Abstract
The reassignment of stop codons is common among many ciliate species. For example, Tetrahymena species recognize only UGA as a stop codon, while Euplotes species recognize only UAA and UAG as stop codons. Recent studies have shown that domain 1 of the translation termination factor eRF1 mediates stop codon recognition. While it is commonly assumed that changes in domain 1 of ciliate eRF1s are responsible for altered stop codon recognition, this has never been demonstrated in vivo. To carry out such an analysis, we made hybrid proteins that contained eRF1 domain 1 from either Tetrahymena thermophila or Euplotes octocarinatus fused to eRF1 domains 2 and 3 from Saccharomyces cerevisiae. We found that the Tetrahymena hybrid eRF1 efficiently terminated at all three stop codons when expressed in yeast cells, indicating that domain 1 is not the sole determinant of stop codon recognition in Tetrahymena species. In contrast, the Euplotes hybrid facilitated efficient translation termination at UAA and UAG codons but not at the UGA codon. Together, these results indicate that while domain 1 facilitates stop codon recognition, other factors can influence this process. Our findings also indicate that these two ciliate species used distinct approaches to diverge from the universal genetic code.
The near-universal nature of the standard genetic code implies that a barrier prevents organisms from easily evolving new coding strategies. However, exceptions to the standard code exist in mitochondria, ciliates, Mycoplasma, Candida, and other species (24). Among the ciliates, Tetrahymena species recognize UGA as a stop codon but have reassigned UAA and UAG to function as glutamine codons (16). Similarly, Euplotes species continue to recognize UAA and UAG as stop codons but have reassigned UGA to function as a cysteine codon (28). The existence of these alternate codes, which frequently include reassignment of the standard stop codons, raises obvious questions about how codon reassignment is carried out.
In eukaryotes, the release factors eRF1 and eRF3 are required for translation termination (43, 45). Normally, all three stop codons are bound and decoded by eRF1, which is a class I release factor with three functional domains (10, 42). Domain 1 binds to the stop codon and initiates the termination process (1, 5, 9, 20, 40). Domain 2 interacts with the peptidyl transferase center of the ribosome and mediates release of the completed polypeptide chain from the peptidyl-tRNA molecule in the ribosomal P site (12, 15). Domain 3 mediates an interaction between eRF1 and its functional partner, eRF3 (7, 8, 19, 27). eRF3 is a class II release factor that contains a GTPase domain. GTP hydrolysis by eRF3 stimulates both polypeptide chain release and proper stop codon recognition by eRF1 (11, 35).
The eRF1 proteins from ciliates have the same basic domain structure as eRF1s from other eukaryotic species and also share significant sequence homology with them. For example, both Tetrahymena thermophila eRF1 and Euplotes octocarinatus eRF1 share ∼56% overall amino acid sequence identity with Saccharomyces cerevisiae eRF1 and have ∼51% amino acid sequence identity with each other. Similar levels of sequence identity are found upon comparison of domain 1 sequences from these species. Since domain 1 of eRF1 is thought to mediate stop codon recognition, a number of studies have sought to use bioinformatic approaches to identify the key residues within domain 1 that mediate stop codon recognition (18, 23, 25, 26, 30). These and other mutational studies of eRF1 proteins have identified two highly conserved sequence motifs in domain 1, the NIKS motif (residues 58 to 61 of S. cerevisiae eRF1) (5, 9, 20) and the YXCXXXF motif (residues 122 to 128 of S. cerevisiae eRF1) (40). The fact that some residues in these motifs have diverged from the consensus sequence in variant-code species has led to considerable speculation that these changes are directly responsible for the altered stop codon specificities of these organisms. However, there has been only limited success in confirming that these specific residues actually mediate stop codon recognition.
In the current study, we asked whether domain 1 from Tetrahymena and that from Euplotes are sufficient to provide the variant stop codon recognition pattern used by these two ciliate species. To do this, we made hybrid molecules that contained eRF1 domain 1 from T. thermophila or E. octocarinatus fused to eRF1 domains 2 and 3 from S. cerevisiae. When these hybrid eRF1 molecules were expressed in S. cerevisiae cells that lacked the endogenous eRF1 gene, we found that the Tetrahymena hybrid eRF1 retained the ability to support growth, while the Euplotes hybrid did not. In vivo translation termination assays revealed that the Tetrahymena hybrid eRF1 promoted efficient termination at all three stop codons, indicating that eRF1 domain 1 alone is not capable of mediating the stop codon specificity observed in that species. In contrast, the Euplotes hybrid eRF1 facilitated efficient translation termination at UAA and UAG codons but not at the UGA codon, indicating that Euplotes domain 1 alone is sufficient for recapitulation of the stop codon specificity from that organism. Our results indicate that these organisms used different approaches to acquire changes in their genetic codes and suggest that the evolution of stop codon reassignment may be more straightforward than previously thought.
MATERIALS AND METHODS
Strain.
S. cerevisiae strain YDB447 (MATα ura3-52 leu2-3,112 ade1-14 lys2− trp1− his3− sup45::HIS3 [psi−]) was used in all experiments.
Hybrid gene constructions.
A clone (Eo-eRF1a) containing the coding region of Euplotes octocarinatus eRF1a was kindly provided by Aihua Liang (Shanxi University, China). For Tetrahymena thermophila eRF1, the entire eRF1 coding region (1,308 bp) was PCR amplified from a full-length T. thermophila cDNA library (6). The resulting PCR product was then cloned into the pCR2.1-TOPO vector using a TOPO TA cloning kit (Invitrogen, Carlsbad, CA) for further manipulations.
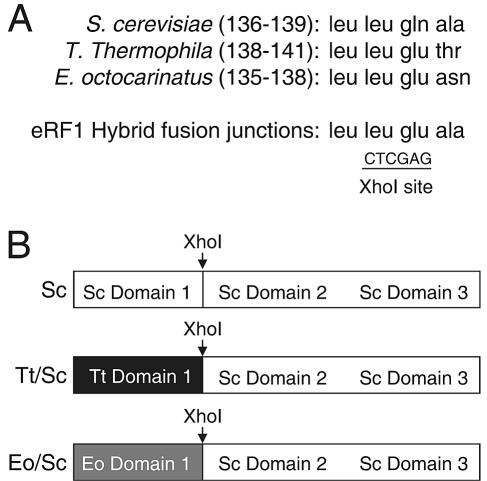
The low-copy-number S. cerevisiae eRF1 expression plasmid (a derivative of pRS315) was pDB800. To make the low-copy-number hybrid eRF1 constructs, the junction between domains 1 and 2 was defined as the hinge region ranging from residue 136 to residue 139 in S. cerevisiae eRF1, from 138 to 141 in T. thermophila eRF1, and from 135 to 138 in E. octocarinatus eRF1 (Fig. 1). To make the E. octocarinatus/S. cerevisiae hybrid eRF1 expression plasmid, domain 1 of E. octocarinatus eRF1 was PCR amplified using E. octocarinatus eRF1/pGBKT7 as the template. Domains 2 and 3 from S. cerevisiae eRF1, together with its endogenous transcription terminator, were PCR amplified using pUKC802 as the template. The PCR products containing E. octocarinatus eRF1 domain 1 and S. cerevisiae domains 2 and 3 were subcloned into pDB788 (SUP45 promoter YCplac111) to yield the final E. octocarinatus/S. cerevisiae eRF1 expression plasmid pDB948.
FIG. 1.
Strategy to construct hybrid eRF1 proteins. (A) Fusion junctions used to make eRF1 hybrid proteins. (B) Schematic showing the T. thermophila/S. cerevisiae (Tt/Sc) and E. octocarinatus/S. cerevisiae (Eo/Sc) hybrid eRF1 proteins and the wild-type S. cerevisiae (Sc) eRF1 control.
To make the T. thermophila/S. cerevisiae hybrid eRF1 expression plasmid, domain 1 of T. thermophila eRF1 was PCR amplified using T. thermophila eRF1/pCR2.1-TOPO as the template. Three in-frame UAA stop codons present in domain 1 of T. thermophila eRF1 at positions 8, 43, and 68 were changed to CAA codons. This T. thermophila eRF1 domain fragment was then fused to a fragment encoding domains 2 and 3 from S. cerevisiae eRF1 (see above) in pDB781 (SUP45 promoter YCplac22), yielding the T. thermophila/S. cerevisiae eRF1 expression plasmid pDB955. Alternatively, the T. thermophila eRF1 domain 1 fragment was fused to a fragment encoding domains 2 and 3 from S. cerevisiae eRF1 in pDB788 (SUP45 promoter YCplac111), yielding the T. thermophila/S. cerevisiae eRF1 expression plasmid pDB960.
