Abstract
Rec2 is the single Rad51 paralog in Ustilago maydis. Here, we find that Rec2 is required for radiation-induced Rad51 nuclear focus formation but that Rec2 foci form independently of Rad51 and Brh2. Brh2 foci also form in the absence of Rad51 and Rec2. By coprecipitation from cleared extracts prepared from Escherichia coli cells expressing the proteins, we found that Rec2 interacts physically not only with Rad51 and itself but also with Brh2. Transgenic expression of Brh2 in rec2 mutants can effectively restore radiation resistance, but the frequencies of spontaneous Rad51 focus formation and allelic recombination are elevated. The Dss1-independent Brh2-RPA70 fusion protein is also active in restoring radiation sensitivity of rec2 but is hyperactive to an extreme degree in allelic recombination and in suppressing the meiotic block of rec2. However, the high frequency of chromosome missegregation in meiotic products is an indicator of a corrupted process. The results demonstrate that the importance of Rec2 function is not only in stimulating recombination activity but also in ensuring that recombination is properly controlled.
Repair of DNA damaged by double-strand breakage or replication fork collapse can take advantage of a homologous sequence for use as a template in directing accurate correction. Rad51 provides the essential homologous pairing and DNA strand exchange activity required for homology-directed or recombinational repair (53). In mitotic cells, Rad51 alone is sufficient to power homologous pairing (47), while in meiotic cells, Dmc1 can contribute through its own innate homologous-pairing activity (6, 43). Accumulating evidence points to a choreographed interaction between Rad51 and BRCA2 as a critical mechanism governing recombinational repair (25, 40, 48). Assembly of Rad51 into its catalytically active form, the nucleoprotein filament generated through Rad51 polymerization on single-stranded DNA, appears to be regulated both positively and negatively by BRCA2. There is evidence for control at three levels of Rad51 filament dynamics. Biochemical analyses using Brh2, the BRCA2-related protein from Ustilago maydis, demonstrate that it can function to nucleate Rad51 assembly at the site of a double-strand/single-strand DNA junction, the prerequisite structure for recombinational repair arising from resection of a double-strand DNA end to reveal a protruding 3′ single-stranded tail (60). On the other hand, molecular genetic experimentation using U. maydis (31) as well as biochemical studies using synthetic peptides modeling BRC elements from the human BRCA2 (21) suggest a role in organizing or stabilizing Rad51 filaments. In contrast, work with BRC peptides has also provided evidence for a role in filament disassembly (14), interference with Rad51 focus formation (11), and inhibition of recombination and repair (51), although these effects could have resulted from the very high levels of BRC peptides used.
Regulated assembly of the filament appears balanced on the one hand by the interaction of Rad51 with BRCA2's BRC-containing domain, located in the central third of the primary sequence (5, 7, 12), and on the other hand with a second domain located at the extreme C terminus of BRCA2 (CRE [C-terminal Rad51-interacting element]) (see below) (44). The BRC domain comprises eight reiterated sequences of about 30 amino acids each whose structure has been proposed to mimic an element in Rad51 that provides a critical determinant at the polymerization interface gluing Rad51 molecules into a chain (39, 46). Rad51 interaction with the C-terminal domain is controlled by phosphorylation of a key BRCA2 residue that is targeted by cyclin-dependent kinases (15). Proper Rad51 filament assembly at DNA sites of repair requires a precisely coordinated interplay between BRCA2's Rad51-interacting domains and its DNA/DSS1 binding domain. The latter consists of a tandem array of oligonucleotide/oligosaccharide binding (OB) folds, with a double-helical tower emerging from one topped by a helix-turn-helix, and a helical domain that is laced to the adjacent OB folds by the intertwining small acidic protein DSS1 (59). The recent reports that BRCA2 from Arabidopsis thaliana interacts with Dmc1 as well as with Rad51 (49), together with the observation that an N-terminal fragment of the BRCA2-related protein CeBRC-2 from Caenorhabditis elegans interacts with the Rad51 paralog RFS-1 (36), raise the notion that the regulatory circuitry of BRCA2 in recombinational repair extends more broadly in scope than previously considered.
The Ustilago maydis Brh2 protein is a streamlined version of the mammalian BRCA2 protein exhibiting a similar modular arrangement (28). There is only a single BRC element and a more circumscribed DNA/Dss1 binding domain. A C-terminal Rad51-interacting domain is present but has not yet been characterized (Q. Zhou, M. Kojic, and W. K. Holloman, unpublished observations). The Rad51 ortholog of U. maydis, on the other hand, is highly conserved and is very similar in sequence and size to the mammalian counterparts (17). But peculiar to U. maydis is Rec2, the single but very divergent Rad51 paralog (42). In most systems that have been examined in detail, for instance, vertebrates, plants, flies, budding and fission yeasts, and other fungi whose genomes have been sequenced, the Rad51-related proteins are more elaborated in number. There are exceptions, such C. elegans, in which there appears to be only a single Rad51-related gene, in this case of unknown function (8), but in general, it seems that there are multiple mitotically expressed paralogs plus a meiosis-specific Dmc1-like protein. Thus, it might be speculated that the single paralog represented by Rec2 is an indicator of a prototypic or more primal recombinational repair system in U. maydis (see the annotated genome at http://mips.gsf.de/genre/proj/ustilago/). In any event, inactivation of any of the above-described proteins, Rec2, Rad51, or Brh2, results in a similar phenotype of profound loss of resistance to DNA clastogens, deficiency in mitotic recombination and mutation avoidance, and failure to complete meiosis, and furthermore, epistasis analysis has indicated a common pathway of operation (3, 17, 23, 28).
In line with the BRCA2 paradigm developed in higher organisms, interaction between U. maydis Brh2 and Rad51 has been established by a combination of biochemical studies involving copurification of the proteins and affinity pull-down procedures (60). Evidence for a physical interaction between Rec2 and Rad51 has also been obtained by yeast two-hybrid analysis, which again is in accord with the emerging paradigm of Rad51 interplay with paralogs (29). Thus, Rad51, as the ultimate executor in catalyzing DNA strand exchange, appears to be organized and directed to an appropriate and active condition for homologous pairing through interplay with Brh2 and Rec2. In other systems, the meiotically expressed Dmc1 has been shown to be capable of supporting strand invasion in an ATP-dependent manner (43), but to date, there is no evidence that any of the other mitotically or somatically expressed paralogs can promote this reaction (32, 33). In contrast, biochemical studies have revealed that Rec2 itself can catalyze homologous pairing and strand exchange reactions with cofactor requirements similar to that of Rad51 (4), thus raising questions about the nature of their hierarchical arrangement and their functional dedication.
An intriguing overlap was observed in the phenotype of the brh2 mutant and the dominant-negative rec2-197 allele with respect to meiotic chromosome segregation. Using heterozygous crosses involving the brh2 null mutant, the meiotic progeny exhibited extremely high frequencies of aneuploidy (28). This was reminiscent of the earlier observations made with heterozygous crosses involving rec2-197, which was considered to exert its dominant-negative effect through an interaction between an N-terminal amino acid stretch of 174 residues expressed by this allele and one or more presumed members of the Rec2 interactive network (29). Rad51 was the first interacting partner of Rec2 to be identified, and preliminary testing of it with the Rec2 N-terminal domain by yeast two-hybrid analysis provided evidence for the interaction as well (29). Unfortunately, determining whether there was an interaction with Brh2 or establishing the identity of any additional Rec2-interacting partners proved to be problematical by two-hybrid methodology, as there was elevated transcriptional activation of the reporter gene caused by fusion of the N-terminal Rec2 polypeptide with the Gal4 DNA binding domain.
