Abstract
RNA editing inserts and deletes uridylates (U's) in kinetoplastid mitochondrial pre-mRNAs by a series of enzymatic steps. Small guide RNAs (gRNAs) specify the edited sequence. Editing, though sometimes extensive, is precise. The effects of mutating pre-mRNA and gRNA sequences in, around, and upstream of the editing site on the specificity and efficiency of in vitro insertion editing were examined. U's could be added opposite guiding pyrimidines, but guiding purines, particularly A's, were required for efficient ligation. A base pair between mRNA and gRNA immediately upstream of the editing site was not required for insertion editing, although it greatly enhanced its efficiency and accuracy. In addition, a gRNA/mRNA duplex upstream of the editing site enhanced insertion editing when it was close to the editing site, but prevented cleavage, and hence editing, when immediately adjacent to the editing site. Thus, several aspects of mRNA-gRNA interaction, as well as gRNA base pairing with added U's, optimize editing efficiency, although they are not required for insertion editing.
In kinetoplastid RNA editing, uridylates (U's) are inserted and removed at specific sites in mitochondrial (mt) premRNAs by a series of enzymatic steps to form mature, functional mRNAs (for recent reviews, see references 13, 17, and 37). Small guide RNAs (gRNAs) specify the edited sequence, which is complementary to the gRNAs by G · U and Watson-Crick base pairing (4). Editing is initiated by an endoribonucleolytic cleavage of the pre-mRNA at the editing site (ES) that is 5" to the anchor duplex between the 5" region of gRNA and the pre-mRNA downstream of the sequence that becomes edited. The U's are added or removed at the 3" end of the 5" pre-mRNA fragment that results from the cleavage (10, 18, 34).
A 3"-terminal uridylyl transferase (TUTase) adds the U's, while a U-specific 3" exonuclease (exoUase) removes U's (2, 10). An RNA ligase rejoins the cleavage fragments after the U addition and removal. Overall, editing extends the gRNA/pre-mRNA duplex in the 5" direction. The enzymatic activities that are required for editing are contained within a large multiprotein complex (1, 9, 29, 31), which sediments in glycerol gradients at 20S (9, 31) and is approximately 1,600 kDa in size (27).
Progress has been made in elucidating how the edited sequence is specified by gRNA, but many questions remain unanswered. The cleavage step contributes to the specificity of editing by selecting the ES (4, 10, 18, 34). In deletion editing, this cleavage occurs downstream of the U's that will be removed (10, 34), and they are removed by the U-specific 3" exonuclease until the first non-U nucleotide is encountered (2, 11, 16a, 20a). Hence, selection of the ES by the endonuclease and the U specificity of the exonuclease may be more important than the sequence upstream of the ES for accurate editing at a deletion site.
In insertion editing, cleavage occurs at an ES where the 3" nucleotide of the 5" fragment can base pair with the gRNA nucleotide adjacent to the purines that guide the U insertions. The U specificity of TUTase must play an important role in insertion editing, but gRNA/mRNA interactions upstream of the ES and the number of guiding purines also appear important to specificity in insertion editing (7, 16). The sequence of the gRNA appears to contribute to both latter sources of specificity (4, 8). The gRNA/pre-mRNA base pair immediately 5" to the ES may help specify the number of inserted U's. It may present the 5" fragment as a substrate for TUTase, help to stabilize the added U's by base pairing with the gRNA purines, or position the 5" fragment in register with gRNA so that the 5" fragment with the correct number of added U's is properly aligned for ligation. In addition, the strong bias for purine ribonucleotides upstream of insertion ESs (6) may reflect selection against base pairing with guiding nucleotides or adjacent purines in gRNA that do not specify insertion of U's.
We used the precleaved insertion editing system (16) to examine the effects of gRNA and pre-mRNA mutations on the U addition and ligation steps of editing to test the above possibilities. We used this system because it does not require prior cleavage and allows mRNA and gRNA sequence changes that would normally disrupt cleavage or change the location of the ES. Where possible, we used an uncleaved substrate in vitro to examine a full round of insertion editing to test the effects of these changes on endonucleolytic cleavage and on the series of editing steps. We found that sequence elements of gRNA and pre-mRNA in and around the ES contribute to specification of the edited mRNA sequence in insertion editing. We also found that no editing occurs in in vitro substrates with a pyrimidine that is upstream and immediately adjacent to the ES. Furthermore, a stable pre-mRNA/gRNA duplex upstream of the ES enhanced editing efficiency except when immediately adjacent to the editing site.
MATERIALS AND METHODS
Preparation and labeling of RNAs.