The final hemagglutinin (HA)-tagged S. cerevisiae eRF1 expression plasmid pDB950 and the E. octocarinatus/S. cerevisiae eRF1 expression plasmid pDB954 each produced the indicated eRF1 protein with an N-terminal HA tag. Finally, the GAL1 promoter HA-S. cerevisiae eRF1 plasmid (pDB967) used in the S. cerevisiae eRF1 depletion experiment was constructed by subcloning a SalI/SphI fragment from pDB950 into GAL1 promoter YCplac22 (pDB451).
Construction of suppressor tRNAs.
Plasmids encoding the UAA suppressor tRNAGln and wild-type (CAA) tRNAGln were generous gifts from Yury Chernoff. The original UAA suppressor tRNAGln (41) has a G36A mutation in the tRNAGln gene that changes the tRNA anticodon from 5′-TTG-3′ (decodes CAA Gln codon) to 5′-TTA-3′ (decodes UAA stop codon). Additional suppressor tRNAs were made by mutating the anticodon to 5′-CTA-3′ (decodes UAG stop codon) and 5′-TCA-3′ (decodes UGA stop codon).
Viability assays.
A plasmid shuffle technique was used to assess whether the T. thermophila/S. cerevisiae eRF1 and E. octocarinatus/S. cerevisiae eRF1 constructs could support viability as the only source of eRF1 in the cell. For this purpose, pDB955 or pDB948 was transformed into a sup45Δ yeast strain (YDB447) that carried plasmid pUKC802 (SUP45-YEp24) to support viability. The strains were streaked on plates containing 5-fluoroorotic acid (5-FOA), which inhibits the growth of cells expressing the URA3 gene but allows the growth of cells that lost pUKC802 (as long as pDB955 or pDB948 support viability as the only source of eRF1). YDB447/pUKC802 transformed with wild-type SUP45-pRS315 (pDB800) was used as a positive control for growth on 5-FOA, while YDB447/pUKC802 transformed with the pRS315 vector alone was used as a negative control.
Dual luciferase readthrough assays.
The dual luciferase assays were carried out as previously described (13, 21). This system monitors readthrough of a stop codon by measuring firefly luciferase activity and allows the normalization of readthrough activity by comparing firefly luciferase activity to the level of upstream Renilla luciferase activity expressed in the same open reading frame before the stop codon. To determine the percent readthrough, assays were also done with reporter that contained a sense codon in place of the stop codon to obtain a value for maximum (100%) readthrough.
Determination of protein expression levels.
Trichloroacetic acid precipitation, protein extraction, and Western blotting were performed as previously described (35). For sodium dodecyl sulfate-polyacrylamide gel electrophoresis, 25 μg of total protein was loaded per lane.
S. cerevisiae eRF1 depletion experiments.
Cultures of strain YDB447 carrying GAL1 promoter HA-S. cerevisiae eRF1-YCplac22 (pDB967) with or without pDB948 were grown in synthetic minimal (SM) medium with galactose as the carbon source for several generations. During the mid-log stage of growth, the cells were harvested, spun down, and resuspended in SM medium with glucose as the carbon source to a cell density that would allow at least six cell doublings without nutrient depletion. After six generations, the cells were harvested for luciferase assays or Western blot analysis. As a control to determine the wild-type (basal) level of readthrough, assays were performed in a sup45Δ strain (YDB447) carrying a plasmid with the yeast SUP45 gene under the control of its own promoter (pDB800).
RESULTS
The objective of this study was to examine the in vivo stop codon recognition mediated by hybrid eRF1 proteins that contained domains 1 from the ciliates Tetrahymena and Euplotes. To do this, we made constructs that expressed two hybrid proteins (Fig. 1). The first fusion protein contained Tetrahymena eRF1 domain 1 joined to S. cerevisiae eRF1 domains 2 and 3 (referred to as T. thermophila/S. cerevisiae eRF1). The second contained Euplotes eRF1a domain 1 fused to S. cerevisiae eRF1 domains 2 and 3 (referred to as E. octocarinatus/S. cerevisiae eRF1). The junction for each hybrid protein corresponded to amino acids 137 and 138 of yeast eRF1, which are located in a hinge region between domains 1 and 2. A control plasmid expressing the intact S. cerevisiae eRF1 was also included in all subsequent experiments.
Since UAA and UAG encode glutamine in Tetrahymena species and UGA encodes cysteine in Euplotes species, we first had to make sure any reassigned stop codons within the ciliate genes were converted back to the universal code. No in-frame stop codons were present in the open reading frame encoding Euplotes domain 1. However, the Tetrahymena domain 1 open reading frame contained three in-frame UAA codons at positions 8, 43, and 68. Site-directed mutagenesis was used to change these to CAA (glutamine) codons to allow the fusion proteins to be expressed in yeast cells.
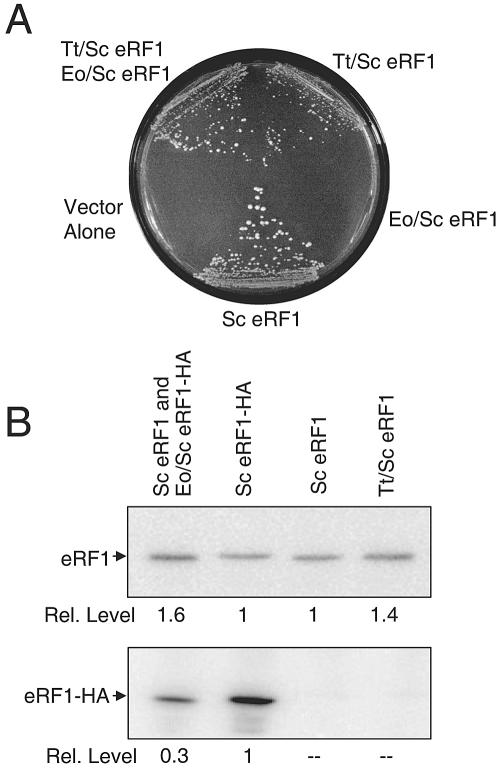
To monitor the function of these hybrid eRF1 proteins, we used a yeast strain with a deletion/disruption of the genomic eRF1 gene (sup45Δ). Since the SUP45 gene is essential, the viability of this strain was maintained by expressing the wild-type SUP45 gene from a low-copy-number plasmid that carried a URA3 selectable marker. Plasmids expressing each hybrid eRF1 gene under SUP45 promoter control were then introduced, and a plasmid shuffle method was used to assess whether the T. thermophila/S. cerevisiae eRF1 and E. octocarinatus/S. cerevisiae eRF1 plasmids could support cell viability as the only source of eRF1. To do this, the strains are plated on a medium supplemented with 5-FOA, a uracil analogue that allows the growth of only those colonies that have lost the original SUP45 plasmid with the URA3 marker (2). As can be seen in Fig. 2A, both S. cerevisiae eRF1 and T. thermophila/S. cerevisiae eRF1 complemented the sup45Δ mutation at 30°C, although the strain carrying the T. thermophila/S. cerevisiae eRF1 formed slightly smaller colonies. In contrast, the E. octocarinatus/S. cerevisiae eRF1 was unable to support growth when present as the only form of eRF1 in the cell. This lack of suppression was observed even when E. octocarinatus/S. cerevisiae eRF1 was expressed from a multicopy plasmid under SUP45 promoter control or when E. octocarinatus/S. cerevisiae eRF1 was expressed under the control of the strong GAL1 promoter (data not shown).
FIG. 2.
Initial functional characterization of hybrid eRF1 proteins. (A) Plasmid shuffling to test the ability of hybrid eRF1 proteins to support cell viability. Strains were streaked on SM glucose plates supplemented with 5-FOA to select colonies that had lost the URA3-based plasmids carrying wild-type eRF1. Growth indicates that the hybrid eRF1 can support cell viability. (B) Western blot confirming the expression of hybrid eRF1 proteins. (Top) Blot probed with polyclonal antibodies to domains 2 and 3 of yeast eRF1 that should recognize all forms of eRF1; (bottom) blot probed with antibodies to the HA epitope tag. Tt, T. thermophila; Sc, S. cerevisiae; Eo, E. octocarinatus.
