses (in kilodaltons) are indicated on the right. (B) hA3G-mediated restriction of HIV and Δvif viruses, with or without FV Bet. Viral supernatants from transfected 293T cells described in the legend to panel A were normalized for Gag p24 release and used to infect target P4 cells, which express β-Gal upon HIV infection. The infectivities of HIV and Δvif in the absence of hA3G were set at 100%. Data are means ± SD of two independent experiments.
hA3G induces G-to-A mutations in SFV genome.
We examined whether hA3G induces G-to-A substitutions in SFV genomic DNA. To this end, DNA from HeLa and HeLa-hA3G cells infected with T1FV or T2FV was prepared and amplified by PCR with FV-specific primers. The DNA products were cloned and sequenced. The sequences corresponding to a 166-bp fragment of the integrase gene, from 10 to 12 independent clones, are shown in Fig. 4. In the absence of hA3G, very few substitutions were identified, none of which were G-to-A substitutions. In sharp contrast, in the presence of hA3G, numerous G-to-A substitutions were observed, both at day 6 (not shown) and day 16 (Fig. 4) postinfection. Deamination hotspots were visible, and GG dinucleotides were preferentially mutated (the substituted G being underlined) as expected (14, 18, 33, 39, 62, 72). Thus, the antiviral activity of hA3G on FV is associated with editing of viral DNA.
FIG. 4.
hA3G triggers G-to-A mutations in the FV genome. HeLa and HeLa-hA3G were infected with two primary FV isolates, T1FV (top) and T2FV (bottom). At day 16 p.i., FV genomes present in cell extracts were amplified by PCR, cloned, and sequenced. For points from HeLa-hA3G cells, individual sequences from 10 to 12 different clones with a high number of G-to-A substitutions were selected and aligned with the consensus FV sequence (corresponding to the major sequence obtained from HeLa cells). A representative fragment of the integrase gene is shown. Similar results were observed at an earlier time point (day 6 p.i.; data not shown).
Antiviral effect of other members of the APOBEC family.
The ability of Vif to block the antiviral activity of APOBEC3G is species specific (6, 28, 40, 41, 53). HIV and simian immunodeficiency virus (SIV) Vif block the APOBEC3G of the species from which they are derived but generally fail to inhibit those of other species. The FV clone 13 strain originated from a chimpanzee. We thus examined the sensitivity of FV to APOBEC3G from its natural host (CpzA3G). Interestingly, this molecule efficiently inhibited WT FV and Δbet (Fig. 5), and no significant difference in the sensitivity of each virus was observed, even at lower CpzA3G:virus ratios (not shown). Therefore, FV is similarly susceptible to restriction by human and chimpanzee APOBEC3G.
FIG. 5.
Effect of various APOBEC proteins on FV infectivity. 293T cells were transiently transfected with WT or Δbet FV (clone 13 strain), along with expression plasmids for the indicated APOBEC proteins. Viral stocks were prepared, and their infectivity was tested on FAB indicator cells. β-Gal levels obtained with FV and Δbet produced in the absence of any APOBEC proteins were set at 100%. The FV:APOBEC plasmid ratio was 1:0.2 or 1:0.5 (for CpzA3G). Similar behavior in WT and Δbet FV was observed at an FV:CpzA3G plasmid ratio of 1:0.05 (not shown). Data are means ± SD of triplicates and are representative of two to three independent experiments.