Given the biochemical evidence that Rec2 resembles Rad51 in possessing an innate homologous-pairing activity plus the genetic evidence for an epistatic relationship between the structural genes in DNA repair, together with the idea that chromosome disjunction during meiosis might present a point of intersection of both Brh2 and Rec2 function, we were curious to learn more about the interplay between the two. Here, we have analyzed molecular genetic and physical relationships between Rec2 and Brh2. We find that Rec2 interacts directly with Brh2 as well as with Rad51 and that the requirement for Rec2 in repair and recombination can be partially bypassed by increasing the level of expression of Brh2. The findings suggest that Rec2 functions in establishing the assembly of the Rad51 filament.
MATERIALS AND METHODS
U. maydis strains and methods.
Manipulations with U. maydis, culture methods, gene transfer procedures, and survival after irradiation have been described previously (19, 24, 28). Haploid strains utilized for testing survival after irradiation or in crosses to measure meiotic recombination included UCM54 (rec2-1 pan1-1 nar1-1 a1b1), UCM350 (pan1-1 nar1-6 a1b1), UCM565 (Δbrh2 pan1-1 nar1-6 a1b1) UCM567 (nar1-1 a2b2), UCM626 (rec2-53 nar1-6 a2b2), and UCM628 (Δrad51 pan1-1 nar1-6 a1b1). Mitotic allelic recombination was measured in UCM96 (ino1-4/ino1-5 pan1-1/+ met1-2/+ nic1-1/+ ade1-1/+)- and UCM110 (ino1-4/ino1-5 rec2-1/rec2-1 pan1-1/+ met1-2/+ nic1-1/+ ade1-1/+)-transformed derivatives as Ino+ prototroph formation after plating cultures onto inositol-free medium. The rec2 mutant alleles rec2-1 and rec2-53 are both deletions resulting in a complete loss of function and have been described previously (29). Meiotic allelic recombination was measured after germinating teliospores on rich medium (1% yeast extract, 2% peptone, 2% sucrose [YEPS]) for 36 h at 30°C. Cells were collected and spread for random-products analysis on minimal medium with supplements and containing nitrate as the sole source of nitrogen. Nar+ recombinants were identified after incubation for 5 days. ino, met, nic, pan, nar, and ab indicate auxotrophic requirements for inositol, methionine, nicotinic acid, and pantothenic acid, inability to metabolize nitrate, and mating type loci, respectively. Green fluorescent protein (GFP) fluorescence microscopy and differential interference contrast imaging of live cells were performed on cells transformed with a set of self-replicating plasmids expressing GFP-Rad51 (pCM1001), GFP-Rec2 (pCM1049), or GFP-Brh2 (pCM1053) as previously described (34). In each case, the same vector containing the gap promoter for driving expression of the GFP-tagged protein and harboring a gene for hygromycin resistance for selection was used. Cells were irradiated with UV by a germicidal lamp emitting at 254-nm or by gamma rays delivered from a 60Co source. Approximately 200 cells of each genotype were inspected for focus formation before and at each time point after irradiation.
Plasmid construction and methods.
Plasmid pMAL-C2 (New England Biolabs) derivatives were engineered to express target proteins in Escherichia coli as C-terminal fusions of maltose binding protein (MBP). For expression of MBP-Brh2, the fusion was from Brh2 amino acid residues 106 to 1075, (Brh2106-1075). For expression of MBP-BRC, the fusion included a 91-residue peptide, Brh2260-350. Point mutations F294A and T296A were introduced into the BRC peptide by site-directed mutagenesis using oligonucleotides and standard procedures. For expression of MBP-Rec2, the fusion utilized the complete open reading frame Rec21-781 derived from pCM538 (4). The open reading frame for Rec2 was introduced into pET28a (Novagen) for expression of the protein with an N-terminal His tag. Rec2-NT, the Rec2 N-terminal fragment (Rec21-174), was expressed in pET28a as a His tag fusion. Full-length Rad511-339 was expressed without a tag in pACYC-Duet (Novagen) or with a His tag in pET28a. Several plasmids used for protein expression in U. maydis have been described previously. Self-replicating pUC-based vectors contain a U. maydis ARS, the glyceraldehyde-3-phosphate dehydrogenase promoter (gap) for driving expression, and a gene expressing hygromycin resistance as a selectable marker. These plasmids and the proteins expressed (in parentheses) are as follows: pCM955 (empty vector), pCM973 (Brh2), pCM1023 (Brh2-RPA70), pCM1030 (Rad51), pCM1031 (Rec2), and pCM1040 (RPA70). Derivatives expressing Rad51, Rec2, and Brh2 as GFP fusion proteins were constructed by inserting a cassette to fuse GFP to the N terminus of each of these proteins. A set of plasmids derived from the nonreplicating vector pCM691 (27) was used for the construction of strains with an integrated transgene expressing the same proteins described above. These plasmids contained the gap promoter for driving expression of the target protein and a selectable marker for resistance to carboxin. The level of protein expressed from one of the self-replicating plasmids was virtually the same as that from an integrated transgene judged by the almost-equal complementation of the radiation sensitivity of any of the tester strains. For simplicity in nomenclature, U. maydis strains expressing a protein from a transgene, whether on a self-replicating plasmid or integrated into the genome, will be denoted as mutant/protein expressed, e.g., rec2/Rad51, indicating rec2 mutant cells expressing Rad51 without any tag, or rec2/GFP-Rec2, indicating rec2 mutant cells expressing GFP-tagged Rec2, etc.
Coprecipitation/pull-down analysis.
E. coli strain BL21(DE3) (Novagen) cells cotransformed with pairs of plasmids expressing proteins with MBP or hexahistidine affinity tags were grown in Luria broth with appropriate antibiotics to mid-log phase, induced with isopropyl-β-d-galactoside, and cultured at 16 or 23°C to maximize solubility of expressed proteins. Harvested cells were crushed by passage through a French press in a solution of 50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 0.5% NP-40, 2 mM MgCl2, benzonase nuclease (10 units; Novagen), and 20% glycerol and centrifuged at 40,000 × g for 30 min. A slurry of affinity resin, either nitrilotriacetic acid-agarose (QIAGEN) charged with Ni2+ or amylose-agarose beads (100 μl; New England Biolabs), was mixed with 500 μl cell extract, and the beads were collected, washed, and then eluted with a solution of 80 μl 50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, and 0.5% NP-40 containing either 10 mM maltose or 250 mM imidazole. Western blotting was performed after electrophoretic transfer of proteins to polyvinylidene difluoride membranes with anti-MBP antiserum (New England Biolabs), His-tagged monoclonal antibody (Novagen), or affinity-purified rabbit polyclonal anti-Rad51 antibodies raised against U. maydis Rad51 protein. Complexes were visualized by chemiluminescence with horseradish peroxidase-coupled secondary antibodies (Amersham Biosciences).
RESULTS
DNA damage-induced focus formation.
It has been well established in a number of experimental systems that Rad51 assembles into intranuclear foci along with other recombinational repair proteins in response to DNA damage. GFP-tagged versions of the corresponding proteins have been found to respond similarly (16, 34, 61), supporting the notion that the intracellular behavior of the tagged proteins accurately depicts the true biological activity. The foci are thought to mark sites of active recombinational repair and are also observed at a low frequency in undamaged cells, likely reflecting the recruitment of the repair proteins to spontaneously damaged sites, such as those arising from replication fork collapse. As in the cases of mammalian and yeast systems (16, 34, 61), modification of U. maydis Rad51 by GFP tagging has an effect on the ability to complement the radiation sensitivity of rad51 mutant cells (FIG 1). However, significant biological activity remains, and there is no dominant-negative interference when GFP-Rad51 is expressed in wild-type cells, supporting the general consensus that the behavior of the fusion protein is biologically relevant. Previously, we observed that GFP-Rad51 focus formation after DNA damage by ionizing radiation was eliminated in both brh2 and dss1 mutants (31). Since Brh2 serves to nucleate Rad51 filament formation during DNA strand exchange reactions in vitro (60) and since Dss1 forms a tight complex with Brh2 and is required for its activation (30, 31), the implication is that the physical interaction between Brh2 and Rad51 is important for recruiting Rad51 to sites of DNA damage.
