Abstract
We find that during embryogenesis the expression of HMGN1, a nuclear protein that binds to nucleosomes and reduces the compaction of the chromatin fiber, is progressively down-regulated throughout the entire embryo, except in committed but continuously renewing cell types, such as the basal layer of the epithelium. In the developing limb bud, the expression of HMGN1 is complementary to Sox9, a master regulator of the chondrocyte lineage. In limb bud micromass cultures, which faithfully mimic in vivo chondrogenic differentiation, loss of HMGN1 accelerates differentiation. Expression of wild-type HMGN1, but not of a mutant HMGN1 that does not bind to chromatin, in Hmgn1−/− micromass cultures inhibits Sox9 expression and retards differentiation. Chromatin immunoprecipitation analysis reveals that HMGN1 binds to Sox9 chromatin in cells that are poised to express Sox9. Loss of HMGN1 elevates the amount of HMGN2 bound to Sox9, suggesting functional redundancy among these proteins. These findings suggest a role for HMGN1 in chromatin remodeling during embryogenesis and in the activation of Sox9 during chondrogenesis.
During organogenesis of the vertebrate embryo, multipotent progenitor cells undergo a complex process of differentiation according to their fate. Cellular differentiation involves the execution of a preprogrammed, orderly process that involves multiple changes in gene expression. It is well documented that chromatin structure plays a key role in regulating gene expression and that the chromatin structure of specific genes is remodeled during the differentiation. It is therefore possible that nuclear proteins such as the high mobility group N (HMGN), which affect chromatin structure (4), histone modifications (17, 18) and transcription rates (11, 30), may play a role in differentiation processes.
The HMG superfamily of proteins consists of three families, HMGA, HMGB, and HMGN, all of which have been shown to modulate the structure and activity of the chromatin fiber (5). Members of the HMGN family bind specifically to the building block of the chromatin fiber, the nucleosome core particle, without any known specificity for the underlying DNA sequence (28). The interaction of the proteins with chromatin is dynamic, and HMGN proteins continuously exchange among nucleosomes (7, 25). The binding of HMGN to nucleosomes reduces the compaction of chromatin fiber and enhances transcription from chromatin templates (11, 22, 30). HMGNs modulate the levels of posttranslational modifications in the histone tails (17), most likely because their presence on nucleosomes affects the ability of nucleosome remodeling complexes to reach their targets. These findings and additional studies suggest that HMGN proteins modulate chromatin-related activities, including transcription (4).
The expression level of Hmgn genes is related to cellular differentiation processes, such as erythropoiesis, myogenesis, osteoblast differentiation, kidney organogenesis, preimplantation development of early mouse embryo, and Xenopus embryogenesis (1, 9, 15, 16, 20, 23, 27). Transient depletion of HMGN proteins from one- or two-cell mouse embryos slowed the progression of preimplantation development (20). Overexpression of HMGN1 in myoblasts inhibited their differentiation into myotubes (23). HMGN2 plays a role in the activation of genes regulating kidney organogenesis (16). During Xenopus embryogenesis, either enhancement or depletion of HMGN protein levels led to malformed tadpole embryos (15). These results suggest a link between regulated expression of HMGN proteins and cellular differentiation during embryo development.
Here we use in situ hybridization and immunohistochemistry to examine the expression of HMGN1, a major member of the HMGN protein family, during mouse embryogenesis. We find a differentiation-related, global down-regulation of HMGN1 expression throughout the entire embryo; however, in specific cell types the levels of this protein remain high, even in fully differentiated organs. We focus on the developmental down-regulation of HMGN protein during limb bud development and demonstrate that expression of the protein is related to chondrocyte differentiation. Chondrocyte differentiation is a multistep genetic program, regulated by combinatorial signaling of various growth and differentiation factor networks (29). Sox9 is a transactivator and master regulator of chondrogenic fate that is required for initiating chondrogenesis and subsequent chondrocyte differentiation (10). We find that the expression patterns of Hmgn1 and Sox9 are complementary both in vivo and in vitro. We demonstrate that in micromass cultures of differentiating limb bud cells, the expression of Sox9 and the rate of differentiation are modulated by the levels of HMGN1 protein and that HMGN1 is present in Sox9 chromatin of chondrogenic cells and absent from this gene in nonchondrogenic cells. We demonstrate that the close homologue HMGN2 is also associated with Sox9 chromatin and that loss of HMGN1 increases the amount of HMGN2 on the Sox9 gene. Taken together with previous studies, our results suggest that HMGN proteins affect chromatin remodeling and cellular differentiation during embryogenesis.
MATERIALS AND METHODS
Mouse strains.
Hmgn1−/− mice were previously described (3).
Whole-mount in situ hybridization.
Whole-mount in situ hybridizations were done according to the method of Wilkinson (34) with minor modifications: punctured, and proteinase K-digested embryos were rinsed with phosphate-buffered saline (PBS) containing 0.1% Tween 20 and immediately fixed with 4% paraformaldehyde (PFA). Blocking reagent (Roche Diagnostics) was used at a concentration of 1.5%. For the Hmgn1 in situ hybridization probe, the 1.2-kb mouse Hmgn1 cDNA cloned in pBluescriptII KS (Stratagene) was linearized by HindIII and XmaI to synthesize sense and antisense probes, respectively. For the Sox9 probe, 500 bp of sequence 3′ to the DNA binding domain of mouse Sox9 cDNA was generated by reverse transcription-PCR using the oligonucleotides 5′-ACCAATACTTGCCACCCAAC-3′ and 5′-TAGGAGCCGGAGTTCTGATG-3′, cloned in pCR2.1-TOPO (Invitrogen), and linearized with BamH I to synthesize the antisense probe. Sense and antisense RNA probes were prepared by transcription of the linearized plasmids using T3 and T7 RNA polymerases (Stratagene) with digoxigenin-11-UTP (Roche Diagnostics).
Cell culture and transfections.
Embryonic day 10.5 (E10.5) limb buds were collected in Dulbecco's modified PBS (GIBCO) at 4°C. Mesenchymal cells were dissociated in Dulbecco's modified PBS containing 0.1% trypsin, 0.4 mM EDTA, and 0.1% collagenase at 37°C for 10 min, resuspended in Dulbecco's modified Eagle's medium-F12 medium (GIBCO) with 10% fetal bovine serum, 50 U/ml penicillin, and 50 mg/ml streptomycin, at 2 × 107 cells/ml, and a 10-μl drop of cell suspension was placed in the center of a well in a standard 24-well polystyrene tissue culture dish or on Labteck chamber slides. Cells were allowed to adhere for 1 h at 37°C and 5% CO2, and 1 ml of medium was added to the culture. Medium was changed every 2 days. Alcian blue staining and quantification were performed as previously described (12). For transient expression of the DNA in micromass cultures, 8 × 106 limb bud mesenchymal cells were resuspended in the 0.4 ml of ice-cold PBS with 20 μg of plasmid DNA in 4-mm wide electroporation cuvettes and electroporated with a BTX T820 electroporator (Genetronics Inc.) using a single 225-V square pulse at 50 ms. HMGN1-YFP and HMNG1(S20,24E)_YFP (where YFP is yellow fluorescent protein) expression vectors were as previously described (26). Transfection efficiency was assessed by YFP after 2 days of culture, and chondrocyte differentiation was assessed by Alcian blue staining (12) or immunostaining for Sox9 after another 4 days of culture. For Alcian blue quantification, the experiments were repeated three times. Mouse embryonic fibroblasts (MEFs) were prepared as previously described (3).
