Abstract
The hematopoietic zinc finger protein, Hzf, is induced in response to genotoxic and oncogenic stress. The Hzf protein is encoded by a p53-responsive gene, and its overexpression, either in cells retaining or lacking functional 53, halts their proliferation. Enforced expression of Hzf led to the appearance of tetraploid cells with supernumerary centrosomes and, ultimately, to cell death. Eliminating Hzf mRNA expression by use of short hairpin (sh) RNAs had no overt effect on unstressed cells but inhibited the maintenance of G2 phase arrest following ionizing radiation (IR), thereby sensitizing cells to DNA damage. Canonical p53-responsive gene products such as p21Cip1 and Mdm2 were induced by IR in cells treated with Hzf shRNA. However, the reduction in the level of Hzf protein was accompanied by increased polyubiquitination and turnover of p21Cip1, an inhibitor of cyclin-dependent kinases whose expression contributes to maintaining the duration of the G2 checkpoint in cells that have sustained DNA damage. Thus, two p53-inducible gene products, Hzf and p21Cip1, act concomitantly to enforce the G2 checkpoint.
The p53 transcription factor is the most frequently inactivated tumor suppressor in human cancer (19). Stabilization and activation of p53 in response to various oncogenic stress signals trigger a program of gene expression that inhibits cell proliferation or, more drastically, induces apoptosis, thereby eliminating incipient tumor cells (36, 49). Cancer-derived p53 mutations compromise the transcriptional activity of p53, enabling cells that have sustained oncogenic damage to persist and eroding their genome integrity. Many p53-responsive genes have been identified that encode proteins such as p21Cip1, a potent cyclin-dependent kinase (CDK) inhibitor that arrests progression through the cell division cycle (16, 20, 59); Bax and Puma, which induce cell death by regulating a mitochondrial-based apoptotic cascade (46, 47, 61); and Mdm2, an E3 ubiquitin protein ligase that degrades p53 and acts in a feedback circuit to terminate the p53 response (21, 31). Although the functions of these canonical p53-induced proteins are relatively well understood, the roles of many other p53 target genes have yet to be determined.
The best-characterized mechanism for initiating the p53 response is an intrinsic signal transduction cascade triggered by genotoxic stress. Different forms of DNA damage resulting from either single- or double-strand breaks, stalled replication forks, or exposure to cytotoxic drugs that affect DNA structure activate protein kinases of the ATM/ATR family (27). These, in turn, induce p53 phosphorylation both directly and through the agency of other kinases, such as Chk1 and Chk2, that are themselves activated through ATM/ATR phosphorylation (2). Phosphorylation of p53 at several sites near its N terminus not only interferes with its association with its negative regulator Mdm2 but also facilitates interactions between p53 and other transcriptional coactivators on p53-responsive promoters (27). Complexity stems from the fact that the p53 protein can undergo many posttranslational modifications including acetylation, sumoylation, and phosphorylation by many different kinases, all of which may contribute to its stability, activation state, and target specificity (5). Apart from the DNA damage response, aberrant growth-promoting signals emanating from oncogenes can also activate p53 by inducing the tumor suppressor, p19Arf (p14ARF in humans), which binds to Mdm2 and antagonizes its E3 ubiquitin ligase activity (40). Several other proteins, including the ribosomal proteins L11 and L23 and the nucleolar protein nucleophosmin (NPM/B23), have been reported to similarly contribute to the p53 response by inhibiting Mdm2 function (25, 34, 39, 62) and, like p53, Mdm2 can itself undergo regulatory phosphorylation that influences its interaction with p53 following different forms of oncogenic stress (44, 50).
In studying the genome-wide transcriptional response to p19Arf induction (33), we noted that the so-called hematopoietic zinc finger gene (Hzf) was induced with kinetics similar to Mdm2 and Cip1. Hzf was originally identified as a gene whose expression is induced in hematopoietic progenitor cells derived from differentiating embryonic stem cells in vitro (23). Although its elimination in the mouse germ line suggested a possible role for Hzf in late megakaryocytic differentiation, mice lacking the gene seemed to be otherwise normal (29). Here we show that Hzf is a bona fide transcriptional target of p53 that contributes to the maintenance of checkpoint arrest in cells responding to DNA damage.
MATERIALS AND METHODS
Cells and culture conditions.
NIH 3T3 and MT-Arf cells (33) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 2 mM glutamine, and 100 U/ml penicillin and streptomycin. Primary mouse embryonic fibroblasts (MEFs) were cultured at passages 3 to 5 on a 3T3 protocol in medium supplemented with 0.1 mM nonessential amino acids, 55 μM 2-mercaptoethanol, and 10 μg/ml gentamicin instead of penicillin and streptomycin (64). For clonogenic survival assays, 104 cells were seeded into 10-cm diameter culture dishes and subjected to irradiation with a 137Cs source at a rate of approximately 1 Gy/min for a total dose of 15 Gy. After 10 days, cultured cells were stained with HEMA-QUICK (Fisher, Middletown VA), and the number of colonies per dish was counted.
Virus production and infection.
293T cells were transfected with retroviral expression plasmids expressing the designated genes of interest together with either green fluorescent protein or drug resistance markers in cis as previously described (64). Replication-defective viruses rescued with plasmids encoding requisite helper functions were harvested 24 to 60 h after transfection, pooled, and stored on ice. Exponentially growing cells in 10-cm-diameter culture dishes were infected with 3 ml of fresh virus-containing supernatant in complete medium containing 8 μg/ml polybrene (Sigma Chemicals, St. Louis, MO). Infection was confirmed either by flow cytometric assay for green fluorescent protein expression or by selection for drug resistance.
Hzf expression plasmids.
Mouse Hzf cDNA was cloned from a mouse embryo (embryonic day 12 [E12]) cDNA library by PCR using the following primers: sense, 5′-AAAGAATTCATCCTCGGCAGCCTGAGCCG-3′; antisense, 5′-AAACTCGAGTCAGTAGGGGGAGAAGAGGA-3′. The PCR product, recloned and verified by nucleotide sequence analysis, was digested with EcoRI and XhoI and cloned into an mouse stem cell virus vector 3′ to sequences specifying a Flag epitope (4). Hzf deletion mutant cDNAs were obtained by PCR using full-length Hzf cDNA as a template. Four PCRs were performed as follows: fragment A, Hzf sense and dZn-1AS (5′-CTCTTCGGGCGGAAATGACTGGCCGCTTGG-3′); fragment B, dZn-1S (5′-AGTCATTTCCGCCCGAAGAGTCAAAGGCAT-3′) and Hzf antisense; fragment C, dZn-1S and dZn-2AS (5′-CGCCATCTCGGTAGAGCAGACGCTTAGCCT-3′); and fragment D, dZn-2S (5′-TCTGCTCTACCGAGATGGCGTGGCTGGGAA-3′) and Hzf antisense. Purified PCR products were mixed (fragments A and B for the d2 mutant and fragments A, C, and D for the d3 mutant) and subjected to a second PCR using Hzf sense and antisense primers. For the knock-down of endogenous Hzf, annealed oligonucleotides (GATCCCCAGCCATGTCCATCCAACCCTTCAAGAGAGGGTTGGATGGACATGGCTTTTTTGGAAA and AGCTTTTCCAAAAAAGCCATGTCCATCCAACCCTCTCTTGAAGGGTTGGATGGACATGGCTGGG) were cloned into a pSUPERretro vector plasmid (Oligoengine, Seattle, WA) in accordance with the manufacturer's instructions.
Northern blotting.
Total RNA prepared from NIH 3T3 and MT-Arf cells was separated on a 1% formaldehyde-agarose gel (20 μg/lane) and transferred to a Hybond-N+ membrane. Prehybridization, hybridization, and washing were performed as previously described (11). For detection by reverse transcription-PCR (RT-PCR), total RNA was reverse transcribed using a random oligohexamer primer and murine leukemia virus reverse transcriptase (Stratagene, La Jolla, CA). Reverse-transcribed products were aliquoted and subjected to PCR using the following primers: Hzf-sense, 5′-CCATCAAAGCTTACCCTCGG-3′; Hzf-antisense, 5′-AGAAGAGGATGGGTCCGTGC-3′; HPRT-sense, 5′-CCAGCAAGCCTTGCAACCTTAACCA-3′; and HPRT-antisense, 5′-GTAATGATCAGTCAACGGGGGAC-3′ (HPRT is hypoxanthine phosphoribosyltransferase). Each of 25 cycles consisted of denaturation at 94°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 1 min. The PCR products were separated on agarose gels, visualized by ethidium bromide, and subjected to Southern blotting using a full-length Hzf cDNA coding sequence as the probe.
