Abstract
Doublesex (dsx) is a transcription factor in Drosophila that regulates somatic sexual differentiation. Male- and female-specific splicing isoforms of DSX share a novel DNA-binding domain, designated the DM motif. Broadly conserved among metazoan sex-determining factors, the DM domain contains a nonclassical zinc module and binds in the DNA minor groove. Here, we characterize the DM motif by site-directed and random mutagenesis using a yeast one-hybrid (Y1H) system and extend this analysis by chemogenetic complementation in vitro. The Y1H system is based on a sex-specific Drosophila enhancer element and validated through studies of intersexual dsx mutations. We demonstrate that the eight motif-specific histidines and cysteines engaged in zinc coordination are each critical and cannot be interchanged; folding also requires conserved aliphatic side chains in the hydrophobic core. Mutations that impair DNA binding tend to occur at conserved positions, whereas neutral substitutions occur at nonconserved sites. Evidence for a specific salt bridge between a conserved lysine and the DNA backbone is obtained through the synthesis of nonstandard protein and DNA analogs. Together, these results provide molecular links between the structure of the DM domain and its function in the regulation of sexual dimorphism.
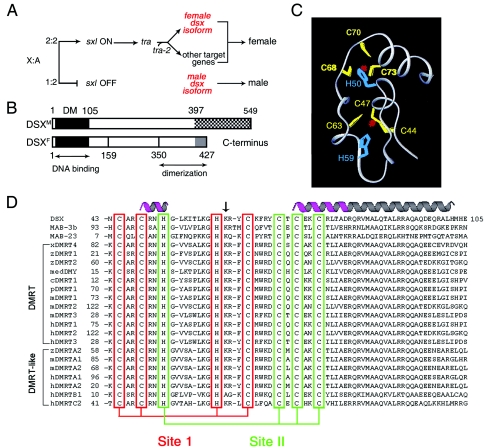
Sexual dimorphism is regulated by diverse molecular mechanisms (18). In mammals, male development is initiated by a gene on the Y chromosome that is designated Sry (sex-determining region of the Y chromosome) (7, 30, 41, 42). Alternative genetic mechanisms operate in Drosophila melanogaster (and Caenorhabditis elegans), wherein sex is determined by the X-to-autosome ratio, a process linked to X-dosage compensation (Fig. 1A) (19, 59). The sex-determining hierarchy of Drosophila defines an RNA-splicing cascade through the actions of Sex Lethal (SXL) and the TRA factors (18). Doublesex (dsx) functions downstream of this cascade to define one branch of a ramifying pathway (Fig. 1A) (29, 59). The sex-specific isoforms of DSX (DSXM and DSXF; Fig. 1B) regulate most aspects of somatic sexual dimorphism in D. melanogaster. These isoforms share an N-terminal DNA-binding domain, designated the DM motif (Fig. 1C). In this article, we present a mutational analysis of the DSX DM domain to identify key determinants of folding and DNA recognition.
FIG. 1.
Sex-determining hierarchy of D. melanogaster and DM motif. (A) X-to-autosome ratio regulates a sex-specific RNA-splicing cascade (50). Not shown are Intersex (IX), a putative transcriptional coactivator that acts with DSXF to promote female development, and a divergent fruitless branch downstream of tra factors. (B) Domain organization of DSX isoforms N-terminal DM and C-terminal dimerization domains. DSXM and DSXF differ at the extreme C terminus (cross-hatched and gray regions). (C) Ribbon model of DM domain with intertwined CCHC and HCCC Zn2+-binding sites (DSX residues 41 to 81) (68). (D) Metazoan DM sequences. Cysteines and histidines that coordinate Zn2+ are aligned (red and green boxes). Stable α-helical elements are highlighted in magenta; the nascent helical tail is shown in gray. The arrow highlights residue K60, conserved on the surface of the Zn module and proposed to contact DNA.
The Wilkins-Wolf hypothesis posits that sex-determining pathways have evolved in a reverse order from bottom (most broadly conserved) to top (least conserved) (65, 66). This viewpoint is supported by the ubiquity of DM genes among metazoan sex-determining pathways (35, 59). Such genes encode a newly recognized class of transcription factors (for a review, see reference 59). DM sequences are broadly conserved in metazoans, including nematodes, fish, reptiles, birds, and mammals (Fig. 1D). The deletion of human DM genes on chromosome 9p is associated with intersexual development (6, 27, 31, 49, 55, 59, 63). Although originally described in relation to sex-determining pathways (67), an extended family of DM genes is proposed to function in general aspects of developmental patterning in vertebrates (35).
The DSX DM domain, a prototype for this class of DNA-binding motifs, binds as a dimer to a pseudopalindromic DNA site (13, 26, 68). DSXM and DSXF contain identical DM domains (25) and exhibit similar DNA-binding properties (15, 17). Minor-groove targeting of the DSX DM domain is proposed to allow simultaneous binding of major-groove factors to composite DNA enhancer elements (68). A paradigm is provided by the well-characterized fat body enhancer (fbe), which regulates the tissue- and sex-specific expression of yolk proteins (3, 4). The fbe contains overlapping DNA target sites for DSX and a bZIP transcription factor (3). Genetic interactions between dsx and classical homeotic pathways have also been described previously (1, 12). Although biochemical mechanisms underlying such interactions have not been characterized, these regulatory relationships raise the possibility of coordinate DNA recognition by DSX and homeodomains. The DSX isoforms may thus function not only as terminal differentiation factors, but also as integrators of sex-, lineage-, and position-specific signals in development (11).
The structure of the DSX DM domain contains two zinc ions and so defines a distinct class of nonclassical zinc fingers (Protein Data Bank code 1LPV) (12, 25). Nuclear magnetic resonance studies of the free domain have demonstrated that the zinc ions are coordinated within a single hydrophobic core (Fig. 1C). Tetrahedral coordination is effected in each metal-binding site by three cysteine residues and one histidine (Fig. 1D, sites I and II). These eight ligands, invariant among DM sequences, are intertwined in the sequence of the protein. As expected in a globular protein, other positions also exhibit conserved sequence preferences, presumably due to structural requirements of folding and/or DNA binding. The Zn module is extended by a flexible C-terminal tail, which forms an α-helix on DNA binding (52, 68). Mutations in dsx that are associated with intersexual development have been found in both the Zn module and tail (25), suggesting a bipartite mechanism of DNA recognition. This model is supported by observations that truncation or mutation of the tail impairs specific DNA binding but not Zn-dependent folding (52). Although the structure of a DM-DNA complex has not been determined, minor-groove targeting has been established through studies of DNA analogs (68). Unlike SRY and related HMG boxes (8, 28), binding of DSX does not induce sharp DNA bending (68).
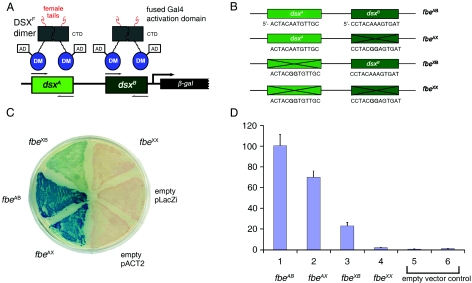
In this article, we investigate sequence determinants of metal-dependent folding and DNA recognition by the DSX DM domain. To enable efficient mutational analysis, a yeast one-hybrid (Y1H) system (47, 54) is designed based on prior molecular-genetic analysis of yolk protein 1 (yp1) (2-4) gene expression and validated through control studies of intersexual mutations in dsx (25). The expression of a reporter gene (lacZ encoding β-galactosidase) is regulated by two DSX-binding sites derived from the fbe gene of Drosophila: transcription is activated on binding of a fusion protein containing DSX and a Saccharomyces cerevisiae transcriptional activation domain (AD) (Fig. 2A). Random and site-directed mutations in the DM domain are characterized in relation to reporter gene expression. A correlation between β-galactosidase expression and specific DNA affinity is established through studies of alanine-scanning variants that were previously characterized in vitro (52). Although exceptions occur, a trend is observed wherein mutations that block transcriptional activation (and so presumably specific DNA binding) occur at conserved sites, whereas neutral substitutions occur at nonconserved sites. The stringent conservation of the eight motif-specific cysteines and histidines is highlighted by the inactivity of “interchange” variants containing Cys→His or His→Cys substitutions. Analysis of allowed and disallowed substitutions in the DM domain suggests possible sites of protein-DNA interaction. Chemical evidence for the function of one such site, a conserved lysine on the surface of the Zn module (K60) (Fig. 1D, arrow), is obtained by an in vitro complementation test: replacement of a putative salt bridge to the DNA backbone by a novel “hydrophobic bridge” between a variant domain and a nonstandard DNA analog containing a neutral phosphodiester modification (10, 44). Together, the dual use of mutational and chemogenetic approaches suggests a general strategy for the characterization of nucleic-acid-binding domains. Application to DSX provides molecular links between the structure of the DM domain and its function in the regulation of sexual dimorphism.
FIG. 2.