The RNA 3"CL13pp and other 3" fragments were purchased as oligoribonucleotides from Oligos, Etc. Unless otherwise specified, RNAs described below were transcribed using T7 polymerase (Promega). The 3" ends of 5" cleavage fragments were verified by ligation to 3"CL13pp and subsequent partial digestion with RNase T1 to ensure correct sequence length after T7 transcription (25).
Transcription templates for variants of 5"CL18 (Fig. 1A) with altered 3" termini were prepared by PCR of 5"CL18-Tmp1 (16) using oligonucleotide primer EcoRI T7 (16) and variants of primer 5"CL18-3" (5"-NCTACGTCTCATACTTCCTATAG-3"), where N is complementary to the desired 3"-terminal nucleotide. Transcription templates for derivatives of gPCA6-1A with various guiding nucleotides (Fig. 1) were synthesized by PCR of gPCA6-3A-Tmp1 (16) with EcoRI T7 and primers gPCA6-N (5"-GGAAGTATGAGACGTAGGNATCGGAG-3"), where N is complementary to the desired guiding nucleotide. Similarly, templates for transcription of gPCA6-2A derivatives with two guiding nucleotide were synthesized by PCR of gPCA6-3A-Tmp1 with EcoRI T7 and primers of sequence 5"-GGAAGTATGAGACGTAGGN1N2ATCGGAG-3", where N1N2 is the complement of the two guiding nucleotides read 5"→3" with respect to gRNA (i.e., right to left in Fig. 3).
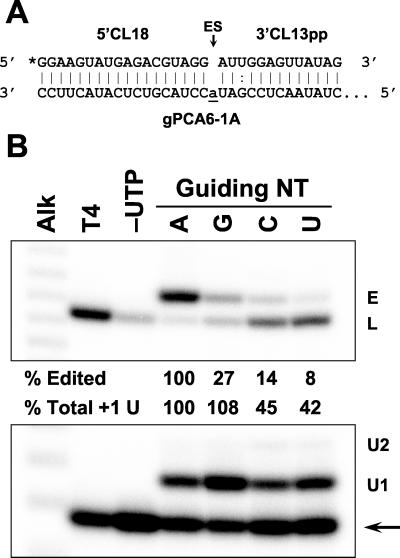
FIG. 1.
Substitution effects on precleaved insertion editing with a single guiding nucleotide (NT). (A) Precleaved insertion editing substrate fragments 5"CL18 and 3"CL13pp aligned with gRNA gPCA6-1A (16). The asterisk indicates a 5" 32P label. Monophosphates are present on the 5" and 3" ends of the 3" substrate fragment but are not shown. (B) Autoradiogram showing the 5" substrate fragment (arrow) and the expected positions of products with one or two added U's (U1 and U2), ligated 5" + 3" substrate fragments (L), and edited RNA with a single inserted U (E). Nucleotides substituted for the wild-type A are indicated above the lanes. Substrate fragments ligated by T4 RNA ligase (T4), reaction with UTP omitted (−UTP), and the alkaline hydrolysis sizing ladder (Alk) are indicated. The numbers indicate the abundance of edited (% Edited) and +1 U addition products (ligated plus nonligated) (% +1 U) relative to the wild type.
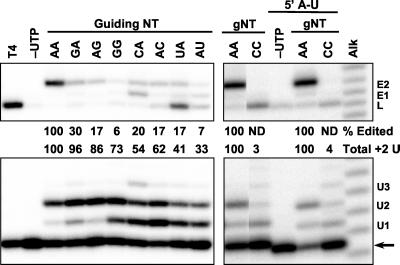
FIG. 3.
Substitution effects on precleaved insertion editing with two guiding nucleotides (gNT). Autoradiograms show the 5"CL18 substrate fragment (arrow) and the expected positions of products with one to three added U's (U1, U2, and U3), ligated 5" + 3" substrate fragments (L), and edited RNA with one or two inserted U's (E1 and E2). Nucleotides substituted for the A's in wild-type gPCA6-2A are indicated as guiding NT or gNT above each lane, with the 3"-most nucleotides (relative to gRNA) on the bottom. The 5" A-U indicates a 5" substrate with a 3" A plus gPCA6-2A with a U substituted to base pair with this A. Substrates ligated by T4 RNA ligase (T4), reaction with UTP omitted (−UTP), and the alkaline hydrolysis sizing ladder (Alk) are indicated. The numbers indicate the abundance of accurately edited and +2 U addition products (ligated plus nonligated) expressed as a percentage of the wild-type value.
