Abstract
Studies in tissue culture cells have demonstrated a role for the Ras-like GTPase Rap1 in the regulation of integrin-mediated cell-matrix and cadherin-mediated cell-cell contacts. To analyze the function of Rap1 in vivo, we have disrupted the Rap1A gene by homologous recombination. Mice homozygous for the deletion allele are viable and fertile. However, primary hematopoietic cells isolated from spleen or thymus have a diminished adhesive capacity on ICAM and fibronectin substrates. In addition, polarization of T cells from Rap1−/− cells after CD3 stimulation was impaired compared to that of wild-type cells. Despite this, these defects did not result in hematopoietic or cell homing abnormalities. Although it is possible that the relatively mild phenotype is a consequence of functional complementation by the Rap1B gene, our genetic studies confirm a role for Rap1A in the regulation of integrins.
Rap1 belongs to the family of Ras-like GTPases, and it is now well established that one of its main biological functions is the regulation of cell-matrix and cell-cell adhesion. First, it was shown that interfering in Rap1 activity via overexpression of the Rap1-specific GAP Spa1 blocks adhesion of HeLa and 32D cells to the extracellular matrix, which depends on integrin activity (32). In addition, constitutively active Rap1 was found to enhance integrin-mediated adhesion of T cells and macrophages (3, 15, 27). Also, in transgenic mice, Rap1V12 under the control of a T-cell specific promoter has shown to affect cell adhesion of T cells (28). Rap1 appeared to act on a variety of different integrins, such as LFA-1, VLA4, and VLA5, but not all integrins (8). More recently it was shown that Rap1 also enhances cell-cell contact formation via cadherins (13, 25). To a great extent, these studies have relied on overexpression of GTPase activating proteins (GAPs) for Rap1, isolated Rap1-binding domains from putative effector molecules (RBDs), or constitutively active or dominant-negative mutants of Rap1. Evidence for a role of endogenous Rap1 has come from studies in which a very specific activator of the Rap-specific GEF EPAC was used (26). Further support for the role of Rap1 has come from genetic disruption of GEFs for Rap1. Targeted disruption of C3G, the first GEF for Rap1 to be cloned (31), results in embryonic lethality (20). In the same study it was shown that deletion of C3G from mouse embryonic fibroblasts (MEFs) results in defects in cell adhesion and migration. Disruption of RapGEF, CD-GEFI, revealed that it was required for Rap1 activation in platelets and integrin-mediated coagulation (6). Although all of these studies taken together provide a large body of evidence in favor for a role of Rap1 in cell adhesion via integrins, many of them still do not discriminate between the action of Rap1A, Rap1B, or any of the Rap2 isoforms. For example, the RapGAPs, Rap1GAP Spa1, will decrease the levels of both GTP-bound Rap1 and Rap2. Obviously, the use of isolated RBDs as an inhibitory tool has an even a higher risk of intervening in the action of various Ras-like GTPases. Furthermore, the above-mentioned GEFs are not specific for a single GTPase. For example, Epac will activate both Rap1 and Rap2, CDGEFIII will activate Rap1, Ras, and R-Ras (18, 34), and C3G will activate Rap1 and R-Ras (10, 11, 21, 22). Indeed, the enhanced migration of MEFs lacking C3G could be suppressed by overexpression of active versions of Rap1, Rap2, and R-Ras. To make picture even more complicated, a role for Rap2 in B-cell adhesion has been put forward by McLeod et al. (19).
In order to directly assess the function of one of the Rap1 isoforms, Rap1A, we generated knockout mice by homologous recombination. Analysis of these mice reveals that Rap1A is dispensable for viability or fertility. The major defect observed thus far is in cell adhesive properties of cells from the immune system. Despite the reduced activity in cell adhesion assays, the immune system does not show any obvious defects in differentiation or maturation of lymphoid cells.
MATERIALS AND METHODS
Targeting construct and microinjections.
In order to inactivate the Rap1A gene, a targeting construct was made by replacing a 420-bp region encoding the N-terminal 19 amino acids of Rap1A, which include conserved residues and part of the GTP-binding motif, by an internal ribosome entry site (IRES)/LacZ/neomycin cassette (23). The homology arms at the 5′ and 3′ ends were 1.3 and 4.5 kb in length, respectively (Fig. 1A). A XhoI-linearized construct was electroporated into HM-1 embryonic stem line. A total of 200 clones were picked, and 4 were identified as positive by PCR and Southern blotting. The HM-1 embryonic stem line has an OlA/126 genetic background. Positive cells were microinjected into blastocysts of a C57BL/6 genetic background. Outcrossing was done with C57BL/6 mice.
FIG. 1.