Design and validation of Y1H system. (A) Specific DSX-DNA recognition regulates the expression of lacZ via fusion protein containing an N-terminal Gal4 AD and a C-terminal DSX domain. A 48-bp Drosophila promoter fragment derived from fbe was inserted upstream of lacZ to provide DSX-binding sites dsxA and dsxB. CTD, C-terminal dimerization domain of DSX. (B) Wild-type DNA-binding sites and inactive variants fbeAB, containing two wild-type sites and control elements (respectively designated fbeAX, fbeXB, and fbeXX) in which one, the other, or both sites are inactivated by twin A→G transitions at center of target sites (bold). (C) Colonies on an X-gal indicator plate in the presence of wild-type or variant enhancer elements. (D) Histogram describing β-galactosidase activity in the presence of wild-type or variant DNA sites (columns 1 to 4), empty reporter vector (column 5) or reporter (column 6) plasmid lacking fbe sites.
MATERIALS AND METHODS
Y1H reporter plasmid.
DSX-binding sites were synthesized by PCR using the template 5′-GCACAACTACAATGTTGCAATCAGCGGAGCCTACAAAGTGATTACAAA-3′ (bold indicates core binding sites). The following oligonucleotides were used as PCR primers (restriction sites for EcoRI or XhoI are underlined): YK-f, 5′-CTGAATTCGCACAACT-3′, and YK-r, 5′-CTCCGCTCGAGTTTGTAAT-3′. Amplifications were performed as follows: denaturation (5 min, 95°C), cycling profile (95°C, 30 s; 55°C, 30 s; 72°C, 10 s; 35 cycles), and final extension (2 min at 72°C). Products were inserted into pLacZi digested with compatible pairs of restriction enzymes to create reporter pLacZi-fbeAB. Three control reporter plasmids were made using the QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) to disrupt one or both binding sites (Fig. 2B). Constructions were verified by DNA sequencing.
DSX deletions and mutations.
Deletions (Fig. 3A) were introduced by PCR and subcloned into pACT2 via SmaI and EcoRI sites. Substitutions were introduced by PCR-based two-stage overlap extension mutagenesis. A polyalanine variant was generated by PCR using a specific 3′ primer encoding eight alanines (residues 98 to 105). Coding regions were sequenced in each case to confirm mutations and exclude PCR errors.
FIG. 3.
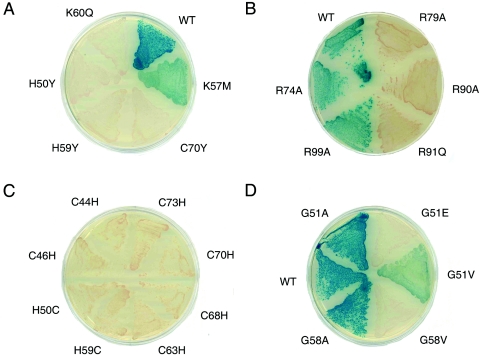
Y1H analysis of variant DM domains. (A) Intersexual mutations H50Y, H59Y, and C70Y inactivate DSX as does K60Q. K57M is light blue, which is consistent with partial activity of isosteric norleucine analog (52). (B) Alanine substitutions previously shown to significantly impair specific DNA binding (R79A and R90A) (52) impair β-galactosidase expression (white colonies), whereas substitutions compatible with high-affinity DNA binding (R74A and R99A) are associated with wild-type β-galactosidase expression (blue). Intersexual mutation R91Q (25) is white. (C) Interchange of histidine and cysteine within the metal-binding sites in each case impairs β-galactosidase expression (white colonies). (D) Variable phenotypes G51A and G58A (blue), G51V (light blue), and G51E and G58V (white). G51 and G58 participate in turns.
Y1H screening.
Random mutations were generated by error-prone PCR, generating a 354-bp DNA fragment spanning the DSX DM domain. The mutation frequency was ∼5 per 1,000 bp. The protocol employed 30 cycles of amplification with Mutazyme II DNA polymerase (Stratagene, La Jolla, CA); four independent reactions were combined to limit founder bias. Fragments were inserted into pACT2 at SmaI and EcoRI sites. Ligated products were transformed into Escherichia coli XL10-Gold ultracompetent cells (Stratagene, La Jolla, CA). Approximately 1 × 105 independent clones were obtained, and plasmids were prepared using an EndoFree plasmid maxi kit (QIAGEN, Valencia, CA). The resulting library encoded fusion proteins with N-terminal Gal4 AD and C-terminal DSX fragments. Yeast strain YM4271 was cotransformed with pLacZi-fbeAB (lacZ reporter vector), and the plasmid library was transformed by the lithium acetate-polyethylene glycol method. Cells were plated on synthetic dropout-Leu-Ura minimal medium supplemented with 80 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-gal). Plates were incubated at 30°C for 7 days. DM mutations were recovered from white colonies by PCR and sequenced. A second plasmid library with a higher mutational frequency (ca. 15 per 1,000 bp) was likewise prepared and utilized for sequencing DM variants from blue colonies.
Yeast colony PCR.
DNA was prepared by boiling (48). A colony was picked with a sterile toothpick and rinsed with 10 μl of an incubation solution (1.2 M sorbitol, 100 mM sodium phosphate [pH 7.4], and 1.5 units/μl Zymolyase; Zymo Research, Orange, CA). The mixture was incubated at 37°C for 15 min and boiled for 3 min; a total of 5 μl was used in a 100-μl PCR. The DM sequence was amplified as a ca. 500-bp fragment using the Expand high-fidelity PCR system (Roche Applied Sciences, Indianapolis, IN). Fragments were separated on 1.2% agarose gels and purified using the QIAquick gel extraction kit (QIAGEN, Valencia, CA).
β-Galactosidase assay.
The MATCHMAKER 1H system (BD Clontech, Palo Alto, CA) was used to analyze lacZ expression. Strain YM4271 and an o-nitrophenyl-β-d-galactose assay were used to measure β-galactosidase activity (54). Cells were grown in synthetic dropout-Leu-Ura selective medium to maintain plasmids. Data represent means (plus or minus standard deviation) of triplicate determinations in Miller's β-galactosidase units. One unit is defined as the amount that hydrolyzes 1 μmol of o-nitrophenyl-β-d-galactose to o-nitrophenol and d-galactose per min per cell.
Methylphosphonate interference.
Methylphosphonate interference (10, 53) was evaluated by gel mobility-shift assay (GMSA) in a 29-bp DNA duplex (5′-GTGCACAACTACAATGTTGCAATCAGCGG-3′ and its complement; bold indicates the dsxA site). Possible stereospecific interference at selected sites was evaluated following high-performance liquid chromatography (HPLC) resolution of single-strand diastereomers (44). DNA-binding reactions were conducted in 20 mM Tris-HCl (pH 7.4), 150 mM KCl, 5 mM MgCl2, 0.1 mM ZnCl2, 5% glycerol, 33 μg/ml bovine serum albumin, and 0.08 μg/μl poly(dI-dC) competitor DNA. The concentration of 33P-labeled DNA was <1.5 nM. Gels (10% acrylamide with 29:1 bis-acrylamide) were run in 45 mM Tris-borate (pH 8.0). Quantification was obtained using a PhosphorImager (Amersham Biosciences, Piscataway, NJ). Dissociation constants in the limit of strongly cooperative binding were estimated as described previously (52).
Chemical synthesis of DSX DM domains.
DM domains were prepared by solid-phase peptide synthesis and native fragment ligation (20, 68). Ligation products were purified by reverse-phase HPLC as described previously (52). Purity was >98% as assessed by analytical reverse-phase HPLC; fidelity of synthesis was verified in each case by mass spectrometry.
Molecular modeling.
Structures were visualized using InsightII (Biosym, Inc., San Diego, CA) implemented in a Silicon Graphics workstation.
RESULTS
Our results are presented in three parts. In the first part, we describe a Y1H system for screening of mutations in DSX. The design is based on molecular-genetic studies of a well-characterized DSX target gene in D. melanogaster (3, 4). Validation of this system is provided by control studies of variant promoter elements (26) and DSX mutations associated with intersexual development (25, 26). In the second part, we utilize the Y1H system to investigate structure-function relationships through deletion analysis, site-directed substitutions, and random mutagenesis. The third part focuses on a potential DNA-binding residue on the surface of the DM domain. Evidence for a salt bridge between a conserved lysine and a specific DNA phosphodiester linkage is obtained by a chemogenetic strategy.
DSX is a modular protein containing an N-terminal DNA-binding domain (the DM motif; residues 39 to 105) and a C-terminal dimerization domain (residues 350 to 412) (2, 5). Although monomeric in solution, the isolated DM domain binds as a dimer to specific DNA target sites (68). Such binding is strengthened by strong C-terminal dimer contacts (16). An AD-DM fusion protein exhibits significant Y1H activity in accord with its autonomous high-affinity DNA-binding activity (15, 52, 68). Because its expression is less toxic to Saccharomyces cerevisiae than was an intact AD-DSXF fusion protein, the AD-DM construct is employed as a template for mutagenesis. The C-terminal domain of DSX is not conserved among vertebrate DM transcription factors.
Design and characterization of a DSX-regulated Y1H system.