Confocal microscopy.
Micromass cultures were grown on Lab-Tek chambered cover slides (Nalgen), and immunostaining was performed as previously described (26) except that the micromass cultures were fixed with 4% PFA in PBS for 10 min, washed with PBS, and permeabilized for 40 min in PBS containing 1% Triton X-100 and blocked overnight in PBS containing 1% fetal bovine serum. Micromass cultures were incubated 6 h with the primary antibody (anti-Sox9) (H-90; Santa Cruz) at a 1:100 dilution and with anti-HMGN1 at a 1:200 dilution. The micromass was washed in PBS three times for 20 min each time, incubated for 2 h with the secondary antibody labeled with either AlexaFluor 488 or AlexaFluor 594 (Molecular Probes). DNA was stained with Hoechst 33258 at 0.5 μg/ml in PBS for 10 min.
Microscopy was performed on a Zeiss LSM 510 confocal setting, using a 63× differential interference contrast objective (1.4 numeric aperture). Stacks (57 μm thick) were collected through the entire micromass.
Histology.
Embryos were fixed in 4% PFA, dehydrated and embedded in paraffin, and sectioned at a thickness of 5 μm. Sections were deparaffinized and subjected to antigen retrieval by microwaving for 10 min in 10 mM citric acid buffer (pH 6.0). After endogenous peroxidase activities were quenched by treatment with 1% H 2O 2 in PBS for 30 min, sections were blocked with 5% goat serum-PBS for 30 min, incubated overnight with primary antibody at 4°C, washed three times with PBS, incubated with biotinylated secondary antibodies for 1 h at room temperature, and stained with the Vectastain ABC Elite kit (Vector Laboratories) using diaminobenzidine as substrate. Sections were counter-stained with hematoxylin and mounted in Permount (Fisher Scientific). For immunofluorescence, the procedure was identical except that AlexaFluor 488-labeled goat anti-rabbit (Invitrogen) was used to visualize the primary antibody.
ChIP assay, RNA analysis, and real-time reverse transcription-PCR.
Chromatin was prepared from E10.5 mouse limb buds and MEFs and immunoprecipitated with a chromatin immunoprecipitation (ChIP) assay kit (Upstate Biotechnology) (17) using either anti-HMGN1, mouse immunoglobulin G (Santa Cruz) or no antibody as a negative control. Primer sets numbered 1 to 21 (see Fig. 7) used for PCR amplification of the Sox9 gene are described in Table S1 of the supplemental material. Differences in specific DNA enrichment were determined in duplicate by real-time quantitative PCR using an ABI Prism 7900HT sequence detector (Applied Biosystems). Beta-globin served as a reference gene. For RNA quantification, 50 ng of purified RNA isolated from limb bud cells or culture cells with TRIZOL (Invitrogen) was subjected to reverse transcription with SuperscriptII (Invitrogen) and amplified with primers specific for either Hmgn1 (forward, 5′-TTGTAGCCATGAGTGACGTTGAA-3′; reverse, 5′-ATGACCCAGAACATTAGCCAGG-3′) or Sox9 (forward, 5′-CGGCTCCAGCAAGAACAAG-3′; reverse, 5′-TTGTGCAGATGCGGGTACTG-3′). The level was normalized using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers (Applied Biosystems).
FIG. 7.
HMGN1 binds to Sox9 chromatin. (A and B) ChIP analysis (with anti HMGN1) of mouse MEFs and E10.5 limb bud cells. (A) Schematic diagram of Sox9 gene, with the positions of primer sets used to amplify the immunoprecipitated DNAs are indicated at the bottom. The arrow indicates transcription start site. (B) ChIP assay for HMGN1 in MEFs and limb bud cells. Enrichment of each DNA sequence in the HMGN1 immunoprecipitate relative to the input DNA was normalized and plotted as the position of the PCR primer pair within the Sox9 gene locus. Note that HMGN1 is associated with Sox9 chromatin in limb bud cells but not in MEFs. (C) Expression levels of Hmgn1 and Sox9 mRNA in MEFs and limb bud cells. Note that while HMGN1 is expressed in both cell types, Sox9 is expressed only in limb bud cells. (D) Western analysis of HMGN1 and Sox9 proteins in limb bud cells and MEFs. Coomassie blue staining of histones indicates equal loading of extracts from MEFs and limb bud cells. (E) Enhanced DNase I sensitivity of Sox9 chromatin in limb bud cells. Nuclei isolated from MEF or E10.5 limb bud cells were digested with the indicated concentrations of DNase I, the DNA from the digested nuclei purified and amplified with forward primer of primer pair 3 and reverse primer of primer pair 4. The level of amplification is inversely related to degree of digestion. Each point is an average from three experiments, with a new preparation of cells each time. (F) Similar rate of DNase I sensitivity of the beta-globin gene in chromatin of limb bud cells and MEFs. Primers used for amplification are described in Materials and Methods. LB, limb bud cells.
RNA dot blot analysis was performed as previously described (33) with a mouse RNA master dot blot (BD-Clonetech). In these blots the amount of RNA spotted from each tissue is normalized to that of eight housekeeping genes (see manufacturer's instructions).
DNase I digestion assay.
To prepare nuclei, E10.5 limb bud cells or MEFs were suspended and swollen in ice-cold lysis buffer (10 mM Tris [pH 7.4], 3 mM CaCl2, 2 mM Mg Cl2); the swollen cells were resuspended in an equal volume of buffer containing 10 mM Tris, pH 7.4, 10 mM NaCl, 3 mM Mg Cl2, and 0.5% NP-40 and then homogenized with a Dounce homogenizer and centrifuged at 800 × g for 10 min at 4°C. Nuclear pellets were stored in 25% glycerol, 5 mM Mg acetate, 50 mM Tris (pH 8.0), 0.1 mM EDTA, and 12 mM 2-mercaptoethanol at −70°C. Nuclei (1 μg of DNA) were digested with DNase I in buffer containing 50 mM Tris (pH 7.4), 100 mM NaCl, 10 mM MgCl2, 5% glycerol, and 1 mM dithiothreitol at 37°C for 15 min. Purified genomic DNA was digested as controls. Digestion was terminated by adding proteinase K buffer (20 mM Tris [pH 8.0], 100 mM KCl, 50 mM Mg Cl2, 1% Tween 20, 1% NP-40, 1 mg/ml proteinase K) at 55°C for 3 h. The control primer for beta-globin spans exon 2 of the beta-globin gene: forward, 5′-TGAAGGCCCATGGCAAGA-3′; reverse, 5′-GCCCTTGAGGCTGTCCAA.
RESULTS
Progressive down-regulation of Hmgn1 expression during mouse embryogenesis.
Whole-mount in situ hybridization and immunostaining analyses revealed that the expression of Hmgn1 is selectively down-regulated during mouse embryogenesis. In 7.5-day-old mouse embryos (E7.5), the expression of Hmgn1 was high in the epiblast, weak in the extra-embryonic ectoderm, and absent from the ectoplacental cone (Fig. 1A, frame a). At E8.5 and E9.5, high Hmgn1 expression was observed throughout the entire embryo, except in the heart (Fig. 1A, frames b and c), an organ known to begin differentiating early (14). At E10.5, fairly strong expression of Hmgn1 was widespread through the embryo and was prominent in the forelimb and hind limb buds, branchial arches, and tail bud (Fig. 1A, frames d to f) but was totally absent from the heart. At subsequent developmental stages, the expression level of Hmgn1 selectively declines, coincident with the onset of differentiation in many organs, as more easily seen in tissue sections (see below).
