Abstract
Transcription factors of the POU family govern cell fate through combinatorial interactions with coactivators and corepressors. The POU factor Oct-3/4 can define differentiation, dedifferentation, or self-renewal of pluripotent embryonic stem (ES) cells in a sensitive, dose-dependent manner (H. Niwa, J.-I. Miyazali, and A. G. Smith, Nat. Genet. 24:372-376, 2000). Here we have developed a complementation assay based on the ability of Oct-3/4 transgenes to rescue self-renewal in conditionally null ES cells and used this to define which domains of Oct-3/4 are required to sustain the undifferentiated stem cell phenotype. Surprisingly, we found that molecules lacking either the N-terminal or C-terminal transactivation domain, though not both, can effectively replace full-length Oct-3/4. Furthermore, a fusion of the heterologous transactivation domain of Oct-2 to the Oct-3/4 POU domain can also sustain self-renewal. Thus, the unique function of Oct-3/4 in ES cell propagation resides in combination of the specific POU domain with a generic proline-rich transactivation domain. Interestingly, however, Oct-3/4 target gene expression elicited by the N- and C-terminal transactivation domains is not identical, indicating that at least one class of genes activated by Oct-3/4 is not required for ES cell propagation.
Transcription factors regulate expression of specific genes to control cellular phenotype. This is achieved via binding to target DNA sequences, followed by interaction in cis with components of the basal transcriptional machinery. These two capacities are often separated into structurally and functionally distinct DNA-binding and regulatory domains. The DNA-binding domain provides for nucleotide sequence-directed localization to target genes. The regulatory domain(s) interacts with one or more components of the basal transcription complex, either directly or indirectly via bridging cofactors, to mediate activation or repression of gene expression (23, 34).
A regulatory domain that can activate expression is designated a transactivation domain. Many transactivation domains have been identified in various transcription factors and are grouped into five different classes based on their characteristic amino acid composition: (i) acidic, (ii) glutamine-rich, (iii) proline-rich, (iv) serine/threonine-rich, and (v) unclassified (23, 46). The activities of transactivation domains are commonly defined in artificial systems based on transient transfection of a transcription factor plus reporter construct into heterologous cells. Although such systems have detected activities of all classes of transactivation domains and revealed different specificities (46), it is difficult to establish physiological distinctions by this method.
The POU family transcription factor Oct-3/4 (also termed Oct-3 or Oct-4, encoded by Pou5f1) (31, 36, 44) is expressed in totipotent and pluripotent cells, including oocytes, early-cleavage-stage embryos, the inner cell mass (ICM) of the blastocyst, epiblast, and germ cells (32, 42). Oct-3/4 is also present in cultured pluripotent embryonic stem (ES) cells, embryonal carcinoma cells, and embryonic germ cells (18, 43, 56) but is absent from all differentiated somatic cell types in vitro and in vivo.
The essential role of this transcription factor in mouse development has been revealed by targeted gene deletion. Oct-3/4-deficient embryos fail to initiate fetal development because the prospective founder cells of the ICM do not acquire pluripotency and become diverted into the extraembryonic trophectoderm lineage (25). Further investigation via conditional repression and expression in ES cells revealed that the precise level of Oct-3/4 governs developmental fate. ES cells require a critical level of Oct-3/4 to maintain their stem cell character and pluripotency. A less than twofold increase causes differentiation into endoderm and mesoderm, whereas reduction to less than 50% of the normal expression level triggers dedifferentiation into trophectoderm (29). These results accord with observations of subtle differences of Oct-3/4 levels in different cell types in the early mouse embryo (32). Therefore, Oct-3/4 can be regarded as a candidate master regulator for initiation, maintenance, and differentiation of pluripotent cells (27).
Oct-3/4 consists of the bipartite DNA-binding POU domain that is diagnostic of POU family members and has both amino-terminal (N-TD) and carboxy-terminal (C-TD) transactivation domains (16, 31, 55). The N-TD is classified as a proline-rich transactivation domain. The C-TD is a serine/threonine-rich transactivation domain, though it also has a high proline content. Several reports have provided evidence for a distinction between N-TD and C-TD activity in heterologous transfection assays (2, 6, 7, 16, 55). It remains unclear whether the two transactivation domains have separate specificities that are significant for the physiological functions of Oct-3/4.
In this report, we have analyzed the capacity of variant forms of Oct-3/4 to sustain ES cell identity. Since the study was carried out in the cellular context in which Oct-3/4 normally operates, the results should reflect the relevant physiological contribution of each domain.
MATERIALS AND METHODS
Cell culture and transfection.
All ES cells were cultured in the absence of feeder cells in Glasgow minimal essential medium (GMEM; Sigma, St. Louis, Mo.) supplemented with 10% fetal calf serum (Biowhittaker, Walkersville, Md.), 1 mM sodium pyruvate, 10−4 M 2-mercaptoethanol, 1× nonessential amino acids, and 1,000 U of leukemia inhibitory factor (LIF) per ml on gelatin-coated dishes (48). In the rescue experiments, 107 ZHBTc4 ES cells (29) were electroporated with 50 μg of linearized plasmid DNA at 800 V and 3 μF in a 0.4-cm cuvette using a Gene Pulser (Bio-Rad, Hercules, Calif.), followed by culture with or without 1 μg of tetracycline (Tc) per ml for 2 days. Then these cells were cultured in the presence of 1 μg of puromycin per ml with or without Tc for 7 days. Under this condition, single-copy integration of the vector DNA into the host genome can be achieved in 80% of drug-resistant colonies. The resulting stem cell colonies were scored by staining with Leishman stain or pooled as the stable transfectants for preparing RNA and protein samples and for transfection of reporter constructs.
For the selection of high expressers, transfectants were cultured in the presence of 9 μg of puromycin per ml in the first 3 days. For supertransfection of episomal vectors, 5 × 104 MG 1.19 ES cells (13) were transfected with 2 μg of supercoiled plasmid DNA using Lipofectamine 2000 (Invitrogen, Groningen, The Netherlands), and then replated and cultured in the presence of 1 μg of puromycin per ml for 8 days. This method allows efficient production of stable transfectants, reaching 50% of treated cells, so differentiation events induced by overexpression of transgenes can be easily confirmed by morphology of primary transfectants (28, 29). EB3 is a germ line-competent ES cell line derived from E14tg2a and has one allele of Pou5f1 inactivated by targeted integration of an IRESBSDpA cassette (H. Niwa, unpublished).
BMT-10 cells (14) were cultured in GMEM supplemented with 10% fetal calf serum. For transient transfection, 105 BMT10 cells were transfected with 4 μg of supercoiled plasmid DNA using Lipofectamine 2000.
Construction of expression vectors and reporter plasmids.
The pacbGHpA cassette from pPGKpurobGHpA (54) was amplified by PCR with oligonucleotide primers 5"-AAGCTTATCATGACCGAGTACAAGC-3" and 5"-GAGCCCCTGCAGGTTCTTTCCGCC-3" using Pfu polymerase (Promega, Madison, Wis.). The product was digested with BspHI and PstI and ligated to the PstI-NotI backbone and the NotI-NcoI internal ribosome entry site (IRES) fragments of pCAG-IZ (28) to give pCAG-IP. cDNAs were introduced between the XhoI and NotI or BstXI sites of pCAG-IP.
To construct the mutant Oct-3/4 expression vectors, the full-length cDNA of mouse Oct-3/4 subcloned between the HincII and SmaI sites of pBluescript KS(−) (Stratagene, La Jolla, Calif.), designated pBl-Oct-3/4, was used as a template for PCR amplification using high-fidelity KOD (Toyobo, Osaka, Japan) or Pfx (Invitrogen, Groningen, The Netherlands) DNA polymerase. Wild-type, ΔN, ΔC, ΔNΔC, Δ2-62, and Δ2-62ΔC Oct-3/4 fragments were generated with the appropriate primer pairs listed in Table 1 following introduction of the resulting PCR product into BstXI sites of pCAG-IP with BstXI adapters (5"-GGCCGCACTGGCCACA-3" and 5"-GCCAGTGCGGCC-3").
TABLE 1.