DSX isoforms regulate the sex- and tissue-specific expression of yp1 via the well-characterized fbe (3, 4). A Y1H reporter plasmid was therefore constructed using a 48-bp enhancer element containing two DSX-binding sites (designated dsxA and dsxB) (Fig. 2A) placed upstream of lacZ. To effect transcriptional regulation in S. cerevisiae, DSXF or fragments containing the DM domain (residues 1 to 118) were subcloned into expression vector pACT2. The resulting constructs produced fusion proteins consisting of N-terminal Gal4 AD and C-terminal DSX sequences (Fig. 2A). The Y1H system yields DSX-dependent expression of β-galactosidase (as monitored by X-gal indicator plates and as measured in extracts) relative to empty vector controls. The fidelity of the Y1H system was validated by the control studies described below. Accompanying figures contain photomicrographs of yeast colonies, wherein the extent of β-galactosidase activity is indicated by a blue indicator dye.
DNA variants that impair specific DSX binding block lacZ expression.
Three variant enhancer elements were constructed to impair DSX binding (Fig. 2B). Paired G→A transitions were thus introduced at the centers of dsxA and/or dsxB based on previous analysis of the sequence specificity of the DSX DM domain (25, 68). Mutations in either DNA site (dsxA or dsxB) impair DSX-dependent expression of lacZ, whereas mutations in both sites essentially eliminate expression (Fig. 2C and D).
Mutations that perturb metal-dependent protein folding block lacZ expression.
Previous studies have demonstrated that the specific DNA-binding activity of DSX requires zinc ions and is blocked by mutations in the invariant cysteines and histidines (25, 68). Three such mutations (H50Y, H59Y, and C70Y) are associated with intersexual phenotype, suggesting that folding of the Zn module is required for sex-specific gene regulation (25). These substitutions block detectable β-galactosidase activity (Fig. 3A). Western blots established that such decrements are not a result of impaired expression of the variant fusion proteins.
DM mutations that impair specific DNA binding block lacZ expression.
An intersexual mutation in the C-terminal tail of the DM domain (R91Q) permits Zn-dependent folding but impairs specific DNA binding by at least 100-fold (25, 68). The corresponding substitution in the AD-DM fusion protein leads to loss of detectable β-galactosidase activity (Fig. 3B). Further, a broad correlation between the extent of impaired DNA binding and Y1H transcriptional activation is obtained by analysis of a set of Ala substitutions previously characterized in the tail (52). Whereas R74A and R99A affect neither specific DNA binding nor Y1H activity, for example, substitutions R79A and R90A exhibit significant impairment in both assays (Fig. 3B). These control studies suggest that the Y1H assay correlates with relative specific DNA-binding activities. Mutations associated with white colonies impair specific DNA binding by at least fivefold and/or cause a commensurate decrease in the folding or stability of the fusion protein.
Deletion analysis and mutagenesis.
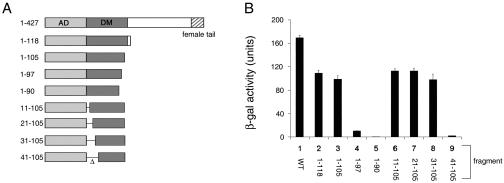
Segments of DSXF that are necessary for Y1H activity were mapped by deletion analysis, defining N- and C-terminal boundaries of a minimal DNA-binding domain (Fig. 4A). C-terminal deletion (yielding DM fragment 1-97) leads to a >10-fold reduction in activity. The deletion of seven additional residues (yielding DM fragment 1-90) leads to loss of detectable lacZ expression (Fig. 4B). By contrast, β-galactosidase activity is not affected following the deletion of the N-terminal 30 residues (constructs 11 to 105, 21 to 105, and 31 to 105 in Fig. 4A). The deletion of residues 1 to 40 abolishes expression of β-galactosidase (Fig. 4B). These results are in accord with the extent of sequence conservation among DM-related genes. The polypeptide segment immediately N terminal to the first zinc ligand (C44) is presumably required to stabilize the Zn module.
FIG. 4.
Deletion analysis of DM domain. (A) Set of DSX fragments of different lengths employed in AD fusion proteins (Fig. 2A). The DM domain, dimerization domain, and C-terminal sex-specific tail are shown in schematic form. (B) Corresponding Y1H activities of fusion proteins quantified by liquid β-galactoside assays.
Invariance of motif-specific cysteines and histidines.
In the classical Zn finger, the last His can sometimes be functionally substituted by other residues (61). Whereas the Zn finger contains a single CCHH metal-binding site, the DSX DM domain is composed of intertwined CCHC and HCCC modules. To investigate the interchangeability of cysteine and histidine in the DM domain, we constructed eight variants: C44H, C47H, H50C, H59C, C63H, C68H, C70H, and C73H. Each interchange yielded white colonies, indicating perturbed folding and/or specific DNA binding (Fig. 3C). One substitution (C63H) is associated with white colonies of smaller size, implying that the C63H AD-DM fusion protein is deleterious to growth, perhaps due to toxic misfolding. The function of the DM motif thus requires a specific pattern of cysteines and histidines in accord with its broad sequence invariance. These results do not exclude the possibility that His→Cys or Cys→His substitutions can be compensated by mutations elsewhere in the core, leading to an alternative scheme.
Random mutagenesis of the DM domain.
To determine the importance of individual residues, PCR-based random mutagenesis was employed to generate libraries of DM variants. These enabled identification of disallowed sequences, mutations with impaired function associated with white colonies, and allowed sequences, substitutions compatible with native Y1H function. Occasional light-blue colonies were observed (S9C, G51V, K57M, K57A, K57N, Y62A, L75P, T76K, and R81G), presumably corresponding to partial impairment of expression, folding, or DNA binding. The partial activity of K57M is in accord with prior GMSA studies of a related variant (52). Representative white, blue, and light blue variant strains are illustrated in Fig. 3D. A total of 200 white colonies were analyzed by DNA sequencing. Among these, ca. 40% yielded single amino acid substitutions; the remainder either had a stop codon (leading to truncated fragments) or contained two or more mutations. The latter were reconstructed to test the individual mutations. Certain mutations were obtained multiple times, presumably reflecting founder bias in initial PCR cycles. In total, 45 loss-of-function mutations were identified at 26 positions (Fig. 5) spanning the internal Zn-binding sites, the surface of the Zn module, and the tail. Because PCR mutagenesis is effectively limited to single-base-pair changes within codons, our mutational screen is likely to be far from saturation.
FIG. 5.
Summary of allowed and disallowed substitutions. Mutations that are highlighted in red impair reporter expression, whereas substitutions that are shown in blue are well tolerated. Azure indicates light-blue colonies and hence partial impairment. For residues in ordered portion of structure (residues 41 to 86), mutations at solvent-exposed sites are listed above the DM sequence, whereas those below the DM sequence indicate buried sites (side chain solvent accessibility, <40%). The polyalanine variant (residues 98 to 105) is indicated by a black bar.
The screen for allowed substitutions employed a library with higher mutational frequency to reduce wild-type background. A total of 200 blue colonies were sequenced, among which ca. 35% yielded single amino acid substitutions. Sequences containing two substitutions where the possibility of intragenic complementation appears low are included in Fig. 5. In addition to the PCR-generated sets of random variants, codons 1 to 8 were each substituted by degenerate oligonucleotides; substitutions at these and other N-terminal sites are well tolerated. Together, these approaches led to the identification of 131 allowed substitutions (Fig. 5). As among loss-of-function mutations, these included sites within the core, at the surface, and in the tail. In some cases, allowed and disallowed substitutions are identified at the same position. Not surprisingly, loss-of-function mutations tend to occur at sites conserved among DM sequences (Fig. 1D). Three findings nonetheless seem anomalous: (i) although conserved glycines G51 and G58 exhibit D-like Ramachandran conformations ordinarily unfavorable to l-amino acids (the [φ, ψ] angles of G51 and G58 are 114 ± 3, 32 ± 3 and 49 ± 2, 58 ± 5, respectively [66]), substitutions G51A and G58A are well tolerated (Fig. 3D); (ii) a conserved aromatic side chain that packs between metal-binding sites I and II (F65; residue 27 of the DM consensus) may functionally be substituted by nonaromatic side chains of variable shape and size (Ala, Val, Cys, or Lys); and (iii) although the deletion of residues 98 to 105 impairs β-galactosidase activity by 10-fold, diverse amino-acid substitutions in this segment are allowed. To resolve this apparent discrepancy, block replacement of the distal tail with polyalanine (i.e., eight successive Ala substitutions) was constructed (Fig. 5, bar). This variant exhibits unimpaired Y1H activity. These results suggest that the length of the distal tail, but not its sequence, contributes to the stability of the specific protein-DNA complex. Implications of these and other mutations for structure-function relationships are considered in the Discussion.
Previous studies have demonstrated that the substitution of certain basic side chains on the surface of the DM domain by norleucine (herein designated Z), an isostere of methionine, can cause a range of decrements in specific DNA binding: substitutions K57Z and K60Z impair specific DNA binding by 1.5- and 10-fold, respectively. In the Y1H indicator system, corresponding mutation K57M is light blue (Fig. 3A), indicating substantial but incomplete activation of reporter expression. Variant K60M is inactive in accord with the impaired DNA binding of the K60Z analog. Mutations K60Q and K60H likewise lead to white colonies (Fig. 3A). By contrast, K60R is well tolerated, suggesting that a positive charge at this site is required for high-affinity DNA recognition. Positions of K60 and other surface side chains are shown in Fig. 6; sites of inactivating mutations are indicated. We suggest that these surfaces either contact DNA or mediate DNA-dependent dimerization of the DM domain. Since K60Z does not perturb cooperativity (51), K60 is a likely DNA contact site.
