Abstract
Foamy viruses (FV) are the oldest known genus of retroviruses and have persisted in nonhuman primates for over 60 million years. FV are efficiently transmitted, leading to a lifelong nonpathogenic infection. Transmission is thought to occur through saliva, but the detailed mechanism is unknown. Interestingly, this persistent infection contrasts with the rapid cytopathicity caused by FV in vitro, suggesting a host defense against FV. To better understand the tissue specificity of FV replication and host immunologic defense against FV cytopathicity, we quantified FV in tissues of healthy rhesus macaques (RM) and those severely immunosuppressed by simian immunodeficiency virus (SIV). Contrary to earlier findings, we find that all immunocompetent animals consistently have high levels of viral RNA in oral tissues but not in other tissues examined, including the small intestine. Strikingly, abundant viral transcripts were detected in the small intestine of all of the SIV-infected RM, which has been shown to be a major site of SIV (and human immunodeficiency virus)-induced CD4+ T-cell depletion. In contrast, there was a trend to lower viral RNA levels in oropharyngeal tissues of SIV-infected animals. The expansion of FV replication to the small intestine but not to other CD4+ T-cell-depleted tissues suggests that factors other than T-cell depletion, such as dysregulation of the jejunal microenvironment after SIV infection, likely account for the expanded tissue tropism of FV replication.
Foamy viruses (FV) are ubiquitous retroviruses of nonhuman primates (NHP), cats, cows, and horses that establish lifelong persistent infections. Thus far, there is no credible evidence that FV infections lead to pathologies in any host, natural or accidental (reviewed in reference 26). FV have cospeciated with NHP for an estimated 60 million years (39). This unusually long period of cospeciation has allowed foamy viruses to be particularly well adapted to their hosts. It is not known why foamy viruses are not pathogenic, while infection by their closest viral relatives, the complex retroviruses such as human immunodeficiency virus (HIV) and human T-cell leukemia virus, and hepatitis B virus, can be deadly. In contrast to the seemingly benign course of infection, FV generally induce cytopathicity in vitro and replicate to high levels in primary cells and cell lines, although stable persistently infected cell lines can be derived (24). Hematopoietic cells obtained from FV+ animals do not produce virus in vivo. However, when such cells are stimulated to divide in tissue culture, they produce virus which spreads to new cells and eventually kills the culture. When primate FV replicate to high titer in tissue culture cells, the cells are invariably killed (27). The ability of FV to rapidly kill many primary cell types in tissue culture, but not in vivo, raises the possibility that the host immune system could play a role in containing viral infection and/or replication in vivo. Many opportunistic viruses are limited by the host's immune system and become associated with pathologies only after immunosuppression. Whether or not this is true for FV has not been determined.
Foamy viruses have a unique replication pathway, aspects of which are related to both retroviruses and hepadnaviruses (reviewed in reference 23). For the most part, FV behave like the retroviruses, in that viral RNA genomes are reverse transcribed into cDNA, which is then stably integrated into the host genome as a provirus. However, FV reverse transcription occurs during viral assembly and/or budding so that the actual genome is DNA rather than RNA, as in the case of hepadnaviruses, such as hepatitis B virus. Thus, the presence of FV DNA in cells is indicative of both proviruses (mostly latent) and also cell-associated virions with DNA genomes.
Foamy virus-host dynamics have been only cursorily examined. Previous studies of naturally infected NHP characterized FV infection as primarily latent. In a single study of naturally infected African green monkeys, a very low copy number of proviral DNA was detected in most tissues (8). In contrast, FV RNA, indicative of viral gene expression and replication, was found exclusively, albeit sporadically and at a low level, in the oral mucosa (8). FV have a high transmission rate, which is at odds with this characterization of FV infection as mostly latent. FV seroprevalence is >90% among NHP in captivity (3, 26), with the majority of NHP seroconverting as juveniles, by the age of 2 years (M. Axthelm, unpublished data). Quarantine of FV-free animals in specific-pathogen-free facilities is required to prevent infection. While humans are the only primates who are not naturally infected with FV, some human populations, such as monkey caretakers, veterinarians, and people in contact with monkeys in natural settings, such as monkey temple workers or bush meat hunters, are zoonotically infected with FV, with an incidence of 1 to 4% (15, 38, 44). Only a small number of FV+ humans have been identified, and there is as yet no evidence for horizontal transmission between humans, or pathogenic consequences (4, 6). It has been speculated that primate FV are transmitted via saliva, based on documentation of humans zoonotically infected after sustaining monkey bites and the observation that virus can consistently be cocultured from saliva of infected monkeys (3). The high FV transmission rate and the difficulty in detecting viral RNA in the presumptive site of viral transmission have presented a paradox. Further, there has not as yet been detailed examination of FV replication in the most common host for retroviral research, the macaque.
One approach taken to study immune control of viral infection in NHP has been examination of immunosuppressed hosts for pathologies or increases in viral replication. In macaques, immunosuppression after infection by simian immunodeficiency virus (SIV) has been used to examine immune control of viruses including cytomegalovirus (CMV) (16, 17) and other herpesviruses. Depletion of CD4+ T cells after infection with HIV or SIV is most pronounced at mucosal surfaces and occurs rapidly, within 10 to 17 days after infection, well before CD4+ T-cell loss is observed in blood (reviewed in reference 7). There is a selective loss of memory CD4+ T cells (cells that specifically recognize previously encountered antigens) from multiple tissues (25). In the gastrointestinally associated lymphoid tissue of the colon (20) and small intestine (40), the lung (30), and the vagina (41), 60 to 80% of memory T cells are depleted early after infection. The culmination of the subsequent chronic phase of immunodeficiency virus infection is associated with susceptibility to opportunistic infections.
Rhesus macaques (RM) infected with SIV are used as a model for AIDS. As most RM are naturally infected with a simian foamy virus (SFVmac), we were able to study the effect of SIV-induced immunosuppression on FV levels using animals enrolled in ongoing SIV studies. We developed sensitive and quantitative PCR (qPCR) for analysis of both RNA and DNA sequences and used these to examine the tissue specificity of FV infection (DNA) and replication (RNA) in rhesus macaques. These were compared to the patterns of FV replication in late stages of SIV infection when animals were severely depleted of CD4+ T cells in mucosal tissues. Our results indicate that FV replication is readily detectable in oral swabs and several oral tissues in both SIV− and SIV+ animals. Most interestingly, SIV-induced immunosuppression expands the sites of FV replication to include the small intestinal jejunum, a site of CD4+ T-cell depletion, but not other mucosal sites that are significantly depleted of CD4+ T cells.
MATERIALS AND METHODS
Animals and virus.