Oligonucleotides used for generating Oct-3/4 mutantsa
| Category and primer | Position | Sequence |
|---|---|---|
| Mouse Oct-3/4 | ||
| wt forward | 39-60 | 5"-CCACCTTCCCCATGGCTGGACA-3" |
| wt reverse | 1101-1120 | 5"-AGGGCTGGTGCCTCAGTTTG-3" |
| ΔN forward | 435-454 | 5"-ACCTTCCCCATGGCCCAGGACATGAAAGCCCTG-3" |
| ΔC reverse | 875-895 | 5"-AGTTAACTTGATCTTTTGCCCTTCTG-3" |
| Δ1-62 forward | 236-254 | 5"-ACCTTCCCCATGGAGTTCTGCGGAGGGATGG-3" |
| Δ1-62 reverse | 217-235 | 5"-AAATGCATGTATGCGGGCGGACATGGG-3" |
| 267V/P forward | 845-871 | 5"-GTGCCCCGGGTATGGTTCTGTAACCGG-3" |
| 267V/P reverse | 830-857 | 5"-ATACCCGGGGCACATCCTTCTCTAGCCC-3" |
| Human Oct-2 | ||
| CTD forward | 5"-ATCCATGGCTTCCATCCCCTCTGTCACTC-3" | |
| CTD reverse | 5"-ATAGTACTAAGGCTGGTAAGGGGCAGG-3" | |
| HA epitope | ||
| HA forward | 5"-TGTACCCATACGACGTGCCAGACTACGC-3" | |
| HA reverse | 5"-TGGCGTAGTCTGGCACGTAGTATGGGTA-3" | |
| EGFP | ||
| EGFP forward | 5"-CCACCATGGTGAGCAAGGGG-3" | |
| EGFP reverse | 5"-AAGGCCTAGGTCTCGCATGCTCTTGTACAGCTCGTCCATGC-3" |
In primers for mouse Oct-3/4, the underlined bases indicate mismatches, and positions are shown based on the reported cDNA sequence (GenBank no. M34381), which contains one base deletion of G at position 934, and the correct sequence results in a length of 1,325 bp.
An in-frame hemagglutinin (HA) epitope was introduced by insertion of double-stranded oligonucleotides shown in Table 1 encoding the HA epitope (YPYDVPDYA) (11) into the NcoI site overlapping the start codon by the partial fill-in method. A point mutation in amino acid 267, valine to proline (267V/P), was introduced into pBl-Oct-3/4 using the pairwise primers in Table 1. To put the Oct-3/4 N-TD or Oct-2 C-TD flanking the C terminus of the Oct-3/4 POU domain, the indicated region was amplified with the appropriate primer pairs listed in Table 1 and plasmid pBl-Oct-3/4 or pOEV1 (24) as a template. To construct the enhanced green fluorescent protein (EGFP) fusion expression vector, pEGFP (Clonetech, Palo Alto, Calif.) was used as a template for PCR with the pairwise primers in Table 1, and the resulting product was introduced into the wild-type, ΔN, or ΔNΔC expression vector.
Luciferase reporter plasmids were constructed by introduction of the regulatory elements listed in Table 2 into pGL3-Basic (Promega, Madison, Wis.).
TABLE 2.
Regulatory elements used for generating luciferase reporter plasmids
| Element | Origin |
|---|---|
| PGK | 507-bp EcoRI-TaqI fragment containing mouse phosphoglycerate kinase (PGK-1) promoter (1) |
| tk | 168-bp BamHI-XhoI fragment derived from pBLCAT2 (19) containing herpes simplex virus thymidine kinase (tk) promoter |
| 6Wtk | 6W enhancer containing 6 copies of oligonucleotides with octamer-binding motif from the mouse immunoglobulin heavy chain gene enhancer (40) upstream of the tk promoter |
| Fgf-4 5" | 460-bp PCR-amplified genomic DNA carrying the Fgf-4 enhancer (57) placed upstream of the tk-luc cassette in plasmid ptk-luc |
| Fgf-4 3" | Fgf-4 enhancer placed downstream of the tk-luc cassette in plasmid ptk-luc |
| 052 | 9.5-kb mouse retrotransposon 052 enhancer/promoter (4) |
| Rex-1 | PCR-amplified 307-bp mouse Zfp42/Rex1 promoter (3) |
Luciferase reporter assay.
For the luciferase reporter assay in ES cells, 2 μg of supercoiled DNA of reporter plasmids was cotransfected into 3 × 104 cells with 0.02 μg of pRL-CMV (Promega, Madison, Wis.) using Lipofectoamine 2000. Two days after transfection, luciferase activities were measured by the dual luciferase assay system (Promega, Madison, Wis.).
RNA analysis.
For Northern blot analysis, 4 μg of total RNA was analyzed by nonradioactive filter hybridization (Gene Image; Amersham Pharmacia, Buckinghamshire, United Kingdom) with Oct-3/4, Sox2, Fgf4, Utf1, Zfp42/Rex1, Upp, Ebaf/Lefty1, Otx1, 226/Tera, and Gap cDNA probes.
EMSA analysis.
Nuclear extracts were prepared from ZHBTc4 ES cells and their derivatives as described by Schreiber et al. (45). Electrophoretic mobility shift assays (EMSAs) were performed with the DIG gel shift kit (Roche, Basel, Switzerland) per the manufacturer's protocol using the digoxigenin (DIG)-labeled double-stranded oligonucleotide containing the typical octamer binding site (ATGCAAAT) attached as a control oligonucleotide in this kit, and with the indodicarbocyanine (Cy5)-labeled double-stranded oligonucleotide with the same sequence as above followed by the detection of fluorescent signal using a Fluorimager (Typhoon 8600; Amersham Pharmacia, Buckinghamshire, United Kingdom).
Immunoblotting.
Ten-microliter aliquots of nuclear extracts or whole-cell lysates with radioimmunoprecipitation assay buffer were fractionated on a sodium dodecyl sulfate SDS-10% polyacrylamide gel and electroblotted onto a polyvinylidene difluoride membrane (Immobilon; Nihon Millipore, Yonezawa, Japan). After treatment in blocking buffer (1× TTBS [10 mM Tris HCl {pH 7.4}, 137 mM NaCl, 2.7 mM KCl, 0.1% Tween 20] plus 3% skimmed milk), membranes were sequentially probed with the anti-HA (11) (clone 12CA5; Roche, Basel, Switzerland), anti-Oct3/4N (47) (provided by H. Hamada), or anti-EGFP (MBL, Nagoya, Japan) and then horseradish peroxidase-coupled anti-mouse immunoglobulin G (IgG) or anti-rabbit IgG (Zymed Laboratories, South San Francisco, Calif.), and developed using ECL reagents (Amersham Pharmacia, Buckinghamshire, United Kingdom).
RESULTS
ES cell-based complementation assay for Oct-3/4 function.
We have previously described how conditional repression of Oct-3/4 in ES cells using the Tc regulatory system (15) induces dedifferentiation into the trophectoderm lineage (29). ZHBTc4 ES cells harbor the Tc-dependent transactivator Oct-3/4 transgene and have had both endogenous alleles of Oct-3/4 inactivated by gene targeting. These cells can be propagated as ES cells in the absence of Tc, in which condition the Oct-3/4 transgene is active, but not in the presence of Tc, which represses the transgene. We reasoned that this system could provide a complementation assay to delineate which elements of Oct-3/4 are required to sustain ES cell identity. The basis of the assay is suppression of dedifferentiation and formation of ES cell colonies in the presence of Tc.
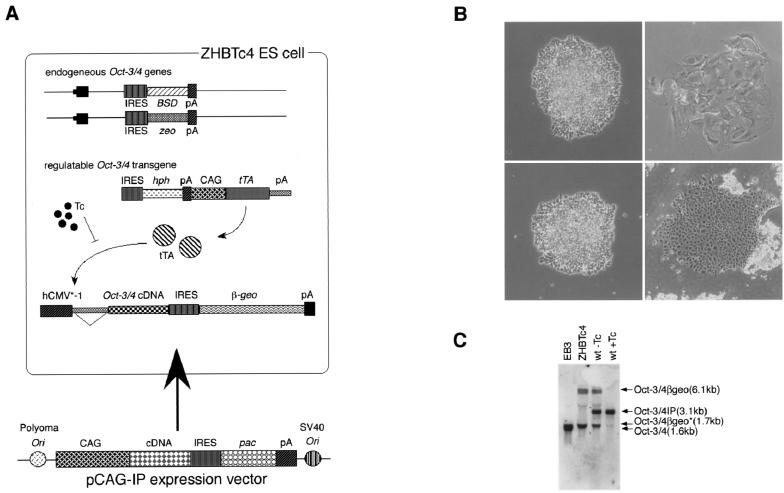
To test this approach, the wild-type Oct-3/4 cDNA under the control of the constitutive CAG expression unit (30) was transfected into ZHBTc4 ES cells (Fig. 1A). ES cell colonies were obtained in the presence of Tc, whereas no stem cell colonies were produced in untransfected controls (without selection) or after transfection of the empty expression vector (Fig. 1B and Table 3). This result indicates that the stem cell phenotype can be rescued by the appropriate level of expression of Oct-3/4 from an introduced transgene. The rescue event was substantiated by Northern hybridization analysis. Expression of the Oct-3/4-IRES-pac transgene was evident, but neither the Tc-regulatable Oct-3/4-IRES-βgeo nor any endogenous Oct-3/4 mRNAs were detected (Fig. 1C).