The human APOBEC family includes activation-induced deaminase (AID), APOBEC1 (hA1), APOBEC2 (hA2), eight APOBEC3 (hA3A to hA3H), and APOBEC4 members (1, 19, 26). The same genes expressing these proteins are found in chimpanzees and probably in other monkeys (19, 67). In mice, only one APOBEC3 (mA3) gene is present, along with APOBEC1 (mA1) and APOBEC2 (mA2). hA3G was the first cytidine deaminase demonstrated to have antiviral activity. hA3F was then shown to be active against HIV and a target of Vif (2, 17, 33, 68, 77). hA3B modestly affects HIV and SIV, whereas hA3C has antiviral activity only against SIV (2) (71). In parallel to hA3G, simian A3G and mA3 display a potent antiretroviral activity (6, 40, 41, 53, 70). We have investigated the capacity of diverse APOBEC proteins to restrict FV replication. WT FV virus was produced in the presence of either AID, hA1, hA2, hA3B, hA3C, hA3F, hA3G, mA1, mA2, mA3, or AGM A3G and used in single-cycle assays of viral replication. We distinguished three groups of APOBEC proteins according to their anti-FV activity. The murine, AGM, and Cpz counterparts of hA3G potently inhibited FV infectivity, as was also the case for hA3F (Fig. 5). The second group included hA3B, with only a modest antiviral effect of <50% of reduction of infectivity. The third group corresponded to noninhibitory proteins (AID, hA1, hA2, hA3C, mA1, and mA2). Of note, similar profiles were obtained with WT and Δbet FV (Fig. 5), supporting the notion that none of the cytidine deaminases were significantly sensitive to Bet.
Genetic analysis of naturally occurring FV isolates in macaques.
SFV replication in animals is associated with a minimal genetic variation (57, 63). Given the high susceptibility of FV to APOBEC restriction, we sought edited viral DNAs in naturally infected macaques. Uncultured PBMCs from seven FV-seropositive animals were analyzed. Using a classical PCR approach of amplification and cloning, we did not detect significant differences when comparing a locus in the integrase gene within each of these animals (not shown). We then screened the same samples by using a sensitive PCR-based method, 3DPCR, capable of preferentially amplifying G-to-A hypermutated genomes present at a frequency at least 10−4 (meaning 1 hypermutated sequence per 10,000 nonmutated genomes) (60, 61) This method relies on the fact that the DNA of an AT-rich variant melts at a slightly lower temperature than parental DNA. By lowering the denaturation temperature, edited genomes are selectively amplified (61). A 425-bp DNA fragment corresponding to the integrase gene was first amplified at 95°C. A second round was performed with nested primers at lower denaturation temperatures until there was no product amplification. DNA from the last positive amplification was cloned and sequenced. As can be seen from the aligned sequences (see Fig. S1 in the supplemental material), animals were infected with two close FV strains, one that was found in four animals, whereas the other was detected in three monkeys. We were able to detect a low level of genetic heterogeneity among each macaque, reflecting the presence of minor variants. However, nothing encoding an excess of G-to-A substitutions, the hallmarks of APOBEC3-edited DNA, was found.
Genetic analysis of naturally occurring FV isolates in infected humans.
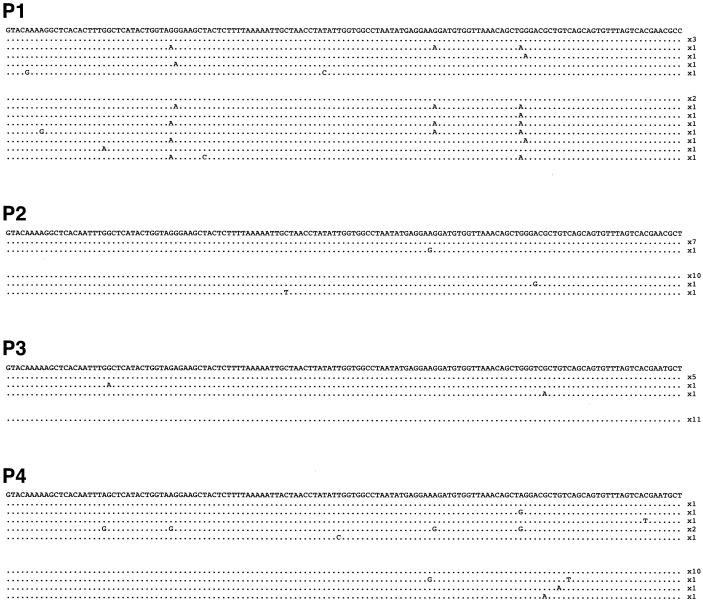
Simian FV infection has been reported in a small number of humans exposed to monkeys, such as animal handlers and hunters (8, 22, 23, 69). We examined the variability of FV sequences in such individuals. In the course of an epidemiological analysis of FV transmission to human populations in southern Cameroon (Calatini et al., manuscript in preparation), we identified four hunters infected by FVs from gorillas, likely through severe bites that occurred between 10 to 30 years ago. In these individuals, viral loads in PBMCs were particularly low, and a nested-PCR technique was required to amplify viral genetic material. Samples were screened by regular PCR and 3DPCR. The sequences, all closely related to known gorilla FV sequences, were aligned to the major sequence from each individual (Fig. 6). Interestingly, we observed several G-to-A transitions in samples from these persons. Multiple mechanisms generate mutations and may explain this variability. However, we noticed that some of the mutations were in the GG and GA contexts, which may be compatible with the existence of an APOBEC3G or APOBEC3F activity that targeted the viruses in their hosts.
