Abstract
Serine/threonine kinase Akt/protein kinase B, the cellular homologue of the transforming viral oncogene v-Akt, plays a central role in the regulation of cell survival and proliferation. We have previously demonstrated that the proto-oncogene TCL1 is an Akt kinase coactivator. TCL1 binds to Akt and mediates the formation of oligomeric TCL1-Akt high-molecular-weight protein complexes in vivo. Within these protein complexes, Akt is preferentially phosphorylated and activated. The MTCP1/TCL1/TCL1b oncogene activation is the hallmark of human T-cell prolymphocytic leukemia (T-PLL), a form of adult leukemia. In the present study, using a PCR-generated random TCL1 library combined with a yeast two-hybrid screening detecting loss of interaction, we identified D16 and I74 as amino acid residues mediating the association of TCL1 with Akt. Based on molecular modeling, we determined that the βC-sheet of TCL1 is essential for TCL1 homodimerization. Studies with mammalian overexpression systems demonstrated that both Akt association and oligomerization domains of TCL1 are distinct functional domains. In vitro kinase assays and overexpression experiments in mammalian cells demonstrated that both TCL1-Akt interaction and oligomerization of TCL1 were required for TCL1-induced Akt activation and substrate phosphorylation. Assays for mitochondrial permeability transition, nuclear translocation, and cell recovery demonstrated that both Akt association and homodimerization of TCL1 are similarly needed for the full function of TCL1 as an Akt kinase coactivator in vivo. The results demonstrate the structural basis of TCL1-induced activation of Akt, which causes human T-PLL.
The TCL1/MTCP1/TCL1b oncogene activation is the hallmark of human T-cell prolymphocytic leukemia (T-PLL). The disease is due to chromosomal translocations involving a T-cell receptor gene and either the 14q32.1 or the Xq28 region. Oncogenes defining a gene family have been identified in these regions, namely, MTCP1 in Xq28 and TCL1 as well as TCL1b in 14q32.1 (34, 43, 49). The oncogenic properties of MTCP1 and TCL1 were further demonstrated by the fact that transgenic mice overexpressing these molecules develop a murine form of T-PLL (17, 48).
TCL1 and TCL1b are highly expressed at early developmental stages. In adults the expression of all TCL1 family members is mostly restricted to the lymphoid compartment. TCL1 expression is limited to immature thymocytes in the T-cell lineage and from pre-B to mature B cells in the B-cell lineage (34, 43). In human diseases, in addition to T-PLL, TCL1 is overexpressed in Burkitt's lymphoma cell lines (49), the majority of AIDS-related non-Hodgkin’s lymphoma-designated immunoblastic lymphoma plasmacytoids (45), lymphoblastic lymphoma, chronic lymphocytic leukemia, mantle cell lymphoma, follicular lymphoma, diffuse large B-cell lymphoma, and primary cutaneous B-cell lymphoma (30). It is also implicated in the development of hematopoietic abnormalities in patients with ataxia-telangiectasia (44).
We have previously found in a yeast two-hybrid screening that TCL1 binds to the serine/threonine kinase Akt (28, 33, 35). In functional assays, all three members of the TCL1 family (TCL1, MTCP1, and TCL1b) were able to interact with Akt1 in mammalian cells and enhance Akt kinase activity. Further, TCL1 promoted the formation of oligomeric TCL1-Akt protein complexes in which the kinase was preferentially phosphorylated and activated (28). Recently, several binding molecules that bind to Akt and modulate its kinase activity have been identified (28, 29, 35, 39).
Akt has a central role in the regulation of several signaling pathways controlling cell survival and proliferation. To date, three mammalian isoforms (Akt1/protein kinase Bα [PKBα], Akt2/PKBβ, and Akt3/PKBγ) of the kinase have been identified. These isoforms share a high degree of structural homology with human Akt1, having 81 and 83% amino acid identity with Akt2 and Akt3, respectively. The analysis of Akt1- and Akt2-knockout mice has suggested a distinct role of each of the isoforms (7, 8). The kinases have two distinct functional domains: an N-terminal pleckstrin homology (PH) domain mediating protein-protein and protein-lipid interactions and a C-terminal catalytic domain (6, 9, 16).
The basic activation process of the Akt isoforms appears to be uniform (47). After the binding of growth factors to their cell surface receptors, Akt is translocated to the plasma membrane secondary to binding of the PH domain with products of the phosphoinositide-3-kinase pathway—phosphatidylinositide (PI)-3,4-bisphosphate and PI-3,4,5-triphosphate (2, 6, 16). The activation of Akt is then regulated by phosphorylation on two regulatory sites, threonine 308/309/305 and serine 473/474/472 (Akt1/2/3, respectively), with phosphorylation of both required for maximal kinase activity (4). Phosphorylation of the threonine residue occurs through the activity of phosphoinositide-dependent kinase 1 (PDK1) (9, 16, 42). The exact mechanisms for the phosphorylation of Ser-473/474/472 remain to be determined (1, 12, 22). After activation, Akt has been shown elsewhere to phosphorylate and inactivate a number of pivotal proapoptotic molecules including BAD, Nur77, and forkhead transcription factor (FHKR) (5, 11, 21, 22).
We hypothesized that TCL1 has two distinct binding sites, one required for the interaction with Akt and the other essential for TCL1 oligomerization, which might explain the molecular mechanism of TCL1-induced Akt activation. Using a PCR-based random TCL1 mutation library combined with a yeast two-hybrid system to screen for loss of interaction as well as molecular modeling, we identified these domains in TCL1. We show that both Akt association and TCL1 dimerization are required for full function of TCL1 as an Akt kinase coactivator, which may contribute to the development of human malignancies associated with TCL1 overexpression.
MATERIALS AND METHODS
PCR-generated random TCL1 library.
Human full-length TCL1 in pGAD424 (Clontech) was amplified with primers flanking the TCL1 sequence, 5"-CCACCAAACCCAAAAAAAGAGATCGAATTCATG and 5"-ATTCATAGATCTCTGCAGGTCGACGGATCCTCA; the EcoRI and BamHI sites next to TCL1 cDNA are underlined. A reaction mixture of 100 μl contained 10 mM Tris-HCl (pH 9.0), 1.0% Triton X-100, 4.2 mM MgCl2, 0.5 mM MnCl2, 3.4 mM enforcing deoxynucleoside triphosphate (dNTP), 0.2 to 0.4 mM (other) dNTPs (15), 2 nM template (wild-type TCL1), 5 μM (each) primer, and 2.5 U of Taq polymerase (Promega). The reaction mixture was put in a Gene Amp 2400 Thermocycler (Perkin-Elmer) at 94°C for 5 min and subjected to 25 cycles of 1 min at 91°C, 1 min at 51°C, and 3 min at 72°C followed by a final extension at 72°C for 10 min. PCR products were subcloned into pGAD424 (Clontech). Nucleotide sequences from the resulting random library were analyzed to determine the frequency of nucleotide substitutions.
Yeast two-hybrid screening for loss of interaction.
Y190 cells (Clontech) were cotransformed by the lithium acetate method with a human Akt2 (Akt2/PAS2-1) and the TCL1 random library as described previously (28, 31). Approximately 104 clones from the cDNA library were screened in the absence of 3-amino-1,2,4-triazole (Sigma). His+ colonies were evaluated for β-galactosidase (β-Gal) activity using a filter-lift assay. Yeast clones were classified according to β-Gal intensity into three categories: 3-h-positive (++), 8-h-positive (+), and 24-h-negative (−) clones. From each category 10 clones were picked for nucleotide sequencing.
Quantitative β-Gal assay.
Y190 cells (Clontech) were cotransformed with Akt2/PAS2-1 together with wild-type, D16G, K30M, Q46R, I74V, and M106V TCL1 in pGAD424 (Clontech). TCL1 mutants were generated by PCR-based site-directed mutagenesis and/or with the Quikchange kit (Stratagene). The liquid β-Gal assay was performed using ONPG (O-nitrophenyl-β-d-galactopyranoside; Sigma) as described previously (28). The values shown were normalized by the expression of each yeast transformant as determined by Western blot analysis (GAL4 activation domain antibody [Ab]; Clontech).
Expression vectors.
Mammalian expression constructs (pCMV6-HA-Akt1, pCMV6-HA-Akt2, pCMV2-Flag-TCL1 [wild type and substitution mutations of TCL1], pME-18S-HA-TCL1 [wild type and substitution mutations of TCL1]) and prokaryotic expression vectors for the generation of glutathione S-transferase (GST) fusion proteins (pGEX4T2-TCL1 [wild type and substitution mutations of TCL1]) have been previously described (28). All nucleotide sequences were verified before performing the experiments.
Site-directed mutagenesis of TCL1.
