Abstract
The triplex of herpesvirus capsids is a unique structural element. In herpes simplex virus type 1 (HSV-1), one molecule of VP19C and two of VP23 form a three-pronged structure that acts to stabilize the capsid shell through interactions with adjacent VP5 molecules. The interaction between VP19C and VP23 was inferred by yeast cryoelectron microscopy studies and subsequently confirmed by the two-hybrid assay. In order to define the functional domains of VP19C and VP23, a Tn7-based transposon was used to randomly insert 15 bp into the coding regions of these two proteins. The mutants were initially screened for interaction in the yeast two-hybrid assay to identify the domains important for triplex formation. Using genetic complementation assays in HSV-1-infected cells, the domains of each protein required for virus replication were similarly uncovered. The same mutations that abolish interaction between these two proteins in the yeast two-hybrid assay similarly failed to complement the growth of the VP23- and VP19C-null mutant viruses in the genetic complementation assay. Some of these mutants were transferred into recombinant baculoviruses to analyze the effect of the mutations on herpesvirus capsid assembly in insect cells. The mutations that abolished the interaction in the yeast two-hybrid assay also abolished capsid assembly in insect cells. The outcome of these experiments showed that insertions in at least four regions and especially the amino terminus of VP23 abolished function, whereas the amino terminus of VP19C can tolerate transposon insertions. A novel finding of these studies was the ability to assemble herpesvirus capsids in insect cells using VP5 and VP19C that contained a histidine handle at their amino terminus.
The genetic information of viruses is enclosed in the capsid shell, a stable protein coat whose function is to protect the nucleic acid and to aid in the infectious process. The herpes simplex virus type 1 (HSV-1) capsid is comprised of seven proteins that together form a large complex assembly. The intimate association of these proteins to create a protective shell around the virus genome requires multiple protein-protein interactions. These interactions drive the self-assembly of this structure.
The HSV-1 capsid is composed of seven proteins, VP5, VP19C, 21, 22a, VP23, VP24, and VP26 (VP stands for virion protein), and the genes that encode these proteins are UL19, UL38, UL26, UL26.5, UL18, UL26, and UL35, respectively (19, 21, 24). Because 21 and 22a are not present in the virion, the prefix VP is not used for them. Capsid assembly is a nuclear event resulting in the production of three types of capsids, designated A, B, and C, corresponding to increasing distance sedimented through sucrose gradients (10). B capsids contain internal scaffold proteins (pre-22a and pre-21), the viral protease (VP24), and the capsid shell proteins (VP5, VP19C, VP23, and VP26). For C capsids, genomic DNA replaces the scaffold proteins. A capsids are devoid of any internal composition, that is, they are empty capsids. VP5, the major capsid protein, in association with scaffold molecules, forms the hexons and pentons present in capsid shells (16, 30, 31). A complex of VP19C and VP23 stabilizes the capsid shell structure; if either is absent, capsid shells are not formed (5, 18). VP26 is located at the outer surface of VP5 hexons (29, 35). A single open reading frame (ORF), UL26, encodes the remaining components of B capsid 21, 22a, and VP24. VP24 encodes the protease, which cleaves the scaffold proteins so that they can be released upon DNA packaging (reviewed in references 19, 21, and 24). When the HSV-1 capsid proteins were expressed by recombinant baculoviruses in insect cells, fully assembled capsids were observed (25, 26). The proteins that are essential for this in vitro assembly process are VP5, VP19C, VP23, and the scaffold protein (25, 26).
VP19C (465 amino acids) and VP23 (318 amino acids) form a three-pronged structure comprised of one molecule of VP19C and two molecules of VP23 (16, 27, 34). There are approximately 320 hetero-trimeric triplex molecules per capsid, which corresponds to one interaction site per VP5 capsomere. The function of the complex is to promote assembly of the capsid shell through interactions with adjacent VP5 molecules. A molecular interaction between the two molecules was discovered using the yeast two-hybrid (Y2H) system (7). The VP19C-VP23 complex has been detected for HSV-1 using sedimentation techniques following coexpression of the two proteins in the baculovirus system (23). Cell localization studies have also demonstrated the requirement of VP19C for the nuclear localization of VP23 (20). Thus, triplex structures are assembled in the cytoplasm and transported to the nucleus, where higher-order capsid shells are synthesized.
The goal of this study was to identify the domains in VP23 that are crucial for the association with VP19C and those in VP19C that are important for binding to VP23 during the formation of the functional triplex structures. Transposition mutations in the genes encoding VP19C and VP23 were generated by a linker-scanning mutagenesis method. The resulting VP19C-VP23 interactions were investigated in vitro by the Gal4 yeast two-hybrid assay and in vivo by recombinant baculovirus infection of insect cells (Sf9) and genetic complementation assays of the VP23 and VP19C null mutants.
MATERIALS AND METHODS
Cell lines and antibodies.
Spodoptera frugiperda (Sf9) cells were grown in Grace's insect cell medium, supplemented with 10% fetal calf serum (Gibco-Invitrogen). Sf9 cells were grown in spinner flasks at 27°C with constant agitation (85 rpm). Cells were seeded at 0.5 × 106 cells/ml and harvested when the cell density reached 2 × 106 cells/ml. Cell viability was in the 90 to 98% range. Vero cells and transformed Vero cell lines were grown in minimum essential medium-alpha medium supplemented with 10% fetal calf serum (Gibco-Invitrogen) and passaged as described by Desai et al. (6). Virus stocks of KOS (HSV-1) and the mutant viruses were prepared as previously described (6). The rabbit polyclonal antiserum R2421 (5) was raised to VP23 isolated from capsids and purified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The R2421 serum also displayed cross-reactivity to VP19C. The UL26-C rabbit antibody was made to a C-terminal (VDVDTARAADLFVSQMMGAR) peptide of UL26 (pre-22a) spanning amino acids 616 to 635, and the UL38C rabbit antiserum was raised against a C-terminal peptide (VILEGVVWRPGEWRA) spanning amino acids 449 to 463.
Plasmids.
For this study the VP19C (UL38) and VP23 (UL18) ORFs were obtained from the yeast two-hybrid vectors that have been previously described (7). The UL38 polypeptide sequence of KOS differs from that of strain 17 (14) by two amino acids (G48 and P366 in strain 17 are V48 and Q366 in KOS). The UL18 amino acid sequence of KOS is identical to that of strain 17 (14). The UL38 ORF was derived as an EcoR1-BamH1 fragment from pGAD424-19C (7). This fragment was cloned into the same respective sites of pBSKII (Stratagene). This plasmid, designated pBS19C, was used for subsequent mutagenesis experiments. The UL18 gene was derived as an EcoR1-BamH1 fragment from pGBT9-23 (7) and was cloned into pGEM3Z (Promega) and designated pGEM23.