Immunofluorescence and immunoblotting.
An Hzf C-terminal peptide (LHPAPGPIRTAHGPILFSPY) conjugated to keyhole limpet hemocyanin was used to immunize rabbits, and antibodies were affinity purified, all as previously described (14). Cells on coverslips were fixed with 4% paraformaldehyde in phosphate-buffered saline for 10 min at room temperature and stained with affinity-purified antibodies to the Hzf C terminus for 1 h at room temperature. Washed coverslips were incubated with Texas Red-conjugated antibodies to rabbit immunoglobulin G (Amersham Biosciences, Piscataway, NJ). Cell lysates prepared in radioimmunoprecipitation assay buffer (1.25% NP-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 150 mM NaCl, 10 mM sodium phosphate pH 7.2, 2 mM EDTA, 50 mM sodium fluoride, 0.2 mM sodium vanadate, 100 U/ml aprotinin) were separated by electrophoresis on polyacrylamide gels containing SDS and transferred to polyvinylidene difluoride membranes (Millipore, Bedford, MA). Proteins were detected using antibodies to the Hzf C terminus, p53 (Ab-7; Oncogene Research Products, Boston, MA), p21Cip1 (C-19; Santa Cruz Biotechnology, Santa Cruz, CA), CDK4 (C-22; Santa Cruz), and Mdm2 (2A10). Sites of antibody binding were detected using horseradish peroxidase-conjugated antibodies to mouse or rabbit immunoglobulin G as previously described (64).
Luciferase reporter assay.
The annealed oligonucleotides (wild-type sense, 5′-TCTCCGCAGCCATCCTGCCCCGACACGCCGGGACCTGCCCCTCT CTCCCTGCGCCGGCT-3′; wild-type antisense, 5′-GATCAGCCGGCGCAGGGAGAGAGGGGCAGGTCCCGGCGTGTCGGGGCAGGATGGCTGCGGAGAGTAC-3′; mutant sense, 5′-TCTCCGCAGCCATCCTGCCCCGACACTAAGGGACCTTAACCTCTCTCCCTGCGCCGGCT-3′; and mutant antisense, 5′-GATCAGCCGGCGCAGGGAGAGAGGTTAAGGTCCCTTAGTGTCGGGGCAGGATGGCTGCGGAGAGTAC-3′) were inserted into KpnI/BglII sites of a pGl2-Promoter vector (Promega, Madison, WI) and transfected into NIH 3T3 cells (5 × 104 cells per 3.5-cm diameter culture dish) together with pRL-SV40 (where SV40 is simian virus 40; Promega) in the presence or absence of a p19Arf expression plasmid. Cell lysates were prepared 48 h later, and luciferase activities were measured using a dual luciferase reporter assay system (Promega). For the measurement of ionizing radiation (IR)-induced luciferase activity, cells were irradiated 12 h before lysate preparation. Luciferase (firefly) activity in each sample was normalized by Renilla luciferase.
Electrophoretic mobility shift assay (EMSA).
Annealed oligonucleotides (wild-type sense, 5′-TTTCATCCTGCCCCGACACGCCGGGACCTGCCCCTCTCTCCCT-3′; wild-type antisense, 5′-TTTAGGGAGAGAGGGGCAGGTCCCGGCGTGTCGGGGCAGGATG-3′; mutant sense, 5′-TTTCATCCTGCCCCGACACTAAGGGACCTTAACCTCTCTCCCT-3′; and mutant antisense, 5′-TTTAGGGAGAGAGGTTAAGGTCCCTTAGTGTCGGGGCAGGATG-3′) were radiolabeled using Klenow fragment (New England Biolabs, Beverly, MA) and [α-32P]dATP (6,000 Ci/mmol; Perkin Elmer Life Sciences, Boston, MA). p53 protein was produced by in vitro transcription/translation using a rabbit reticulocyte lysate system (Promega) and preincubated at room temperature for 15 min in buffer containing 20 mM HEPES (pH 7.9), 50 mM KCl, 5 mM MgCl2, 10 mM ZnSO4, 0.2% NP-40, 10% glycerol, 1 mM spermidine, 0.1 mg/ml bovine serum albumin, 0.5 mM dithiothreitol, and 10 ng/μl poly(dI-dC) in the presence or absence of 1 μg of p53 antibody (PAb421 [Oncogene Research Products] and DO-1 [Santa Cruz], respectively). Radiolabeled probe (104 cpm per sample) was then added and incubated for 15 min at room temperature; samples were electrophoretically separated on 4% polyacrylamide gels containing 22.5 mM Tris-borate and 0.5 mM EDTA, and complexes were detected by autoradiography.
ChIP.
Chromatin immunoprecipitation (ChIP) was performed by a modified protocol (22) using p53-null MEFs infected with control or human p53-expressing retroviruses. A total of 107 cells used for each immunoprecipitation were cross-linked in 1% formaldehyde for 10 min and then neutralized with 0.125 M glycine. Cells were scraped, washed with phosphate-buffered saline, resuspended in cell lysis buffer I (10 mM HEPES, pH 6.5, 10 mM EDTA, 0.5 mM EGTA, 0.25% Triton X-100, and protease inhibitors), and incubated on ice for 10 min. Cells were then collected by centrifugation and resuspended in cell lysis buffer II (10 mM HEPES, pH 6.5, 1 mM EDTA, 0.5 mM EGTA, 0.2 M NaCl, and protease inhibitors). Nuclei were precipitated and suspended (0.3 ml for each immunoprecipitation reaction) in nuclei lysis buffer (50 mM Tris-HCl, pH 8.1, 10 mM EDTA, 1% SDS, and protease inhibitors) and sonicated 10 times for 15 s to yield genomic DNA 200 to 600 base pairs in length. Sonicated chromatin was diluted to 1 ml with immunoprecipitation dilution buffer (1% Triton X-100, 2 mM EDTA, 20 mM Tris-Cl, pH 8.1, and 150 mM NaCl) and precleared with salmon sperm DNA-coated protein A beads (Upstate, Charlottesville, CA) twice for 90 min. Precleared lysates were then immunoprecipitated overnight at 4°C with 1 μg of anti-p53 antibody FL-393 (Santa Cruz) or PAb421. Immune complexes recovered after incubation with protein A beads for 3 h were washed seven times with immunoprecipitation wash buffer (50 mM HEPES, pH 7.6, 1 mM EDTA, 0.7% deoxycholic acid, 0.5 M LiCl, 1% NP-40, and protease inhibitors) and once with Tris-EDTA buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA). Beads were rotated for 3 min at 4°C between each washing step. Immune complexes were eluted twice for 15 min with 0.1 ml of elution buffer (0.1 M NaHCO3 and 1% SDS) and incubated with 10 μg of RNase A for 60 min at 37°C, followed by treatment with 4 μg of proteinase K at 42°C for 150 min. Immune complexes were incubated at 65°C overnight to reverse formaldehyde cross-links, and genomic DNA was isolated with a PCR purification kit (QIAGEN, Valencia, CA) and subjected to PCR (35 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min) using the following Hzf primers: sense, 5′-AGATGCTGGGTGTAGCTTCG-3′; antisense, 5′-CCACTCTGACTTTCCTTCTC-3′). As a positive control, the Cip1 gene was similarly amplified using the following primers: sense, 5′-CCAGAGGATACCTTGCAAGGC-3′; and antisense, (5′-TCTCTGTCTCCATTCATGCTCCTCC-3′.
Fluorescence-activated cell sorting (FACS) analysis.
Cells seeded at subconfluent density were pulse labeled with 10 μM bromodeoxyuridine (BrdU) for 30 min and exposed to 15 Gy of gamma irradiation. Irradiated cells were collected at the indicated times and stained with fluorescein isothiocyanate-conjugated antibody to BrdU and 7-amino-actinomycin D using a fluorescein isothiocyanate-BrdU Flow kit (BD Biosciences, San Diego, CA). Stained cells were analyzed using a FACSCaliber sorter (BD Biosciences).