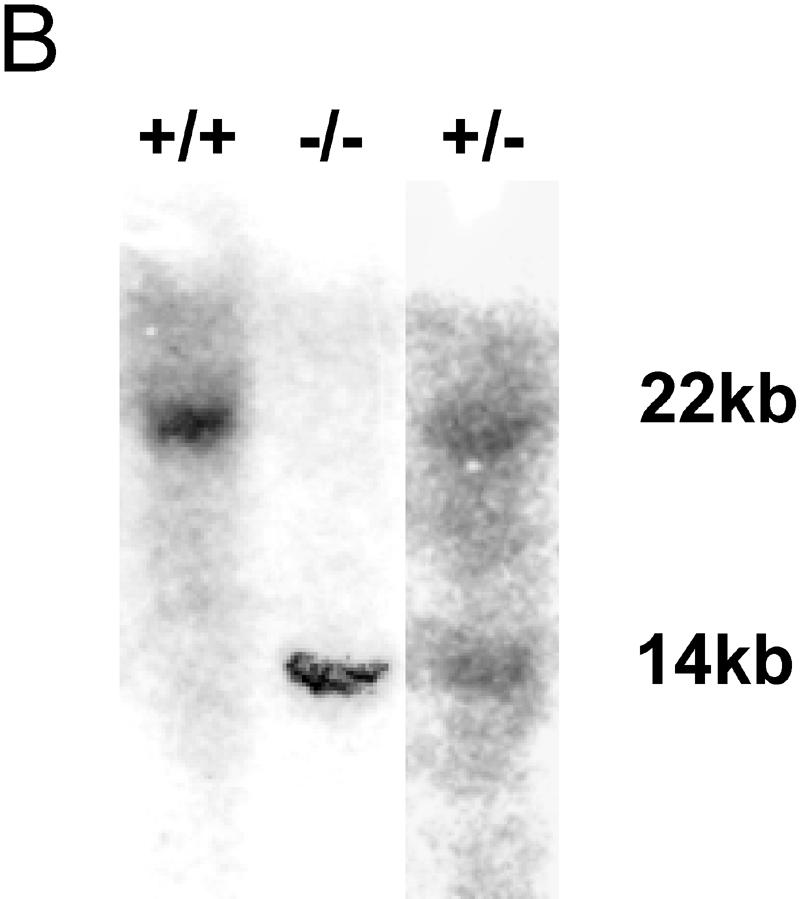

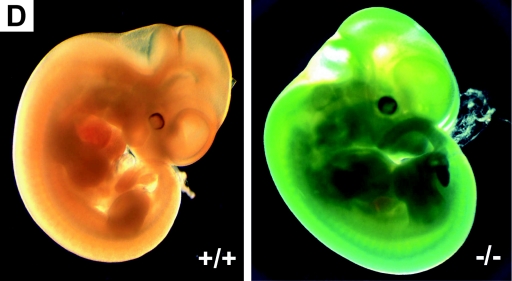
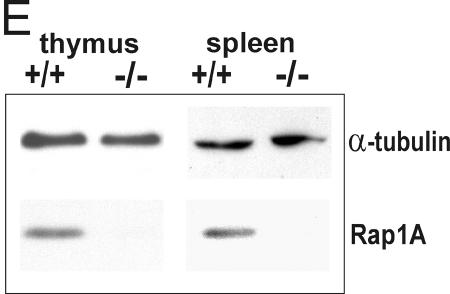
Generation of Rap1A knockout mice. (A) Schematic map of the Rap1A gene locus (exons 2 to 4 are shown), targeting construct, and recombinant allele. The black rectangles represent coding exons. The 5-kb targeting cassette contains an IRES sequence, β-galactosidase gene (LacZ), and the neomycin gene (NEO). The 3′ external probe used for probing the genomic blot is shown in panel B as a solid black box under the targeted allele. Restriction enzyme sites: B, BamHI; H, HindIII; P, PstI. (B) Southern blot analysis of the Rap1A gene. DNA was prepared from mouse tails, digested by the ApaI restriction enzyme, and hybridized with 3′ external probe. The wild-type allele generated a 22-kb band, and the targeted allele generated a 14-kb band. (C) PCR-based genotyping of Rap1A knockout. ForPrimer1 and RevPrimer1 generated a 1.2-kb band from the wild-type allele. ForPrimer2 and RevPrimer2 produced 0.6-kb band from targeted allele. (D) β-Galactosidase staining in whole-mount embryonic day 12.5 −/− embryo. (E) Western blot for Rap1A protein. Proteins extracts from thymus and spleen were analyzed by Western blotting with Rap1A-specific antibodies. Rap1A band is absent in protein extracts isolated from −/− mice. α-Tubulin was used as a loading control.
β-Galactosidase staining of murine embryos.
Embryos at embryonic day 12.5 were isolated and fixed in 4% paraformaldehyde for 15 to 20 min at 4°C. Embryos were washed three times, each for 15 min, in washing buffer (5 mM EGTA, 0.01% deoxycholate, 0.02% NP-40, 2 mM MgCl2, phosphate-buffered saline [PBS]). Staining solution [5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 5 mM EGTA, 0.01% deoxycholate, 0.02% NP-40, 2 mM MgCl2, PBS] was added, and embryos were incubated overnight at 37°C in the dark. The staining was stopped by fixation in 4% paraformaldehyde overnight at 4°C.
Mouse genotyping-Southern blotting and PCR.
Genotyping was performed on DNA extracts prepared from mouse tails. DNA was digested with ApaI and analyzed by Southern blotting. Using a 3′ Rap1A external probe, the recombinant allele is detected as a 14-kb band, whereas the wild-type allele is 22 kb. In addition, mice were genotyped by PCR with ForPrimer (5′-GTTACTCCATATCAACCATTG-3′) and RevPrimer1 (5′-CCACCTCACTTTCTCCCAC-3′) to produce a 1.2-kp product specific for the wild-type allele or ForPrimer and RevPrimer2 (5′-CCAAGGACTACTAGCTTGTACTCACG-3′) to generate a 0.6-kp product specific for the targeted allele.
RT of Rap1A mRNA.
RNA was isolated from mouse organs by using RNA isolation kit (QIAGEN). RNA was treated with DNase I to remove DNA. RNA was reverse transcribed into cDNA by using reverse transcriptase (Gibco). Standard PCR was carried out on a cDNA template. Two primers were used: 5′Primer (5′-GCGGGATTGTCAATATTTAAAC-3′) and 3′Primer (5′-GCCATAGAAATCAGTTATCCC-3′), which generated a product of 1,186 bp. Reverse transcription-PCR (RT-PCR) products were cloned into TOPO cloning vector (Invitrogen) and sequenced by using vector primers (Invitrogen).
Flow cytometric analysis.
Single cell suspensions were prepared from the thymus, spleen, and peripheral lymph nodes. The cells were first blocked with CD16/CD32 blocking antibodies and later stained with specific antibodies against surface markers labeled with fluorescein isothiocyanate (FITC), APC, phycoerythrin (PE), or PER-CY5. Staining was performed for 15 min at 4°C and subsequently washed two times with PBS. Fluorescence was measured in a DakoCytomation FACS Analyzer (CYAN). Antibodies to the following surface markers were used in these studies: CD4, CD8, CD3, B220, CD45, CD44, CD25, αβTCR, CD69, CD5, ScaI, LFA-1(CD 11a), and VLA-4(CD 49d). All antibodies were purchased from Pharmingen.
GST pull-down assay.
About 2 × 107 of thymus or spleen cells were left untreated or stimulated with TPA (12-O-tetradecanoylphorbol-13-acetate; 50 ng/ml) for 5 min at 37°C in RPMI medium. Stimulation was interrupted by adding of 5 volumes of ice-cold lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 0.5% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 2 μg of aprotinin/ml, 2 μg of leupeptin/ml, 0.1 mM Na3VO4, 10 mM NaF). Lysis was performed for 45 min on ice. The extracts were centrifuged at 15,000 × g for 15 min at 4°C. Supernatants were collected, and 5 μg of RalGDSRBD-glutathione S-transferase (GST), precoupled to glutathione-agarose beads, was added. Samples were mixed with slow agitation for 1 h at 4°C. The beads were washed three times with lysis buffer, and 30 μl of 1× Laemmli buffer was added. Proteins isolated in pull-down assays or present in total lysates were separated by sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes, and detected with antibodies to α-tubulin and GAPDH (glyceraldehyde-3-phosphate dehydrogenase; both purchased from Santa Cruz Biotechnology, Santa Cruz, CA), both Rap1 isoforms (SC-65; Santa Cruz, California), or Rap1A (kindly provided by V. Pizon). The signal was detected by enhanced luminescence (Amersham Biotechnologies).