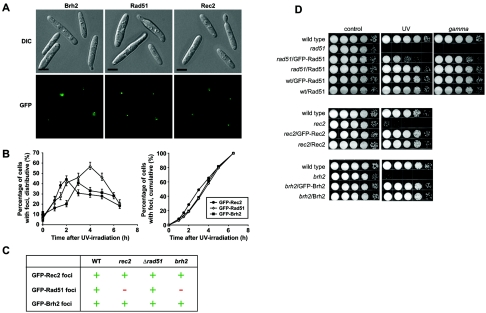
FIG. 1.
Focus formation. Wild-type (WT) (UCM350) cells expressing GFP-Rad51, Rec2, or Brh2 were viewed by differential interference contrast (DIC) imaging and fluorescence microscopy without fixation. (A) Cells were examined for focus formation 4 h after irradiation with UV (30 J/m2). Cells with representative Brh2, Rad51, or Rec2 foci are shown. Typically, cells had one to two foci. Bar indicates 3 μm. (B) The fraction of cells with foci at each time point was determined after counting approximately 200 cells. The cumulative representations of each distribution are shown on the right. (C) rec2 (UCM54), brh2 (UCM565), or rad51 (UCM628) mutant cells expressing GFP-Rad51, Rec2, or Brh2 were examined for foci at 2 and 4 h as described above. +, competent in focus formation; −, no focus formation. (D) Survival of rad51, rec2, and brh2 mutant strains expressing GFP-tagged or untagged Rad51, Rec2, or Brh2, respectively, after irradiation with UV (120 J/m2) and, in the case of rad51, with gamma rays (400 Gy). Serial 10-fold dilutions of cell suspensions were spotted from left to right as shown.
While Brh2 (and Dss1) is necessary for Rad51 focus formation, we were interested to learn if it was sufficient. Following DNA damage induced with UV irradiation, cells with Rad51 foci accumulated over a period of several hours, with the maximal fraction of focus-containing cells in the population occurring approximately 4 h posttreatment (Fig. 1). These kinetics are somewhat delayed with respect to the time course observed previously with Rad51 foci formed following gamma irradiation (31), probably reflecting differences in the nature of the DNA damage and in the cellular response to the different types of damage. No Rad51 foci were observed in the rec2 mutant following irradiation with UV. As the foci represent aggregates of Rad51 that presumably reflect the formation of nucleoprotein filaments at sites of DNA damage or replication fork collapse, this result could suggest a role for Rec2 in promoting the establishment of Rad51 filaments.
By this same logic, if one assumes that the physical interaction between Rad51 and Rec2, observed previously by two-hybrid analysis, is functionally important, then it might be predicted that recruitment of Rec2 to damage-induced foci would be dependent on Rad51 and, in turn, ultimately dependent on Brh2. To investigate this notion, we monitored Rec2 focus formation in cells expressing the GFP-tagged protein, which is fully active in complementing the UV sensitivity of the rec2 mutant (data not shown). As in the case of Rad51, in the absence of exogenous DNA damage, cells expressing GFP-Rec2 exhibited diffuse fluorescence within the nucleus. Rec2 foci were evident only in a small fraction of cells in the population in normal growth, but following DNA damage by UV light, the number of cells with Rec2 foci increased substantially. By visual comparison, the Rec2 foci were less intense than Rad51 foci. The time course of cells accumulating Rec2 foci was not completely coincident with that of Rad51 but appeared slightly advanced by comparison (Fig. 1B). After converting the data to a cumulative form to enable higher precision (38), it also seemed apparent that there was displacement of the fraction of cells with Rec2 foci to an earlier stage relative to cells with Rad51 foci. This observation could be interpreted to mean that Rec2 is recruited to sites of damage slightly earlier than Rad51, although other interpretations are possible. By the same token, the cumulative curve of Brh2 focus formation was almost coincident with that of Rad51. As it is likely that Rad51 is more abundant than Brh2, given Brh2's activity in nucleating assembly of Rad51 filaments, we tend to favor the view that Rec2 has some priority in recruitment to damaged sites.
When the genetic requirements were examined, it was found that Rec2 focus formation was independent not only of Rad51 but also of Brh2, as focus formation was completely competent in the deletion mutants following radiation damage (Fig. 1C). Similarly, Brh2 foci formed in the absence of Rad51 as well as in the absence of Rec2. Again, the GFP modification had no effect on the ability to complement the UV sensitivity of the brh2 mutant. Thus, Rad51 is dependent on Rec2 as well as on Brh2 for focus formation, but neither Rec2 nor Brh2 has a requirement for Rad51. These findings imply that both Rec2 and Brh2 can access sites in DNA requiring repair in an autonomous fashion. A similar observation was made with the BRCA2-related protein CeBRC-2 from C. elegans, which was also found to form foci after DNA damage independently of Rad51 (36).
Rec2 interaction with Brh2.
A question of interest was whether Rec2 can associate with Brh2. We initially tested for an interaction between Brh2 and Rec2 using yeast two-hybrid methodology. However, an interpretation of the findings was not straightforward due to the problematical behavior of the Gal4 fusion proteins. Therefore, we turned to coprecipitation or pull-down procedures to test for an interaction between Brh2 and Rec2 directly. We were unable to obtain clear, reproducible signals from affinity-tagged proteins expressed in U. maydis, likely due to the high levels of vicious proteases released when U. maydis cells were ruptured, and so we switched our efforts to expressing the proteins in E. coli. The procedure was to coexpress the proteins in E. coli, capture appropriately tagged proteins on affinity beads after opening the cells, and then identify interacting proteins by Western blotting. As a validation control, we tested the system by checking for interactions between Rec2 and Rad51, since we had previously determined that these two proteins interact using the two-hybrid procedure (29). In this case, Rec2 was tagged with a hexahistidine leader sequence (His-Rec2), while Rad51 was untagged. Previous work had already established that the His tag had no negative effect on the DNA repair activity of Rec2 (4). Therefore, extracts were prepared from cells expressing the proteins and spun at high speed to remove insoluble material. His-Rec2 was selectively captured using nitrilotriacetic acid-agarose charged with Ni2+, bound material was then eluted using imidazole, and proteins were separated by gel electrophoresis. By Western blotting with anti-Rad51 antibodies, it was evident that Rad51 associated with Rec2 (Fig. 2A). In addition, Rad51 was found to associate with the Rec2 N-terminal domain when expressed separately as a His fusion (His-Rec2 NT), although based on the relative intensities of the bands, it would appear that the interaction between Rad51 and the Rec2 N-terminal domain is considerably weaker than that with full-length Rec2. To eliminate any complications in interpretation as a result of mutual, adventitious binding to DNA, all preparations were treated with benzonase nuclease to hydrolyze nucleic acids, but we observed no difference in pull-down patterns regardless of the benzonase treatment. The results obtained conform to the yeast two-hybrid results and thus confirm the Rec2-Rad51 interaction.
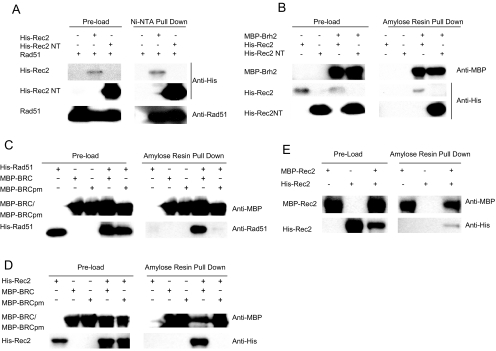
FIG. 2.
