Abstract
Biogenesis of the small and large ribosomal subunits requires modification, processing, and folding of pre-rRNA to yield mature rRNA. Here, we report that efficient biogenesis of both small- and large-subunit rRNAs requires the DEAH box ATPase Prp43p, a pre-mRNA splicing factor. By steady-state analysis, a cold-sensitive prp43 mutant accumulates 35S pre-rRNA and depletes 20S, 27S, and 7S pre-rRNAs, precursors to the small- and large-subunit rRNAs. By pulse-chase analysis, the prp43 mutant is defective in the formation of 20S and 27S pre-rRNAs and in the accumulation of 18S and 25S mature rRNAs. Wild-type Prp43p immunoprecipitates pre-rRNAs and mature rRNAs, indicating a direct role in ribosome biogenesis. The Prp43p-Q423N mutant immunoprecipitates 27SA2 pre-rRNA threefold more efficiently than the wild type, suggesting a critical role for Prp43p at the earliest stages of large-subunit biogenesis. Consistent with an early role for Prp43p in ribosome biogenesis, Prp43p immunoprecipitates the majority of snoRNAs; further, compared to the wild type, the prp43 mutant generally immunoprecipitates the snoRNAs more efficiently. In the prp43 mutant, the snoRNA snR64 fails to methylate residue C2337 in 27S pre-rRNA, suggesting a role in snoRNA function. We propose that Prp43p promotes recycling of snoRNAs and biogenesis factors during pre-rRNA processing, similar to its recycling role in pre-mRNA splicing. The dual function for Prp43p in the cell raises the possibility that ribosome biogenesis and pre-mRNA splicing may be coordinately regulated.
Ribosome biogenesis is an elaborate process involving rRNA transcription, modification, processing, and folding, as well as ribonucleoprotein (RNP) assembly and export (10, 20, 41). Ribosome biogenesis occurs largely in the nucleolus where RNA polymerase I transcribes a large rRNA precursor (pre-rRNA) that is processed to mature 5.8S, 18S, and 25S rRNAs. In parallel, RNA polymerase III transcribes a precursor of mature 5S rRNA in the nucleolus. The large precursor is cleaved by Rnt1p and trimmed at the 3′ end to yield the 35S pre-rRNA (Fig. 1A). Before processing of 35S pre-rRNA, >70 box C/D and H/ACA small nucleolar RNAs (snoRNAs) bind and modify target sequences through 2′-O-methylation or pseudouridinylation, respectively, in the 18S and 25S regions (6).
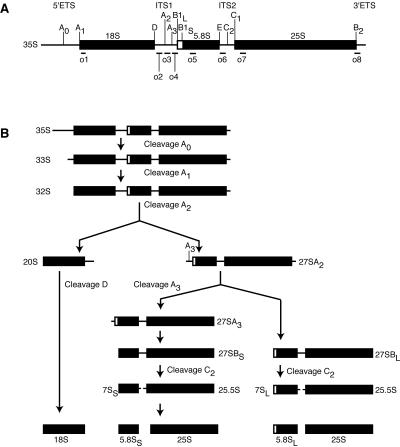
FIG. 1.
The pre-rRNA processing pathway in Saccharomyces cerevisiae. (A) Schematic of the 35S pre-RNA precursor. Mature rRNA sequences are indicated by black boxes; the 5′ external transcribed sequence (5′ETS) and 3′ETS and internal transcribed sequence 1 (ITS1) and ITS2 are indicated by lines. Processing sites (A0 to E) are indicated above. Binding sites for oligonucleotide probes (o1 to o8) are indicated below. (B) Schematic of the pre-rRNA processing pathway. 27SA2 is processed primarily by the left branch. 23S pre-RNA (not shown) results from premature cleavage of 35S at site A3.
The processing of 35S pre-rRNA initiates with cleavages at sites A0, A1, and A2, which yield the small-subunit (SSU) precursor 20S and the large-subunit precursor 27SA2 (Fig. 1B) (41). The 20S pre-rRNA is exported to the cytoplasm, where it is further modified and trimmed to yield mature 18S rRNA, the sole rRNA component of the small subunit. The 27SA2 pre-rRNA is cleaved and trimmed to generate mature 5.8S and 25S rRNAs (Fig. 1B), which then associate with 5S rRNA to form the large subunit. Although small- and large-subunit rRNAs are cotranscribed, these rRNAs are independently processed and assembled into RNPs (10).
Ribosome biogenesis requires at least 18 members of the ubiquitous DExD/H box family of proteins, which generally hydrolyze ATP in an RNA-dependent manner (10). Other members of this family promote numerous RNA-dependent processes, including transcription, pre-mRNA splicing, mRNA export, translation, and RNA degradation (43). The family is defined by eight conserved motifs that function in RNA binding, ATP hydrolysis, coupling RNA binding to ATP hydrolysis, and (in some cases) RNA unwinding. These proteins may function as general chaperones; for example, the DExD/H box protein Cyt19 promotes folding of a group I intron by unwinding nonnative base pairing (37). These proteins may also function as specific unwindases; for example, Prp28p promotes dissociation of U1 small nuclear RNA (snRNA) from the 5′ splice site of a pre-mRNA (9, 49). Additionally, DExD/H box proteins may function to remodel RNPs; for example, Sub2p has been implicated in displacing Mud2p from single-stranded pre-mRNA (31). Thus, in ribosome biogenesis DExD/H box ATPases likely promote the folding, unwinding, and remodeling of pre-rRNA.
One DEAH box ATPase that has recently been implicated in ribosome biogenesis is Prp43p, a protein originally identified as a nuclear pre-mRNA splicing factor (2). Pre-mRNA splicing is catalyzed by the spliceosome, a macromolecular machine composed of five snRNAs (U1, U2, U4, U5, and U6) and roughly 100 proteins (29, 48). The spliceosome catalyzes splicing in two steps. First, the 2′ hydroxyl of a conserved intronic adenosine attacks the 5′ splice site, producing a lariat intermediate and a cleaved 5′ exon. Then, the 3′ hydroxyl of the 5′ exon attacks the 3′ splice site, yielding ligated mRNA and excised lariat intron (48). After ligating the exons, the spliceosome first releases the mRNA for export and then releases the excised intron for debranching and degradation. Intron release is promoted by Prp43p (2, 36), as the dissociation and subsequent degradation of the excised intron are hindered in vivo by the temperature-sensitive mutant prp43-1 (2) and in vitro by recombinant dominant negative Prp43p mutants (36). In addition, hPrp43 associates with the 12S U2 small nuclear ribonucleoprotein particle (snRNP) (57) and with spliceosome assembly complexes (21, 35), suggesting that Prp43p performs additional roles in splicing.
Several observations have implicated Prp43p in ribosome biogenesis. First, Prp43p localizes to the nucleolus (1, 26). This localization is unexpected for a splicing factor; in budding yeast, the only other splicing factor known to localize to the nucleolus is Snu13p, a component of both the U4 snRNP and the box C/D small nucleolar ribonucleoprotein particles (snoRNPs) (56). Second, Prp43p copurifies with Pwp2p (17, 19) and interacts by two-hybrid with Sas10p (27); Pwp2p and Sas10p are components of the SSU processome, suggesting a role in small-subunit processing. Third, Prp43p copurifies with the large-subunit factors Npa1p (11), Nsa3p (38), and Erb1p (24), suggesting an additional role in large-subunit biogenesis. Fourth, Prp43p copurifies with additional factors implicated in ribosome biogenesis, including Asc1p (17, 24), Nop6p (24), and Bud20p (24). Fifth, Prp43p copurifies with Nop1p (17), the putative methyltransferase of the box C/D snoRNPs, and Nop58p (19), a core box C/D snoRNP component. Finally, Prp43p is transcriptionally coregulated with a number of 60S ribosome biogenesis factors in budding yeast, including Erb1p, Nmd3p, Noc3p, Nug1p, and Rrb1p (33); furthermore, coregulation of Prp43p with the 60S ribosome biogenesis factors Noc3p and Nmd3p is conserved across species (50). Although these studies have linked Prp43p to factors associated with ribosome biogenesis, direct evidence that Prp43p functions in ribosome biogenesis has been lacking.