Adhesion assay.
The adhesion assay was based on previous reports (28). Maxisorb 96-well plates (Nunc) were coated with the indicated concentrations of fibronectin (Sigma) or murine ICAM-FC (R&D Systems) in PBS at 4°C. Wells were washed three times with PBS and blocked with 2% bovine serum albumin (BSA) in Hanks balanced salt solution (HBSS; Invitrogen) for 1 h at 37°C. Fresh thymus or spleen cells were incubated 10 min in erythrocyte lysis buffer (Stem Cell Technology) to remove erythrocytes and then washed in RPMI and stained in 2.5 μM BCEF-AM (Calbiochem)-HBSS for 30 min at 37°C. After staining, the cells were washed three times in HBSS medium and resuspended in RPMI medium-0.5% BSA. The blocked 96-well plates were washed three times with 0.5% BSA-HBSS. A total of 106 cells were loaded in one well in a volume of 100 μl. Then, 50 μl of TPA was added to obtain a final concentration of 50 ng/ml. The cells were left to adhere for 30 min at 37°C. Nonadherent cells were removed by three washing steps with warm 0.5% BSA-RPMI. Adhesion was measured in spectrofluorometer at an excitation of 485 nm and an emission of 530 nm. The adhesion was counted as a percentage of total cell input (measured before incubation) added to the well.
Proliferation assay.
T cells were isolated from spleen by using Pan T-cell kit (MACS) by magnetic sorting. We coated 96-well plates with 1 μg of murine anti-CD3 (145-2C11) or 1 μg of anti-CD3 and anti-CD28/ml. A total of 105 cells were plated per well, and stimulation was performed for 48 h at 37°C. As a positive control, cells were stimulated with 30 ng of TPA/ml plus 100 ng of ionomycin/ml. After the stimulation, 1 μCi of [3H]thymidine (Amersham Biosciences) was added, and cells were further incubated for 18 h. Labeled DNA from cells was collected on GSC filters. Radioactivity was measured in a microplate scintillation counter (Packard).
IL-2 measurements.
Cell medium was collected from CD3 and CD3/CD28- or TPA-stimulated cells after 48 h. The interleukin-2 (IL-2) concentration was determined by using a BD OptEIATM Mouse IL-2 ELISA set (BD Biosciences).
Immunofluorescence staining.
To assess T-cell polarization, T cells were first incubated with anti-CD3 (10 μg/ml) for a half an hour at 4°C and then washed two times with cold RPMI incubated with anti-American hamster immunoglobulin G (10 μg/ml) for 30 min on ice. After two washing steps in cold RPMI, the cells were stimulated at 37°C for 3 min or left unstimulated at 4°C. The cells were fixed in suspension in 3.3% paraformaldehyde and mounted on poly-l-lysine-coated slides and then blocked with1% BSA. The cells were stained with anti-LFA-1 (1:1,500; Pharmingen). The fluorescence was visualized on Zeiss microscope using ×63 magnification. The cells were assessed for polarization of LFA-1 as described previously (12). Briefly, photos of 10 fields, each comprising 40 to 50 cells, were obtained for each treatment. Cells with LFA-1 polarized to one side of cell were regarded as polarized; those showing equal distribution of LFA-1 were considered not polarized. The percentage of polarized cells in each photographed field was counted.
Statistical methods.
Analysis of proliferation and IL-2 production ratios were done by fitting a two-factor log-linear model to each of the datasets and performing an analysis of variance. As factors we used the type of treatment (CD3, CD3/CD28, and TPA/ionomycin) and the type of mouse (knockout and wild type).
RESULTS
Targeted disruption of Rap1A.
HM-1 embryonic stem cells were electroporated with a targeting vector for the Rap1A gene, in which the second exon had been replaced by an IRES/LacZ/neomycin cassette. Correct insertion would delete the 19 N-terminal amino acids of Rap1A. Four recombinant clones were identified by PCR and Southern blotting. Sequencing of PCR products confirmed a precise incorporation of the IRES/LacZ/neomycin cassette in exon 2. Chimeric mice were generated with two clones, both of which gave germ line transmission. DNA analysis, either by Southern blotting or by PCR, of offspring from interbred heterozygous Rap1A mutants (Fig. 1B and C) revealed that homozygous Rap1A−/− mice were born in Mendelian proportions (i.e., of the 132 first-generation [F1] mice, 28 were +/+ and 27 were −/−).
Northern blotting of mRNA from tissues of homozygous knockout mice, using a probe against the complete coding sequence, did not reveal the expected lack of Rap1A RNA but instead revealed the presence of a slightly shorter message (data not shown). We therefore cloned and sequenced cDNA of Rap1A from homozygous mutant mice. The cDNAs were derived from shortened WT mRNA, lacking the first 19 codons, which resulted from insertion of the targeting vector. However, the IRES/LacZ/neomycin cassette was absent due to the usage of a cryptic splice site at the 5′ region of the cassette. Nevertheless, we were able to detect LacZ mRNA by RT-PCR, but we could not detect a fusion mRNA of IRES/LacZ/neomycin with the 5′ end of Rap1A. Staining of Rap1A−/− embryos at day 12.5 post coitus for β-galactosidase (Fig. 1D) also confirmed the stability of the LacZ transcript. In conclusion, mRNA from the targeted Rap1A allele lacks the first 19 codons, whereas eventual translation from a further downstream located ATG would result in 51-amino-acid-shorter protein product.
To establish that the Rap1A gene had been successfully targeted a Western blot with lysates from embryonic thymus and spleen was probed with an antibody raised against Rap1A (kindly provided by V. Pizon). This antibody is almost specific for Rap1A and only weakly recognizes overexpressed Rap1B (unpublished data). As can be seen in Fig. 1E, no Rap1A protein was detected in these lysates. From these results we conclude that the Rap1A gene has been successfully targeted.
General phenotype of Rap1A-deficient mice.
Rap1A-deficient mice appeared to be fully viable and fertile. Growth rates of pups as determined by measuring their weight between weeks 2 and 8 after birth were not different from that of wild-type littermates (data not shown). The life span of Rap1A mutant mice did not differ from that of wild-type littermates. In addition, no obvious behavioral abnormalities were noted in Rap1A mutants.
Immunological phenotype of Rap1A knockout mice.