Rec2 interactions. E. coli strains expressing the indicated proteins were processed as described in Materials and Methods. After specific elution from affinity resins, samples were electrophoresed in 10% polyacrylamide gels containing sodium dodecyl sulfate and analyzed by Western blotting. (A) His-Rec2 and His-Rec2 NT (Rec21-174) interact with Rad51. NTA, nitrilotriacetic acid. (B) MBP-Brh2 interacts with His-Rec2 and His-Rec2 NT (Rec21-174). (C) His-Rad51 interacts with MBP-BRC (Brh2260-330) but not with MBP-BRCpm (point mutations FT294 and 296AA). (D) His-Rec2 interacts with MBP-BRC (Brh2260-330) but not with MBP-BRCpm (point mutations FT294 and 296AA). (E) His-Rec2 interacts with MBP-Rec2.
A general problem with this approach is the lack of uniformity in expression of the various proteins. Therefore, the interpretation of the results should be taken only in the qualitative sense without attempting to draw any specific conclusions concerning stoichiometries. With this proviso in mind, we addressed the question of whether there is a physical interaction between Rec2 and Brh2 using Brh2 tagged with MBP to pull down His-tagged Rec2. Previously, we established that the MBP tag had little or no negative effect on the DNA repair activity of Brh2 (31). Thus, the procedure devised was to prepare extracts from cells expressing the tagged proteins, mix the soluble supernatant with amylose beads, and then specifically elute MBP-Brh2 complexes from the beads using buffer containing maltose. The identity of the proteins was determined in Western blots using antibodies directed against the MBP tag or His tag. In controls, MBP-Brh2 protein expressed alone was found highly enriched in the amylose bead eluate, whereas His-Rec2 was absent in amylose bead eluates when expressed by itself. When extracts from cells coexpressing MBP-Brh2 and His-Rec2 were tested, His-Rec2 was pulled down with MBP-Brh2, indicating a direct interaction between Brh2 and Rec2 (Fig. 2B). The Rec2 N-terminal domain was also found to interact with Brh2. The His-Rec2 NT was expressed better by comparison to the full-length Rec2, which most likely accounts for why there appears to be more His-Rec2 NT in the pull down with MBP-Brh2 than is the case with the full-length His-Rec2.
Structural studies of Rad51 have identified a key motif in the linker region joining the N-terminal and ATPase domains that serves at the interface between neighboring subunits (1, 13, 39, 46, 58). The motif forms an element that is thought to zip onto the adjacent Rad51 molecule through docking of a phenylalanine residue in the former with a hydrophobic pocket in the latter. A similar ball-and-socket coupling is proposed to be mimicked by the BRC-Rad51 interaction. However, no corresponding docking site is evident by sequence alignment of Rec2, so the basis for the Brh2-Rec2 interaction is not clear. Nevertheless, to determine whether an analogous pocket might be present in Rec2, in spite of the absence of any sequence motif, we tested whether BRC by itself could interact with Rec2. We isolated the BRC motif to an 80-amino-acid stretch of the Brh2 N-terminal region and expressed this as a fusion protein with MBP. In the control, Rad51 was efficiently pulled down by MBP-BRC, although there was a low level of background signal from Rad51 (Fig. 2C). The basis for this is unknown but seems more likely due to Rad51 self-aggregation than any genuine association of Rad51 with the amylose resin. In the experimental case, it was clear that Rec2 could be efficiently pulled down by MBP-BRC although at a somewhat lower efficiency than with Rad51 (Fig. 2D). As a control, we tested whether the BRC element modified by mutations that are known to abrogate the interaction with Rad51 in other systems would lose the ability to associate with Rec2. The phenylalanine and threonine residues F294 and T296, corresponding to amino acids essential in BRC's interaction with Rad51 (11, 14, 39), were changed to alanine. The MBP-BRC fusion with these two point mutations (BRCpm) was severely reduced in its ability to interact with Rad51, as expected (Fig. 2C), and concomitantly was also abrogated in the interaction with Rec2 (Fig. 2D). These findings suggest that a structural feature similar to the Rad51 oligomerization motif docking site could be present in Rec2.
Finally, we wished to confirm our earlier findings, based on two-hybrid analysis, that Rec2 could interact with itself (29). Using MBP-Rec2 in the pull down, it was clear that His-Rec2 could be captured (Fig. 2E). These data provide additional support for the notion that Rec2 is capable of self-interaction.
Suppression of rec2 radiation sensitivity.
In light of the physical interaction between Rec2 and Brh2 plus Rad51's requirement for both Rec2 and Brh2 in DNA damage-induced focus formation, it might be supposed that Rec2 and Brh2 cooperate with each other in some overlapping manner to enable Rad51. If their action is to work as interacting partners, then one might compensate for the loss of the other when present at high levels. Therefore, a question of interest was whether overexpression of Rec2 could suppress the radiation sensitivity of the brh2 mutant and vice versa. For these determinations, we introduced the appropriate gene driven by a strong constitutive promoter into the strain to be tested via a self-replicating plasmid or by integration into the genome. We emphasize that in this set of experiments, it was the native, untagged protein being expressed. Unfortunately, using polyclonal antibodies raised against the recombinant proteins, we were unable to unequivocally detect any of the proteins of interest in crude cell extracts by Western blotting, and so we could not make an assessment of the level of overexpression in any case. However, introduction of each transgene into its corresponding mutant partner completely complemented the phenotype as determined by recovery of radiation resistance (Fig. 3A). Therefore, we were able to conclude that expression of the transgenes is strong enough to provide a level of protein sufficient for normal DNA repair activity.
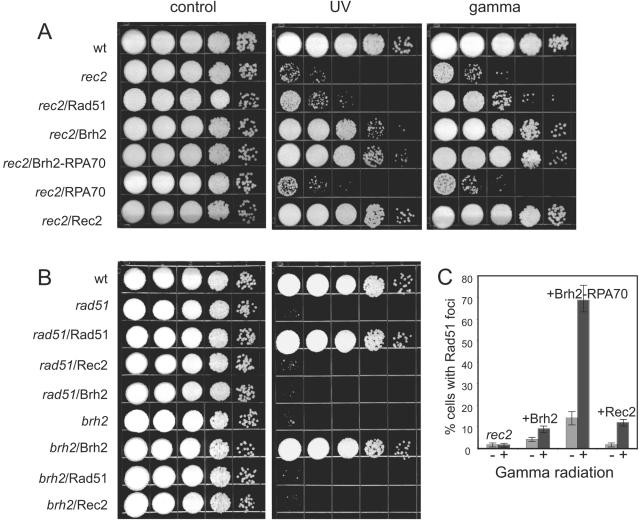
FIG. 3.
Suppression of rec2 radiation sensitivity and restoration of Rad51 focus formation. (A) rec2 strains (UCM54) expressing Rad51, Brh2, Brh2-RPA70, RPA70, or Rec2 were tested for survival after irradiation with UV (120 J/m2) or gamma rays (400 Gy). Serial 10-fold dilutions of cell suspensions were spotted from left to right as shown. wt, wild type. (B) rad51 (UCM628) or brh2 (UCM565) strains expressing untagged Rad51, Rec2, or Brh2 were tested for survival after irradiation with UV as described above. (C) GFP-Rad51 was expressed in rec2 strains with integrated transgenes expressing Brh2, Brh2-RPA70, or Rec2. Cells were monitored for Rad51 focus formation 60 min after administering a 40-Gy dose of gamma rays.
The transgene expressing Rec2 introduced into the brh2 mutant failed to suppress the radiation sensitivity (Fig. 3B). Similarly, no suppression of radiation sensitivity was observed when the transgenes expressing Rec2 were expressed in the rad51 mutant. In contrast, when a transgene expressing Brh2 was introduced in the rec2 mutant, there was substantial recovery of both UV and gamma radiation resistance (Fig. 3A and B). Rad51 was partially active in restoring resistance to gamma radiation when introduced as a transgene in rec2 but weakly active in restoring resistance to UV by comparison with Brh2. With regard to this latter observation, promoter strength may come into play here, since no suppression of rec2's UV sensitivity was noted in a previous study when Rad51 was expressed from its natural promoter (17). In addition, Brh2-RPA70, the Dss1-independent Brh2 variant with the natural DNA binding domain replaced by that from the RPA70 subunit, was very active in suppressing both UV and gamma sensitivity of rec2. This Brh2 fusion was previously reported to be able to complement the radiation sensitivity of brh2 as well as dss1 with remarkable proficiency and to be hyperactive in promoting recombination (31).