Amino acid substitution mutants of TCL1 (D16G, K30M, Q46E, I74V, M106V, 36-38A, or 36A/38Δ) were generated by PCR using the primers listed below, and wild type and mutants were subcloned into pGAD424 (Clontech) and pME18SHA (28) or pCMV Flag (Kodak) expression vectors. pGEX4T2-D16G TCL1 and pGEX4T2-36-38A TCL1 were generated by subcloning the corresponding cDNA from pGAD424. All nucleotide sequences were verified before the indicated experiments. Mutated codons are underlined in the primer sequences. Primers were the following: for D16G, 5"-ATG GCC GAG TGC CCG ACA CTC GGG GAG GCA GTC ACC GAC CAC CCG GGC CGC CTG TGG GCC; for K30M, 5"-GTG TAT TTG GAC GAG ATG CAG CAC GCC TGG CTG; for Q46R, 5"-G ATA AAG GAT AGG TTA CGG TTA CGG GTG CTC TTG; for I74V, 5"-CCA AGC CTG CTG CCT GTC ATG TGG CAG CTC TAC; for M106V, 5"-ATC ATC GGA TCC TCA GTC ATC TGG CAG CAG CTC GAG AAG CAC GTC CTC C; for 36-38A, 5"-CAG CAC GCC TGG CTG GCC GCG GCC ATC GAG ATA AAG GAT and reverse complement; and for 38A/38Δ, 5"-GCC TGG CTG GCC TTA ATC GAG ATA and reverse complement.
Coimmunoprecipitation assays.
293T cells (American Type Culture Collection) were cotransfected with a total of 10 μg of indicated plasmids: pCMV-Flag-TCL1 (wild type and indicated mutated forms) with pCMV-HA-Akt1, pCMV-HA-Akt2, pCMV-HA-Akt3, pCMV-PH-Akt1 or pCMV-C-terminal Akt1 (41), and pME18S-HA-TCL1 (28). For the TCL1 dimerization experiment, Flag-tagged TCL1 and hemagglutinin (HA)-tagged TCL1 were transfected. Seventy-two hours after transfection cells were washed twice with ice-cold phosphate-buffered saline (PBS) and lysed with ice-cold Brij 97 lysis buffer. Lysates were precleaned with a protein G-protein A mixture (50% [vol/vol]; Pharmacia) for 1 h; immunoprecipitated with anti-HA, anti-myc, or anti-Flag Ab with anti-mouse immunoglobulin G as a control; run on a sodium dodecyl sulfate (SDS)-polyacrylamide gel (4 to 20% Tris-glycine gel; Novex); and immunoblotted with anti-HA Ab (3F10; Boehringer Mannheim).
In vitro Akt kinase assays.
GST-TCL1 and control GST proteins were generated as described previously (28). Akt1 was immobilized with anti-HA or anti-green fluorescent protein (GFP) monoclonal Ab (MAb) (Clontech) from 293T cells transfected with pCMV6-HA-Akt1. In vitro kinase assays were performed with the Akt kinase assay kit (Cell Signaling) using baculovirus K179D Akt1 (28) by incubating a total of 20 μl of the indicated immobilized GST fusion proteins (0 to 15 μg) in the presence of 2 μl (5 ng) of immobilized Akt for 7 min at 30°C. The total amount of GST protein in the reaction mixtures was normalized to 15 μg by adding GST control fusion protein into the reaction samples. The reactions were terminated by adding SDS sample buffer and analyzed on SDS gels (Invitrogen).
GST pull-down assays.
In vitro translation of indicated wild-type and mutant forms of TCL1 (in PSG5 vector; Stratagene) was performed using the TNT-coupled reticulocyte lysate system (Promega), [35S]methionine (Amersham Pharmacia Biotech), and T7 polymerase. In vitro-translated TCL1 protein product was mixed with 6 μg of each GST fusion protein and 20 μl of glutathione-Sepharose 4B beads (Amersham Pharmacia Biotech) (wild-type TCL1 and mutant TCL1 in Fig. 3B) in 400 μl of NETN buffer (20 mM Tris [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5% NP-40), incubated for 3 h at 4°C, and washed three times with NETN buffer. After the final wash 20 μl of 2× SDS sample buffer was added to each reaction mixture, and the samples were resolved on an SDS gel (Invitrogen) and autoradiographed.
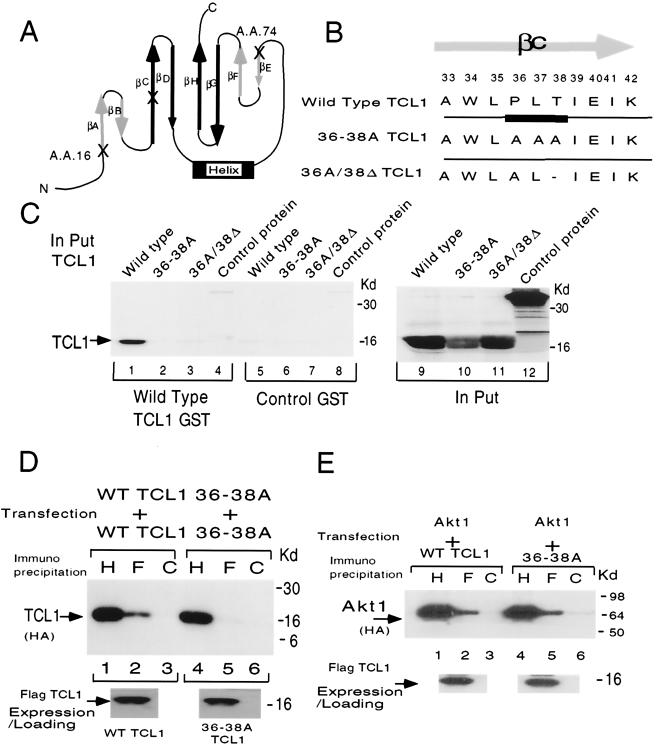
FIG. 3.
βC-sheet mediates the homodimerization of TCL1. (A) The alignment of amino acid positions D16 and I74 within the topology of the eight-strand antiparallel β-sheets of TCL1 (23). Amino acids D16 and I74 are symmetrically located at the very beginning of two β-sheets (βA and βE, respectively), forming a single surface of the TCL1 barrel structure (Fig. 6A). (B) The mutation forms of TCL1 targeting the βC-sheet used in this study. In order to mutate the βC-sheet, we created P36A/L37A/T38A TCL1 (designated 36-38A TCL1, second row) and P36A/T38Δ TCL1 (designated 36A/38Δ, third row). (C) Pull-down assays were performed using an in vitro-translated full-length GST fusion protein of TCL1 and its mutant forms. Only wild-type TCL1 (lane 1), not 36-38A TCL1 (lane 2), P36A/T38Δ (lane 3), or control protein (luciferase, lane 4), could interact and thus dimerize with TCL1. Control GST showed no background retention (lanes 5 to 8), demonstrating the specificity of TCL1 binding. The in vitro-translated protein products utilized are shown in the right panel (lanes 9 to 12, wild-type TCL1, 36-38A TCL1, P36A/T38Δ, and control luciferase protein, respectively). (D) Coimmunoprecipitation experiments in mammalian overexpression systems confirmed that 36-38A TCL1 had lost the capacity for homodimerization (compare lane 2 with lane 5, wild-type and 36-38A TCL1, respectively). The equal amounts of loading and the expression of TCL1 in this experimental system are shown below (anti-HA [H], anti-Flag [F], and control [C] Abs were used for immunoprecipitation in panels D and E). (E) In a mammalian overexpression system in 293 cells, 36-38A TCL1 retained the capacity to associate with Akt1 (panel D, compare lanes 2 and 5). The results were similar with Akt2 and Akt3 (data not shown). The expression levels of the Flag constructs were comparable (data not shown). The results (C to E) were consistent in a separate set of experiments. WT, wild type.
In vivo phosphorylation of FKHR and BAD.
In vivo levels of phosphorylated FKHR and BAD were assessed using the phospho-FKHR Ab or anti-phospho-BAD Ab (Cell Signaling) with the following modifications. 293 cells were transfected with 2 μg of the indicated form of HA-tagged TCL1 or control vector per six-well dish by the calcium phosphate method (28). For BAD, 293 cells were transfected with 3 μg of pEBG-MBAD with a total of 7 μg of the indicated plasmids: sham (vector control) and TCL1 (1 μg). Forty-eight hours after transfection, the cells were serum starved for 24 h before harvesting. The cells were lysed, and an equal amount of protein was loaded into individual lanes and Western blotted using the indicated Abs.
Mitochondrial transmembrane potential (MTP) measurement.
MTP measurement using rhodamine 123 was performed as described previously (28). Briefly, transfected cells were stimulated with 50 ng of human tumor necrosis factor alpha (PeproTech)/ml for 4 h at 37°C, stained with 2.5 μM rhodamine 123 (Molecular Probes) for 15 min and with 2 μg of propidium iodide (Boehringer Mannheim)/ml, and analyzed using fluorescence-activated cell sorting (FACS).
Cell recovery assays.
Cell recovery assay after cytokine withdrawal was performed essentially as described previously (24). Internal ribosomal entry site GFP retroviruses of wild-type and indicated mutant forms of TCL1 were generated as described previously (28). Splenic CD4+ T cells were purified from DO10 T-cell-receptor-transgenic mice by using T-cell enrichment columns (R&D Systems). After CD4+ T cells were stimulated with anti-CD3 MAb plus anti-CD28 MAb for 48 h, cells were washed with PBS and incubated with 500 μl of the retroviruses in the presence of interleukin 2 (IL-2; 20 U/ml) in a 24-well plate precoated with recombinant fibronectin fragment (CH-296, RetroNectin, 27 mg/ml; TaKaRa). After 4 h of infection, 500 μl of fresh medium containing IL-2 and IL-4 (20 ng/ml) was added and cells were allowed to grow for an additional 60 h. Under these conditions, the transduction efficiency to CD4+ T cells was 15 to 20% as assessed by counting GFP-positive cells by FACS. The cells were washed twice with PBS and cultured in the presence or absence of IL-4 and IL-2 for an additional 72 h. The number of live infected cells (GFP-positive, propidium iodide-negative cells) was determined by FACS (24, 31). The percent cell recovery was calculated as (number of viable cells without cytokines/number of viable cells with cytokines) × 100. The data presented are relative % cell recovery compared to the wild-type TCL1 transduction = 100 × {[% viable cells gating on transduced cells (GFP positive) of mutant form of TCL1] − [% viable cells in sham transduction (40.4%)]}/{[% viable cells gating on transduced cells (GFP positive) of wild-type TCL1 (80.3%)] − [% viable cells in sham transduction (40.4%)]}.