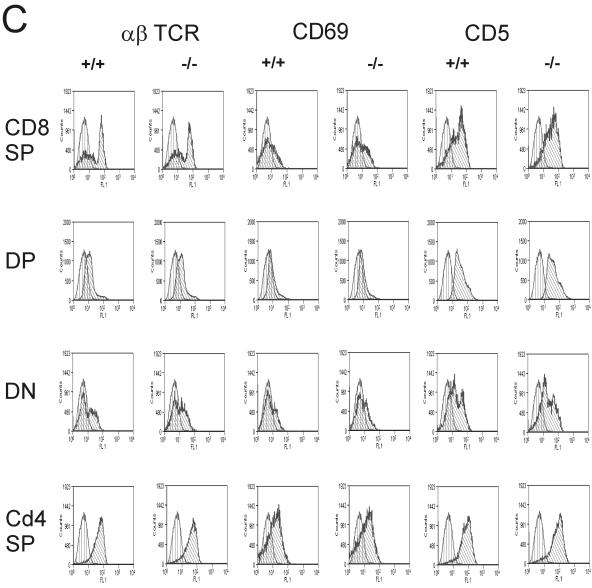
Given the fact that genetic disruption of the RapGAP Spa1 resulted in T-cell anergy and altered memory responses (14), we set out to investigate the different lymphoid compartments in Rap1A mutant mice. T-cell maturation, as measured by the relative fraction of CD4/8 double-negative (DN), single-positive (SP), and double-positive (DP) cells in the thymus, appeared to be normal (Fig. 2A). Furthermore, expression profiles for TCRαβ, CD5, or CD69 in SP, DP, and DN populations of Rap1A−/− thymocytes appeared to be indistinguishable from that of wild-type mice and demonstrated unperturbed positive selection in Rap1A−/− mouse (Fig. 2B).
FIG. 2.
Normal differentiation in thymuses of wild-type and Rap1A−/− mice. (A) Expression pattern for CD4 and CD8. (B) CD44 and CD25 expression in CD4− CD8−population. Thymocytes were four-color stained for CD4, CD8, CD44, and CD25 markers. The results are representative for one of 10 sets of wild-type and Rap1A mutant mice. (C) Positive selection in the thymus is intact in Rap1A−/− mice. The thymus was three-color stained for CD4 PE, CD8 APC, and FITC-labeled αβTCR, CD69, or CD5. Expression profiles for each FITC marker were measured in each CD4/CD8 population.
Likewise, analysis of a very early thymic population CD4− CD8− (DN) for expression of CD44 and CD25 (alpha-domain of the IL-2 receptor) showed no abnormalities (Fig. 2C). Profiling of thymic cells by fluorescence-activated cell sorting (FACS) analysis using either a combination of CD3 and CD45 or a ScaI antibody also did not reveal any difference (data not shown). Therefore, we conclude that development and differentiation of thymus is not altered in Rap1A−/− mouse.
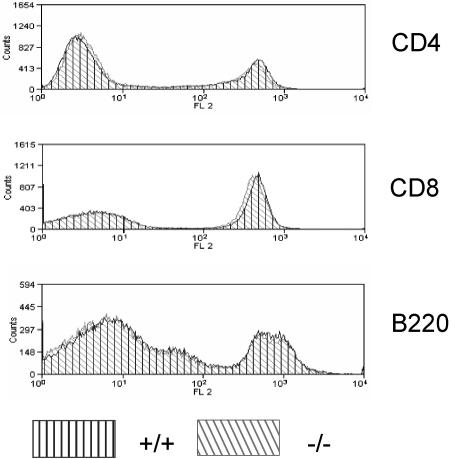
Also, separation of spleen cells by FACS stained with CD4/CD8, CD3/CD45, or B220/CD45 gave identical patterns for Rap1A mutant and wild-type cells (data not shown). Homing of peripheral T cells into lymph nodes, as investigated by studying the expression of B220, CD4, and CD8 on cells isolated from lymph nodes, was not affected (Fig. 3). In the bone marrow, FACS analysis with anti-B220, Mac-1, NK-1, GR-1, and SCA-1 did not reveal differences between Rap1 and wild-type mice (data not shown). Moreover, we did not find any abnormalities in the absolute number of thymus and spleen cells. Neither lymphocyte nor thrombocyte numbers in blood were altered in Rap1A−/− mice (data not shown). We conclude from this analysis that the immune system of Rap1A mutant mouse is largely intact. We do realize, however, that certain defects, e.g., in thymocyte selection or homing, may have gone undetected in our FACS analysis.
FIG. 3.
Homing of T and B cells into peripheral lymph nodes is unperturbed in Rap1A−/− mice. Overlay histograms show CD4, CD8 and B220 expressions in peripheral lymph nodes of wild-type mice and −/− mice.
Adhesion and Rap1 activity in spleen and thymus.
Since Rap1 has been established to regulate cell adhesion via integrins, we decided to investigate adhesive properties of cells isolated from the spleen or thymus derived from Rap1A mutant mice.
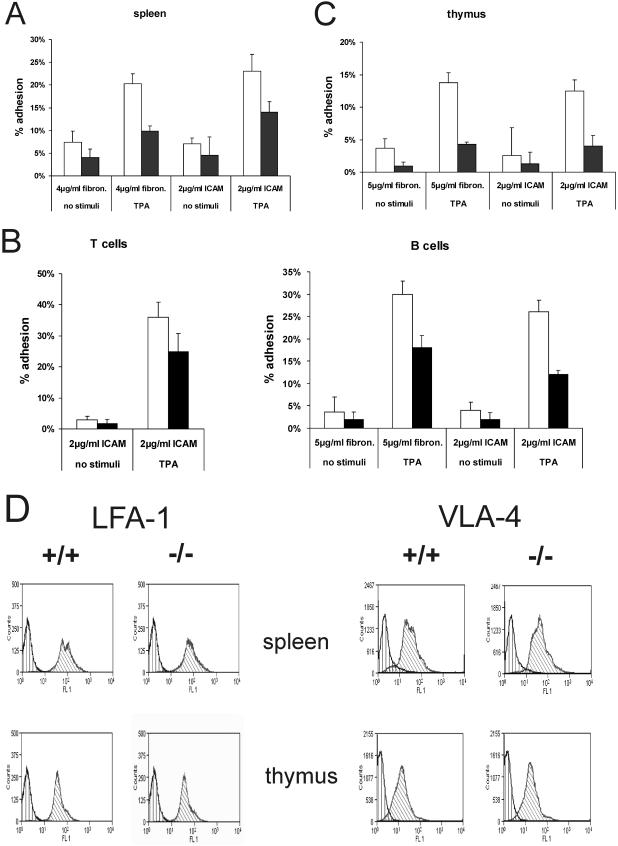
Spleen cells from wild-type mice clearly adhered better to fibronectin, a ligand for α4β1 integrins (VLA-4), than cells isolated from Rap1A mutant mice. If spleen cells were allowed to adhere for 30 min, about two times as many wild-type cells were bound to fibronectin than Rap1A mutant cells (Fig. 4A). Upon stimulation with 50 ng of TPA/ml, a twofold increase in adhesion was seen for both cell populations, meaning that a significant difference was still seen. Adherence to ICAM, which is mediated via LFA-1, was slightly decreased in the case of Rap1A mutant cells. However, the difference in the number of adherent cells was more pronounced (about twofold), after stimulation with TPA (Fig. 4A). Separated T and B cells from spleen showed that both cell types from Rap1A knockout mice had an impaired capacity to bind to ICAM-FC, although adhesion could still be increased by TPA treatment (Fig. 4B). Furthermore, thymus cells from wild-type mice adhered better to either fibronectin or ICAM-FC (Fig. 4C). Stimulation of both wild-type and mutant cell populations with TPA increased adhesion, but again the number of adherent cells from Rap1A mutant mice remained significantly decreased. Importantly, disruption of Rap1A did not change the level of LFA-1 cell surface expression or VLA-4, as shown by FACS analysis for thymus and spleen (Fig. 4D).