The capsid proteins were expressed in recombinant baculoviruses using the BAC-to-BAC system (Invitrogen) (13). The transfer vector pFastBacHta (pFBHta) was used to express VP5, VP19C, and UL80HSVCT (a chimera of the human cytomegalovirus [HCMV] scaffold protein which lacks the C-terminal tail that interacts with the CMV major capsid protein fused instead to the HSV-1 scaffold C-terminal 25 amino acids). In this vector the proteins encode a six-histidine handle at the N terminus. In addition, a tobacco etch virus cleavage site between the histidine handle and the foreign protein allows for subsequent removal of the histidine residues. The VP5 (UL19) ORF was derived from pGBT95 (7) as an EcoR1-Sal1 fragment (partial Sal1 digest) and cloned into the same sites of pFBHta and then designated pFBHtaUL19. Similarly, VP19C (UL38) was derived from the yeast two-hybrid vector (7) and cloned into pFBHta as an EcoR1-Sal1 fragment. This plasmid was designated pFBHtaUL38. Some of the HSV-1 capsid ORFs for baculovirus expression were generated by PCRs using Pfu Turbo (Stratagene). The UL18 (VP23) gene was PCR amplified using primer pairs F (GGAATTCAAACCATGCTGGCGGACGGCTTTGAAACTGAC) and R (GCTCGAGTTAGGGATAGCGTATAACGGGGGC). The template used for this reaction was pGBT9-23. The PCR product was digested with EcoR1 and Xho1 and cloned into the same restriction sites of pFastBac1 (pFB), and the resultant plasmid was designated pFBUL18. This vector does not express a histidine handle. The strategy to generate the HCMV scaffold protein chimera was similar to that developed by Oien et al. (17). The UL80 scaffold protein was amplified using F (GGAATTCATGTCGCACCCTCTGAGTGCTGCGGTT) and R (GCTCGAGGCGTTCACCACGCCGGCCTGAGCGCG) primers, with pEB11 as the template. This fragment was cloned into pFastBac1 using the EcoR1-Xho1 sites. This plasmid was designated pFB80. Oligonucleotides, once annealed, that create the C-terminal tail of HSV-1 scaffold were made and cloned into the Xho1-HindIII sites of pFB80 to give pFB80HSVCT. The chimera protein was amplified using the same forward primer used to amplify UL80 and the R primer (GGAAGCTTTCAGCGGGCCCCCATCATCTG), digested with EcoR1 and HindIII, cloned into pFastBacHta, and designated pFBHta80HSVCT. All PCR-generated constructs were sequenced for authentic amplification.
The UL18 and UL38 TN mutants were also cloned into pcDNA 3.1(-). The EcoR1-BamH1 fragments were derived from the respective yeast two-hybrid vectors and ligated to similar sites in pcDNA 3.1.
Transposition mutagenesis.
The transposition reaction was done following the GPS-LS protocol (NEB) (1, 2, 4). The UL18 and UL38 ORFs isolated as EcoRI-to-BamHI fragments were used as the templates for the transposition. Following transposition, the fragments were ligated to pGBT9 (UL18) or pGAD424 (UL38) and plated on selective plates containing kanamycin and ampicillin. Plasmid DNA was extracted from the pooled colonies, and this DNA was digested with PmeI to remove the transposon element. The DNA was purified, religated, and transformed into Escherichia coli. DNA extracted from individual colonies was analyzed using restriction enzymes (PmeI) to map the site of the transposition. All of the mutants were sequenced to confirm the positions and residues inserted as a result of the transposition.
Yeast two-hybrid assays.
The yeast two-hybrid assays were performed essentially as described by Desai and Person (7). Plasmid pGBT9-23, in which VP23 was fused to the Gal4 DNA binding domain, was cotransformed into SFY526 cells together with pGAD424-19C, in which VP19C was fused to the Gal4 transactivation domain (7). The mutant clones were cotransformed with respective wild-type interactive protein into SFY526, and the transformants were allowed to grow on filter paper on SD (leu-/trp-) plates in order to screen each mutation, using the 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) filter assay (3).
Complementation assay.
Vero cells (1 × 106 per 35-mm dish) were transfected with 1 μg of plasmid DNA and 6 μl of Lipofectamine reagent (Invitrogen) diluted in 200 μl of serum-free medium. The DNA-lipid complexes were allowed to form at room temperature for 30 min and incubated with the cells for 5 h. At 24 h after the addition of the complexes to the culture, the cells were infected with either K23Z (VP23 null mutant) (5) or KΔ19C (VP19C null mutant) (18) at a multiplicity of infection (MOI) of 5 PFU/cell. The following day (24 h later), the infected cells were harvested following freeze-thaw and sonicated, and the virus titers were determined on both C32 (18) and Vero cell monolayers.
Generation of recombinant baculoviruses using the BAC-to-BAC system.
Introduction of the various capsid ORFs into recombinant baculoviruses was achieved by transposition reactions in E. coli using the DH10BAC host cell line. This cell line harbors the baculovirus genome cloned into a bacterial artificial chromosome (13), bacmid. The transfer plasmids were transformed into this host cell, and the transposition was carried out according to the manufacturer's protocol (Invitrogen). Positive clones were identified by the disruption of the lacZ gene and confirmed by PCR analysis of the bacmid DNA. The bacmid DNA was transfected into low-passage Sf9 cells as described below to reconstitute the infectious virus.
Baculovirus transfection.
Sf9 cells (9 × 105 cells in 35-mm dishes) were transfected with bacmid DNA using CELLFECTIN reagent (Invitrogen). A solution of 1 μg baculovirus DNA in 100 μl Grace's insect cell medium was mixed with a solution containing 9 μl CELLFECTIN reagent diluted into 100 μl Grace's insect cell medium. The mixture was incubated for 30 min. The insect cells were then washed with 2 ml Grace's medium and overlaid with 0.8 ml Grace's medium, which was first added to the tube containing the lipid-DNA complexes. The Sf9 transfected cells were incubated in a 27°C incubator for 5 h. The transfection mixtures were then removed, and 2 ml Grace's medium containing antibiotics and serum (10%) was added to the cells, which were then incubated for 72 h in a 27°C incubator. The supernatant of the transfected cells which contained the recombinant virus was harvested and clarified, and this was designated a P1 stock.
Baculovirus amplification.
Viruses were amplified by infection of 6 × 106 Sf9 cells in 100-mm dishes using 100 to 150 μl of the transfection stock (P1 stock). Supernatants were harvested 5 days after infection and clarified, and the titer of the viruses was determined by plaque assay on Sf9 cell monolayers. These stocks were typically 1 × 108 PFU/ml and were designated P2 stocks. P3 stocks were made by infection of Sf9 cells in spinner flasks (1 × 106 cells/ml) at an MOI of 0.1 PFU/cell. When greater than 90% of the cells were dead, the culture supernatant containing the virus was harvested and the titer was determined. P3 stocks were also generally 108 PFU/ml.
Radiolabeling and SDS-PAGE.
Radiolabeling of infected cells was performed at 24, 48, 72, and 96 h postinfection. Cells were washed with phosphate-buffered saline (PBS) and overlaid with PBS for 2 h. Subsequently, 100 μCi of [35S]methionine (NEN-Dupont) was added in fresh PBS, and the cells were pulse-labeled with this mixture for 2 h. At the end of this period, the cells were washed and harvested for analysis. SDS-PAGE analysis was performed as described by Person and Desai (18).
Western blot analysis.
Unlabeled infected cell extracts were resolved by SDS-PAGE and transferred to Immobilon-P membranes (Millipore) in Tris-glycine buffer using a Bio-Rad mini-transblot apparatus. Transfer buffer and procedures were according to the manufacturer's protocol. The membrane was incubated in 5% bovine serum albumin (BSA) in TN buffer (10 mM Tris [pH 7.4], 150 mM NaCl) overnight at 4°C to block nonspecific reactivity. Filters were then incubated with an appropriate dilution of antibody in 5% BSA-TN for 90 min at room temperature. Following antibody reactivity, the membranes were washed three times for 7 min each at room temperature in TN followed by two washes in TN containing 0.5% NP-40 (10 min each) and finally three washes in TN buffer (7 min each). Antigen detection was performed by incubation with 125I-labeled protein A (NEN-DuPont) diluted 1:1,000 in 5% BSA-TN for 2 h at room temperature. The filters were washed as described above and dried prior to autoradiography.
Electron microscopy.
Sf9 cells (1 × 107) in 100-mm dishes were infected at an MOI of 5 PFU/cell. The samples were processed for transmission electron microscopy (TEM) essentially as described by Hendricks et al. (11). Briefly, the cells were fixed for 1 h at room temperature in a solution containing 2.0% glutaraldehyde in 100 mM cacodylate (pH 7.4) and 2.5% sucrose. The cells were then lifted off the dish, pelleted and washed, and subsequently stained with OsO4 (1 h at 4°C). The pellet was then washed three times in 100 mM cacodylate, pH 7.4, washed three times in double-distilled H2O, and incubated overnight in Kellenberger's uranyl acetate (11). The pellet was then dehydrated through a graded series of ethanol and embedded in EMBED-812. Sections were cut on a Leica Ultracut UCT ultramicrotome, collected onto 400 mesh nickel grids, poststained in uranyl acetate and lead citrate, and observed in a Philips EM420 TEM equipped with a Soft Imaging System's MegaView III digital camera and analySIS software.