Nuclear translocation of Akt.
GFP fusion Akt3 (in pEGFP vector [Clontech], 0.5 μg) was cotransfected with 2.5 μg of TCL1 (wild type, D16G/I74V, 36-38A, or sham control vector) per six-well dish into 293 cells by the calcium phosphate method. Sixteen hours after transfection, cells were serum starved for an additional 6 h, fixed with 4% paraformaldehyde, and visualized under an immunofluorescence microscope.
Abs utilized in this study.
Anti-HA Ab (3F10; Boehringer Mannheim); anti-Flag Ab (M2; Kodak); anti-phospho-Akt (Ser-473, Thr-308); anti-phospho-glycogen synthase kinase 3α (GSK-3α); anti-Akt; FKHR; phospho-FKHR (S-256; Cell Signaling); 27D6 anti-TCL1 Ab (generous gift from R. Bichi); isoform-specific anti-Akt1, anti-Akt2, and anti-Akt3 Ab (Upstate Biotechnologies); GAL4 activation domain Ab (Clontech); and GFP MAb (Clontech) were used in this study.
Statistical analysis.
Data were subjected to a one-way analysis of variance followed by Dunnett multiple comparison tests. P < 0.05 was considered to be significant.
Quantitation of the kinase assays.
All kinase assays were quantitated by using the NIH Image (version 1.62) program calculating fold increase compared to control.
RESULTS
TCL1 random mutation library screening for loss of Akt-TCL1 interaction revealed accumulation of amino acid substitutions at positions D16, K30, Q46, I74, and M106.
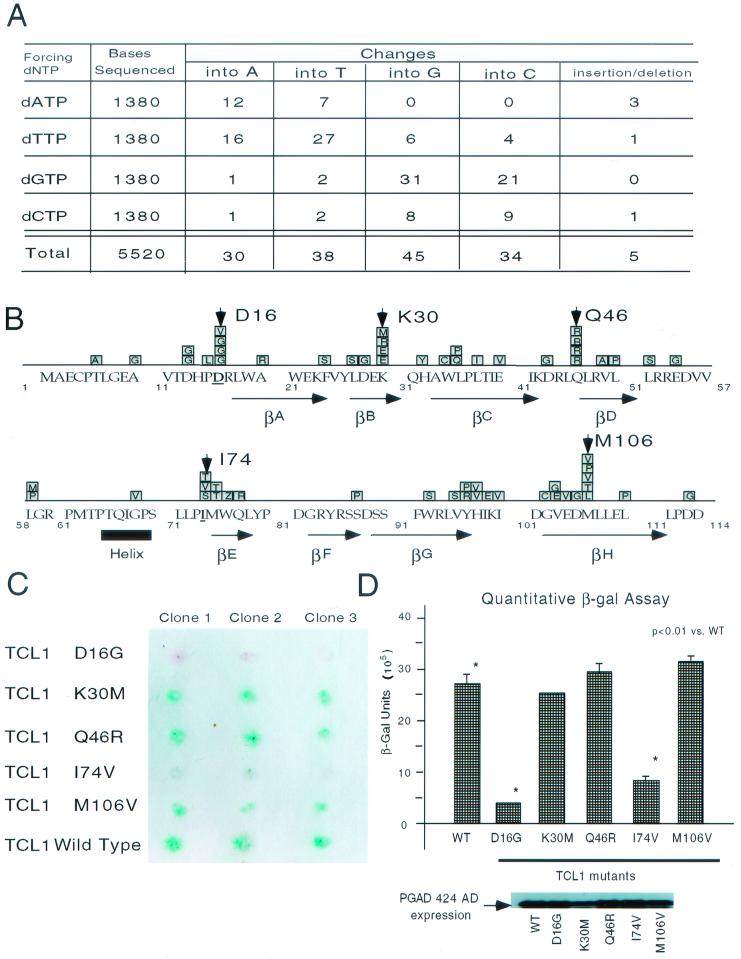
In order to define amino acid residues which mediate the Akt-TCL1 interaction, we generated a random TCL1 library by PCR-mediated random mutagenesis. Enforced dATP resulted in 1.4%, dTTP resulted in 3.8%, dGTP resulted in 4.0%, and dCTP resulted in 1.4% substitution incidence. The total frequency of nucleotide substitutions in this library was 2.7% with a 0.09% insertion-deletion rate (Fig. 1A). The size of the library was approximately 2.5 × 104 bp. The substitutions were distributed over more than 90% of the whole molecule in the 25 sampling clones sequenced (data not shown).
FIG. 1.
Yeast two-hybrid screening of random TCL1 library for loss of interaction. (A) The nucleotide change and deletion-insertion rates for the enforced nucleotide in each PCR for generation of the random TCL1 library are shown. Enforced dATP resulted in a 1.4% (12 + 7 of 1,380) frequency of nucleotide substitutions, dTTP resulted in a 3.8% (16 + 27 + 6 + 4 of 1,380) frequency, dGTP resulted in a 4.0% (1 + 2 + 31 + 21 of 1,380) frequency, and dCTP resulted in a 1.4% (1 + 2 + 8 + 9 of 1,380) frequency. The total incidence of nucleotide substitution in this library was 2.7% (30 + 38 + 45 + 34 of 5,520) with an 0.09% (5 of 5,520) insertion-deletion rate. (B) Amino acid substitutions observed in clones interacting weakly with Akt (8 h positive [+]) are shown aligned with the structure of TCL1 (23). Accumulation of substitutions for amino acids D16, K30, Q46, I74, and M106 was observed. (C) The β-Gal lifting assay was repeated using the wild-type and individual mutant forms of TCL1 (D16G, K30M, Q46R, I74V, and M106V). Three independent D16G TCL1 and I74V TCL1 yeast clones demonstrated dramatically reduced interaction with Akt as shown by color reaction in the β-Gal lifting assay. The transformation efficiency of the Y190 yeast strain (Clontech) and the protein expression of the individual constructs were comparable (data not shown). (D) D16G and I74V TCL1 showed reduced Akt interaction in quantitative β-Gal assay. The data presented were normalized by the level of expression of each TCL1 mutant. Yeast transformation efficiencies (data not shown) as well as the expression of each mutant TCL1 (shown in inset below) were comparable. WT, wild type.
We then performed a yeast two-hybrid assay to study the interaction of individual clones with Akt2. The surviving yeast clones were classified into three categories based on the intensity of blue color reaction in the β-Gal lifting assay (β-Gal positive at 3 h [++], β-Gal positive at 8 h [+], and negative [−]). The nucleotide sequences of 10 clones of each category were determined. The ++ clones contained wild-type TCL1 or mutations at positions P5, P15, D43, L45, P61, M75, and D88, which did not affect β-Gal activity, indicating that the residues were not responsible for Akt interaction. The amino acid substitutions observed in the 10 clones showing reduced interaction with Akt (8 h positive [+]) are shown aligned with the amino acid sequence of TCL1 (Fig. 1B). Among these clones, there was an apparent accumulation of substitutions at specific residues. In 9 out of the 10 clones, we found substitutions of at least one of the amino acids at D16, K30, Q46, I74, or M106. Negative clones (−) contained no insert, inserts with massive deletions, frameshifts, and/or multiple mutations and were therefore excluded from further analysis.
We hypothesized that the clones, which showed reduced Akt interaction, might include the crucial residue(s) mediating Akt-TCL1 interaction. Therefore, we reintroduced the individual mutations (D16G, K30M, Q46R, I74V, or M106V) into TCL1 by site-directed mutagenesis. The substitutions D16G and I74V resulted in dramatically decreased binding with Akt as shown by β-Gal lifting assay (Fig. 1C) and quantitative liquid β-Gal assay (Fig. 1D).
D16 and I74 mediate Akt interaction.
Consistently, in coimmunoprecipitation experiments in a mammalian overexpression system, D16G TCL1 showed markedly reduced interaction with Akt2. I74V TCL1 showed a more modest, but reproducible, reduction of interaction with Akt2 (Fig. 2A and B). Similar to D16G TCL1, D16G/I74V TCL1 showed reduced interaction with Akt2 in coimmunoprecipitation assays (data not shown). Further, D16G TCL1 exhibited decreased binding not only with Akt2 but also with Akt1 and Akt3 (Fig. 2C and data not shown). Both D16G and double mutant D16G/I74V TCL1 retained the ability to form homodimers in the overexpression system, indicating that their overall structure and folding remained intact (Fig. 2D and data not shown).