We next carried out Western blot analysis to confirm that each of the hybrid constructs was expressed (Fig. 2B). This experiment was done in two ways. First, a polyclonal antibody raised to residues 236 to 437 of S. cerevisiae eRF1 (located in domains 2 and 3) was used to determine the total amount of eRF1 in each strain. Second, an HA epitope-specific monoclonal antibody was used to detect the Euplotes hybrid eRF1 (E. octocarinatus/S. cerevisiae eRF1-HA) and an HA-tagged wild-type eRF1 control (S. cerevisiae eRF1-HA). As shown in Fig. 2B, each of the hybrid eRF1 constructs was expressed. By comparing each blot to that of the control expressing only HA-tagged S. cerevisiae eRF1 (lane 2), it can be calculated that the abundance of the E. octocarinatus/S. cerevisiae eRF1 is ∼0.5× (0.3 × 1.6 = 0.5) the level of the S. cerevisiae eRF1 control. This level of eRF1 should support cell growth if this hybrid eRF1 is functional, since it was previously shown that as little as 10% of the normal level of eRF1 can support yeast cell viability (29). We conclude that T. thermophila/S. cerevisiae eRF1 is capable of supporting growth as the only source of eRF1 in the cell, while E. octocarinatus/S. cerevisiae eRF1 is unable to support cell viability when present as the only source of eRF1.
Tetrahymena domain 1 efficiently recognizes all three stop codons in vivo.
While the ability to support cell viability suggests that T. thermophila/S. cerevisiae eRF1 can mediate translation termination at all three stop codons, we wanted to more accurately determine its termination efficiency at each stop codon. To do this, we utilized a dual luciferase readthrough reporter system to measure the efficiency of stop codon recognition by T. thermophila/S. cerevisiae eRF1 in yeast cells grown at 30°C. This basic reporter system has been used to measure the efficiency of stop codon recognition in several previous studies (13, 17, 21, 34, 35) (Fig. 3A). Reporter plasmids were introduced into strains carrying either S. cerevisiae eRF1 or T. thermophila/S. cerevisiae eRF1 as the sole source of eRF1, and the level of readthrough at each stop codon was measured by determining the firefly luciferase activity. The firefly luciferase activity was then normalized to the Renilla luciferase activity, which serves as an internal control for mRNA abundance and any possible changes in the rate of translation initiation.
FIG. 3.
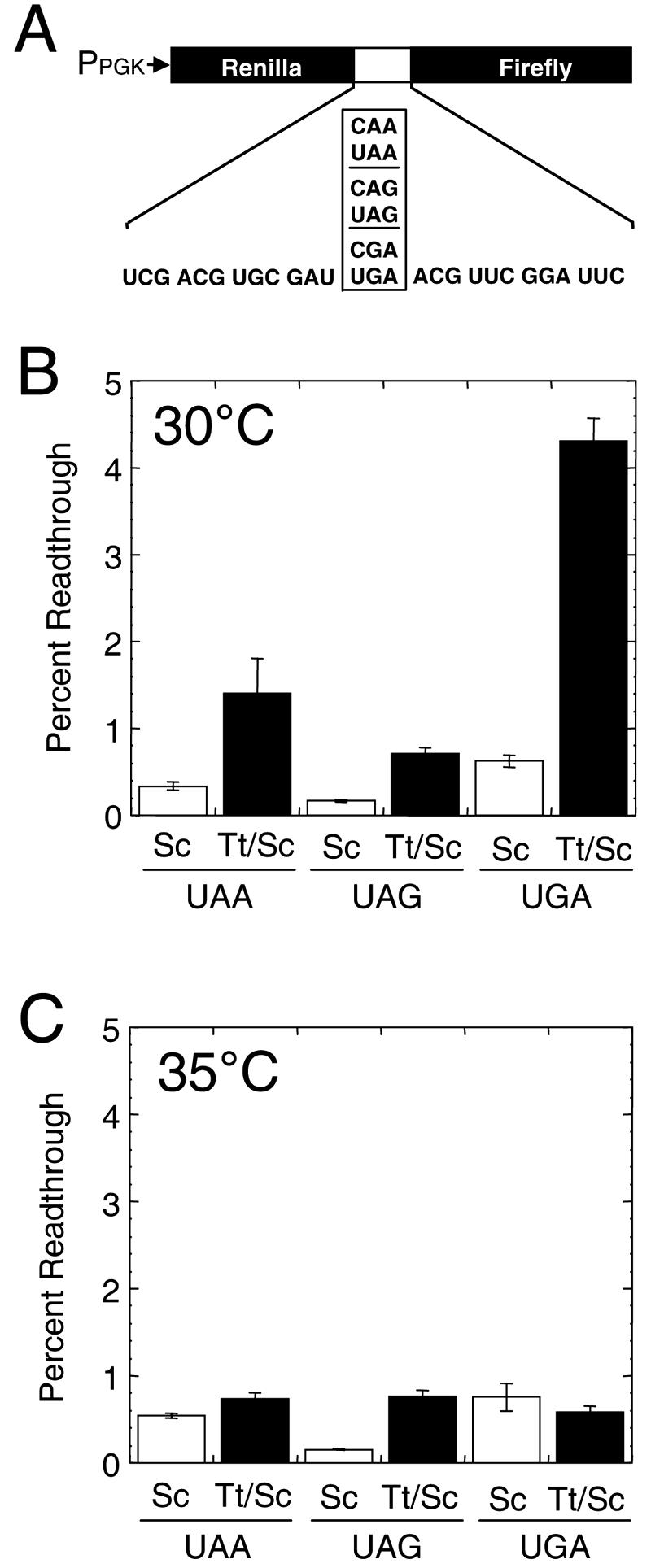
Measurements of stop codon readthrough measured in cells expressing hybrid eRF1 proteins. (A) Dual luciferase readthrough reporter plasmids used to monitor translation termination. (B) Stop codon readthrough measured at UAA, UAG, and UGA codons in cells expressing S. cerevisiae eRF1 (Sc) or T. thermophila/S. cerevisiae eRF1 (Tt/Sc) that were grown at 30°C. (C) Stop codon readthrough measured at UAA, UAG, and UGA codons in cells expressing S. cerevisiae eRF1 or T. thermophila/S. cerevisiae eRF1 in cells grown at 35°C.
We found that the T. thermophila/S. cerevisiae eRF1 exhibited a relatively low level of readthrough at all three stop codons in cells grown at 30°C, although the level of readthrough observed at each stop codon was higher than that of the readthrough mediated by S. cerevisiae eRF1 (Fig. 3B). Since the genetic code of Tetrahymena recognizes only UGA as a stop codon, it was surprising that readthrough at the UGA codon was the highest of all three stop codons (4.3%) and also represented the largest increase (6.9-fold) over the readthrough level mediated by S. cerevisiae eRF1 (0.6%). In contrast, the levels of readthrough mediated at the UAA codon (1.4%) and at the UAG codon (0.7%) by T. thermophila/S. cerevisiae eRF1 were remarkably low. These values were only 4.1-fold and 4.3-fold higher, respectively, than the level of readthrough allowed by S. cerevisiae eRF1.
Since Tetrahymena thermophila exhibits optimal growth rates at 35°C (31), we next measured readthrough levels at this temperature (Fig. 3C). Readthrough at the UAG codon did not change significantly in cells expressing either S. cerevisiae eRF1 or T. thermophila/S. cerevisiae eRF1 from the level measured at 30°C. In contrast, growth at this higher temperature caused readthrough at the UAA and UGA codons to increase slightly in cells expressing S. cerevisiae eRF1 (0.5% at UAA and 0.8% at UGA), indicating a slight decrease in the efficiency of termination at these codons by S. cerevisiae eRF1 at 35°C. However, readthrough was significantly lower in cells expressing T. thermophila/S. cerevisiae eRF1 at 35°C (0.7% at UAA and 0.6% at UGA) than at 30°C. The readthrough allowed by T. thermophila/S. cerevisiae eRF1 at each of these two stop codons was similar to the levels observed with S. cerevisiae eRF1. When taken together, these results indicate that the T. thermophila/S. cerevisiae eRF1 efficiently recognizes all three stop codons when expressed in yeast cells.