Ubiquitination assay for p21Cip1 protein.
NIH 3T3 cells were transfected with 0.1 μg of Flag-p21, 1 μg of shHzf, and 2 μg of His-ubiquitin expression plasmids using Fugene 6 (Roche, Indianapolis, IN). His-ubiquitinated proteins were purified under denaturing conditions as described previously (32). Cells were suspended in lysis buffer containing 8 M urea, 0.1 M NaHPO4, 10 mM Tris-Cl, pH 8.0, 10 mM imidazole, 10% glycerol, 0.1% Triton X-100, 0.5 M NaCl, and 10 mM β-mercaptoethanol and sonicated for 15 s, and lysates were cleared by centrifugation. Cleared lysates were incubated with Ni-nitrilotriacetic acid agarose beads (QIAGEN) at room temperature for 6 h. Beads were washed five times with lysis buffer and boiled in denaturing gel sample buffer containing 0.2 M imidazole, and eluted proteins were separated on denaturing gels and subjected to immunoblotting.
RESULTS
Hzf is a direct p53 target gene.
Microarray studies performed with an NIH 3T3 cell line engineered to express an inducible Arf gene identified numerous p19Arf-responsive genes that were temporally regulated in either a p53-dependent or -independent manner (33). Among these, the Hzf gene (gene identifier 7305176) encoding a previously described “hematopoietic zinc finger” (Hzf) protein (23) was rapidly induced with kinetics similar to that of the bona fide p53-regulated target gene, Mdm2. The failure of exogenously expressed p19Arf to increase Hzf mRNA levels in p53-null MEFs indicated that Hzf expression is p53 dependent. Using PCR and a cDNA library from mouse mid-gestation embryos (E12), we cloned and sequenced an Hzf cDNA containing an open reading frame encoding the predicted 44-kDa protein that includes three zinc finger motifs. Although we noted some mismatches between this Hzf nucleotide sequence and that determined earlier (23), the mouse expressed sequence tag sequences in the NCBI database corresponding to Hzf, as well as the cDNA reconstructed from the mouse genomic DNA sequence database, are identical to that of the cDNA that we recovered.
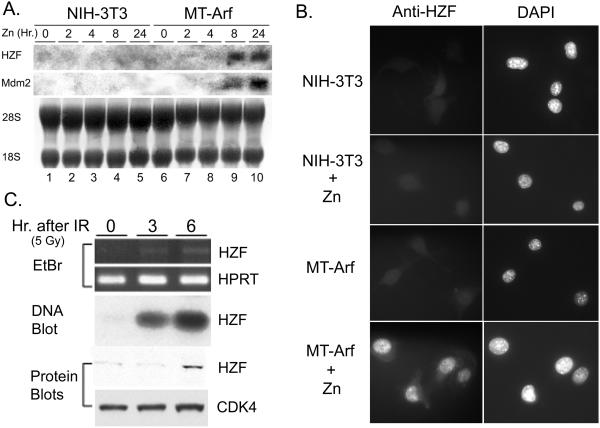
To confirm that Hzf mRNA is induced by p19Arf, RNAs prepared from control Arf-null NIH 3T3 cells and from a zinc-inducible derivative in which the metallothionein promoter regulates an Arf transgene (MT-Arf) were analyzed by Northern blotting using the Hzf cDNA as a probe. In this inducible system, p19Arf protein is first detected within 4 h after the addition of zinc sulfate to the medium, after which p53 and p53-induced proteins such as Mdm2 or p21Cip1 accumulate; the cells withdraw completely from the division cycle within 24 h, arresting in both the G1 and G2 phases (33). Hzf and Mdm2 mRNAs accumulated de novo in MT-Arf cells, but not in control NIH 3T3 cells, within 8 h after the addition of zinc sulfate (Fig. 1A). Using antibodies directed to the predicted C terminus of Hzf, we observed accumulation of the protein predominantly within nuclei of induced MT-Arf cells (Fig. 1B). Since p53 can be activated in a p19Arf-independent manner by many genotoxic stresses, we exposed NIH 3T3 cells to IR and determined that both Hzf mRNA and protein were again rapidly induced (Fig. 1C). Together, these results raised the possibility that Hzf might be a direct target of p53-mediated transcription.
FIG. 1.
Hzf is induced by p19Arf or IR. (A) NIH 3T3 and MT-Arf cells were treated with 80 μM zinc sulfate for the indicated periods, and Hzf and Mdm2 mRNAs were detected by Northern blotting. 28S and 18S rRNAs in the same membrane were stained with methylene blue before hybridization, confirming that equal amounts of RNA were loaded in each lane. (B) NIH 3T3 and MT-Arf cells treated for 24 h with zinc sulfate as indicated were stained with purified rabbit polyclonal antibodies directed to a C-terminal peptide of Hzf (left panels). Cells were counterstained with DAPI (4′,6′-diamidino-2-phenylindole) (right panels). (C) NIH 3T3 cells were exposed to 5 Gy of IR, and RNAs prepared at the indicated times were amplified by RT-PCR (top two panels). RT-PCR products were subjected to Southern blotting using Hzf cDNA as a probe (DNA blot, middle panel). Lysates were prepared from the same samples after IR, and Hzf protein was detected by immunoblotting using Cdk4 protein as a loading control (bottom two panels).
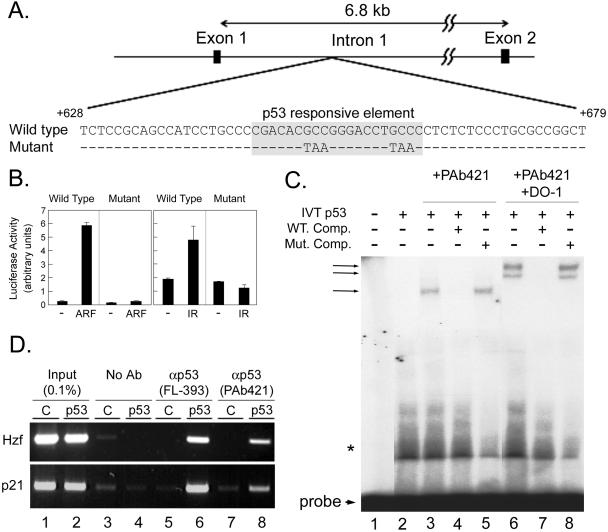
We therefore searched for potential p53-responsive elements (p53RE) within the Hzf gene in the NCBI mouse genomic DNA sequence database (http://www.ncbi.nih.gov/genome/guide/mouse/). Although we could not find a p53RE in the promoter region of Hzf, there is one potential p53RE within its first intron (Fig. 2A). Intronic p53-binding sites are not unusual and have been found in the p53-responsive Mdm2, Bax, Puma, and Pml genes (13, 26, 47, 55, 61). Based on these observations, an Hzf genomic fragment including the putative wild-type or mutated p53RE (Fig. 2A) was inserted into a luciferase reporter plasmid containing the SV40 minimal promoter, which was cotransfected into NIH 3T3 cells with a control or p19Arf expression plasmid. Expression of p19Arf led to an approximately 20-fold induction of luciferase activity, whereas no effect was observed when the p53RE was mutated (Fig. 2B, left two panels). Luciferase activity was also induced by IR and again strictly depended upon the wild-type p53RE enhancer element (Fig. 2B, right two panels).
FIG. 2.
Hzf is a transcriptional target of p53. (A) The schematic indicates a potential p53-responsive element in Hzf intron 1. The designated mutated sequence was also utilized in the following experiments. (B) Empty or p19Arf expression plasmids were cotransfected into NIH 3T3 cells together with a luciferase reporter plasmid containing a wild-type or mutant p53 response element. Alternatively, transfected cells were exposed to 5 Gy of IR, and luciferase activities were measured after 12 h. Bars indicate the standard errors for each group. (C) An EMSA was performed using the Hzf-derived p53 response element as a probe. PAb421 was used to stabilize DNA-p53 complexes, and DO-1 was added to supershift the complexes. Nonradiolabeled DNA fragments (either wild type or mutant, added at a 600-fold molar excess to the radiolabeled probe) were used to compete for binding. The bottom arrow designates the position of unbound probe; arrows at the top indicate the mobilities of complexes containing p53. The asterisk indicates nonspecifically bound proteins from the reticulocyte lysate. (D) ChIP using antibodies to p53. p53 complexes were immunoprecipitated from p53-null MEFs infected with control (C) or p53 retroviruses. The Hzf intron 1 sequence and the region of the p21Cip1 promoter were amplified by PCR.