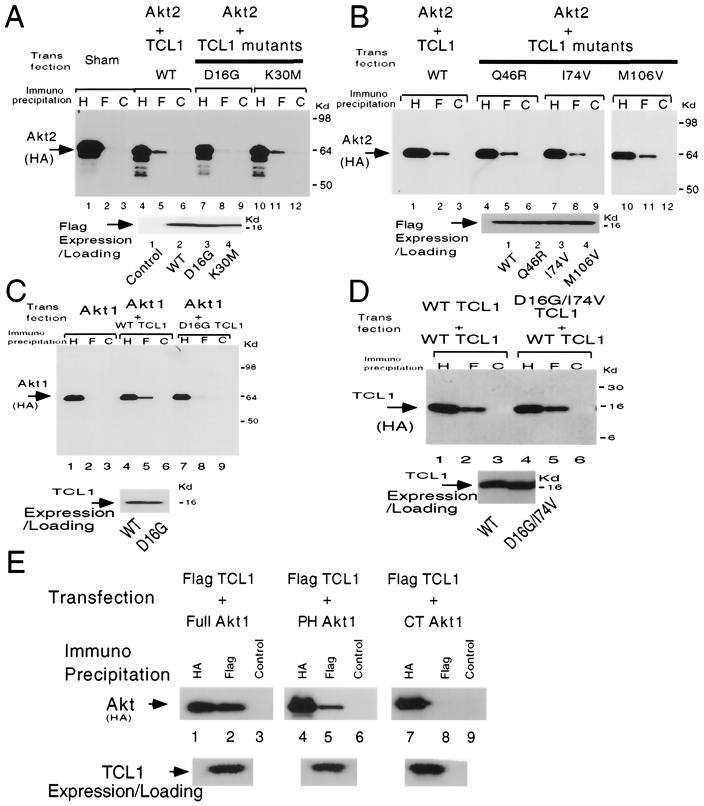
FIG. 2.
Identification of D16 and I74 as the putative binding sites of Akt in mammalian cells. (A and B) In coimmunoprecipitation experiments in a mammalian overexpression system, D16G TCL1 (panel A, lanes 4 to 6 with 7 to 9, wild-type TCL1 and D16G TCL1, respectively) showed markedly reduced interaction with Akt2. The interaction of I74V with Akt2 (panel B, lanes 7 to 9) was modest but reproducibly reduced. The equal amount of loading and the expression of the indicated TCL1 constructs were similar when analyzed by Flag Western blotting (anti-HA [H], anti-Flag [F], and control [C] Abs were used for immunoprecipitation in all panels). (C) In a mammalian overexpression experiment in 293 cells, D16G TCL1 showed reduced interaction with Akt1 (compare lane 5 with lane 8) compared with that for wild-type TCL1. D16G TCL1 also could not associate with two other isoforms of Akt (Akt2 and Akt3) in a coimmunoprecipitation assay (data not shown). (D) In a mammalian overexpression experiment using 293 cells D16G/I74V (double substitution mutation) TCL1 retained the capacity for homodimerization in coimmunoprecipitation assays compared to wild-type TCL1 (wild-type TCL1, lanes 1 to 3; D16G/I74V, lanes 4 to 6). Similarly, D16G TCL1 retained the capacity for homodimerization (data not shown). (E) TCL1 interacts with the PH domain of Akt1 in a mammalian overexpression system (lanes 1 to 3, 4 to 6, and 7 to 9, full-length Akt1, PH domain of Akt, and C-terminal Akt1, respectively). The results were consistent in three independent experiments. WT, wild type.
These observations strongly suggest that D16 and I74 are residues mediating the Akt-TCL1 interaction and that they are distinct from the TCL1 homodimerization domain.
D16 and I74 mediate Akt interaction.
In a mammalian overexpression system we then determined that the interaction between Akt1 and TCL1 is mediated through the Akt1 PH domain, since TCL1 coimmunoprecipitated with both full-length Akt1 and the PH domain of Akt1 (lanes 1 to 6) but not with C terminus Akt (Fig. 2E).
The βC-sheet of TCL1 is the functional domain required for homodimerization.
Based on the X-ray structure, TCL1 is predicted to form a symmetrical β-barrel structure (23). Amino acids D16 and I74 are localized at symmetrical positions at the very beginning of the first and fifth β-sheet (βA and βE, D16 and I74, respectively) of TCL1. Since TCL1 is composed of 114 amino acids, position 74 is almost identical to position 16 (17th amino acid from the beginning of the second groove [Fig. 3A]). Moreover, in the X-ray crystal structure, both positions are located on the same surface of TCL1, composed of βA and βE (see also Fig. 7A). Based on the X-ray analysis (23), we hypothesized that one of the β-sheets located on the opposite side of the surface mediating Akt interaction might be required for homodimerization. Therefore, we mutated the βC-sheet of TCL1, generating P36A/L37A/T38A TCL1 (36-38A TCL1) and P36A/T38Δ TCL1 (36A/38Δ TCL1), and tested whether these mutant forms of TCL1 were capable of dimerizing (Fig. 3B).
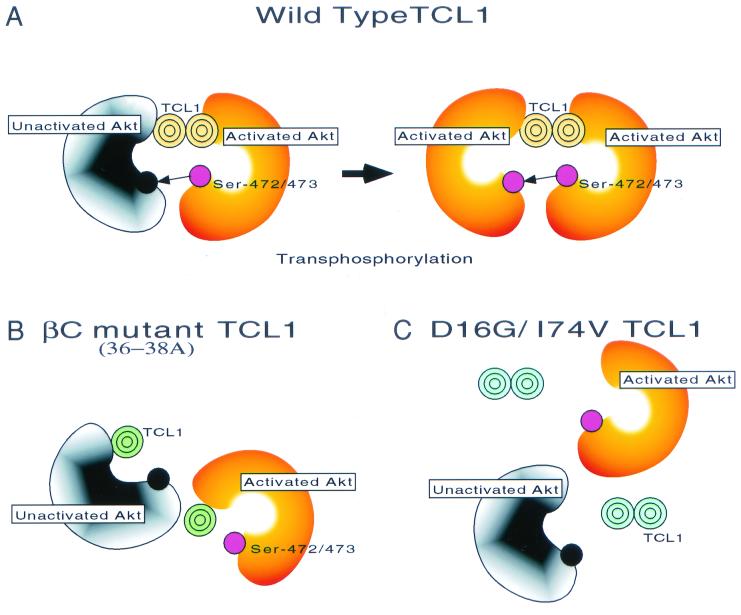
FIG. 7.
Model of TCL1-dependent Akt kinase activation. Based on the findings presented in this study, both TCL1 dimerization (B) (36-38A mutant TCL1) and the TCL1-Akt interaction domain (C) (D16/I74V TCL1) are required for the recruitment of individual Akt molecules into the same high-molecular-weight protein complex, bringing Akt molecules close to each other. Thus, both domains are needed for the full function of TCL1 as an Akt coactivator in vitro and in vivo (A).
Using GST pull-down assays with in vitro-translated TCL1, we demonstrated that wild-type TCL1, but not the mutant forms of TCL1 (36-38A and 36A/38Δ), could homodimerize (Fig. 3C). The results were consistent in coimmunoprecipitation experiments in a mammalian overexpression system in which 36-38A TCL1 and 36A/38Δ TCL1 could not homodimerize (Fig. 3D and data not shown). Similarly, 36-38A TCL1 did not interact with wild-type TCL1 in coimmunoprecipitation assays (data not shown). However, 36-38A TCL1 was able to interact with Akt1, Akt2, and Akt3 (Fig. 3E and data not shown). The results indicate that the overall folding of 36-38A TCL1 remained intact and show that mutating the βC-sheet specifically resulted in the loss of homodimerization while Akt binding capability remained intact.
Both association with Akt and TCL1 homodimerization are required for TCL1-induced Akt activation in vitro and in vivo.
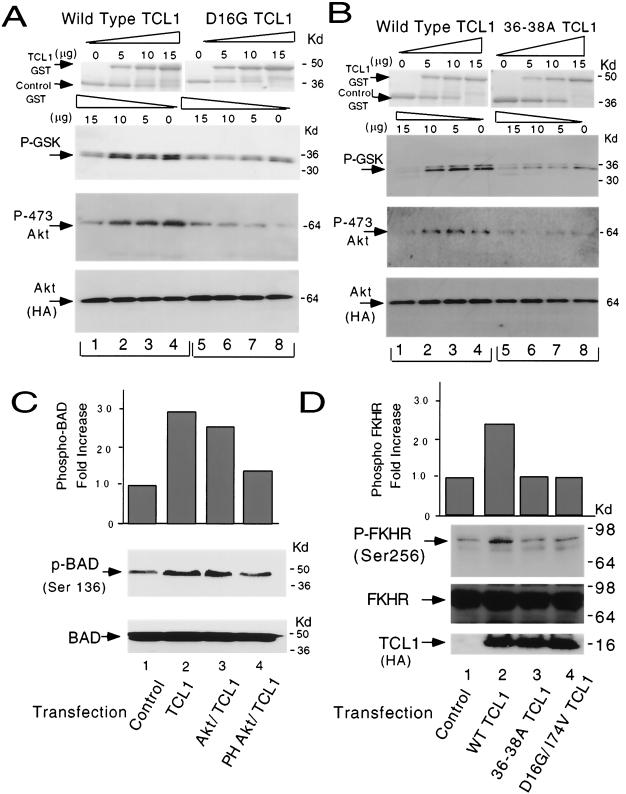
In in vitro Akt kinase assays, wild-type TCL1 enhanced Akt kinase activity, which correlated well with increased Ser-473 phosphorylation of Akt. However, D16G TCL1 had no effect on Akt kinase activity in a dose escalation experiment (Fig. 4A). Notably, while D16G TCL1 was not able to enhance Akt kinase activity as examined by GSK-3α phosphorylation, it also could not induce Akt Ser-473 phosphorylation.