Tetrahymena domain 1 is not more susceptible to increased readthrough induced by suppressor tRNAs.
A previous study reported that Tetrahymena encodes two tRNAGln species that contain anticodons complementary to the UAG and UAA stop codons (37). More recently, The Institute for Genome Research has released a draft of the Tetrahymena macronuclear genome (http://www.tigr.org/tdb/e2k1/ttg/), and their preliminary gene annotations indicate that there are 52 tRNAGln genes in the genome, of which 39 are expected to be UAA or UAG suppressors (see http://www.ciliate.org/). Since the results presented above show that Tetrahymena domain 1 retains the ability to efficiently recognize UAA and UAG codons, we hypothesized that Tetrahymena domain 1 may facilitate UGA-specific termination by competing less efficiently with UAA- and UAG-specific suppressor tRNAs than the eRF1 domain 1 from universal-code organisms.
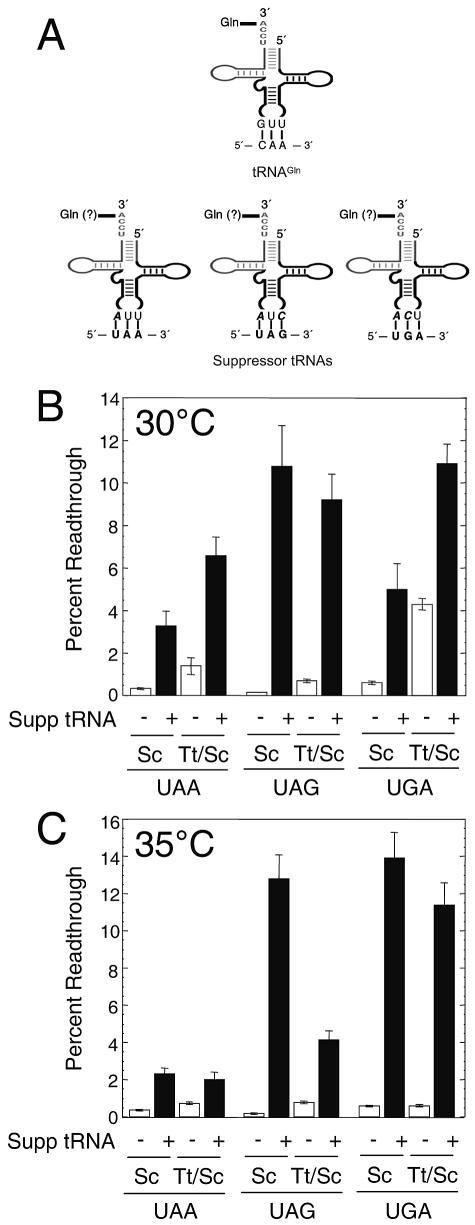
To test this hypothesis, we designed variants of a previously isolated UAA suppressor tRNAGln (41). These new suppressor tRNAGln species were made with anticodons complementary to the UAA, UAG, and UGA stop codons via standard Watson-Crick base pairing (Fig. 4A). A multicopy plasmid encoding each suppressor tRNA was transformed into yeast cells expressing S. cerevisiae eRF1 or T. thermophila/S. cerevisiae eRF1 along with a readthrough reporter plasmid for the stop codon corresponding to each suppressor tRNA.
FIG. 4.
Susceptibilities of S. cerevisiae eRF1 and T. thermophila/S. cerevisiae eRF1 to competition by suppressor tRNAs. (A) tRNAGln and UAA-, UAG-, and UGA-specific nonsense suppressor tRNAs that were constructed. (Top) Gln tRNA with its anticodon domain oriented toward the bottom. The anticodon base pairs with the codon in the mRNA as indicated; (bottom) suppressor tRNAs with mutated residues in the anticodon (bold italic) to allow Watson-Crick base pairing with stop codons (bold) in the mRNA. The amino acids attached to these suppressor tRNAs are indicated as Gln (?) because we have not confirmed that they continue to be charged with glutamine. (B) Stop codon readthrough measured in the presence (black bars) and the absence (white bars) of suppressor (Supp) tRNAs in cells expressing S. cerevisiae eRF1 (Sc) or T. thermophila/S. cerevisiae eRF1 (Tt/Sc) at 30°C. (C) Stop codon readthrough measured in the presence and the absence of suppressor tRNAs in cells expressing S. cerevisiae eRF1 or T. thermophila/S. cerevisiae eRF1 at 35°C.
We first carried out readthrough assays with cells harboring these suppressor tRNAs at 30°C (Fig. 4B). We found that each suppressor tRNA species significantly increased readthrough at the corresponding stop codon in cells expressing either S. cerevisiae eRF1 or T. thermophila/S. cerevisiae eRF1. Readthrough at the UAG codon was similar in both strains expressing the UAG suppressor tRNA (10.8% readthrough for S. cerevisiae eRF1 and 9.2% readthrough for T. thermophila/S. cerevisiae eRF1). The level of readthrough at the UAA codon was roughly twofold higher in the strain expressing T. thermophila/S. cerevisiae eRF1 (3.3% readthrough for S. cerevisiae eRF1 and 6.6% for T. thermophila/S. cerevisiae eRF1). A similar twofold difference in readthrough levels was also observed at the UGA stop codon (5.0% readthrough for S. cerevisiae eRF1 and 10.9% for T. thermophila/S. cerevisiae eRF1). We next examined the effects of suppressor tRNAs on readthrough in cells grown at 35°C (Fig. 4C). In cells expressing either S. cerevisiae eRF1 or T. thermophila/S. cerevisiae eRF1, readthrough levels were similar both at the UAA codon (2.3% readthrough for S. cerevisiae eRF1 and 2.0% for T. thermophila/S. cerevisiae eRF1) and at the UGA codon (13.9% readthrough for S. cerevisiae eRF1 and 11.4% for T. thermophila/S. cerevisiae eRF1), while the readthrough level at the UAG codon was threefold lower in cells expressing the T. thermophila/S. cerevisiae eRF1 (12.8% readthrough for S. cerevisiae eRF1 and 4.2% for T. thermophila/S. cerevisiae eRF1). When taken together, these results indicate that the T. thermophila/S. cerevisiae eRF1 is not more susceptible to readthrough at the UAA and UAG codons than the S. cerevisiae eRF1 when suppressor tRNAs are present.
Coexpression of E. octocarinatus/S. cerevisiae eRF1 influences readthrough in cells expressing T. thermophila/S. cerevisiae eRF1.
Since E. octocarinatus/S. cerevisiae eRF1 was unable to support cell growth when it was present as the only form of eRF1, we were unable to measure the efficiency of translation termination after evicting the S. cerevisiae eRF1 plasmid. To gain more insight into the stop codon recognition of domain 1 from Euplotes eRF1, we next constructed strains that expressed both E. octocarinatus/S. cerevisiae eRF1 and T. thermophila/S. cerevisiae eRF1 under SUP45 promoter control and carried out assays with our dual luciferase readthrough reporters (Fig. 5). We found that coexpression of E. octocarinatus/S. cerevisiae eRF1 and T. thermophila/S. cerevisiae eRF1 reduced the level of readthrough 2.3-fold at the UAA codon compared to that seen with a strain expressing T. thermophila/S. cerevisiae eRF1 alone. Similarly, when both the E. octocarinatus/S. cerevisiae and T. thermophila/S. cerevisiae hybrid release factors were present, a 1.8-fold decrease in readthrough relative to that seen with cells expressing T. thermophila/S. cerevisiae eRF1 alone was observed at the UAG codon. In contrast, expression of both E. octocarinatus/S. cerevisiae eRF1 and T. thermophila/S. cerevisiae eRF1 increased the level of readthrough 2.2-fold over that observed with the T. thermophila/S. cerevisiae eRF1 alone at the UGA stop codon. These results suggest that E. octocarinatus/S. cerevisiae eRF1 is capable of recognizing UAA and UAG stop codons but not the UGA codon.
FIG. 5.