To determine whether induction of Hzf by p19Arf or IR could be mediated by the direct binding of p53 to the intronic p53RE element, an EMSA was performed using the Hzf p53RE as a probe (Fig. 2C). The human p53 protein, produced by in vitro translation in a rabbit reticulocyte lysate, was mixed with a radiolabeled fragment containing the p53RE. To activate p53 and stabilize the DNA-p53 protein complex in vitro (18), we utilized an antibody (PAb421) that binds to the p53 C terminus. As expected, p53 stably associated with the probe only in the presence of PAb421 (Fig. 2C, lane 3 versus lane 2), and no such complex was observed using control lysates lacking p53 (not shown). An excess amount of a nonradiolabeled wild-type Hzf p53RE fragment competed with the probe for binding (lane 4), whereas the mutant fragment did not (lane 5). In agreement with this result, when radiolabeled, the mutated oligonucleotide was unable to bind to p53 (data not shown). To confirm that the high-molecular-mass complexes seen in lanes 3 and 5 contained the p53 protein, we performed EMSAs in which we added yet another antibody to p53 (DO-1) that recognizes a p53 epitope distinct from that bound by PAb421. The addition of PAb421 and DO-1 together further slowed the mobility of the complexes (lanes 6 and 8), indicating that they are due to direct binding of p53 to the probe. We next performed a ChIP assay to verify that p53 binds to the Hzf locus in vivo (Fig. 2D). MEFs null for p53 were infected with a retrovirus encoding human p53, and cross-linked p53 complexes were immunoprecipitated using two different antibodies to p53. Hzf genomic DNA (intron 1) was amplified only from immunoprecipitated complexes recovered from cells expressing p53 (Fig. 2D, lanes 6 and 8). As expected, sequences from the p53-responsive Cip1 promoter could be amplified from the same samples (Fig. 2D), further confirming the specificity of the analysis. Taken together, these results indicate that Hzf is a direct transcriptional target of p53.
Hzf overexpression leads to tetraploidization, checkpoint failure, and cell death.
To address the biological activity of Hzf, we used retroviral vectors to enforce expression of the exogenous Hzf protein or of mutants lacking two (d2) or three (d3) zinc fingers (Fig. 3A). Overexpression of Hzf inhibited colony formation in both NIH 3T3 cells and in p53-null MEFs, suggesting that Hzf itself has p53-independent antiproliferative activity (Fig. 3B), whereas the mutants were without effect (data not shown). When analyzed 3 days after infection, many Hzf-transduced cells exhibited a 4N DNA content; in contrast, cells infected with vectors encoding the two Hzf deletion mutants were not significantly affected (Fig. 3C). Microscopic inspection of fixed and stained cells indicated that many of those transduced with wild-type Hzf became binucleate (Fig. 3D); in turn, staining with antibodies to γ-tubulin revealed that more than half of this population contained greater than two centrosomes (Fig. 3E). Despite the increased number of cells with 4N DNA content, subsequent biochemical analyses indicated that the Hzf-transduced population exhibited very low levels of mitotic cyclins A and B and cyclin A-dependent histone H1 kinase activity, whereas cyclins D1 and E were readily detected (data not shown). Together, these data imply that cells engineered to overexpress Hzf undergo an abortive mitosis, return to a G1 state without undergoing cytokinesis, and thereby become tetraploid and exhibit additional centrosomes. If their p53-dependent checkpoint is intact, such cells should arrest in G1 (35, 43, 45, 48). However, we noted that many such cells continued to incorporate BrdU for several more days (Fig. 3G), and this concomitantly led to cell death (Fig. 3H). Immunofluorescence staining with antibodies to γ-tubulin (Fig. 3F) and centriolin (not shown) further revealed that many such cells exhibited more than four centrosomes despite the absence of detectable 8N DNA content, indicating that the centrosome and DNA replication cycles had been uncoupled. Therefore, although NIH 3T3 cells retain wild-type p53 and can mount a robust p53 transcriptional response (see above), p53-dependent checkpoints fail when Hzf expression is enforced, and the cells form extranumerary centrosomes, reenter S phase, and ultimately die.
FIG. 3.
Hzf overexpression in fibroblasts causes the inhibition of cell proliferation. (A) Schematic representations of Hzf and mutant proteins. Full-length Hzf protein (WT) contains three C2H2-type zinc finger domains (black bars). (B) Colony formation assay. NIH 3T3 or p53-null fibroblasts were infected with control or Hzf retroviruses. After 3 days of infection, 104 cells were seeded per plate, cultured for another 10 days, and stained with Giemsa. (C) NIH 3T3 cells were infected with indicated retroviruses and analyzed 3 days later for DNA content by FACS. (D) Cells were fixed and stained with Giemsa, and binucleate cells were counted at the indicated times postinfection. (E) After 3 days of infection, cells were stained using anti-γ-tubulin antibody, and the numbers of centrosomes per cell were counted. (F) Extranumerary centrosomes are present in cells 4 days after infection with an Hzf-expressing vector versus a vector control. (G) NIH 3T3 cells infected with the indicated retroviruses were pulse-labeled with BrdU for 30 min and stained with anti-BrdU antibody. (H) The same cells as shown in panel G were stained with annexin V and analyzed by FACS.
Hzf contributes to prolonged G2 phase arrest and survival after DNA damage.
As an alternative approach, the levels of endogenous Hzf induced in response to p53 were “knocked down” using short hairpin (sh) RNAs. The shHzf RNA used for these studies was designed to target the 3′ untranslated region of Hzf mRNA and efficiently limited endogenous Hzf expression at both the mRNA and protein level (see Fig. 5A). Because we suspected that cells lacking Hzf protein might exhibit checkpoint defects following DNA damage, wild-type primary MEFs infected for 48 h with control or shHzf-encoding retroviral vectors were pulsed for 30 min with BrdU to label cells in S phase. These were then exposed to IR (Fig. 4A). BrdU-positive cells were gated by flow cytometry, and their DNA content was analyzed at various times following IR treatment. In these analyses, a nonirradiated BrdU-positive population (S phase cells) will enter G2 (4N DNA content) and complete mitosis, thereby giving rise to a G1 population (2N DNA). However, IR should transiently arrest the BrdU-labeled cells in G2 phase, so that the subsequent formation of G1 phase cells will be retarded as long as the G2 checkpoint is maintained. In nonirradiated cells infected with the control vector, BrdU-positive cells (Fig. 4A, gated trapezoids) exhibited a DNA content between 2N and 4N, as expected for cells in S phase (Fig. 4A, control, second row, 0 h). By 12 h after IR, the majority of these cells were arrested in G2, although some had reentered G1. A significant percentage (55%) of irradiated cells remained in G2 even 24 h after exposure to IR (Fig. 4A, control, second row, 24 h). G2 phase arrest was similarly observed in the shHzf-treated cells 12 h after IR treatment. However, these cells did not efficiently maintain the G2 arrest, since many fewer BrdU-positive cells (36%) remained in G2 phase 24 h after IR (Fig. 4A, fourth row, 24 h). Therefore, although Hzf-deficient cells are able to appropriately initiate a G2 arrest in response to IR, Hzf is required for maintenance of the G2 checkpoint.
FIG. 5.
IR-induced induction of p21Cip1 is impaired in the absence of Hzf. (A) Expression of endogenous Hzf was knocked down in NIH 3T3 cells using an shHzf retrovirus. Hzf protein was then restored by exogenous Hzf protein (Flag-tagged). Cells were irradiated (15 Gy) and lysates were prepared at the indicated times after IR. Expression of Hzf, p53, p21Cip1, Mdm2, and CDK4 (as loading control) was analyzed by immunoblotting. Antibody raised against the C-terminal portion of Hzf protein recognizes both the endogenous and exogenous Flag-tagged proteins. (B) Similar experiments were performed using the Flag-Hzf d2 mutant.
FIG. 4.