FIG.1.
Expression patterns of Hmgn1 during mouse embryogenesis. (A) Whole-mount in situ hybridization with digoxigenen-labeled Hmgn1 antisense probes. At E7.5 strong expression is observed in the embryonic region (arrowhead). No signal is observed in the ectoplacental cone (arrow). At E8.5 and E9.5, strong expression of Hmgn1 is visible throughout the embryo except in the heart (arrowhead). At E10.5, HMGN1 is most strongly expressed in the distal portion of forelimb and hind limb mesenchyme (filled arrowhead), in branchial arches (arrow), and in tail bud (open arrowhead). After stage E11.5 Hmgn1 expression remains strong mostly in the developing forelimbs and hind limbs and in the tail. As the embryos develops, Hmgn1 becomes progressively more localized to the interdigit mesenchyme of the limbs and to the tail. (B) Whole-mount in situ hybridization with Hmgn1 sense probes. (C) Bar graph depicting the relative expression levels of Hmgn1 transcripts at the developmental stage indicated or in the tissue indicated, determined by quantitative dot blot analysis with RNA master blots (Clonetech), relative to 7-day embryos, which are considered as 100%.
Quantitative RNA dot blot analysis fully supports the conclusion of the in situ analyses (Fig. 1C). In these blots, the amount of RNA in each spot is normalized to the transcription levels of eight housekeeping genes; therefore, the intensity of the spot indicates the relative mRNA abundance in a tissue. The relative levels of Hmgn1 RNA were highest in 7-day-old embryos and then gradually decreased; the RNA levels in 17-day-old embryo were only 50% of those of the 7-day-old embryos. In differentiated tissues such as the adult heart and spleen, the levels of Hmgn1 RNA are only about 10% of the level present in young embryos (Fig. 1C).
To discern the developmental pattern of Hmgn1 expression in the internal organs of the embryos, we analyzed the presence of HMGN1 protein in multiple serial sections by immunohistochemistry using affinity-purified anti-mouse HMGN1 antibody. Representative results from several tissues (Fig. 2) (images are best viewed at large magnifications; see Fig. S1 to S3 in the supplemental material) are described below.
FIG. 2.
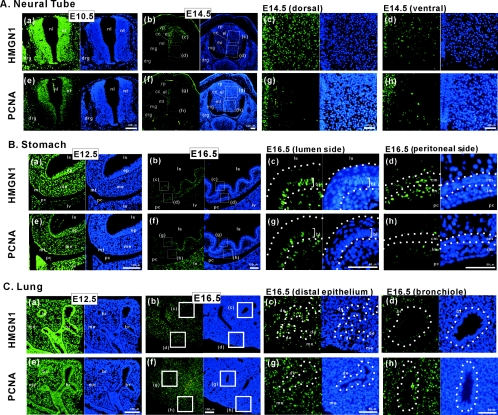
HMGN1 expression correlates with cellular differentiation rather than cell proliferation. The localization of HMGN1 and PCNA in developing mouse tissues was detected by immunofluorescence. Images at higher magnifications are presented in Fig. S1 to S3 of the supplemental material. Green staining visualizes either HMGN1 or PCNA, as indicated in the figure. Cell nuclei are counterstained blue with DAPI (4′,6′-diamidino-2-phenylindole). (A) Sections through the neural tube. (a) At E10.5, HMGN1 was detected in the neural tube (nt), dorsal root ganglia (drg), and the mesenchymal cells. nl, neural lumen. (b) At E14.5, HMGN1 was localized to the roof plate (rp) and ependymal layer (el) of the neural tube but down-regulated in the mantle layer (ml) and absent from marginal layer (mg). cc, central canal. (c and d) Higher magnification of dorsal and ventral regions of E14.5 neural tube indicated in the boxed areas of frames b are shown. (B) Sections through the developing mouse stomach at E12.5 (a and e) and E16.5 (b and f). At E12.5, HMGN1 was detected both in the epithelial (ep) and mesenchymal layer (me); however at E16.5, its expression was localized to basal layer (bl) of gastric mucosa and longitudinal muscle layer (lm). mt, mesothelium; ls, lumen of stomach; pc, peritoneal cavity; lv, liver; cm, circular muscle layer. Frames c and d show higher magnifications of E16.5 stomach wall lumenal side and peritoneal cavity side indicated by boxed areas in frames b. (C) Section through the lung. (a) At E12.5, HMGN1 was detected in both of the bronchi (br) and surrounding mesenchymal cells (me). (b) At E16.5, bronchial expression of HMGN1 was down-regulated in the proximal region (br) and decreased in the mesenchymal region (me) but remained strong in the distal region (de). (c and d) Higher magnification of distal bronchi and proximal bronchi of E16.5 lung section indicated by boxed areas in frames b. Corresponding PCNA staining is shown in adjacent sections of all panels.
Neural tube.
In E10.5 transverse sections, HMGN1 protein is visible throughout the embryo, including the surface ectoderm, neural tube, dorsal root ganglia, and the mesenchymal cells (Fig. 2A). The neural tube is already differentiated into an outer marginal layer, an intermediate mantle layer, and inner ependymal layer; HMGN1 is strongly expressed in all the three layers. By E14.5 HMGN1 expression is localized primarily to the dorsal half of the mantle layer (Fig. 2A, frames c), which expresses higher HMGN1 levels than the ventral half of the mantle layer (Fig. 2A, frames d), an indication that HMGN1 expression correlates with neural tube differentiation (19, 36).
Stomach.
At E12.5, HMGN1 was detected in both the epithelial and mesenchymal layers of the developing stomach (Fig. 2B), most prominently in the outermost mesothelium. At E16.5, when the stomach underwent additional differentiation, HMGN1 expression in the epithelium was most prominent in the relatively undifferentiated basal layer (Fig. 2B, bl in frames c).
Lung.
At E12.5 HMGN1 was detected throughout the developing lung, both in the epithelial and in the dense mesenchymal cells (Fig. 2C, me) surrounding the bronchioles (Fig. 2C). However at E16.5, when the bronchioles are fully differentiated but the distal epithelium Fig. 2C, de still forms alveoli, expression of HMGN1 is localized to the developing distal epithelium and is greatly decreased in the bronchiols (Fig. 2C, br in frames d).
To examine whether the HMGN1 expression levels are related to cellular proliferation, we compared the expression patterns of HMGN1 with that of PCNA. Comparison of the PCNA-stained panels with the HMGN1-stained panels reveals that the expression of HMGN1 and PCNA do not always overlap, which is an indication that in the developing embryo the levels of HMGN1 are not linked to cellular proliferation per se, a finding consistent with previous observations in other experimental systems (16, 24).
Our analyses revealed that in every tissue examined, the expression of HMGN1 was down-regulated as organ differentiation proceeded during development. However, the levels of the protein remained high in progenitor cells, such as the basal skin layer cells that are committed and poised for further differentiation and renewal. Thus, the dot blot RNA analysis, the in situ hybridization, and the immunofluorescence analyses indicate a differentiation-related down-regulation of HMGN1 expression.
Reciprocal expression patterns of Hmgn1 and Sox9 in the developing limb bud.