All animals used in these studies were rhesus macaques (Macaca mulatta) of Indian origin bred and housed at the Oregon National Primate Research Center. All studies were conducted in accordance with the standards of the Center's Animal Use and Care Committee and the Guide for the Care and Use of Laboratory Animals-approved protocols (13). Animals were euthanized in accordance with the Panel on Euthanasia of the American Veterinary Medical Association (2). The RM were naturally infected with SFV, through interaction with SFV-infected animals in the facility, but the time since infection with FV was unknown for any individual animal. Both colony and specific-pathogen-free RM were identified as SFV+ or SFV− by screening for the presence of serum antibodies against the SFVmac Gag antigen, as detected by a quantitative enzyme-linked immunosorbent assay (ELISA) as described below. Animals were infected with either SIVmac239, a CCR5-tropic virus, or SIVmac155T3, a CXCR4-tropic virus, as previously described (30). SIV plasma loads were determined as previously described (22). Animals were considered immunosuppressed by the presence of a simian AIDS-related pathology. In all cases, there was a >90% CD4+ T-cell depletion from mucosal sites, as measured by phenotypic analysis of lymphocytes obtained by bronchoalveolar lavage (BAL) and also, in some cases, the jejunum, as described further below. Absolute blood CD4+ T-cell counts were determined by obtaining total lymphocyte counts from whole blood and measuring the percentage of CD4+ T cells in peripheral blood mononuclear cells (PBMC), as described below. Necropsy tissues were obtained from SIV-immunosuppressed SFV+ animals or healthy SFV+ animals that were sacrificed for reasons unrelated to viral infections. The SIV-infected cohort was all male, with an average age of 5.1 years, while the SIV− cohort included three males and two females, with an average age of 3.2 years. Additional animals were used to obtain PBMC, blood, and buccal swabs. In these cases, all SIV− animals were in good health, and SIV+ animals were sampled >70 days postinfection with pathogenic SIV.
Sample collection.
A set of tissues for each macaque was obtained at necropsy, and sections were preserved in RNA stabilization solution, RNAlater (Ambion), according to the manufacturer's instructions, and stored at −20°C until nucleic acid isolation, as described below. Buccal swabs, composed of saliva and cellular material obtained by scraping the tongue, cheek, and gums, were collected with a polyester fiber-tipped swab (Fisher). PBMC were isolated by standard density gradient centrifugation of whole blood. BAL cells, a source of T cells from the lung, and jejunal lamina propria lymphocytes were isolated as previously described (31).
Analysis of CD4+ T cells.
In order to determine CD4+ T-cell percentages in tissues and PBMC, bronchoalveolar lymphocytes, jejunal lamina propria lymphocytes, and PBMC were phenotyped by flow cytometry. The lymphocyte preparations were stained with antibodies to the lymphocyte-specific marker CD3 and to the CD4 and/or CD8 T-cell subset-specific molecules. The specific monoclonal antibody clones used were α-CD3 (SP34), α-CD4 (L200), and α-CD8β (2ST8.5h7), which had been conjugated to one of the following fluorochromes: Pacific Blue, fluorescein isothiocyanate, phycoerythrin-Texas Red, Texas Red, allophycocyanin, allophycocyanin-Cy7, or Am-Cyan (all obtained from BD Biosciences, Pharmingen, San Diego, Calif.). Data were collected using a three-laser BD LSR flow cytometer (Becton Dickinson Immunocytometry Systems, San Diego, Calif.) and analyzed with FlowJo v. 6.1 (TreeStar, San Carlos, Calif).
FV Gag ELISA.
The SFV-1 viral clone (29) was kindly provided by A. Mergia (University of Florida). The N terminus of the SFV-1 Gag protein (amino acids 1 to 193) was produced as a glutathione S-transferase (GST) fusion protein by cloning bases 1736 to 2318 of SFV-1 into the pGEX-2X vector (Promega). The protein was expressed in the Escherichia coli strain PLys and extracted by standard methods. SFV-1 Gag1-193-GST was titrated and determined to be optimal at 500 ng/ml in the ELISA. The ELISA was done as previously described for SIV antigens (14), with the following changes. Briefly, 96-well Immunosorp plates (Nalge Nunc) were coated with 100 μl/well of the SFV-1 Gag1-193-GST antigen or the GST control antigen alone, diluted to 500 ng/ml in a carbonate-bicarbonate buffer (10 mM Na2CO3, 40 mM NaHCO3, pH 9.6) at 4°C overnight. Heat-inactivated plasma was serially diluted in twofold dilutions that ranged from 1:100 to 1:51,200. The previously described ELISA protocol for SIV antigens was followed except that midpoint titers were obtained by determining the reciprocal of the plasma dilution at the maximum optical density/2. Every individual assay included standard plasma derived from an RM with high levels of anti-FV Gag antibodies, in order to normalize for interassay variability, and all titers were normalized to this plasma standard. Additionally, all assays included plasma from an RM previously identified as FV−. In every assay, this serum gave a value that was at or below background.
Isolation of DNA and RNA from PBMC, buccal swabs, and tissues.
Buccal swabs were placed directly into RLT buffer (QIAGEN) containing 1% beta-mercaptoethanol (Sigma). The tube was briefly vortexed, the swab was removed from the solution, and the solution was then frozen at −80°C. The swab solution was thawed at 37°C for 15 min immediately prior to RNA extraction. Total RNA was extracted according to a standard protocol using the RNeasy Mini kit (QIAGEN). Tissues preserved in RNA stabilization solution were removed from the solution, minced, and placed in either RLT buffer (QIAGEN) with 1% beta-mercaptoethanol for RNA extraction or ATL buffer (QIAGEN) for DNA extraction. For DNA and RNA isolation, tissues were homogenized in the presence of 2.5-mm zirconia beads (Biospec, Inc.). Homogenization was performed using a Mini Bead Beater-8 (Biospec, Inc.) for three homogenization cycles at 3 min per cycle. Tissue homogenates were then removed from the beads. Total DNA and RNA were extracted from the tissue homogenates as well as from PBMC using the DNA Mini (QIAGEN) and RNeasy Mini (QIAGEN) kits, respectively. DNA and RNA were quantified by spectrophotometry. DNA preparations were treated with RNase A according to the manufacturer's instructions (QIAGEN). RNA preparations were treated with RNase-free DNase (Promega) at 1 μl/μg for 2 h at 37°C and then for 15 min at 65°C to heat inactivate the enzyme.
Primary foamy virus isolation and sequencing of the FV gag gene.