FIG. 1.
Oct-3/4 complementation assay. (A) Schematic of complementation assay system. ZHBTc4 ES cells (29) contain a Tc-regulated Oct-3/4 transgene and both alleles of endogenous Oct-3/4 have been inactivated by targeted integration of IRESzeopA and IRESBSDpA cassettes. The Oct-3/4 transgene is repressed by addition of Tc, so stem cell colonies can form in the presence of Tc only when an introduced transgene in the pCAG-IP vector can replicate the function of Oct-3/4 and sustain stem cell renewal. Abbreviations: BSD, Aspergillus blasticidin S deaminase; zeo, zeocin resistance gene; IRES, encephalomyocarditis virus internal ribosome entry site; hph, hygromycin phosphotransferase; CAG, CAG expression unit; tTA, Tc-dependent transactivator; β-geo, fusion gene between the Escherichia coli genes for β-galactosidase and neomycin phosphotransferase; pac, puromycin acetyltransferase; Ori, DNA replication origin. (B) Photomicrographs of ZHBTc4 ES cells cultured in the absence of Tc (left upper, undifferentiated), ZHBTc4 cells cultured in the presence of Tc (right upper, differentiated), ZHBTc4 cells transfected with the Oct3/4wt expression vector cultured in the presence of Tc (left lower, undifferentiated), and the rescued ZHBTc4 Oct-3/4wt transfectants transferred to medium lacking Tc (right lower, differentiated). (C) Expression of Oct-3/4 transgenes in ZHBTc4 ES cells transfected with the wild-type (wt) expression vector. ZHBTc4 cells maintained in the absence of Tc express full-length and truncated mRNAs derived from the Oct-3/4βgeo transgene (29) and lack endogenous Oct-3/4 transcripts. Oct-3/4wt transfectants grown with Tc lack transcripts derived from the Oct-3/4βgeo transgene and express the Oct-3/4IP transgene only. Oct-3/4wt transfectants grown without Tc express transcripts derived from both transgenes. EB3 was maintained by the endogenous Oct-3/4.
TABLE 3.
Generation of stem cell colonies of ZHBTc4 ES cells in the presence and absence of Tc by transfection of 4 various expression vectorsa
| Gene | No. of stem cell colonies per 107 treated cells
|
Rescue index (+Tc/−Tc) | |
|---|---|---|---|
| +Tc | −Tc | ||
| Oct-3/4 | 84 | 83 | 1.01 |
| Oct-1 | 0 | 21 | 0 |
| Oct-2 | 0 | 271 | 0 |
| Oct-6 | 0 | 45 | 0 |
| Sox2 | 0 | 138 | 0 |
| None | 0 | 321 | 0 |
107 ZHBTc4 ES cells were transfected with 50 μg of linearlized vector DNAs by electroporation. After 7 days of selection with puromycin in the presence or absence of Tc, the number of stem cell colonies was scored. Rescue index = number of stem cell colonies in the presence of Tc/number of stem cell colonies in the absence of Tc.
The rescued clones are expected to undergo differentiation on induction of the Tc-regulatable Oct-3/4 transgene due to overproduction of Oct-3/4 beyond the upper threshold level. Consistent with this, transfer of rescued clones to culture medium lacking Tc resulted in differentiation (Fig. 1B) in 40% of colonies, in a manner identical to that observed for overexpression of Oct-3/4 in heterozygous ZHTc6 cells (29). Conversely, transfectants isolated in the absence of Tc are anticipated to express only low levels of the transgene. Indeed most of these cells differentiated into trophectodermal phenotypes on addition of Tc (data not shown), indicative of inadequate levels of Oct-3/4.
To investigate the specificity of this rescue assay, constructs for expression of other POU factors were introduced into the ZHBTc4 cells. In contrast to Oct-3/4, expression of Oct-1, Oct-2, or Oct-6 did not produce any stem cell colonies in the presence of Tc (Table 3). Either Oct-1 or Oct-6 overexpression gave fewer stem cell colonies in the absence of Tc, as we observed in the episomal expression system (29) without induction of differentiation, indicating that a moderate level of overexpression of these POU factors still provides some strange toxicity in ES cells. The Sry-related transcription factor Sox2, which is a partner of Oct-3/4 (57) in activation of specific genes such as fibroblast growth factor (Fgf-4) and Utf-1, also cannot substitute for Oct-3/4 function in ZHBTc4 ES cells. These results confirm the specific requirement for Oct-3/4 function to maintain the undifferentiated self-renewing phenotype of ES cells.
DNA-binding ability is essential for maintaining stem cell renewal.
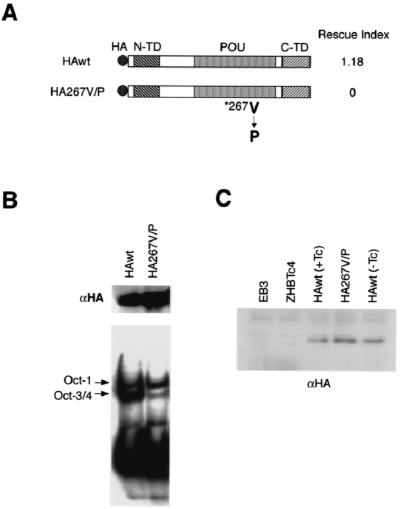
Oct-3/4 contains three functionally characterized domains, the transcriptional activation domains N-TD and C-TD and the POU DNA-binding domain. First, we tested whether DNA binding by Oct-3/4 was required to maintain stem cell renewal. We inactivated Oct-3/4 DNA binding by changing a single amino acid (Val267 to Pro) in the recognition helix of the POU homeodomain (HA267V/P) (55). The mutated protein was efficiently produced in transiently transfected cells and, as expected, showed dramatically reduced binding to the octamer DNA probe (Fig. 2B). When the mutant Oct-3/4 expression vector was introduced into ZHBTc4 ES cells, it could not rescue stem cell renewal in the presence of Tc (Fig. 2A) although stable production of the HA-tagged mutant protein was confirmed by Western blot analysis of transfectants isolated in the presence of Tc (Fig. 2C). These data indicate that the high-affinity DNA-binding ability of Oct-3/4 is essential for its proper function in ES cells.
FIG. 2.
Rescue of stem cell self-renewal by wild-type Oct-3/4 and the 267V/P mutant lacking DNA-binding ability. (A) Diagram of wild-type and mutant gene products. N-TD, C-TD, and the POU domain are represented as boxes. The point mutation in the HA267V/P mutant is positioned in the POU homeodomain. Both cDNAs have an in-frame HA tag upstream of the start codon. Rescue index is scored as in Table 3. (B) Western blot and EMSA analysis of nuclear extracts prepared from BMT10 cells transfected with the HAwt or HA267V/P expression vector. Production of comparable amounts of transgene products was confirmed in each transfectant by immunoblotting with anti-HA antibody (upper panel). EMSA analysis of the same nuclear extracts with the DIG-labeled probe confirmed that HA267V/P binds to octamer motif with much lower efficiency than Oct3/4wt (lower panel). Equal loading of nuclear extracts was confirmed by the intensity of shift bands corresponding to endogenous Oct-1. (C) Western blot analysis of nuclear extracts prepared from the rescued and unrescued ES cells transfected with HAwt or HA267V/P. Similar amounts of transgene product with HA tag were detected by anti-HA antibody.
Functional assay of N- and C-terminal regions.