FIG. 4.
Both Akt association and TCL1 homodimerization are required for TCL1-induced Akt kinase activation. (A) In in vitro kinase assays increasing amounts (0 to 15 μg) of D16G TCL1 GST fusion protein did not enhance Akt1 activity. Fold increases in GSK-3α phosphorylation were 1.0, 2.3, 3.5, and 4.7 for wild-type TCL1 (lanes 1 to 4) compared with 1.0, 0.9, 1.1, and 1.1 for D16G TCL1 (lanes 5 to 8) as detected by phospho-GSK-3α immunoblotting (top panel). Fold increases in Akt Ser-473 phosphorylation were 1.0, 2.9, 3.5, and 4.8 for wild-type TCL1 and 1.0, 1.2, 0.9, and 0.7 for D16G TCL1 as analyzed by anti-phospho-Ser-473 immunoblotting (middle panel). Equal amounts of Akt were used in the reaction mixtures as shown by anti-HA immunoblotting panels (bottom panel). (B) Dimerization-deficient 36-38A TCL1-GST was not able to enhance Akt1 activity. Fold increases in GSK-3α phosphorylation were 1.0, 3.8, 5.3, and 4.5 for wild-type TCL1 (lanes 1 to 4) compared with 1.0, 1.2, 0.9, and 0.7 for 36-38A TCL1 (lanes 5 to 8) as analyzed by phospho-GSK-3α blotting (top panel). Fold increases of Akt Ser-473 phosphorylation were 1.0, 2.9, 3.5, and 4.8 for wild-type TCL1 (lanes 1 to 4) and 1.0, 1.2, 0.9, and 0.7 for 36-38A TCL1 (lanes 5 to 8) in phospho-Ser-473 Akt immunoblotting (middle panel). Equal amounts of Akt were used in the reaction mixtures as shown by anti-HA immunoblotting panels (bottom panel). Similar results were obtained using Akt2 and Akt3 (panels A and B and data not shown). The results were consistent in a separate set of experiments. (C) Overexpression of TCL1 in cells enhanced BAD phosphorylation at Ser-136 (top panel, 2.9-fold increase compared to the control). The increased BAD phosphorylation was compromised in the presence of PH Akt (1.3-fold compared to the control), but not in the presence of wild-type Akt (2.5-fold compared to the control). Equal amounts of BAD were transfected as verified by anti-BAD Western blotting (bottom panel). The results were consistent in two separate independent experiments. (D) Overexpression of TCL1 in cells enhanced FKHR phosphorylation on Ser-256 (top panel, 2.4-fold increase compared to the control). Equal amounts of nonphosphorylated FKHR and indicated forms of TCL1 were expressed and loaded in this experiment as verified by Western blotting (middle and bottom panels, FKHR and anti-HA blotting, respectively). The results were consistent in two separate independent experiments. WT, wild type.
Similarly, 36-38A TCL1, which bound to Akt but could not homodimerize, was unable to enhance Akt kinase activity in in vitro kinase assays as shown by stable levels of phosphorylated GSK-3α and Ser-473 Akt (Fig. 4B). Analogous to Akt1, TCL1 enhanced Akt2-dependent GSK-3α phosphorylation in vitro (data not shown). The results indicate that both interaction of TCL1 with Akt and TCL1 homodimerization are required for TCL1 to function as an Akt kinase coactivator in vitro.
In an intact cell context, transient transfection of wild-type TCL1 enhanced BAD phosphorylation at Ser-136, which could be compromised by the cointroduction of pleckstrin homology (pH) of Akt (Fig. 4C). Moreover, transient transfection of wild-type TCL1 enhanced the phosphorylation of FKHR (5) in 293 cells. The increased FKHR phosphorylation could not be achieved by mutant forms of TCL1 which lack the capacity for dimerization (36-38A TCL1) or Akt association (D16G/I74V TCL1) (Fig. 4D).
Both Akt association and TCL1 dimerization are required for TCL1-mediated antiapoptosis and nuclear translocation of Akt.
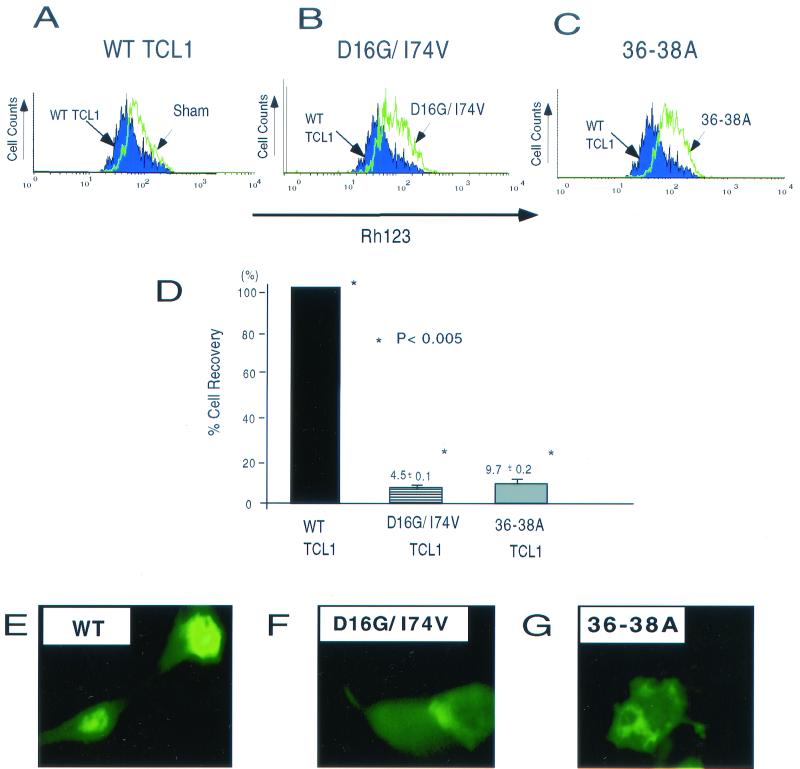
MTP has a key role in the regulation of cell death machinery (18), and Akt activation regulates this process (25). Consistent with our previous report (28), overexpression of wild-type TCL1 stabilized the mitochondrial membranous potential (Fig. 5A). The stabilization of the MTP was not achieved by introduction of double mutant D16G/I74V or 36-38A TCL1 (Fig. 5B and C).
FIG. 5.
Both Akt association and TCL1 dimerization are required for TCL1-mediated stabilization of mitochondrial membranous potential, cell survival, and nuclear translocation in vivo. (A to C) In a mammalian overexpression system using 293 cells, wild-type TCL1 stabilized the mitochondrial membranous potential after tumor necrosis factor treatment (A) (compared with overlaid sham transfection). The stabilization could not be achieved by the introduction of double mutant D16G/I74V TCL1 (B) or 36-38A TCL1 (C). The expression levels of TCL1 constructs used in this experiment were comparable as verified by Western blotting (data not shown). (D) Cell recovery assay after IL-2-IL-4 withdrawal of activated CD4+ T cells was performed by retroviral transduction of wild-type and mutant forms of TCL1. Percent cell recovery (mean ± standard deviation) compared to the wild type (100%) was 4.5% ± 0.1% in D16G/I74V and 9.7% ± 0.2% in 36-38A TCL1. Percent cell recovery enhanced by wild-type TCL1 compared to the sham transduction was approximately 40% in these experiments. (E to G) Wild-type TCL1 (E) but not D16G/I74V TCL1 (F) or 36-38A TCL1 (G) facilitated the nuclear translocation of Akt in a mammalian overexpression system. Equal expression of each construct was confirmed by Western blotting (data not shown). WT, wild type.
TCL1 also enhanced cell recovery after cytokine withdrawal in activated CD4+ T cells. Cell recovery was enhanced by wild-type TCL1. The enhancement was not observed with either D16G/I74V TCL1 or 36-38A TCL1 (4.5% ± 0.1% [D16G/I74V TCL1] or 9.7% ± 0.2% [36-38A TCL1] compared to the wild-type TCL1). The results demonstrate that both Akt association and TCL1 dimerization are required for the complete function of TCL1 in vivo (Fig. 5D).
TCL1 has been reported previously to enhance nuclear translocation of Akt (35). We further examined the functional requirement of Akt association and oligomerization of TCL1 for the nuclear translocation of Akt. Consistent with a previous report (35), wild-type TCL1 facilitated the nuclear translocation of Akt, which could not be achieved by D16G/I74V TCL1 or 36-38A TCL1 (Fig. 5E to G). These results establish that both TCL1 oligomerization and association with Akt are required for the complete function of TCL1 in vivo.