We now report that Prp43p functions in ribosome biogenesis. In the cold-sensitive mutant prp43-Q423N at the nonpermissive temperature, the steady-state levels of 35S pre-rRNA increased and the steady-state levels of downstream pre- rRNAs decreased, suggesting that Prp43p is required for pre-rRNA processing. Supporting a role in ribosome biogenesis, pulse-chase analysis of pre-rRNA processing in the mutant revealed a requirement for Prp43p in the formation of 20S and 27S pre-rRNAs, as well as in the formation of 18S and 25S mature rRNAs. Supporting a direct role in ribosome biogenesis, Prp43p coimmunoprecipitated both small- and large-subunit pre-rRNAs, as well as mature rRNAs. Further, the Prp43p-Q423N mutant coimmunoprecipitated a greater fraction of 27SA2 pre-rRNA, in particular, indicating that the mutant stalls on pre-rRNA during an early step in large-subunit biogenesis. Supporting a role for Prp43p in both small- and large-subunit biogenesis, Prp43p also coimmunoprecipitated most snoRNAs. Suggesting a requirement for Prp43p in snoRNA function, in the prp43-Q423N mutant the box C/D snoRNA snR64 failed to methylate residue C2337 in 27S pre-rRNA. These results demonstrate that ribosome biogenesis requires Prp43p and establish a second role for Prp43p in the cell.
MATERIALS AND METHODS
Plasmids and strains.
PRP43 was subcloned from pRSJA1 (2) into pRS315 using SacI and XhoI sites and into pRS316 using HindIII and SacII sites, yielding pJPS298 and pJPS261, respectively. To tandem affinity purification (TAP) tag PRP43, the tag was amplified from pKG1810 (51) using primers Prp43-TAPF and Prp43-TAPR and then integrated into pJPS261 in yJPS549, yielding pJPS1162. The prp43-Q423N mutation was introduced by QuikChange (Stratagene) into pJPS298, yielding pJPS643; into pJPS1162, yielding pJPS1163; and into pGALWT (2), yielding pGALQ423N.
To generate yJPS549 (MATa his3Δ leu2Δ LYS2 met15Δ0 ura3Δ prp43::KanMX4 [pJPS261]), BY4743 (Invitrogen) was transformed with pJPS261 and sporulated. PRP43 (yJPS568) and prp43-Q423N (yJPS575) were generated by transforming yJPS549 with pJPS298 and pJPS643, respectively, and shuffling out pJPS261 using 5-fluoroorotic acid (7). The TAP-tagged PRP43 (yJPS797) and prp43-Q423N (yJPS798) strains were generated by transformation of pJPS1162 and pJPS1163, respectively, into yJPS728, which was generated by replacement of KanMX4 in yJPS549 with HIS3. Prp43p overexpression strains were constructed by transforming BY4741 (Open Biosystems) with pGALWT and pGALQ423N. The TAP-tagged PRP22 strain (yJPS773) was constructed by integrative transformation of BY4741 (Open Biosystems) with DNA amplified from pKG1810 (51) using primers Prp22-TAPF and Prp22-TAPR. The TAP-tagged DBP2, DHH1, SUB2, and TIF1 strains were obtained from Open Biosystems.
Immunoprecipitations.
Immunoprecipitations were performed essentially as previously described (42). One liter of yeast was grown in rich media at 30°C to an optical density at 600 nm of 0.3 to 0.8 and shifted to 15°C for 2 h. Cells were harvested by centrifugation, resuspended in IPP150 (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% NP-40) and protease inhibitors [1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride, 1-μg/ml leupeptin, 1-μg/ml aprotinin, 1 mM benzamidine], and lysed by bead beating. After centrifugation at 2,000 to 10,000 × g for 10 min, lysates were incubated with 350 μl immunoglobulin G-Sepharose beads (Amersham), equilibrated in IPP150 for 5 h at 4°C, and washed three times with 50 ml IPP450 (10 mM Tris-HCl [pH 8.0], 450 mM NaCl, 0.1% NP-40) and once with 50 ml of IPP150. RNA was extracted from the beads by hot phenol-chloroform-isoamyl alcohol (4) and analyzed by microarray or Northern analysis. Alternatively, in the analysis of the bound snRNAs, the pellets were washed with buffer D (20 mM HEPES [pH 7.9], 0.2 mM EDTA, 50 mM KCl, 20% glycerol) and analyzed by primer extension.
Microarray analysis.
RNA extracted from immunoprecipitates was treated with RNase-free DNase I. A total of 10 μg of immunoprecipitated RNA was labeled with Alexa Fluor 647 (Molecular Probes Ulysis kit). Total RNA was isolated from wild-type yeast cells by hot acidic phenol extraction (40), and 10 μg was labeled with Alexa Fluor 546. Labeled RNAs were ethanol precipitated and dissolved in hybridization buffer containing 33% formamide (23). Labeled RNAs were hybridized to arrays in a rotating incubator at 42°C for 16 to 20 h and washed (25). Arrays were scanned on an Axon 4000B. Images were quantitated with GenePix (Axon Instruments). Individual channels were spatially detrended, and dye bias was corrected (23). Normalized intensities were converted to log 2 ratios of Prp43p-associated RNA relative to total RNA. The average ratio of all probes across individual RNAs was calculated for each of the three samples (see Fig. 4). To improve signal-to-noise values, probes with intensities of <99% of the 200 negative control spots were not included.
FIG. 4.
Prp43p coimmunoprecipitates introns and snRNAs as well as pre-rRNAs and many snoRNAs. RNA was coimmunoprecipitated from TAP-tagged PRP43, prp43-Q423N, or PRP22 strains after a 2-h shift to 15°C. Bound RNA was assayed by microarray. Quantitation of the enrichment (bound versus whole-cell RNA) is shown for the snoRNAs, snRNAs, (pre-)rRNAs, and three introns that are enriched in Prp43p and/or Prp43p-Q423N coimmunoprecipitates. The darkness of a bar reflects the degree of enrichment, according to the linear scale above, with the exception of the darkest bar, which reflects enrichment ranging from 8 to 20 fold. Due to normalization (23), only relative differences among 5.8S, 18S, and 25S rRNAs could be detected. RNA classes are labeled on the left; probe targets are labeled on the right. In addition, the genomic context for each snoRNA is indicated as monocistronic (mono), polycistronic (poly), or intronic (intron) (5, 44). Finally, the 18S and/or 25S rRNA targets are indicated, where known (34, 44, 45, 52). U3a, U3b, and snR30 do not modify rRNA but do promote rRNA processing, as do U14 and snR10 (6). snR4 and snR45 are not implicated in either rRNA modification or processing. Since the snoRNAs snR39, snR41, snR44, snR58, snR59, snR65, and snR78 were not detected in wild-type cells above background, these snoRNAs were not included in the figure. The representative data derive from a single experiment; the bound RNAs were similarly enriched in an independent coimmunoprecipitation. Note that Northern analysis failed to confirm the enrichment of snR66 in the Prp22p coimmunoprecipitation (data not shown); in contrast, Northern analysis has confirmed enrichment of snR66 in the Prp43p coimmunoprecipitation. The intensity ratios used in the figure and the raw data set are available online (http://hugheslab.med.utoronto.ca/Leeds).
Northern and primer extension analysis.
For large RNAs, 5 to 10 μg of RNA was treated with glyoxal (4), separated on a 1.25% agarose gel in 10 mM sodium phosphate (pH 7.0), and transferred by capillary action to HybondN (Amersham). For small RNAs, 5 to 10 μg of RNA was separated on a 6% denaturing polyacrylamide gel and transferred semidry to HybondN. Membranes were probed with 32P-labeled oligonucleotides or random-primed probes in Rapid-Hyb buffer (Amersham) and washed two or three times with 2× to 6× SSC-0.1% sodium dodecyl sulfate (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Primer extensions were performed as previously described (49). Northern blots and primer extension gels were exposed to a phosphor screen, developed with a Storm PhosphorImager (Molecular Dynamics), and quantitated with ImageQuant (Molecular Dynamics).
Pulse-chase analysis.
yJPS797 and yJPS798 were grown in 100 ml of synthetic defined medium without uracil at 30°C to an optical density at 600 nm of 0.2 to 0.4. Cells were pelleted and resuspended in 6 ml of synthetic defined medium without uracil at room temperature. Cultures were then pulsed with 100 μCi [3H]uracil (60 Ci/mmol; Amersham) for 2 min and then chased with 300 μg of cold uracil. Aliquots (each, 1 ml) were taken at each time point during the chase; the 0-min aliquot was taken immediately before the chase. Following lysis of the cells and extraction of the RNA by phenol-chloroform-isoamyl alcohol (4), 25,000 cpm of labeled RNA was treated with glyoxal (4), resolved on a 1.25% agarose gel in 10 mM sodium phosphate (pH 7.0), transferred by capillary action to HybondN, sprayed with EN3HANCE (Perkin-Elmer), and exposed to BIOMAX MS film (Kodak).