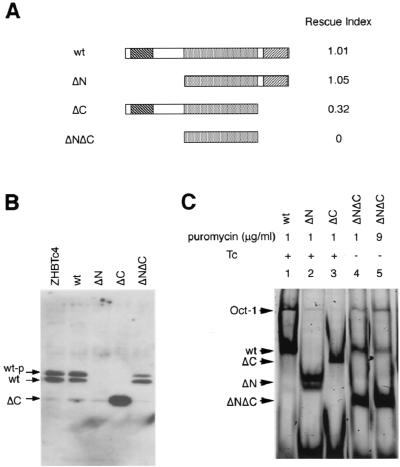
As the next step, Oct-3/4 deletion mutants lacking either N-terminal amino acids (aa) 3 to 129 (ΔN), C-terminal aa 283 to 352 (ΔC), or both (ΔNΔC) were generated, and their ability to rescue stem cell renewal was assayed. Transfection with the ΔNΔC (POU domain only) mutant resulted in differentiated cells and no stem cell colonies. However, both ΔN and ΔC mutants yielded high numbers of undifferentiated ES cell colonies (Fig. 3A). The ES cell identity of rescued cells was established by the following criteria: (i) characteristic colony morphology, (ii) contact-independent proliferation, (iii) zeocin resistance, reflecting activity of the endogenous Oct-3/4 promoter carrying a knock-in of zeo, (iv) expression of the Oct-3/4-independent stem cell marker gene Sox2, and (v) differentiation on withdrawal of LIF. The latter assay demonstrates that self-renewal remains cytokine dependent and that the cells retain the capacity for differentiation.
FIG. 3.
Rescue of stem cell renewal by deletion mutants. (A) Diagram of wild-type and mutant gene products. ΔN and ΔC lack aa 3 to 129 and 283 to 352, respectively, and ΔNΔC consists of aa 130 to 282. The rescue index was determined as in Table 3. (B) Western blot analysis of nuclear extracts prepared from rescued transfectants with wild-type, ΔN, and ΔC grown in the presence of Tc and unrescued transfectants with ΔNΔC grown in the absence of Tc with anti-Oct3/4N antibody. The rescued cells with ΔN or ΔC lack wild-type Oct-3/4 protein. Mutant protein of ΔC but not ΔN and ΔNΔC can be detected by anti-Oct3/4N antibody. Two different sizes of bands are occasionally detected with anti-Oct3/4N antibody. The upper band appears to represent the phosphorylated form, because this signal can be intensified by preparation of nuclear extracts in the presence of phosphatase inhibitors (PI). We always detected a single ΔC band, because the phosphorylation sites of Oct-3/4 are located in the C terminus (6, 35). (C) EMSA analysis of nuclear extracts analyzed in panel B with the Cy5-labeled probe. All mutants can bind to the octamer motif with comparable intensity to the wild-type Oct-3/4. The signal for ΔNΔC is weaker in lane 4, but it expressed an amount comparable to the others in unrescued cells selected with 9 μg of puromycin per ml in lane 5.
This finding implied that, in combination with the POU domain, either the N or the C terminus is sufficient to maintain stem cell phenotype. The absence of wild-type Oct-3/4 protein in the transfectants grown in the presence of Tc was confirmed by immunoblotting with polyclonal anti-Oct3/4N antibody, recognizing the N terminus of Oct-3/4 (47) (Fig. 3B). EMSAs revealed that the ΔNΔC mutant protein is present in unrescued ZHBTc4 ES cells maintained in the absence of Tc and that only the mutant proteins were present in the ΔN- and ΔC-rescued ES cells (Fig. 3C).
The reduced level of expression of the ΔNΔC mutant protein selected in normal conditions might be due to preparation of nuclear lysate from unrescued ES cells (lane 4 in Fig. 3C), in which there is no selection pressure in the expression level of transgene to rescue ES self-renewal. Indeed, higher amounts of the ΔNΔC protein can be observed in nonrescued cells selected with a high dose of puromycin (lane 5 in Fig. 3C). These data suggest that the two separated transactivation domains have equivalent functionality in ES self-renewal. This is somewhat surprising given that they represent different classes of transactivation domain (6), each with a relatively high degree of conservation of characteristic amino acid composition, and that assays in heterologous cells have pointed to distinctions in activity (see Discussion).
POU domain alone is not sufficient for maintaining stem cell renewal.
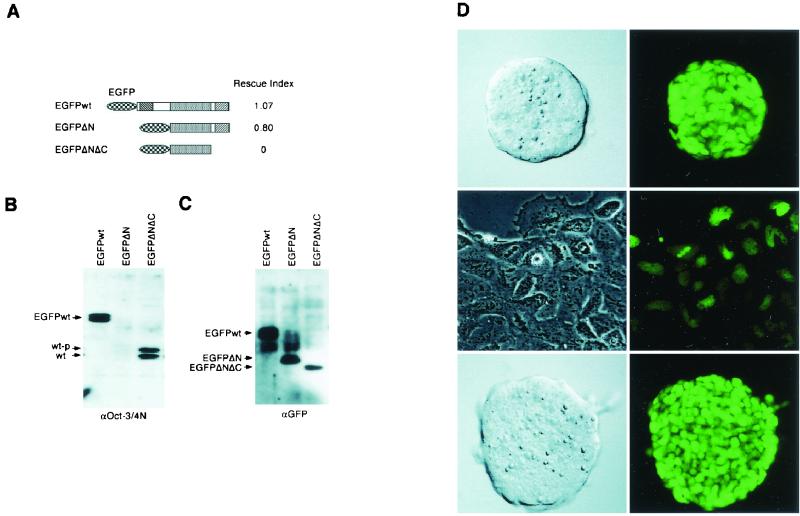
The previous experiments suggested that the Oct-3/4 POU domain alone is necessary but not sufficient to support stem cell renewal. However, since the expression level of Oct-3/4 is critical for maintaining stem cell phenotype (29), it is necessary to examine nuclear protein in order to distinguish between a true qualitative difference and a quantitative effect arising from inappropriate intracellular localization or instability. For this purpose, we generated constructs in which the EGFP coding sequence is fused in-frame to Oct-3/4 sequences.
N-terminal GFP fusions with full-length Oct-3/4 (EGFPwt) or with ΔN Oct-3/4 (EGFPΔN) could rescue stem cell renewal efficiently when introduced into ZHBTc4 cells (Fig. 4A). Immunoblotting confirmed that these fusion proteins maintain stem cell renewal in the absence of endogenous Oct-3/4 (Fig. 4B). However, an EGFP fusion with ΔNΔC (EGFPΔNΔC) did not generate any rescued stem cell colonies in the presence of Tc (Fig. 4A), although a significant amount of the EGFPΔNΔC mutant protein was expressed in transfectants (Fig. 4C). The signal of EGFPΔNΔC might be weaker than the others because the nuclear extract was prepared from unrescued ES cells, as mentioned above. By fluorescence microscopy, the EGFP signals can be detected at similar intensity in the nuclei of cells transfected with each of the EGFP fusion proteins (Fig. 4D).
FIG. 4.
Rescue of stem cell renewal by EGFP fusion constructs. (A) Diagram of EGFP fusion constructs with wild-type and mutant gene products. The rescue index was determined as in Table 3. (B) Western blot analysis of nuclear extracts prepared from rescued transfectants with EGFPwt and EGFPΔN grown in the presence of Tc and unrescued transfectants with EGFPΔNΔC grown in the absence of Tc. Anti-Oct3/4N antibody detects wild-type Oct-3/4 in the latter but not the former, confirming the rescue events in EGFPwt and EGFPΔN transfectants. (C) Western blot analysis of the blot used in panel B with anti-GFP antibody. Proper production of EGFP fusion proteins was confirmed. (D) Photomicrographs of phase-contrast and fluorescent views of a stem cell colony rescued by the expression of EGFPwt (top) and of nucleus-localized EGFPΔNΔC in a stem cell colony grown in the absence of Tc (bottom). Nuclear fluorescence from EGFPwt is retained in differentiated cells produced after withdrawal of LIF (middle).
We previously reported that maintenance of Oct-3/4 expression cannot prevent differentiation induced by withdrawal of LIF (29). This finding was confirmed by removal of LIF from ES cells rescued by the expression of EGFPwt. These cells readily differentiated while retaining Oct-3/4 activity, as revealed by EGFP fluorescence (Fig. 4D).
The subcellular localization of the EGFP fusions indicates that the nuclear localization signal of Oct-3/4 is present in the POU domain, as previously reported for Oct-6 (50). However, the proper production and nuclear localization of EGFPΔNΔC fusion protein confirm that the POU binding domain alone lacks an essential function to maintain stem cell renewal.