FIG. 6.
Analysis of naturally occurring viral sequences from FV-infected humans. Uncultured PBMCs from four humans (P1 to P4) accidentally infected by gorilla FVs were analyzed for the presence of FV sequences. A total of 16 to 20 different PCR-derived clones are depicted for each individual. Analysis was performed using either regular PCR (95°C denaturation temperature; top part of the sequences for each individual) or 3DPCR (85°C denaturation temperature; bottom part of the sequences). Sequences are aligned to the major sequence of each individual, which served as a reference. The frequency of occurrence of each unique sequence is depicted on the right. A fragment of the integrase gene is shown.
DISCUSSION
Nonhuman primates become infected by FVs as young adults, probably via saliva, but do not develop any evident pathology (36). We demonstrate here that multiple APOBEC proteins have the capacity to restrict FV replication in cell culture experiments. hA3G protein efficiently inhibits replication of molecularly cloned viruses, as well as viral strains isolated from naturally infected animals, and edits their genomes. Murine (mA3) and simian (AgmA3G and CpzA3G) homologues are also potently active, as is hA3F. In contrast, hA3B displays a modest inhibitory effect, whereas other family members tested (AID, hA1, hA2, hA3C, mA1, and mA2) are not active against FV.
That some APOBEC proteins neutralize FV replication was not totally unexpected, due to their known activity on HIV, HBV, murine leukemia virus, and endogenous retroviruses (14, 58, 64). However, not all retroviruses are sensitive to these enzymes, and species-specific antiviral activities of APOBEC proteins have been reported for HIV and murine leukemia virus (1, 19, 28, 41). Our study revealed original features of the relations between FV and APOBEC. We show that these viruses are targeted by murine, simian, or human molecules, suggesting that there is no significant species-specific sensitivity of FVs to APOBEC.
Primate lentiviruses have evolved a viral protein, Vif, to degrade APOBECs and thus to overcome them (58, 76). This is apparently not the case for FV, since primary isolates were restricted by hA3G. FVs possess various regulatory proteins, whose functions are not fully understood. It has been recently reported that Bet may counteract the antiviral activity of APOBEC (37, 49). Using feline FVs, Löchelt et al. observed that Bet-deficient but not WT viruses are susceptible to feline APOBEC3 editing (37). Russell et al. reported that simian, human, and murine APOBEC3 proteins inhibit the infectivity of a simian FV-based viral vector (49). A partial rescue of infectivity was observed when Bet was overexpressed in trans (in the presence of up to 12-fold-more Bet than APOBEC expression plasmids) (49). Both reports concluded that FV Bet, in contrast to HIV Vif, does not induce APOBEC degradation. In both cases, experiments were performed in single-cycle assays of infectivity, using cloned viruses or viral vectors. We examined here whether Bet may act as a functional homologue of Vif. Several lines of evidence argue against this hypothesis. First, WT and Δbet isogenic strains appeared to be similarly sensitive to the inhibitory effect of hA3G and other deaminases. Second, Bet was unable to rescue Vif-defective HIV infectivity in the presence of hA3G. Third, naturally occurring viral isolates were sensitive to hA3G. Differences between these two reports and our own experiments may be due to the use of different FV strains, to the relative levels of expression in APOBEC and viral proteins, and to infectivity assays. To reconcile both views, it seems that Bet, when overexpressed, might bind APOBEC3, without inducing its degradation, thus exhibiting a protective effect only when small amounts of cytidine deaminase are present within the cell. Moreover, although we cannot formally rule out that all FV strains tested in our study carry defective Bet proteins, our results suggest that FVs are not equipped with a protein whose task would be to efficiently neutralize APOBEC proteins.