Hzf knockdown causes a defect in sustained cell cycle arrest and increased radiosensitivity. (A) Wild-type MEFs infected with control or shHzf-expressing retroviruses as indicated at the right were pulse labeled for 30 min with BrdU and exposed to IR (15 Gy). At the indicated times after irradiation, the DNA content of cells was analyzed by FACS. Trapezoids in the top panels define the distribution of BrdU-positive cells, whose DNA contents are displayed in the lower panels. (B) Clonogenic survival assays after IR treatment. Control and Hzf-knockdown NIH 3T3 cells were treated with the indicated doses of IR (abscissa). After 10 days, the numbers of colonies in each plate were enumerated, and their survival was compared to those of nonirradiated control cultures. Standard errors are indicated by bars.
After genotoxic stress, cell cycle arrest induced by p53 is important for allowing cells time to repair damaged DNA strands, but the failure to do so can reprogram cells to commit suicide. Since knockdown of Hzf expression by shRNA caused a defect in maintaining G2 arrest, we checked whether or not the viability of cells lacking Hzf was compromised after IR treatment. NIH 3T3 cells, infected with vectors expressing shHzf RNA or not, were subjected to different doses of IR, and cell survival rates were determined by clonogenic assays. As expected, the number of colonies decreased with increasing IR in a dose-dependent manner, but there were always fewer colonies recovered from Hzf knockdown cells (Fig. 4B).
Hzf is required for accumulation of the p21Cip1 protein after DNA damage.
To explain the effects of Hzf loss on the cellular response to IR, we surveyed the expression of several cell cycle regulators that have been implicated in maintaining the G2 checkpoint (9, 58). Chief among these are the Cdc25 family proteins required for mitotic entry that are down-regulated by several DNA damage signaling molecules, including p53 (41, 54). We first compared the protein levels of Cdc25A, Cdc25B, and Cdc25C in control and Hzf knockdown cells after IR but did not note significant effects of Hzf loss of function on their expression. Whereas Cdc25B levels remained constant, Cdc25A and Cdc25C were reduced to undetectable levels after IR, whether cells were exposed to shHzf RNA or not (data not shown), in agreement with previous observations that their turnover is increased in response to DNA damage (2).
By contrast, induction of p21Cip1 by IR was significantly diminished in shHzf knockdown cells (Fig. 5A, lanes 9 to 12) versus its levels in cells infected with a control virus (lanes 1 to 4). Note that in response to IR treatment, the Hzf and p53 proteins were induced prior to the accumulation of p21Cip1 (lanes 1 to 4), whereas introduction of shRNA to Hzf reduced the levels of the Hzf and p21Cip1 proteins without affecting that of p53 (lanes 9 to 12). Moreover, shHzf RNA did not affect the p53-mediated induction of Mdm2, so p53 activity per se was not attenuated in Hzf knockdown cells after DNA damage.
To verify that the defect in p21Cip1 induction could be attributed to the loss of Hzf protein but not to off-target effects caused by the shHzf RNA, Flag-tagged Hzf proteins were reintroduced into the knockdown cells. Under the conditions used, the reprogrammed level of the exogenous Flag-tagged Hzf protein was comparable to that of the endogenous protein; due to the absence of the 3′ untranslated region target sequence in the Flag-Hzf construct, its expression was not affected by the shHzf interfering RNA (Fig. 5A, top panel, lanes 5 to 8 and 13 to 16). By contrast, shHzf RNA again reduced the level of the endogenous Hzf protein, which migrated faster than Flag-Hzf on the denaturing gels. The presence of the Flag-tagged Hzf protein did not affect basal p21Cip1 expression in nonirradiated control cells or markedly increase its level after radiation treatment (compare lanes 1 to 4 and 5 to 8). However, introduction of the Flag-Hzf protein restored IR induction of p21Cip1 in cells expressing shHzf (lanes 13 to 16 versus lanes 9 to 12). We next tested whether the Hzf d2 mutant previously shown to be defective in inducing Hzf-mediated phenotypes would rescue the inhibitory effects of shHzf on IR induction of p21Cip1. While in this case the levels of enforced expression of Flag-tagged Hzf proteins eclipsed that of endogenous Hzf, the Flag-Hzf d2 mutant was unable to restore Hzf function (Fig. 5B, lanes 16 to 18 versus lanes 13 to 15). Therefore, Hzf is required for the efficient accumulation of p21Cip1 following IR exposure. These results are consistent with a previous demonstration that the p53-p21Cip1 pathway is required for sustained G2 arrest after DNA damage, although it is not essential for its initiation (7, 58).
Because p53-dependent expression of p21Cip1 can prevent irradiated G1 phase cells from entering S phase (6, 12, 16, 20, 59), we further explored the effects of shHzf on irradiated cells stimulated to reenter the division cycle from quiescence. Confluent cultures of shHzf knockdown cells or of cells expressing the control vector were starved of serum. After 24 h, the cells were trypsinized, replated at a lower density, and stimulated with serum to reenter G1 phase synchronously. Stimulated cells were exposed to various doses of ionizing radiation, and their DNA content was measured both 12 and 18 h after mitogenic stimulation. Although shHzf treatment significantly diminished the levels of p21Cip1 induced in response to all doses of IR tested (see Fig. S1A in the supplemental material), both control and shHzf-treated cells retained an intact G1 checkpoint response (see Fig. S1B in the supplemental material). Evidently, the residual amount of p21Cip1 expressed in irradiated shHzf-treated cells was still adequate to inhibit entry into S phase.
Hzf loss does not affect the transcription of Cip1 but accelerates p21Cip1 turnover.
Expression of p21Cip1 is regulated at both the transcriptional and posttranslational level. Cip1 mRNA levels, as well as those of other canonical p53 target genes including cyclin G1 and Mdm2, were increased after IR in both control and Hzf knockdown cells, indicating that Hzf loss does not interfere at a detectable level with p53-mediated transcriptional activation of these target genes (Fig. 6A). The half-life of the p21Cip1 protein was also similar in nonirradiated control NIH 3T3 and Hzf knockdown cells, respectively (Fig. 6B and C). Whereas IR treatment did not alter p21Cip1 protein stability in control cells, its half-life was markedly reduced in irradiated Hzf knockdown cells (Fig. 6B and C). Therefore, the defect in p21Cip1 protein accumulation after IR in Hzf knockdown cells is due to its increased turnover.
FIG. 6.
Hzf knock-down destabilizes p21Cip1 after IR exposure. (A) Expression of p21Cip1, cyclin G1, and Mdm2 mRNA was determined by Northern blotting in nonirradiated (−) or irradiated (+) NIH 3T3 cells (15 Gy) infected with control or shHzf-expressing retroviruses. Methylene blue staining of 28S and 18S rRNAs indicated that equal amounts of RNA were loaded in each lane. (B) The same cells were treated with 100 μg/ml of cycloheximide (CHX) for the indicated periods. Proteins from cell lysates were separated on denaturing gels and subjected to immunoblotting with antibody to p21Cip1. (C) The intensities of the p21Cip1 signals in panel B were quantitated by densitometry to calculate the half-life of the protein under each condition.
Despite the fact that p21Cip1 can undergo ubiquitination, its basal turnover is generally thought to be regulated by ubiquitin-independent proteasomal degradation (3, 52). Native p21Cip1 has a disordered structure (30) and can be degraded by the 20S proteasome in vitro in the absence of polyubiquitination (38). However, ubiquitin-dependent p21Cip1 degradation has been observed in cells subjected to low doses of UV irradiation (3), implying that different modes of regulation can govern the protein's stability. As expected, treatment of cells with the proteasome inhibitor MG-132 increased p21Cip1 protein levels in control and Hzf knockdown cells, whether they were irradiated or not (Fig. 7A, bottom panel). When immunoblots were overexposed (Fig. 7A, top panel), we observed the accumulation of higher-molecular-weight species of p21Cip1 in irradiated Hzf knockdown cells (Fig. 7A, lane 8, brackets), which we thought might represent ubiquitinated forms of the protein.