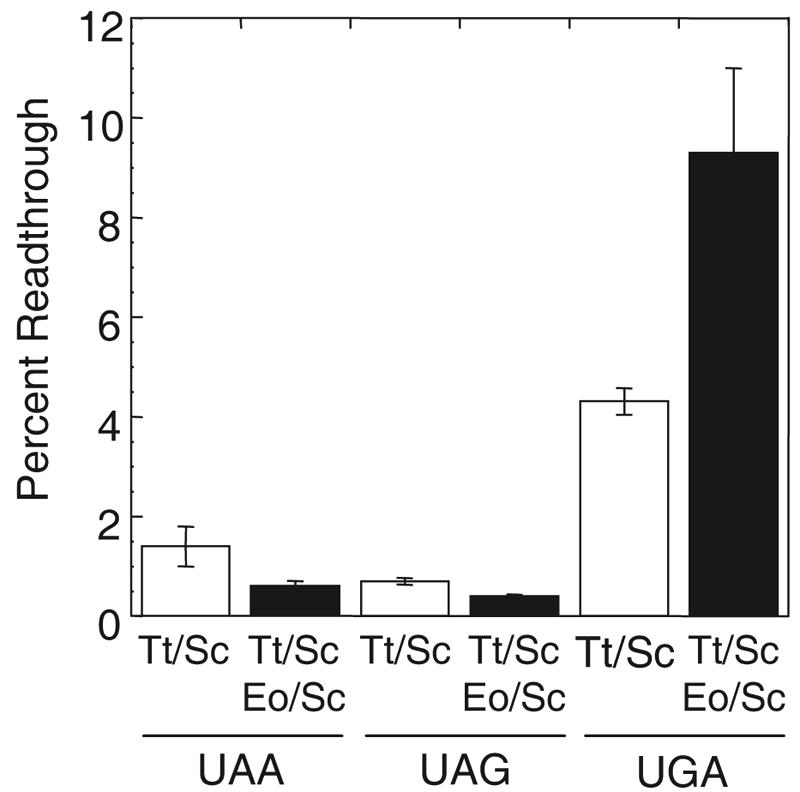
Stop codon readthrough measured in cells expressing either T. thermophila/S. cerevisiae eRF1 only (Tt/Sc) or both T. thermophila/S. cerevisiae eRF1 and E. octocarinatus/S. cerevisiae eRF1 (Eo/Sc). Cultures were grown at 30°C in SM glucose medium.
Direct evidence that Euplotes domain 1 recognizes UAA and UAG stop codons but not the UGA stop codon.
To directly examine the level of readthrough allowed by E. octocarinatus/S. cerevisiae eRF1 at each stop codon, we set up a dual expression system where S. cerevisiae eRF1 was expressed from the regulated GAL1 promoter while E. octocarinatus/S. cerevisiae eRF1 was expressed from the constitutive SUP45 promoter. In this system, S. cerevisiae eRF1 expression was initially maintained by growing the cells in a medium with galactose as the carbon source. After shifting to a medium with glucose as the carbon source to inhibit expression from the GAL1 promoter, the preexisting S. cerevisiae eRF1 was diluted out in subsequent cell divisions, while expression of the E. octocarinatus/S. cerevisiae eRF1 remained constant. In a control strain that carried only the plasmid encoding S. cerevisiae eRF1 under GAL promoter control, cell growth slowed considerably after six generations under these conditions (data not shown), suggesting that a significant depletion of eRF1 had occurred.
To confirm that S. cerevisiae eRF1 was depleted following the carbon source shift, we carried out Western blotting to examine the steady-state level of HA-tagged S. cerevisiae eRF1 in cell extracts prepared from cultures grown with galactose as the carbon source or after shifting the cultures from galactose to glucose and continuing cultivation for six generations (Fig. 6A). In galactose-grown cultures, we found that the level of HA-tagged S. cerevisiae eRF1 expressed from the GAL promoter was fourfold higher than the level expressed from the SUP45 promoter. After the cultures were grown for six generations with glucose as the carbon source, the level of S. cerevisiae eRF1 expressed from the GAL promoter was reduced to roughly 1/10 the control level, indicating that efficient depletion had occurred.
FIG. 6.
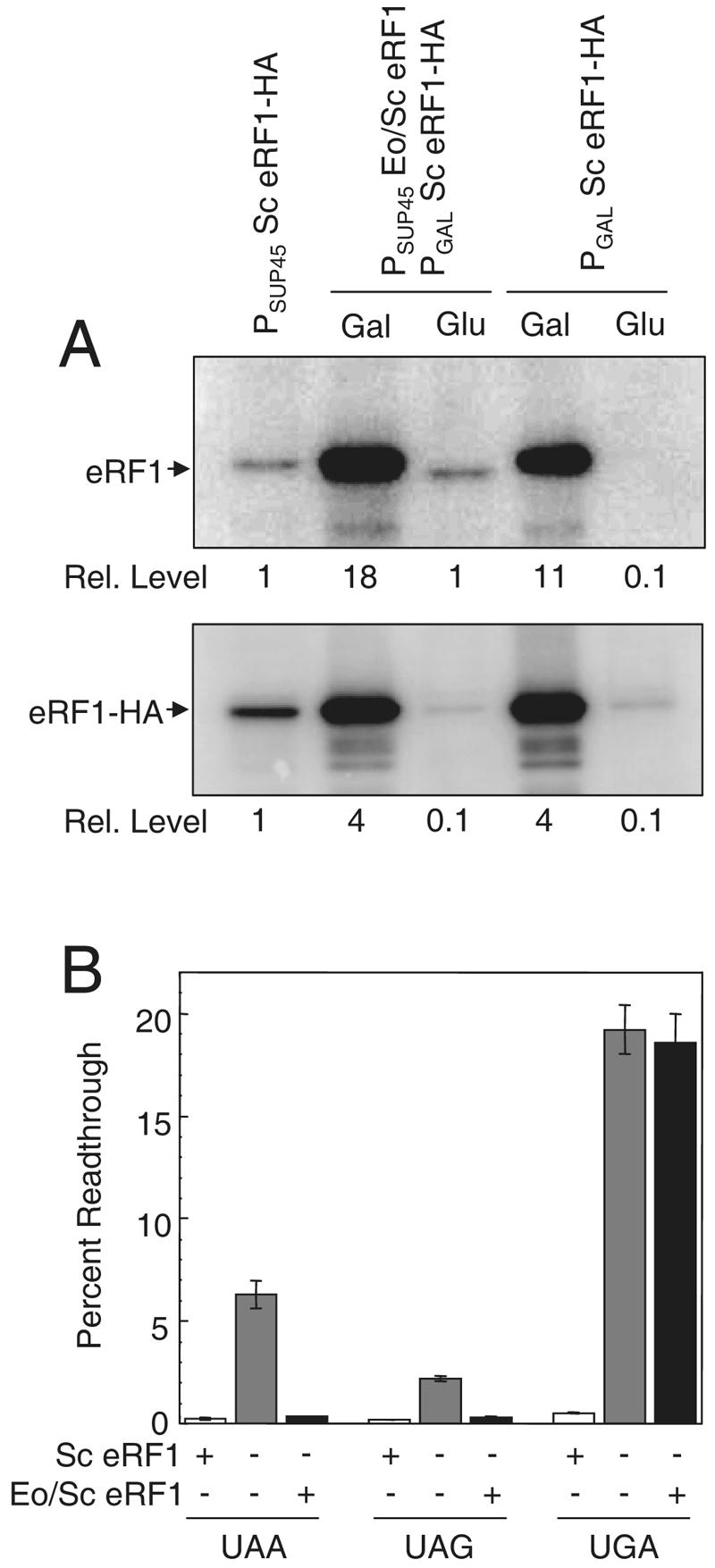
Stop codon readthrough mediated by E. octocarinatus/S. cerevisiae (Eo/Sc) eRF1 measured after depletion of S. cerevisiae (Sc) eRF1. (A) Western blot confirming the relative levels of eRF1 proteins. (Top) Blot probed with polyclonal antibodies to domains 2 and 3 of yeast eRF1 that should recognize all forms of eRF1; (bottom) blot probed with antibodies to the HA epitope tag. Cultures were grown in SM medium with either glucose or galactose as the carbon source to regulate the expression of S. cerevisiae eRF1 under GAL1 promoter control. (B) Stop codon readthrough measured in cells expressing S. cerevisiae eRF1, no eRF1, or E. octocarinatus/S. cerevisiae eRF1. Cultures were grown at 30°C.