To gain additional insights into the role of HMGN1 in developmental processes, we focused on its possible involvement in the differentiation of the limb bud, where Hmgn1 expression correlated inversely with mesenchymal chondrogenic differentiation. Whole-mount in situ hybridization demonstrates clearly that the expression of Hmgn1 is down-regulated during development and is complementary to that of Sox9, a major transactivator involved in the initiation and propagation of chondrogenesis (Fig. 3). Thus, in the E10.5 forelimb bud (and hind bud; data not shown), Hmgn1 was strongly expressed in a broad anterior and distal region and down-regulated in the proximal region (Fig. 3A), where mesenchymal cells condense and differentiate into chondrocytes, as evidenced by the onset of Sox9 expression (Fig. 3B). As chondrogenesis initiated in the more distal digit region over time, expression of Hmgn1 was localized to the future interdigit region and down-regulated in the future digit region (Fig. 3C), coinciding with activation of Sox9 expression (Fig. 3D). In the E12.5 forelimb bud, Hmgn1 expression is most prominent in the interdigit mesenchyme (Fig. 3E), while Sox9 transcripts are most abundant in future digit regions (Fig. 3F). In the E13.5 and E14.5 forelimb buds, Hmgn1 expression is most prominent in the interdigital mesenchyme (Fig. 3G and I), while Sox9 transcripts are most abundant in the cartilage of phalanges (Fig. 3H and J). A similar pattern of reciprocal Hmgn1 and Sox9 expression was observed during hind limb bud development (data not shown). Thus, during the development of both limbs, the decline of Hmgn1 expression preceded that of Sox9, the latter being expressed in regions with significantly diminished levels of HMGN1 (Fig. 3E to J).
FIG. 3.
Reciprocal expression patterns of Hmgn1 and Sox9 during limb bud development. (A to J) Whole-mount in situ hybridization analysis of Hmgn1 and Sox9 expression in E10.5 to E14.5 forelimbs. At E10.5 Hmgn1 is strongly expressed in the distal region of limb bud (arrowhead in A) while weak Sox9 expression commences in the proximal region where Hmgn1 is down-regulated (arrowhead in B). In the E11.5 limb bud, the expression of Hmgn1 starts to be restricted to the interdigit mesenchyme (arrowheads in C), whereas Sox9 expression is observed in the early digit condensation (arrowheads in D). At E12.5 Hmgn1 is expressed in the interdigit mesenchyme (arrowheads in E), while Sox9 is expressed in digit chondrogenic condensations (arrowheads F). In the E13.5 forelimb bud the expression of Hmgn1 in the interdigit mesenchyme begins to weaken (arrowheads in G) and is restricted to perichondrium while the expression of Sox9 is strongest in the distal regions of the digit cartilage (arrowheads in H). At E14.5 Hmgn1 remains localized to the perichondrium while Sox9 is expressed mainly in the distal digit cartilage (I and J). (K to R) Immunohistochemical staining of HMGN1 and Sox9 protein in the developing forelimb bud. Higher magnifications are shown in supplemental Fig. S4. At E10.5, HMGN1 is strongly expressed in distal mesenchyme (K, arrow) and in the epithelium (K, arrowhead), while Sox9 is strongly expressed in the proximal mesenchyme (L, arrow). At E12.5, HMGN1 protein expression is seen through most of the limb bud but is relatively weak in the differentiating digit chondrocytes (M, arrows) where Sox9 protein is strongly expressed (N). At E14.5, the relative levels of HMGN1 protein (Q) are highest in the skin (sk) and perichondrium (pc) and lowest in the digit cartilage (dc). Conversely, Sox9 protein is strongly expressed in the digit cartilage (dc).
More detailed immunohistochemical analyses of sections from the developing limb bud region verified the reciprocal expression of Hmgn1 and Sox9. The expression of Hmgn1 decreased while that of Sox9 increased as the prechondrogenic mesenchyme condensed in the cartilage primordia. Thus, in the E10.5 and E12.5 forelimb buds, Sox9 protein was observed in the proximal region (Fig. 3L) and digit cartilages (Fig. 3N), while HMGN1 protein was expressed in the surface ectoderm and throughout the distal mesenchyme region but absent from the prechondrogenic mesenchyme (Fig. 3K and M). In the more fully differentiated E14.5 forelimb, HMGN1 protein was abundant in the perichondrium of the phalanges but depleted from the cartilage, where Sox9 protein was expressed (Fig. 3). At higher magnification the difference in the expression patterns of HMGN1 and Sox9 in the developing phalanges was very clear: the expression of Sox9 was confined to the cartilage, while HMGN1 was expressed in the surrounding perichondrium but not in cartilage (Fig. 3Q and 3R; see Fig. S4 in the supplemental material).
Reciprocal expression patterns of Hmgn1 and Sox9 in micromass cultures.
To gain additional insights into the role of HMGN1 in chondrocyte differentiation, we analyzed the pattern of Hmgn1 and Sox9 expression in a micromass culture system (Fig. 4A), which has been commonly used as a model to study in vitro chondrogenesis. When plated at high density, mouse limb bud mesenchymal cells differentiate into chondrocytes and form cartilage nodules (Fig. 4A). The progress of differentiation can be assessed by Alcian blue staining, which is most prominent in differentiated nodules (Fig. 4B). Under our growth conditions, nodules of differentiated chondrocytes can be seen within 3 days of plating (Fig. 4B, +) and distinct, strongly staining nodules containing differentiated chondrocytes are clearly visible after 5 days of plating. In situ hybridization with digoxigenin-labeled RNA probes indicates that the levels of Hmgn1 transcripts decrease while those of Sox9 increase during differentiation. Thus, 2 days after plating, Hmgn1 was expressed throughout most of the monolayer cells. At day 3, Hmgn1 expression was down-regulated in the regions in which the cells condensed into the evolving nodules (Fig. 4B, +), and by day 5, Hmgn1 transcripts were highly depleted and practically absent in the differentiated cartilage nodule (Fig. 4C, top panels). In contrast, Sox9 expression was first observed in the small cell aggregates detectable after 2 days of culturing. In the evolving nodules, the levels of Sox9 RNA gradually increases and is prominent in the fully differentiated, 5-day-old nodules (Fig. 4C, bottom panels).
FIG. 4.
Reciprocal expression patterns of Hmgn1 and Sox9 during chondrogenesis in vitro in micromass cultures. (A) Schematic diagram of micromass culture method. Mesenchymal limb bud cells obtained from E10.5 embryos are dissociated by trypsin-collagenase treatment, collected and concentrated by centrifugation, and then resuspended in the culture medium at high density (2.0 × 107 cells/ml). Cells are plated as a 10-μl spot and flooded with medium after 1 h of incubation and adhesion. (B) Detection of chondrocyte differentiation by Alcian blue staining. Mesenchymal cells plated at high density differentiate into chondrocytes after 3 to 5 days in culture. (C) Reciprocal expression of Hmgn1 and Sox9 transcripts during mesenchymal differentiation into nodules. Plus symbols indicate developing nodules. After 5 days, Hmgn1 expression is down-regulated in the fully differentiated region of the chondrogenic nodule, where Sox9 expression is strongly expressed. (D) Confocal immunofluorescence analysis of HMGN1 and SOX9 expression in 5-day-old differentiated nodules. (a) Low magnification. HMGN1 protein is absent from the chondrocytic nodule, while SOX9 protein is detected only within the nodule. (b) Higher magnification of the images shown in part a centered on a differentiating nodule. Note that in the DNA-Sox9 merge, the center of the nodule is mostly green (high Sox protein levels) and the surrounding cells are red (low Sox protein; DNA stain prevails), while in the DNA-HMGN1 confocal merge the center is red due to low HMGN1 (light green) and the surrounding cells are green due to relatively high levels of HMGN1. Proteins are shown in green, and DNA (Hoechst) is shown in red. Phase-contrast images are used to visualize node morphology. Scale bar, 50 μm.