Data and figure preparation.
For figure preparation, autoradiographs were scanned at 300 dots per inch in Adobe Photoshop. Electron micrographs were captured as 16-bit images, exported as 16-bit tiff files, and were linearly adjusted for brightness/contrast and resized in Adobe Photoshop. Figures were compiled using Adobe Illustrator.
RESULTS
Transposition mutagenesis of the genes encoding the triplex proteins (VP19C and VP23).
In order to define the functional domains of VP19C and VP23, we employed the GPS linker-scanning mutagenesis method (NEB), which randomly inserts 15-bp “linkers” throughout the target DNA (4). The system employs a Tn7-based transprimer that encodes an antibiotic marker (kanamycin) for positive selection of transposition events. The transposition was carried out in vitro rather than in a host cell, and only one transprimer was inserted per molecule (1, 2); transposition was performed on a purified fragment encoding UL38 (VP19C) or UL18 (VP23) so that insertions occurred in the gene rather than plasmid sequences. The kanamycin gene was removed from the plasmid by digestion with a unique Pme1 site, leaving a 15-bp insertion (10 bp of restriction site sequence and 5 bp of duplicated sequence of the target).
Purified DNA fragments that encode the UL38 and UL18 ORFs were subjected to this mutagenesis procedure. The UL38 fragments that carried the 15-bp transprimer were cloned into the yeast two-hybrid plasmid, pGAD424, which encodes the Gal4 transcriptional activation domain. The UL18 fragments were cloned into pGBT9, which contains the Gal4 DNA binding domain (9). We isolated and analyzed 82 mutants for UL38 and 60 mutants for UL18. The mutations as judged by restriction enzyme analysis using Pme1 appeared to have randomly integrated throughout the UL38 and UL18 genes (Fig. 1A and B). Using double enzyme digests, we were able to crudely map the location of the Pme1 site in the sequence. Subsequently, we sequenced the region of the transposition insertion to determine the nature of the mutation.
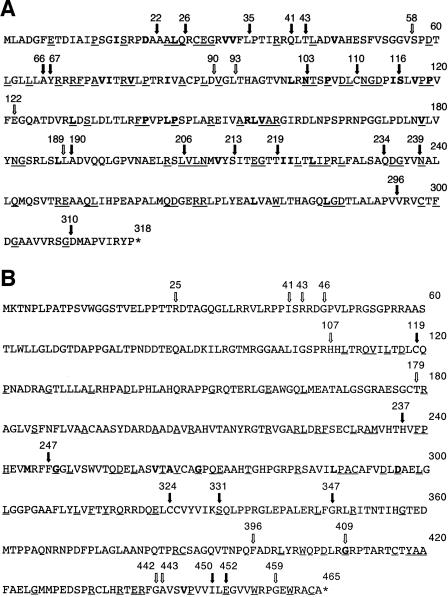
FIG. 1.
Transposition mutants of the triplex proteins. The figure shows the amino acid sequences of the VP23 (A) and VP19C (B) proteins. The arrows show the sites of the transposition insertions. The filled arrow indicates that the insertion abolished the interaction between VP19C and VP23 in the yeast two-hybrid assay, whereas, the nonfilled arrow indicates that the two proteins were still capable of interaction. Amino acids that are conserved in the alphaherpesviruses are underlined, and those in bold indicate amino acids or side chain groups that are conserved in all three herpesvirus families. Amino acids of VP23 (N102, P105, P119, P140, and P144) and VP19C (G248, G268, D296, and G409) are absolutely conserved in all three families.
The Y2H assay (9) was used to rapidly screen mutants for loss or decrease of interaction (Tables 1 and 2). This in vitro assay allowed us to screen through the numerous mutants in a very short time, with the appearance of blue color of the yeast colonies in the presence of X-Gal as a result of β-galactosidase gene expression being indicative of protein-protein interaction. The UL38 mutants were cotransformed into yeast cells together with the plasmid encoding wild-type VP23 and the UL18 mutants with wild-type VP19C-expressing plasmid. Analysis of the transformants revealed many mutants that abolished the interaction, some that weakened the interaction, and others that did not affect the interaction, as indicated by the variable expression of the β-galactosidase reporter gene (Tables 1 and 2; Fig. 1A and B). Due to the A-T-rich sequence of the Pme1 site, there were many translation termination codons inserted into the genes, depending on the insertion frame. For the UL38 TN mutants, 42 mutants contained a translation termination codon: 7 of these occurred prior to the 100th codon, and the others were distributed throughout the downstream portion of the sequence. Since the most-C-terminal translation termination in VP19C was at codon 452, and this mutation abolished the interaction (Table 2 and Fig. 1B), the remaining translation termination mutants are not shown in the table. Similarly for UL18, 27 mutants encoded a stop codon at the transprimer insertion site. Of these, 11 mutants resulted in translation termination before the 50th codon, and the most-C-terminal translation termination mutation that also abolished interaction was at residue 309 (Fig. 1A). All of the translation termination mutants of VP23 and VP19C did not interact with the wild-type protein partner in the Y2H assay.
TABLE 1.
VP23 transposition mutants: insertion site, amino acids inserted, and the yeast two-hybrid resultb
| Wild-type interaction | Weak interaction | No interaction |
|---|---|---|
| 90-CLNMD | 58-LFKHS | 22-VFKHA |
| 93-MFKHL | 122-VFKQF | 26-CLNMQ |
| 189-CLNTL | 35-LLFKH | |
| 41-LMFKQ | ||
| 43-LVFKQ | ||
| 66-FKQLL | ||
| 67-VFKQA | ||
| 103-CLNSN | ||
| 110-CLNIC | ||
| 116-CLNIS | ||
| 190-FKQLL | ||
| 206-CLNTL | ||
| 213-CLNNS | ||
| 219-CLNTT | ||
| 234-VFKHQ | ||
| 239-CLNMN | ||
| 296-V*a | ||
| 310-V* |
*, stop codon.
In the X-Gal filter assay the time it took for the appearance of blue colonies was determined. For the wild-type interaction, colonies turned blue within 45 min of the start of the assay. “Weak interaction” indicates it took greater than 60 min for the appearance of blue color and in most cases the color was faint in intensity. “No interaction” indicates that the colonies remained clear even after prolonged incubation. The data for the X-Gal filter were derived from two to three independent transformations.
TABLE 2.
VP19C transposition mutants: insertion site, amino acids inserted, and the yeast two-hybrid resultb
| Wild-type interaction | Weak interaction | No interaction |
|---|---|---|
| 25-LFKHT | 396-CLNTQ | 119-LFKHL |
| 41-MFKHP | 409-CLNMR | 237-LFKHT |
| 43-CLNIS | 442-VFKHF | 247-LFKQF |
| 46-CLNSD | 443-CLNIG | 324-FKQEL |
| 107-MFKQR | 331-CLNIK | |
| 179-CLNRC | 347-VFKQF | |
| 459-CLNSP | 452-VFKHL | |
| 450-V*a | ||
| 452-DV* |
*, stop codon.
In the X-Gal filter assay the time it took for the appearance of blue colonies was determined. For the wild-type interaction, colonies turned blue within 45 min of the start of the assay. “Weak interaction” indicates it took greater than 60 min for the appearance of blue color and in most cases the color was faint in intensity. “No interaction” indicates that the colonies remained clear even after prolonged incubation. The data for the X-Gal filter were derived from two to three independent transformations.