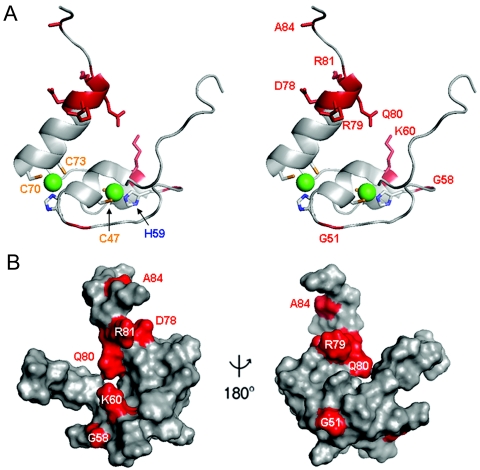
FIG. 6.
Surface of DSX DM domain. (A) Ribbon model of DM domain (residues 35 to 86, stereo pair). Cys and His side chains are shown; Zn2+ atoms are shown as green spheres. The proximal portion of the C-terminal tail is shown at the top. Mutations presumed to impair specific DNA binding are highlighted in red. (B) Corresponding molecular surface shown with unaffected DNA binding (light gray) and decreased DNA binding (red). Left and right panels are related by a 180o rotation of the vertical axis.
Analysis of a candidate protein-DNA contact by methylphosphonate complementation.
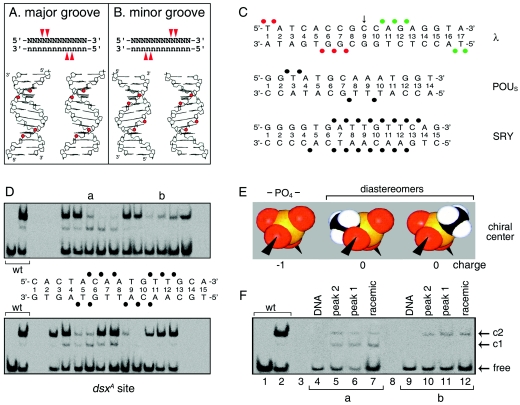
Genetic evidence for base-specific contacts can be obtained through the isolation of mutations conferring altered or relaxed specificity (21, 34). A powerful approach (described as chemogenetics) (60) likewise exploits base analogs to modify individual donors or acceptors in the major groove. Although the minor groove of DNA lacks the distinctive “signposts” of hydrogen-bond donors and acceptors characteristic of the major groove, minor-groove-specific DNA-binding proteins often exhibit a motif-specific pattern of salt bridges to the DNA backbone. We thus sought to generalize chemogenetics to screen variant DNA sites containing DNA backbone modifications. Binding of small globular domains to one face of canonical B DNA is associated with near mirror-image backbone footprints, depending on whether such binding occurs in the major or minor DNA groove (Fig. 7A and B). Classical examples of major-groove footprints are provided by λ repressor and the POU domain (Fig. 7C) (10, 36, 37).
FIG. 7.
Methylphosphonate footprint of DSX DM domain. (A and B) Classical backbone footprints of major- and minor-groove DNA-binding proteins. Binding of a small globular domain to one face of B DNA yields a staggered pattern of phosphate contacts (red arrowheads) whose orientation depends on which groove is occupied (14, 68). (C) Patterns of protein contacts to DNA phosphodiester groups; sites of interference are depicted as filled circles. Major-groove patterns are observed in λ repressor (top) (36, 37) and human Oct-2 POUS domains (middle) (10). The SRY HMG box (bottom) exhibits a nonclassical pattern due to DNA bending and unwinding (51, 58). Contacts by symmetry-related protomers in λ complex are shown in red and green. (D) GMSA methylphosphonate interference. Sites of interference are indicated by filled circles. Phosphodiester positions across 15-bp DNA site in complementary strands are designated by a common number based on nucleoside position in the upper strand. Base pairs are numbered 1 to 15 from left. (E and F) Test for stereospecific interference following HPLC resolution of Sp and Rp diastereomers. (E) Corey-Pauling-Koltun (CPK) models of negatively charged phosphodiester linkage (PO4−, left) and neutral isomers (center and right). Phosphorus is shown in orange, oxygen in red, and the methyl group in black and white. (F) Representative gels showing absence of stereospecific interference at two sites of partial interference (a and b).
We employed methylphosphonate as a neutral substitution for the negatively charged phosphodiester linkage in the DNA backbone (53). The methylphosphonate moiety is similar to that of the native phosphodiester bridge (Fig. 7E) (32). Racemic mixtures of Rp and Sp diastereomers were initially employed. The DSX DM domain binds to a pseudopalindromic DNA site (dsxA) to form a specific 2:1 complex as detected by GMSA (Fig. 7D). Systematic DNA-binding studies of modified DNA sites provide an interference footprint (Fig. 8).
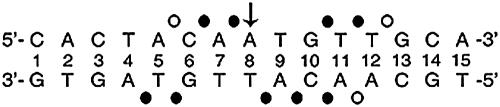
FIG. 8.
Interference footprint by systematic DNA-binding studies of modified DNA sites. Sites of strong or weak interference (filled or open circles, respectively) are nearly symmetric about the central base pair (arrow).
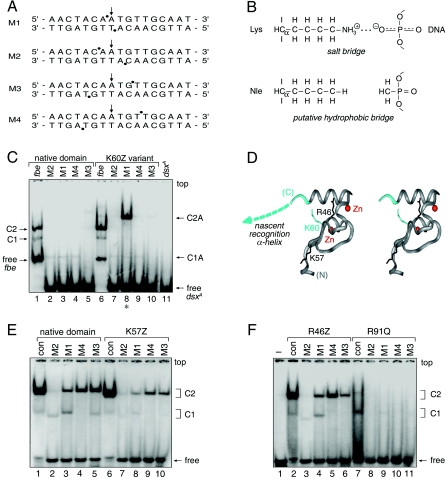
Sites of strong or weak interference are nearly symmetric about the central base pair. Their orientation is opposite to that of classical major-groove footprints, reflecting the minor-groove localization of the DM domain (68). Stereospecific interference was not observed at representative sites of partial interference (Fig. 7D, top panel, lanes a and b, and F). Assuming that a DM protomer makes nearly equivalent half-site interactions, the footprint defines four sets of DNA probes (M1 to M4, see below), each containing two substitutions (Fig. 9A). The DSX domain binds only weakly to these modified oligonucleotides (Fig. 9C, lanes 2 to 5). To test whether specific binding might be rescued by a neutral substitution of a basic side chain (i.e., replacement of a salt bridge by a “hydrophobic bridge” [Fig. 9B]), potential complementation was investigated using R46Z, K57Z, and K60Z variant domains. These substitutions (Fig. 9D) impair specific DNA binding by 2-, 1.5-, and 10-fold, respectively (52).
FIG. 9.
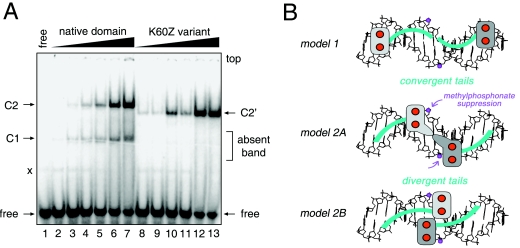
Chemogenetic analysis of putative protein-DNA contact. (A) DNA probes defined by the DSX DM domain footprint. Sites of interference (filled circles) are nearly symmetric about the central base pair (arrow). (B) Chemogenetic strategy envisages substitution of salt bridge at protein-DNA interface (upper panel) by hydrophobic bridge (lower panel). (C) GMSA autoradiogram results for specific binding of native protein (lanes 1 to 5) or K60Z variant (lanes 6 to 10) to 33P-labeled DNA sites. Native and variant protein concentrations were 24 nM and 500 nM, respectively. Control lanes 1 and 6 demonstrate binding to unmodified dsxA site within 29-bp fat body enhancer (fbe) duplex. Native 1:1 and 2:1 complexes are respectively labeled C1 and C2 (fbe) and C1A and C2A (M1 to M4). Binding of native protein to modified dsxA probes M1 to M4 (lanes 2 to 5) yields only a weak 1:1 complex. Whereas binding of K60Z variant to probes M2 to M4 is likewise perturbed, formation of a 2:1 complex is rescued by modification M1 (lane 8, asterisk). Note accidental similarity of motilities between free fbe probe in lanes 1 and 6 (29 bp) and the C1 complex containing dsxA (15 bp; lanes 2 to 5). (D) Ribbon model (stereo pair) of DSX Zn module (DSX residues 35 to 78; gray) and tail (dashed line; azure). Sites of norleucine substitution and zinc ions (red spheres) are shown. (E and F) GMSA screening of DSX analogs against methylphosphonate DNA probes M1 to M4. No specific pattern of second-site compensation is observed for native domain and K57Z (E) or R46Z and R91Q (F) variant proteins. The native domain concentration was 120 nM, whereas the concentrations of the DSX analogs were in each case 520 nM. The control lanes (con, lane 1 in panel E and lane 2 in panel F) employ a 15-bp dsxA duplex site containing a methylphosphonate modification outside of the footprint (i.e., at a noninterfering site). Respective percentages of DNA probe shifted to the C1 and C2 forms for the native domain were as follows: control, 2% and 94%; M2, 5% and 3%; M1, 11% and 23%; M4, nondetectable and 43%; and M3, 5% and 50%. Respective percentages for the K57Z variant were as follows: control, 1% and 91%; M2, <1% and 3%; M1, 2% and 9%; M4, nondetectable and 18%; and M3, nondetectable and 17%. Respective percentages for the R46Z variant were as follows: control, 3% and 78%; M2, 1% and nondetectable; M1, 7% and 18%; M4, nondetectable and 34%; and M3, nondetectable and 14%. Respective percentages for the R91Q variant were as follows: control, 10% and nondetectable; M2, both nondetectable; M1, 1% and nondetectable; M4, both nondetectable; and M3, <0.5% and nondetectable.