PBMC isolated from eight FV+ rhesus macaques were cocultured with TF cells, an SFV− rhesus macaque fibroblast cell line which was generated by expression of the human telomerase protein in neonatal RM fibroblasts (19). The PBMC were treated with interleukin-2 as previously described (35), prior to coculture. Infected TF cells were maintained until significant cytopathic effects were observed. The cells were used to obtain virus as previously described (45). Cell-free viral preparations were used to infect fresh TF cells, until 80 to 90% cytopathic effect was observed. Cells were then lysed, and genomic DNA was isolated using the DNA Mini kit (QIAGEN). The complete 1.9-kb gag gene was PCR amplified from DNA, and primary isolates were sequenced using eight primers spanning the gag gene. A contiguous gag sequence for each isolate was generated with Sequencher v. 4.5 (Applied Biosystems), and the complete gag nucleic acid and predicted protein sequences were aligned using Clustal X, version 1.83. At the nucleic acid level, three of the eight viral gag genes were completely homologous, while the remaining five were between 90 and 98% homologous (GenBank accession no. DQ120930 to DQ120937). All of the primary isolates were between 84 and 85% identical to the gag gene of SFV-1 (GenBank accession no. X58484).
Quantitation of FV DNA and RNA.
Quantitative PCR assays for DNA (qPCR) and reverse transcription-PCR (RT-PCR) for RNA (qRT-PCR) were performed using the ABI Prism 7700 sequence detection system (Applied Biosystems Inc.). Primers and probes were designed to detect an 80-bp highly conserved nucleic acid binding region of the gag gene of the primary isolates (37, 46). This region was completely conserved among the eight primary isolates whose sequences were obtained. The primers used were gag1759F, ACGAACATCTGGTGCGGG, and gag1835R, CTGCGTTTCCACCAGCTGA, and the probe was gag1789 6-carboxyfluorescein (FAM)-AGGAAGAGGGAACCAAAACCGAAACCA-6-carboxyltetramethylrhodamine (TAMRA). The qPCR conditions used were as follows: 95°C for 1 min, 95°C for 15 s, and 60°C for 1 min for 45 cycles. The qRT-PCR conditions used were as follows: 48°C for 30 min, 95°C for 10 min, 95°C for 15 s, and 60°C for 1 min for 40 cycles.
To standardize the qPCR, a 125-bp region that included the PCR target sequence from one primary isolate was cloned into a PCR cloning vector, TOPO-TA (Invitrogen). Known amounts of the target sequence (from 101 to 107 copies) were added to 500 ng of human genomic DNA that was quantified by the manufacturer (Promega) to generate DNA standard curves in every individual assay. In addition to a standard curve, each PCR run included a buffer-only and an SFV-negative control. DNA derived from PBMC or tissue samples was used at 500 ng (105 cell equivalents) per PCR. The lower limit of detection of the qPCR was 10 copies per 105 cell equivalents.
A c-myc cellular DNA qPCR was done for each sample in every individual assay to normalize for cellular DNA content (c-mycF1, GCCCCTCAACGTTAGCTTCA; c-mycR1, CGCAGTAGAAATACGGCTGCA; probe, FAM-CAACAGGAACTATGACCTCGACTACGACTCG-TAMRA). All assays included a c-myc standard curve generated by adding 10-fold dilutions of human genomic DNA to the PCR mixtures, ranging from 105 to 10 copies.
Each PBMC DNA sample was tested in duplicate, in two or more independent PCR assays. Each tissue DNA sample was tested in duplicate, in three independent assays, and in all cases, standard deviations were calculated. We found that, for the qPCR assays, >90% of the samples tested in duplicate had less than twofold variability, while the maximum variability was 4.4-fold. All qPCR standard curves had correlation coefficients of ≥0.95.
To generate standards for the qRT-PCR, the PCR target sequence was subcloned from the TOPO-TA vector into the pNEB vector (New England Biolabs) with a T7 promoter inserted upstream of the cloning site so that RNA could be transcribed using T7 RNA polymerase. The in vitro transcription reactions were done using the Riboprobe Combination System according to standard protocols (Promega). Tenfold dilutions (ranging from 107 to 10 copies) of the viral RNA sequence were added to 50 ng (104 cell equivalents) of RNA isolated from TF cells. Control reaction mixtures lacking reverse transcriptase were included, in order to ensure that there was no DNA contamination in the RNA preparations, and SFV-negative RNA was used as a control in every individual assay. The lower limit of detection of the qRT-PCR was 50 copies per 104 cell equivalents.
RNA input was normalized by qRT-PCR using an 18S RNA standard that was included in every assay, using a commercially available primer-probe set (Applied Biosystems). An 18S standard curve was generated in every assay by adding 10-fold dilutions (ranging from 104 to 10 cell equivalents) of RNA, quantified using spectrophotometry, to the 18S qRT-PCR mixtures.
Buccal swab RNA was tested in duplicate, in two independent assays. Tissue RNA was tested in duplicate, in three independent assays, and standard deviations were calculated for each sample set. Animals previously identified as SFV− by ELISA tested negative for SFV sequences by qRT-PCR. For the FV gag qRT-PCR assays, we found maximum variation in 12 replicates to be 1.5-fold, and for the 18S qRT-PCR, for samples run in 24 replicates, we found maximum variation to be 1.7-fold. The correlation coefficient of all qRT-PCR standard curves was ≥0.95.
Statistical analyses.
Between-group differences in FV RNA levels were analyzed using the Mann-Whitney U test, with the exception of the FV RNA levels in the tongue and DNA levels in the jejunum, which were evaluated using log-transformed data and an independent sample t test. Prism v. 4 (GraphPad Software) was used to perform statistical analyses.
Nucleotide sequence accession numbers.
The gag gene sequences of the eight isolates were deposited in GenBank under accession numbers DQ120930 to DQ120937.
RESULTS
SFV DNA is present at a low copy number in PBMC and tissues from healthy and immunosuppressed animals.
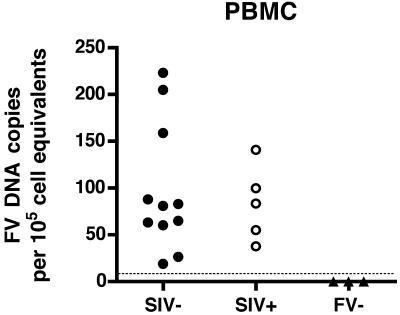
Initially, we measured foamy virus DNA in PBMC from 11 SIV-negative, immunocompetent RM by qPCR (Fig. 1). We found levels ranging from 19 to 223 SFV DNA copies per 105 cell equivalents of DNA (mean = 98). PBMC obtained from five SIV-infected SFV+ RM had levels ranging from 38 to 141 SFV DNA copies (mean = 83). This indicates that there are no significant differences in PBMC SFV DNA levels between the SIV-uninfected and SIV-infected macaques. Animals were screened for SFV antibodies using an SFVmac Gag ELISA, and, as expected, all seronegative animals were negative for SFV DNA sequences (Fig. 1).
FIG. 1.
Foamy virus DNA loads in peripheral blood mononuclear cells of SIV− and SIV+ RM. Each symbol indicates the mean normalized SFV DNA level from two independent assays run in duplicate from an individual animal. The lower limit of detection of the assay is shown by the dotted line. All animals which were negative by ELISA had DNA levels below the level of detection.