Using this system, we assayed the level of readthrough at each stop codon in strains that had undergone the shift protocol and were left either with S. cerevisiae eRF1 primarily, with no eRF1, or with E. octocarinatus/S. cerevisiae eRF1 primarily (Fig. 6B). At the UAA codon, we observed 6.3% readthrough following S. cerevisiae eRF1 depletion in the absence of any other eRF1 species. In contrast, the cells that retained S. cerevisiae eRF1 or E. octocarinatus/S. cerevisiae eRF1 expression showed much less UAA readthrough (0.26% and 0.35% readthrough, respectively). At the UAG codon, we observed 2.2% readthrough following S. cerevisiae eRF1 depletion in the absence of other eRF1 species, while 0.19% and 0.28% readthroughs were measured in cells with primarily S. cerevisiae eRF1 and primarily E. octocarinatus/S. cerevisiae eRF1, respectively. When taken together, these results indicate that E. octocarinatus/S. cerevisiae eRF1 can recognize UAA and UAG codons at a level that is only slightly less efficient than that of their recognition by S. cerevisiae eRF1.
Significantly different results were obtained when readthrough was measured at the UGA stop codon. We observed 19.2% readthrough following S. cerevisiae eRF1 depletion when no other eRF1 species was present. In cells that retained constitutive S. cerevisiae eRF1 expression, we observed only 0.51% readthrough. In cells that retained E. octocarinatus/S. cerevisiae eRF1 expression after the shift, we found 18.6% readthrough at the UGA stop codon. Since the increase in readthrough with E. octocarinatus/S. cerevisiae eRF1 was similar to the increase observed in the complete absence of eRF1, we conclude that the Euplotes domain 1 is largely unable to recognize the UGA stop codon.
DISCUSSION
Domain 1 of eRF1 is thought to be solely responsible for stop codon recognition in eukaryotes. In the current study, we tested this hypothesis by asking whether hybrid eRF1 proteins with domains 1 from the ciliates Tetrahymena and Euplotes could recapitulate the variant stop codon recognition of those organisms. We found that the E. octocarinatus/S. cerevisiae hybrid eRF1 containing Euplotes domain 1 efficiently recognized UAA and UAG codons but not the UGA stop codon. This pattern of stop codon recognition accurately reflected the stop codon usage observed in Euplotes species, indicating that the presence of this domain alone is sufficient for their variant stop codon recognition.
Only a few studies have heretofore addressed stop codon recognition in Euplotes. In one, in vitro fMet release assays were carried out with Euplotes aediculatus eRF1 (22). It was shown that the Euplotes protein facilitated termination at UAA and UAG stop codons but not at a UGA stop codon. This provided the first evidence that Euplotes eRF1 exhibits the same stop codon specificity in a heterologous release assay as that observed in the intact organism. Another assay system took advantage of an in vitro cross-linking assay to monitor the domains of eRF1 required for proper stop codon recognition (3, 5). It was shown that human eRF1 could be cross-linked to an mRNA that contained the photoactivated uridine analog 4-thiouridine in the first position of the stop codon. By use of this system, human eRF1 could be cross-linked to mRNAs that contain any of the three stop codons (UAA, UAG, and UGA). Hybrid eRF1 proteins that contained Euplotes aediculatus domain 1 fused to domains 2 and 3 of human eRF1 were also examined. It was found that hybrid proteins with domain 1 from Euplotes gave efficient cross-linking at the UAA and UAG stop codons but not at the UGA codon (4). These in vitro results suggested that domain 1 was sufficient to confer the Euplotes stop codon specificity to the termination process and are entirely consistent with our finding that the Euplotes domain 1 is sufficient to mediate efficient in vivo termination at the UAA and UAG codons but not at the UGA stop codon.
In contrast to our finding that Euplotes domain 1 contains all the information required for UAG- and UAA-specific stop codon recognition, our results indicate that Tetrahymena domain 1 retains the ability to efficiently recognize all three stop codons in vivo. This demonstrates that domain 1 from Tetrahymena eRF1 is not sufficient to recapitulate the altered stop codon specificity observed in Tetrahymena species. These results are only partially consistent with a previous study that used a hybrid eRF1 with Tetrahymena thermophila domain 1 and Schizosaccharomyces pombe domains 2 and 3 (T. thermophila/S. pombe eRF1) (20). In that study, an in vitro release assay showed that the T. thermophila/S. pombe hybrid eRF1 stimulated release at the UGA stop codon but not at the UAA or UAG stop codons. These results suggested that domain 1 of Tetrahymena eRF1 was sufficient to confer the stop codon specificity of Tetrahymena to translation termination. However, when the T. thermophila/S. pombe eRF1 was expressed in an S. cerevisiae strain in which the endogenous yeast eRF1 gene was deleted (sup45Δ), the Tetrahymena hybrid eRF1 fully complemented the sup45Δ mutation and supported cell growth. This result suggested that Tetrahymena eRF1 domain 1 retained the ability to properly recognize each of the three stop codons and terminate translation. However, the observation that this yeast strain was temperature sensitive for growth led the authors to speculate that the function of domain 1 from Tetrahymena eRF1 is optimized for UGA-only termination at 37°C and that the growth observed at 30°C was a nonphysiological response of the hybrid eRF1 toward stop codons due to alterations in affinity and/or rate constants of the molecules interacting with eRF1 in vivo (20). Given our data that the T. thermophila/S. cerevisiae eRF1 allows very little stop codon readthrough at 35°C, we think this discrepancy between the two studies may be attributable to the fact that Ito et al. studied Tetrahymena domain 1 in the context of domains 2 and 3 from S. pombe, while we used domains 2 and 3 from S. cerevisiae. Since each study was done with an S. cerevisiae strain that lacked its endogenous eRF1, it is possible that the large evolutionary distance between S. pombe and S. cerevisiae, rather than Tetrahymena domain 1, was responsible for the temperature-sensitive nature of the strain expressing the T. thermophila/S. pombe eRF1 protein. Since in vivo readthrough assays were not carried out in their study, we directly tested this temperature sensitivity model of stop codon recognition by using our T. thermophila/S. cerevisiae eRF1 system. We found that T. thermophila/S. cerevisiae eRF1 exhibited minimal readthrough at all three stop codons in yeast cells grown at either 30°C and 35°C, indicating that domain 1 of Tetrahymena eRF1 alone was not sufficient to confer the stop codon specificity observed in that organism.
Three possibilities could explain why UGA-specific termination was not observed in yeast expressing T. thermophila/S. cerevisiae eRF1. First, interactions with residues in domains 2 or 3 of Tetrahymena eRF1 may be required to properly modulate the variant stop codon recognition of domain 1. Such amino acid changes that restrict stop codon recognition would not be present in domains 2 and 3 of S. cerevisiae eRF1. Second, another component of the termination machinery such as eRF3 may be necessary to faithfully mediate UGA-specific stop codon recognition by Tetrahymena eRF1. If unique changes in Tetrahymena eRF3 were required to implement the variant genetic code through its action on eRF1, they would not be present in S. cerevisiae eRF3. Finally, Tetrahymena species have been shown to possess natural glutaminyl tRNAs with anticodons complementary to UAA and UAG codons (37) that may provide a level of readthrough of these stop codons sufficient to support viability. We found that the T. thermophila/S. cerevisiae eRF1 allowed as much as 11% readthrough in the presence of artificially constructed suppressor tRNAs. The natural suppressor tRNAs present in Tetrahymena species may have evolved to facilitate even higher levels of readthrough. In this regard, these suppressor tRNAs should be quite abundant, since the Tetrahymena thermophila macronuclear genome database indicates that 39/52 total tRNAGln genes encode suppressor tRNAs.
The possibility that eRF3 influences stop codon recognition by Tetrahymena eRF1 is particularly intriguing, since the UGA-specific termination mediated by a T. thermophila/S. pombe hybrid eRF1 in a previous study was demonstrated using only an in vitro fMet release assay that did not contain eRF3 (20). This could have a significant impact on stop codon recognition, since eRF3 was previously shown to greatly stimulate in vitro fMet release assays (11), and a recent in vivo study found that eRF3 modulates stop codon recognition by eRF1 in yeast (35). Based on these observations, UGA-specific stop codon recognition by Tetrahymena eRF1 may result from fundamental differences in the mechanisms by which eRF3 modulates its stop codon recognition. Such a mechanism would have negated the need for this organism to alter its eRF1 protein away from the universal genetic code. Importantly, an eRF3-specific component to stop codon recognition could result in termination at all stop codons when eRF3 from a universal-code organism is present (as observed in our in vivo experiments) but only in UGA-specific termination when Tetrahymena eRF3 (or no eRF3) is present. Further studies will be required to test this hypothesis.