Transactivation domain is required for stem cell renewal of ES cells.
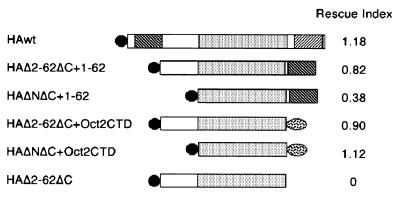
Deletion of the entire N- or C-terminal sequence suggested that the transactivation domains in these regions are necessary for stem cell renewal. To test the requirement for transactivation domains more precisely, we made various mutant forms of Oct-3/4 and examined their function in our rescue system. A deletion of aa 2 to 62 of the minimal proline-rich N-TD (aa 13 to 60) (16) in combination with the ΔC truncation abolished the ability to rescue stem cell renewal (HAΔ2-62ΔC in Fig. 5). This confirms the requirement specifically for a transactivation domain.
FIG. 5.
Rescue of stem cell renewal by combination of the POU domain with transactivation domains. Diagram of mutant gene products and the rescue index for them are shown. The rescue index was determined as in Table 3.
Interestingly, when aa 1 to 62 containing the N-TD was transferred to the C terminus of the Δ2-62ΔC mutant, the resulting Oct-3/4 (HAΔ2-62ΔC+1-62) rescued stem cell renewal. Therefore, the orientation of the transactivation domain relative to the POU domain is not critical. Moreover, addition of these residues to the C terminus of the POU domain only also yielded a version of Oct-3/4 (HAΔNΔC+1-62) competent to support the propagation of ES cells (Fig. 5). These data show that combination of the minimal N-TD with the POU domain is sufficient to maintain stem cell self-renewal in vitro.
Both N-TD and C-TD are proline-rich, although the latter is also serine/threonine-rich. The similarity in amino acid composition might be the basis for their interchangeable role in sustaining stem cell renewal. To test this idea, we assayed the function of a heterologous transactivation domain. Oct-2 cannot substitute for Oct-3/4 function in ES cells (Table 1). However, the transactivation domain at the C terminus of Oct-2 (Oct2CTD) is a well-characterized proline-rich transactivation domain (52). We combined the Oct2CTD with the Oct-3/4 POU domain and introduced the resultant fusion (HAΔ2-62ΔC+Oct2CTD or HAΔNΔC+Oct2CTD) into ZHBTc4 ES cells. In contrast to full-length Oct-2, the chimeric molecule could sustain the undifferentiated state (Fig. 5). This finding indicates that the transactivation domains in Oct-3/4 do not have any unique character in their function and can be replaced with other transactivation domains.
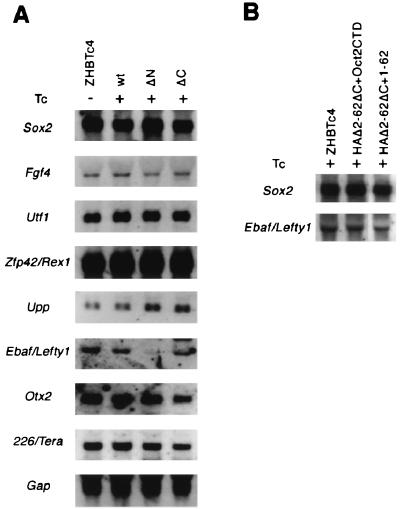
Gene expression in ES cells rescued by mutants.
Pools of colonies rescued by transfection of wild-type, ΔN, or ΔC Oct-3/4 expression vectors were expanded in the presence of Tc for further characterization. To interrogate the function of the mutant Oct-3/4 proteins in these rescued ES cells, transcription of seven Oct-3/4 target genes was examined by Northern blot analysis (Fig. 6A). All seven genes are downregulated in Tc-treated ZHBTc4 ES cells with kinetics parallel to the disappearance of Oct-3/4 transcripts (29) (data not shown on Utf-1 and 226/Tera). Expression of six of the seven was restored in the rescued ES cells. Expression of one gene, Ebaf/lefty1, however, was appreciable in cells rescued by full-length Oct-3/4 and ΔC, but was barely detectable in ES cells rescued by ΔN. This result was consistent with three different RNA preparations. These data suggest either that N-TD possesses specific activity to direct expression of Ebaf/Lefty1 or that C-TD specifically lacks this ability.
FIG. 6.
Comparison of gene expression pattern in rescued ES cells. (A) Expression of Oct-3/4 target genes in ZHBTc4 ES cells and rescued ES cells with wild-type, ΔN, or ΔC. Total RNAs prepared from these ES cells were blotted on nylon membranes, and replicate filters were analyzed by nonradioactive filter hybridization with the indicated cDNA probes. Gap was used as a loading control. (B) Expression of Ebaf/Lefty1 in ES cells rescued by Δ2-62ΔC+Oct2CTD and Δ2-62ΔC+1-62 mutant. Sox2 was used as a loading control.
To distinguish these two possibilities, we next assayed Ebaf/Lefty1 expression in ES cells rescued by other mutants. Normal levels of Ebaf/Lefty1 expression were detected in ES cells rescued by HAΔ2-62ΔC+1-62 and HAΔ2-62ΔC+Oct2CTD. The finding that Oct2CTD can substitute for N-TD and activate Ebaf/Lefty1 expression implies that the Oct-3/4 C-TD is specifically deficient in this capacity.
Comparative analysis of transcriptional activation by mutant Oct-3/4 proteins.
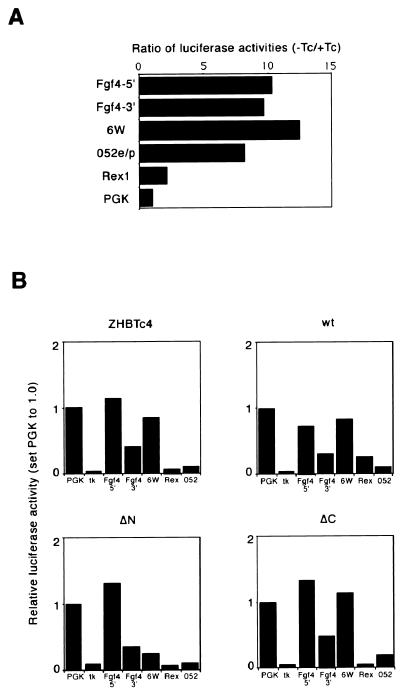
To characterize further the difference between N-TD and C-TD in activating particular target genes, we employed luciferase reporter assays. Five constructs containing transcriptional regulatory elements activated by Oct-3/4 in ES cells were prepared (Table 3). They are known to be activated in different manners by Oct-3/4 and cofactors, such as the Fgf-4 enhancer activated by Oct-3/4 and Sox2 (57), the 052 promoter and enhancer activated by Oct-3/4 and adenovirus E1A (4, 47), the Rex1 promoter activated by Oct-3/4 and Rox1 (3), and 6Wtk activated by Oct-3/4 alone (41). For all reporter plasmids, downregulation of the luciferase activity on repression of Oct-3/4 was confirmed by measuring luciferase activities in ZHBTc4 ES cells with or without Tc (Fig. 7A). When transfected into rescued ES cells, each of these reporters was activated by different Oct-3/4 variants to a similar extent, except for the 6W construct (Fig. 7B). 6W luciferase activity in the presence of ΔC was comparable to that induced by full-length Oct-3/4, and in both cases significantly greater than that in the presence of ΔN. N-TD is therefore required for full transactivation in this instance. These data also suggested that there is no cooperativity between N-TD and C-TD, because reporter activities were comparable to those observed in wild-type ES cells or ZHBTc4 cells rescued with Oct-3/4wt (Fig. 7B).
FIG. 7.
Transactivation ability of mutant proteins. (A) Oct-3/4-dependent activation of reporter constructs. Luciferase reporter plasmids carrying various regulatory elements were transfected into ZHBTc4 ES cells in the presence or absence of Tc. The data were calibrated by both pRL-CMV and PGK-luc, and the ratio for PGK was set at 1.0. Fgf4-5", Fgf4-3", 6W, 052, and Rex1 showed severalfold-higher ratios, confirming their Oct-3/4-dependent activation in ES cells. (B) Expression of luciferase reporters in rescued ES cells. Reporter plasmids tested in A were transfected into ES cells rescued by wild-type, ΔN, or ΔC, and their activities were estimated. The data were calibrated by both pRL-CMV and PGK-luc, and the ratio for PGK was set at 1.0 as above. Note the lower magnitude of activation of 6W in ΔN than in the others.