DISCUSSION
The use of a random mutation library combined with a functional assay system is a powerful strategy for structural-functional analysis (32, 51). We used this approach combined with a yeast two-hybrid assay in order to identify the residues of TCL1 required for Akt binding. Enforced dNTP-driven PCR amplification was utilized to generate a random TCL1 library as described previously (15). Since TCL1 cDNA consists of only 342 nucleotides, the approximately 3% substitution frequency resulted on the average in nine nucleotide (and one to three amino acid) substitutions per single TCL1 molecule, making further analysis by library screening feasible.
Although the yeast two-hybrid system was originally developed to identify protein-protein interaction (13), our results indicate that it can be used to screen for loss of interaction. This was made possible by the very high β-Gal activity induced by the wild-type TCL1-Akt interaction in the system. It enabled mutant forms of TCL1 with very weak Akt interactions to grow on Trp-Leu-His-deficient selection plates and be identified using the β-Gal lifting assay. The screening was also relatively specific and possibly applicable to other molecules, since, of the five residues showing accumulation of mutations, two were found to be functionally relevant.
Analysis of the X-ray structure has revealed that TCL1 forms a closed symmetrical β-barrel structure, consisting of eight antiparallel β-strands. The barrel consists of two similar four-stranded β-meander motifs: motif 1, composed of strands A to D, and motif 2, composed of strands E to H. A long loop following strand D inverts the second motif by juxtaposing strand E to strand A. Each β-sheet of TCL1 contains two strands from each motif (23). One sheet is composed of two pairs of the shorter strands (A-B and E-F), whereas the other is formed by the longer strands (C-D and G-H). The loop contains one helical turn (Pro-64-Gly-68) (Fig. 6A).
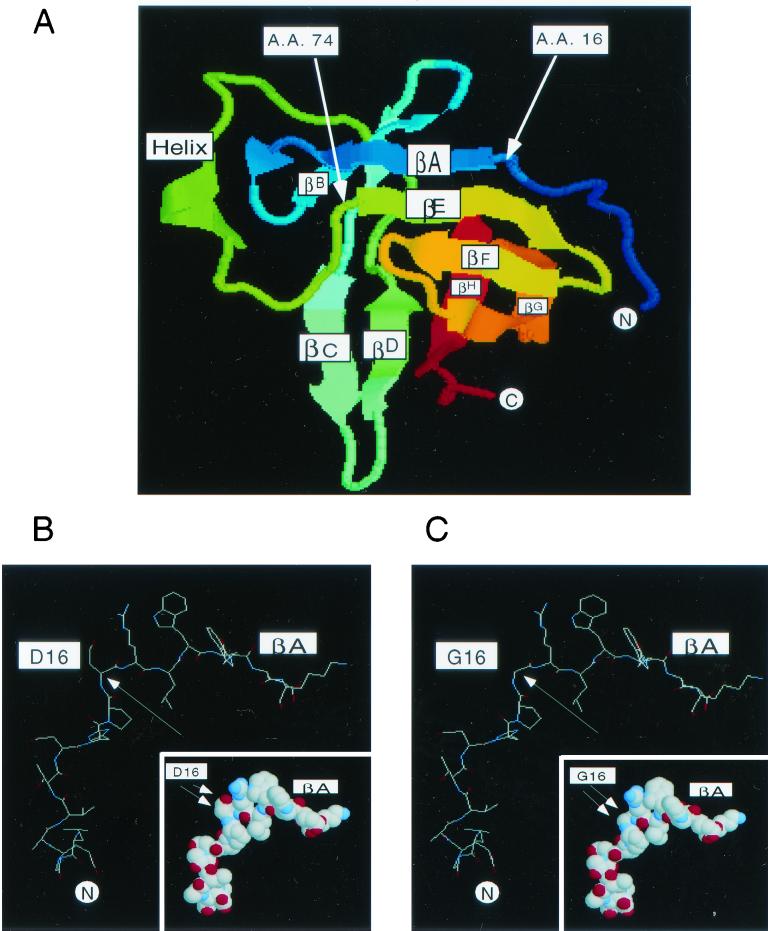
FIG. 6.
Alignment of amino acid positions D16 and I74 within the topology of the eight-strand antiparallel β-sheets of TCL1. (A) The crystal structure of TCL1 (23) and the amino acid positions of D16 and I74 are shown. Amino acids D16 and I74 are located at the very beginning of two β-sheets (βA and βE, respectively). βA and βE form the top surface of the TCL1 barrel structure. The crystal study suggested that the βC-sheet of TCL1 serves as a dimerization domain (19, 23). Since the Akt-TCL1 interaction is mediated by the surface formed by βA and βE, it is logical that TCL1 dimerization can be mediated by the surface directly on the opposite side of the molecule, the βC-sheet. The arrows indicate the positions of amino acids D16 and I74. The Swiss-Prot accession no. of human TCL1 is P56279. (B and C) Molecular modeling of wild-type (B) and D16G (C) TCL1 based on the Swiss-pdbViewer (version 3.7b2) program is shown. D16, which is hydrophilic and accessible to solvent, is likely to contribute to the stabilization of βA to βE through electrostatic interactions together with surrounding amino acids including a cluster of basic side chains of R17, E29, and Q77. Molecular modeling predicts that the substitution of glycine (G16) for aspartic acid (D16) changes the angle between amino acids 15 and 17 (PDR to PGR) from 88.45 to 71.62o, altering the alignment of βA to βE. In addition, the absence of a side chain towards the βE surface in the glycine mutation might also have a role in destabilizing the interaction. The changing angle between amino acids 15 and 17 is indicated by a single arrow. Note that the amino acid substitution of G16 for D16 results in the loss of the side chain as shown in the inset by double arrows.
The results obtained in the screening of the random TCL1 mutation library and their functional verification demonstrated that amino acids D16 and, to a lesser extent, I74 mediate Akt binding. These residues are localized at very similar positions at the very beginning of the first β-strand of each of the two β-meander motifs of TCL1 (βA and βE, D16 and I74, respectively [Fig. 3A]). Moreover, they reside on the same flat surface, formed by βA and βE, of mature TCL1 (Fig. 6A). βA and βE are linked together in an antiparallel fashion by CO-to-NH hydrogen bonds (23).
D16, which is hydrophilic and accessible to solvent, is likely to contribute to the stabilization of βA to βE through electrostatic interaction together with surrounding amino acids including a cluster of basic side chains of R17, E29, and Q77. Molecular modeling predicts that the substitution of glycine (G16) for aspartic acid (D16) changes the angle between amino acids 15 and 17 (PDR to PGR) from 88.45 to 71.62°, altering the alignment of βA to βE. In addition, the absence of a side chain towards the βE surface in glycine mutation (Fig. 6B and C , inset) might also have a role in destabilizing the interaction.
In contrast to D16, the role of I74 in Akt interaction seems less clear. Although in the yeast system, I74V mutation clearly disrupted Akt binding, its effects in mammalian cells and functional assays were less evident. I74 is located at the beginning of the βE-sheet, which is well conserved among the TCL1 family proteins. The residue appears to be poorly solvent accessible. It may participate in forming a long loop structure composed of amino acids 53 to 73 via a hydrophobic interaction with P64 and contribute to maintaining a local hydrophobic surface.
It is possible that Akt binds to TCL1 through the βA- to βE-sheets via two distinct sites: D16, conceivably together with the surrounding conserved amino acids, and the hydrophobic contact region P64/I74. Detailed molecular characterization of the interaction awaits the elucidation of the cocrystal structure of the Akt-TCL1 complex.
We have demonstrated elsewhere that TCL1 forms dimers as predicted by X-ray crystallographic studies (23, 28). The crystal study suggested that the βC-sheet of TCL1 acts as a dimerization domain (19, 23). Since the Akt-TCL1 interaction is mediated by the surface formed by βA and βE, it is logical that TCL1 dimerization can be mediated by the surface directly on the opposite side of the molecule and therefore targets the βC-sheet. Our data demonstrate that 36-38A and 36A/38Δ mutations, which presumably destroy the βC-sheet, eliminate the homodimerization of TCL1 but have no effect on Akt association. Based on this, the TCL1-TCL1 binding is conferred by interaction between the βC-sheets of two symmetrical TCL1 molecules.
How does the presence of TCL1 lead to enhanced Akt phosphorylation and kinase activation? It is well documented elsewhere that dimerization can lead to activation of surface receptor kinases (e.g., vascular endothelial growth factor receptor and platelet-derived growth factor receptor) and regulate their intracellular responses (3, 20, 40). It is unclear whether intracellular nontransmembrane kinases can be activated through dimerization. It has been reported elsewhere that TEL-JAK2 fusion protein causes human leukemia due to oligomerization and constitutive kinase activation (27). The Akt PH domain has been suggested previously to mediate dimerization (6, 10). Further, a conditionally activated Akt fused to the hormone binding domain of estrogen receptor was able to stimulate PHAS-1 phosphorylation (26), suggesting that Akt dimerization may promote kinase activity.