We next surveyed necropsy tissues from healthy and SIV-infected RM (Table 1) for SFV DNA. In the SIV-infected RM, severe CD4+ T-cell depletion was evident in mucosal sites, such as the lung, as measured using BAL samples. CD4+ T-cell depletion was >90% in the SIV-infected RM compared to immunocompetent animals, where BAL CD4+ T cells are typically 25 to 60% of total lymphocytes (30). CD4+ T cells in the peripheral blood were not as depleted, with the exception of RM SIV+ 4, which had been infected for the longest time with SIV (Table 1). CD4+ T-cell numbers in the blood of healthy RM range between ∼600 and 2,000 per μl of blood, with a mean of 1,330 (33), whereas we found CD4+ T-cell numbers ranging from 70 to 415, with a mean of 323. Most importantly, all the SIV-infected RM were severely immunosuppressed based on the presence of at least one simian AIDS-defining opportunistic infection (Table 1).
TABLE 1.
Status of SIV+ animals at necropsy
| Animal | SIV strain | DPIa | SIV loadb | CD4 (blood)c | % CD4 in:
|
Opportunistic infection(s) | |
|---|---|---|---|---|---|---|---|
| BALd | LPLe | ||||||
| SIV+ 1 | mac239 | 554 | 5 × 106 | 415 | 0.25 | 0.28 | CMV, cryptosporidium |
| SIV+ 2 | mac239 | 491 | 3 × 106 | 399 | 0.4 | NDf | CMV |
| SIV+ 3 | mac155T3 | 380 | 6 × 105 | 407 | 0.42 | 1.23 | CMV, pneumocystis |
| SIV+ 4 | mac155T3 | 730 | 6 × 105 | 70 | 0.1 | ND | CMV, cryptosporidium |
DPI, days postinfection.
The SIV load is given as SIV gag RNA copies per ml of plasma.
Absolute CD4+ T-cell counts per μl in the blood.
The percentage of CD4+ cells of total CD3+ bronchoalveolar lavage lymphocytes.
The percentage of CD4+ cells of total CD3+ jejunal lamina propria lymphocytes.
ND, not determined.
SFV DNA was present in the majority of tissues examined from healthy animals, similar to a previous finding in African green monkeys (8). We found levels ranging from 50 copies per 105 cell equivalents of DNA in the parotid salivary gland to 104 copies per 105 cell equivalents in the tonsil and tongue (data not shown). In immunosuppressed RM, we saw a similar prevalence of SFV DNA. In our evaluation of SFV DNA we did not specifically isolate high-molecular-weight chromosomal DNA. Therefore, DNA extracted from infected cells could contain both integrated proviral DNA and viral genomic DNA. Our findings indicate that SFV DNA, in either a proviral or a genomic form, is present in PBMC and diverse tissues in both immunocompetent and immunosuppressed RM.
SFV RNA is consistently detected at high levels in the oral cavity.
The difficulty in detecting infectious virus in SFV-infected animals suggests that most tissues harbor only latent proviruses. In order to determine which, if any, of the tissues containing SFV DNA are permissive for SFV replication, we used qRT-PCR for viral gag RNA expression as an indication of viral replication. Since we used primers in the gag gene, our measurements include both genomic RNA and gag transcripts. Using this assay, we examined blood, buccal swabs, and some necropsy tissues of healthy and SIV-infected RM. First, we measured SFV RNA in PBMC from nine immunocompetent and nine SIV-infected RM. SFV RNA was not detected in PBMC of any of the animals (data not shown). This finding is not unexpected (43) and confirms that blood is a site of SFV persistence but not of replication.
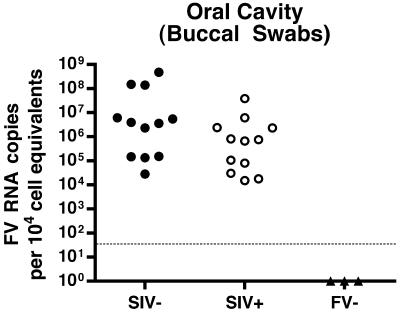
A previous study showed that SFV infection of NHP leads to only very limited replication, even in the oral mucosa, which is not consistent with the observed efficient transmission. To determine levels of SFV RNA in the oral cavity of the RM, we used oral swabs, a noninvasive, accessible source of saliva and cellular material. We evaluated 12 SIV− and 12 SIV+ SFV-seropositive RM and found SFV RNA in all of the SFV+ animals tested but not in the animals which were SFV− by ELISA. Viral RNA levels in the oral cavity ranged from 1.4 × 104 to 4.7 × 108 copies per 104 cell equivalents of RNA (Fig. 2). Such consistent detection of high levels of SFV RNA in the oral cavity is consistent with the high rate of SFV transmission among NHP.
FIG. 2.
Foamy virus RNA loads in the oral cavity of SIV− and SIV+ RM. RNA levels from buccal swabs were normalized to cell equivalents using a qRT-PCR for 18S RNA. Each symbol indicates the SFV RNA mean value from two independent assays, each run in duplicate from an individual animal. The lower limit of detection of the assay is shown by the dotted line. All animals which were negative by ELISA had RNA levels below the level of detection.
If CD4+ T-cell-dependent mechanisms limit SFV replication, then viral RNA levels should increase following SIV-induced immunosuppression. However, the SFV load in the SIV− population (mean = 7.1 × 107) was more than a log greater than that of the SIV+ population (mean = 4.3 × 106). Thus, there is a trend toward lower RNA levels in immunosuppressed animals (P = 0.088; Fig. 2), which is not consistent with a role for CD4+ T cells in limiting viral replication in the oral cavity. However, other factors may confound our interpretation; for example, CD4+ T cells could be a permissive cell type for SFV replication, or they could stimulate other cell types which are permissive for viral replication, such as macrophages.
Multiple oropharyngeal tissues are permissive for SFV replication in both healthy and SIV-immunosuppressed hosts.