RNase H protection assay.
A total of 5 μg of whole-cell RNA, extracted from cells shifted to 15°C for 2 h, was annealed with 300 ng of a chimeric 2′-O-methyl oligonucleotide, RNase H (Roche) digested, phenol-chloroform-isoamyl alcohol extracted, and separated on an agarose gel; Northern analysis was then carried out (58).
Oligonucleotides.
Oligonucleotides for Northern analysis were as follows: o1, 5′CATGGCTTAATCTTTGAGAC; o2, 5′GCTCTTTGCTCTTGCC; o3, 5′ATGAAAACTCCACAGTG; o4, 5′CCAGTTACGAAAATTCTTG; o5, 5′CGCATTTCGCTGCGTTCTTCATCG; o6, 5′GGCCAGCAATTTCAAGT; o7, 5′GCCCGTTCCCTTGGCTGTG; o8, 5′GAAATAAAAAACAAATCAGAC; 5S, 5′GCGTATGGTCACCCACTAC; U3A, 5′CCAAGTTGGATTCAGTGGCTC; U14, 5′TCCACGGTAGGAGTACGCTTACGAACCCATCGT; U18, 5′CTCTTTTGTCACTCATATCGGGGGTCTTACTTCCCAT; U24, 5′CCATCTGAAGTAGCAAATATG; snR3, 5′GCAATCCACGAGTTCTTATA; snR10, 5′GAAAGACTGTTGCACCCAAGATAG; snR30, 5′GAAACTGCTCGTAGTCTGACG; snR37, 5′CGCAATTCTCTAAAAACGCGAAC; snR47, 5′GGACGAAGAAATTCATGTTGTTGTTG; snR74, 5′CATGAAGATCAGACATATGCTTGTC; U6, 5′AAAACGAAATAAATCTCTTTG; Sup56, 5′AGGTATCGTAAGATGCAAGAGTTCGAATCTCTTAGC; RPL10, 5′TTAAGCTTGAGCAGCAAAGTATTCTGGGAATTCTC; and U3A intron, 5′CAAAAGCTGCTGCAATGG. An actin intron random prime probe was generated by PCR of genomic DNA using ACT1iF (5′GTATGTTCTAGCGCTTGCACCATC) and ACT1iR (5′CTAAACATATAATATAGCAACAAAAA). Chimeric 2′O-methyl oligonucleotides for RNase H protection assays (Dharmacon) were as follows: C2337, 5′mGmAmUmAmGdGdGdAdCmAmGmUmGmGmGmAmAmUmCmUmC mG; and U68, 5′mUmUmCmAdCdAdTdTmAmCmGmUmAmUmCmGmCmAmUmUmU.
Oligonucleotides for snRNA primer extensions were as follows: U2 (ssU2M5), 5′AAAGTCTCTTCCCGTCCATTTTATTA; and U5 (ssU5-125), 5′GGAGACAACACCCGGATGGTTCTGGTA. U1, U4, and U6 primers were described previously (49). Oligonucleotides for TAP tagging PRP43 were as follows: PRP43-TAPF, 5′TTGAAACAAGGTAAAAACAAAAAGAAGAGTAAGCACTCCAAGAAAATGAAGCGACGATGGAAAAAGAATTTC; and PRP43-TAPR, 5′AAATTTATATAAATCTATTTTTTTTTTTTTTTCGACACAAAATGTTACGACTCACTATAGGGA. Oligonucleotides for TAP tagging PRP22 were as follows: Prp22-TAPF, 5′CTAAGCTCAATAAGGCAGTCAAGGGAAAGGGCATTAGGTATCAAGAGGATGAAGCGACGATGGAAA; and Prp22-TAPR, 5′ATAGGTCTATAAAACTCGATAATTATAATGCATAAAAAGCTAACAATGTACGACTCACTATAGGG.
RESULTS
The cold-sensitive mutant prp43-Q423N is defective for pre-mRNA splicing.
To investigate the function of Prp43p, we engineered the mutation Q423N in motif VI (QRAGRAGR) of the DExD/H box domain to disrupt the catalytic activity of Prp43p. In the crystal structure of a related DExD/H box ATPase, the hepatitis C virus protein NS3, the equivalent residue Q460 forms a hydrogen bond with H293 in the Walker B ATPase motif, implicating the glutamine in ATPase activity (30). Indeed, in Prp43p the lethal mutation Q423A inhibits its ATPase activity 10 fold (36). As mutations in the DExD/H box domains of related proteins disrupt catalytic activity without disrupting recruitment to a substrate (14, 55), we anticipated that the prp43-Q423N mutation disrupts catalysis without disrupting association with its substrates.
The prp43-Q423N mutation confers a cold-sensitive growth phenotype.
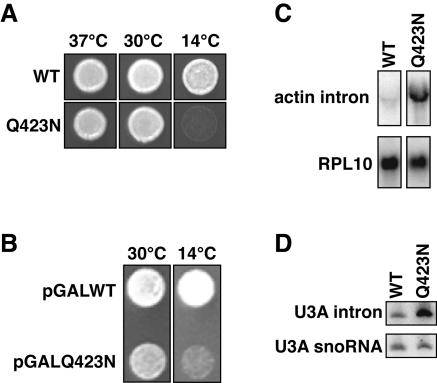
The mutant grew as well as the wild type at both 30°C and 37°C but grew poorly at 14°C (Fig. 2A). When overexpressed in the presence of wild-type Prp43p, Prp43p-Q423N conferred a dominant negative phenotype (Fig. 2B). These observations suggest that Prp43p-Q423N is indeed impaired for catalysis but not for recruitment to its substrate and that Prp43p-Q423N provides an experimental tool to identify the substrates for Prp43p (see below) (22).
FIG. 2.
The prp43-Q423N mutant is cold sensitive, dominant, and defective in splicing. (A) Growth of PRP43 (yJPS568) and prp43-Q423N (yJPS575) on rich media at various temperatures. (B) Growth of cells containing both a chromosomal copy of PRP43 and a galactose-driven, plasmid-borne copy of either PRP43 (pGALWT) or prp43-Q423N (pGALQ423N). Overexpression of the plasmid-borne copy was induced on selective galactose-containing media. (C and D) Northern analysis of excised intron levels from actin (C) or U3A (D) in PRP43 (yJPS568) and prp43-Q423N (yJPS575). mRNA from the intronless gene RPL10 (C) or U3A snoRNA (D) were probed for comparison. Whole-cell RNA was harvested from cells after a 2-h shift to 15°C in rich media.
The prp43-Q423N mutation impedes the splicing cycle.
In vivo, prp43-Q423N cells accumulated intron excised from actin and U3A precursors at 30°C (data not shown); this accumulation increased to >7 fold after a 2-h shift to 15°C (Fig. 2C and D). Spliced actin mRNA and U3A snoRNA levels did not change after the temperature shift (Fig. 2D and data not shown), as expected (2, 36). In vitro, prp43-Q423N extracts similarly accumulated excised actin intron (data not shown). Thus, consistent with previous findings (2), the Prp43p-Q423N mutant impeded degradation of the excised lariat intron. These observations were also consistent with the finding that Prp43p promotes release of the excised intron from the spliceosome (2, 36), a prerequisite for intron degradation.
The cold-sensitive mutant prp43-Q423N is also defective for pre-rRNA processing.