FIG. 4.
LFA-1- and VLA-4-dependent adhesion is impaired in Rap1A−/−cells from the thymus and spleen. □, adhesion of wild-type mice; ▪, adhesion of Rap1A−/− mice. Each bar represents an average value of four independently performed experiments. (A) Adhesion of spleen cells on fibronectin and ICAM. (B) Adhesion of separated T and B cells from spleens. The adhesion of T on ICAM and of B cells on ICAM and fibronectin is shown. (C) Adhesion of thymus cells on ICAM and fibronectin. (D) Equal expression of LFA-1 and VLA-4 in wild-type and Rap1A−/− mouse. The isotype control is shown as a vertical-line histogram. Diagonal-line histograms represent cells stained with anti-LFA-1 or anti-VLA-4.
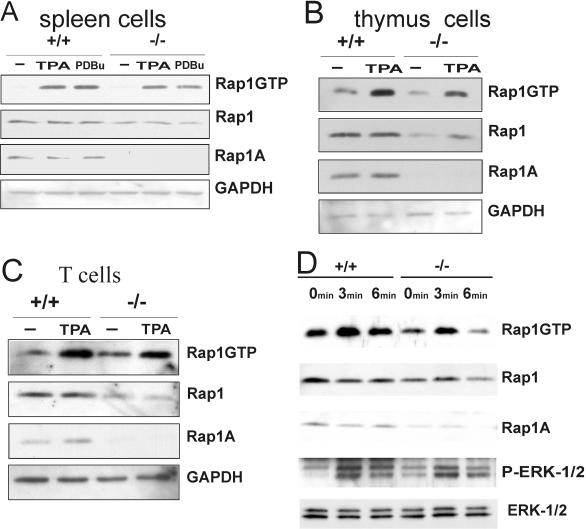
In parallel experiments, Rap1 activity was measured for all cells used in adhesion experiments. Pull-down assays performed on total cell populations isolated from the spleen showed a modest but consistent decrease in the fraction of GTP-bound Rap1. This was seen before and after stimulation with either TPA or PDBu (Fig. 5). A similar decrease was seen in the total amount of Rap1 expressed in these cells. Rap1A, on the other hand, was completely absent in cells from Rap1A knockout mice. A stronger decline in the fraction GTP-bound Rap1 and total amount of Rap1 was observed for thymus cells, as well as for T and B cells separated from spleen (Fig. 5C and data not shown). Likewise, stimulation of T cells from knockout mice with CD3 also shows a decreased GTP loading of Rap1 (Fig. 5D). This indicates that the response of T cells to physiological stimuli is also affected. In all cases, Rap1A could readily be detected in total lysates of wild type but not in that of mutant mice. Together, these data show that Rap1A deficiency results in an impairment of adhesion with a lymphoid origin, which is consistent with a decrease in GTP-bound Rap1.
FIG. 5.
Activity of Rap1 (Rap1GTP) in cell extracts derived from the spleen and thymus is decreased in Rap1-deficient mice. (A) GTP-bound Rap1 was isolated by using RalGDS-RBD from spleen cells before and after stimulation with TPA and detected by Western blotting with a Rap1 antibody (upper panel). Total Rap1 and Rap1A were detected in total lysates (middle two panels) by using an antibody that detects both Rap1 isoforms and one that is specific for Rap1A. Equal loading was confirmed by probing for GAPDH. (B) Detection of GTP-bound Rap1A in thymus cells as described for panel A for spleen cells. (C) Detection of GTP-bound Rap1A in T cells isolated from spleen as described for panel A for spleen cells. (D) T cells were induced with anti-CD3 antibodies for various periods of time. The cell extracts were subjected to Rap1 GST pull-down assay, and then the samples were analyzed by Western blotting. Rap1 activity (Rap1GTP) and total Rap1 (Rap1) was detected by using anti-Rap1 antibodies. In addition, the cell extracts were tested for activation of ERK1 and ERK2 (p-ERK1/2) by using anti-phospho ERK antibodies.
Proliferation potential of T cells is decreased in Rap1A−/− mice.
Recent reports have presented several lines of evidence for involvement of Rap1 protein in signaling pathways leading to cell activation and proliferation (14, 16). Therefore, we measured T-cell proliferative responses after T-cell-receptor (TCR) stimulation. The cells were stimulated with anti-CD3, anti-CD3/anti-CD28, or TPA. It was found that proliferation after stimulation with only CD3 was substantially decreased in −/− mice (Fig. 6A). In these mice, the T-cell proliferative response to CD3/CD28 stimulation was also impaired, although to a lower extent than that observed for CD3 alone. Furthermore, in parallel to a weaker proliferative response, Rap1A-negative T cells stimulated with CD3 produced significantly less IL-2 when measured 48 h after stimulation (Fig. 6B). Strikingly, impairment in IL-2 production in response to CD3/CD28, although significant, was not as drastic as for CD3 alone. Importantly, no detectable differences were found in the level of CD25 or CD69 cell surface expression (data not shown). We therefore conclude that Rap1A−/− mice displayed a weaker CD3 response, accompanied by a weaker IL-2 expression.