The most frequent amino acid insertions were CLNXX, LFKXX, and VFKXX. In addition, insertions MFKXX and FKQXX were detected in some of the transpositions (Tables 1 and 2). The transprimer and restriction enzyme sequence TGTTTAAACA would encode amino acids CLN (first frame), V* (* is the stop codon) (second frame), and FK (third frame). Due to the GC-rich sequence of HSV-1, LFK and VFK were more commonly observed in the third frame. Amino acid insertions that disrupted a key functional site or structural domain abolished interaction with the corresponding interactive protein, and this was seen by the absence of blue color in the colonies assayed (no-interaction columns in Tables 1 and 2). There were mutant insertions that resulted in a weak interaction in the Y2H assay, as judged by the late development of blue color of the colonies and the intensity of the blue color (weak interaction columns in Tables 1 and 2). Finally, some of the insertions did not affect the VP19C-VP23 interaction in this assay, that is, the kinetics of appearance of the blue color in the colonies and the intensity of the color were similar to the wild-type proteins assayed at the same time (wild-type interaction columns in Tables 1 and 2). The locations of the transposition inserts in the VP23 and VP19C polypeptides are shown in Fig. 1A and B, as are the effects of the insertions on the interaction between VP19C and VP23.
Complementation assay.
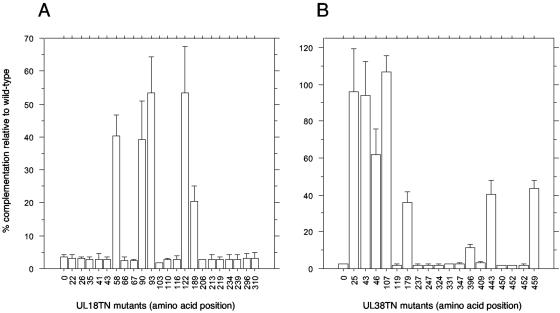
In order to correlate the in vitro results from the yeast two-hybrid assay with the ability of the VP23 and VP19C TN mutants to support virus growth, it was decided to test these mutants using the classical genetic complementation assay. Most of the TN mutants were cloned into pcDNA3.1, in which the CMV promoter drove the expression of the mutant and wild-type proteins. Vero cells were transfected with plasmids encoding the wild-type protein or no protein (empty vector) and with the different TN mutants. The following day the cells were infected with either the VP19C or VP23 null mutant viruses (5, 18). Following another day of incubation, the virus progeny yield was determined by plating the lysates on the complementing C32 cell line (18). The level of background virus (rescued virus) was estimated by the results of plating the lysates on Vero cells. As shown in Fig. 2A and B, the complementation assay was able to distinguish between mutants that support the replication of the null mutant (complementation of 20 to 100% relative to wild-type protein) and those that do not (complementation of 2 to 5% relative to wild type; this level was similar to the number seen with empty vector). In general there was very good correlation between the ability of the mutants to participate in a molecular interaction (Y2H assay) and to complement the growth of the null mutant virus. Even the mutants that gave a weak interaction in the yeast assay were able to support the replication of the null mutant except for the VP19C TN mutant at residue 409, which gave a level of complementation similar to the empty vector.
FIG. 2.
Complementation assays for the VP23 and VP19C TN mutants. The VP23 and VP19C TN mutants were examined for their ability to complement the growth of K23Z (A) and KΔ19C (B), respectively. The data are represented as percent complementation relative to the wild-type protein. The numbers on the x axis refer to the amino acid position of the insertion. The wild-type and vector-alone (0) controls were included in each transfection assay. All assays were done twice for each mutant and three times for mutants containing insertions at amino acids 58, 90, 93, 122, and 189 (A) and 25, 43, 46, 107, 179, 442, and 459 (B). Error bars are indicated.
Assembly of HSV-1 capsids using histidine-tagged capsid proteins.
The yeast two-hybrid assay was a very fast and informative assay for the analysis of the multitude of mutants generated. In addition, the genetic complementation assay was a rapid transient assay to determine the ability of the mutants to support virus growth. In order to correlate the findings from both these assays with the effects of these mutations on capsid assembly, we decided to generate recombinant baculoviruses, a system that has greatly contributed to our understanding of the assembly process of HSV capsids (25, 26). In this in vitro system, the capsid proteins are expressed in insect cells and subsequently assayed for capsid assembly. As the baculovirus genome has been cloned into a mini-F plasmid, it can be propagated and manipulated in E. coli (13).
The ORFs of the capsid proteins were derived using PCR assays or from previously described expression vectors. The ORFs for VP5, VP19C, and a chimera scaffold protein consisting of the human cytomegalovirus UL80 sequences of the mature assembly protein fused to the C-terminal 25 amino acids of HSV (17) were cloned into the transfer plasmid pFastBacHta. In this plasmid the proteins were expressed under the regulation of the polyhedrin promoter, with a histidine (six-histidine) handle fused to the N termini of these proteins. This was desirable for the purification of the proteins from infected cell lysates using affinity chromatography. A tobacco etch virus protease cleavage site provided an enzymatic method for releasing the protein from the handle. The reason for using the HCMV/HSV chimera scaffold protein derived from reported observations by Newcomb et al. (15) that demonstrated the superiority of this scaffold protein over the HSV scaffold protein for assembly reactions in vitro. The VP23 ORF was cloned into pFastBac, which produces the native protein, that is, no histidine tag. The reason for this was the ability to use affinity chromatography to purify the triplex complex using the histidine-tagged VP19C.
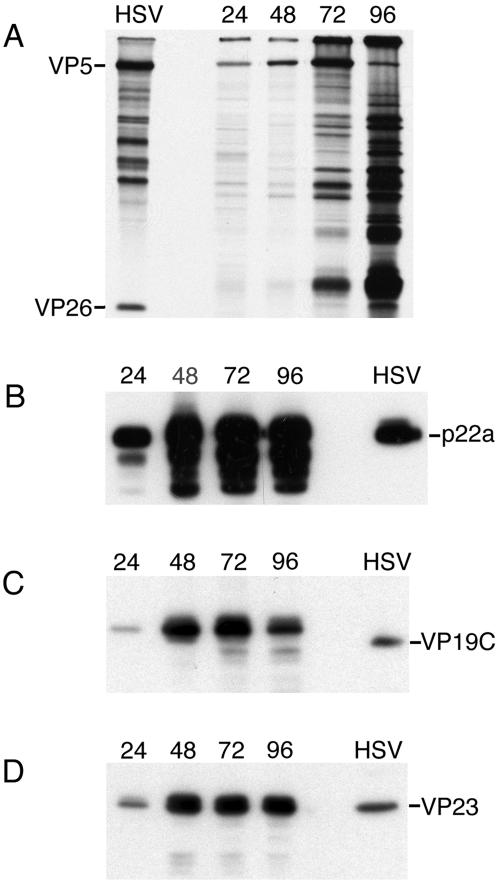
Sf9 cells were infected with baculoviruses expressing the UL80HSVCT, VP19C, and VP23 proteins, and lysates harvested at different times after infection were analyzed by SDS-PAGE followed by Western blot assays for the expression of the capsid proteins (Fig. 3B to D). To detect VP5 expression, the cells were pulse-labeled with [35S]methionine at the indicated times after infection (Fig. 3A). All the proteins were expressed in insect cells as judged by reactivity of the proteins to the specific antisera or by the presence of radioactivity in the gel. Accumulation of the proteins increased with time, especially from 24 to 48 h after infection (Fig. 3). Thereafter, there was stable accumulation of the proteins. In addition, we were able to detect the accumulation of these proteins in infected cells using Coomassie blue-stained protein gels (data not shown). While there does appear to be some degradation of the protein at late times after infection, especially for VP5 (panel A) and UL80HSVCT (panel B), the predominant polypeptide species in most cases was the full-length species. Additionally, we also observed a double polypeptide species for VP23 (panel D) similar to that observed in HSV-infected cells (5). The mobility of the recombinant VP5 and VP19C proteins was decreased (more evident for VP19C) (panel C) compared to that observed in lysates derived from KOS-infected cells due to the histidine and protease cleavage tag.
FIG. 3.