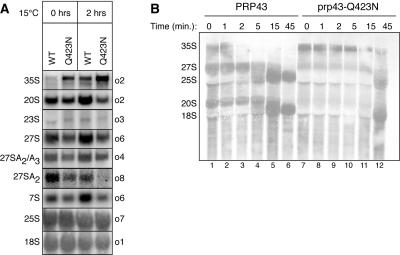
Consistent with the nucleolar localization of Prp43p (26), we found evidence that Prp43p functions in pre-rRNA processing (Fig. 3). By Northern analysis of whole-cell RNA, the prp43-Q423N mutant accumulated 35S pre-rRNA at 30°C (Fig. 3A). Although the prp43-Q423N mutant showed no growth defect at 30°C (Fig. 2A), the accumulation of 35S pre-rRNA at 30°C paralleled the accumulation of excised intron at this temperature (data not shown). Similarly, 35S pre-rRNA accumulated to levels ∼3-fold higher in the mutant than in the wild type after a 2-h shift to 15°C (Fig. 3A), paralleling the accumulation of excised intron in the mutant after a 2-h shift to 15°C (Fig. 2C and D). Although 35S accumulation is often accompanied by premature A3 cleavage and increased formation of aberrant 23S pre-rRNA (53), we did not observe increased formation of 23S pre-rRNA in the mutant. As 23S accumulation is characteristic of mutants deficient in the SSU processome (53), the prp43-Q423N mutant did not appear deficient in SSU processome functions. Suggesting a broader role in ribosome biogenesis, the accumulation of 35S pre-rRNA in the prp43-Q423N mutant was accompanied by a general decrease in both the small- and large-subunit pre-rRNAs; specifically, the 20S, 27S, and 7S pre-rRNAs were lower at 30°C and decreased ∼3 fold relative to the wild type after a 2-h shift to 15°C (Fig. 3A). In contrast, the mature rRNAs did not decrease, as expected for such a short temperature shift (Fig. 3A; data not shown). Consistent with the interaction of Prp43p with both small- and large-subunit biogenesis factors (11, 17, 19, 24, 27, 38), our data suggest that Prp43p promotes biogenesis of both small- and large-subunit rRNAs.
FIG. 3.
The prp43-Q423N mutant is defective in pre-rRNA processing. (A) Northern analysis of steady-state pre-rRNA and rRNA levels in PRP43 (yJPS568) and prp43-Q423N (yJPS575). RNA was harvested from cells before and after a 2-h shift to 15°C (see Materials and Methods). Probes are indicated on the right; pre-rRNA and rRNA species are indicated on the left. The 27S species include all 27SA and 27SB intermediates. The 27SA2/A3 species include 27SA2 and 27SA3. Note that the 27SA2 species is detected at the 3′ end. (B) [3H]uracil pulse-chase analysis of pre-rRNA processing. yJPS797 and yJPS798 were pulsed with [3H]uracil for 2 min and then chased with unlabeled uracil for the times indicated above. The zero time point was taken immediately prior to the addition of unlabeled uracil.
A broad role for Prp43p in the biogenesis of both small- and large-subunit rRNAs is supported by [3H]uracil pulse-chase analysis of pre-rRNA processing (Fig. 3B). In particular, the analysis identifies a role for Prp43p in processing 35S pre-rRNA to 20S and 27S pre-rRNAs. At the beginning of the chase, 35S pre-rRNA levels were higher in the mutant than in the wild type. Further, whereas 35S pre-rRNA chased by 5 min in the wild type, 35S pre-RNA persisted 10-fold longer in the mutant. Although this persistence could be accounted for by either an increase in stability or a delay in processing of 35S pre-rRNA, the 20S and 27S pre-rRNAs levels argued strongly for a delay in 35S pre-rRNA processing; the levels of 20S and 27S pre-rRNAs were reduced in the mutant, compared to the wild type, at the beginning of the chase and throughout the chase. Finally, the formation of both 18S and 25S mature rRNAs was delayed nearly 10 fold in the mutant; while these mature rRNAs appeared in the wild type after 5 min, neither of these mature rRNAs appeared in the mutant until 45 min. These data identified a central role for Prp43p early in ribosome biogenesis. Further, these data indicated a requirement for Prp43p in the biogenesis of both small- and large-subunit rRNAs. Given that most ribosome biogenesis factors promote either small- or large-subunit biogenesis (53), Prp43p performs an unusually broad role in ribosome biogenesis.
Prp43p associates with intronic RNAs, snRNAs, rRNAs, and snoRNAs.
To determine whether the pre-rRNA processing defects in the prp43-Q423N mutant cells are direct or indirect, we investigated the RNAs that bind to Prp43p and Prp43p-Q423N by microarray analysis (40). For comparison, we also investigated the RNAs that bind to the DEAH box splicing factor Prp22p, which dissociates the ligated mRNA from the spliceosome before Prp43p dissociates the excised intron (46, 54). Using the TAP tag (42), we immunoprecipitated each protein after cells were shifted for 2 h at 15°C and then analyzed the bound RNAs by microarray. We limited our analysis of the array data to groups of RNAs rather than individual RNAs, given that noise and cross-hybridization can complicate interpretation of specific signals on the array. As expected, Prp43p, Prp43p-Q423N, and Prp22p all bound U2, U5, and U6 snRNAs, as evidenced by the 3- to 20-fold enrichment of these snRNAs in the immunoprecipitates relative to whole-cell RNA (Fig. 4); in contrast, these proteins did not detectably bind U1 and U4 snRNAs, as these RNAs were not enriched in the coimmunoprecipitates, as expected. Both Prp43p and Prp43p-Q423N also bound detectably to a few introns, which were enriched two- to threefold (Fig. 4), but binding to the rest of the introns, although expected, was below the level of detection. This failure to detect many bound introns, despite the detection of bound snRNAs, presumably reflects the fact that any given intron would be bound to only a small fraction of the total spliceosomes and consequently be much more difficult to detect than the snRNAs. Similar to Prp43p, Prp22p was expected to bind introns, but none were detected. The binding of snRNAs and a few introns was consistent with the known roles of Prp43p and Prp22p in pre-mRNA splicing and thereby validated the microarrays as a tool to identify Prp43p substrates. An average twofold increase in the binding of Prp43p-Q423N, relative to wild-type protein, to snRNAs and introns confirmed that the mutated protein is functional for recruitment to the spliceosome but defective for a subsequent function, such as the catalysis of RNA unwinding, that is required for release of Prp43p from the spliceosome.
In addition, Prp43p and Prp43p-Q423N bound pre-rRNAs, which were enriched from 3 to 14 fold (Fig. 4). Coimmunoprecipitated RNAs hybridized to probes to the 5′-A0, A0-A1, and/or D-A2 external and internal transcribed sequences (Fig. 1A), consistent with Prp43p binding to small-subunit pre- rRNAs, such as 35S and 20S; in addition, coimmunoprecipitated RNAs hybridized to probes to the A2-A3, A3-B1, E-C2, and/or C2-C1 internal transcribed sequences (Fig. 1A), consistent with Prp43p binding to small- and/or large-subunit precursors, such as 27S pre-rRNA. In contrast, the coimmunoprecipitated RNAs did not hybridize to probes to 3′ external transcribed sequences specific to the primary pre-rRNA transcript, a precursor to 35S pre-rRNA, indicating that Prp43p and Prp43p-Q423N do not interact detectably with the primary transcript. The RNAs that interacted with Prp43p were further enriched two- to fourfold in the Prp43p-Q423N immunoprecipitates, suggesting that the mutated protein stalled on these RNAs and that these RNAs corresponded to genuine substrates. By comparison with the pre-rRNAs, the mature rRNAs did not interact detectably with Prp43p and Prp43p-Q423N, as the ratios of bound to total RNA, ranging from 0.9 to 1.3, were well below the practical cutoff for significance (2.0); however, by Northern analysis, which is more sensitive, we were able to detect binding of mature rRNAs (see below). In contrast to Prp43p, Prp22p did not bind to any of the pre-rRNAs, further demonstrating the specificity of pre-rRNA binding by Prp43p (Fig. 4). These data confirm a direct role for Prp43p in pre-rRNA processing in addition to its established role in pre-mRNA splicing.
Prp43p and Prp43p-Q423N associated with a majority of snoRNAs (Fig. 4), providing additional evidence that Prp43p functions in ribosome biogenesis. Of the 60 analyzed snoRNAs, 47 were enriched from 2 to 11 fold in the wild-type and/or mutant immunoprecipitates, relative to whole-cell RNA. We observed association with all subclasses of snoRNAs, indicating that Prp43p does not function exclusively with a subclass of snoRNAs. Specifically, the associated snoRNAs derived from the three classes of transcripts—monocistronic, polycistronic and intronic. Further, both box C/D and H/ACA snoRNAs associated with Prp43p and Prp43p-Q423N. These snoRNAs facilitated processing and/or modification of both pre-18S and pre-25S rRNAs. For example, the essential box C/D snoRNAs U3 and U14 (snR128) and the nonessential box H/ACA snoRNA snR30 are required for processing 35S to 20S pre-rRNA (Fig. 1B); the snoRNAs snR53, snR71, snR74, and snR76 are required for 2′-O-methylation of 18S; and the snoRNAs snR3 and snR37 are required for pseudouridinylation of 25S (6). The binding of box C/D snoRNAs is consistent with the association of Prp43p with the box C/D factors Nop1p (17) and Nop58p (19). Intriguingly, the enrichment of 41 of the 60 analyzed snoRNAs increased two- to fivefold in the mutant Prp43p-Q423N immunoprecipitates, relative to wild type, suggesting that the mutant DEAH box ATPase stalled on snoRNAs and/or snoRNA-bound substrates, such as pre-rRNAs. These data support a general role for Prp43p in ribosome biogenesis.