As another approach to measure activity in suppressing the rec2 repair deficiency, we examined GFP-Rad51 focus formation in strains expressing Brh2 and Brh2-RPA70 (Fig. 3C). Expression of Brh2 in the rec2 mutant elevated the spontaneous level of GFP-Rad51 focus formation severalfold higher than in a control strain expressing Rec2, while expression of Brh2-RPA70 had a more pronounced effect. After a 40-Gy dose of ionizing radiation, the frequency of cells with GFP-Rad51 foci in the strain expressing Bh2 was similar to that of the Rec2 control when sampled 1 h after irradiation. However, the frequency was much higher when Brh2-RPA70 was expressed, indicating an imbalanced recovery in function. Rec2 expressed in the brh2 mutant had no ability to restore GFP-Rad51 focus formation. These findings suggest that Brh2 and Rec2 are not interchangeable cofactors governing Rad51 but have specific nonoverlapping functions.
Suppression of rec2 recombination deficiency.
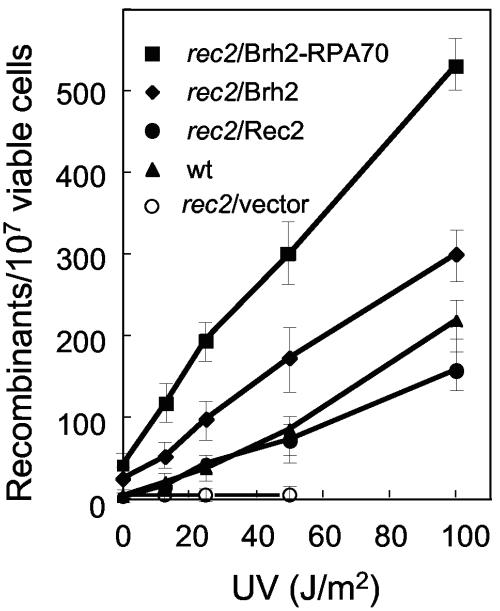
Notwithstanding the increased resistance to radiation and the recovery of damaged-induced Rad51 focus formation afforded to the rec2 mutant by transgene expression of Brh2, it would not necessarily be expected that the integrity of the repair process would be restored to normal in the absence of Rec2. To explore this issue in more detail, we investigated recombination activity in rec2 strains expressing Brh2 or the hyperactive variant Brh2-RPA70. From the early studies conducted by Holliday, it has been known that the rec2 mutant exhibits a nearly normal level of spontaneous recombination between heteroalleles (23). However, unlike the case in the wild type, there is no large increase in recombination following DNA damage in the rec2 mutant cells that survive. When a transgene expressing Rec2 was introduced into a rec2 diploid strain, there was complete complementation of the recombination deficiency as evidenced by the virtually identical dose-dependent responses in recombination following UV irradiation (Fig. 4). In a rec2 strain expressing Brh2, and to an even greater extent in a strain expressing Brh2-RPA70, spontaneous and UV-induced recombination were markedly higher than in the wild type.
FIG. 4.
Hyperactive allelic recombination. rec2/rec2 homozygous diploid strains (UCM110) heteroallelic at the ino1 locus and expressing Rec2, Brh2, or Brh2-RPA70 from a transgene, plus a wild-type control strain (UCM96), were irradiated with increasing doses of UV light and plated onto minimal medium to score Ino+ recombinants or onto YEPS to monitor cell viability. Five independent cultures of each strain were tested. The mean frequencies and standard deviations are shown. wt, wild type.
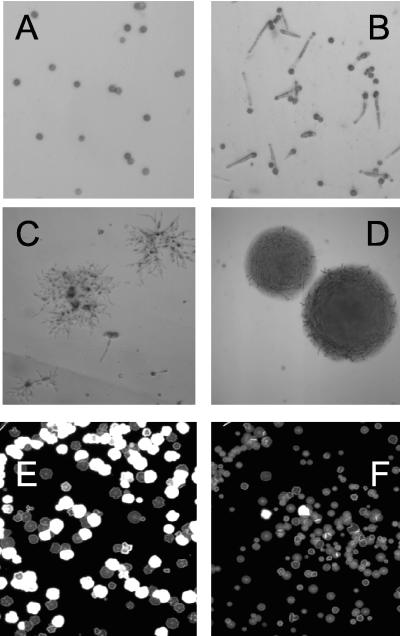
During rec2 meiosis, the process is aborted, presumably due to a failure in repair of Spo11-induced double-strand DNA breaks. Teliospores are produced from matings between compatible haploid strains, and normally, these germinate after about 18 h, with the development of a promycelium or metabasidium that septates into compartments housing each of the four uninucleate meiotic products (37). Haploid basidiospores with an unlimited capacity for growth arise after a postmeiotic division by budding off from the individual metabasidium compartments. In rec2 homozygous crosses, teliospores germinate to form a promycelium, but subsequent processes cease, as evidenced by the lack of septation and the complete failure in formation of viable basidiospores when plated on rich medium.
In matings between rec2 strains in which one parent contained a transgene expressing Brh2, there was no change in the ability of the teliospores to progress further in the meiotic cycle (Table 1). However, expression of Brh2-RPA70 resulted in a significant improvement in meiotic development as measured by basidiospore formation and colony-forming activity, approaching a third of the wild-type level. However, it was clear from the extremely ragged and irregular colony morphology that meiosis in rec2 × rec2/Brh2-RPA70, while enabled, was probably disturbed (Fig. 5). This was evident by an assay for chromosome missegregation that measures heterozygosity at the mating type loci (2). In wild-type crosses, >99% of the meiotic progeny are monosomic at the mating type loci as determined by the infrequent formation of white fuzzy colonies which represent disomes heterozygous at the a and b mating type loci. By contrast, almost half of the viable cells arising from the rec2 × rec2/Brh2-RPA70 cross were disomic for the mating type loci, indicating a significant failure in proper chromosome distribution (compare Fig. 5E and F). We also measured allelic recombination in these meioses but found that the frequency was almost identical to that of the wild type (Table 1). This suggests that in meiosis, Spo11-initiated events are limiting in terms of the extent of recombination regardless of the hyperactive state of Brh2. Thus, recombination can be facilitated in the absence of Rec2 to a considerable extent by Brh2, even more so by the hyperactive variant Brh2-RPA70, and exclusively so by Brh2-RPA70 in meiosis, but the quality of these processes appears corrupted.
TABLE 1.
Suppression of rec2 meiotic deficiency by Brh2-RPA70
| Crossa | Basidiospore viabilityb (%) | Fuz+ progenyc (%) | Recombination frequency (10−4)d |
|---|---|---|---|
| wt × wt | 45 ± 10 | 0.8 | 7.8 |
| rec2 × rec2 | <10−3 | ||
| rec2 × rec2/Brh2 | <10−3 | ||
| rec2 × rec2/Brh2-RPA70 | 15 ± 5 | 46 | 7.6 |
Strains were as follows: wild type (wt) × wt, UCM350 × UCM567; rec2 × rec2, UCM54 × UCM626.
Teliospores were spread on YEPS plates and incubated for 5 days. Colony formation was taken as a measurement of viable basidiospore generation.
A total of 200 to 300 random meiotic products obtained after germinating teliospores for 36 h and then dispersing microcolonies into single cells were spread onto medium containing charcoal. The percentage of colonies that turned white and fuzzy was determined after incubation for 3 days.