FIG. 7.
p21Cip1 ubiquitination is increased in Hzf knockdown cells. (A) NIH 3T3 cells infected with control or shHzf-expressing retroviruses were exposed to IR (15 Gy). After 12 h, some cells were treated as indicated at the top of the panel with 10 μM MG-132 for an additional 4 h. Proteins from cell lysates were then separated on denaturing gels and immunoblotted with antibody to p21Cip1. The bracket at the right indicates the positions of modified forms of the protein. (B) Cells were transiently transfected with vectors expressing Flag-tagged p21Cip1 together with His-tagged ubiquitin and/or shHzf as indicated, and some were then exposed to IR as indicated at the top of the panel. Cells treated 12 h later with MG-132 for an additional 4 h were lysed in buffer containing 8 M urea. His-tagged proteins isolated by affinity chromatography on nickel agarose were separated on denaturing gels and subjected to immunoblotting using antibody to the Flag epitope. The levels of expression of Flag-p21Cip1 in the total cell lysates (lower panel) are compared to the relative recoveries of the affinity-purified His-tagged ubiquitinated species (top panel). (C) Nonirradiated cells were transfected with vectors encoding His-ubiquitin and Flag-tagged p21Cip1 as described for panel B (+ lanes) or with 10 times more of the p21Cip1 expression plasmid (++ lanes), and affinity-purified ubiquitinated proteins were blotted with antibodies to the Flag epitope. The levels of Flag-p21Cip1 in the total lysates (lower panel) were compared to the levels of ubiquitinated species (upper panel) as described for panel B.
To test whether or not ubiquitination of the p21Cip1 protein is accelerated in Hzf knockdown cells after IR treatment, cells were transiently transfected with expression plasmids encoding Flag-tagged p21Cip1 and His-tagged ubiquitin. His-ubiquitinated proteins were affinity purified on nickel agarose under denaturing conditions in buffer containing 8 M urea, and the recovered proteins were separated on gels and blotted with antibody to the Flag epitope (Fig. 7B). In control cells, the level of p21Cip1 ubiquitination did not change after IR exposure (Fig. 7B, lanes 3 and 4). In shHzf-transfected cells, however, we observed a significant increase of His-ubiquitinated p21Cip1 protein after IR treatment (lane 8 versus lane 7). Enforced expression of Flag-tagged p21Cip1 at much higher levels revealed ubiquitinated forms of the protein even in the absence of IR treatment (Fig. 7C, lane 3). shHzf expression further enhanced this effect, indicating that interference with basal levels of Hzf expression can also influence p21Cip1 ubiquitination (Fig. 7C, lane 7). Although we had assumed that induction of Hzf by IR might be a prerequisite in revealing any subsequent effects of shHzf on p21Cip1 ubiquitination and turnover (Fig. 6), this need not be the case. Taken together, these results strongly suggest that Hzf protects p21Cip1 protein from ubiquitination and accelerated turnover and that this contributes to maintenance of G2 checkpoint arrest after p53 activation.
DISCUSSION
In the adult mouse, the Hzf gene is predominantly expressed in megakaryocytes, and its targeted disruption in the mouse germ line has no reported effects on cell cycle dynamics, including the endoreduplication of DNA that occurs in such cells. Instead, Hzf inactivation results only in subtle defects affecting the maturation of megakaryocyte α-granules, and the animals otherwise appear to be overtly normal (23, 29). The limited role of Hzf in hematopoiesis is likely to be independent of p53, since no such phenotype has been described in p53-null mice, which have been scrupulously analyzed by many laboratories. The Hzf protein localizes to the nucleoplasm and contains three typical C2H2-type zinc finger domains that can potentially serve as nucleic acid-binding motifs. However, Rzf, the human orthologue of Hzf, does not bind to single- or double-stranded nucleic acids in vitro and appears not to act as a transcription factor (51). Thus, the biochemical function of the Hzf protein remains unknown.
We have now found that Hzf is a direct transcriptional target of the p53 tumor suppressor. Hzf is induced by ionizing radiation or via the Arf tumor suppressor pathway with kinetics similar to those of other p53-responsive genes, whereas its induction by these stimuli did not occur in cells lacking functional p53. A promoter-reporter plasmid containing a putative p53 enhancer sequence derived from the first intron of the Hzf gene responded to p53. EMSAs revealed that p53 could bind directly to the putative enhancer element but not to mutated consensus sequences, and ChIP experiments confirmed that p53 can bind directly to this intronic sequence in living cells. Conversely, shRNA-mediated reduction of Hzf did not affect the p53-dependent transcriptional activation of Cip1 or Mdm2 after DNA damage, so Hzf functions “downstream” of p53 and is itself not involved in regulating p53 activity per se. Together, these findings imply that Hzf can act to regulate p53-directed responses to DNA damage and oncogene activation, which can adversely affect genome integrity.
Several lines of evidence suggest that Hzf plays a role in cell cycle checkpoint control. Overexpression of the Hzf protein led to an accumulation of tetraploid cells, some of which were binuclear; these cells contained extranumerary centrosomes, continued to cycle, and ultimately died. Similar events have been observed when functions of key regulators of mitotic entry or exit are compromised, resulting from debilitating mutations affecting a wide spectrum of genes that regulate G2 checkpoint arrest, mitotic progression, the spindle checkpoint, kinetochore and centrosome integrity, and cytokinesis (8, 15, 17, 24, 28, 35, 37, 42, 48, 53, 57, 58, 60, 63). Like cells that overexpress Hzf, those that fail to undergo mitosis for other reasons become tetraploid, contain twice their normal complement of centrosomes, and revert to a G1 state in which the mitotic cyclins A and B are no longer expressed and are replaced by cyclins D and E. Unless p53 function is disrupted, such cells arrest in G1 phase (1, 15, 35, 43, 48). Possibly, alterations in centrosome composition coupled with a p53-dependent reduction in cyclin E-dependent Cdk activity prevent further centrosome duplication as well as the reinitiation of DNA synthesis (15). However, if p53 function is disabled, such cells reenter S phase, acquire even more centrosomes, and advance into mitosis with multipolar spindles, leading to abortive divisions and cell death. Notably, Hzf can prevent colony formation both in p53-positive fibroblasts as well as in primary p53-null MEFs, so its effects must be at least partially p53 independent. How overexpression of Hzf triggers the accumulation of tetraploid cells that bypass the p53-dependent G1 checkpoint and continue to cycle remains a mystery.
We obtained some mechanistic insights from loss-of-function experiments performed using shRNAs directed to Hzf. Knock-down of endogenous Hzf by shRNA limited the duration of G2 phase arrest following DNA damage, and these cells exhibited increased radiosensitivity. Cdc25A and Cdc25C levels fell dramatically after shHzf-treated cells were exposed to ionizing radiation, but p21Cip1 did not efficiently accumulate to expected levels. This suggested that Hzf regulates the level of p21Cip1 following irradiation of cells, in agreement with observations that Cip1-null cells are defective in maintaining G2 cell cycle arrest triggered by DNA damage and are more sensitive to DNA-damaging agents (7, 9, 58). Therefore, we reason that the failure of p21Cip1 to accumulate under these conditions could contribute to the impaired G2 arrest and to the increased radiosensitivity of Hzf knock-down cells. In contrast, when Hzf knock-down cells were stimulated to synchronously enter the cell cycle from quiescence and were then exposed to IR early in G1 phase, the observed reduction in p21Cip1 levels was not sufficient to allow entry into S phase. We cannot exclude the possibility, however, that an shHzf-mediated reduction of p21Cip1 in continuously cycling cells might blunt G1 arrest.
In unstressed cells “basal” ubiquitination of p21Cip1 seems not to be required for its proteasome-dependent degradation (3, 10, 52, 56), as nonubiquitinated p21Cip1 is degraded by the proteasome as efficiently both in vitro (38) and in vivo (52). Other investigators have reported that UV irradiation regulates ubiquitin-dependent degradation of p21Cip1 (3). In turn, our findings revealed that Hzf suppression can increase the ubiquitination and turnover of p21Cip1. Therefore, we presume that p53-induced Hzf can protect the p21Cip1 protein from ubiquitination and facilitate its accumulation in response to DNA damage.
However, we think it likely that Hzf affects the activity of proteins other than p21Cip1. First, Hzf overexpression causes phenotypic changes without affecting p21Cip1 expression at a detectable level, and it also does so in p53-null cells that express very low levels of p21Cip1. Second, when shRNAs directed to p21Cip1 were used, the attenuation of G2 arrest following IR treatment was less than that observed in cells treated with shHzf (data not shown), whereas inactivation of p21Cip1 more strongly compromises the G1 checkpoint (12, 20, 59). The simplest idea is that Hzf's effects might be mediated through the ubiquitin-proteasome pathway and involve several targets, p21Cip1 among them.