What might then be the strategy of FVs to escape this antiviral barrier and to persist in their hosts? Like endogenous retroviruses, FVs are able to retrotranspose within the infected cell (20, 21). However, it is unlikely that this mode of propagation provides a means to escape neutralization by cytidine deaminases, since murine and human APOBEC molecules potently inhibit retrotransposition of endogenous retroviruses (13, 14, 54).
One clue may reside in the low viral loads observed in infected animals, which suggest that FVs replicate at only low levels in vivo. A significant diversity is observed among FVs from different simian species, suggesting that cross-species infection was associated with adaptative changes (7, 15). However, within each monkey species, viral persistence is associated with strong genetic stability. “Molecular clock” analysis revealed an extremely low rate of SFV evolution (63). A long-term longitudinal study of infected African green monkeys revealed high homology (>99.5%) within the env gene (57). Our study extends these findings. We did not detect any significant nucleotide diversity in the predominant viral sequences isolated from seven naturally infected animals. Furthermore, no hyperedited viral genomes were detected in PBMC DNA. The absence of imprints of an APOBEC activity may seem paradoxical at first sight. However, it is possible that hypermutated, defective genomes are counterselected and thus below detection thresholds in the low viral loads obtained from PBMC samples. It will be worth looking for such mutated genomes in other tissues or fluids (saliva, plasma) where FVs may replicate more actively. The viral reservoir of FV in vivo is not known, but one can also hypothesize that this virus replicates essentially in APOBEC-negative cells or that the restriction is not absolute in some APOBEC-positive cells. On the other hand, editing-independent antiviral effects of APOBEC have been previously described (9, 45) and may also be operative in FV-infected primates, lowering viral loads. The sensitivity of a given retrovirus to APOBEC proteins also likely depends on its kinetics of replication in vivo. The lifestyle of FVs, characterized by small amounts of replicative cycles, may facilitate escape from cytidine deaminases.
A small fraction (<2%) of humans in contact with monkeys are infected by FVs (8, 22, 23, 69). Therefore, FVs are actively crossing into human populations, despite the presence of APOBEC proteins. However, viral loads are extremely low in infected humans: a sensitive nested-PCR method is required to isolate FV sequences from blood, and no virus has been detected in the saliva (23). Consistently, no secondary human infections have been reported so far, despite decades of human infection by SFVs (23). We have analyzed here viral sequences from four humans infected by gorilla FVs. We report the presence of a few viral genomes bearing G-to-A substitutions in these individuals. Numerous mechanisms may explain the presence of these mutants (for example, natural polymorphism of the viral strains, reverse transcriptase errors, and genome editing). As for monkeys, it is possible that heavily mutated and hence defective genomes are constantly counterselected. It is thus tempting to speculate that in FV-infected humans and monkeys, APOBEC family members are involved in the control of viral spread.
Supplementary Material
Acknowledgments
We thank Maxine Linial, Rolf Flügel, Allan Hance, Ned Landau, Klaus Strebel, and the NIH AIDS Research and Reference Reagent Program for the kind gift of reagents; Françoise Porrot for excellent technical help; and N. Herrenschmidt and F. Wanert for access to the troop of M. tonkeana macaques at the primatology center (Strasbourg, France).