Teliospores from UCM350 × UCM567 (wt × wt) and UCM54 × UCM626 (rec2 × rec2/Brh2-RPA70) were germinated on YEPS for 38 h. Microcolonies were collected and dispersed into single cells which were spread onto supplemented minimal medium containing nitrate as the sole nitrogen source to determine recombination and on YEPS to determine viability. Nar+ recombinants were identified as colonies appearing on nitrate minimal medium after 5 days.
FIG. 5.
Meiotic rescue. Teliospores obtained from wild-type and rec2/rec2 homozygous crosses with or without an integrated transgene expressing Brh2-RPA70 were germinated on YEPS at 30°C for 38 h. (A) Teliospores (wild type) before germination viewed at ×1,000 magnification. (B) rec2/rec2 teliospores at 38 h after germination, arrested after promycelium formation. (C) rec2/rec2/Brh2-RPA70 microcolonies at 38 h. (D) Wild-type microcolonies at 38 h. Microcolonies from rec2/rec2/Brh2-RPA70 (E) and the wild type (F) were collected and dispersed to single cells which were spread onto medium containing charcoal to enable fuzz formation. Colonies appearing after incubation for 3 days were recorded. A section from petri dishes with Fuz+ (white) and Fuz− (gray) colonies is shown.
DISCUSSION
As the single Rad51 paralog present in U. maydis, Rec2 provides an integral function to the recombinational repair system powered by Rad51 and regulated by Brh2 and Dss1. Expressed by itself, the N-terminal region of Rec2 interferes with functions normally processed by Brh2 and attendant components such as promoting survival after radiation-induced damage (29). This disturbance is particularly striking in meiotic chromosome disjunction, where the frequency of missegregation caused by the rec2 allele expressing the N-terminal domain is comparable to that in crosses heterozygous for brh2 (28). These genetic interactions have suggested the possibility of some more physical interplay between Rec2 and components of the recombinational repair system. Here, we have found that Rec2, in addition to Rad51, interacts directly with Brh2. Moreover, coprecipitation experiments in this study show that the Rec2 N-terminal domain by itself has the capacity to interact with Brh2, thus providing a mechanistic link rationalizing earlier observations on its ability to interfere with functions performed by Brh2. By sequence comparison, no obvious run of residues resembling the Rad51 polymerization motif is evident in Rec2. Furthermore, since the Rad51 domain(s) important in the interplay with the extreme C-terminal region of BRCA2 (CRE) is not yet defined, it is premature to know whether a related element is present in Rec2. It is possible that structural elements resembling the BRC or the CRE docking site are present in Rec2 and are suitable as binding interfaces but that these are simply unrecognizable from the primary sequence. However, recent analysis of BRC peptides binding to Rad51 filaments by electron microscopy image reconstruction provide evidence for interaction through an undefined site in the N-terminal domain of Rad51 that is distinct from the polymerization interface (21). Perhaps a related region is present in the N-terminal domain of Rec2, and it is this that mediates the interplay with Brh2.
A key point for experimentation in this study was whether there might be functional overlap or redundancy between Brh2 and Rec2. This issue was raised since both Brh2 and Rec2 physically interact with Rad51 and since both are necessary for the formation of DNA damage-induced Rad51 nuclear foci but are able to form foci independently of each other and in the absence of Rad51. The results show that Brh2 and Rec2 are not interchangeable. There is no reciprocity in their ability to compensate for each other. Rather, there is a fixed hierarchy in their interaction with Rad51 to the effect that in the absence of Rec2, cells can survive radiation damage and recover a substantial degree of proficiency in recombination if the level of Brh2 is increased. These observations suggest that Brh2, not Rad51, is limiting in promoting repair and that its role is to enable Rad51 to promote repair while that of Rec2 is to contribute to establishing the quality or integrity of the Rad51 filament. The hyperactivity of the Brh2-RPA70 fusion in mitosis and meiosis, which is partly a consequence of its freedom from Dss1 regulation, is an indication of Rad51's innate promiscuity in promoting recombination once it gains access to DNA.
Our observations are reminiscent of previous studies on phenotype suppression in Saccharomyces cerevisiae rad55/rad57 and dmc1 mutants and in chicken DT40 cell lines with a deletion of the Rad51 paralogs. In S. cerevisiae, the two Rad51 paralogs, Rad55 and Rad57, are required for resistance to ionizing radiation and for proficiency in DNA double-strand-break-induced homologous recombination, and in the vertebrate system, all five Rad51 paralogs, Xrcc2, Xrcc3, Rad51B, Rad51C, and Rad51D, are required for DNA repair and recombination proficiency. Overexpression of Rad51 partially corrects the sensitivity to DNA-damaging agents of the yeast and chicken paralog mutants (22, 26, 55, 56). In addition, overexpression of Rad51 largely suppresses the meiotic defects of dmc1 (57). No information is available yet as to whether overexpressing BRCA2 might suppress the repair deficiency of the paralog mutants in DT40 cell lines. However, it should be noted that overexpression of Rad52 had only a marginal effect on suppressing X-ray sensitivity of a rad55 or rad57 mutant when expressed by itself but could improve survival substantially when overexpressed together with Rad51 (22). Paradoxically, Rad52 was reported to be toxic when overexpressed in xrcc3 mutant DT40 cells (20).
Until recently, the paralogs were thought to act during the early stages of DNA repair by facilitating loading and stabilization of RAD51 onto single-stranded DNA. In yeast, this notion was supported by the genetic studies on phenotype suppression noted above and by the observation that certain rad51 mutant alleles encoding proteins that bind DNA more tightly could bypass the requirement for Rad55 and Rad57 in recombination and repair proficiency (18, 22, 26). In the higher eukaryotes, the notion was supported by the observations that formation of damage-induced RAD51 foci, which are thought to correspond to sites of DNA repair, is dependent on the paralogs and that overexpression of RAD51 partially suppresses the DNA repair deficiency in paralog mutant cells (55, 56). In addition, in vitro studies with purified proteins have shown that certain paralogs can serve as mediators enhancing Rad51-catalyzed DNA strand exchange by overcoming the inhibitory effects of RPA (50, 54) or by maintaining Rad51 in an ATP-bound state (45). On the other hand, recent evidence has implicated the paralogs functioning at a stage after the loading of Rad51 onto single-stranded DNA. In yeast, by use of chromatin immunoprecipitation analysis, it was concluded that Rad55 was required for formation of strand invasion intermediates (52), while in mammalian cells, accumulating evidence for certain paralogs has suggested roles in stabilizing heteroduplex DNA formed in the wake of Rad51-promoted strand exchange (9) and also in Holliday junction processing (35). How and when Rec2 functions is still an open question. The possibility that Rad51 filament formation is a dynamic process required throughout recombination and potentiated during the entire course by Rec2 cannot be excluded.
From the studies reported here, it seems clear that Rec2, Brh2, and Rad51 form an axis of function about which the recombinational repair process revolves and depends upon for execution and control. While Rad51 provides the catalytic muscle to power recombination, Rec2 and Brh2 provide the governance and regulation. The importance of Rec2 in the process is evident, even without any knowledge of molecular interplay, from the sensitivity to radiation, loss of recombination proficiency, and abortive meiosis that result from its absence. Assembly of Rad51 into damage-induced nuclear foci fails in the absence of Rec2. Yet Rec2 itself has a measure of autonomy in its action, being independent of Rad51 and Brh2 in its own ability to assemble into foci following DNA damage. The activity of Rec2 in recombination is likely to be in conjunction with that of Brh2, given their physical interaction and Brh2's capacity to compensate for the loss of Rec2. This latter attribute is particularly striking in the case of the hyperactive Brh2-RPA70 fusion, which can rescue meiosis and promote an elevated frequency of allelic recombination. Thus, there is a dualism in Rec2, one arm serving to activate recombination function and the other arm serving to temper it.