Prp43p associates with precursors to both small- and large-subunit RNAs.
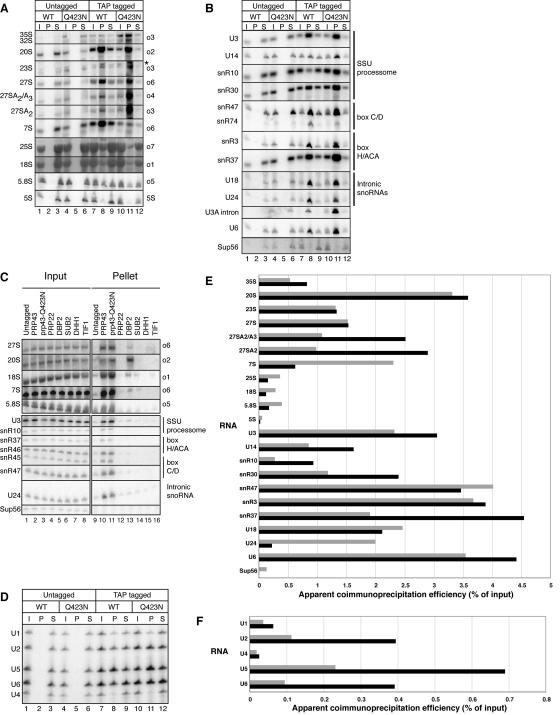
Prp43p and Prp43p-Q423N associate with pre-rRNA intermediates and, less efficiently, with mature rRNAs (Fig. 5A, C, and E). To determine specifically which pre- rRNAs bound to Prp43p and Prp43p-Q423N in the microarray experiment (Fig. 4), we analyzed the coimmunoprecipitated pre-RNAs directly by Northern analysis. Consistent with a direct role in 35S processing, both Prp43p and Prp43p-Q423N interacted with 35S pre-rRNA, coimmunoprecipitating ∼0.5% of the input, a level that is ∼20 fold above the undetectable background binding of 35S in control coimmunoprecipitates from an untagged strain (Fig. 5A and E). Both Prp43p and Prp43p-Q423N associated most strongly with the 20S pre-rRNA, coimmunoprecipitating ∼3% of the input in the tagged strains, compared to ∼0.03% or less in the untagged strains (Fig. 5A and E). Although this binding efficiency is low, it is comparable to the efficiency of binding U6 snRNA (Fig. 5B); only ∼4% of U6 snRNA was coimmunoprecipitated in this experiment, as expected given the transient binding of Prp43p to a subpopulation of spliceosomes (28). This efficiency of binding is within twofold of the efficiency of binding 20S pre-rRNA by the nuclear DExD/H-box ATPase Dbp2p (Fig. 5C), a factor known to interact with 20S pre-rRNA (32). In contrast, Prp22p coimmunoprecipitated 20S pre-rRNA more than 20-fold-less efficiently than Prp43p (Fig. 5C), consistent with the specificity observed by microarray. Given that the abundance of Prp22p is 19-fold lower than Prp43p (18), we tested whether the association of 20S pre-rRNA was general to highly expressed factors or specific to Prp43p. Three DEAD-box ATPases expressed two- to sixfold more highly than Prp43p (18) also coimmunoprecipitated 20S pre-rRNA less efficiently. Specifically, the nuclear ATPase Sub2p and the two cytoplasmic ATPases Tif1p and Dhh1p coimmunoprecipitated 20S pre-rRNA >15-fold less efficiently than Prp43p (Fig. 5C). Thus, the association of Prp43p with 20S pre-rRNA is specific and suggests a direct role for Prp43p in small-subunit biogenesis.
FIG. 5.
Prp43p coimmunoprecipitates precursors to both small- and large-subunit rRNAs, as well as the mature rRNAs themselves. (A to C) Northern analysis of pre-rRNAs and mature rRNAs (A and C) or snoRNAs (B and C) that coimmunoprecipitate with Prp43p and Prp43p-Q423N or the controls Prp22p, Dbp2p, Sub2p, Dhh1p, and Tif1p. To increase stringency, cell lysates analyzed that are shown in panel C were cleared at 10,000 × g, rather than 2,000 × g (A and B). (D) Primer extension analysis of the snRNAs that coimmunoprecipitated. (E) Quantitation of the coimmunoprecipitation efficiency for RNAs shown in panels A and B. Gray bars, RNA bound to Prp43p; black bars, RNA bound to Prp43p-Q423N. snR47 and the U3a intron were omitted, given undetectable levels of these RNAs in the input for wild-type PRP43. (F) Quantitation of the coimmunoprecipitation efficiency for RNAs shown in panel D. Bars are as defined for panel E. RNA was coimmunoprecipitated from TAP-tagged strains and from isogenic untagged strains after a 2-h shift to 15°C. RNA corresponding to approximately 0.15% (5 μg) of the total input (I), 25% of the total pellet (P) and 0.15% (5 μg) of the total supernatant (S) was analyzed in each experiment (A, B, C, and D). Note that the RNA analyzed in the results shown in panel D derived from an independent experiment in which the pellets were washed in buffer D rather than IPP450 (see Materials and Methods). In panels A to C, RNA species are indicated to the left and Northern probes or RNA classes are shown on the right. In panel D, snRNA species are indicated on the left. In panel A, the asterisk marks nonspecific hybridization.
Suggesting a parallel role in large-subunit biogenesis, Prp43p and/or Prp43p-Q423N also associate with the general population of 27S pre-rRNAs (Fig. 5A). Both Prp43p and Prp43p-Q423N bound the 27S population with the same efficiency, roughly 1.5% (Fig. 5E). In contrast, by probing coimmunoprecipitated RNA between sites A3 and B1L (Fig. 1A), we observed that the Prp43p-Q423N mutant bound the 27SA2/A3 subpopulation ∼3-fold more efficiently than the wild type (Fig. 5A and E). Further, by probing between sites A2 and A3, we discovered that Prp43p-Q423N binds 27SA2 more efficiently than wild-type Prp43p (Fig. 5A and E). Prp43-Q423N coimmunoprecipitated 27SA2 pre-rRNA as efficiently as 20S pre-rRNA (Fig. 5A and E). This increased association of 27SA2 pre-rRNA with Prp43p-Q423N suggests that the Prp43p-Q423N mutant stalls on this pre-rRNA, implicating a direct role for wild-type Prp43p at the 27SA2 pre-rRNA stage of ribosome biogenesis.
Prp43p also binds the large-subunit pre-rRNA 7S. Wild-type Prp43p bound ∼2% of 7S pre-rRNA, whereas Prp43p-Q423N bound two- to fourfold less (Fig. 5A and E). This binding is specific, as Prp22p bound 7S pre-rRNA 100-fold-less efficiently (Fig. 5C), consistent with the microarray data (Fig. 4). Further demonstrating specificity, the more highly expressed DExD/H box ATPases Dbp2p, Sub2p, Dhh1p and Tif1p (18) all bound 7S with at least 45-fold-lower efficiency (Fig. 5C). The association of Prp43p with 7S pre-rRNA suggests additional roles for Prp43p downstream of 27SA2 pre-rRNA.