This work was supported by grants from the Institut Pasteur, ANRS, Sidaction, CNRS, INSERM, and the European Community. F.D. is a fellow of Sidaction, R.S. is the recipient of a Boehringer-Ingelheim Fonds fellowship, and S.C. is the recipient of a Virus Cancer Prevention Association fellowship.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Bieniasz, P. D. 2004. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 5:1109-1115. [DOI] [PubMed] [Google Scholar]
- 2.Bishop, K. N., R. K. Holmes, A. M. Sheehy, N. O. Davidson, S. J. Cho, and M. H. Malim. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392-1396. [DOI] [PubMed] [Google Scholar]
- 3.Bishop, K. N., R. K. Holmes, A. M. Sheehy, and M. H. Malim. 2004. APOBEC-mediated editing of viral RNA. Science 305:645. [DOI] [PubMed] [Google Scholar]
- 4.Bock, M., M. Heinkelein, D. Lindemann, and A. Rethwilm. 1998. Cells expressing the human foamy virus (HFV) accessory Bet protein are resistant to productive HFV superinfection. Virology 250:194-204. [DOI] [PubMed] [Google Scholar]
- 5.Boeke, J. D., and J. Stoye. 1997. Retrotransposons, endogenous retroviruses, and the evolution of retroelements, p. 343-436. In J. M. Coffin, S. H. Hugues, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 6.Bogerd, H. P., B. P. Doehle, H. L. Wiegand, and B. R. Cullen. 2004. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl. Acad. Sci. USA 101:3770-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calattini, S., E. Nerrienet, P. Mauclere, M. C. Georges-Courbot, A. Saib, and A. Gessain. 2004. Natural simian foamy virus infection in wild-caught gorillas, mandrills and drills from Cameroon and Gabon. J. Gen. Virol. 85:3313-3317. [DOI] [PubMed] [Google Scholar]
- 8.Callahan, M. E., W. M. Switzer, A. L. Matthews, B. D. Roberts, W. Heneine, T. M. Folks, and P. A. Sandstrom. 1999. Persistent zoonotic infection of a human with simian foamy virus in the absence of an intact orf-2 accessory gene. J. Virol. 73:9619-9624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu, Y. L., V. B. Soros, J. F. Kreisberg, K. Stopak, W. Yonemoto, and W. C. Greene. 2005. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 435:108-114. [DOI] [PubMed] [Google Scholar]
- 10.Conticello, S. G., R. S. Harris, and M. S. Neuberger. 2003. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13:2009-2013. [DOI] [PubMed] [Google Scholar]
- 11.Delelis, O., J. Lehmann-Che, and A. Saib. 2004. Foamy viruses—a world apart. Curr. Opin. Microbiol. 7:400-406. [DOI] [PubMed] [Google Scholar]
- 12.Delelis, O., A. Saib, and P. Sonigo. 2003. Biphasic DNA synthesis in spumaviruses. J. Virol. 77:8141-8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dutko, J. A., A. Schafer, A. E. Kenny, B. R. Cullen, and M. J. Curcio. 2005. Inhibition of a yeast LTR retrotransposon by human APOBEC3 cytidine deaminases. Curr. Biol. 15:661-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esnault, C., O. Heidmann, F. Delebecque, M. Dewannieux, D. Ribet, A. J. Hance, T. Heidmann, and O. Schwartz. 2005. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433:430-433. [DOI] [PubMed] [Google Scholar]
- 15.Falcone, V., M. Schweizer, and D. Neumann-Haefelin. 2003. Replication of primate foamy viruses in natural and experimental hosts. Curr. Top. Microbiol. Immunol. 277:161-180. [DOI] [PubMed] [Google Scholar]
- 16.Giron, M.-L., H. de Thé, and A. Saïb. 1998. An evolutionarily conserved splice generates a secreted Env-Bet fusion protein during human foamy virus infection. J. Virol. 72:4906-4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hache, G., M. T. Liddament, and R. S. Harris. 2005. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J. Biol. Chem. 280:10920-10924. [DOI] [PubMed] [Google Scholar]
- 18.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 19.Harris, R. S., and M. T. Liddament. 2004. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 4:868-877. [DOI] [PubMed] [Google Scholar]