PDK1 is required for phosphorylating Akt at Thr-305/308 (37, 42) and for activation of Akt, which is thought to be the prerequisite for subsequent Ser-473 phosphorylation (3). However, recent studies using PDK1−/− cells suggest that phosphorylation of Ser-473 may be independent from phosphorylation of Thr-308 (50). Supporting this notion, Ser-473 of Akt was recently shown to be regulated by integrin-linked kinase (36). Since wortmannin treatment can block the TCL1-dependent Akt activation, some degree of basal Akt activity seems to be required for the TCL1-dependent Akt kinase activation to occur (28, 35). This observation suggests not only that TCL1 functions within the intracellular phosphoinositide 3-kinase (wortmannin-sensitive) pathway but also that association of Akt with PIs and/or a certain preactivation of Akt, sensitive to wortmannin, may be prerequisite for the TCL1-induced Akt activation. Our observations also suggest that the effect of TCL1 on the phosphorylation status and activation of Akt seems to be additive to, and possibly at least partially independent of, the effects of PDK1. However, in our experimental system of in vitro kinase assays, we cannot completely exclude the potential contamination of immobilized Akt by PDK1 or other functionally equivalent kinases during the immobilization process from transfected cells.
The Akt PH domain can directly bind to phospholipids including PI-3,4-bisphosphate and PI-3,4,5-triphosphate, thereby facilitating the translocation of the protein to the plasma membrane (14, 38). As the molecular masses of phospholipids are relatively small (less than 5 kDa), it is feasible for the PH domain of Akt, which is composed of over 100 amino acids, to interact with PIs as well as TCL1.
As demonstrated by coimmunoprecipitation assays, our data show that TCL1 binds to Akt and facilitates the formation of Akt-TCL1 hetero-oligomers in vivo. Thereby, Akt molecules come into close physical proximity with each other. We have recently demonstrated that, in the presence of TCL1, Akt Ser-473 phosphorylation can result from transphosphorylation by other Akt molecules in the oligomers (28a). In this scenario, PDK1 is required for triggering the Akt kinase activation and TCL1 further enhances Akt kinase activity. These data are in accordance with reports demonstrating the requirement of Akt kinase activity for Ser-473 phosphorylation in vitro (46).
Therefore, although it is likely that unidentified kinases capable of Akt Ser-473 phosphorylation exist, at least under certain circumstances Ser-473 phosphorylation can occur through oligomerization-induced Akt-Akt transphosphorylation (Fig. 7). Based on the findings of the present study, it seems likely that the TCL1-Akt interaction domain is required for the recruitment of individual Akt molecules into the oligomeric complexes and that the TCL1 dimerization domain is essential for bringing several Akt molecules into the same complex. Thus, both domains are needed for the full function of TCL1 as an Akt coactivator in vitro and in vivo (Fig. 7).
The physiological expression of TCL1 is relatively abundant at early embryonic stages (34) and during CD3-negative stages of T-cell development (49), periods when external growth factor stimuli may be limited. TCL1 family proteins may function as a structural amplification loop in the phosphoinositide 3-kinase-Akt pathway, providing a survival advantage.
Acknowledgments
We thank B. Hemmings (Friedrich Miescher Institute) for Akt3 cDNA; T. Franke (Columbia University) for Akt expression vectors; J. Testa (FCCC) for Akt2 cDNA; A. Bellacosa (FCCC), T. Kitamura (Tokyo University), E. Minet and P. Silver (DFCI), and J. Chiorirni (NIH) for providing reagents and technical advice; L. Cantley (BIDMC), J. O'Shea (NIH), J. P. Kinet (BIDMC), and colleagues for their valuable suggestions; F. Pechenick for secretarial assistance; and T. B. Strom (BIDMC) for generous support for the project.
G.P. is supported by a grant from the Ministère de l'Enseignement National et de la Recherche. M.N. is supported by a Cancer Research Institute Investigator Award. J.L. is supported by grants from the National Kidney Foundation, the Academy of Finland, and The Foundation for Pediatric Research. G.K. is supported by the Emmy Noether Program of the Deutsche Forschungsgemeinschaft (DFG). C.R. and F.H. are supported by the Association pour la Recherche contre le Cancer (ARC).
REFERENCES
- 1.Alessi, D. R., S. R. James, C. P. Downes, A. B. Holmes, P. R. Gaffney, C. B. Reese, and P. Cohen. 1997. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Ba. Curr. Biol. 7:261-269. [DOI] [PubMed] [Google Scholar]
- 2.Andjelkovic, M., D. R. Alessi, R. Meier, A. Fernandez, N. J. Lamb, M. Frech, P. Cron, P. Cohen, J. M. Lucocq, and B. A. Hemmings. 1997. Role of translocation in the activation and function of protein kinase B. J. Biol. Chem. 272:31515-31524. [DOI] [PubMed] [Google Scholar]
- 3.Blume-Jensen, P., and T. Hunter. 2000. Oncogenic kinase signaling. Nature 411:355-365. [DOI] [PubMed] [Google Scholar]
- 4.Brodbeck, D., P. Cron, and B. A. Hemmings. 1999. A human protein kinase Bg with regulatory phosphorylation sites in the activation loop and in the C-terminal hydrophobic domain. J. Biol. Chem. 274:9133-9136. [DOI] [PubMed] [Google Scholar]
- 5.Brunet, A., A. Bonni, M. J. Zigmond, M. Z. Lin, P. Juo, L. S. Hu, M. J. Anderson, K. C. Arden, J. Blenis, and M. E. Greenberg. 1999. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96:857-868. [DOI] [PubMed] [Google Scholar]
- 6.Chan, T. O., S. E. Rittenhouse, and P. N. Tsichlis. 1999. AKT/PKB and other D3 phosphoinositide-regulated kinases: kinase activation by phosphoinositide-dependent phosphorylation. Annu. Rev. Biochem. 68:965-1014. [DOI] [PubMed] [Google Scholar]
- 7.Chen, W. S., P. Z. Xu, K. Gottlob, M. L. Chen, K. Sokol, T. Shiyanova, I. Roninson, W. Weng, R. Suzuki, K. Tobe, T. Kadowaki, and N. Hay. 2001. Growth retardation and increased apoptosis in mice with homozygous disruption of the akt1 gene. Genes Dev. 15:2203-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho, H., J. Mu, J. K. Kim, J. L. Thorvaldsen, Q. Chu, E. B. Crenshaw III, K. H. Kaestner, M. S. Bartolomei, G. I. Shulman, and M. J. Birnbaum. 2001. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB beta). Science 292:1728-1731. [DOI] [PubMed] [Google Scholar]
- 9.Coffer, P. J., J. Jin, and J. R. Woodgett. 1998. Protein kinase B (c-Akt): a multifunctional mediator of phosphatidylinositol 3-kinase activation. Biochem. J. 335:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datta, K., T. F. Franke, T. O. Chan, A. Makris, S. I. Yang, D. R. Kaplan, D. K. Morrison, E. A. Golemis, and P. N. Tsichlis. 1995. AH/PH domain-mediated interaction between Akt molecules and its potential role in Akt regulation. Mol. Cell. Biol. 15:2304-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datta, S. R., A. Brunet, and M. E. Greenberg. 1999. Cellular survival: a play in three Akts. Genes Dev. 13:2905-2927. [DOI] [PubMed] [Google Scholar]
- 12.Delcommenne, M., C. Tan, V. Gray, L. Rue, J. Woodgett, and S. Dedhar. 1998. Phosphoinositide-3-OH kinase-dependent regulation of glycogen synthase kinase 3 and protein kinase B/AKT by the integrin-linked kinase. Proc. Natl. Acad. Sci. USA 95:11211-11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245-246. [DOI] [PubMed] [Google Scholar]
- 14.Franke, T. F., D. R. Kaplan, L. C. Cantley, and A. Toker. 1997. Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science 275:665-668. [DOI] [PubMed] [Google Scholar]
- 15.Fromant, M., S. Blanquet, and P. Plateau. 1995. Direct random mutagenesis of gene-sized DNA fragments using polymerase chain reaction. Anal. Biochem. 224:347-353. [DOI] [PubMed] [Google Scholar]
- 16.Fruman, D. A., R. E. Meyers, and L. C. Cantley. 1998. Phosphoinositide kinases. Annu. Rev. Biochem. 67:481-507. [DOI] [PubMed] [Google Scholar]
- 17.Gritti, C., H. Dastot, J. Soulier, A. Janin, M. T. Daniel, A. Madani, G. Grimber, P. Briand, F. Sigaux, and M. H. Stern. 1998. Transgenic mice for MTCP1 develop T-cell prolymphocytic leukemia. Blood 92:368-373. [PubMed] [Google Scholar]
- 18.Gross, A., J. M. McDonnell, and S. J. Korsmeyer. 1999. BCL-2 family members and the mitochondria in apoptosis. Genes Dev. 13:1899-1911. [DOI] [PubMed] [Google Scholar]