Prp43p also interacts with the mature rRNAs. Wild-type Prp43p immunoprecipitated ∼0.4% of 5.8S, 18S, and 25S rRNAs and the Prp43p-Q423N mutant immunoprecipitated ∼0.2% (Fig. 5A and E). By comparison, Prp22p, Dbp2p, Sub2p, Dhh1p, and Tif1p bound at least 10-fold less of the 18S and 5.8S rRNAs (Fig. 5C). Further, although the binding of mature rRNAs by Prp43p was ∼10-fold lower than for 20S pre-rRNA, for example (Fig. 5E), binding was higher than for tRNALeu (Sup56). Specifically, wild-type Prp43p bound only ∼0.1% of tRNALeu (Sup56), and the mutant bound undetectable levels (Fig. 5B and E). Prp43p did not bind 5S rRNA as significantly as the other mature rRNAs; the wild type bound only ∼0.05% of 5S rRNA and the mutant bound only ∼0.02% (Fig. 5A and E). The binding of mature rRNAs by Prp43p suggests an association of Prp43p with mature rRNAs at late stages of ribosome assembly. In total, these Northern data further support a broad role for Prp43p in both small- and large-subunit biogenesis.
Northern analysis also confirmed that both Prp43p and Prp43p-Q423N specifically associated with a wide range of snoRNAs (Fig. 5B, C, and E). Prp43p and Prp43p-Q423N bound both box C/D and H/ACA snoRNAs that processed and/or modified 18S and/or 25S rRNAs. The bound snoRNAs also derive from all three classes of pre-snoRNA transcripts. The snoRNAs bound as efficiently as the pre-rRNAs; the efficiency of binding ranged from ∼0.5% to ∼5% (Fig. 5E). As observed by microarray (Fig. 4), a number of snoRNAs were enriched in the mutant compared to the wild-type immunoprecipitates (Fig. 5B and E). Specifically, U14, snR30, and snR37 were enriched twofold, and snR10 was enriched threefold (Fig. 5E). Although the relative enrichment of a few snoRNAs (e.g., snR37 and snR47) appeared to vary between the array and the Northern (Fig. 4 and 5), we attributed these differences to limitations inherent to microarray experiments—noise and cross-hybridization that can confound the analysis of specific RNAs. Further, in every case tested, we found that snoRNAs not detected as enriched by array were detected as enriched by Northern analysis. These snoRNAs included snR4, snR10, snR42, snR45, and snR46 (Fig. 5C and E; data not shown); the remainder of the snoRNAs were not tested by Northern analysis. Compared to Prp43p, Prp22p coimmunoprecipitated U3, U24, snR45, snR46, and snR47 at least 10-fold-less efficiently (Fig. 5C), consistent with the specificity observed by microarray. Additionally, Prp22p coimmunoprecipitated snR10 and snR37 roughly fivefold-less efficiently than Prp43p. Although the abundance of Prp22p was 19-fold lower than Prp43p (18), the highly expressed factors Dbp2p, Sub2p, Dhh1p, and Tif1p coimmunoprecipitated U3, U24, snR45, snR46, and snR47 less efficiently than Prp43p by at least 10 fold and coimmunoprecipitated snR10 and snR37 roughly 5-fold-less efficiently than Prp43p. Thus, the association of Prp43p with snoRNAs was specific. These data support early roles for Prp43p in pre-rRNA processing and suggest roles for Prp43p in promoting the binding and/or dissociation of snoRNAs.
Northern and primer extension analysis also confirmed that Prp43p and Prp43p-Q423N bound introns and snRNAs. By Northern analysis, Prp43p and Prp43p-Q423N bound the excised U3A intron and U6 snRNA (Fig. 5B). By primer extension analysis, Prp43p and Prp43p-Q423N bound primarily U2, U5, and U6, the snRNAs associated with active spliceosomes (Fig. 5D and F). Prp43p bound these snRNAs with low efficiency, ranging from ∼0.2% to ∼0.4% for wild-type Prp43p (Fig. 5F); low efficiency was expected, given that Prp43p binds the spliceosome transiently (28). The efficiency of binding snRNAs increased three- to fourfold in the mutant (Fig. 5F), consistent with a defect in dissociating the intron and releasing the spliceosome. This analysis of complexes derived from living cells reaffirms the role of Prp43p in splicing.
The prp43-Q423N mutant is defective for methylation of 27S pre-rRNA by a box C/D snoRNA.
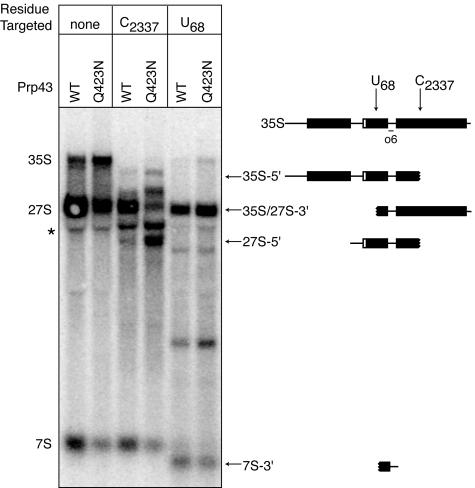
To examine the efficiency of box C/D snoRNA-mediated methylation in the prp43-Q423N mutant, we probed for methylation of C2337 in 25S rRNA by the monocistronic, nonintronic snoRNA snR64 (34). We probed for methylation at different stages of processing using a site-directed RNase H cleavage protection assay (58). We chose C2337 for probing because RNase H cleavage at this site in 35S or 27S pre-rRNA yields products that can be resolved by agarose gel electrophoresis. In both wild-type and mutant cells, RNase H cleaved C2337 in 35S, as shown by depletion of 35S pre-rRNA and formation of the 5′ cleavage product (Fig. 6), indicating that C2337 was largely unmodified at the 35S stage in both the wild type and the mutant. At the 27S stage, C2337 was largely protected from RNase H cleavage in the wild type (Fig. 6). However, this site was cleaved in the mutant, as illustrated by the depletion of 27S pre-rRNA and the formation of the 5′ cleavage product (Fig. 6). By Northern analysis, the levels of snR64 were unchanged in the prp43-Q423N mutant (data not shown). These observations indicated that snR64 fails to direct methylation of C2337 in 27S pre-rRNA in the prp43 mutant. In a positive control, RNase H cleaved the unmodified 5.8S residue U68 in 35S, 27S, and 7S pre-rRNAs in both wild type and mutant (Fig. 6). Our results demonstrate a requirement for Prp43p in the methylation of C2337, suggesting a role for Prp43p in snoRNA binding, snoRNA-mediated methylation, and/or snoRNA recycling.
FIG. 6.
Prp43p is required for snoRNA-mediated methylation of C2337 in 25S rRNA at the 27S pre-rRNA stage. 2′-O-methylation by the box C/D snoRNA snR64 at C2337 was assessed by a site-specific RNase H cleavage protection assay (58). The unmodified 5.8S residue U68 was assayed as a positive control for cleavage. The first two lanes represent negative controls. RNA from PRP43 or prp43-Q423N strains, shifted 2 h at 15°C, was RNase H digested and analyzed by Northern using probe o6. The schematic shown at the top indicates the position of C2337, U68, and the o6 probe in 35S; the other schematics symbolize the cleavage products, marked by arrows and labeled with the pre-rRNA from which it derives. The asterisk marks nonspecific hybridization.
DISCUSSION
We find that the essential DEAH box protein Prp43p, a known pre-mRNA splicing factor, plays a novel role in ribosome biogenesis. First, the prp43-Q423N mutant is defective in 35S pre-rRNA processing (Fig. 3). Second, Prp43p associates with pre-rRNAs (Fig. 4 and 5A). Third, Prp43p associates with all classes of snoRNAs (Fig. 4, 5B, and 5C). Fourth, the prp43-Q423N mutant inhibits 2′-O-methylation at site C2337 in 27S rRNA (Fig. 6). Consistent with the interaction of Prp43p with both small- and large-subunit biogenesis factors (11, 17, 19, 24, 27, 38), our analysis reveals a requirement for Prp43p in both small- and large-subunit biogenesis, indicating a global role for Prp43p in ribosome biogenesis. Stevens and coworkers and Henry and coworkers have similarly discovered a role for Prp43p in the biogenesis of both small and large ribosomal subunits (32a). We conclude that Prp43p plays a dual role in pre-mRNA splicing and ribosome biogenesis. Given the association of hPrp43 with the spliceosome (21, 35, 57) and its residence in the nucleolus (1), this dual role is likely conserved from yeast to humans. These results have important implications for understanding the mechanism of Prp43p function.