- 20.Heinkelein, M., T. Pietschmann, G. Jarmy, M. Dressler, H. Imrich, J. Thurow, D. Lindemann, M. Bock, A. Moebes, J. Roy, O. Herchenroder, and A. Rethwilm. 2000. Efficient intracellular retrotransposition of an exogenous primate retrovirus genome. EMBO J. 19:3436-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinkelein, M., M. Rammling, T. Juretzek, D. Lindemann, and A. Rethwilm. 2003. Retrotransposition and cell-to-cell transfer of foamy viruses. J. Virol. 77:11855-11858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heneine, W., M. Schweizer, P. Sandstrom, and T. Folks. 2003. Human infection with foamy viruses. Curr. Top. Microbiol. Immunol. 277:181-196. [DOI] [PubMed] [Google Scholar]
- 23.Heneine, W., W. M. Switzer, P. Sandstrom, J. Brown, S. Vedapuri, C. A. Schable, A. S. Khan, N. W. Lerche, M. Schweizer, D. Neumann-Haefelin, L. E. Chapman, and T. M. Folks. 1998. Identification of a human population infected with simian foamy viruses. Nat. Med. 4:403-407. [DOI] [PubMed] [Google Scholar]
- 24.Hill, C. L., P. D. Bieniasz, and M. O. McClure. 1999. Properties of human foamy virus relevant to its development as a vector for gene therapy. J. Gen. Virol. 80:2003-2009. [DOI] [PubMed] [Google Scholar]
- 25.Ibrahim, F., G. de Thé, and A. Gessain. 1995. Isolation and characterization of a new simian T-cell leukemia virus type 1 from naturally infected celebes macaques (Macaca tonkeana): complete nucleotide sequence and phylogenetic relationship with the Australo-Melanesian human T-cell leukemia virus type 1. J. Virol. 69:6980-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, and N. Navaratnam. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79:285-296. [DOI] [PubMed] [Google Scholar]
- 27.Kao, S., M. A. Khan, E. Miyagi, R. Plishka, A. Buckler-White, and K. Strebel. 2003. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 77:11398-11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi, M., A. Takaori-Kondo, K. Shindo, A. Abudu, K. Fukunaga, and T. Uchiyama. 2004. APOBEC3G targets specific virus species. J. Virol. 78:8238-8244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lecellier, C. H., P. Dunoyer, K. Arar, J. Lehmann-Che, S. Eyquem, C. Himber, A. Saib, and O. Voinnet. 2005. A cellular MicroRNA mediates antiviral defense in human cells. Science 308:557-560. [DOI] [PubMed] [Google Scholar]
- 30.Lecellier, C. H., W. Vermeulen, F. Bachelerie, M.-L. Giron, and A. Saïb. 2002. Intra- and intercellular trafficking of the foamy virus auxiliary bet protein. J. Virol. 76:3388-3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 32.Lehmann-Che, J., M.-L. Giron, O. Delelis, M. Löchelt, P. Bittoun, J. Tobaly-Tapiero, H. de Thé, and A. Saïb. 2005. Protease-dependent uncoating of a complex retrovirus. J. Virol. 79:9244-9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liddament, M. T., W. L. Brown, A. J. Schumacher, and R. S. Harris. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 14:1385-1391. [DOI] [PubMed] [Google Scholar]
- 34.Lindemann, D., and A. Rethwilm. 1998. Characterization of a human foamy virus 170-kilodalton Env-Bet fusion protein generated by alternative splicing. J. Virol. 72:4088-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linial, M. 2000. Why aren't foamy viruses pathogenic? Trends Microbiol. 8:284-289. [DOI] [PubMed] [Google Scholar]
- 36.Linial, M. L. 1999. Foamy viruses are unconventional retroviruses. J. Virol. 73:1747-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lochelt, M., F. Romen, P. Bastone, H. Muckenfuss, N. Kirchner, Y. B. Kim, U. Truyen, U. Rosler, M. Battenberg, A. Saib, E. Flory, K. Cichutek, and C. Munk. 2005. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc. Natl. Acad. Sci. USA 102:7982-7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lochelt, M., H. Zentgraf, and R. M. Flugel. 1991. Construction of an infectious DNA clone of the full-length human spumaretrovirus genome and mutagenesis of the bel 1 gene. Virology 184:43-54. [DOI] [PubMed] [Google Scholar]