Probes M1 to M4 were screened against the norleucine analogs to test for second-site suppression. Remarkably, M1 binding is selectively rescued by K60Z (Fig. 9D, lane 8) but not by the other substitutions. The K57Z and R46Z domains bind to the M1 to M4 probes with the same order of affinity as that of the wild-type domain (Fig. 9E and F); there is no site-specific change or relaxation of chemical specificity. Intersexual mutation R91Q blocks binding to all sites tested (Fig. 9F, lanes 8 to 11). Native and K60Z domains bind with near-equal affinity to the M1 site (ΔΔG, <0.2 kcal/mol as inferred from the relationship ΔΔG = RTln [K/K′] where K and K′ are the respective wild-type and variant dissociation constants; ΔΔG indicates the change in respective free energies of binding, and ln indicates the natural logarithm) (Fig. 10A). By contrast, the M1 modifications impair binding of the native domain by 10-fold (ΔΔG, ∼1.3 kcal/mol). The K60Z substitution thus leads to “relaxed specificity” for the M1 DNA modifications (rather than altered specificity), as the variant protein does not effectively discriminate between unmodified and M1-modified DNA. Suppression is specific for the M1 probe: the native domain binds to sites M2 to M4 at least fivefold more strongly than does the K60Z variant (ΔΔG, >1 kcal/mol). Similar relaxation of specificity is observed in GMSA studies of variant sites singly containing either the upper- or the lower-strand M1 modifications (see the supplemental material). 1H-nuclear magnetic resonance and CD spectra of the K60Z domain are essentially identical to those of the native domain (52), suggesting that the substitution's effects on M1 DNA binding are due to the local absence of a positive charge. These data suggest that, in the native complex, dimer-related K60 side chains interact with the two DNA phosphodiester groups at the sites of M1 modification. Tolerance of the K60R substitution suggests that the detailed molecular structure of the positive charge (ε-amino group of Lys versus guanidinium group of Arg) does not matter. Intolerance of the K60H and K60Q substitutions conversely suggests that intermolecular hydrogen bonding at this site is not in itself sufficient.
FIG. 10.
Relaxed sequence specificity of K60Z domain and structural implications. (A) Native DSX domain and K60Z variant domain exhibit similar specific affinities for methylphosphonate probe M1 (band C2 and C2′, specific 2:1 complex). The free DNA duplex probe M1 is shown in lane 1; native domain is shown in lanes 2 to 7; and variant domain is shown in lanes 8 to 13. The K60Z substitution destabilizes the 1:1 band (C1 at left; indicated by “absent band”). Because the substitution does not significantly perturb the cooperativity of binding to the unmodified DNA probe (see Fig. S2 in the supplemental material), we presume that the variant C1 complex is too kinetically unstable to be detected by GMSA. Native protein concentrations in lanes 2 to 7 are 24 nM, 48 nM, 96 nM, 144 nM, 240 nM, and 500 nM, respectively. Concentrations of K60Z variant domain in lanes 8 to 13 are the same, respectively, as those for native protein. Respective percentages of DNA probe shifted to the C1 and C2 forms for the native domain are as follows: lane 2, <0.5% and <0.5%; lane 3, 1% and <0.5%; lane 4, 1% and 1%; lane 5, 3% and 5%; lane 6, 5% and 16%; and lane 7, 10%, and 36%. Percentages of DNA probe shifted to the C2′ form for the K60Z variant are as follows: lane 8, 1%; lane 9, 1%; lane 10, 10%; lane 11, 4%; lane 12, 29%; and lane 13, 33%. The faint band “x” is a minor contaminant of the free DNA. (B) Alternative models of protein-DNA complex. In model 1, Zn modules bind at ends of DNA site and tails (shown in blue) converge within the minor groove. In model 2A, Zn modules bind near center; tails diverge to follow the minor groove. In model 2B, Zn modules bind farther apart; tails diverge. Zinc ions are shown as red spheres; one module is shaded in dark gray, the other in light gray. Methylphosphonates are shown in lilac (arrows). The DNA (16 bp) is shown as B DNA (black). DNA-dependent dimerization of DM domain may be mediated by protein-protein contacts near the center of the DNA site.
DISCUSSION
The DM domain defines a newly recognized family of minor-groove transcription factors (59, 68). Such proteins participate in metazoan sex-determining pathways and general developmental patterning (35). A prototype DM factor is provided by DSX (59), a classical target of the Drosophila sex-determining hierarchy (29). Although the sex-specific isoforms of DSX have long been regarded as terminal differentiation factors (18), recent genetic evidence suggests that DSXF and DSXM also function to integrate sex-specific and positional information in morphogenesis (1, 12). Of particular interest are potential interactions between homeotic proteins (such as Abdominal A and Abdominal B) (9, 38, 39, 43) in the joint regulation of segmental identity (1). Because the homeodomain contains a helix-turn-helix (HTH) motif that binds within the DNA major groove (40), it is possible that coordinate gene regulation is effected through the approximation of major- and minor-groove DNA-binding sites within a sex-specific multiprotein-DNA complex. Juxtaposition of major- and minor-groove DNA control sites has been observed in the fbe control site: DSX target sites overlap a putative bZIP site, which in principle may allow or preclude simultaneous occupancy (3). The helical arms of bZIP transcription factors (like the homeodomain HTH) bind within the major groove (23).
The DM domain contains a novel Zn module and nascent helical tail (68). The helical propensity of the tail, intrinsic to its sequence, depends on specific DNA binding for its realization. Such induced fit is unrelated to metal ion binding. How these elements bind DNA is not well understood. As a first step toward their functional characterization, we sought to delimit domain boundaries by deletion analysis and identify key side chains by mutagenesis. To this end, a rapid and efficient genetic screen in S. cerevisiae was constructed based on a DSXF-regulated Y1H system. Its design recapitulates the physiological regulation of yp1 gene expression by specific DSX target sites in the fbe (3, 4). Control studies of intersexual dsx mutations and nonconsensus base-pair substitutions in the fbe sites (26) established the validity of this model. As expected (25), DM-regulated expression of the Y1H reporter gene requires intact metal-binding sites in the Zn module. Functional boundaries of the DM domain span DSX residues 31 to 105 in accord with its evolutionary consensus (59) and previous DNA-binding studies (25, 68). Surprisingly, although deletion of the C-terminal segment (residues 98 to 105) leads to a 10-fold decrease in reporter gene activation, none of the 18 substitutions in this segment are deleterious, and the segment itself may functionally be replaced by polyalanine. These results suggest that a critical parameter is provided by the length of the distal tail but not its sequence. Since the tail folds on DNA binding to form one or more α-helical segments (52, 68), we imagine that a minimum C-terminal length is required for segmental helical stability. Similar findings have been described previously in studies of adaptive RNA binding by helical arginine-rich ARM peptides (62).
Contributions of individual side chains were probed by a combination of site-directed and random mutagenesis. Site-directed mutations established a correlation with previous effects of alanine substitutions in the tail as assayed by GMSA (52), verified the dispensability of N-terminal residues 1 to 31, and demonstrated that cysteines and histidines are not interchangeable in the Zn module. Random mutagenesis provided an overview of allowed and disallowed substitutions. The majority of substitutions do not perturb reporter gene activation; these are distributed throughout the DM domain. Indeed, some residues tolerate diverse substitutions, indicating that such side chains are required neither for folding nor for DNA binding. Loss-of-function mutations are by contrast confined to a limited region of the DM domain, defining sites that are critical to folding or predicted to interact with DNA (52).
Folding determinants of the DM Zn module.
The core of the DM Zn module contains conserved aliphatic and aromatic side chains in addition to the immediate metal-binding ligands. Substitutions of internal side chains I54 and L56 result in white colonies, indicating that formation of a hydrophobic core is essential. Because the side chain of L56 packs against C44, H59, and K60, loss-of-function substitutions by Ala, Pro, or Gln would be expected to introduce, respectively, a destabilizing cavity (L56A), conformational perturbation (L56P), or an unfavorable buried polar group (L56Q). The side chain of I54 likewise packs against C44, L52, and H59, wherein substitutions would introduce a similar spectrum of perturbations. Substitutions at N49 are also deleterious. This internal polar side chain packs between the two metal-binding sites, enabling the side chain carboxamide to participate in a network of hydrogen bonds. The asymmetric distribution of partial charges near the S-Zn2+ bonds may be stabilized by this network. In addition, N49 has long-range contacts with the aliphatic portions of K60 and R79 (which may in turn contact DNA; see below). Many sites at the surface of the DM domain or in the tail are tolerant of substitutions (residues P41, P42, K53, T55, R61, K64, R66, Y67, T69, E71, K72, R74, L75, V82, M83, L85, and Q86). Such findings are reminiscent of a pioneering structure-based analysis of allowed and disallowed sequences in the HTH domain of phage λ repressor by Bowie, Hecht, and coworkers (11, 33).