The emerging picture of the Rad51 filament features a dynamic structure subject to the opposing forces of assembly and dissociation (10, 41). Promoting the formation of the structure, stabilizing it, maintaining the ATP-bound state, and promoting the transition to disassembly are all possible stages for imposing regulation. At what level Rec2 acts and how it works together with Brh2 to promote and potentiate the assembly of the Rad51 filament await discovery. We expect that the application of chromatin immunoprecipitation methodology to ordering the molecular events of filament establishment together with experimentation on the combined actions of Rec2, Brh2, and Rad51 using purified proteins will provide a revealing picture of this molecular triangle.
Acknowledgments
We thank Lorraine Symington and Robin Holliday for helpful comments on the manuscript and Haijuan Yang, Nikola Pavletich, and Maria Jasin for continuing interest and enthusiastic discussions.
W.K.H. gratefully acknowledges financial support for this work from National Institutes of Health grant GM42482, the Department of Defense Breast Cancer Research Program grant DAMD17-3-1-0234, and the William Randolph Hearst Foundation.
REFERENCES
- 1.Ariza, A., D. J. Richard, M. F. White, and C. S. Bond. 2005. Conformational flexibility revealed by the crystal structure of a crenarchaeal RadA. Nucleic Acids Res. 33:1465-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banuett, F., and I. Herskowitz. 1989. Different alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl. Acad. Sci. USA 86:5878-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauchwitz, R., and W. K. Holloman. 1990. Isolation of the REC2 gene controlling recombination in Ustilago maydis. Gene 96:285-288. [DOI] [PubMed] [Google Scholar]
- 4.Bennett, R. L., and W. K. Holloman. 2001. A RecA homologue in Ustilago maydis that is distinct and evolutionarily distant from Rad51 actively promotes DNA pairing reactions in the absence of auxiliary factors. Biochemistry 38:14379-14386. [DOI] [PubMed] [Google Scholar]
- 5.Bignell, G., G. Micklem, M. R. Stratton, A. Ashworth, and R. Wooster. 1997. The BRC repeats are conserved in mammalian BRCA2 proteins. Hum. Mol. Genet. 6:53-58. [DOI] [PubMed] [Google Scholar]
- 6.Bishop, D. K., D. Park, L. Xu, and N. Kleckner. 1992. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69:439-456. [DOI] [PubMed] [Google Scholar]
- 7.Bork, P., N. Blomberg, and M. Nilges. 1996. Internal repeats in the BRCA2 protein sequence. Nat. Genet. 13:22-23. [DOI] [PubMed] [Google Scholar]
- 8.Boulton, S. J., A. Gartner, J. Reboul, P. Vaglio, N. Dyson, D. E. Hill, and M. Vidal. 2002. Combined functional genomic maps of the C. elegans DNA damage response. Science 295:127-131. [DOI] [PubMed] [Google Scholar]
- 9.Brenneman, M. A., B. M. Wagener, C. A. Miller, C. Allen, and J. A. Nickoloff. 2002. XRCC3 controls the fidelity of homologous recombination: roles for XRCC3 in late stages of recombination. Mol. Cell 10:387-395. [DOI] [PubMed] [Google Scholar]
- 10.Bugreev, D. V., and A. V. Mazin. 2004. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl. Acad. Sci. USA 101:9988-9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, C. F., P. L. Chen, Q. Zhong, Z. D. Sharp, and W. H. Lee. 1999. Expression of BRC repeats in breast cancer cells disrupts the BRCA2-Rad51 complex and leads to radiation hypersensitivity and loss of G(2)/M checkpoint control. J. Biol. Chem. 274:32931-32935. [DOI] [PubMed] [Google Scholar]
- 12.Chen, P. L., C. F. Chen, Y. Chen, J. Xiao, Z. D. Sharp, and W. H. Lee. 1998. The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc. Natl. Acad. Sci. USA 95:5287-5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conway, A. B., T. W. Lynch, Y. Zhang, G. S. Fortin, C. W. Fung, L. S. Symington, and P. A. Rice. 2004. Crystal structure of a Rad51 filament. Nat. Struct. Mol. Biol. 11:791-796. [DOI] [PubMed] [Google Scholar]
- 14.Davies, A. A., J. Y. Masson, M. J. McIlwraith, A. Z. Stasiak, A. Stasiak, A. R. Venkitaraman, and S. C. West. 2001. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell 7:273-282. [DOI] [PubMed] [Google Scholar]
- 15.Esashi, F., N. Christ, J. Gannon, Y. Liu, T. Hunt, M. Jasin, and S. C. West. 2005. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature 434:598-604. [DOI] [PubMed] [Google Scholar]
- 16.Essers, J., A. B. Houtsmuller, L. van Veelen, C. Paulusma, A. L. Nigg, A. Pastink, W. Vermeulen, J. H. Hoeijmakers, and R. Kanaar. 2002. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J. 21:2030-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferguson, D. O., M. C. Rice, M. H. Rendi, H. Kotani, E. B. Kmiec, and W. K. Holloman. 1997. Interaction between Ustilago maydis REC2 and RAD51 genes in DNA repair and mitotic recombination. Genetics 145:243-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fortin, G. S., and L. S. Symington. 2002. Mutations in yeast Rad51 that partially bypass the requirement for Rad55 and Rad57 in DNA repair by increasing the stability of Rad51-DNA complexes. EMBO J. 21:3160-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fotheringham, S., and W. K. Holloman. 1989. Cloning and disruption of Ustilago maydis genes. Mol. Cell. Biol. 9:4052-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujimori, A., S. Tachiiri, E. Sonoda, L. H. Thompson, P. K. Dhar, M. Hiraoka, S. Takeda, Y. Zhang, M. Reth, and M. Takata. 2001. Rad52 partially substitutes for the Rad51 paralog XRCC3 in maintaining chromosomal integrity in vertebrate cells. EMBO J. 20:5513-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galkin, V. E., F. Esashi, X. Yu, S. Yang, S. C. West, and E. H. Egelman. 2005. BRCA2 BRC motifs bind RAD51-DNA filaments. Proc. Natl. Acad. Sci. USA 102:8537-8542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hays, S. L., A. A. Firmenich, and P. Berg. 1995. Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc. Natl. Acad. Sci. USA 92:6925-6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holliday, R. 1967. Altered recombination frequencies in radiation sensitive strains of Ustilago. Mutat. Res. 4:275-288. [DOI] [PubMed] [Google Scholar]
- 24.Holliday, R. 1974. Ustilago maydis, p. 575-595. In R. C. King (ed.), Handbook of genetics, vol. 1. Plenum Press, New York, N.Y. [Google Scholar]
- 25.Jasin, M. 2002. Homologous repair of DNA damage and tumorigenesis: the BRCA connection. Oncogene 21:8981-8993. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, R. D., and L. S. Symington. 1995. Functional differences and interactions among the putative RecA homologs Rad51, Rad55, and Rad57. Mol. Cell. Biol. 15:4843-4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kojic, M., and W. K. Holloman. 2000. Shuttle vectors for genetic manipulations in Ustilago maydis. Can. J. Microbiol. 46:333-338. [DOI] [PubMed] [Google Scholar]
- 28.Kojic, M., C. F. Kostrub, A. R. Buchman, and W. K. Holloman. 2002. BRCA2 homolog required for proficiency in DNA repair, recombination, and genome stability in Ustilago maydis. Mol. Cell 10:683-691. [DOI] [PubMed] [Google Scholar]