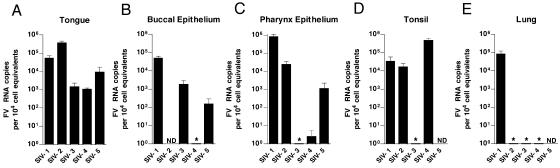
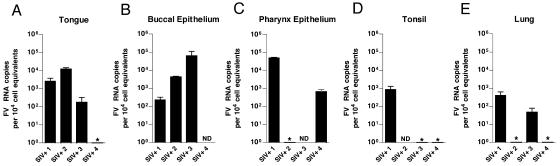
We next screened a large number of tissues obtained at necropsy from five healthy RM and the four SIV-infected, immunocompromised RM described in Table 1. In samples from the SIV− animals, SFV RNA was consistently found in multiple oropharyngeally associated tissues, such as the pharyngeal epithelium, tongue, and tonsils. All five SIV− animals had detectable SFV RNA in at least one of the oral tissues, although there was variability in the specific site of viral replication. Figure 3 shows the results of these analyses. The tongue was positive in all of the animals (Fig. 3A), and the buccal epithelium (Fig. 3B), pharyngeal epithelium (Fig. 3C), and tonsil (Fig. 3D) were positive in most of the animals. Only one animal had detectable RNA in the lung (Fig. 3E). Interestingly, SFV RNA was undetectable in both the parotid and submaxillary salivary glands (data not shown). This indicates that SFV enters saliva distal from the site of its secretion. All other tissues examined (Table 2) had undetectable SFV RNA. A similar pattern was seen in the four SIV-immunosuppressed animals (Fig. 4). In these animals, the buccal epithelium (Fig. 4A) and tongue (Fig. 4B) were most consistently permissive, with two animals having RNA in the pharyngeal epithelium (Fig. 4C) and lung (Fig. 4E) and only one having SFV RNA in the tonsil (Fig. 4D). All of the SIV-immunosuppressed RM also had viral RNA in at least one oropharyngeal tissue. When we compared the SFV RNA levels in oropharyngeally associated tissues from SIV− and SIV+ animals, we also observed a trend towards lower SFV RNA loads in the SIV+ RM. This trend was most evident in the tongue, with a P value of 0.13 (data not shown).
FIG. 3.
Foamy virus RNA levels in permissive tissues of healthy RM. Results of qRT-PCR for SFV RNA in tissues from five SIV− RM are shown. Each bar represents the mean viral load from three independent qRT-PCRs for one animal. Viral loads were normalized to cell equivalents by qRT-PCR for 18S RNA. Error bars represent the standard deviations. All SFV-seronegative RM were PCR negative for SFV RNA in all tissues examined. ND, not determined. *, undetectable.
TABLE 2.
Tissue distribution of SFV gag RNA in tissues from healthy or SIV+ immunosuppressed animals
| Tissue | FV RNAa
|
|
|---|---|---|
| SIV− | SIV+ | |
| Buccal epithelium | + | + |
| Pharyngeal epithelium | + | + |
| Tongue | + | + |
| Tonsil | + | + |
| Lung | + | + |
| Small intestine | − | + |
| Mesenteric lymph node | − | + |
| Parotid salivary gland | − | − |
| Colon | − | − |
| PBMC | − | − |
RNAs were analyzed by qRT-PCR as described in Materials and Methods. The lower limit of the assay was 50 to 100 SFV gag copies per 104 cell equivalents of RNA. This table summarizes the data for the four SIV− animals in shown in Fig. 3 and the five SIV+ animals shown in Fig. 4, with the exception of PBMC, which were evaluated from eight healthy and eight SIV-infected RM. A + indicates that at least one animal in the group had significant levels of RNA in the tissue.
FIG. 4.
Foamy virus RNA levels in permissive oropharyngeal tissues of SIV-immunosuppressed RM. Results of qRT-PCR for SFV RNA in tissues from four SIV+ immunosuppressed RM are shown. Analysis was done as in Fig. 3. Error bars represent the standard deviations. All SFV-seronegative RM were PCR negative for SFV RNA in all tissues examined. ND, not determined. *, undetectable.
SFV replication is extended to the small intestine with SIV-induced immunosuppression.
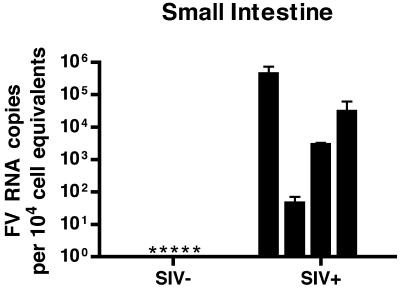
A striking finding in these analyses was that the small intestine from all four of the SIV+ RM contained detectable levels of SFV RNA (Fig. 5; between 50 and 4.3 × 105 SFV RNA copies per 104 cell equivalents), in contrast to small intestine from all of the five immunocompetent RM, where SFV RNA was below the level of detection (<50 copies). This result is statistically significant (P = 0.016).
FIG. 5.
Foamy virus replication is expanded to the small intestine of SIV-immunosuppressed RM. Results of qRT-PCR for SFV+ and SFV− RNA are shown. Each bar represents the mean viral load from three independent qRT-PCRs for one animal. Viral loads were normalized to cell equivalents by a qRT-PCR for 18S RNA. Error bars represent the standard deviations. *, undetectable.
The jejunum samples that we analyzed came from the middle region of the jejunum (jejunum 2). To determine whether or not FV RNA is absent from all regions of the jejunum in healthy animals, we examined all regions (jejunum 1, 2, and 3) obtained from one SIV− animal and detected no FV RNA in any region (data not shown). We evaluated the CD4+ T-cell percentages in the jejunum of two of the SIV+ RM and found, in contrast to 12% in a healthy young adult animal, 0.28% and 1.23% CD4+ T cells (Table 1). In other reports, normal levels were found to be between 5% and 45% in juvenile RM and between 30% and 50% in mature adult RM (42). Therefore, these animals are significantly depleted of CD4+ T cells in the jejunum, having ≤10% of the normal levels. Additionally, we found detectable SFV RNA, 1.8 × 103 copies per 104 cell equivalents, in the mesenteric lymph node of one SIV+ animal (data not shown). Interestingly, this was observed in the RM that had the lowest levels of CD4+ T cells, SIV+ 4 (Table 1).
DISCUSSION
We have performed the first quantitative and systematic evaluation of the tissue specificity of FV replication in nonhuman primates and have found that FV infection is characterized by high viral RNA levels (up to 104 copies/cell) in the oral cavity. These results are consistent with the high rate of FV transmission between natural hosts and the frequent acquisition of FV by humans through monkey bites. However, the actual number of virions that the FV RNA copy number represents is unknown, since the copy number that we determined includes gag mRNAs in addition to viral genomic RNA. Our studies revealed that the tongue and oral secretions were most consistently permissive for FV replication. Oral swabs containing superficial cells and saliva had the highest levels. The lack of FV RNA in the salivary glands is consistent with FV entering saliva from infected cells in the oral cavity. Tissue culture studies have shown that FV are highly cell associated and that high levels of replication invariably lead to cell death (47, 48). Extrapolating from the in vitro studies to the in vivo situation leads us to hypothesize that FV also kill their in vivo target cells which would be shed into saliva in the oral cavity. Since replication is not associated with pathogenesis, it is reasonable to assume that the permissive cells in vivo, containing cell-associated virus, have a naturally high turnover rate and even when not infected are sloughed off into saliva.