- 39.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 40.Mangeat, B., P. Turelli, S. Liao, and D. Trono. 2004. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 279:14481-14483. [DOI] [PubMed] [Google Scholar]
- 41.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 42.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 43.Mikovits, J. A., P. M. Hoffman, A. Rethwilm, and F. W. Ruscetti. 1996. In vitro infection of primary and retrovirus-infected human leukocytes by human foamy virus. J. Virol. 70:2774-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neumann-Haefelin, D., U. Fleps, R. Renne, and M. Schweizer. 1993. Foamy viruses. Intervirology 35:196-207. [DOI] [PubMed] [Google Scholar]
- 45.Newman, E. N., R. K. Holmes, H. M. Craig, K. C. Klein, J. R. Lingappa, M. H. Malim, and A. M. Sheehy. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15:166-170. [DOI] [PubMed] [Google Scholar]
- 46.Paccaud, M. 1957. Study of foamy virus. II. Study of lesions in monkey and human kidney cells, monkey testicular cells and carcinomatous HeLa cells caused by foamy virus (strains FV I, FV II, and FV III). Ann. Inst. Pasteur (Paris) 92:481-488. (In French.) [PubMed] [Google Scholar]
- 47.Petit, C., O. Schwartz, and F. Mammano. 1999. Oligomerization within virions and subcellular localization of human immunodeficiency virus type 1 integrase. J. Virol. 73:5079-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rethwilm, A. 2003. The replication strategy of foamy viruses. Curr. Top. Microbiol. Immunol. 277:1-26. [DOI] [PubMed] [Google Scholar]
- 49.Russell, R. A., H. L. Wiegand, M. D. Moore, A. Schafer, M. O. McClure, and B. R. Cullen. 2005. Foamy virus Bet proteins function as novel inhibitors of the APOBEC3 family of innate antiretroviral defense factors. J. Virol. 79:8724-8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saib, A., M. H. Koken, P. van der Spek, J. Peries, and H. de Thé. 1995. Involvement of a spliced and defective human foamy virus in the establishment of chronic infection. J. Virol. 69:5261-5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schiffer, C., C.-H. Lecellier, A. Mannioui, N. Felix, E. Nelson, J. Lehmann-Che, M.-L. Giron, J. C. Gluckman, A. Saib, and B. Canque. 2004. Persistent infection with primate foamy virus type 1 increases human immunodeficiency virus type 1 cell binding via a Bet-independent mechanism. J. Virol. 78:11405-11410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmidt, M., and A. Rethwilm. 1995. Replicating foamy virus-based vectors directing high level expression of foreign genes. Virology 210:167-178. [DOI] [PubMed] [Google Scholar]
- 53.Schrofelbauer, B., D. Chen, and N. R. Landau. 2004. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Natl. Acad. Sci. USA 101:3927-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schumacher, A. J., D. V. Nissley, and R. S. Harris. 2005. APOBEC3G hypermutates genomic DNA and inhibits Ty1 retrotransposition in yeast. Proc. Natl. Acad. Sci. USA 102:9854-9859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwartz, O., V. Maréchal, O. Danos, and J. M. Heard. 1995. Human immunodeficiency virus type 1 Nef increases the efficiency of reverse transcription in the infected cell. J. Virol. 69:4053-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schwartz, O., V. Maréchal, B. Friguet, F. Arenzana-Seisdedos, and J.-M. Heard. 1998. Antiviral activity of the proteasome on incoming human immunodeficiency virus type 1. J. Virol. 72:3845-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schweizer, M., H. Schleer, M. Pietrek, J. Liegibel, V. Falcone, and D. Neumann-Haefelin. 1999. Genetic stability of foamy viruses: long-term study in an African green monkey population. J. Virol. 73:9256-9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 59.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12:591-601. [DOI] [PubMed] [Google Scholar]