Confocal immunofluorescence analysis of the fully formed 5-day-old nodules revealed reciprocity in HMGN1 and Sox9 protein levels at the single-cell resolution. The fully differentiated nodules present after 5 days of growth in micromass culture were clearly depleted of HMGN1 protein, while the undifferentiated cells surrounding the nodules expressed high levels of HMGN1 protein (Fig. 4D, frame a). In contrast, Sox9 levels were high in the nodules but undetectable in the undifferentiated cells surrounding the nodules. Although higher magnifications revealed some residual HMGN1 protein in the nodule, a merge of the DNA and HMGN1 confocal images clearly demonstrates a reduced level of HMGN1 protein in the cells at the center of the nodules. In contrast, Sox9 levels are highest in the cells at the center of the nodule and gradually decrease toward the periphery of the nodule; Sox9 is absent from the cells surrounding the nodules. In the DNA-Sox9 image merge, the center of the nodule is green (high protein levels) and the surrounding cells are red (high DNA, low protein), while in the DNA-HMGN1 merge the opposite is visible: the center is red due to low HMGN1 (light green), while the surrounding cells are green due to relatively high levels of HMGN1 (Fig. 4D, frame b).
Thus, the expression levels of Hmgn1 and Sox9 in the micromass cultures faithfully reproduce the Hmgn1 and Sox9 expression patterns observed in mouse embryos. Chondrocyte differentiation is associated with decreased levels of HMGN1 protein and increased levels of Sox9. Taken together, the data demonstrate reciprocity in the expression of Hmgn1 and Sox9.
HMGN1 modulates chondrocyte differentiation.
The reciprocal expression of HMGN1 and Sox9 raises the possibility that HMGN1 modulates the rate of chondrocyte differentiation. To test this possibility, we first compared the rate of nodule formation in micromass cultures prepared from cells derived from the limb buds of wild-type and Hmgn1−/− E10.5 embryos. On the basis of Alcian blue staining, Hmgn1−/− cultures were found to be differentiated faster than Hmgn1+/+ cultures at all time points examined (Fig. 5A). Quantification of Alcian blue staining confirmed that the rate of differentiation of the micromass prepared from Hmgn1−/− limb bud cells was significantly faster than that prepared from Hmgn1+/+ limb buds. After 4 days in culture, the amount of Alcian blue stain recovered from the Hmgn1−/− cultures was almost twice that obtained from Hmgn1+/+ cultures, and after 5 days it was still more than 1.5 times higher (Fig. 5B). Thus, genetic inactivation of Hmgn1 accelerated differentiation.
FIG.5.
Loss of HMGN1 enhances the rate of differentiation. (A) Alcian blue staining of micromass cultures derived from E10.5 limb bud mesenchymal cells of Hmgn1−/− or Hmgn1+/+ embryos after different lengths of time in culture. (B) Quantification of Alcian blue-stained cells. Alcian blue-stained cultures were lysed (12), centrifuged, and the A595 values of the supernatants were determined.
To further test this possibility, we examined the effect of HMGN1 on cartilage nodule formation by reexpressing HMGN1 protein in Hmgn1−/− cultures. To this end, E10.5 limb bud mesenchymal cells prepared from Hmgn1−/− embryo were transiently transfected with vectors expressing either YFP (control) or the HMGN1-YFP fusion protein. Fluorescent analysis of the cells verified that the expression level of the control plasmid transcribing YFP was similar to that of the plasmid transcribing the HMGN1-YFP fusion protein (Fig. 6A, frames a and b). The number and staining intensity of the Alcian blue-positive nodule in the cultures expressing the HMGN1-YFP fusion protein were significantly lower than those in the cultures expressing the control YFP protein (compare Fig. 6A, frames d and e, and B). Likewise, the levels of Sox9 in cells expressing HMGN1-YFP were lower than in those expressing only YFP (Fig. 6A, frames g and h). Thus, overexpression of HMGN1 inhibited the formation of differentiated nodules and the expression of Sox9, suggesting that down-regulation of HMGN1 is required for chondrocyte differentiation.
FIG. 6.
HMGN1 reduces the rate of chondrocyte differentiation. (A) E10.5 limb bud mesenchyme cells derived from Hmgn1−/− embryos were transfected with plasmids expressing either YFP (Control), HMGN1-YFP (HMGN1) fusion protein or HMGN1(S20,24E)-YFP (HMGN1 S20,24E), a mutant that cannot bind to nucleosomes. Transfection efficiency was assessed by YFP after 2 days of culture, and chondrocyte differentiation was assessed by Alcian blue staining or immunostaining for Sox9, as indicated, after 4 days of culture. Note that the numbers and the sizes of Alcian blue- or Sox9-positive nodules (arrows point to representative nodules) were lower in the HMGN-transfected micromass (e and h), whereas the mutant HMGN1(S20,24E) had no effect (f and i). (B) Representative high-magnification micrographs of fields taken from different plates of Alcian blue-stained 8-day-old micromass cultures transfected with vectors expressing the protein indicated at the top of the fields. (C) Quantification of Alcian blue staining by measuring absorbance at 595 nm.
To test whether the HMGN1-induced inhibition of nodule formation is related to the ability of the protein to bind to chromatin, we expressed the double point mutant HMGN1(S20,24E)-YFP fusion protein, rather than the wild-type fusion protein, in the Hmgn1−/− E10.5 limb bud cells. The HMGN1(S20,24E) mutant enters the nucleus but does not bind nucleosomes (26). In contrast to the wild-type HMGN1, this mutant did not affect Sox9 expression (Fig. 6A, frames f and i), and the rate of nodule formation was comparable to that observed in cells transfected with the control plasmid expressing YFP (Fig. 6A, compare frames d and g to f and i, and B). Quantification of the effect by measuring the Alcian blue staining in the cultures (Fig. 6B) revealed that the amount of stain in the cultures expressing wild-type HMGN1 was 60% of that in control cells transfected with plasmids expressing either YFP or the HMGN1(S20,24E) mutant (Fig. 6C). Thus, reexpression of wild-type but not mutant HMGN1 in Hmgn1−/− limb bud cells reduced the rate of Sox9 expression and chondrocyte differentiation. We therefore conclude that the interaction of HMGN1 with chromatin affects the rate of chondrocyte differentiation by modulating the expression levels of chondrogenic factors such as Sox9.
Specific binding of HMGN1 to the Sox9 gene.
The interrelationship between HMGN1 and Sox9 expression raised the possibility that HMGN1 protein is directly involved in the regulation of Sox9 gene expression. We therefore tested whether HMGN1 protein was directly associated with the Sox9 gene by using a ChIP assay with affinity-purified antibodies against mouse HMGN1. The ChIP analyses were performed with chromatin isolated either from nonchondrogenic MEFs or from E10.5 limb bud cells, which contain both prechondrogenic cells and chondrocytes (Fig. 3A and B). The relative amounts of Hmgn1 transcripts and protein in the MEFs and limb bud cells are similar, while those of Sox9 are almost 10 times higher in the limb bud cells (Fig. 7C and D). However, although Sox9 expression is detected in the E10.5 limb bud, most of the cells are only poised to express the gene; high-level Sox9 expression occurs at later developmental stages, when HMGN1 is significantly down-regulated (Fig. 3).