Contrary to expectation, we saw a small decrease in FV RNA levels in the oral pharyngeal tissues in late stages of SIV immunosuppression, and this finding is inconsistent with T-cell immune control of FV infection in these tissues. This decrease could be caused by SIV induction of innate immunity, such as alpha/beta interferon (IFN-α/β) production (1, 5), or adaptive immunity, such as production of IFN-γ. FV infection does not normally induce IFN production, although FV are sensitive to IFN-α, -β, and -γ (9, 32). Alternatively, this decrease could be due to SIV-induced loss of a cell type that normally supports FV replication. The permissive cell type in vivo is not as of yet known. Primate FV infects all cell types in vitro and is most cytopathic in cells of epithelial and fibroblast origin. However, in leukocyte-derived human cell lines it is generally persistent, with very low levels of virus produced. For example, the titer of FV produced by clonal lines of infected Jurkat T cells is less than 102 infectious virions/2 × 106 cells (28), and because so few cells are productively infected, no cytopathic effects are seen. The small amount of virus detected is likely to be produced after spontaneous activation of very small numbers of cells. Therefore, it is possible that in vivo CD4+ T cells, or another cell type lost after SIV infection, are persistently infected with FV and that activation of a subset leads to the levels of viral RNA seen. Until the target cell is known, detailed understanding of the mechanism is impossible.
Our most striking finding is the expansion of FV replication to the jejunum of SIV-immunosuppressed animals. For two SIV+ animals examined, the level of CD4+ T cells in the jejunal lamina propria was significantly depressed, less than 10% of those observed in healthy RM (Table 1). However, the small intestine is only one of many mucosal tissues that are severely depleted of CD4+ T cells after SIV infection. For example, although significant CD4+ T-cell depletion has been demonstrated in the colon of SIV-immunosuppressed RM (20), we did not observe FV RNA in this tissue. Thus, our results are not consistent with general control of viral replication by CD4+ T cells but suggest that SIV-induced changes that are unique to the small intestine account for the extended tissue tropism of FV replication.
A question of interest is whether or not there is increased migration of FV-infected cells to the jejunum from permissive tissues after SIV infection, which would be consistent with the lowered FV RNA levels that we find in the oral mucosa. For retroviruses, one way to address this is by measuring FV DNA levels. We did, in fact, quantify FV DNA loads in the jejunum of the SIV− and SIV+ animals and found significantly higher levels of FV DNA in the SIV-infected macaques (data not shown). However, in the case of foamy viruses, the complication is that the functional genome in virions is DNA (49), so that proviral DNA cannot be distinguished from viral genomic DNA using PCR. Therefore, it is likely that the higher DNA levels in the jejunum result from increased viral transcription leading to cell-associated virions containing genomic DNA.
SIV infection is known to induce dysregulation of intestinal cytokines and growth factors, inducing hyperinflammation, maladsorption, and changes in the mucosal barrier in the small intestine (10). This altered tissue microenvironment could lead to induction of latent proviruses and drive FV replication at this ectopic site. While the Tas transactivator protein of PFV (prototype FV of chimpanzee origin) has been shown to transactivate genes under the control of the HIV type 1 long terminal repeat (18), transactivation of the FV long terminal repeat by HIV Tat has not been examined. CMV, which establishes an opportunistic infection associated with AIDS-related immunosuppression, is present in the intestine of HIV-infected people and induces expression of latent HIV genomes in this tissue (36). Recently, rhesus macaque CMV (RhCMV) has been described in the gastrointestinal tract of SIV-infected macaques (12), and all of the SIV-immunosuppressed RM in our study had an RhCMV-induced pathology (Table 1). Thus, it is possible that RhCMV is involved in transactivation of latent FV proviruses in the intestine.
A current model of HIV and SIV pathogenesis proposes that, subsequent to the massive mucosal memory CD4+ T-cell depletion, there is chronic immune activation, and opportunistic agents may contribute to this state. With immunosuppression, persistent viruses such as CMV are transformed into opportunistic pathogens. Foamy virus, while apparently a- pathogenic in immunocompetent hosts, has now been shown to expand its replication to include the small intestine in immunosuppressed hosts. In this context, it is interesting that “generalized giant cell disease” was observed in a number of cynomolgus macaques infected with SIV (21). Giant cells were seen with multiple nuclei in various tissues, including the small intestine. These were attributed to macrophages but also resemble FV-induced syncytia, although it is not known whether these cells are producing FV. It is possible that FV plays a role in the pathology in the gut seen during progression to simian AIDS and contributes to dysregulation of the immune system. Finally, it is interesting that FV-persistently infected T cells in culture are more permissive to HIV infection than uninfected cells (34). The differences in disease progression after SIV infection of FV+ and FV− animals have not as yet been evaluated. Because SIV- and SHIV (SIV-HIV hybrid viruses)-infected monkeys are widely used as a model for pathogenesis of HIV, and a majority of these animals are also infected with foamy viruses, the contribution of foamy viruses to simian AIDS needs to be more carefully examined.
Some humans have been identified who are infected with primate foamy viruses through various types of contacts with NHP. Thus far, there is no evidence for human-to-human transmission in the small number of cases examined (11). It is an outstanding question as to whether this lack of transmission is due to differences in patterns of FV replication in the natural host compared to zoonotically infected humans or to behavioral differences. It is not known whether foamy virus replication is as robust in the oral mucosa of human hosts. Further, there are no reports of FV infection in immunocompromised people, but the status of FV in such individuals would also be of interest.
Acknowledgments
We thank Lyn Daly, John Edgar, Shoko Hagen, Anne Lewis, Richard Lum, Elizabeth Majors, Ed Reed-Inderbitzen, Bill Sutton, Andrew Sylwester, Cara Taormina, and Joshua Walker for technical advice and assistance and Al Legasse and Shannon Planer for animal care. Thanks to Barbra Richardson and Dina Lauman for assistance with statistical analysis and to Nancy Haigwood for critical reading of the manuscript.
This work was funded by NIH grant CA 81297 to M.L.L. S.M.M. was partially supported by NIH training grant T32 CA 09229. L.J.P. was supported by NIH grant AI 54292. M.K.A. was supported by NIH grants U42 RR016025 and U24 RR018107.