FIG. 6.
Analysis of Rap1a, knockout/wild-type ratios (KO/WT) of proliferation responses and IL-2 production to CD3, CD3/CD28, and TPA/ionomycin stimuli in T cells. The asterisks above the error bars indicate the significance of the results: ✽✽✽, P < 0.001; ✽✽, P < 0.01). (A) Relative proliferation changes in T cells. The estimated KO/WT proliferation ratios lie at 0.78 for CD3 and at 0.9 for CD3/CD28. (B) Relative IL-2 production in Rap1A−/− T cells measured 48 h after stimulation. The estimated KO/WT IL-2 production ratios lie at 0.28 for CD3 and at 0.81 for CD3/CD28.
CD-3 induced LFA-1 polarization on the surface of T cells.
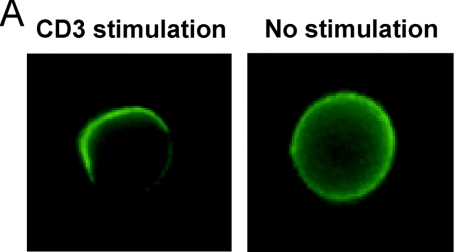
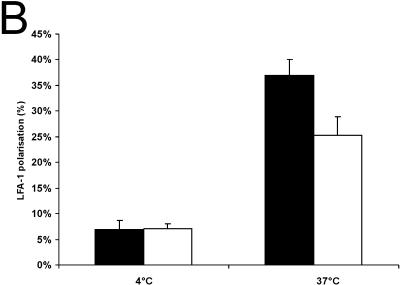
The mechanism by which Rap1 activates integrins is not understood in detail but, based on experiments in which Rap1V12 is overexpressed in T cells, clustering of integrins on the surface (28) or relocalization to one side of the cell as a consequence of polarization may be involved (29). Since TCR stimulation activates Rap1 (15) (Fig. 5D), we investigated the localization of LFA-1 on T cells after cross-linking with CD3. As can be seen in Fig. 7, we observed a clear polarization effect after 3 min of stimulation. Quantification of the number of polarized cells revealed that in Rap1A knockout cells the fraction of polarized cells is significantly decreased (Fig. 7).
FIG. 7.
CD3 induced LFA-1 polarization on the surface of T cells. (A) Examples of polarized and nonpolarized T cells. (B) LFA-1 polarization was assessed by determining the percentage of polarized cells in the fields of about 50 cells. Ten fields were counted for each treatment. Similar results were obtained in three independent experiments. Results for wild-type (□) and Rap1A−/− (▪) mice are shown.
DISCUSSION
A wide variety of functions, ranging from inhibition of the extracellular signal-regulated kinase pathway, to exocytosis, to control of proliferation to regulation of adhesion and migration, have been ascribed to the Ras-like GTPases Rap1A and Rap1B. Rap1A and Rap1B are paralogs and differ by 8 amino acids, mostly located in the C-terminal tail region. It is unknown whether there is a functional difference between the two proteins, which, apart from differences in primary structure, could be brought about by distinct subcellular locations or tissue-specific expression. It is also unclear to what extent the functions of Rap1 overlap with those of the highly related Rap2A, -B, and -C GTPases. Indeed, regulatory proteins such as GEFs and GAPs rarely discriminate between the Rap1 and Rap2 proteins. Direct measurement of GTP loading of Rap1 and Rap2 proteins has shown that both proteins are coregulated with Rap1 activity peaking before that of Rap2 (21, 22). Evidence for distinct functions of highly related GTPases has come, for example, from the functional analysis of the Ras-like GTPases RalA and RalB. Here, the RalA protein was found to contribute to anchorage-independent cell proliferation, whereas the RalB gene was found to be more important for protection against apoptosis (4). Furthermore, RalA but not RalB was shown to increase the transport of E-cadherin to the plasma membrane (30).
To probe for Rap1A-specific functions, we created a null mutation by removal of the N-terminal coding regions from the Rap1A locus. Homozygous mutant mice appear viable and fertile without any gross morphological or behavioral changes. Also, an extensive survey of hematopoietic characteristics did not reveal any striking phenotypes. For example, analysis of thymocytes showed the presence of all expected classes of DN, DP, and SP CD4/CD8 cells in very similar ratios, whereas also the distribution of markers such as TCRαβ, CD5, or CD69 appeared to be unaffected. However, when the adhesive properties of T and B cells were tested by using fibronectin- and ICAM-coated surfaces, a clear decrease in the adhesion of cells derived from Rap1A knockout mice was seen. Reduced binding was not a consequence of lowered expression of α4β1 integrins (VLA-4), the receptors for fibronectin or LFA-1, which binds ICAM. Binding of Rap1A−/− cells to two different substrates was not completely abrogated, which is most likely due to the presence of Rap1B in these cells. Indeed, Western blotting with an anti-Rap1A antibody, which only slightly cross-reacts with Rap1B, confirmed the absence of Rap1A, but an antibody that recognizes both Rap1A and B (SC-65) showed the residual presence of Rap1 in these cells. Also, the fraction of GTP-bound Rap1 was lowered, but a clear signal from Rap1B could still be detected. Interestingly, while the present study was in review, Chrzanowska-Wodnicka et al. reported that in Rap1B knockout mice platelet aggregation, which is dependent on αIIbβ3 integrin activation, is defective (5). In addition, bleeding defects cause lethality of more then 40% of the embryos. This latter effect may not be caused by impaired platelet function but results from endothelial defects. Indeed, the permeability of endothelial cells has been described as under the control of the cyclic AMP-regulated, Rap-specific GEF Epac (7, 9, 33). Given the different defects in integrin functioning seen in Rap1A and 1B knockout mice, it will be interesting to see what the compound phenotype is. Apart from Rap1B, Rap2 isoforms may function redundantly with Rap1A. Indeed, in most tissue culture experiments reported, all of these isoforms are likely blocked by overexpression of RapGAPs or Ras-binding domains. Furthermore, Rap1-independent cell adhesion mechanisms may exist. For example, in studies performed in T cells from lymph nodes overexpressing the RapGAP Spa1, an effect of cell adhesion after stimulation with SCF was seen but not after treatment with TPA (29). To what extent the adhesion phenotype is caused by a less efficient polarization of these cells is currently unclear. However, it should be noted that T cells expressing Rap1V12 display both increased adhesiveness and a polarized phenotype (29). Notably, not all primary cells derived from knockout mice display a diminished adhesive capacity on fibronectin and ICAM. MEFs isolated from wild-type and knockout mice behave identically in an adhesion assay (unpublished observation). This may be a consequence of a relatively higher level of Rap1B in these cells, but it is also possible that the higher basal level of adhesion obscures differences between both cell populations.