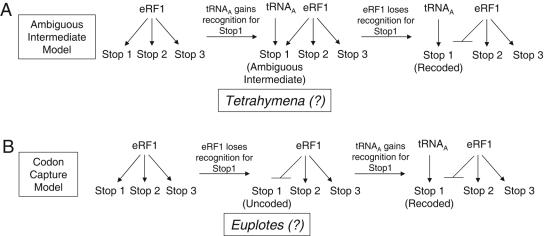
Two major models explaining how various species diverged from the universal genetic code have been proposed (for reviews, see references 24 and 36) (Fig. 7). In the codon capture hypothesis, codon reassignment occurs by a two-step mechanism (32, 33). Recognition of a codon by either a cognate tRNA (for sense codons) or a release factor (for stop codons) must first be lost in order to make the codon available for reassignment. The codon specificity of another tRNA must then be altered to capture the available codon. In the ambiguous intermediate hypothesis, a mutation in a tRNA gene leads to the concurrent recognition of a codon by two different tRNAs (at a sense codon) or by a tRNA and a release factor (in the case of a stop codon) (38, 39). Ambiguities of either type ultimately could be resolved by loss of codon recognition by the original tRNA or release factor, thus fixing the reassignment event. In support of this mechanism, codon-specific ambiguous decoding has been demonstrated in certain Candida species (44). A key difference in these two mechanisms is that the ambiguous intermediate mechanism does not require codon disappearance as a prerequisite for codon reassignment.
FIG. 7.
Two evolutionary models for stop codon reassignment. (A) The ambiguous intermediate model. (B) The codon capture model. The states of stop codon reassignment that are most consistent with our data are indicated for Euplotes and Tetrahymena.
Our results indicate that Tetrahymena eRF1 domain 1 retains the inherent ability to efficiently decode the three universal stop codons. However, this organism evolved a set of suppressor tRNAs that can efficiently decode the UAA and UAG stop codons as glutamine codons (37). While it remains a possibility that other domains of eRF1 or other factors such as eRF3 are required to faithfully recapitulate the stop codon specificity found in Tetrahymena species, the present data are consistent with an ambiguous intermediate mechanism where these natural suppressor tRNAs alone mediate stop codon reassignment in this organism in competition with termination at the same codon by eRF1 (Fig. 7A). Thus, Tetrahymena appears to have arrested at an intermediate stage of the codon reassignment process, as proposed in the ambiguous intermediate hypothesis.
In contrast, we found that the E. octocarinatus/S. cerevisiae eRF1 is significantly compromised in its ability to recognize the UGA stop codon. This resulted in a high level of readthrough of the UGA stop codon in the absence of a suppressor tRNA, highlighting the fact that the termination process normally represents a competition between stop codon recognition by eRF1 and near-cognate tRNAs. In the absence of UGA recognition by eRF1, near-cognate mispairing can be quite efficient. This demonstrates that reassignment possibly could occur by losing the ability to recognize a stop codon without the appearance of a cognate tRNA for that codon. Since Euplotes species use UGA as a cysteine codon, it is interesting to note that a previous search failed to find any UGA-specific tRNACys genes in the Euplotes genome (14). If this finding is confirmed, it would indicate that stop codon reassignment in Euplotes has taken place by a simple one-step mechanism that consisted solely of losing UGA codon recognition by eRF1 (Fig. 7B). This result would suggest that stop codon recoding had occurred even though the process had arrested at an intermediate step in the codon capture hypothesis, an outcome that had not previously been predicted to represent a stable reassigned state. Alternatively, if UGA-specific suppressor tRNACys species are ultimately found in the Euplotes genome, it would indicate that this organism had lost UGA recognition by eRF1 and gained UGA recognition by one or more tRNACys species. Such an outcome could have arisen either by a codon capture mechanism or by completion of an ambiguous intermediate mechanism (with eRF1 recognition lost after acquisition of the UGA-specific tRNACys species).
Our results provide strong evidence that distinct paths were taken by Tetrahymena and Euplotes to diverge from the universal genetic code. The question of whether these different approaches were necessitated by fundamental differences between the mechanism of decoding the UAA and UAG stop codons and that of decoding the UGA stop codon remains to be determined. However, an increased understanding of these differences may provide important clues about the mechanism of eukaryotic stop codon recognition. For example, it may be possible to completely lose UGA recognition (as we observed with the E. octocarinatus/S. cerevisiae hybrid eRF1), but it may not be possible to lose UAA and UAG recognition while retaining UGA recognition. Such a restriction on stop codon recognition patterns may be the most likely reason why Tetrahymena would retain recognition of UAA and UAG codons throughout the millions of years since it adopted a variant genetic code with UGA-specific termination. In this regard, it appears that UAA- and UAG-specific species evolved these patterns of stop codon recognition independently at least two times, while the UGA-specific species evolved them at least three times (23). Analysis of the mechanisms of stop codon reassignment in species evolving from each of these independent events should provide additional valuable information about how reassignment can occur.
Acknowledgments
We thank Yury Chernoff and Mick Tuite for strains and plasmids and Kim Keeling for critically reading the manuscript.
This work was supported by NIH grants RO1 GM 68854 (D.M.B.), and RO1 GM 29480 (P.J.F.) and by NSF grant MCB-0343813 (L.A.K.).
REFERENCES
- 1.Bertram, G., H. A. Bell, D. W. Ritchie, G. Fullerton, and I. Stansfield. 2000. Terminating eukaryote translation: domain 1 of release factor eRF1 functions in stop codon recognition. RNA 6:1236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345-346. [DOI] [PubMed] [Google Scholar]
- 3.Chavatte, L., L. Frolova, L. Kisselev, and A. Favre. 2001. The polypeptide chain release factor eRF1 specifically contacts the s(4)UGA stop codon located in the A site of eukaryotic ribosomes. Eur. J. Biochem. 268:2896-2904. [DOI] [PubMed] [Google Scholar]
- 4.Chavatte, L., S. Kervestin, A. Favre, and O. Jean-Jean. 2003. Stop codon selection in eukaryotic translation termination: comparison of the discriminating potential between human and ciliate eRF1s. EMBO J. 22:1644-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chavatte, L., A. Seit-Nebi, V. Dubovaya, and A. Favre. 2002. The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. EMBO J. 21:5302-5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chilcoat, N. D., N. C. Elde, and A. P. Turkewitz. 2001. An antisense approach to phenotype-based gene cloning in Tetrahymena. Proc. Natl. Acad. Sci. USA 98:8709-8713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebihara, K., and Y. Nakamura. 1999. C-terminal interaction of translational release factors eRF1 and eRF3 of fission yeast: G-domain uncoupled binding and the role of conserved amino acids. RNA 5:739-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eurwilaichitr, L., F. M. Graves, I. Stansfield, and M. F. Tuite. 1999. The C-terminus of eRF1 defines a functionally important domain for translation termination in Saccharomyces cerevisiae. Mol. Microbiol. 32:485-496. [DOI] [PubMed] [Google Scholar]
- 9.Frolova, L., A. Seit-Nebi, and L. Kisselev. 2002. Highly conserved NIKS tetrapeptide is functionally essential in eukaryotic translation termination factor eRF1. RNA 8:129-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frolova, L. Y., T. I. Merkulova, and L. L. Kisselev. 2000. Translation termination in eukaryotes: polypeptide release factor eRF1 is composed of functionally and structurally distinct domains. RNA 6:381-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frolova, L. Y., J. L. Simonsen, T. I. Merkulova, D. Y. Litvinov, P. M. Martensen, V. O. Rechinsky, J. H. Camonis, L. L. Kisselev, and J. Justesen. 1998. Functional expression of eukaryotic polypeptide chain release factors 1 and 3 by means of baculovirus/insect cells and complex formation between the factors. Eur. J. Biochem. 256:36-44. [DOI] [PubMed] [Google Scholar]