Induction of differentiation by overexpression of mutant Oct-3/4 proteins.
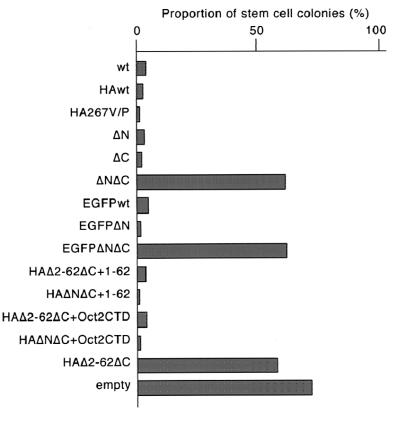
We previously showed that upregulation of Oct-3/4 in ES cells provokes endodermal and mesodermal differentiation. To investigate which part of Oct-3/4 is responsible for this phenomenon, expression vectors carrying mutant Oct3/4 transgenes were introduced into ES cells by episomal supertransfection (13, 28). Supertransfection of full-length Oct-3/4 produced many differentiated colonies to endoderm-like cells as previously. All mutant expression vectors that rescued stem cell renewal similarly generated differentiated colonies (Fig. 8). In contrast, transfection with inactive ΔNΔC, EGFPΔNΔC, and HAΔ2-62ΔC constructs produced stem cell colonies. Thus, the minimal combination of POU domain plus transactivation domain is sufficient and necessary to induce differentiation when overexpressed. Unexpectedly, however, expression of the DNA-binding mutant HA267V/P blocked self-renewal and generated differentiated colonies. Thus, Oct-3/4 DNA-binding ability, which is required to sustain self-renewal, is not essential to induce differentiation upon elevated expression.
FIG. 8.
Episomal expression of various mutants of Oct-3/4 in MG1.19 ES cells. The bar chart shows the proportion of undifferentiated colonies obtained after supertransfection of episomal vectors for expression of the indicated mutants and selection for 8 days in puromycin. Supertransfection of Oct-3/4wt and most mutants gave predominantly differentiated colonies. Mutants lacking both transactivation domains failed to induce significant differentiation.
DISCUSSION
The mammalian fetus is generated from a pluripotent founder population, the epiblast that develops in the blastocyst. This pluripotent stage of development can be continuously propagated in culture in the form of mouse ES cells (9, 20). The mechanisms by which pluripotency is established and sustained remain obscure (33). However, it appears that the POU factor Oct-3/4 plays a central role in both processes (27). Oct-3/4 activity is essential for the initial formation of pluripotent cells in the mouse blastocyst (25) and for their maintenance in culture as ES cells (29). The present study was aimed at clarifying the contribution of different functional domains of Oct-3/4 protein to these effects. The findings demonstrate that the POU domain of Oct-3/4 is uniquely required and, in combination with a generic proline-rich transactivation domain, is sufficient to maintain the undifferentiated stem cell phenotype.
Oct-3/4 possesses two separate transactivation domains that have been reported to show distinct activities in different assay systems. Imagawa et al. reported that they detected transactivation ability only in the N terminus by an assay in which the N and C termini of Oct-3/4 were fused to the c-Jun DNA-binding domain and activity was tested in P19 EC cells using a reporter construct with multiple c-Jun-binding sites (16). In contrast, Vigano' and Staudt identified both N-TD and C-TD activities by transactivation of a reporter plasmid containing six copies of the octamer motif upstream of the β-globin promoter in HeLa cells. They found stronger activity from C-TD than N-TD and also that an Oct-3/4 mutant unable to bind the octamer target could activate transcription in an octamer-dependent manner (55).
However, these heterologous assay systems may lead to misreading of native function. Thus, Brehm et al. revealed that the activity of C-TD is modulated in a cell type-dependent manner (6). Oct-3/4 can directly activate transcription of target genes when the cis binding site is positioned proximal to the promoter (41). More commonly, however, Oct-3/4 appears to bind to distal elements, in which case gene activation (or repression) requires cooperation with other factors. Adenovirus E1A can serve as a bridging factor between Oct-3/4 and the basal transcription machinery and this function appears to be reproduced by a cellular factor(s) in pluripotent cells (41).
Several nuclear proteins seem to interact with Oct-3/4, including high-mobility group and bromodomain proteins (8), and the Sry-related transcription factor Sox2. The last participates with Oct-3/4 in cooperative enhancer binding and synergistic transcriptional activation of the Fgf-4 (57) and Utf-1 genes (26). In reporter assays, the Oct-3/4 POU domain plus N-TD is sufficient to stimulate transcription from the Fgf-4 enhancer in the absence of Sox2. The C-TD, in contrast, is latent in this assay and becomes active only by cooperation with Sox2, resulting in the synergistic enhancement of transcription (2). Therefore, the activities of the two transactivation domains can be differentially affected by protein-protein interaction.
In this study we have found that in the relevant physiological context of ES cells, N-TD and C-TD function redundantly to maintain self-renewal. We found that the presence of either N-TD or C-TD was sufficient, provided the Oct-3/4 POU domain was intact. The requirement for a transactivation domain demonstrates that the critical function of Oct-3/4 does involve target gene activation and cannot be attributed entirely to gene repression. The data indicate, however, that the two Oct-3/4 transactivation domains have overlapping specificity and that neither makes a unique contribution to ES cell propagation.
Yet the activity of these two transactivation domains is not identical in ES cells. Analysis of expression of Oct-3/4 target genes revealed that Ebaf/Lefty1 expression is significantly reduced in ES cells rescued by the expression of the ΔN mutant. This mutant also failed to activate the 6Wtkluc reporter, which can be activated by Oct-3/4 alone in HeLa cells (41). This suggests that the N-TD has a specific function to transactivate a suite of target genes independently of interaction with partners such as Sox2 (57) or cellular E1A-like factors (41). However, it might be better to regard this phenomenon as mediated by a specific defect of the C-TD to transactivate a particular type of target promoter, because the Oct-2 C-TD possesses a function similar to that of the N-TD. The data also imply that expression of a subset of Oct-3/4 target genes represented by Ebaf/Lefty-1 is not necessary for self-renewal of ES cells in vitro.
We recently identified the Oct-3/4-dependent enhancer element in the upstream region of the Ebaf/Lefty-1 promoter (N. Fukui and H. Niwa, unpublished data), confirming that this gene is one of the direct targets of Oct-3/4. It should be noted, however, that an ES cell in culture is not experiencing the same environment as ICM or epiblast cells in the embryo (49). Thus, it is quite possible that N-TD-specific targets that appear redundant in ES cells might function in vivo at earlier or later stages of development to induce or maintain stem cell populations or to direct lineage commitment. Ebaf/Lefty1 encodes a transforming growth factor beta superfamily protein (38) that antagonizes the function of activin and nodal and acts as a midline barrier to establish left-right asymmetry (21, 22). Conceivably, it might act in stem cells to protect them from the effect of differentiation inducers during development. Therefore, this class of Oct-3/4 target genes might be particularly important for ordered differentiation of the epiblast and/or for maintaining the identity of primordial germ cells during gastrulation.
The capacity for integration into the developing embryo and full developmental repertoire have not been examined in this study. Unfortunately, the ZHBTc4 system does not lend itself to such analyses because expression of the variant forms of Oct-3/4 is under the control of the constitutively active CAG expression unit and would show unregulated expression after blastocyst injection. Generation of mice carrying the ΔN or ΔC deletion introduced by gene targeting is therefore under way to enable the function of N-TD-dependent target genes to be determined in vivo.
The rescue effect of the ΔN mutant was weaker than that of the wild type and ΔC mutant. It might be partly due to the lack of N-TD-dependent gene expression. In addition, it was recently reported that Ets-2-induced transactivation of the tau interferon promoter is repressed by Oct-3/4 (10). In this case, full repression required both the N-terminal and POU domains, so the ΔN mutant might have a weaker ability to repress such trophectoderm-specific genes. However, the repressor function should not depend completely on the N terminus because ΔN can rescue ES self-renewal and we could not detect any ectopic expression of trophectoderm-specific genes such as Hand-1 and Cdx-2 (29) in the rescued ES cells (data not shown). It is also possible that the difference between ΔN and ΔC is based on the slightly different amino acid composition of N-TD and C-TD. We need to investigate whether various types of transactivation domains except the proline-rich type can substitute for the function of transactivation domains in Oct-3/4 in the future.