REFERENCES
- 1.Ahmed, R. K., G. Biberfeld, and R. Thorstensson. 2005. Innate immunity in experimental SIV infection and vaccination. Mol. Immunol. 42:251-258. [DOI] [PubMed] [Google Scholar]
- 2.AVMA Panel on Euthanasia. American Veterinary Medical Association. 2001. 2000 report of the AVMA Panel on Euthanasia. J. Am. Vet. Med. Assoc. 218:669-696. [DOI] [PubMed] [Google Scholar]
- 3.Blewett, E. L., D. H. Black, N. W. Lerche, G. White, and R. Eberle. 2000. Simian foamy virus infections in a baboon breeding colony. Virology 278:183-193. [DOI] [PubMed] [Google Scholar]
- 4.Boneva, R. S., A. J. Grindon, S. L. Orton, W. M. Switzer, V. Shanmugam, A. I. Hussain, V. B. Bhullar, M. E. Chamberland, W. Heneine, T. M. Folks, and L. E. Chapman. 2002. Simian foamy virus infection in a blood donor. Transfusion 42:886-891. [DOI] [PubMed] [Google Scholar]
- 5.Bosinger, S. E., K. A. Hosiawa, M. J. Cameron, D. Persad, L. Ran, L. Xu, M. R. Boulassel, M. Parenteau, J. Fournier, E. W. Rud, and D. J. Kelvin. 2004. Gene expression profiling of host response in models of acute HIV infection. J. Immunol. 173:6858-6863. [DOI] [PubMed] [Google Scholar]
- 6.Brooks, J. I., E. W. Rud, R. G. Pilon, J. M. Smith, W. M. Switzer, and P. A. Sandstrom. 2002. Cross-species retroviral transmission from macaques to human beings. Lancet 360:387-388. [DOI] [PubMed] [Google Scholar]
- 7.Douek, D. C., L. J. Picker, and R. A. Koup. 2003. T cell dynamics in HIV-1 infection. Annu. Rev. Immunol. 21:265-304. [DOI] [PubMed] [Google Scholar]
- 8.Falcone, V., J. Leupold, J. Clotten, E. Urbanyi, O. Herchenroder, W. Spatz, B. Volk, N. Bohm, A. Toniolo, D. Neumann-Haefelin, and M. Schweizer. 1999. Sites of simian foamy virus persistence in naturally infected African green monkeys: latent provirus is ubiquitous, whereas viral replication is restricted to the oral mucosa. Virology 257:7-14. [DOI] [PubMed] [Google Scholar]
- 9.Falcone, V., M. Schweizer, A. Toniolo, D. Neumann-Haefelin, and A. Meyerhans. 1999. Gamma interferon is a major suppressive factor produced by activated human peripheral blood lymphocytes that is able to inhibit foamy virus-induced cytopathic effects. J. Virol. 73:1724-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.George, M. D., E. Reay, S. Sankaran, and S. Dandekar. 2005. Early antiretroviral therapy for simian immunodeficiency virus infection leads to mucosal CD4+ T-cell restoration and enhanced gene expression regulating mucosal repair and regeneration. J. Virol. 79:2709-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heneine, W., W. M. Switzer, P. Sandstrom, J. Brown, S. Vedapuri, C. A. Schable, A. S. Khan, N. W. Lerche, M. Schweizer, D. Neumann-Haefelin, L. E. Chapman, and T. M. Folks. 1998. Identification of a human population infected with simian foamy viruses. Nat. Med. 4:403-407. [DOI] [PubMed] [Google Scholar]
- 12.Hutto, E. H., D. C. Anderson, and K. G. Mansfield. 2004. Cytomegalovirus-associated discrete gastrointestinal masses in macaques infected with the simian immunodeficiency virus. Vet. Pathol. 41:691-695. [DOI] [PubMed] [Google Scholar]
- 13.Institute for Laboratory Animal Research. 1996. Guide for the care and use of laboratory animals. National Academic Press, Washington, D.C.
- 14.Jayaraman, P., D. Mohan, P. Polacino, L. Kuller, N. Sheikh, H. Bielefeldt-Ohmann, B. Richardson, D. Anderson, S. L. Hu, and N. L. Haigwood. 2004. Perinatal transmission of SHIV-SF162P3 in Macaca nemestrina. J. Med. Primatol. 33:243-250. [DOI] [PubMed] [Google Scholar]
- 15.Jones-Engel, L., G. A. Engel, M. A. Schillaci, A. Rompis, A. Putra, K. G. Suaryana, A. Fuentes, B. Beer, S. Hicks, R. White, B. Wilson, and J. S. Allan. 2005. Primate-to-human retroviral transmission in Asia. Emerg. Infect. Dis. 11:1028-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaur, A., C. L. Hale, B. Noren, N. Kassis, M. A. Simon, and R. P. Johnson. 2002. Decreased frequency of cytomegalovirus (CMV)-specific CD4+ T lymphocytes in simian immunodeficiency virus-infected rhesus macaques: inverse relationship with CMV viremia. J. Virol. 76:3646-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaur, A., N. Kassis, C. L. Hale, M. Simon, M. Elliott, A. Gomez-Yafal, J. D. Lifson, R. C. Desrosiers, F. Wang, P. Barry, M. Mach, and R. P. Johnson. 2003. Direct relationship between suppression of virus-specific immunity and emergence of cytomegalovirus disease in simian AIDS. J. Virol. 77:5749-5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keller, A., E. D. Garrett, and B. R. Cullen. 1992. The Bel-1 protein of human foamy virus activates human immunodeficiency virus type 1 gene expression via a novel DNA target site. J. Virol. 66:3946-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirchoff, V., S. Wong, S. St. Jeor, and G. S. Pari. 2002. Generation of a life-expanded rhesus monkey fibroblast cell line for the growth of rhesus rhadinovirus (RRV). Arch. Virol. 147:321-333. [DOI] [PubMed] [Google Scholar]
- 20.Li, Q., L. Duan, J. D. Estes, Z. M. Ma, T. Rourke, Y. Wang, C. Reilly, J. Carlis, C. J. Miller, and A. T. Haase. 2005. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature 434:1148-1152. [DOI] [PubMed] [Google Scholar]
- 21.Li, S. L., E. E. Kaaya, H. Feichtinger, P. Putkonen, C. Parravicini, D. Bottiger, G. Biberfeld, and P. Biberfeld. 1991. Monocyte/macrophage giant cell disease in SIV-infected cynomolgus monkeys. Res. Virol. 142:173-182. [DOI] [PubMed] [Google Scholar]
- 22.Lifson, J. D., J. L. Rossio, M. Piatak, Jr., T. Parks, L. Li, R. Kiser, V. Coalter, B. Fisher, B. M. Flynn, S. Czajak, V. M. Hirsch, K. A. Reimann, J. E. Schmitz, J. Ghrayeb, N. Bischofberger, M. A. Nowak, R. C. Desrosiers, and D. Wodarz. 2001. Role of CD8+ lymphocytes in control of simian immunodeficiency virus infection and resistance to rechallenge after transient early antiretroviral treatment. J. Virol. 75:10187-10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linial, M. L. 1999. Foamy viruses are unconventional retroviruses. J. Virol. 73:1747-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linial, M. L. 2000. Why aren't foamy viruses pathogenic? Trends Microbiol. 8:284-289. [DOI] [PubMed] [Google Scholar]