As stated above, Rap1A deficiency appears not to result in defective immune functions. This clearly contrasts with findings in mice in which the Rap1 effector RAPL has been disrupted (17). Here, both cell trafficking and homing of lymphocytes are severely affected. Also, in LFA-I knockout mice these functions are compromised (1).
One of the other pathways in which Rap1 is involved is triggered by activation of the TCR and leads to the stimulation of proliferation of T cells. Our in vitro studies revealed a significant defect in proliferation after CD3 stimulation of Rap1A−/− cells. Strikingly, this effect was not as pronounced if T cells were stimulated simultaneously with CD3 and CD28 (Fig. 6A). Previously, T-cell anergy has been claimed to rely on a block in IL-2 production as a result of activated Rap1 (2). However, in an in vivo model it was shown that expression of activated Rap1 resulted in an enhanced immune response with no detectable proliferative defects (28). On the other hand, examples of knockouts of ADAP genes working down from the TCR, such as Fyb/Slap (12) or Slap-130Fyb (24), show that defects in integrin activation correlate with impaired IL-2 production and cell proliferation. It is possible that weaker proliferation responses of Rap1−/− T cells are a secondary consequence of adhesion defect.
In summary, our studies define a role for Rap1A in integrin-mediated adhesion, but the lack of Rap1A does not lead to severe immunological phenotypes. Since it is likely that Rap1B successfully substitutes for Rap1A in many cells, generation of Rap1 double mutants will help to further define the physiological role of these Ras-like GTPases.
Acknowledgments
J.S. acknowledges support received from the Deutsche Krebshilfe grant 10-1872. T.Z. acknowledges support received from BioSapiens Network of Excellence funded by the European Commission within its FP6 Programme under the thematic area “Life Sciences, Genomics, and Biotechnology for Health” (contract number LHSG-CT-2003-503265).
We thank Johanes L. Bos for valuable discussions and a critical reading of the manuscript. We thank Véronique Pizon for providing Rap1A-specific antibody. M.D. thanks Reinchard Marks for providing CD3 and CD28 antibodies. We thank Claudia Orelio for helpful comments on the analysis of hematopoietic cells.
REFERENCES
- 1.Berlin-Rufenach, C., F. Otto, M. Mathies, J. Westermann, M. J. Owen, A. Hamann, and N. Hogg. 1999. Lymphocyte migration in lymphocyte function-associated antigen (LFA)-1-deficient mice. J. Exp. Med. 189:1467-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boussiotis, V. A., G. J. Freeman, A. Berezovskaya, D. L. Barber, and L. M. Nadler. 1997. Maintenance of human T-cell anergy: blocking of IL-2 gene transcription by activated Rap1. Science 278:124-128. [DOI] [PubMed] [Google Scholar]
- 3.Caron, E., A. J. Self, and A. Hall. 2000. The GTPase Rap1 controls functional activation of macrophage integrin αMβ2 by LPS and other inflammatory mediators. Curr. Biol. 10:974-978. [DOI] [PubMed] [Google Scholar]
- 4.Chien, Y., and M. A. White. 2003. RAL GTPases are linchpin modulators of human tumour-cell proliferation and survival. EMBO Rep. 4:800-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chrzanowska-Wodnicka, M., S. S. Smyth, S. M. Schoenwaelder, T. H. Fischer, and G. C. White II. 2005. Rap1b is required for normal platelet function and hemostasis in mice. J. Clin. Investig. 115:680-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crittenden, J. R., W. Bergmeier, Y. Zhang, C. L. Piffath, Y. Liang, D. D. Wagner, D. E. Housman, and A. M. Graybiel. 2004. CalDAG-GEFI integrates signaling for platelet aggregation and thrombus formation. Nat. Med. 10:982-986. [DOI] [PubMed] [Google Scholar]
- 7.Cullere, X., S. K. Shaw, L. Andersson, J. Hirahashi, F. W. Luscinskas, and T. N. Mayadas. 2005. Regulation of vascular endothelial barrier function by Epac, a cAMP-activated exchange factor for Rap GTPase. Blood 105:1950-1955. [DOI] [PubMed] [Google Scholar]
- 8.Enserink, J. M., L. S. Price, T. Methi, M. Mahic, A. Sonnenberg, J. L. Bos, and K. Tasken. 2004. The cAMP-Epac-Rap1 pathway regulates cell spreading and cell adhesion to laminin-5 through the α3β1 integrin but not the α6β4 integrin. J. Biol. Chem. 279:44889-44896. [DOI] [PubMed] [Google Scholar]
- 9.Fukuhara, S., A. Sakurai, H. Sano, A. Yamagishi, S. Somekawa, N. Takakura, Y. Saito, K. Kangawa, and N. Mochizuki. 2005. Cyclic AMP potentiates vascular endothelial cadherin-mediated cell-cell contact to enhance endothelial barrier function through an Epac-Rap1 signaling pathway. Mol. Cell. Biol. 25:136-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gotoh, T., S. Hattori, et al. 1995. Identification of Rap1 as a target for the Crk SH3 domain-binding guanine nucleotide-releasing factor C3G. Mol. Cell. Biol. 15:6746-6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gotoh, T., Y. Niino, M. Tokuda, O. Hatase, S. Nakamura, M. Matsuda, and S. Hattori. 1997. Activation of R-Ras by Ras-guanine nucleotide-releasing factor. J. Biol. Chem. 272:18602-18607. [DOI] [PubMed] [Google Scholar]
- 12.Griffiths, E. K., C. Krawczyk, Y. Y. Kong, M. Raab, S. J. Hyduk, D. Bouchard, V. S. Chan, I. Kozieradzki, A. J. Oliveira-Dos-Santos, A. Wakeham, P. S. Ohashi, M. I. Cybulsky, C. E. Rudd, and J. M. Penninger. 2001. Positive regulation of T-cell activation and integrin adhesion by the adapter Fyb/Slap. Science 293:2260-2263. [DOI] [PubMed] [Google Scholar]