Our data confirm and extend the role for Prp43p in splicing. Consistent with previous results (2, 36), we find that the prp43-Q423N mutant impedes degradation of excised introns (Fig. 2C and D). Further, Prp43p binds in vivo to both excised introns and to a U2/U6.U5 spliceosome complex, mostly likely in a postsplicing configuration. The dominant negative Prp43p-Q423N mutant interacts with these species to a greater extent (Fig. 4, 5D, and 5F), suggesting that the mutant stalls on an intron-bound spliceosome. Indeed, we observe a requirement for Prp43p in vitro in dissociating the U3A intron and recycling the activated spliceosome (E. C. Small and J. P. Staley, unpublished observations), consistent with previous findings (2, 36). However, since Prp43p performs a second function in ribosome biogenesis, we cannot currently determine if Prp43p is essential for splicing in vivo.
Prp43p performs a unique and broad role in pre-rRNA processing, functioning in both small- and large-subunit biogenesis. Mutations in almost all known ribosome biogenesis factors lead to defects in the biogenesis of either small- or large-subunit processing (16). In stark contrast, the prp43-Q423N mutation leads to defects in the biogenesis of both subunits, as reflected by the accumulation of 35S pre-rRNA and the reduction in 18S and 25S mature rRNAs, as observed by pulse-chase analysis (Fig. 3B). A broad role for Prp43p is further supported by a steady-state analysis that reveals an accumulation of 35S pre-rRNA and a reduction of 20S and 7S pre-rRNAs (Fig. 3A). In addition, Prp43p associates with both small- and large-subunit precursors, including 35S, 27S, 20S, and 7S pre-rRNAs (Fig. 4 and 5A). To our knowledge, only two other ribosome biogenesis factors, Rrp5p and Rrp12p, clearly function in both small- and large-subunit biogenesis. These factors function distinctly from Prp43p—Rrp5p is required for cleavage at A0, A1, A2 and A3 but not at C2, (15), and Rrp12p is required for the export of both large and small subunits (39). Thus, our data suggest a novel and general role for Prp43p in ribosome biogenesis.
Our data indicate that Prp43p plays a key role at an early stage in ribosome biogenesis. By steady-state and pulse-chase analysis, 35S pre-rRNA accumulates in the mutant and downstream pre-rRNAs decrease (Fig. 3). Further, Prp43p binds 35S pre-rRNA (Fig. 5A and E). Finally, Prp43p associates broadly with snoRNAs (Fig. 4, 5B, 5C, and 5E), which are thought to bind and to modify 35S pre-rRNA either co- or posttranscriptionally (6). These observations could be explained by a requirement for Prp43p in dissociating the snoRNPs from 35S pre-rRNA. Such a function could also be required at later stages of ribosome biogenesis and provide a rationalization for interactions of Prp43p with downstream pre-rRNAs. For example, both box C/D and H/ACA snoRNAs remain associated with 27SA2 pre-rRNA (11), the major intermediate upon which Prp43p-Q423N stalls (Fig. 5A). Moreover, a requirement for Prp43p in promoting recycling of the snoRNAs would be consistent with the 27S hypomethylation defect at C2337 in the prp43-Q423N mutant (Fig. 6). Finally, a role in recycling snoRNPs could extend to include a role in recycling biogenesis factors, thereby rationalizing the association of Prp43p with all pre-rRNAs and mature rRNAs. Such a recycling role would parallel the role of Prp43p in promoting recycling of snRNPs and splicing factors in pre-mRNA splicing.
Prp43p joins an expanding group of DExD/H box proteins that play multiple roles in the cell (8, 47). Prp43p also joins a specialized subset of ribosome biogenesis factors that play a dual role in the cell, providing a link between ribosome biogenesis and other key cellular functions. For example, the ribosome biogenesis factors Yhp1p/Nop7p and Noc3p perform a second role in DNA replication (13, 59). Similarly, a number of proteins play a dual role in ribosome biogenesis and the cell cycle (12). As ribosome biogenesis factors are highly regulated, the dual role for these proteins provides a link to coordinately regulate ribosome biogenesis with DNA replication and the cell cycle (12). The dual role for Prp43p may similarly provide a link to coordinately regulate ribosome biogenesis and pre-mRNA splicing.
Acknowledgments
We thank Scott Stevens, Susan Baserga, and Staley laboratory members for helpful discussions; Angela Hilliker, Hiroshi Maita, Rabiah Mayas, Melissa Mefford, and Joseph Piccirilli for critical reading of the manuscript; Yves Henry and Scott Stevens for sharing results before publication; John Abelson and Kathy Gould for strains and/or plasmids; the Turkewitz laboratory for assistance with tritium; and Channon Jordan, Martha Norman, and Valerie Shaw for technical assistance.
This work was supported by a Canadian Institute for Health Research (CIHR) postdoctoral fellowship to S.L.H., a Canadian Foundation for Innovation grant and a CIHR grant to T.R.H., and an NIH grant (GM62264) and a Packard Foundation Fellowship (19057) to J.P.S.
REFERENCES
- 1.Andersen, J. S., C. E. Lyon, A. H. Fox, A. K. Leung, Y. W. Lam, H. Steen, M. Mann, and A. I. Lamond. 2002. Directed proteomic analysis of the human nucleolus. Curr. Biol. 12:1-11. [DOI] [PubMed] [Google Scholar]
- 2.Arenas, J. E., and J. N. Abelson. 1997. Prp43: an RNA helicase-like factor involved in spliceosome disassembly. Proc. Natl. Acad. Sci. USA 94:11798-11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reference deleted.
- 4.Ausubel, F. M., R. E. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1993. Current protocols in molecular biology, vol. 2. John Wiley & Sons, New York, N.Y.
- 5.Badis, G., M. Fromont-Racine, and A. Jacquier. 2003. A snoRNA that guides the two most conserved pseudouridine modifications within rRNA confers a growth advantage in yeast. RNA 9:771-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertrand, E., and M. J. Fournier. 2004. The snoRNPs and related machines: ancient devices that mediate maturation of rRNA and other RNAs, p. 223-257. In M. O. J. Olson (ed.), The nucleolus. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 7.Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345-346. [DOI] [PubMed] [Google Scholar]
- 8.Burckin, T., R. Nagel, Y. Mandel-Gutfreund, L. Shiue, T. A. Clark, J. L. Chong, T. H. Chang, S. Squazzo, G. Hartzog, and M. Ares, Jr. 2005. Exploring functional relationships between components of the gene expression machinery. Nat. Struct. Mol. Biol. 12:175-182. [DOI] [PubMed] [Google Scholar]
- 9.Chen, J. Y., L. Stands, J. P. Staley, R. R. Jackups, Jr., L. J. Latus, and T. H. Chang. 2001. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol. Cell 7:227-232. [DOI] [PubMed] [Google Scholar]
- 10.de la Cruz, J., D. Kressler, and P. Linder. 2004. Ribosomal subunit assembly, p. 258-285. In M. O. J. Olson (ed.), The nucleolus. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 11.Dez, C., C. Froment, J. Noaillac-Depeyre, B. Monsarrat, M. Caizergues-Ferrer, and Y. Henry. 2004. Npa1p, a component of very early pre-60S ribosomal particles, associates with a subset of small nucleolar RNPs required for peptidyl transferase center modification. Mol. Cell. Biol. 24:6324-6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dez, C., and D. Tollervey. 2004. Ribosome synthesis meets the cell cycle. Curr. Opin. Microbiol. 7:631-637. [DOI] [PubMed] [Google Scholar]
- 13.Du, Y. C., and B. Stillman. 2002. Yph1p, an ORC-interacting protein: potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell 109:835-848. [DOI] [PubMed] [Google Scholar]
- 14.Edwalds-Gilbert, G., D. H. Kim, E. Silverman, and R. J. Lin. 2004. Definition of a spliceosome interaction domain in yeast Prp2 ATPase. RNA 10:210-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eppens, N. A., S. Rensen, S. Granneman, H. A. Raué, and J. Venema. 1999. The roles of Rrp5p in the synthesis of yeast 18S and 5.8S rRNA can be functionally and physically separated. RNA 5:779-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313-318. [DOI] [PubMed] [Google Scholar]
- 17.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 18.Ghaemmaghami, S., W. K. Huh, K. Bower, R. W. Howson, A. Belle, N. Dephoure, E. K. O'Shea, and J. S. Weissman. 2003. Global analysis of protein expression in yeast. Nature 425:737-741. [DOI] [PubMed] [Google Scholar]