The DNA purified from the immunoprecipitated chromatin was amplified with 21 primer sets spanning an approximately 10-kb long genomic region encompassing the Sox9 gene and its 4.4-kb 5′ and 5.7-kb 3′ flanking regions. In limb bud cells, but not in MEFs, these analyses identified three regions in the Sox9 chromatin as enriched in HMGN1: a region 2 kb upstream of the promoter, exon 2, and exon 3 (Fig. 7A and B). The DNA sequences of the three regions do not contain any common motifs (data not shown), supporting previous findings that the interaction of HMGN1 with chromatin is not regulated by the sequence of the nucleosomal DNA (28).
To test whether the presence of HMGN1 is associated with changes in chromatin structure, we first focused on the region spanned by primer set 3 (Fig. 7A), since its location was in the 5′ region of the gene and may be involved in the regulation of Sox9 gene expression. We digested nuclei isolated from E10.5 limb bud cells or MEFs with different amounts of DNase I and used PCR to determine the amount of undigested DNA in a 748-bp long region amplified by forward primer 3 and reverse primer 4. The yield of the resulting amplified fragment is a measure of the relative rate of DNA digestion in chromatin. A DNase I-hypersensitive site between primer sets 3 and 4 in the Sox9 promoter in nuclei prepared from E10.5 limb bud cells was digested faster than in MEF nuclei (Fig. 7E), suggesting that the chromatin structure of Sox9 promoter in limb bud cells, which are poised to begin Sox9 expression, is less compact than the corresponding region in MEFs. Similar results were obtained by analysis of other regions of the Sox9 gene, suggesting that the HMGN1-containing gene is more susceptible to DNase I digestion. Similar analysis of the DNase I sensitivity of the beta-globin gene, which is not expressed in either MEFs or limb bud cells, showed only minor differences between the two cell types (Fig. 7F). Our finding both in vivo and in vitro that the expression of Hmgn1 precedes that of Sox9 and that HMGN1 is associated with Sox9 chromatin in limb bud but not in MEFs argues for a role for HMGN1 in Sox9 gene regulation during chondrocyte development.
Enhanced binding of HMGN2 on the Sox9 gene in Hmgn1−/− mice.
The lack of phenotype in Hmgn1−/− mice raises the possibility that homeostatic mechanisms, perhaps involving HMGN2, compensate for loss of HMGN1 protein. The levels of HMGN2 RNA and protein in Hmgn1−/− mice is the same as in their Hmgn1+/+ littermates (not shown). ChIP analysis with affinity pure antibodies to HMGN2 revealed the levels of this protein in the 10.5-day limb bud Sox9 chromatin are higher than in the Sox9 chromatin of MEFs (Fig. 8). Interestingly, while HMGN1 was enriched in distinct regions of the gene (Fig. 7B), HMGN2 protein was enriched along the entire length of the gene (Fig. 8A). Significantly, in the Sox9 chromatin derived from the limb bud of Hmgn1−/− mice, the level of HMGN2 was enriched in several regions, especially those spanning primer sets 6 to 10 and 17 to 19 (Fig. 8B). The increased level of HMGN2 in the Sox9 chromatin of the Hmgn1−/− limb bud suggests functional redundancy among HMGN proteins.
FIG. 8.
Enhanced binding of HMGN2 on the Sox9 gene in Hmgn1−/− mice. (A) ChIP assay for HMGN2 on Sox9 in MEF and limb bud cells. Enrichment of each DNA sequence in the HMGN2 immunoprecipitate relative to the input DNA was normalized to the expression of the beta-globin gene and plotted as the position of the PCR primer pair within the Sox9 gene locus (as described in the legend of Fig. 7). (B) ChIP assay for HMGN2 on Sox9 chromatin in limb bud cells obtained from Hmgn1−/− mice (KO-LB). The results are shown together with the plots of HMGN2 and HMGN1 in Hmgn1+/+ mice, superimposed for comparison. Each point is an average from three experiments, with a new preparation of each time limb bud cells. LB, limb bud cells.
DISCUSSION
The widespread occurrence of HMGN proteins in vertebrate nuclei and their ability to alter the structure of chromatin and modulate access to nucleosomes raise the possibility that they play a role in gene expression and cellular differentiation. To gain insights into the possible role of HMGN in vertebrate development, we first analyzed the pattern of HMGN1 expression during mouse embryogenesis and then focused on the possible role of this protein in modulating the expression of Sox9, a master regulator of chondrocyte differentiation.
A role for HMGN1 in Sox9 expression.
Numerous types of experiments suggest a role for HMGN1 in transcription from chromatin templates (4); however, it is still not clear whether HMGN proteins act indiscriminately as general facilitators or whether they play a role in specific gene expression. DNA array analysis of the expression pattern of UV-treated Hmgn1+/+ and Hmgn1−/− fibroblasts (3) and of fibroblasts expressing increased amounts of HMGN3 (32) linked the HMGN proteins to changes in the transcript levels of specific genes. Likewise, studies on Xenopus development suggest that altered expression levels of HMGNs alter the expression of specific genes such as Xbra and chordin (15). These findings and additional studies (17) suggest that HMGN proteins may affect the transcription of specific genes rather than act only as general coregulators of transcription from chromatin. Our present findings that HMGN1 is associated with the Sox9 chromatin of limb bud cells but not of nonchondrogenic MEFs (Fig. 7) provides evidence for the involvement of HMGN1 in tissue-specific gene regulation. The ChIP analyses with HMGN2 indicate a moderate enrichment of HMGN2 along the entire length of the Sox9 gene, suggesting that this protein is also involved in the transcription of this gene. Our finding that the Sox9 chromatin in Hmgn1−/− is enriched in HMGN2 is particularly significant. Loss of HMGN1 does not change the cellular levels of HMGN2, and the expression of HMGN2 during limb development is indistinguishable from that of HMGN1 (not shown). The results suggest that HMGN2 replaces the missing HMGN1, the first direct experimental evidence for functional redundancy among members of the HMGN protein family.
The regulation of Sox9 transcription is not fully understood. In human SOX9, regulatory elements are scattered through a 350-kb region upstream of the start of transcription (35). The enhancer of the human SOX9 is located in the first intron, while the promoter of the mouse Sox9 gene is located 193 bp upstream of the transcriptional start site (13, 21). The association of HMGNs with the chromatin of E10.5 limb bud cells may serve to generate a chromatin structure that poises the Sox9 gene for transcription at the proper developmental stage. This view is in agreement with the original suggestion that the binding of HMGNs to chromatin potentiates genes for transcription (31) and with subsequent studies of in vitro reconstituted chromatin templates (reviewed in reference 4). At later developmental stages, when cells are committed to a differentiation pathway, HMGNs may still play a positive role in Sox9 expression; however, high levels of HMGNs may interfere with proper development and Sox9 expression. This view is in agreement with our present observations that ectopic expression of HMGN1 in Hmgn1−/− micromass cultures inhibits the rate of differentiation and Sox9 expression and with previous studies in Xenopus (15) and mice (20) indicating that either depletion or overexpression of HMGN protein interferes with proper embryonic development.