gPCA6-2A RNAs with single-base substitutions 5" of the ES (with respect to pre-mRNA) (see Fig. 5A) were transcribed from templates amplified by PCR of gPCA6-3A-Tmp1 with primers EcoRIT7 and 5"-GGAAGTATGAGACGTAG NTTATCGGAG-3", where N is the complement of the substituted nucleotide. Variants of gPCA6-2A forming mismatches with 5"CL18 or 3"CL13pp (see Fig. 6A) were prepared by transcription from a template prepared by PCR of gPCA6-3A-Tmp1 with primer EcoRI T7 and 3" primers as follows: one 5" mismatch, 5"-GGAAGTATGAGACGTAGCTTATCGGAG-3"; two 5" mismatches, 5"-GGAAGTATGAGACGTACCTTATCGGAG-3"; three 5" mismatches, 5"-GGAAGTATGAGACGTCCCTTATCGGAG-3"; one 3" mismatch, 5"-GGAAGTATGAGACGTAGGTTGTCGGAGT-3"; two 3" mismatches, 5"-GGAAGTATGAGACGTAGGTTGGCGGAGTT-3"; and three 3" mismatches, 5"-GGAAGTATGAGACGTAGGTTGGGGGAGTTA-3".
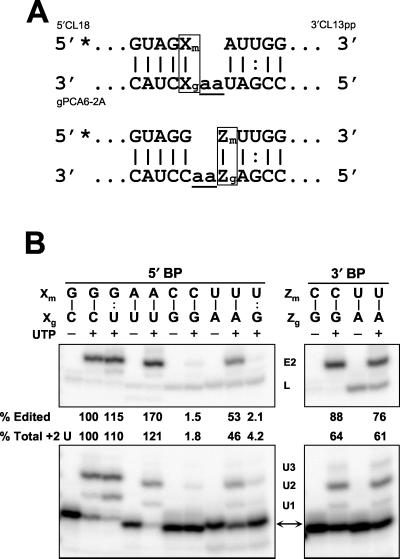
FIG. 5.
Effects on precleaved insertion editing of various base pairs flanking the ES. (A) Diagram of 5"CL18 and 3"CL13pp precleaved insertion substrate mRNAs aligned with gPCA6-2A gRNA. Xm and Zm indicate mRNA nucleotides and Xg and Zg indicate gRNA nucleotides that flank the ES where nucleotide substitutions were made. Wild-type base pairs upstream and downstream adjacent to ES2 in A6 pre-mRNA are Xm · Xg = G · C and Zm · Zg = A · U, respectively. Substitutions to the RNAs used in panel B changed the sequence of substrate and gRNAs but preserved the fully duplexed structure shown. (B) Precleaved insertion editing with wild-type and other Xm · Xg base pairs upstream of the ES (left panel) and with pyrimidine-purine Zm · Zg base pairs downstream of the ES (right panel). Reactions were performed in the absence of UTP (−) or presence of 100 μM UTP (+). Products of U addition and editing are indicated as in Fig. 3, and abundances of edited and total +2 U addition products (see legend to Fig. 3) are reported relative to wild-type sequences.
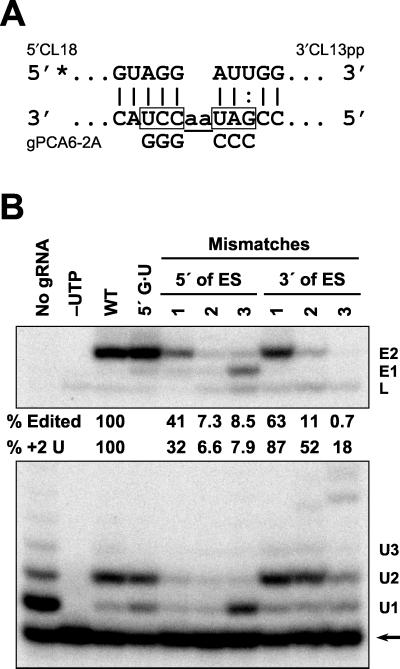
FIG. 6.
The effects on precleaved insertion editing of mismatches upstream or downstream of the ES. (A) Diagram showing the creation of mismatches with 5"CL18 or 3"CL13pp by nucleotide replacement with G's or C's in gPCA6-2A upstream and downstream of the ES, respectively. The asterisk indicates 5" 32P labeling. (B) Autoradiogram of products formed during precleaved editing with mismatches adjoining the ES. Substrates, products, and quantification are as in Fig. 2. Mismatches 5" to the ES are 3" to 5", while those 3" to the ES are 5" to 3". Lanes with no gRNA, no UTP, wild-type gPCA6-2A (WT), and with a G · U rather than G · C base pair 5" to the ES are labeled.