How does the absence of Hzf increase p21Cip1 ubiquitination and degradation? We have not detected a direct physical interaction between Hzf and p21Cip1 in cells, nor have we observed an effect of Hzf loss on the localization of p21Cip1 in cells before or after DNA damage. A yeast two-hybrid interactive screen using Hzf as bait identified APC5, a component of the anaphase-promoting (E3 ligase) complex as a protein that can physically interact with Hzf but not with the Hzf d2 mutant (M. Sugimoto, unpublished). However, we have so far been unable to implicate Hzf as a bona fide regulator of the anaphase-promoting complex in vivo. Whatever the exact mechanisms, our results indicate that Hzf and p21Cip1, both p53 targets, act in concert to coordinate the p53 response upon DNA damage.
Supplementary Material
Acknowledgments
We thank the members of the Sherr-Roussel laboratory for advice and criticism throughout the course of this work, Kenji Tago for help with the luciferase assay, Elijahu Berkovich for assistance with the ChIP protocol, and Masatoshi Takagi for annexin V staining and FACS analysis.
This work was supported in part by NCI Cancer Center Core Grant CA-21765 and by ALSAC of St. Jude Children's Research Hospital. C.J.S. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Andreassen, P. R., O. D. Lohez, F. B. Lacroix, and R. L. Margolis. 2001. Tetraploid state induces p53-dependent arrest of nontransformed mammalian cells in G1. Mol. Biol. Cell 12:1315-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartek, J., and J. Lukas. 2003. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 3:421-429. [DOI] [PubMed] [Google Scholar]
- 3.Bendjennat, M., J. Boulaire, T. Jascur, H. Brickner, V. Barbier, A. Sarasin, A. Fotedar, and R. Fotedar. 2003. UV irradiation triggers ubiquitin-dependent degradation of p21(WAF1) to promote DNA repair. Cell 114:599-610. [DOI] [PubMed] [Google Scholar]
- 4.Bertwistle, D., M. Sugimoto, and C. J. Sherr. 2004. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol. Cell. Biol. 24:985-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooks, C. L., and W. Gu. 2003. Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr. Opin. Cell Biol. 15:164-171. [DOI] [PubMed] [Google Scholar]
- 6.Brugarolas, J., C. Chandrasekaran, J. I. Gordon, D. Beach, T. Jacks, and G. J. Hannon. 1995. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature 377:552-557. [DOI] [PubMed] [Google Scholar]
- 7.Bunz, F., A. Dutriaux, C. Lengauer, T. Waldman, S. Zhou, J. P. Brown, J. M. Sedivy, K. W. Kinzler, and B. Vogelstein. 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282:1497-1501. [DOI] [PubMed] [Google Scholar]
- 8.Carroll, P. E., M. Okuda, H. F. Horn, P. Biddinger, P. J. Stambrook, L. L. Gleich, Y.-Q. Li, P. Tarapore, and K. Fukasawa. 1999. Centrosome hyperamplification in human cancer: chromosome instability induced by p53 mutation and/or Mdm2 overexpression. Oncogene 18:1935-1944. [DOI] [PubMed] [Google Scholar]
- 9.Chan, T. A., P. M. Hwang, H. Hermeking, K. W. Kinzler, and B. Vogelstein. 2000. Cooperative effects of genes controlling the G2/M checkpoint. Genes Dev. 14:1584-1588. [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, X., Y. Chi, A. Bloecher, R. Aebersold, B. E. Clurman, and J. M. Roberts. 2004. N-acetylation and ubiquitin-independent proteasomal degradation of p21(Cip1). Mol. Cell 16:839-847. [DOI] [PubMed] [Google Scholar]
- 11.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng, C., P. Zhang, J. W. Harper, S. J. Elledge, and P. Leder. 1995. Mice lacking p21Cip1/Waf1 undergo normal development, but are defective in G1 checkpoint control. Cell 82:675-684. [DOI] [PubMed] [Google Scholar]
- 13.De Stanchina, E., E. Querido, M. Narita, R. V. Davuluri, P. P. Pandolfi, G. Ferbeyre, and S. W. Lowe. 2004. PML is a direct p53 target that modulates p53 effector functions. Mol. Cell 13:523-535. [DOI] [PubMed] [Google Scholar]
- 14.Downing, J. R., S. A. Shurtleff, and C. J. Sherr. 1991. Peptide antisera to human colony-stimulating factor 1 receptor detect ligand-induced conformational changes and a binding site for phosphatidylinositol 3-kinase. Mol. Cell. Biol. 11:2489-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doxsey, S., W. Zimmerman, and K. Mikule. 2005. Centrosome control of the cell cycle. Trends Cell Biol. 15:303-311. [DOI] [PubMed] [Google Scholar]
- 16.El-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 17.Fukusawa, K., T. Choi, R. Kuriyama, S. Rulong, and G. F. Vande Woude. 1996. Abnormal centrosome amplification in the absence of p53. Science 271:1744-1747. [DOI] [PubMed] [Google Scholar]
- 18.Funk, W. D., D. T. Pak, R. H. Karas, W. E. Wright, and J. W. Shay. 1992. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol. Cell. Biol. 12:2866-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hainaut, P., T. Soussi, B. Shomer, M. Hollstein, M. Greenblatt, E. Hovig, C. C. Harris, and R. Montesano. 1997. Database of p53 gene somatic mutations in human tumors and cell lines: updated compilation and future prospects. Nucleic Acids Res. 25:151-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harper, J. W., G. R. Adami, N. Wei, K. Keyomarsi, and S. J. Elledge. 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75:805-816. [DOI] [PubMed] [Google Scholar]
- 21.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296-299. [DOI] [PubMed] [Google Scholar]
- 22.Hershko, T., and D. Ginsberg. 2004. Up-regulation of Bcl-2 homology 3 (BH3)-only proteins by E2F1 mediates apoptosis. J. Biol. Chem. 279:8627-8634. [DOI] [PubMed] [Google Scholar]
- 23.Hidaka, M., G. Caruana, W. L. Stanford, M. Sam, P. H. Correll, and A. Bernstein. 2000. Gene trapping of two novel genes, Hzf and Hh1, expressed in hematopoietic cells. Mech. Dev. 90:3-15. [DOI] [PubMed] [Google Scholar]
- 24.Hollander, M. C., M. S. Sheikh, D. V. Bulavin, K. Lundgren, L. Augeri-Henmueller, R. Shehee, T. A. Molinaro, K. E. Kim, E. Tolosa, J. D. Ashwell, M. P. Ronsenberg, Q. Zhan, P. M. Fernandez-Salguero, W. F. Morgan, C. X. Deng, and A. J. Jr. Fornace. 1999. Genomic instability in Gadd45a-deficient mice. Nat. Genet. 23:176-184. [DOI] [PubMed] [Google Scholar]
- 25.Jin, A., K. Itahana, K. O'Keefe, and Y. Zhang. 2004. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol. Cell. Biol. 24:7669-7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Juven, T., Y. Barak, A. Zauberman, D. L. George, and M. Oren. 1993. Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the Mdm2 gene. Oncogene 8:3411-3416. [PubMed] [Google Scholar]
- 27.Kastan, M. B., and J. Bartek. 2004. Cell cycle checkpoints and cancer. Nature 432:316-323. [DOI] [PubMed] [Google Scholar]
- 28.Katayama, H., W. R. Brinkley, and S. Sen. 2003. The aurora kinases: role in cell transformation and tumorigenesis. Cancer Metastasis Rev. 22:451-464. [DOI] [PubMed] [Google Scholar]
- 29.Kimura, Y., A. Hart, M. Hirashima, C. Wang, D. Holmyard, J. Pittman, X. L. Pang, C. W. Jackson, and A. Bernstein. 2002. Zinc finger protein, Hzf, is required for megakaryocyte development and hemostasis. J. Exp. Med. 195:941-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kriwacki, R. W., L. Hengst, L. Tennant, S. I. Reed, and P. E. Wright. 1996. Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: conformational disorder mediates binding diversity. Proc. Natl. Acad. Sci. USA 93:11504-11509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kubbutat, M. H., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature 387:299-303. [DOI] [PubMed] [Google Scholar]