- 60.Suspene, R., D. Guetard, M. Henry, P. Sommer, S. Wain-Hobson, and J. P. Vartanian. 2005. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc. Natl. Acad. Sci. USA 102:8321-8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suspene, R., M. Henry, S. Guillot, S. Wain-Hobson, and J. P. Vartanian. 2005. Recovery of APOBEC3-edited human immunodeficiency virus G→A hypermutants by differential DNA denaturation PCR. J. Gen. Virol. 86:125-129. [DOI] [PubMed] [Google Scholar]
- 62.Suspene, R., P. Sommer, M. Henry, S. Ferris, D. Guetard, S. Pochet, A. Chester, N. Navaratnam, S. Wain-Hobson, and J. P. Vartanian. 2004. APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase. Nucleic Acids Res. 32:2421-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Switzer, W. M., M. Salemi, V. Shanmugam, F. Gao, M. E. Cong, C. Kuiken, V. Bhullar, B. E. Beer, D. Vallet, A. Gautier-Hion, Z. Tooze, F. Villinger, E. C. Holmes, and W. Heneine. 2005. Ancient co-speciation of simian foamy viruses and primates. Nature 434:376-380. [DOI] [PubMed] [Google Scholar]
- 64.Turelli, P., and D. Trono. 2005. Editing at the crossroad of innate and adaptive immunity. Science 307:1061-1065. [DOI] [PubMed] [Google Scholar]
- 65.Turelli, P., S. Vianin, and D. Trono. 2004. The innate antiretroviral factor APOBEC3G does not affect human LINE-1 retrotransposition in a cell culture assay. J. Biol. Chem. 279:43371-43373. [DOI] [PubMed] [Google Scholar]
- 66.von Laer, D., D. Neumann-Haefelin, J. L. Heeney, and M. Schweizer. 1996. Lymphocytes are the major reservoir for foamy viruses in peripheral blood. Virology 221:240-244. [DOI] [PubMed] [Google Scholar]
- 67.Wedekind, J. E., G. S. Dance, M. P. Sowden, and H. C. Smith. 2003. Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends Genet. 19:207-216. [DOI] [PubMed] [Google Scholar]
- 68.Wiegand, H. L., B. P. Doehle, H. P. Bogerd, and B. R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23:2451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wolfe, N. D., W. M. Switzer, J. K. Carr, V. B. Bhullar, V. Shanmugam, U. Tamoufe, A. T. Prosser, J. N. Torimiro, A. Wright, E. Mpoudi-Ngole, F. E. McCutchan, D. L. Birx, T. M. Folks, D. S. Burke, and W. Heneine. 2004. Naturally acquired simian retrovirus infections in central African hunters. Lancet 363:932-937. [DOI] [PubMed] [Google Scholar]
- 70.Xu, H., E. S. Svarovskaia, R. Barr, Y. Zhang, M. A. Khan, K. Strebel, and V. K. Pathak. 2004. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc. Natl. Acad. Sci. USA 101:5652-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu, Q., D. Chen, R. Konig, R. Mariani, D. Unutmaz, and N. R. Landau. 2004. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 279:53379-53386. [DOI] [PubMed] [Google Scholar]
- 72.Yu, Q., R. Konig, S. Pillai, K. Chiles, M. Kearney, S. Palmer, D. Richman, J. M. Coffin, and N. R. Landau. 2004. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 11:435-442. [DOI] [PubMed] [Google Scholar]
- 73.Yu, S. F., D. N. Baldwin, S. R. Gwynn, S. Yendapalli, and M. L. Linial. 1996. Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science 271:1579-1582. [DOI] [PubMed] [Google Scholar]
- 74.Yu, S. F., and M. L. Linial. 1993. Analysis of the role of the bel and bet open reading frames of human foamy virus by using a new quantitative assay. J. Virol. 67:6618-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu, S. F., J. Stone, and M. L. Linial. 1996. Productive persistent infection of hematopoietic cells by human foamy virus. J. Virol. 70:1250-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 77.Zheng, Y. H., D. Irwin, T. Kurosu, K. Tokunaga, T. Sata, and B. M. Peterlin. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 78:6073-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.