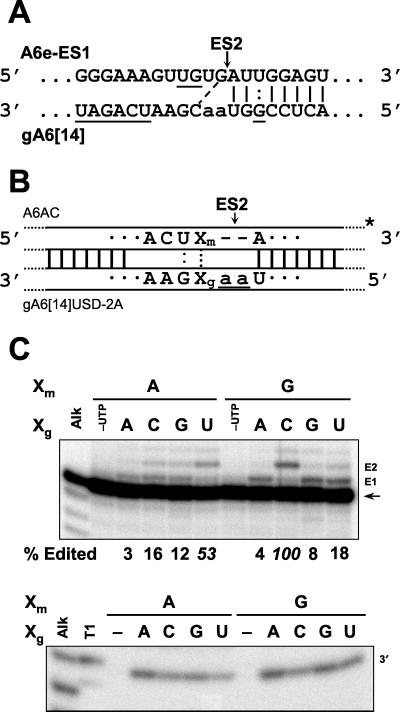
Pre-mRNA A6AC was prepared as described previously (16). Transcription templates for its variants were prepared in the same manner, except that 3" olignucleotide primers A6AC N40 (5"-CTATAACTCCAATNAGTACTTTC-3") were used, where N is complementary to the nucleotide upstream of ES2, as shown in Fig. 7B. gA6[14]USD-3A was prepared as described (16). Templates for transcription of its derivatives with pyrimidine guiding nucleotides were prepared likewise but used 3" primers 5"-GAAGAAAGGGAAAACTTCG N1N2N3ATTGGAGTT-3", where N1N2N3 is the complement of the three guiding nucleotides read 5"→3" with respect to gRNA (i.e., read right to left in Fig. 4B). Variants of gA6[14]USD-2A (16) with substitutions at position Xg, as shown in Fig. 7B, were prepared using 3" primers 5"-AAAGAAAGGGAAAACTTC NTTATTGGAGTT-3", where N is complementary to Xg.
FIG. 7.
Consequences of varying the upstream base pair to full-round insertion editing. (A) Sequence of A6e-ES1 and gA6[14] RNAs in the vicinity of ES2 (18). A dashed line indicates the potential base pair that may direct insertion of two U's at ES2. Underlining indicates residues which have been changed in A6AC and gA6[14]USD. (B) Diagram of A6AC substrate aligned with gA6[14]USD-2A gRNA, showing positions Xm and Xg upstream of ES2 in mRNA and gRNA, respectively, that were mutated. (C) Autoradiogram showing 3" cleavage (bottom panel) and edited (top panel) products of insertion editing reactions using 3" radiolabeled substrate A6AC mRNA. Nucleotides at the Xm and Xg positions in the mRNA and gRNA, respectively, upstream of the ES are indicated, and other labeling is as in Fig. 3. Negative controls shown lacked UTP (−UTP) and gRNA (Xg = −) in the top and bottom panels, respectively. T1 indicates partial digestion of A6AC with RNase T1 in the bottom panel (omitted from top panel) showing cleavage at ES2 (3").
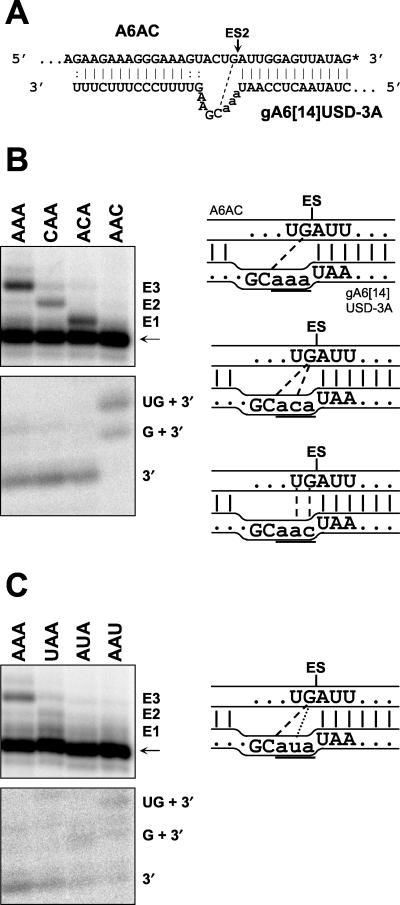
FIG. 4.