Expression of the HSV-1 capsid proteins in insect cells. Sf9 cells (1 × 106 cells in 35-mm dishes) were infected with recombinant baculoviruses at an MOI of 5 PFU/cell. The cells were harvested at 24, 48, 72, and 96 h after infection for Western blot analysis (B to D) or pulse-labeled with [35S]methionine for 2 h (A). Lysates were analyzed by SDS-PAGE followed by autoradiography for VP5 (A) or transferred to membranes followed by incubation using antibodies to the different capsid proteins (UL26C [B], UL38C [C], and R2421 [D]). Lane HSV, HSV-1-infected cell extracts (B to D) or radiolabeled B capsids (A).
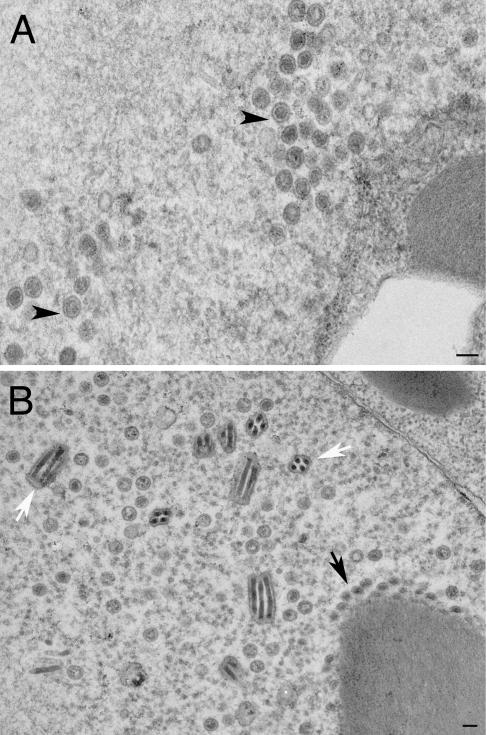
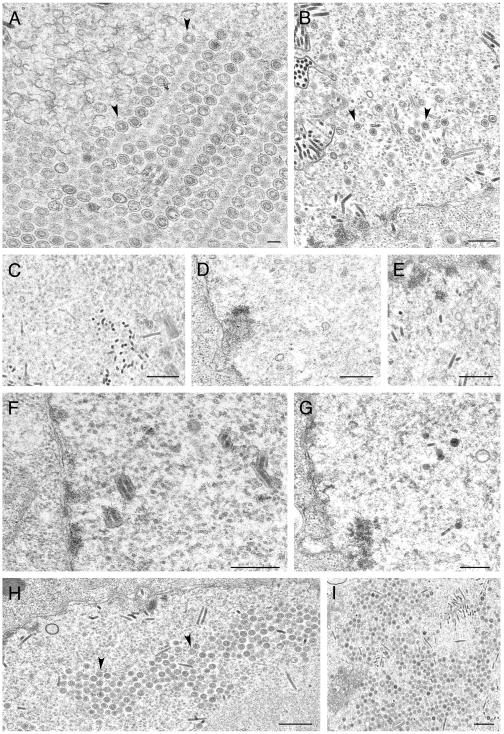
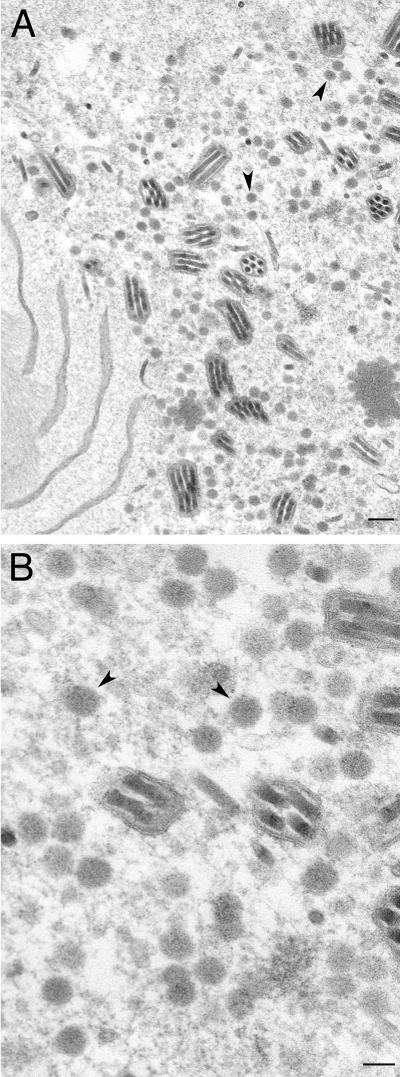
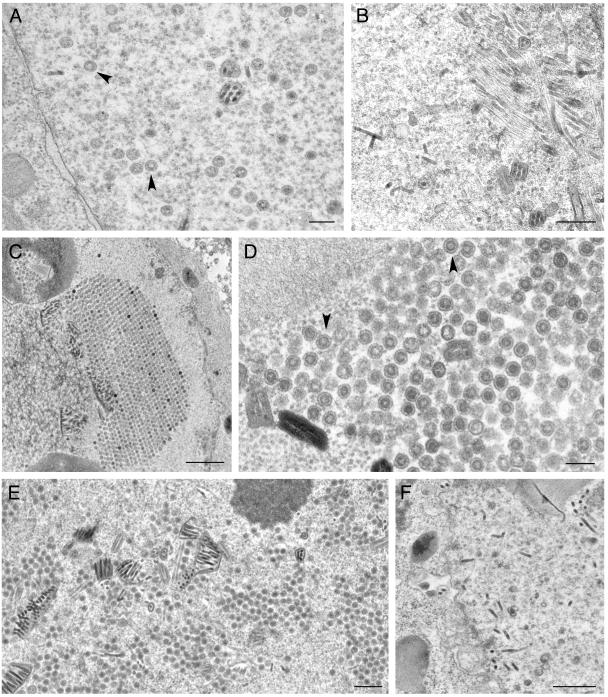
We decided to test these viruses for their ability to form capsids in infected cells. When insect cells were coinfected with these four viruses, fully assembled capsids were readily observed by conventional electron microscopy of the infected cells (Fig. 4A). This indicated that histidine tags at the N termini of VP5 and VP19C did not preclude assembly of a closed capsid shell. In some cells, large numbers of capsids were observed in the nuclei of the insect cells, indicating the efficiency of this reaction in these cells using these recombinant proteins (see Fig. 7A, below). In Fig. 4B, we observed partial capsid shells that were synthesized from an electron-dense region, which was presumably a region of highly concentrated protein. We decided to proceed with these viruses for subsequent analysis. It has previously been reported that coinfection of insect cells with baculoviruses expressing VP5 and VP19C produced small 88-nm particles (20, 22). We carried out similar infections to determine if these particles also formed in the presence of histidine tags at the N-terminal ends of these proteins. As shown in Fig. 5, the same small particles were readily observed in coinfected cells.
FIG. 4.
Capsid assembly in insect cells using histidine-tagged HSV-1 capsid proteins. Sf9 cells in 100-mm dishes (10 × 106 cells) were coinfected with recombinant baculoviruses expressing VP5, VP19C, scaffold protein, and VP23 at an MOI of 5 PFU/cell. The cells were fixed and processed for conventional EM 68 h after infection. HSV-assembled capsids marked by black arrowheads were evident in the nuclei of these cells (A). The white arrows in panel B indicate baculovirus capsids. The black arrow indicates an area where apparently partial shells were assembled from an electron-dense region. Bar, 0.1 μm.
FIG. 7.
Ultrastructural analysis of insect cells infected with viruses expressing VP23 TN mutants. Sf9 cells were infected as described in the legend of Fig. 4 and processed for conventional EM. Assembled capsids marked by arrows are shown for cells infected with the viruses expressing the wild-type proteins (A and B) as well as for those with a VP23 TN insertion at amino acid 90 (H and I). Capsids were not detected in sections examined for cells infected with viruses expressing transpositions at amino acids 296 (C), 239 (D), 22 (E), 219 (F), and 234 (G). Bars, 0.1 μm (A) and 0.5 μm (B to I).
FIG. 5.
Assembly of 88-nm VP5-VP19C particles in insect cells. Sf9 cells were coinfected with baculoviruses expressing the histidine-tagged VP5 and VP19C proteins as described in the Fig. 4 legend. The cells were processed for conventional EM 68 h after infection. The 80-nm particles are indicated with black arrowheads. Bars, 0.5 μm (A) and 0.1 μm (B).