- 29.Kojic, M., C. W. Thompson, and W. K. Holloman. 2001. Disruptions of the Ustilago maydis REC2 gene identify a protein domain important in directing recombinational repair of DNA. Mol. Microbiol. 40:1415-1426. [DOI] [PubMed] [Google Scholar]
- 30.Kojic, M., H. Yang, C. F. Kostrub, N. P. Pavletich, and W. K. Holloman. 2003. The BRCA2-interacting protein DSS1 is vital for DNA repair, recombination, and genome stability in Ustilago maydis. Mol. Cell 12:1043-1049. [DOI] [PubMed] [Google Scholar]
- 31.Kojic, M., Q. Zhou, M. Lisby, and W. K. Holloman. 2005. Brh2-Dss1 interplay enables properly controlled recombination in Ustilago maydis. Mol. Cell. Biol. 25:2547-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurumizaka, H., S. Ikawa, M. Nakada, K. Eda, W. Kagawa, M. Takata, S. Takeda, S. Yokoyama, and T. Shibata. 2001. Homologous-pairing activity of the human DNA-repair proteins Xrcc3.Rad51C. Proc. Natl. Acad. Sci. USA 98:5538-5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lio, Y. C., A. V. Mazin, S. C. Kowalczykowski, and D. J. Chen. 2003. Complex formation by the human Rad51B and Rad51C DNA repair proteins and their activities in vitro. J. Biol. Chem. 278:2469-2478. [DOI] [PubMed] [Google Scholar]
- 34.Lisby, M., J. H. Barlow, R. C. Burgess, and R. Rothstein. 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118:699-713. [DOI] [PubMed] [Google Scholar]
- 35.Liu, Y., J. Y. Masson, R. Shah, P. O'Regan, and S. C. West. 2004. RAD51C is required for Holliday junction processing in mammalian cells. Science 303:243-246. [DOI] [PubMed] [Google Scholar]
- 36.Martin, J. S., N. Winkelmann, M. I. Petalcorin, M. J. McIlwraith, and S. J. Boulton. 2005. RAD-51-dependent and -independent roles of a Caenorhabditis elegans BRCA2-related protein during DNA double-strand break repair. Mol. Cell. Biol. 25:3127-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Donnell, K. L., and D. J. McLaughlin. 1984. Ultrastructure of meiosis in Ustilago maydis. Mycologia 76:465-485. [Google Scholar]
- 38.Padmore, R., L. Cao, and N. Kleckner. 1991. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell 66:1239-1256. [DOI] [PubMed] [Google Scholar]
- 39.Pellegrini, L., D. S. Yu, T. Lo, S. Anand, M. Lee, T. L. Blundell, and A. R. Venkitaraman. 2002. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature 420:287-293. [DOI] [PubMed] [Google Scholar]
- 40.Powell, S. N., and L. A. Kachnic. 2003. Roles of BRCA1 and BRCA2 in homologous recombination, DNA replication fidelity and the cellular response to ionizing radiation. Oncogene 22:5784-5791. [DOI] [PubMed] [Google Scholar]
- 41.Ristic, D., M. Modesti, T. van der Heijden, J. van Noort, C. Dekker, R. Kanaar, and C. Wyman. 2005. Human Rad51 filaments on double- and single-stranded DNA: correlating regular and irregular forms with recombination function. Nucleic Acids Res. 33:3292-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rubin, B. P., D. O. Ferguson, and W. K. Holloman. 1994. Structure of REC2, a recombinational repair gene of Ustilago maydis, and its function in homologous recombination between plasmid and chromosomal sequences. Mol. Cell. Biol. 14:6287-6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sehorn, M. G., S. Sigurdsson, W. Bussen, V. M. Unger, and P. Sung. 2004. Human meiotic recombinase Dmc1 promotes ATP-dependent homologous DNA strand exchange. Nature 429:433-437. [DOI] [PubMed] [Google Scholar]
- 44.Sharan, S. K., M. Morimatsu, U. Albrecht, D. S. Lim, E. Regel, C. Dinh, A. Sands, G. Eichele, P. Hasty, and A. Bradley. 1997. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature 386:804-810. [DOI] [PubMed] [Google Scholar]
- 45.Shim, K. S., C. Schmutte, G. Tombline, C. D. Heinen, and R. Fishel. 2004. hXRCC2 enhances ADP/ATP processing and strand exchange by hRAD51. J. Biol. Chem. 279:30385-30394. [DOI] [PubMed] [Google Scholar]
- 46.Shin, D. S., L. Pellegrini, D. S. Daniels, B. Yelent, L. Craig, D. Bates, D. S. Yu, M. K. Shivji, C. Hitomi, A. S. Arvai, N. Volkmann, H. Tsuruta, T. L. Blundell, A. R. Venkitaraman, and J. A. Tainer. 2003. Full-length archaeal Rad51 structure and mutants: mechanisms for RAD51 assembly and control by BRCA2. EMBO J. 22:4566-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shinohara, A., H. Ogawa, and T. Ogawa. 1992. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69:457-470. [DOI] [PubMed] [Google Scholar]
- 48.Shivji, M. K., and A. R. Venkitaraman. 2004. DNA recombination, chromosomal stability and carcinogenesis: insights into the role of BRCA2. DNA Repair 3:835-843. [DOI] [PubMed] [Google Scholar]
- 49.Siaud, N., E. Dray, I. Gy, N. Takvorian, and M. P. Doutriaux. 2004. Brca2 is involved in meiosis in Arabidopsis thaliana as suggested by its interaction with Dmc1. EMBO J. 23:1392-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sigurdsson, S., S. Van Komen, W. Bussen, D. Schild, J. S. Albala, and P. Sung. 2001. Mediator function of the human Rad51B-Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev. 15:3308-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stark, J. M., P. Hu, A. J. Pierce, M. E. Moynahan, N. Ellis, and M. Jasin. 2002. ATP hydrolysis by mammalian RAD51 has a key role during homology-directed DNA repair. J. Biol. Chem. 277:20185-20194. [DOI] [PubMed] [Google Scholar]
- 52.Sugawara, N., X. Wang, and J. E. Haber. 2003. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell 12:209-219. [DOI] [PubMed] [Google Scholar]
- 53.Sung, P. 1994. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 265:1241-1243. [DOI] [PubMed] [Google Scholar]
- 54.Sung, P. 1997. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 11:1111-1121. [DOI] [PubMed] [Google Scholar]
- 55.Takata, M., M. S. Sasaki, E. Sonoda, T. Fukushima, C. Morrison, J. S. Albala, S. M. Swagemakers, R. Kanaar, L. H. Thompson, and S. Takeda. 2000. The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol. Cell. Biol. 20:6476-6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takata, M., M. S. Sasaki, S. Tachiiri, T. Fukushima, E. Sonoda, D. Schild, L. H. Thompson, and S. Takeda. 2001. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol. 21:2858-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsubouchi, H., and G. S. Roeder. 2003. The importance of genetic recombination for fidelity of chromosome pairing in meiosis. Dev. Cell 5:915-925. [DOI] [PubMed] [Google Scholar]
- 58.Wu, Y., Y. He, I. A. Moya, X. Qian, and Y. Luo. 2004. Crystal structure of archaeal recombinase RADA: a snapshot of its extended conformation. Mol. Cell 15:423-435. [DOI] [PubMed] [Google Scholar]
- 59.Yang, H., P. D. Jeffrey, J. Miller, E. Kinnucan, Y. Sun, N. H. Thoma, N. Zheng, P. L. Chen, W. H. Lee, and N. P. Pavletich. 2002. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 297:1837-1848. [DOI] [PubMed] [Google Scholar]
- 60.Yang, H., Q. Li, J. Fan, W. K. Holloman, and N. P. Pavletich. 2004. The BRCA2 homolog Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature 433:653-657. [DOI] [PubMed] [Google Scholar]
- 61.Yu, D. S., E. Sonoda, S. Takeda, C. L. H. Huang, L. Pellegrini, T. L. Blundell, and A. R. Venkitaraman. 2003. Dynamic control of Rad51 recombinase by self-association and interaction with BRCA2. Mol. Cell 12:1029-1041. [DOI] [PubMed] [Google Scholar]