- 13.Hogan, C., N. Serpente, P. Cogram, C. R. Hosking, C. U. Bialucha, S. M. Feller, V. M. Braga, W. Birchmeier, and Y. Fujita. 2004. Rap1 regulates the formation of E-cadherin-based cell-cell contacts. Mol. Cell. Biol. 24:6690-6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishida, D., H. Yang, K. Masuda, K. Uesugi, H. Kawamoto, M. Hattori, and N. Minato. 2003. Antigen-driven T-cell anergy and defective memory T-cell response via deregulated Rap1 activation in SPA-1-deficient mice. Proc. Natl. Acad. Sci. USA 100:10919-10924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katagiri, K., M. Hattori, N. Minato, S. Irie, K. Takatsu, and T. Kinashi. 2000. Rap1 is a potent activation signal for leukocyte function-associated antigen 1 distinct from protein kinase C and phosphatidylinositol-3-OH kinase. Mol. Cell. Biol. 20:1956-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katagiri, K., M. Hattori, N. Minato, and T. Kinashi. 2002. Rap1 functions as a key regulator of T-cell and antigen-presenting cell interactions and modulates T-cell responses. Mol. Cell. Biol. 22:1001-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katagiri, K., N. Ohnishi, K. Kabashima, T. Iyoda, N. Takeda, Y. Shinkai, K. Inaba, and T. Kinashi. 2004. Crucial functions of the Rap1 effector molecule RAPL in lymphocyte and dendritic cell trafficking. Nat. Immunol. 5:1045-1051. [DOI] [PubMed] [Google Scholar]
- 18.Kawasaki, H., G. M. Springett, et al. 1998. A Rap guanine nucleotide exchange factor enriched highly in the basal ganglia. Proc. Natl. Acad. Sci. USA 95:13278-13283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLeod, S. J., A. H. Li, R. L. Lee, A. E. Burgess, and M. R. Gold. 2002. The Rap GTPases regulate B-cell migration toward the chemokine stromal cell-derived factor-1 (CXCL12): potential role for Rap2 in promoting B-cell migration. J. Immunol. 169:1365-1371. [DOI] [PubMed] [Google Scholar]
- 20.Ohba, Y., K. Ikuta, A. Ogura, J. Matsuda, N. Mochizuki, K. Nagashima, K. Kurokawa, B. J. Mayer, K. Maki, J. Miyazaki, and M. Matsuda. 2001. Requirement for C3G-dependent Rap1 activation for cell adhesion and embryogenesis. EMBO J. 20:3333-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohba, Y., N. Mochizuki, K. Matsuo, S. Yamashita, M. Nakaya, Y. Hashimoto, M. Hamaguchi, T. Kurata, K. Nagashima, and M. Matsuda. 2000. Rap2 as a slowly responding molecular switch in the Rap1 signaling cascade. Mol. Cell. Biol. 20:6074-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohba, Y., N. Mochizuki, A. Yamashita, M. S. Chan, J. W. Schrader, S. Hattori, K. Nagashima, and M. Matsuda. 2000. Regulatory proteins of R-Ras, TC21/R-Ras2, and M-Ras/R-Ras3. J. Biol. Chem. 275:20020-20026. [DOI] [PubMed] [Google Scholar]
- 23.Otto, F., A. P. Thornell, T. Crompton, A. Denzel, K. C. Gilmour, I. R. Rosewell, G. W. Stamp, R. S. Beddington, S. Mundlos, B. R. Olsen, P. B. Selby, and M. J. Owen. 1997. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89:765-771. [DOI] [PubMed] [Google Scholar]
- 24.Peterson, E. J., M. L. Woods, S. A. Dmowski, G. Derimanov, M. S. Jordan, J. N. Wu, P. S. Myung, Q. H. Liu, J. T. Pribila, B. D. Freedman, Y. Shimizu, and G. A. Koretzky. 2001. Coupling of the TCR to integrin activation by Slap-130/Fyb. Science 293:2263-2265. [DOI] [PubMed] [Google Scholar]
- 25.Price, L. S., A. Hajdo-Milasinovic, J. Zhao, F. J. Zwartkruis, J. G. Collard, and J. L. Bos. 2004. Rap1 regulates E-cadherin-mediated cell-cell adhesion. J. Biol. Chem. 279:35127-35132. [DOI] [PubMed] [Google Scholar]
- 26.Rangarajan, S., J. M. Enserink, H. B. Kuiperij, J. de Rooij, L. S. Price, F. Schwede, and J. L. Bos. 2003. Cyclic AMP induces integrin-mediated cell adhesion through Epac and Rap1 upon stimulation of the β2-adrenergic receptor. J. Cell Biol. 160:487-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reedquist, K. A., E. Ross, E. A. Koop, R. M. Wolthuis, F. J. Zwartkruis, Y. van Kooyk, M. Salmon, C. D. Buckley, and J. L. Bos. 2000. The small GTPase, Rap1, mediates CD31-induced integrin adhesion. J. Cell Biol. 148:1151-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sebzda, E., M. Bracke, T. Tugal, N. Hogg, and D. A. Cantrell. 2002. Rap1A positively regulates T cells via integrin activation rather than inhibiting lymphocyte signaling. Nat. Immunol. 3:251-258. [DOI] [PubMed] [Google Scholar]
- 29.Shimonaka, M., K. Katagiri, T. Nakayama, N. Fujita, T. Tsuruo, O. Yoshie, and T. Kinashi. 2003. Rap1 translates chemokine signals to integrin activation, cell polarization, and motility across vascular endothelium under flow. J. Cell Biol. 161:417-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shipitsin, M., and L. A. Feig. 2004. RalA but not RalB enhances polarized delivery of membrane proteins to the basolateral surface of epithelial cells. Mol. Cell. Biol. 24:5746-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka, S., T. Morishita, Y. Hashimoto, S. Hattori, S. Nakamura, M. Shibuya, K. Matuoka, T. Takenawa, T. Kurata, K. Nagashima, et al. 1994. C3G, a guanine nucleotide-releasing protein expressed ubiquitously, binds to the Src homology 3 domains of CRK and GRB2/ASH proteins. Proc. Natl. Acad. Sci. USA 91:3443-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsukamoto, N., M. Hattori, et al. 1999. Rap1 GTPase-activating protein SPA-1 negatively regulates cell adhesion. J. Biol. Chem. 274:18463-18469. [DOI] [PubMed] [Google Scholar]
- 33.Wittchen, E. S., R. A. Worthylake, P. Kelly, P. J. Casey, L. A. Quilliam, and K. Burridge. 2005. Rap1 GTPase inhibits leukocyte transmigration by promoting endothelial barrier function. J. Biol. Chem. 280:11675-11682. [DOI] [PubMed] [Google Scholar]
- 34.Yamashita, S., N. Mochizuki, et al. 2000. CalDAG-GEFIII activation of Ras, R-ras, and Rap1. J. Biol. Chem. 275:25488-25493. [DOI] [PubMed] [Google Scholar]