- 25.Mattapallil, J. J., D. C. Douek, B. Hill, Y. Nishimura, M. Martin, and M. Roederer. 2005. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature 434:1093-1097. [DOI] [PubMed] [Google Scholar]
- 26.Meiering, C. D., and M. L. Linial. 2001. Historical perspective of foamy virus epidemiology and infection. Clin. Microbiol. Rev. 14:165-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meiering, C. D., and M. L. Linial. 2002. Reactivation of a complex retrovirus is controlled by a molecular switch and is inhibited by a viral protein. Proc. Natl. Acad. Sci. USA 99:15130-15135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meiering, C. D., C. Rubio, C. May, and M. L. Linial. 2001. Cell-type-specific regulation of the two foamy virus promoters. J. Virol. 75:6547-6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mergia, A., and M. Wu. 1998. Characterization of provirus clones of simian foamy virus type 1. J. Virol. 72:817-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Picker, L. J., S. I. Hagen, R. Lum, E. F. Reed-Inderbitzin, L. M. Daly, A. W. Sylwester, J. M. Walker, D. C. Siess, M. Piatak, Jr., C. Wang, D. B. Allison, V. C. Maino, J. D. Lifson, T. Kodama, and M. K. Axthelm. 2004. Insufficient production and tissue delivery of CD4+ memory T cells in rapidly progressive simian immunodeficiency virus infection. J. Exp. Med. 200:1299-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pitcher, C. J., S. I. Hagen, J. M. Walker, R. Lum, B. L. Mitchell, V. C. Maino, M. K. Axthelm, and L. J. Picker. 2002. Development and homeostasis of T cell memory in rhesus macaque. J. Immunol. 168:29-43. [DOI] [PubMed] [Google Scholar]
- 32.Rhodes-Feuillette, A., J. Lasneret, S. Paulien, W. Ogunkolade, J. Peries, and M. Canivet. 1990. Effects of human recombinant alpha and gamma and of highly purified natural beta interferons on simian Spumavirinae prototype (simian foamy virus 1) multiplication in human cells. Res. Virol. 141:31-43. [DOI] [PubMed] [Google Scholar]
- 33.Schenkel, A. R., H. Uno, and C. D. Pauza. 1999. Asymptomatic simian immunodeficiency virus infection decreases blood CD4+ T cells by accumulating recirculating lymphocytes in the lymphoid tissues. J. Virol. 73:601-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schiffer, C., C. H. Lecellier, A. Mannioui, N. Felix, E. Nelson, J. Lehmann-Che, M. L. Giron, J. C. Gluckman, A. Saib, and B. Canque. 2004. Persistent infection with primate foamy virus type 1 increases human immunodeficiency virus type 1 cell binding via a Bet-independent mechanism. J. Virol. 78:11405-11410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schweizer, M., V. Falcone, J. Gange, R. Turek, and D. Neumann-Haefelin. 1997. Simian foamy virus isolated from an accidentally infected human individual. J. Virol. 71:4821-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, P. D., and U. E. Mai. 1992. Immunopathophysiology of gastrointestinal disease in HIV infection. Gastroenterol. Clin. N. Am. 21:331-345. [PubMed] [Google Scholar]
- 37.Stenbak, C. R., and M. L. Linial. 2004. Role of the C terminus of foamy virus Gag in RNA packaging and Pol expression. J. Virol. 78:9423-9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Switzer, W. M., V. Bhullar, V. Shanmugam, M. E. Cong, B. Parekh, N. W. Lerche, J. L. Yee, J. J. Ely, R. Boneva, L. E. Chapman, T. M. Folks, and W. Heneine. 2004. Frequent simian foamy virus infection in persons occupationally exposed to nonhuman primates. J. Virol. 78:2780-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Switzer, W. M., M. Salemi, V. Shanmugam, F. Gao, M. E. Cong, C. Kuiken, V. Bhullar, B. E. Beer, D. Vallet, A. Gautier-Hion, Z. Tooze, F. Villinger, E. C. Holmes, and W. Heneine. 2005. Ancient co-speciation of simian foamy viruses and primates. Nature 434:376-380. [DOI] [PubMed] [Google Scholar]
- 40.Veazey, R. S., M. DeMaria, L. V. Chalifoux, D. E. Shvetz, D. R. Pauley, H. L. Knight, M. Rosenzweig, R. P. Johnson, R. C. Desrosiers, and A. A. Lackner. 1998. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science 280:427-431. [DOI] [PubMed] [Google Scholar]
- 41.Veazey, R. S., P. A. Marx, and A. A. Lackner. 2003. Vaginal CD4+ T cells express high levels of CCR5 and are rapidly depleted in simian immunodeficiency virus infection. J. Infect. Dis. 187:769-776. [DOI] [PubMed] [Google Scholar]
- 42.Veazey, R. S., M. Rosenzweig, D. E. Shvetz, D. R. Pauley, M. DeMaria, L. V. Chalifoux, R. P. Johnson, and A. A. Lackner. 1997. Characterization of gut-associated lymphoid tissue (GALT) of normal rhesus macaques. Clin. Immunol. Immunopathol. 82:230-242. [DOI] [PubMed] [Google Scholar]
- 43.von Laer, D., D. Neumann-Haefelin, J. L. Heeney, and M. Schweizer. 1996. Lymphocytes are the major reservoir for foamy viruses in peripheral blood. Virology 221:240-244. [DOI] [PubMed] [Google Scholar]
- 44.Wolfe, N. D., W. M. Switzer, J. K. Carr, V. B. Bhullar, V. Shanmugam, U. Tamoufe, A. T. Prosser, J. N. Torimiro, A. Wright, E. Mpoudi-Ngole, F. E. McCutchan, D. L. Birx, T. M. Folks, D. S. Burke, and W. Heneine. 2004. Naturally acquired simian retrovirus infections in central African hunters. Lancet 363:932-937. [DOI] [PubMed] [Google Scholar]
- 45.Yu, S. F., D. N. Baldwin, S. R. Gwynn, S. Yendapalli, and M. L. Linial. 1996. Human foamy virus replication—a pathway distinct from that of retroviruses and hepadnaviruses. Science 271:1579-1582. [DOI] [PubMed] [Google Scholar]
- 46.Yu, S. F., K. Edelmann, R. K. Strong, A. Moebes, A. Rethwilm, and M. L. Linial. 1996. The carboxyl terminus of the human foamy virus Gag protein contains separable nucleic acid binding and nuclear transport domains. J. Virol. 70:8255-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu, S. F., and M. L. Linial. 1993. Analysis of the role of the bel and bet open reading frames of human foamy virus by using a new quantitative assay. J. Virol. 67:6618-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu, S. F., J. Stone, and M. L. Linial. 1996. Productive persistent infection of hematopoietic cells by human foamy virus. J. Virol. 70:1250-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu, S. F., M. D. Sullivan, and M. L. Linial. 1999. Evidence that the human foamy virus genome is DNA. J. Virol. 73:1565-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]