Developmental role for HMGN proteins.
Our studies link the expression of HMGN1 protein to cellular differentiation. We observe a widespread, progressive down-regulation of Hmgn1 expression during embryonic development in every tissue examined, only a few types of which are shown in Fig. 2. We also found that these expression patterns were not identical to the PCNA expression pattern, suggesting that the expression level of HMGN1 does not simply reflect the cellular proliferation state. HMGN1 expression is significantly reduced in differentiated tissues but remains robust in undifferentiated cells or cells with regenerative-renewal capacity, such as the basal layer of skin, the inner lumen of stomach, the branching distal epithelium of lung, or the peripheral mesenchymal cells layer of the kidney (Fig. 2 and T. Furusawa, unpublished data).
These results are consistent with tissue culture studies that demonstrated down-regulation of Hmgn1 expression during erythropoiesis, myogenesis, and osteoblast differentiation (8, 23, 27) and with biochemical studies that linked this down-regulation specifically to differentiation rather than a decreased rate of cellular replication (4). Likewise, during kidney organogenesis, the closely related protein HMGN2 is expressed mainly in cells undergoing inductive interactions and differentiation (16) and is down-regulated in terminally differentiated cells. In the developing limb bud, the expression of Hmgn1 was restricted to the undifferentiated mesenchymal regions. The prechondrogenic regions and the differentiated digits contained very little, if any, HMGN1 (Fig. 2). These observations raise the possibility that down-regulation of HMGN1 expression is a prerequisite of proper differentiation and, conversely, that elevated expression of Hmgn1 may retard cellular differentiation. Indeed, overexpression of Hmgn1 reduced the number and size of the differentiated nodules (Fig. 4). Likewise, previous studies with differentiation of C2C12 myoblasts demonstrated that ectopic expression of HMGN1 inhibits myotube formation (23). Thus, down-regulation of Hmgn1 is required for both chondrogenesis and myogenesis in vitro. These findings and previous studies with microinjected one-cell mouse embryos (20) and with Xenopus laevis embryos (15) argue for a role of HMGNs in vertebrate embryonic development.
In X. laevis, misexpression of either of the two major members of the HMGN protein family, HMGN1 or HMGN2, leads to specific, developmentally timed abnormalities in the growing embryos (15). Either overexpression or depletion of HMGNs in Xenopus zygotes caused severe malformations in postblastula development without affecting preblastula development. Significantly, HMGN proteins are expressed only after midblastula transition. The data suggest that once the proteins are synthesized and functional, their levels need to be monitored, and either too much or too little protein leads to developmental abnormalities.
In considering molecular mechanisms whereby HMGN proteins affect differentiation processes, we note that in the in vitro micromass cultures, loss of Hmgn1 resulted in faster chondrogenesis. In this system, reexpression of HMGN1 protein in Hmgn1−/− cells restores the wild-type phenotype and retards the rate of differentiation, whereas reexpression of HMGN1(S20,24E), the double point mutant form of HMGN1 which does not bind to nucleosomes, did not restore the wild-type phenotype (Fig. 5 and 6). Likewise, this mutant protein did not lead to developmental abnormalities in X. laevis (15). In one-cell stage mouse embryos, disruption of the interaction of HMGN with chromatin led to an abnormal rate of development (20). Taken together, these results argue that HMGNs modulate a differentiation-related process by interacting with chromatin.
Since Hmgn1−/− mice seem to develop normally (2), it is likely that homeostatic mechanisms, perhaps involving HMGN2, compensate for the loss of the HMGN1 protein. Indeed, our ChIP analyses indicate an increase of HMGN2 in the Sox9 chromatin obtained from Hmgn1−/− mice, suggesting functional redundancy among these proteins. We also observed that during embryogenesis and during limb bud development, Hmgn2 is expressed in a pattern that is indistinguishable from that of Hmgn1 (T. Furusawa, unpublished data). Furthermore, we already demonstrated that transient depletion of both Hmgn1 and Hmgn2 transcripts in 1-day-old mouse embryos was necessary to retard the progression of preimplantation development. Depletion of only one of the Hmgn transcripts had no significant effect on the rate of development and did not cause an embryonic developmental delay (20), suggesting a functional overlap that compensates for loss of one of the major components of HMGN protein.
In the nucleus, HMGN1 functions as a member of a dynamic network of chromatin binding proteins, involving other HMG proteins and histone H1, which compete for nucleosome binding sites (6, 7). As discussed elsewhere (6), changes in one component of the network may trigger compensatory adjustments in other components of the network and in chromatin, all aimed to optimize the cellular requirement at any given time. Thus, changes in HMGN1 alter the interaction of H1 with chromatin (7) and modify the pattern of posttranslational modification of histones (17, 18). These changes reflect the action of homeostatic mechanisms that allow survival and development of Hmgn1−/− mice, which mostly appear normal. Yet their stress response is significantly impaired, and they are hypersensitive to irradiation by either UV (3) or X ray (2). By analogy, the Hmgn1−/− limb bud cells grown in micromass cultures are placed under conditions in which homeostatic mechanisms operating in the intact embryo are altered, thus leading to changes in Sox9 expression and in differentiation of the micromass.
Supplementary Material
Acknowledgments
We thank Susan H. Garfield and Stephen M. Wincovitch (Laboratory of Experimental Carcinogenesis, Center for Cancer Research, National Cancer Institute) for help with the confocal microscopy and the NCI CCR Fellows Editorial Board for constructive criticisms of the manuscript.