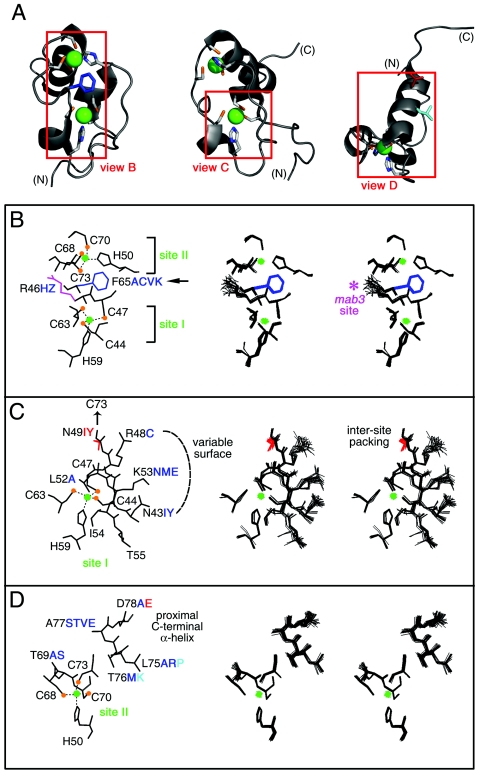
A trend is observed wherein mutations that block transcriptional activation occur at conserved sites, whereas neutral substitutions occur at nonconserved sites. N43 and K53, for example, are well defined on the surface of the Zn module (Fig. 11C) but not conserved (Fig. 1D). Substitutions N43I, N43Y, K53N, K53M, and K53E are well tolerated (Fig. 5). Key exceptions to this trend are noteworthy. (i) Arg is conserved at positions 46 and 48 (positions 12 and 14 of the DM consensus). Indeed, R46 is the site of an intersexual mutation in mab-3 (C. elegans) (59). Uncharged substitutions at these sites (Fig. 11B and C) are nonetheless well tolerated. (ii) Two glycines (G51 and G58) are invariant at sites adjoining metal-binding ligands (H50 and H59, respectively). In the solution structure, the glycines exhibit positive φ angles and so occupy regions of the Ramachandran plot ordinarily unfavorable to l-amino acids. Nonetheless, at each site, Ala is well tolerated, whereas other substitutions are disallowed (Fig. 3D). Modeling suggests that the variant side chains would project into solvent and not disrupt core packing. Tolerance of some substitutions suggests that positive φ angles at positions 51 and 58 are not necessary for metal-dependent folding; alternatively, it is possible that some l-amino acids can adopt unfavorable positive φ angles with only a modest free-energy penalty (∼1 kcal/mol). Intolerance of other substitutions may indicate that neighboring surfaces are close to the DNA or DNA contact sites. (iii) DM sequences contain a conserved aromatic side chain at position 65. In the DSX structure, F65 packs between metal-binding sites (Fig. 11B). Surprisingly, the aromatic side chain may functionally be substituted by Ala, Cys, Lys, or Val, indicating that a broad range of packing schemes is well tolerated. This feature contrasts with the importance of a central aromatic side chain in the classical Zn finger (45, 46). It would be of future interest to purify an F65A variant to assess its structure, stability, and DNA-binding properties.
FIG. 11.
Structural environments of mutation sites. (A) Ribbon models of DSX DM domain. Regions boxed in red are shown in panels B, C, and D, respectively. Zinc ions are shown in bright green, and Zn coordinating cysteines are tipped in orange. The F65 side chain is shown in dark blue (B), that for D78 in red (D), and that for L75 in light blue (D). (B through D) Local environments of selected side chains. In each panel, one representative model is labeled at left; the corresponding DG/SA ensemble is shown in stereo at right. Thiolate-Zn bonds in each panel are shown as dotted lines. Color schemes follow that of panel A. Mutations that impair reporter gene expression are shown in red. Mutations that have no or partial effects are shown in blue and aqua. (B) Zn coordination sites. Residues C44, C47, H59, and C63 comprise site I; residues H50, C68, C70, and C73 comprise site II. Conserved residues R46 and F65 are also shown. Mutation of R46 (side chain shown in magenta) to W in the homologous sequence of mab-3 in C. elegans causes intersexual development of chromosomal male. Residue F65 functions as a bridge between the two Zn-binding sites, participating in an aromatic-aromatic interaction with H50 and additional side chain interactions with several residues near site I (not shown). (C) Packing of ordered residues near site I allows for the formation of a well-defined polar or basic surface. The buried side chain of N49 interacts with C73, promoting the formation of Zn-binding site II. The side chains of residue N43, R48, and K53 form a hydrophilic polar surface. Ordered residues L52, I54, and T55 underlie this surface. (D) Packing of ordered residues near site II. The side chains of α-helical residues in the C-terminal region are also shown. These presumably nucleate helix propagation on DNA binding.
Putative DNA-binding surface of the DM domain.
Of special interest are sites on the protein surface at which substitutions are not tolerated. Because such substitutions would not be expected to impair metal ion binding or protein folding, these sites define putative DNA contacts. Candidates are provided by several tail residues and K60, which defines a basic patch on the surface of the Zn module (Fig. 6). K60 is highly conserved among metazoan DM sequences (arrow in Fig. 1D). Whereas substitutions at K57 (K57M, K57A, and K57N) result in light-blue colonies, several substitutions at K60 (K60E, K60Q, K60M, and K60H) yield white colonies. The inactivity of the K60H and K60Q variants is noteworthy in light of the hydrogen-bonding capability of His and Gln. Interestingly, the K60R variant exhibits wild-type activity, suggesting that the positive charge of K60 contributes to specific DNA binding. The importance of K60 is consistent with the 10-fold decrement in specific DNA binding that was previously observed in GMSA studies of a norleucine analog (52). Further evidence that K60 contacts the DNA backbone is provided by methylphosphonate interference experiments (below). It is possible that K57 also contacts the DNA but that this contact contributes only modestly to the binding free energy.
The disordered tail of the DM domain is proposed to function as a recognition helix (68). Eleven residues are of functional importance; these include two aliphatic side chains (A84 and A92), three polar side chains (Q80, T87, and Q93), four basic side chains (R79, R81, R90, and R91), and two acidic side chains (D78 and E97). Surprisingly, five of these residues (T87, R91, A92, Q93, and E97) exhibit striking divergence among metazoan DM sequences. We speculate that these variable sites contribute to differences among the sequence specificities of DM domains. Although the importance of D78 and E97 may seem surprising, the contribution of acidic side chains to a protein-DNA interface is well established (56). Such side chains may participate in a network of charge-stabilized hydrogen bonds to orient the DNA-binding surface (as in the Asp-Arg recognition element of a classical Zn finger) (57) or contact a base directly (as in the catabolite gene activator protein [CAP]) (22). Although D78A is well tolerated, the analogous D→A substitution in a classical Zn finger allows specific DNA binding but with relaxed specificity (24).
The profound loss of DNA binding due to intersexual mutation R91Q suggests that the distinctive guanidinium group of R91 is integral to a network of base-specific protein-DNA contacts. Such essential arginines have been well described previously in structures of protein-DNA and protein-RNA complexes (56, 64). Although R91 is conserved, a few DM domains contain other residues (Fig. 11). We speculate that such divergence indicates altered sequence specificity. The back surface of the proximal C-terminal α-helix and adjoining surface of metal-binding site II (Fig. 11D) allow diverse substitutions (positions 69, 74, 76, and 77). L75 is conserved at the edge of this surface. We speculate that helicogenic residues at this site contribute to segmental stability: the partial impairment of L75P may thus be due to structural perturbation of the helix.
Chemogenetic evidence for a specific protein-DNA contact.
Insight into the chemical basis of protein-DNA recognition may be obtained by the use of base analogs. In particular, the use of modified DNA sites readily provides a footprint of a protein-DNA complex, enabling the roles of individual functional groups to be resolved. Dissection of chemical specificity at the level of individual functional groups is designated chemogenetics (60). Although application of this strategy to the major groove is widespread, analysis of the minor groove is restricted by its more limited repertoire of functional groups. To circumvent this restriction, we have sought to generalize the chemogenetic approach to the DNA backbone in order to test the role of a key lysine (K60) that is identified above as a potential DNA contact. Our approach exploited the site-specific substitution of the negatively charged phosphodiester linkage by a neutral analog (methylphosphonate). Such modifications can interfere with protein binding and so yield a discrete footprint (10, 44).