- 19.Guignard, L., A. Padilla, J. Mispelter, Y.-S. Yang, M.-H. Stern, J. M. Lhoste, and C. Roumestand. 2000. Backbone dynamics and solution structure refinement of the 15N-labeled human oncogenic protein p13MTCP1: comparison with X-ray data. J. Biomol. NMR 17:215-230. [DOI] [PubMed] [Google Scholar]
- 20.Helden, C.-H. 1995. Dimerization of cell surface receptors in signal transduction. Cell 80:213-223. [DOI] [PubMed] [Google Scholar]
- 21.Hemmings, B. A. 1997. Akt signaling: linking membrane events to life and death decisions. Science 275:628-630. [DOI] [PubMed] [Google Scholar]
- 22.Hill, M. M., M. Andjelkovic, D. P. Brazil, S. Ferrari, D. Fabbro, and B. A. Hemmings. 2001. Insulin-stimulated protein kinase B phosphorylation on Ser-473 is independent of its activity and occurs through a staurosporine-insensitive kinase. J. Biol. Chem. 276:25643-25646. [DOI] [PubMed] [Google Scholar]
- 23.Hoh, F., Y. S. Yang, L. Guignard, A. Padilla, M. H. Stern, J. M. Lhoste, and H. van Tilbeurgh. 1998. Crystal structure of p14TCL1, an oncogene product involved in T-cell prolymphocytic leukemia, reveals a novel beta-barrel topology. Structure 6:147-155. [DOI] [PubMed] [Google Scholar]
- 24.Kagami, S., H. Nakajima, A. Suto, K. Hirose, K. Suzuki, S. Morita, I. Kato, Y. Saito, T. Kitamura, and I. Iwamoto. 2001. Stat5a regulates T helper cell differentiation by several distinct mechanisms. Blood 97:2358-2365. [DOI] [PubMed] [Google Scholar]
- 25.Kennedy, S. G., E. S. Kandel, T. K. Cross, and N. Hay. 1999. Akt/protein kinase B inhibits cell death by preventing the release of cytochrome c from mitochondria. Mol. Cell. Biol. 19:5800-5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohn, A. D., A. Barthel, K. S. Kovacina, A. Boge, B. Wallach, S. A. Summers, M. J. Birnbaum, P. H. Scott, J. C. Lawrence, Jr., and R. A. Roth. 1998. Construction and characterization of a conditionally active version of the serine/threonine kinase Akt. J. Biol. Chem. 273:11937-11943. [DOI] [PubMed] [Google Scholar]
- 27.Lacronique, V., A. Boureux, V. D. Valle, H. Poirel, C. T. Quang, M. Mauchauffe, C. Berthou, M. Lessard, R. Berger, J. Ghysdael, and O. A. Bernard. 1997. A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science 278:1309-1312. [DOI] [PubMed] [Google Scholar]
- 28.Laine, J., G. Kunstle, T. Obata, M. Sha, and M. Noguchi. 2000. The protooncogene TCL1 is an Akt kinase coactivator. Mol. Cell 6:395-407. [DOI] [PubMed] [Google Scholar]
- 28a.Laine, J., G. Künstle, T. Obata, and M. Noguchi. 2002. Differential regulation of Akt kinase isoforms by the members of the TCL1 oncogene family. J. Biol. Chem. 276:3743-3751. [DOI] [PubMed] [Google Scholar]
- 29.Maira, S. M., I. Galetic, D. P. Brazil, S. Kaech, E. Ingley, M. Thelen, and B. A. Hemmings. 2001. Carboxyl-terminal modulator protein (CTMP), a negative regulator of PKB/Akt and v-Akt at the plasma membrane. Science 294:374-380. [DOI] [PubMed] [Google Scholar]
- 30.Narducci, M. G., E. Pescarmona, C. Lazzeri, S. Signoretti, A. M. Lavinia, D. Remotti, E. Scala, C. D. Baroni, A. Stoppacciaro, C. M. Croce, and G. Russo. 2000. Regulation of TCL1 expression in B- and T-cell lymphomas and reactive lymphoid tissues. Cancer Res. 60:2095-2100. [PubMed] [Google Scholar]
- 31.Noguchi, M., A. Sarin, M. J. Aman, H. Nakajima, E. W. Shores, P. A. Henkart, and W. J. Leonard. 1997. Functional cleavage of the common cytokine receptor gamma chain (gammac) by calpain. Proc. Natl. Acad. Sci. USA 94:11534-11539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Onishi, M., T. Nosaka, K. Misawa, A. L. Mui, D. Gorman, M. McMahon, A. Miyajima, and T. Kitamura. 1998. Identification and characterization of a constitutively active STAT5 mutant that promotes cell proliferation. Mol. Cell. Biol. 18:3871-3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pekarsky, Y., C. Hallas, and C. M. Croce. 2001. The role of TCL1 in human T-cell leukemia. Oncogene 20:5638-5643. [DOI] [PubMed] [Google Scholar]
- 34.Pekarsky, Y., C. Hallas, M. Isobe, G. Russo, and C. M. Croce. 1999. Abnormalities at 14q32.1 in T cell malignancies involve two oncogenes. Proc. Natl. Acad. Sci. USA 96:2949-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pekarsky, Y., A. Koval, C. Hallas, R. Bichi, M. Tresini, S. Malstrom, G. Russo, P. Tsichlis, and C. M. Croce. 2000. Tcl1 enhances Akt kinase activity and mediates its nuclear translocation. Proc. Natl. Acad. Sci. USA 97:3028-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Persad, S., S. Attwell, V. Gray, N. Mawji, J. T. Deng, D. Leung, J. Yan, J. Sanghera, M. P. Walsh, and S. Dedhar. 2001. Regulation of protein kinase B/Akt-serine 473 phosphorylation by integrin-linked kinase: critical roles for kinase activity and amino acids arginine 211 and serine 343. J. Biol. Chem. 276:27462-27469. [DOI] [PubMed] [Google Scholar]
- 37.Pullen, N., P. B. Dennis, M. Andjelkovic, A. Dufner, S. C. Kozma, B. A. Hemmings, and G. Thomas. 1998. Phosphorylation and activation of p70s6k by PDK1. Science 279:707-710. [DOI] [PubMed] [Google Scholar]
- 38.Sable, C. L., N. Filippa, C. Filloux, B. A. Hemmings, and E. Van Obberghen. 1998. Involvement of the pleckstrin homology domain in the insulin-stimulated activation of protein kinase B. J. Biol. Chem. 273:29600-29606. [DOI] [PubMed] [Google Scholar]
- 39.Sato, S., N. Fujita, and T. Tsuruo. 2000. Modulation of Akt kinase activity by binding to Hsp90. Proc. Natl. Acad. Sci. USA 97:10832-10837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlessinger, J. 2000. Cell signaling by receptor tyrosine kinases. Cell 103:211-225. [DOI] [PubMed] [Google Scholar]
- 41.Songyang, Z., D. Baltimore, L. C. Cantley, D. R. Kaplan, and T. F. Franke. 1997. Interleukin 3-dependent survival by the Akt protein kinase. Proc. Natl. Acad. Sci. USA 94:11345-11350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stephens, L., K. Anderson, D. Stokoe, H. Erdjument-Bromage, G. F. Painter, A. B. Holmes, P. R. Gaffney, C. B. Reese, F. McCormick, P. Tempst, J. Coadwell, and P. T. Hawkins. 1998. Protein kinase B kinases that mediate phosphatidylinositol 3,4,5-trisphosphate-dependent activation of protein kinase B. Science 279:710-714. [DOI] [PubMed] [Google Scholar]
- 43.Stern, M. H., J. Soulier, M. Rosenzwajg, K. Nakahara, N. Canki-Klain, A. Aurias, F. Sigaux, and I. R. Kirsch. 1993. MTCP-1: a novel gene on the human chromosome Xq28 translocated to the T cell receptor alpha/delta locus in mature T cell proliferations. Oncogene 8:2475-2483. [PubMed] [Google Scholar]
- 44.Taylor, A. M., J. A. Metcalfe, J. Thick, and Y. F. Mak. 1996. Leukemia and lymphoma in ataxia telangiectasia. Blood 87:423-438. [PubMed] [Google Scholar]
- 45.Teitell, M., M. A. Damore, G. G. Sulur, D. E. Turner, M. H. Stern, J. W. Said, C. T. Denny, and R. Wall. 1999. TCL1 oncogene expression in AIDS-related lymphomas and lymphoid tissues. Proc. Natl. Acad. Sci. USA 96:9809-9814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toker, A., and A. C. Newton. 2000. Akt/protein kinase B is regulated by autophosphorylation at the hypothetical PDK-2 site. J. Biol. Chem. 275:8271-8274. [DOI] [PubMed] [Google Scholar]
- 47.Vanhaesebroeck, B., and D. R. Alessi. 2000. The PI3K-PDK1 connection: more than just a road to PKB. Biochem. J. 346:561-576. [PMC free article] [PubMed] [Google Scholar]
- 48.Virgilio, L., C. Lazzeri, R. Bichi, K. Nibu, M. G. Narducci, G. Russo, J. L. Rothstein, and C. M. Croce. 1998. Deregulated expression of TCL1 causes T cell leukemia in mice. Proc. Natl. Acad. Sci. USA 95:3885-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Virgilio, L., M. G. Narducci, M. Isobe, L. G. Billips, M. D. Cooper, C. M. Croce, and G. Russo. 1994. Identification of the TCL1 gene involved in T-cell malignancies. Proc. Natl. Acad. Sci. USA 91:12530-12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams, M. R., J. S. Arthur, A. Balendran, J. van der Kaay, V. Poli, P. Cohen, and D. R. Alessi. 2000. The role of 3-phosphoinositide-dependent protein kinase 1 in activating AGC kinases defined in embryonic stem cells. Curr. Biol. 10:439-448. [DOI] [PubMed] [Google Scholar]
- 51.Yaffe, M. B., K. Rittinger, S. Volinia, P. R. Caron, A. Aitken, H. Leffers, S. J. Gamblin, S. J. Smerdon, and L. C. Cantley. 1997. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 91:961-971. [DOI] [PubMed] [Google Scholar]