T.F. is a recipient of a fellowship from the Japan Society for Promotion of Sciences. This research was supported by the Center for Cancer Research, National Cancer Institute, NIH.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Begum, N., J. M. Pash, and J. S. Bhorjee. 1990. Expression and synthesis of high mobility group chromosomal proteins in different rat skeletal cell lines during myogenesis. J. Biol. Chem. 265:11936-11941. [PubMed] [Google Scholar]
- 2.Birger, Y., F. Catez, T. Furusawa, J. H. Lim, M. Prymakowska-Bosak, K. L. West, Y. V. Postnikov, D. C. Haines, and M. Bustin. 2005. Increased tumorigenicity and sensitivity to ionizing radiation upon loss of chromosomal protein HMGN1. Cancer Res. 65:6711-6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birger, Y., K. L. West, Y. V. Postnikov, J. H. Lim, T. Furusawa, J. P. Wagner, C. S. Laufer, K. H. Kraemer, and M. Bustin. 2003. Chromosomal protein HMGN1 enhances the rate of DNA repair in chromatin. EMBO J. 22:1665-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bustin, M. 2001. Chromatin unfolding and activation by HMGN(*) chromosomal proteins. Trends Biochem. Sci. 26:431-437. [DOI] [PubMed] [Google Scholar]
- 5.Bustin, M. 1999. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 19:5237-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bustin, M., F. Catez, and J. H. Lim. 2005. The dynamics of histone H1 function in chromatin. Mol. Cell 17:617-620. [DOI] [PubMed] [Google Scholar]
- 7.Catez, F., H. Yang, K. J. Tracey, R. Reeves, T. Misteli, and M. Bustin. 2004. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol. Cell. Biol. 24:4321-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crippa, M. P., J. M. Nickol, and M. Bustin. 1991. Developmental changes in the expression of high mobility group chromosomal proteins. J. Biol. Chem. 266:2712-2714. [PubMed] [Google Scholar]
- 9.Crippa, M. P., J. M. Nickol, and M. Bustin. 1991. Differentiation-dependent alteration in the chromatin structure of chromosomal protein HMG-17 gene during erythropoiesis. J. Mol. Biol. 217:75-84. [DOI] [PubMed] [Google Scholar]
- 10.de Crombrugghe, B., V. Lefebvre, and K. Nakashima. 2001. Regulatory mechanisms in the pathways of cartilage and bone formation. Curr. Opin. Cell Biol. 13:721-727. [DOI] [PubMed] [Google Scholar]
- 11.Ding, H. F., S. Rimsky, S. C. Batson, M. Bustin, and U. Hansen. 1994. Stimulation of RNA polymerase II elongation by chromosmal protein HMG-14. Science 265:796-799. [DOI] [PubMed] [Google Scholar]
- 12.Hatakeyama, Y., R. S. Tuan, and L. Shum. 2004. Distinct functions of BMP4 and GDF5 in the regulation of chondrogenesis. J. Cell Biochem. 91:1204-1217. [DOI] [PubMed] [Google Scholar]
- 13.Kanai, Y., and P. Koopman. 1999. Structural and functional characterization of the mouse Sox9 promoter: implications for campomelic dysplasia. Hum. Mol. Genet. 8:691-696. [DOI] [PubMed] [Google Scholar]
- 14.Kelly, R. G., and M. E. Buckingham. 2002. The anterior heart-forming field: voyage to the arterial pole of the heart. Trends Genet. 18:210-216. [DOI] [PubMed] [Google Scholar]
- 15.Korner, U., M. Bustin, U. Scheer, and R. Hock. 2003. Developmental role of HMGN proteins in Xenopus laevis. Mech. Dev. 120:1177-1192. [DOI] [PubMed] [Google Scholar]
- 16.Lehtonen, S., and E. Lehtonen. 2001. HMG-17 is an early marker of inductive interactions in the developing mouse kidney. Differentiation 67:154-163. [DOI] [PubMed] [Google Scholar]
- 17.Lim, J. H., F. Catez, Y. Birger, K. L. West, M. Prymakowska-Bosak, Y. V. Postnikov, and M. Bustin. 2004. Chromosomal protein HMGN1 modulates histone H3 phosphorylation. Mol. Cell 15:573-584. [DOI] [PubMed] [Google Scholar]
- 18.Lim, J. H., K. L. West, Y. Rubinstein, M. Bergel, Y. V. Postnikov, and M. Bustin. 2005. Chromosomal protein HMGN1 enhances the acetylation of lysine 14 in histone H3. EMBO J. [DOI] [PMC free article] [PubMed]
- 19.Megason, S. G., and A. P. McMahon. 2002. A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development 129:2087-2098. [DOI] [PubMed] [Google Scholar]
- 20.Mohamed, O. A., M. Bustin, and H. J. Clarke. 2001. High-mobility group proteins 14 and 17 maintain the timing of early embryonic development in the mouse. Dev. Biol. 229:237-249. [DOI] [PubMed] [Google Scholar]
- 21.Morishita, M., T. Kishino, K. Furukawa, A. Yonekura, Y. Miyazaki, T. Kanematsu, S. Yamashita, and T. Tsukazaki. 2001. A 30-base-pair element in the first intron of SOX9 acts as an enhancer in ATDC5. Biochem. Biophys. Res. Commun. 288:347-355. [DOI] [PubMed] [Google Scholar]
- 22.Paranjape, S. M., A. Krumm, and J. T. Kadonaga. 1995. HMG17 is a chromatin-specific transcriptional coactivator that increases the efficiency of transcription initiation. Genes Dev. 9:1978-1991. [DOI] [PubMed] [Google Scholar]
- 23.Pash, J. M., P. J. Alfonso, and M. Bustin. 1993. Aberrant expression of high mobility group chromosomal protein 14 affects cellular differentiation. J. Biol. Chem. 268:13632-13638. [PubMed] [Google Scholar]
- 24.Pash, J. M., J. S. Bhorjee, B. M. Patterson, and M. Bustin. 1990. Persistence of chromosomal proteins HMG-14/-17 in myotubes following differentiation-dependent reduction of HMG mRNA. J. Biol. Chem. 265:4197-4199. [PubMed] [Google Scholar]
- 25.Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404:604-609. [DOI] [PubMed] [Google Scholar]
- 26.Prymakowska-Bosak, M., T. Misteli, J. E. Herrera, H. Shirakawa, Y. Birger, S. Garfield, and M. Bustin. 2001. Mitotic phosphorylation prevents the binding of HMGN proteins to chromatin. Mol. Cell. Biol. 21:5169-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shakoori, A. R., T. A. Owen, V. Shalhoub, J. L. Stein, M. Bustin, G. S. Stein, and J. B. Lian. 1993. Differential expression of the chromosomal high mobility group proteins 14 and 17 during the onset of differentiation in mammalian osteoblasts and promyelocytic leukemia cells. J. Cell Biochem. 51:479-487. [DOI] [PubMed] [Google Scholar]
- 28.Shirakawa, H., J. E. Herrera, M. Bustin, and Y. Postnikov. 2000. Targeting of high mobility group-14/-17 proteins in chromatin is independent of DNA sequence. J. Biol. Chem. 275:37937-37944. [DOI] [PubMed] [Google Scholar]
- 29.Shum, L., C. M. Coleman, Y. Hatakeyama, and R. S. Tuan. 2003. Morphogenesis and dysmorphogenesis of the appendicular skeleton. Birth Defects Res. C 69:102-122. [DOI] [PubMed] [Google Scholar]
- 30.Trieschmann, L., P. J. Alfonso, M. P. Crippa, A. P. Wolffe, and M. Bustin. 1995. Incorporation of chromosomal proteins HMG-14/-17 into nascent nucleosomes induces an extended chromatin conformation and enhances the utilization of active transcription complexes. EMBO J. 14:1478-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weisbrod, S. 1982. Active chromatin. Nature 27:289-295. [DOI] [PubMed] [Google Scholar]
- 32.West, K. L., M. A. Castellini, M. K. Duncan, and M. Bustin. 2004. Chromosomal proteins HMGN3a and HMGN3b regulate the expression of glycine transporter 1. Mol. Cell. Biol. 24:3747-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.West, K. L., Y. Ito, Y. Birger, Y. Postnikov, H. Shirakawa, and M. Bustin. 2001. HMGN3a and HMGN3b, two protein isoforms with a tissue-specific expression pattern, expand the cellular repertoire of nucleosome-binding proteins. J. Biol. Chem. 276:25959-25969. [DOI] [PubMed] [Google Scholar]
- 34.Wilkinson, D. G. 1992. In situ hybridization, p. 75-83. In D. G. Wilkinson (ed.), In situ hybridization: a practical approach. IRL Press, Oxford University, Oxford, United Kingdom.
- 35.Wunderle, V. M., R. Critcher, N. Hastie, P. N. Goodfellow, and A. Schedl. 1998. Deletion of long-range regulatory elements upstream of SOX9 causes campomelic dysplasia. Proc. Natl. Acad. Sci. USA 95:10649-10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zechner, D., Y. Fujita, J. Hulsken, T. Muller, I. Walther, M. M. Taketo, E. B. Crenshaw, 3rd, W. Birchmeier, and C. Birchmeier. 2003. β-Catenin signals regulate cell growth and the balance between progenitor cell expansion and differentiation in the nervous system. Dev. Biol. 258:406-418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.