Binding of the DSX DM domain to a modified DNA site may be rescued by chemical complementation: substitution of a positively charged side chain in the protein (lysine) by a neutral isostere (norleucine). Such complementation between the DM domain (as modified at residue 60) and the DNA backbone (as modified near the center of the DNA site) thus provides evidence for an intermolecular contact. The results, specific to the respective sites of modification, distinguish between two classes of models of the DM-DNA complex (Fig. 10B). In model 1, respective Zn modules bind at the ends of the DNA site, whereas the tails converge toward the middle. The second model (model 2A or 2B) envisages binding of the Zn module near the center of the pseudopalindromic DNA site; the motif's C-terminal tails diverge outward to follow the minor groove. The dimer-related Zn modules may occupy distinct half sites (model 2A) or cross at the center (model 2B). Since the sites of modification in M1 flank the central base pair (arrows in Fig. 9A and 10B), their chemogenetic interaction with K60 favors model 2A or 2B.
Understanding the molecular-genetic function of dsx in Drosophila development will in the future require biochemical reconstitution of sex-specific transcriptional preinitiation complexes. A central feature of such complexes will be the DM-DNA interface. The present study has utilized prior molecular-genetic characterization of a DSX-responsive enhancer element to construct a Y1H system for mutational analysis of the DM domain. The results suggest sites of protein-DNA interaction and provide insight into the structural requirements of zinc-dependent protein folding. We anticipate that these data will provide a foundation for crystallographic studies of DM-DNA complexes. Integration of structural and mutational studies promises to provide insight into the evolution and function of DM transcription factors.
Supplementary Material
Acknowledgments
This work was supported in part by a grant from the National Institutes of Health to B. Baker (GM037731-20, subcontract GM037731-19S1 to M.A.W.). R.S. was supported in part by an NIH training grant in molecular oncology at the Case School of Medicine (CA059366).
We thank B. Baker for advice and encouragement, N. B. Phillips and W. Yang for discussion, J. Bayrer and K. Huang for assistance with figures, and E. Collins and S. Price for preparation of the manuscript.
This is a contribution from the Cleveland Center for Structural Biology.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ahmad, S. M., and B. S. Baker. 2002. Sex-specific deployment of FGF signaling in Drosophila recruits mesodermal cells into the male genital imaginal disc. Cell 109:651-661. [DOI] [PubMed] [Google Scholar]
- 2.An, W., S. Cho, H. Ishii, and P. C. Wensink. 1996. Sex-specific and non-sex-specific oligomerization domains in both of the doublesex transcription factors from Drosophila melanogaster. Mol. Cell. Biol. 16:3106-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.An, W., and P. C. Wensink. 1995. Integrating sex- and tissue-specific regulation within a single Drosophila enhancer. Genes Dev. 9:256-266. [DOI] [PubMed] [Google Scholar]
- 4.An, W., and P. C. Wensink. 1995. Three protein binding sites form an enhancer that regulates sex- and fat body-specific transcription of Drosophila yolk protein genes. EMBO J. 14:1221-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayrer, J. R., W. Zhang, and M. A. Weiss. 2005. Dimerization of doublesex is mediated by a cryptic UBA-domain fold. Implications for sex-specific gene regulation. J. Biol. Chem. 280:32989-32996. [DOI] [PubMed] [Google Scholar]
- 6.Bennett, C. P., Z. Docherty, S. A. Robb, P. Ramani, J. R. Hawkins, and D. Grant. 1993. Deletion 9p and sex reversal. J. Med. Genet. 30:518-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berta, P., J. R. Hawkins, A. H. Sinclair, A. Taylor, B. L. Griffiths, P. N. Goodfellow, and M. Fellous. 1990. Genetic evidence equating SRY and the testis-determining factor. Nature 348:448-450. [DOI] [PubMed] [Google Scholar]
- 8.Bewley, C. A., A. M. Gronenborn, and G. M. Clore. 1998. Minor groove-binding architectural proteins: structure, function, and DNA recognition. Annu. Rev. Biophys. Biomol. Struct. 27:105-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bopp, D. 2001. Merging sex and position. Bioessays 23:304-306. [DOI] [PubMed] [Google Scholar]
- 10.Botfield, M. C., and M. A. Weiss. 1994. Bipartite DNA recognition by the human Oct-2 POU domain: POUs-specific phosphate contacts are analogous to those of bacteriophage λ repressor. Biochemistry 33:2349-2355. [DOI] [PubMed] [Google Scholar]
- 11.Bowie, J. U., J. F. Reidhaar-Olson, W. A. Lim, and R. T. Sauer. 1990. Deciphering the message in protein sciences: tolerance to amino acid substitutions. Science 247:1306-1310. [DOI] [PubMed] [Google Scholar]
- 12.Burtis, K. C. 2002. Doublesex in the middle. Science 297:1135-1136. [DOI] [PubMed] [Google Scholar]
- 13.Burtis, K. C., K. T. Coschigano, B. S. Baker, and P. C. Wensink. 1991. The doublesex proteins of Drosophila melanogaster bind directly to a sex-specific yolk protein gene enhancer. EMBO J. 10:2577-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bushman, F. D., J. E. Anderson, S. C. Harrison, and M. Ptashne. 1985. Ethylation interference and X-ray crystallography identify similar interactions between 434 repressor and operator. Nature 316:651-653. [DOI] [PubMed] [Google Scholar]
- 15.Cho, S., and P. C. Wensink. 1997. DNA binding by the male and female doublesex proteins of Drosophila melanogaster. J. Biol. Chem. 272:3185-3189. [DOI] [PubMed] [Google Scholar]
- 16.Cho, S., and P. C. Wensink. 1998. Linkage between oligomerization and DNA binding in Drosophila doublesex proteins. Biochemistry 37:11301-11308. [DOI] [PubMed] [Google Scholar]
- 17.Cho, S., and P. C. Wensink. 1996. Purification and physical properties of the male and female double sex proteins of Drosophila. Proc. Natl. Acad. Sci. USA 93:2043-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cline, T. W., and B. J. Meyer. 1996. Vive la difference: males vs females in flies vs worms. Annu. Rev. Genet. 30:637-702. [DOI] [PubMed] [Google Scholar]
- 19.Dawes, H. E., D. S. Berlin, D. M. Lapidus, C. Nusbaum, T. L. Davis, and B. J. Meyer. 1999. Dosage compensation proteins targeted to X chromosomes by a determinant of hermaphrodite fate. Science 284:1800-1804. [DOI] [PubMed] [Google Scholar]
- 20.Dawson, P. E., T. W. Muir, I. Clark-Lewis, and S. B. Kent. 1994. Synthesis of proteins by native chemical ligation. Science 266:776-779. [DOI] [PubMed] [Google Scholar]
- 21.Ebright, R. H., P. Cossart, B. Gicquel-Sanzey, and J. Beckwith. 1984. Mutations that alter the DNA sequence specificity of the catabolite gene activator protein of E. coli. Nature 311:232-235. [DOI] [PubMed] [Google Scholar]
- 22.Ebright, R. H., A. Kolb, H. Buc, T. A. Kunkel, J. S. Krakow, and J. Beckwith. 1987. Role of glutamic acid-181 in DNA-sequence recognition by the catabolite gene activator protein (CAP) of Escherichia coli: Altered DNA-sequence-recognition properties of [Val181]CAP and [Leu181]CAP. Proc. Natl. Acad. Sci. USA 84:6083-6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellenberger, T. E., C. J. Brandl, K. Struhl, and S. C. Harrison. 1992. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell 71:1223-1237. [DOI] [PubMed] [Google Scholar]
- 24.Elrod-Erickson, M., and C. O. Paco. 1999. Binding studies with mutants of Zif268. Contribution of individual side chains to binding affinity and specificity in the Zif268 zinc finger-DNA complex. J. Biol. Chem. 274:19281-19285. [DOI] [PubMed] [Google Scholar]
- 25.Erdman, S. E., and K. C. Burtis. 1993. The Drosophila doublesex proteins share a novel zinc finger related DNA binding domain. EMBO J. 12:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erdman, S. E., H. J. Chen, and K. C. Burtis. 1996. Functional and genetic characterization of the oligomerization and DNA binding properties of the Drosophila doublesex proteins. Genetics 144:1639-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flejter, W. L., J. Fergestad, J. Gorski, T. Varvill, and S. Chandrasekharappa. 1998. A gene involved in XY sex reversal is located on chromosome 9, distal to marker D9S1779. Am. J. Hum. Genet. 63:794-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giese, K., J. Cox, and R. Grosschedl. 1992. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell 69:185-195. [DOI] [PubMed] [Google Scholar]
- 29.Graham, P., J. K. M. Penn, and P. Schedl. 2002. Masters change, slaves remain. Bioessays 25:1-4. [DOI] [PubMed] [Google Scholar]