Overproduction of Oct-3/4 induces differentiation in ES cells and possibly also in the nascent hypoblast in vivo (32). We have proposed that this effect may arise by repression of a subset of target genes due to sequestration of a particular cofactor(s) (27, 29). This model is dependent on protein-protein interactions. In the present study, we found that overexpression of the mutant HA267V/P could induce differentiation even though this amino acid substitution abolishes the DNA-binding ability of the POU domain. In contrast, overproduction of the POU domain alone had no effect on stem cell renewal, although stable production and DNA-binding ability were confirmed. A fusion of the N and C termini of Oct-3/4 with the Oct-2 POU domain also did not induce differentiation when overexpressed (data not shown). These data suggest that the titration out of a relevant partner(s) (41) is achieved by protein interactions that are independent of DNA binding and are mediated by both the POU domain and a proline-rich transactivation domain and that activation of a particular type of target genes which are repressed in this mechanism should be important to maintain ES self-renewal (27).
Oct-3/4 uniquely can maintain ES cell self-renewal. Other POU factors, Oct-1, Oct-2, and Oct-6, that have common DNA-binding site specificity with Oct-3/4 (37), cannot substitute for it. The data indicate that the presence of a transactivation domain is essential but not specific. Therefore, the singular capacity of Oct-3/4 should be conferred by the POU domain. Consistent with this, the POU domain is the most highly conserved region of Oct-3/4 (12). The specific attributes of the Oct-3/4 POU domain should reside in part at the level of target sequence selectivity. POU domains consist of a POU homeodomain (POU-H) and a POU-specific domain (POU-S) separated by a linker sequence (51). The linker facilitates the adoption of different conformations on DNA binding and thereby allows interaction with a range of target sequences (5).
In addition to DNA binding, however, both subdomains of the POU domain can engage in selective interactions with different types of proteins. For example, the POU domain of Oct-1 but not Oct-2 selectively recruits herpes simplex virus transactivator VP16, and the Oct-1 POU-H is critical for this specificity (17). Coactivator Bob-1/OCA-B/OBF-1 makes selective contacts with both POU-H and POU-S of Oct-1 and Oct-2 but does not interact with the POU domains of Oct-3/4, Oct-6, or Pit-1 (39). Oct-3/4 interactions with adenovirus E1A, Sox2, and Ets-2 involve the POU domain (10, 41, 57), and Ambrosetti et al. showed that POU-H plays a critical role in the formation of a functional Sox2-Oct-3/4 complex (2).
Finally, the POU domain mediates Oct-3/4 dimerization, which impacts both transcriptional activation and cooperativity with cofactors (5, 53). The conformation adopted by the POU domain on binding to a particular DNA sequence leads to dimerization and/or selective cofactor recruitment, which in turn may stabilize DNA binding (53). Thus, the emerging picture of the Oct-3/4 POU domain is as a highly plastic transcriptional organizer. Flexible reciprocal interactions with target DNA sequences and protein cofactors result in assembly of gene-specific transcription complexes (3, 5, 26, 57).
Functional analysis of chimeric and mutant POU domains in the ZHBTc4 complementation system should allow further refinement of the molecular features that underlie the maintenance and propagation of pluripotency and may ultimately pinpoint a minimal set of key target genes.
Acknowledgments
We are grateful to Hiroshi Hamada for the 052CAT plasmid, probes for Oct-3/4 target genes, and anti-Oct3/4N antibody, Hans Schöler for the 6W and hOct2A constructs, and Akihiko Okuda for Utf1 cDNA.
This work was supported by the Ministry of Education, Science and Culture of Japan (H.N.) and the Biotechnology and Biological Sciences Research Council of the United Kingdom (I.C. and A.G.S.).
REFERENCES
- 1.Adra, C. N., P. H. Boer, and M. W. McBurney. 1987. Cloning and expression of the mouse pgk-1 gene and the nucleotide sequence of its promoter. Gene 60:65-74. [DOI] [PubMed] [Google Scholar]
- 2.Ambrosetti, D.-C., H. R. Schöler, L. Dailey, and C. Basilico. 2000. Modulation of the activity of multiple transcriptional activation domains by the DNA binding domains mediates the synergistic action of Sox2 and Oct-3 on the fibroblast growth factor-4 enhancer. J. Biol. Chem. 275:23387-23397. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Shushan, E., J. R. Thompson, L. J. Gudas, and Y. Bergman. 1998. Rex-1, a gene encoding a transcription factor expressed in the early embryo, is regulated via Oct-3/4 and Oct-6 binding to an octamer site and a novel protein, Rox-1, binding to an adjacent site. Mol. Cell. Biol. 18:1866-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhat, K., M. W. McBurney, and H. Hamada. 1988. Functional cloning of mouse chromosomal loci specifically active in embryonal carcinoma stem cells. Mol. Cell. Biol. 8:3251-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Botquin, V., H. Hess, G. Fuhrman, C. Anastassiadis, M. K. Gross, G. Vriend, and H. R. Schöler. 1998. New POU dimer configuration mediates antagonistic control of an osteopontin preimplantation enhancer by Oct-4 and Sox-2. Genes Dev. 12:2073-2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brehm, A., K. Ohbo, and H. Schöler. 1997. The carboxy-terminal transactivation domain of Oct-4 acquires cell specificity through the POU domain. Mol. Cell. Biol. 17:154-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brehm, A., K. Ohbo, W. Zwerschke, V. Botquin, P. Jansen-Dürr, and H. R. Schöler. 1999. Synergism with germ line transcription factor Oct-4: viral oncoproteins share the ability to mimic a stem cell-specific activity. Mol. Cell. Biol. 19:2635-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butteroni, C., M. De Felici, H. R. Schöler, and M. Pesce. 2000. Phage display screening reveals an association between germline-specific transcription factor Oct-4 and multiple cellular proteins. J. Mol. Biol. 304:529-540. [DOI] [PubMed] [Google Scholar]
- 9.Evans, M. J., and M. H. Kaufman. 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292:154-156. [DOI] [PubMed] [Google Scholar]
- 10.Ezashi, T., D. Ghosh, and M. Roberts. 2001. Repression of Ets-2-induced transactivation of the tau interferon promoter by Oct-4. Mol. Cell. Biol. 21:7883-7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Field, J., J. Nikawa, D. Broek, B. MacDonald, L. Rodgers, I. A. Wilson, R. A. Lerner, and M. Wigler. 1988. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol. Cell. Biol. 8:2159-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frankenberg, S., D. Tisdall, and L. Selwood. 2001. Identification of a homologue of POU5F1 (OCT3/4) in a marsupial, the brushtail possum. Mol. Reprod. Dev. 58:255-261. [DOI] [PubMed] [Google Scholar]
- 13.Gassmann, M., G. Donoho, and P. Berg. 1995. Maintenance of an extrachromosomal plasmid vector in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 92:1292-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerard, R. D., and Y. Gluzman. 1985. New host cell system for regulated simian virus 40 DNA replication. Mol. Cell. Biol. 5:3231-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imagawa, M., A. Miyamoto, M. Shirakawa, H. Hamada, and M. Muramatsu. 1991. Stringent integrity requirements for both trans-activation and DNA-binding in a trans-activator, Oct3. Nucleic Acids Res. 19:4503-4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai, J.-S., M. A. Cleary, and W. Herr. 1992. A single amino acid exchange transfers VP16-induced positive control from the Oct-1 to the Oct-2 homeodomain. Genes Dev. 6:2058-2065. [DOI] [PubMed] [Google Scholar]
- 18.Lenardo, M. J., L. Staudt, P. Robbins, A. Kuang, R. C. Mulligan, and D. Baltimore. 1989. Repression of the IgH enhancer in teratocarcinoma cells associated with a novel octamer factor. Science 243:544-546. [DOI] [PubMed] [Google Scholar]