- 32.Kuo, M.-L., W. den Besten, D. Bertwistle, M. F. Roussel, and C. J. Sherr. 2004. N-terminal polyubiquitination and degradation of the Arf tumor suppressor. Genes Dev. 18:1862-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuo, M.-L., E. J. Duncavage, R. Mathew, W. den Besten, D. Pie, D. Naeve, T. Yamamoto, C. Cheng, C. J. Sherr, and M. F. Roussel. 2003. Arf induces p53-dependent and independent anti-proliferative genes. Cancer Res. 63:1046-1053. [PubMed] [Google Scholar]
- 34.Kurki, S., K. Peltonen, L. Latonen, T. M. Kiviharju, P. M. Ojala, D. Meek, and M. Laiho. 2004. Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell 5:465-475. [DOI] [PubMed] [Google Scholar]
- 35.Lanni, J. S., and T. Jacks. 1998. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol. Cell. Biol. 18:1055-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levine, A. J. 1997. p53, the cellular gatekeeper for growth and division. Cell 88:323-331. [DOI] [PubMed] [Google Scholar]
- 37.Li, F., E. J. Ackermann, C. F. Bennett, A. L. Rothermel, J. Plescia, S. Tognin, A. Villa, P. C. Marchisio, and D. C. Altieri. 1999. Pleiotropic cell division defects and apoptosis induced by interference with survivin function. Nat. Cell Biol. 1:461-466. [DOI] [PubMed] [Google Scholar]
- 38.Liu, C. W., M. J. Corboy, G. N. DeMartino, and P. J. Thomas. 2003. Endoproteolytic activity of the proteasome. Science 299:408-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lohrum, M. A., R. L. Ludwig, M. H. Kubbutat, M. Hanlon, and K. H. Vousden. 2003. Regulation of HDM2 activity by ribosomal protein L11. Cancer Cell 3:577-587. [DOI] [PubMed] [Google Scholar]
- 40.Lowe, S. W., and C. J. Sherr. 2003. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr. Opin. Genet. Dev. 13:77-83. [DOI] [PubMed] [Google Scholar]
- 41.Mailand, N., J. Falck, C. Lukas, R. G. Syljuasen, M. Welcker, J. Bartek, and J. Lukas. 2000. Rapid destruction of human Cdc25A in response to DNA damage. Science 288:1425-1429. [DOI] [PubMed] [Google Scholar]
- 42.Mantel, C., S. E. Braun, S. Reid, O. Henegariu, L. Liu, G. Hangoc, and H. E. Broxmeyer. 1999. p21(cip-1/waf-1) deficiency causes deformed nuclear architecture, centriole overduplication, polyploidy, and relaxed microtubule damage checkpoints in human hematopoietic cells. Blood 93:1390-1398. [PubMed] [Google Scholar]
- 43.Margolis, R. L., O. D. Lohez, and P. R. Andreassen. 2003. G1 tetraploidy checkpoint and the suppression of tumorigenesis. J. Cell Biochem. 88:673-683. [DOI] [PubMed] [Google Scholar]
- 44.Maya, R., M. Balass, S. T. Kim, D. Shkedy, J. F. Leal, O. Shifman, M. Moas, T. Buschmann, Z. Ronai, Y. Shiloh, M. B. Kastan, E. Katzir, and M. Oren. 2001. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 15:1067-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meraldi, P., R. Honda, and E. A. Nigg. 2002. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53−/− cells. EMBO J. 21:483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyashita, T., and J. C. Reed. 1995. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 80:293-299. [DOI] [PubMed] [Google Scholar]
- 47.Nakano, K., and K. H. Vousden. 2001. PUMA, a novel proapoptotic gene, is induced by p53. Mol. Cell 7:683-694. [DOI] [PubMed] [Google Scholar]
- 48.Nigg, E. A. 2002. Centrosome aberrations: cause or consequence of cancer progression? Nat. Rev. Cancer 2:815-825. [DOI] [PubMed] [Google Scholar]
- 49.Prives, C. 1998. Signaling to p53: breaking the MDM2-p53 circuit. Cell 95:5-8. [DOI] [PubMed] [Google Scholar]
- 50.Ries, S., C. Biederer, D. Woods, O. Shifman, S. Shirasawa, T. Sasazuki, M. McMahon, M. Oren, and F. McCormick. 2000. Opposing effects of Ras on p53: transcriptional activation of mdm2 and induction of p19ARF. Cell 103:321-330. [DOI] [PubMed] [Google Scholar]
- 51.Sharma, S., D. Dimasi, K. Higginson, and N. G. Della. 2004. RZF, a zinc-finger protein in the photoreceptors of human retina. Gene 342:219-229. [DOI] [PubMed] [Google Scholar]
- 52.Sheaff, R. J., J. D. Singer, J. Swanger, M. Smitherman, J. M. Roberts, and B. E. Clurman. 2000. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol. Cell 5:403-410. [DOI] [PubMed] [Google Scholar]
- 53.Smith, L., S. J. Liu, L. Goodrich, D. Jacobson, C. Degnin, N. Bentley, A. Carr, G. Flaggs, K. Keegan, M. Hoekstra, and M. J. Thayer. 1998. Duplication of ATR inhibits MyoD, induces aneuploidy, and eliminates radiation-induced G1 arrest. Nat. Genet. 19:39-46. [DOI] [PubMed] [Google Scholar]
- 54.St. Clair, S., L. Giono, S. Varmeh-Ziaie, L. Resnick-Silverman, W. J. Liu, A. Padi, J. Dastidar, A. DaCosta, M. Mattia, and J. J. Manfredi. 2004. DNA damage-induced downregulation of Cdc25C is mediated by p53 via two independent mechanisms: one involves direct binding to the Cdc25C promoter. Mol. Cell 16:725-736. [DOI] [PubMed] [Google Scholar]
- 55.Thornborrow, E. C., S. Patel, A. E. Mastropietro, E. M. Schwartzfarb, and J. J. Manfredi. 2002. A conserved intronic response element mediates direct p53-dependent transcriptional activation of both the human and murine Bax genes. Oncogene 21:990-999. [DOI] [PubMed] [Google Scholar]
- 56.Touitou, R., J. Richardson, S. Bose, M. Nakanishi, J. Rivett, and M. J. Allday. 2001. A degradation signal located in the C terminus of p21WAF1/CIP1 is a binding site for the C8α-subunit of the 20S proteasome. EMBO J. 20:2367-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tutt, A., A. Gabriel, D. Bertwistle, F. Connor, H. Paterson, J. Peacock, G. Ross, and A. Ashworth. 1999. Absence of Brca2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr. Biol. 9:1107-1110. [DOI] [PubMed] [Google Scholar]
- 58.Waldman, T., C. Lengauer, K. W. Kinzler, and B. Vogelstein. 1996. Uncoupling of S phase and mitosis induced by anticancer agents in cells lacking p21. Nature 381:713-716. [DOI] [PubMed] [Google Scholar]
- 59.Xiong, Y., G. J. Hannon, H. Zhang, D. Casso, R. Kobayashi, and D. Beach. 1993. p21 is a universal inhibitor of cyclin kinases. Nature 366:701-704. [DOI] [PubMed] [Google Scholar]
- 60.Xu, X., Z. Weaver, S. P. Linke, C. Li, J. Gotay, X. W. Wang, C. C. Harris, T. Ried, and C. X. Deng. 1999. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol. Cell 3:389-395. [DOI] [PubMed] [Google Scholar]
- 61.Yu, J., L. Zhang, P. M. Hwang, K. W. Kinzler, and B. Vogelstein. 2001. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol. Cell 7:673-682. [DOI] [PubMed] [Google Scholar]
- 62.Yuan, X., Y. Zhou, E. Casanova, M. Chai, E. Kiss, H. J. Grone, G. Schutz, and I. Grummt. 2005. Genetic inactivation of the transcription factor TIF-1A leads to nucleolar disruption, cell cycle arrest, and p53-mediated apoptosis. Mol. Cell 19:77-87. [DOI] [PubMed] [Google Scholar]
- 63.Zhou, H., J. Kuang, L. Zhong, W. L. Kuo, J. W. Gray, A. Sahin, B. R. Brinkley, and S. Sen. 1998. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy, and transformation. Nat. Genet. 20:189-193. [DOI] [PubMed] [Google Scholar]
- 64.Zindy, F., C. M. Eischen, D. H. Randle, T. Kamijo, J. L. Cleveland, C. J. Sherr, and M. F. Roussel. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 12:2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.