- 30.Gubbay, J., J. Collignon, P. Koopman, B. Capel, A. Economou, A. Munsterberg, N. Vivian, P. Goodfellow, and R. Lovell-Badge. 1990. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature 346:245-250. [DOI] [PubMed] [Google Scholar]
- 31.Guioli, S., K. Schmitt, R. Critcher, M. Bouzyk, N. K. Spurr, T. Ogata, J. J. Hoo, L. Pinsky, G. Gimelli, L. Pasztor, and P. N. Goodfellow. 1998. Molecular analysis of 9p deletions associated with XY sex reversal: refining the localization of a sex-determining gene to the tip of the chromosome. Am. J. Hum. Genet. 63:905-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han, F., W. Watt, D. J. Duchamp, L. Callahan, F. J. Kezdy, and K. Agarwal. 1990. Molecular structure of deoxycytidyl-3′-methylphosphonate (RP) 5′-deoxyguanidine, d[Cp(CH3)G]. A neutral dinucleotide with Watson-Crick base pairing and a right handed helical twist. Nucleic Acids Res. 18:2759-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hecht, M. H., H. C. M. Nelson, and R. T. Sauer. 1983. Mutation in λ repressor's amino-terminal domain: implication for protein stability and DNA binding. Proc. Natl. Acad. Sci. USA 80:2676-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hochschild, A., and M. Ptashne. 1986. Homologous interactions of λ repressor and λ Cro with the λ operator. Cell 44:925-933. [DOI] [PubMed] [Google Scholar]
- 35.Hodgkin, J. 2002. The remarkable ubiquity of DM domain factors as regulator of sexual phenotype: ancestry or aptitude? Gene Dev. 16:2322-2326. [DOI] [PubMed] [Google Scholar]
- 36.Johnson, A. D., B. J. Meyer, and M. Ptashne. 1979. Interactions between DNA-bound repressors govern regulation by the λ phage repressor. Proc. Natl. Acad. Sci. USA 76:5061-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson, A. D., A. R. Poteete, G. Lauer, R. T. Sauer, G. K. Ackers, and M. Ptashne. 1981. λ Repressor and cro—components of an efficient molecular switch. Nature 294:217-223. [DOI] [PubMed] [Google Scholar]
- 38.Keisman, E. L., and B. S. Baker. 2001. The Drosophila sex determination hierarchy modulates wingless and decapentaplegic signaling to deploy dachshund sex-specifically in the genital imaginal disc. Development 128:1643-1656. [DOI] [PubMed] [Google Scholar]
- 39.Keisman, E. L., A. E. Christiansen, and B. S. Baker. 2001. The sex determination gene doublesex regulates the A/P organizer to direct sex-specific patterns of growth in the Drosophila genital imaginal disc. Dev. Cell. 1:215-225. [DOI] [PubMed] [Google Scholar]
- 40.Kissinger, C. R., B. S. Liu, E. Martin-Blanco, T. B. Kornberg, and C. O. Pabo. 1990. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell 63:579-590. [DOI] [PubMed] [Google Scholar]
- 41.Koopman, P., J. Gubbay, N. Vivian, P. Goodfellow, and R. Lovell-Badge. 1991. Male development of chromosomally female mice transgenic for Sry. Nature 351:117-121. [DOI] [PubMed] [Google Scholar]
- 42.Koopman, P., A. Munsterberg, B. Capel, N. Vivian, and R. Lovell-Badge. 1990. Expression of a candidate sex-determining gene during mouse testis differentiation. Nature 348:450-452. [DOI] [PubMed] [Google Scholar]
- 43.Kopp, A., I. Duncan, and S. B. Carroll. 2000. Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature 408:553-559. [DOI] [PubMed] [Google Scholar]
- 44.Labeots, L. A., and M. A. Weiss. 1997. Electrostatics and hydration at the homeodomain-DNA interface: chemical probes of an interfacial water cavity. J. Mol. Biol. 269:113-128. [DOI] [PubMed] [Google Scholar]
- 45.Lachenmann, M. J., J. E. Ladbury, N. B. Phillips, N. Narayana, X. Qian, and M. A. Weiss. 2002. The hidden thermodynamics of a zinc finger. J. Mol. Biol. 316:969-989. [DOI] [PubMed] [Google Scholar]
- 46.Lee, M. S., R. J. Mortishire-Smith, and P. E. Wright. 1992. The zinc finger motif. Conservation of chemical shifts and correlation with structure. FEBS Lett. 309:29-32. [DOI] [PubMed] [Google Scholar]
- 47.Li, J. J., and I. Herskowitz. 1993. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science 262:1870-1874. [DOI] [PubMed] [Google Scholar]
- 48.Ling, M., F. Merante, and B. H. Robinson. 1995. A rapid and relible DNA preparation mechod for screening a large number of yeast clones by polymerase chain reaction. Nucleic Acids Res. 23:4924-4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McDonald, M. T., W. Flejter, S. Sheldon, M. J. Putzi, and J. L. Gorski. 1997. XY sex reversal and gonadal dysgenesis due to 9p24 monosomy. Am. J. Med. Genet. 73:321-326. [PubMed] [Google Scholar]
- 50.Meyer, B. J. 1988. Primary events in C. elegans sex determination and dosage compensation. Trends Genet. 12:337-342. [DOI] [PubMed] [Google Scholar]
- 51.Murphy, E. C., V. B. Zhurkin, J. M. Louis, G. Cornilescu, and G. M. Clore. 2001. Structural basis for SRY-dependent 46-X, Y sex reversal: modulation of DNA bending by a naturally occurring point mutation. J. Mol. Biol. 312:481-499. [DOI] [PubMed] [Google Scholar]
- 52.Narendra, U., L. Zhu, B. Li, J. Wilken, and M. A. Weiss. 2002. Sex-specific gene regulation: the doublesex DM motif is a bipartite DNA-binding domain. J. Biol. Chem. 277:43463-43473. [DOI] [PubMed] [Google Scholar]
- 53.Noble, S. A., E. F. Fisher, and M. H. Caruthers. 1984. Methylphosphonates as probes of protein-nucleic acid interactions. Nucleic Acids Res. 12:3387-3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oh, D. B., Y. G. Kim, and A. Rich. 2002. Z-DNA-binding proteins can act as potent effectors of gene expression in vivo. Proc. Natl. Acad. Sci. USA 99:16666-16671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ounap, K., O. Uibo, R. Zordania, L. Kiho, T. Ilus, E. Oiglane-Shlik, and O. Bartsch. 2004. Three patients with 9P deletions including DMRT1 and DMRT2: a girl with XY complement, bilateral ovotestes, and extreme growth retardation, and two two females with normal pubertal development. Am. J. Med. Genet. 130A:415-423. [DOI] [PubMed] [Google Scholar]
- 56.Pabo, C. O., and R. T. Sauer. 1992. Transcription factors: structural families and principles of DNA recognition. Annu. Rev. Biochem. 61:1053-1095. [DOI] [PubMed] [Google Scholar]
- 57.Pavletich, N. P., and C. O. Pabo. 1991. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science 252:809-817. [DOI] [PubMed] [Google Scholar]
- 58.Phillips, N. B., A. Jancso-Radek, V. Ittah, R. Singh, G. Chan, N. Narayana, E. Haas, and M. A. Weiss. SRY and human sex determination: the basic tail of the HMG box functions as a kinetic clamp to augment DNA bending. Submitted for publication. [DOI] [PubMed]
- 59.Raymond, C. S., C. E. Shamu, M. M. Shen, K. J. Seifert, B. Hirsch, J. Hodgkin, and D. Zarkower. 1998. Evidence for evolutionary conservation of sex-determining genes. Nature 391:691-695. [DOI] [PubMed] [Google Scholar]
- 60.Sigler, P. B. 1994. Chemogenetics. Nat. Struct. Biol. 1:3-4. [DOI] [PubMed] [Google Scholar]
- 61.Simpson, R. J. Y., E. D. Cram, R. Czolij, J. M. Matthews, M. Crossley, and J. P. Mackay. 2003. CCHX zinc finger derivatives retain the ability to bind Zn(II) and mediate protein-DNA interactions. J. Biol. Chem. 278:28011-28018. [DOI] [PubMed] [Google Scholar]
- 62.Tan, R., L. Chen, J. Buettner, D. Hudson, and A. D. Frankel. 1993. RNA recognition by an isolated α helix. Cell 73:1031-1040. [DOI] [PubMed] [Google Scholar]
- 63.Veitia, R. A., M. Nunes, L. Quintana-Murci, R. Rappaport, E. Thibaud, F. Jaubert, M. Fellous, K. McElreavey, J. Goncalves, M. Silva, J. C. Rodrigues, M. Caspurro, F. Boieiro, R. Marques, and J. Lavinha. 1998. Swyer syndrome and 46, XY partial gonadal dysgenesis associated with 9p deletions in the absence of monosomy-9p syndrome. Am. J. Hum. Genet. 63:901-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weiss, M. A., and N. Narayana. 1998. RNA recognition by arginine-rich peptide motifs. Biopolymers 48:167-180. [DOI] [PubMed] [Google Scholar]
- 65.Wilkins, A. S. 1995. Moving up the hierarchy: a hypothesis onthe evolution of a genetic sex-determination pathway. Bioessays 17:71-77. [DOI] [PubMed] [Google Scholar]
- 66.Wolf, U. 1999. Reorganization of sex-determining pathway with the evolution of placentation. Hum. Genet. 105:288-292. [DOI] [PubMed] [Google Scholar]
- 67.Zarkower, D. 2001. Establishing sexual dimorphism: conservation amidst diversity? Nat. Rev. Genet. 2:175-185. [DOI] [PubMed] [Google Scholar]
- 68.Zhu, L., J. Wilken, N. B. Phillips, U. Narendra, G. Chan, S. M. Stratton, S. B. Kent, and M. A. Weiss. 2000. Sexual dimorphism in diverse metazoans is regulated by a novel class of intertwined zinc fingers. Genes Dev. 14:1750-1764. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.