- 19.Luckow, B., and G. Schultz. 1987. CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res. 15:5490.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin, G. R. 1981. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 78:7634-7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meno, C., K. Gritsman, S. Ohishi, Y. Ohfuji, E. Heckscher, K. Mochida, A. Shimono, H. Kondoh, W. S. Talbot, R. J. Robertson, A. F. Schier, and H. Hamada. 1999. Mouse Lefty2 and zebrafish antivin are feedback inhibitors of nodal signaling during vertebrate gastrulation. Mol. Cell 4:287-298. [DOI] [PubMed] [Google Scholar]
- 22.Meno, C., A. Shimono, Y. Saijoh, K. Yashiro, K. Mochida, S. Ohishi, S. Noji, H. Kondoh, and H. Hamada. 1998. Lefty-1 is required for left-right determination as a regulator of lefty-2 and nodal. Cell 94:287-297. [DOI] [PubMed] [Google Scholar]
- 23.Mitchell, P. J., and R. Tjian. 1989. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science 245:371-378. [DOI] [PubMed] [Google Scholar]
- 24.Müller, M. M., S. Ruppert, W. Schaffner, and P. Matthias. 1988. A cloned octamer transcription factor stimulates transcription from lymphoid-specific promoter in non-B cells. Nature 336:544-551. [DOI] [PubMed] [Google Scholar]
- 25.Nichols, J., B. Zevnik, K. Anastassiadis, H. Niwa, D. Klewe-Nebenius, I. Chambers, H. Schöler, and A. Smith. 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95:379-391. [DOI] [PubMed]
- 26.Nishimoto, M., A. Fukushima, A. Okuda, and M. Muramatsu. 1999. The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol. Cell. Biol. 19:5453-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niwa, H. 2001. Molecular mechanism to maintain stem cell renewal of ES cells. Cell Struct. Funct. 26:137-148. [DOI] [PubMed] [Google Scholar]
- 28.Niwa, H., T. Burdon, I. Chambers, and A. Smith. 1998. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 12:2048-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niwa, H., J.-I. Miyazaki, and A. G. Smith. 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24:372-376. [DOI] [PubMed] [Google Scholar]
- 30.Niwa, H., K.-I. Yamamura, and J.-I. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-200. [DOI] [PubMed] [Google Scholar]
- 31.Okamoto, K., H. Okazawa, A. Okuda, M. Sakai, M. Muramatu, and H. Hamada. 1990. A novel octamer binding transcriptional factor is differentially expressed in mouse embryonic cells. Cell 60:461-472. [DOI] [PubMed] [Google Scholar]
- 32.Palmieri, S. L., W. Peter, H. Hess, and H. R. Schöler. 1994. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineage involved in implantation. Dev. Biol. 166:259-267. [DOI] [PubMed] [Google Scholar]
- 33.Pesce, M., and H. R. Schöler. 2000. Oct-4: control of totipotency and germline determination. Mol. Reprod. Dev. 55:452-457. [DOI] [PubMed] [Google Scholar]
- 34.Ptashne, M. 1988. How eukaryotic transcriptional activators work. Nature 335:683-689. [DOI] [PubMed] [Google Scholar]
- 35.Rosfjord, E., B. Scoltz, R. Lewis, and A. Rizzino. 1995. Phosphorylation and DNA-binding of the octamer binding transcription factor Oct-3. Biochem. Biophys. Res. Commun. 212:847-853. [DOI] [PubMed] [Google Scholar]
- 36.Rosner, M. H., M. A. Vigano, K. Ozato, P. M. Timmons, F. Poirier, P. W. J. Rigby, and L. M. Staudt. 1990. A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature 345:686-692. [DOI] [PubMed] [Google Scholar]
- 37.Ryan, A. K., and M. G. Rosenfeld. 1997. POU domain family values: flexibility, partnerships, and developmental codes. Genes Dev. 11:1207-1225. [DOI] [PubMed] [Google Scholar]
- 38.Saijoh, Y., H. Fukii, C. Meno, M. Sato, Y. Hirota, S. Nagamatsu, M. Ikeda, and H. Hamada. 1996. Identification of putative downstream genes of Oct-3, a pluripotent cell-specific transcription factor. Genes Cells 1:239-252. [DOI] [PubMed] [Google Scholar]
- 39.Sauter, P., and P. Matthias. 1998. Coactivator OBF-1 makes selective contacts with both the POU-specific domain and the POU homeodomain and acts as a molecular clamp on DNA. Mol. Cell. Biol. 18:7397-7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schöler, H. R., R. Balling, A. K. Hatzopoulos, N. Suzuki, and P. Gruss. 1989. Octamer binding proteins confer transcriptional activity in early mouse embryogenesis. EMBO J. 8:2551-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schöler, H. R., T. Ciesiolka, and P. Gruss. 1991. A nexus between Oct-4 and E1A: implications for gene regulation in embryonic stem cells. Cell 66:291-304. [DOI] [PubMed] [Google Scholar]
- 42.Schöler, H. R., G. R. Dressler, R. Baling, H. Rohdewohld, and P. Gruss. 1990. Oct-4: a germline-specific transcription factor mapping to the mouse t-complex. EMBO J. 9:2185-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schöler, H. R., A. K. Hatzopoulos, R. Balling, N. Suzuki, and P. Gruss. 1989. A family of octamer-specific proteins present during mouse embryogenesis: evidence for germline-specific expression of an Oct factor. EMBO J. 8:2543-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schöler, H. R., S. Ruppert, N. Suzuki, K. Chowdhury, and P. Gruss. 1990. New type of POU domain in germ line-specific protein Oct-4. Nature 344:435-439. [DOI] [PubMed] [Google Scholar]
- 45.Schreiber, E., P. Matthias, M. M. Müller, and W. Schaffner. 1989. Rapid detection of octamer binding protein with "mini-extracts', prepared from a small number of cells. Nucleic Acids Res. 17:6419.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seipel, K., O. Georgiev, and W. Schaffner. 1992. Different activation domains stimulate transcription from remote ("enhancer') and proximal ("promoter') positions. EMBO J. 11:4961-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shimazaki, T., H. Okazawa, H. Fujii, M. Ikeda, K. Tamai, R. D. G. McKay, M. Muramatsu, and H. Hamada. 1993. Hybrid cell extinction and re-expression of Oct-3 function correlates with differentiation potential. EMBO J. 12:4489-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith, A. G. 1991. Culture and differentiation of embryonic stem cells. J. Tissue Culture Methods 13:89-94. [Google Scholar]
- 49.Smith, A. G. 2001. Embryonic stem cells, p. 205-230. In D. R. Marshak, R. L. Gardner, and D. Gottleib (ed.), Stem cell biology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Sock, E., J. Enderich, M. G. Rosenfeld, and M. Wegner. 1996. Identification of the nuclear localization signal of the POU domain protein Tst-1/Oct-6. J. Biol. Chem. 271:17512-17518. [DOI] [PubMed] [Google Scholar]
- 51.Sturm, R. A., and W. Herr. 1988. The POU domain is a bipartite DNA-binding structure. Nature 336:601-604. [DOI] [PubMed] [Google Scholar]
- 52.Tanaka, M., W. Clouston, and W. Herr. 1994. The Oct-2 glutamine-rich and proline-rich activation domains can synergize with each other or duplicates of themselves to activate transcription. Mol. Cell. Biol. 14:6046-6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tomilin, A., A. Reményi, K. Lins, H. Bak, S. Leidel, G. Vriend, M. Wilmanns, and H. R. Schöler. 2000. Synergism with the coactivator OBF-1 (OCA-B, BOB-1) is mediated by a specific POU dimer configuration. Cell 103:853-864. [DOI] [PubMed] [Google Scholar]
- 54.Tucker, K. L., C. Beard, J. Dausmann, L. Jackson-Grusby, P. W. Laird, H. Lei, E. Li, and R. Jaenisch. 1996. Germ-line passage is required for establishment of methylation and expression patterns of imprinted but not of nonimprinted genes. Genes Dev. 10:1008-1020. [DOI] [PubMed] [Google Scholar]
- 55.Vigano', M. A., and L. M. Staudt. 1996. Transcriptional activation by Oct-3: evidence for a specific role of the POU-specific domain in mediating functional interaction with Oct-1. Nucleic Acids Res. 24:2112-2118. [DOI] [PMC free article] [PubMed]
- 56.Yeom, Y., G. Fuhrmann, C. E. Ovitt, A. Brehm, K. Ohbo, M. Gross, K. Hübner, and H. R. Schöler. 1996. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development 122:881-894. [DOI] [PubMed] [Google Scholar]
- 57.Yuan, H., N. Corbi, C. Basilico, and L. Dailey. 1995. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox-2 and Oct-3. Genes Dev. 9:2635-2645. [DOI] [PubMed] [Google Scholar]