Introduction of the VP23 and VP19C transposition mutants into the baculovirus genome and analysis of capsid assembly.
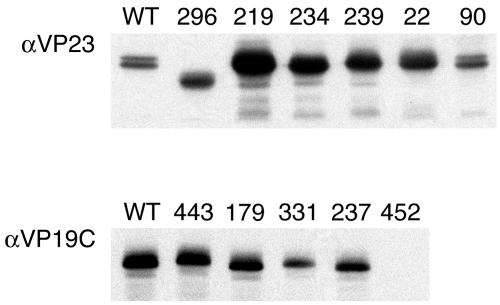
Since we were able to achieve capsid assembly in Sf9 cells using the recombinant baculovirus-expressed capsid proteins, the next step was to introduce selected VP23 and VP19C TN mutants into the baculovirus genome and determine the effects of these mutations on the capsid assembly process. For VP23 we chose to introduce 1822, 1890, 18219, 18234, 18239, and 18296 (Table 1 and Fig. 1A), and for VP19C we chose 38179, 38237, 38331, 38443, and 38452DV* (Table 2 and Fig. 1B). These mutants were chosen for their phenotypes displayed in the yeast two-hybrid assay. The mutant ORFs were introduced into the baculovirus bacmid using the BAC-to-BAC method. The bacmid DNAs were transfected into insect cells to reconstitute the baculovirus. Following amplification, expression for the mutant polypeptide was monitored using Western blot assays. The majority of the TN mutant polypeptides accumulated in infected cells at levels comparable to the wild-type protein (Fig. 6). The 18296 polypeptide displayed a faster mobility than the wild-type protein, because the transposition mutant resulted in polypeptide chain termination after amino acid 296. Some of the mutant polypeptides accumulated at greatly increased levels; the reason for this was not known, although it could be due to the titers of the different virus stocks. The polypeptide encoded by 38452DV* was not detected in this assay using also a polyclonal antibody to the whole protein (data not shown), indicating that this mutant protein was not stable.
FIG. 6.
Expression of the mutant TN polypeptides. Sf9 cells (1 × 106 cells in 35-mm dishes) were infected with the VP23 and VP19C TN mutants at an MOI of 5 PFU/cell. The cells were harvested 3 days after infection, and lysates prepared were analyzed by SDS-PAGE (12% acrylamide) followed by Western blotting. Detection of the mutant proteins was achieved by reacting the filters with VP23 or VP19C antiserum.
All the VP23 and VP19C TN mutants were tested for their ability to form capsids in cells coinfected with the baculoviruses expressing the capsid proteins. Insect cells were coinfected with the viruses and processed for TEM 68 h after infection. The thin sections were examined for the presence of assembled capsids. Capsids were observed in cells coinfected with baculoviruses expressing all the wild-type capsid proteins as shown before (Fig. 7A and B and 8A). In cells infected with the VP23 TN mutants, capsids were only observed in cells infected with 1890 (Fig. 7H and I), which is the only mutant in this group that gave a strong interaction in the Y2H assay. Fully assembled capsids were also observed in cells expressing the VP19C 38179 (Fig. 8C and D) and 38443 (Fig. 8E) polypeptides. In the VP19C 38179-infected cells, we saw a crystalline-like array of capsids (Fig. 8C). These data correlate well with the yeast two-hybrid data. The VP19C 38331 and 38237 mutants could not assemble capsids.
FIG. 8.
Ultrastructural analysis of insect cells infected with viruses expressing VP19C TN mutants. Sf9 cells were infected and the cells were processed for conventional EM as described in the legend of Fig. 4. Assembled capsids were evident in cells infected with the viruses expressing the wild-type proteins, indicated by black arrowheads (A), as well as for those with transpositions at amino acids 179 (C and D) and 443 (E). Capsids were not detected in any of the sections examined for cells infected with viruses expressing transposition insertions at amino acids 331 (B) and 237 (F). Bars, 0.2 μm (A and D), 0.5 μm (B, E, and F), and 1.0 μm (C).
DISCUSSION
The interaction between VP19C and VP23 was originally inferred by cryo-EM studies (16, 35) and subsequently confirmed by the yeast two-hybrid assay (7). VP19C and VP23 form a three-pronged structure that is unique to herpesvirus capsid architecture. The function of the complex is to stabilize the capsid shell through interactions with adjacent VP5 capsomeres. The VP19C-VP23 complex has been detected for HSV-1 using sedimentation techniques during coexpression in the baculovirus system (23) and by coimmunoprecipitation assays (data not shown). Nuclear localization of VP23 is dependent on its interaction with VP19C (20). Self-interaction of the CMV homologue of VP23 (33) has been detected using the yeast two-hybrid assay; for HSV-1, a dimer has been observed using sedimentation assays of lysates derived from Sf9 cells infected with a VP23-expressing baculovirus (23). Data from cryo-EM have shown the triangular-shaped knob exhibits extensive intimate associations of the three protein subunits, revealing the dimer interface between the two VP23 polypeptides and an extensive interaction of each of the two VP23 molecules with VP19C (34). All three subunits also exhibit a triangular symmetrical interaction with the floor domain, which is made up of VP5 (34).
The manner in which a hetero-trimeric protein complex interacts with the same shell protein (VP5) is perplexing. Purified VP23 behaves as a molten globule, which suggests that it only acquires tertiary structure when it has become incorporated into the triplex (12). In the baculovirus expression system, preformed triplexes will assemble into capsids when VP5 and the scaffold protein are supplied in trans (23). It may also play a role in the reconfiguration of the capsid structure during the assembly process. When VP23 was included in the infection of Sf9 cells together with VP5 and VP19C, the 88-nm spherical particles (Fig. 5) did not assemble (22). It has also been reported that VP19C forms one or more covalent disulfide bonds with VP5 (36). In our own chemical cross-linking studies, we detected one complex comprised of VP5, VP19C, and pre-22a and another comprised of VP5, VP19C, and VP23 (8). Thus, the triplex structure displays unique physical associations and configuration.
Prior results from the analysis of truncation mutants of VP19C in recombinant baculovirus-infected cells showed that the N-terminal 105 amino acids and the C-terminal 15 amino acids were dispensable for triplex formation but not capsid assembly in this system (23). There are few mutational analyses of VP23 in the literature. In the yeast two-hybrid assay we were unable to get an interaction of the N- or the C-terminal half of VP23 with VP19C (7). We did try to insert a flu hemagglutinin epitope in the N terminus of VP23 (at codon 11) but were not able to get a recombinant virus that could replicate on Vero cells, thus suggesting that this site was sensitive to insertion sequences. In unpublished observations noted in the manuscript by Spencer et al. (23), using a three-hybrid system consisting of VP23, VP19C, and VP5 they determined that the termini of VP23 were important for interaction.
Since very little information was available about the key domains and residues that mediate interaction of VP23 with VP19C, we decided to approach this problem using a random insertion method. The Tn7 transposon we used for the most part did randomly insert into the UL18 and UL38 sequences, but because of the A-T sequence preference, some of the insertions were not random in nature. However, by analyzing many mutants we were able to identify a number of insertions distributed throughout the ORFs (Fig. 1A and B). The other problem with this transposon was the diagnostic Pme1 restriction site (GTTTAAAC), which tends to generate a high frequency of translation termination mutations; this was especially true for VP19C, and so our data for this protein were not as extensive.
The data from both the genetic complementation assay and capsid assembly in insect cells correlated well with the data obtained in the yeast two-hybrid analysis (Tables 3 and 4). The single exception was a VP19C insertion at amino acid 409, which gave a weak reaction in the yeast two-hybrid assay but failed to complement the growth of the VP19C null mutant. This was a very weak interaction, because the colonies turned faint blue only after overnight incubation. The genetic complementation assay was used to uncover domains of the two proteins that may also play a role in other interactions during assembly and maturation besides triplex formation. Unfortunately, we did not discover mutants that could still form triplexes (Y2H) but failed to complement the null mutant virus.