- 19.Grandi, P., V. Rybin, J. Bassler, E. Petfalski, D. Strauss, M. Marzioch, T. Schafer, B. Kuster, H. Tschochner, D. Tollervey, A. C. Gavin, and E. Hurt. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10:105-115. [DOI] [PubMed] [Google Scholar]
- 20.Granneman, S., and S. J. Baserga. 2004. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res. 296:43-50. [DOI] [PubMed] [Google Scholar]
- 21.Hartmuth, K., H. Urlaub, H. P. Vornlocher, C. L. Will, M. Gentzel, M. Wilm, and R. Lührmann. 2002. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl. Acad. Sci. USA 99:16719-16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herskowitz, I. 1987. Functional inactivation of genes by dominant negative mutations. Nature 329:219-222. [DOI] [PubMed] [Google Scholar]
- 23.Hiley, S. L., J. Jackman, T. Babak, M. Trochesset, Q. D. Morris, E. Phizicky, and T. R. Hughes. 2005. Detection and discovery of RNA modifications using microarrays. Nucleic Acids Res. 33:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 25.Hughes, T. R., M. Mao, A. R. Jones, J. Burchard, M. J. Marton, K. W. Shannon, S. M. Lefkowitz, M. Ziman, J. M. Schelter, M. R. Meyer, S. Kobayashi, C. Davis, H. Dai, Y. D. He, S. B. Stephaniants, G. Cavet, W. L. Walker, A. West, E. Coffey, D. D. Shoemaker, R. Stoughton, A. P. Blanchard, S. H. Friend, and P. S. Linsley. 2001. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 19:342-347. [DOI] [PubMed] [Google Scholar]
- 26.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 27.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.James, S. A., W. Turner, and B. Schwer. 2002. How Slu7 and Prp18 cooperate in the second step of yeast pre-mRNA splicing. RNA 8:1068-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jurica, M. S., and M. J. Moore. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12:5-14. [DOI] [PubMed] [Google Scholar]
- 30.Kim, J. L., K. A. Morgenstern, J. P. Griffith, M. D. Dwyer, J. A. Thomson, M. A. Murcko, C. Lin, and P. R. Caron. 1998. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure 6:89-100. [DOI] [PubMed] [Google Scholar]
- 31.Kistler, A. L., and C. Guthrie. 2001. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes Dev. 15:42-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krogan, N. J., W. T. Peng, G. Cagney, M. D. Robinson, R. Haw, G. Zhong, X. Guo, X. Zhang, V. Canadien, D. P. Richards, B. K. Beattie, A. Lalev, W. Zhang, A. P. Davierwala, S. Mnaimneh, A. Starostine, A. P. Tikuisis, J. Grigull, N. Datta, J. E. Bray, T. R. Hughes, A. Emili, and J. F. Greenblatt. 2004. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell 13:225-239. [DOI] [PubMed] [Google Scholar]
- 32a.Lebaron, S., C. Froment, M. Froment-Racine, J. C. Rain, B. Monsarrat, M. Caizegues-Ferrer, and Y. Henry. 2005. The splicing ATPase Prp43p is a component of multiple preribosomal particles. Mol. Cell. Biol. 25:9269-9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, I., S. V. Date, A. T. Adai, and E. M. Marcotte. 2004. A probabilistic functional network of yeast genes. Science 306:1555-1558. [DOI] [PubMed] [Google Scholar]
- 34.Lowe, T. M., and S. R. Eddy. 1999. A computational screen for methylation guide snoRNAs in yeast. Science 283:1168-1171. [DOI] [PubMed] [Google Scholar]
- 35.Makarov, E. M., O. V. Makarova, H. Urlaub, M. Gentzel, C. L. Will, M. Wilm, and R. Lührmann. 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298:2205-2208. [DOI] [PubMed] [Google Scholar]
- 36.Martin, A., S. Schneider, and B. Schwer. 2002. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 277:17743-17750. [DOI] [PubMed] [Google Scholar]
- 37.Mohr, S., J. M. Stryker, and A. M. Lambowitz. 2002. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell 109:769-779. [DOI] [PubMed] [Google Scholar]
- 38.Nissan, T. A., J. Bassler, E. Petfalski, D. Tollervey, and E. Hurt. 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 15:5539-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oeffinger, M., M. Dlakic, and D. Tollervey. 2004. A pre-ribosome-associated HEAT-repeat protein is required for export of both ribosomal subunits. Genes Dev. 18:196-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng, W. T., M. D. Robinson, S. Mnaimneh, N. J. Krogan, G. Cagney, Q. Morris, A. P. Davierwala, J. Grigull, X. Yang, W. Zhang, N. Mitsakakis, O. W. Ryan, N. Datta, V. Jojic, C. Pal, V. Canadien, D. Richards, B. Beattie, L. F. Wu, S. J. Altschuler, S. Roweis, B. J. Frey, A. Emili, J. F. Greenblatt, and T. R. Hughes. 2003. A panoramic view of yeast noncoding RNA processing. Cell 113:919-933. [DOI] [PubMed] [Google Scholar]
- 41.Raué, H. A. 2004. Pre-ribosomal RNA processing and assembly in Saccharomyces cerevisiae: the machine that makes the machine, p. 199-222. In M. O. J. Olson (ed.), The nucleolus. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 42.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Séraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 43.Rocak, S., and P. Linder. 2004. DEAD-box proteins: the driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 5:232-241. [DOI] [PubMed] [Google Scholar]
- 44.Samarsky, D. A., and M. J. Fournier. 1999. A comprehensive database for the small nucleolar RNAs from Saccharomyces cerevisiae. Nucleic Acids Res. 27:161-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schattner, P., W. A. Decatur, C. A. Davis, M. Ares, Jr., M. J. Fournier, and T. M. Lowe. 2004. Genome-wide searching for pseudouridylation guide snoRNAs: analysis of the Saccharomyces cerevisiae genome. Nucleic Acids Res. 32:4281-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwer, B., and C. H. Gross. 1998. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 17:2086-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silverman, E., G. Edwalds-Gilbert, and R. J. Lin. 2003. DExD/H-box proteins and their partners: helping RNA helicases unwind. Gene 312:1-16. [DOI] [PubMed] [Google Scholar]
- 48.Staley, J. P., and C. Guthrie. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92:315-326. [DOI] [PubMed] [Google Scholar]
- 49.Staley, J. P., and C. Guthrie. 1999. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell 3:55-64. [DOI] [PubMed] [Google Scholar]
- 50.Stuart, J. M., E. Segal, D. Koller, and S. K. Kim. 2003. A gene-coexpression network for global discovery of conserved genetic modules. Science 302:249-255. [DOI] [PubMed] [Google Scholar]
- 51.Tasto, J. J., R. H. Carnahan, W. H. McDonald, and K. L. Gould. 2001. Vectors and gene targeting modules for tandem affinity purification in Schizosaccharomyces pombe. Yeast 18:657-662. [DOI] [PubMed] [Google Scholar]
- 52.Torchet, C., G. Badis, F. Devaux, G. Costanzo, M. Werner, and A. Jacquier. 2005. The complete set of H/ACA snoRNAs that guide rRNA pseudouridylations in Saccharomyces cerevisiae. RNA 11:928-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33:261-311. [DOI] [PubMed] [Google Scholar]
- 54.Wagner, J. D., E. Jankowsky, M. Company, A. M. Pyle, and J. N. Abelson. 1998. The DEAH-box protein Prp22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. EMBO J. 17:2926-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, Y., and C. Guthrie. 1998. PRP16, a DEAH-box RNA helicase, is recruited to the spliceosome primarily via its nonconserved N-terminal domain. RNA 4:1216-1229. [PMC free article] [PubMed] [Google Scholar]
- 56.Watkins, N. J., V. Segault, B. Charpentier, S. Nottrott, P. Fabrizio, A. Bachi, M. Wilm, M. Rosbash, C. Branlant, and R. Lührmann. 2000. A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103:457-466. [DOI] [PubMed] [Google Scholar]
- 57.Will, C. L., H. Urlaub, T. Achsel, M. Gentzel, M. Wilm, and R. Lührmann. 2002. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 21:4978-4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu, Y. T., M. D. Shu, and J. A. Steitz. 1997. A new method for detecting sites of 2′-O-methylation in RNA molecules. RNA 3:324-331. [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, Y., Z. Yu, X. Fu, and C. Liang. 2002. Noc3p, a bHLH protein, plays an integral role in the initiation of DNA replication in budding yeast. Cell 109:849-860. [DOI] [PubMed] [Google Scholar]