- 12.Frolova, L. Y., R. Y. Tsivkovskii, G. F. Sivolobova, N. Y. Oparina, O. I. Serpinsky, V. M. Blinov, S. I. Tatkov, and L. L. Kisselev. 1999. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA 5:1014-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grentzmann, G., J. A. Ingram, P. J. Kelly, R. F. Gesteland, and J. F. Atkins. 1998. A dual-luciferase reporter system for studying recoding signals. RNA 4:479-486. [PMC free article] [PubMed] [Google Scholar]
- 14.Grimm, M., C. Brunen-Nieweler, V. Junker, K. Heckmann, and H. Beier. 1998. The hypotrichous ciliate Euplotes octocarinatus has only one type of tRNACys with GCA anticodon encoded on a single macronuclear DNA molecule. Nucleic Acids Res. 26:4557-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heurgue-Hamard, V., S. Champ, L. Mora, T. Merkoulova-Rainon, L. L. Kisselev, and R. H. Buckingham. 2005. The glutamine residue of the conserved GGQ motif in Saccharomyces cerevisiae release factor eRF1 is methylated by the product of the YDR140w gene. J. Biol. Chem. 280:2439-2445. [DOI] [PubMed] [Google Scholar]
- 16.Horowitz, S., and M. A. Gorovsky. 1985. An unusual genetic code in nuclear genes of Tetrahymena. Proc. Natl. Acad. Sci. USA 82:2452-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howard, M. T., B. H. Shirts, L. M. Petros, K. M. Flanigan, R. F. Gesteland, and J. F. Atkins. 2000. Sequence specificity of aminoglycoside-induced stop codon readthrough: potential implications for treatment of Duchenne muscular dystrophy. Ann. Neurol. 48:164-169. [PubMed] [Google Scholar]
- 18.Inagaki, Y., and W. F. Doolittle. 2001. Class I release factors in ciliates with variant genetic codes. Nucleic Acids Res. 29:921-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito, K., K. Ebihara, and Y. Nakamura. 1998. The stretch of C-terminal acidic amino acids of translational release factor eRF1 is a primary binding site for eRF3 of fission yeast. RNA 4:958-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito, K., L. Frolova, A. Seit-Nebi, A. Karamyshev, L. Kisselev, and Y. Nakamura. 2002. Omnipotent decoding potential resides in eukaryotic translation termination factor eRF1 of variant-code organisms and is modulated by the interactions of amino acid sequences within domain 1. Proc. Natl. Acad. Sci. USA 99:8494-8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keeling, K. M., J. Lanier, M. Du, J. Salas-Marco, L. Gao, A. Kaenjak-Angeletti, and D. M. Bedwell. 2004. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA 10:691-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kervestin, S., L. Frolova, L. Kisselev, and O. Jean-Jean. 2001. Stop codon recognition in ciliates: Euplotes release factor does not respond to reassigned UGA codon. EMBO Rep. 2:680-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, O. T., K. Yura, N. Go, and T. Harumoto. 2005. Newly sequenced eRF1s from ciliates: the diversity of stop codon usage and the molecular surfaces that are important for stop codon interactions. Gene 346:277-286. [DOI] [PubMed] [Google Scholar]
- 24.Knight, R. D., S. J. Freeland, and L. F. Landweber. 2001. Rewiring the keyboard: evolvability of the genetic code. Nat. Rev. Genet. 2:49-58. [DOI] [PubMed] [Google Scholar]
- 25.Liang, H., J. Y. Wong, Q. Bao, A. R. Cavalcanti, and L. F. Landweber. 2005. Decoding the decoding region: analysis of eukaryotic release factor (eRF1) stop codon-binding residues. J. Mol. Evol. 60:337-344. [DOI] [PubMed] [Google Scholar]
- 26.Lozupone, C. A., R. D. Knight, and L. F. Landweber. 2001. The molecular basis of nuclear genetic code change in ciliates. Curr. Biol. 11:65-74. [DOI] [PubMed] [Google Scholar]
- 27.Merkulova, T. I., L. Y. Frolova, M. Lazar, J. Camonis, and L. L. Kisselev. 1999. C-terminal domains of human translation termination factors eRF1 and eRF3 mediate their in vivo interaction. FEBS Lett. 443:41-47. [DOI] [PubMed] [Google Scholar]
- 28.Meyer, F., H. J. Schmidt, E. Plumper, A. Hasilik, G. Mersmann, H. E. Meyer, A. Engstrom, and K. Heckmann. 1991. UGA is translated as cysteine in pheromone 3 of Euplotes octocarinatus. Proc. Natl. Acad. Sci. USA 88:3758-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moskalenko, S. E., S. V. Chabelskaya, S. G. Inge-Vechtomov, M. Philippe, and G. A. Zhouravleva. 2003. Viable nonsense mutants for the essential gene SUP45 of Saccharomyces cerevisiae. BMC Mol. Biol. 4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muramatsu, T., K. Heckmann, C. Kitanaka, and Y. Kuchino. 2001. Molecular mechanism of stop codon recognition by eRF1: a wobble hypothesis for peptide anticodons. FEBS Lett. 488:105-109. [DOI] [PubMed] [Google Scholar]
- 31.Orias, E., E. P. Hamilton, and J. D. Orias. 2000. Tetrahymena as a laboratory organism: useful strains, cell culture, and cell line maintenance. Methods Cell Biol. 62:189-211. [DOI] [PubMed] [Google Scholar]
- 32.Osawa, S., and T. H. Jukes. 1989. Codon reassignment (codon capture) in evolution. J. Mol. Evol. 28:271-278. [DOI] [PubMed] [Google Scholar]
- 33.Osawa, S., and T. H. Jukes. 1995. On codon reassignment. J. Mol. Evol. 41:247-249. [DOI] [PubMed] [Google Scholar]
- 34.Salas-Marco, J., and D. M. Bedwell. 2005. Discrimination between defects in elongation fidelity and termination efficiency provides mechanistic insights into translational readthrough. J. Mol. Biol. 348:801-815. [DOI] [PubMed] [Google Scholar]
- 35.Salas-Marco, J., and D. M. Bedwell. 2004. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol. Cell. Biol. 24:7769-7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santos, M. A., G. Moura, S. E. Massey, and M. F. Tuite. 2004. Driving change: the evolution of alternative genetic codes. Trends Genet. 20:95-102. [DOI] [PubMed] [Google Scholar]
- 37.Schull, C., and H. Beier. 1994. Three Tetrahymena tRNA(Gln) isoacceptors as tools for studying unorthodox codon recognition and codon context effects during protein synthesis in vitro. Nucleic Acids Res. 22:1974-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schultz, D. W., and M. Yarus. 1996. On malleability in the genetic code. J. Mol. Evol. 42:597-601. [DOI] [PubMed] [Google Scholar]
- 39.Schultz, D. W., and M. Yarus. 1994. Transfer RNA mutation and the malleability of the genetic code. J. Mol. Biol. 235:1377-1380. [DOI] [PubMed] [Google Scholar]
- 40.Seit-Nebi, A., L. Frolova, and L. Kisselev. 2002. Conversion of omnipotent translation termination factor eRF1 into ciliate-like UGA-only unipotent eRF1. EMBO Rep. 3:881-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sizonenko, G., I. Chernov, V. Kulikov, T. Karpova, P. Kashkin, I. Pavlov, T. Berkhtereva, E. Sakharova, O. Tikhodeev, and S. Inge-Vechtomova. 1990. A new class of ochre-suppressors in Saccharomyces: mutations in the tRNA-gln gene. Dokl. Akad. Nauk SSSR 310:1480-1484. (In Russian.) [PubMed] [Google Scholar]
- 42.Song, H., P. Mugnier, A. K. Das, H. M. Webb, D. R. Evans, M. F. Tuite, B. A. Hemmings, and D. Barford. 2000. The crystal structure of human eukaryotic release factor eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100:311-321. [DOI] [PubMed] [Google Scholar]
- 43.Stansfield, I., K. M. Jones, V. V. Kushnirov, A. R. Dagkesamanskaya, A. I. Poznyakovski, S. V. Paushkin, C. R. Nierras, B. S. Cox, M. D. Ter-Avanesyan, and M. F. Tuite. 1995. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 14:4365-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki, T., T. Ueda, and K. Watanabe. 1997. The ‘polysemous’ codon—a codon with multiple amino acid assignment caused by dual specificity of tRNA identity. EMBO J. 16:1122-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhouravleva, G., L. Frolova, X. Le Goff, R. Le Guellec, S. Inge-Vechtomov, L. Kisselev, and M. Philippe. 1995. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 14:4065-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]