TABLE 3.
VP23 TN mutants: summary of phenotypes
| Mutant | Y2H result (interaction) | Complementation of K23Z | Capsid assembly in insect cells |
|---|---|---|---|
| 22-VFKHA | None | − | No capsids |
| 26-CLNMQ | None | − | NDb |
| 35-LLFKH | None | − | ND |
| 41-LMFKQ | None | − | ND |
| 43-LVFKQ | None | − | ND |
| 58-LFKHS | Weak | + | ND |
| 66-FKQLL | None | − | ND |
| 67-VFKQA | None | − | ND |
| 90-CLNMD | Wild type | + | Capsids |
| 93-MFKHL | Wild type | + | ND |
| 103-CLNSN | None | − | ND |
| 110-CLNIC | None | − | ND |
| 116-CLNIS | None | − | ND |
| 122-VFKQF | Weak | + | ND |
| 189-CLNTL | Weak | + | ND |
| 190-FKQLL | None | ND | ND |
| 206-CLNTL | None | − | ND |
| 213-CLNNS | None | − | ND |
| 219-CLNTT | None | − | No capsids |
| 234-VFKHQ | None | − | No capsids |
| 239-CLNMN | None | − | No capsids |
| 296-V*a | None | − | No capsids |
| 310-V* | None | − | ND |
*, stop codon.
ND, not done.
TABLE 4.
VP19C TN mutants: summary of phenotypes
| Mutant | Y2H result (interaction) | Complementation of KΔ19C | Capsid assembly in insect cells |
|---|---|---|---|
| 25-LFKHT | Wild type | + | NDb |
| 41-MFKHP | Wild type | ND | ND |
| 43-CLNIS | Wild type | + | ND |
| 46-CLNSD | Wild type | + | ND |
| 107-MFKQR | Wild type | + | ND |
| 119-LFKHL | None | − | ND |
| 179-CLNRC | Wild type | + | Capsids |
| 237-LFKHT | None | − | No capsids |
| 247-LFKQF | None | − | ND |
| 324-FKQEL | None | − | ND |
| 331-CLNIK | None | − | No capsids |
| 347-VFKQF | None | − | ND |
| 396-CLNTQ | Weak | + | ND |
| 409-CLNMR | Weak | − | ND |
| 442-VFKHF | Weak | ND | ND |
| 443-CLNIG | Weak | + | Capsids |
| 450-V*a | None | − | ND |
| 452-DV* | None | − | Unstable protein |
| 452-VFKHL | None | − | ND |
| 459-CLNSP | Wild type | + | ND |
*, stop codon.
ND, not done.
What we discovered is that, unlike VP19C, in VP23 there are relatively few regions of this protein that are not required for this bimolecular interaction. For VP23 only 2 out of the 23 mutants analyzed had wild-type activity, whereas for VP19C 7 out of the 20 mutants had wild-type activity (Fig. 1A and B and Tables 3 and 4). In VP19C there is a large amino-terminal region that is not required for interaction as judged by this assay (Fig. 1B). Insertions at the N terminus of VP23 (after residues 22 and 25) abolished the interaction with VP19C. This was consistent with our inability to isolate a viable virus containing an epitope tag after residue 10 of the protein (Fig. 1A). Translation termination after residue 296 and 310 abolished the interaction, indicating the C-terminal nine residues may also be important for interaction with VP19C (Fig. 1B and Table 4). Some of the internal regions of VP23 were not as sensitive to mutagenesis as the termini. Using a computer program (42) that identifies potential coiled-coil regions, a segment of VP23 between residues 225 to 250 was judged to exhibit this secondary structure. Sequence analysis of VP23 revealed a isoleucine/leucine-rich region between residues 218 and 223. TN insertion at residue 219, which could potentially disrupt this structure, abolished interaction with VP19C (Table 1 and Fig. 1A). If this region is a potential dimerization domain for VP23, the yeast two-hybrid data strongly imply that the biologically active form of VP23 is a dimer. Many of the TN insertions in VP23 that abolish interaction with VP19C were in conserved regions of the protein (Fig. 1A). Insertions at amino acids in which the hydrophobic side chain were conserved in all three families were at L116, I213, and I219. Insertion at positions in which the amino acid was conserved for alpha- and betaherpesviruses was at T103. For VP19C, what is clear is the relatively large region at the N terminus, which appears to tolerate the TN insertions (Fig. 1B and Table 4). This confirms the studies of Spencer et al. (23). The degree of homology between the VP19C homologues of the three herpesvirus families is low relative to the VP5 homologues and is very weak in the N terminus (Fig. 1B) (28). In fact, it appears as if the HSV-1 VP19C specifies a longer N-terminal extension than the other proteins. The majority of the TN mutants of VP19C that do not affect the interaction with VP23 are in nonconserved regions or amino acids, with the only exception being the insertion at position 179, which is not a conserved amino acid but is in a conserved region in the alphaherpesviruses (Fig. 1B). Many of the mutants that weaken or abolish the triplex interaction were at positions that are conserved or in conserved regions of the alphaherpesviruses (C119, H237, F247, S331, and E452), and some (G409) are conserved in members of all three families.
In the study by Spencer et al. (23) the C-terminal truncation of 15 amino acids accumulated a stable polypeptide, unlike 38452DV*, which did not, even though the translation termination occurs in the same region. We therefore concluded that the nature of the insertion (DV*) at amino acid position 452 resulted in an unstable protein (Fig. 1B and Table 4). Their protein in baculovirus still could interact with VP23 to form triplex structures, but it could not assemble capsids. We did not introduce the TN mutant with an insertion at amino acid 450 into virus, and so we were not able to determine whether that mutant, which would be exactly a deletion of 15 amino acids, accumulated a stable polypeptide. That mutant, as well as the TN insertion that terminated translation at amino acid 448, did not interact with VP23. We synthesized and cloned into the yeast two-hybrid vectors UL38 fragments that would encode VP19C translation termination at amino acids 450, 454, and 458. All three polypeptides could not interact with VP23 in the yeast assay. This difference between the two pieces of data could possibly be explained by the different assays used. We did, however, get a TN insertion at 459, which still interacted with VP23, thus indicating that the C terminus could tolerate insertions of five amino acids.
One novel outcome of this study was the ability to assemble capsids with histidine handles on three of the proteins. The handle on the scaffold protein would be expected to not interfere with assembly, because the essential interaction occurs at the C terminus. For VP5, since the scaffold interaction domain has been mapped to the N terminus (spanning amino acids 27 to 71) (32), one would expect that an N-terminal tag could interfere with this interaction. However, this does not appear to be the case and is consistent with the ability to isolate a viable virus in which a hemagglutinin tag was inserted at the 10th amino acid (data not shown). The surprising result was that seen with the VP19C tagged protein, which can still participate in capsid assembly. We have recently isolated an HSV-1 recombinant that also specifies a histidine tag at the N terminus of the protein (data not shown). This virus replicates in Vero cells, and we are currently analyzing these capsids using immuno-EM to determine the location of the N terminus of VP19C in the capsid shell.
Acknowledgments
We thank Augusto Frisancho for help with the complementation assays. Jewell Walters constructed the VP26GFP fusion protein construct. Also, we thank Ed Brignole for plasmid pEB11 as a template for the UL80 PCR. We acknowledge Wade Gibson's support over the many years and his enthusiasm for this work. Both he and, of course, Stanley Person have been great mentors and friends over the years in the Wood Basic Sciences lab. Thanks again to Stan Person for critical review of the manuscript.
This work was supported by NIH PHS grant AI33077 and by National Science Foundation grant NSF-DBI-0099706 to J.M.M.
REFERENCES
- 1.Biery, M. C., M. Lopata, and N. L. Craig. 2000. A minimal system for Tn7 transposition: the transposon-encoded proteins TnsA and TnsB can execute DNA breakage and joining reactions that generate circularized Tn7 species. J. Mol. Biol. 297:25-37. [DOI] [PubMed] [Google Scholar]
- 2.Biery, M. C., F. J. Stewart, A. E. Stellwagen, E. A. Raleigh, and N. L. Craig. 2000. A simple in vitro Tn7-based transposition system with low target site selectivity for genome and gene analysis. Nucleic Acids Res. 28:1067-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breeden, L., and K. Nasmyth. 1985. Regulation of the yeast HO gene. Cold Spring Harbor Symp. Quant. Biol. 50:643-650. [DOI] [PubMed] [Google Scholar]
- 4.Craig, N. L. 1991. Tn7: a target site-specific transposon. Mol. Microbiol. 5:2569-2573. [DOI] [PubMed] [Google Scholar]
- 5.Desai, P., N. A. DeLuca, J. C. Glorioso, and S. Person. 1993. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. J. Virol. 67:1357-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desai, P., N. A. DeLuca, and S. Person. 1998. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology 247:115-124. [DOI] [PubMed] [Google Scholar]
- 7.Desai, P., and S. Person. 1996. Molecular interactions between the HSV-1 capsid proteins as measured by the yeast two-hybrid system. Virology 220:516-521. [DOI] [PubMed] [Google Scholar]
- 8.Desai, P., S. C. Watkins, and S. Person. 1994. The size and symmetry of B capsids of herpes simplex virus type 1 are determined by the gene products of the UL26 open reading frame. J. Virol. 68:5365-5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245-246. [DOI] [PubMed] [Google Scholar]
- 10.Gibson, W., and B. Roizman. 1972. Proteins specified by herpes simplex virus. 8. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol. 10:1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hendricks, L. C., M. McCaffery, G. E. Palade, and M. G. Farquhar. 1993. Disruption of endoplasmic reticulum to Golgi transport leads to the accumulation of large aggregates containing beta-COP in pancreatic acinar cells. Mol. Biol. Cell 4:413-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirkitadze, M. D., P. N. Barlow, N. C. Price, S. M. Kelly, C. J. Boutell, F. J. Rixon, and D. A. McClelland. 1998. The herpes simplex virus triplex protein, VP23, exists as a molten globule. J. Virol. 72:10066-10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luckow, V. A., S. C. Lee, G. F. Barry, and P. O. Olins. 1993. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 67:4566-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 15.Newcomb, W. W., F. L. Homa, D. R. Thomsen, B. L. Trus, N. Cheng, A. Steven, F. Booy, and J. C. Brown. 1999. Assembly of the herpes simplex virus procapsid from purified components and identification of small complexes containing the major capsid and scaffolding proteins. J. Virol. 73:4239-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newcomb, W. W., B. L. Trus, F. P. Booy, A. C. Steven, J. S. Wall, and J. C. Brown. 1993. Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J. Mol. Biol. 232:499-511. [DOI] [PubMed] [Google Scholar]
- 17.Oien, N. L., D. R. Thomsen, M. W. Wathen, W. W. Newcomb, J. C. Brown, and F. L. Homa. 1997. Assembly of herpes simplex virus capsids using the human cytomegalovirus scaffold protein: critical role of the C terminus. J. Virol. 71:1281-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Person, S., and P. Desai. 1998. Capsids are formed in a mutant virus blocked at the maturation site of the UL26 and UL26.5 open reading frames of herpes simplex virus type 1 but are not formed in a null mutant of UL38 (VP19C). Virology 242:193-203. [DOI] [PubMed] [Google Scholar]
- 19.Rixon, F. J. 1993. Structure and assembly of herpesviruses. Semin. Virol. 4:135-144. [Google Scholar]
- 20.Rixon, F. J., C. Addison, A. McGregor, S. J. Macnab, P. Nicholson, V. G. Preston, and J. D. Tatman. 1996. Multiple interactions control the intracellular localization of the herpes simplex virus type 1 capsid proteins. J. Gen. Virol. 77:2251-2260. [DOI] [PubMed] [Google Scholar]
- 21.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, vol. 2. Lippincott, Williams and Wilkins, New York, N.Y.
- 22.Saad, A., Z. H. Zhou, J. Jakana, W. Chiu, and F. J. Rixon. 1999. Roles of triplex and scaffolding proteins in herpes simplex virus type 1 capsid formation suggested by structures of recombinant particles. J. Virol. 73:6821-6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spencer, J. V., W. W. Newcomb, D. R. Thomsen, F. L. Homa, and J. C. Brown. 1998. Assembly of the herpes simplex virus capsid: preformed triplexes bind to the nascent capsid. J. Virol. 72:3944-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steven, A. C., and P. G. Spear. 1996. Herpesvirus capsid assembly and envelopment. Oxford University Press, New York, N.Y.
- 25.Tatman, J. D., V. G. Preston, P. Nicholson, R. M. Elliott, and F. J. Rixon. 1994. Assembly of herpes simplex virus type 1 capsids using a panel of recombinant baculoviruses. J. Gen. Virol. 75:1101-1113. [DOI] [PubMed] [Google Scholar]
- 26.Thomsen, D. R., L. L. Roof, and F. L. Homa. 1994. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J. Virol. 68:2442-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trus, B. L., F. P. Booy, W. W. Newcomb, J. C. Brown, F. L. Homa, D. R. Thomsen, and A. C. Steven. 1996. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J. Mol. Biol. 263:447-462. [DOI] [PubMed] [Google Scholar]
- 28.Trus, B. L., J. B. Heymann, K. Nealon, N. Cheng, W. W. Newcomb, J. C. Brown, D. H. Kedes, and A. C. Steven. 2001. Capsid structure of Kaposi's sarcoma-associated herpesvirus, a gammaherpesvirus, compared to those of an alphaherpesvirus, herpes simplex virus type 1, and a betaherpesvirus, cytomegalovirus. J. Virol. 75:2879-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trus, B. L., F. L. Homa, F. P. Booy, W. W. Newcomb, D. R. Thomsen, N. Cheng, J. C. Brown, and A. C. Steven. 1995. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J. Virol. 69:7362-7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trus, B. L., W. W. Newcomb, F. P. Booy, J. C. Brown, and A. C. Steven. 1992. Distinct monoclonal antibodies separately label the hexons or the pentons of herpes simplex virus capsid. Proc. Natl. Acad. Sci. USA 89:11508-11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vernon, S. K., M. Ponce de Leon, G. H. Cohen, R. J. Eisenberg, and B. A. Rubin. 1981. Morphological components of herpesvirus. III. Localization of herpes simplex virus type 1 nucleocapsid polypeptides by immune electron microscopy. J. Gen. Virol. 54:39-46. [DOI] [PubMed] [Google Scholar]
- 32.Walters, J. N., G. L. Sexton, J. M. McCaffery, and P. Desai. 2003. Mutation of single hydrophobic residue I27, L35, F39, L58, L65, L67, or L71 in the N terminus of VP5 abolishes interaction with the scaffold protein and prevents closure of herpes simplex virus type 1 capsid shells. J. Virol. 77:4043-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wood, L. J., M. K. Baxter, S. M. Plafker, and W. Gibson. 1997. Human cytomegalovirus capsid assembly protein precursor (pUL80.5) interacts with itself and with the major capsid protein (pUL86) through two different domains. J. Virol. 71:179-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou, Z. H., M. Dougherty, J. Jakana, J. He, F. J. Rixon, and W. Chiu. 2000. Seeing the herpesvirus capsid at 8.5 Å. Science 288:877-880. [DOI] [PubMed] [Google Scholar]
- 35.Zhou, Z. H., J. He, J. Jakana, J. D. Tatman, F. J. Rixon, and W. Chiu. 1995. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat. Struct. Biol. 2:1026-1030. [DOI] [PubMed] [Google Scholar]
- 36.Zweig, M., C. J. Heilman, Jr., and B. Hampar. 1979. Identification of disulfide-linked protein complexes in the nucleocapsids of herpes simplex virus type 2. Virology 94:442-450. [DOI] [PubMed] [Google Scholar]