Guiding nucleotide effects on a full round of editing. (A) Alignment of A6AC substrate mRNA with gA6[14]USD-3A gRNA. The asterisk indicates the 32P 3"-end label. (B) Autoradiogram (left) with top panel showing substrate RNA (arrow) and edited RNAs with 1, 2, or 3 inserted U's (E1, E2, and E3) and bottom panel showing the wild-type 3" cleavage product (3") and products of cleavage shifted upstream one or two nucleotides (G + 3" and UG + 3"). The sequences of the 3 guiding nucleotides (all A's in the wild type) are indicated above the lanes, with the 3" nucleotide, relative to gRNA, on the bottom. Diagrams on the right show possible base pairing (dotted lines) between substrate and gRNAs that can account for some products of cleavage and editing. The potential upstream base pairs are not expected to occur until after cleavage. (C) Left, autoradiogram showing products of editing reactions using A6AC substrate and gA6[14]USD-3A with U's substituted for a guiding A (labeling as in B). Right, diagram showing possible base pairing as in B.
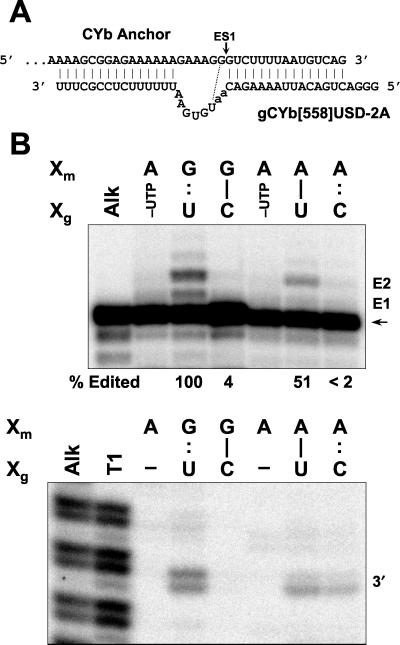
A fragment of apocytochrome b (CYb) pre-mRNA corresponding to nucleotides (nt) 1 to 66 of the transcript (36) was prepared by PCR of plasmid pCYb-pre (12) with oligonucleotides SP6-CYb (5"-ATTTAGGTGACACTATAGTTAAGAATAATGGTTATAAATTTTA-3") and CYb Anchor (5"-CTGACATTAAAAGACCCTTTC-3"), and transcription from this PCR template was performed using SP6 RNA polymerase (Epicentre Technologies). This pre-mRNA includes 51 nt upstream and 15 nt downstream of ES1 (14). CYb Short A51, with an A replacing the G upstream adjacent to ES1 (see Fig. 8A), was prepared similarly but using 3" primer CYb Anchor A51 (5"-CTGACATTAAAAGACTCTTTCTTTTTTC-3").
FIG. 8.
Mutations upstream of the ES diminish full-round insertion editing of CYb mRNA. (A) Diagram showing CYb anchor mRNA aligned with gCYb[558]USD-2A. The potential base pair upstream of ES1 where nucleotide substitutions were made is indicated with the dotted line. (B) Autoradiogram showing cleavage (bottom panel) and edited (top panel) products of the editing reactions. Doubled bands most likely reflect spontaneous formation of 2",3"-cyclic phosphate termini. Nucleotides at the Xm and Xg positions in the mRNA and gRNA, respectively, upstream of the ES are indicated, and other labeling is as in Fig. 7.
The gCYb[558] gRNA (30) was modified to form a stable duplex with CYb Short 6 nt upstream of CYb ES1. The DNA template for PCR of transcription templates for gCYb[558] variants was gCYb-I Tmp (5"-GGAATTCCTAATACGACTCACTATAGGGACTGACATTAAAAGACAATATAAATTT-3" plus complementary sequence). Transcription templates for gCYb[558]USD-2A and gCYb[558]USD-2A C22 were synthesized by PCR of gCYb-I Tmp with 5" primer EcoRI T7 Long (5"-CGGCGGAATTCTGTAATACGACTCACTATAG-3") and either gCYbUSD (5"-AAAGCGGAGAAAAAATTCACATTGTCTTTTAA-3") or gCYbUSD C22 (5"-AAAGCGGAGAAAAAATTCACGTTGTCTTTTAA-3"). gCYb-I gRNA was prepared as previously described (28).
Precleaved substrate RNAs were labeled at the 5" terminus of the 5" cleavage fragment by alkaline phosphatase and polynucleotide kinase treatment, as described (16). Full-round editing substrates were labeled at the 3" terminus by ligation of [5"-32P]pCp (38).
Preparation of mt extract.
Partially purified Trypanosoma brucei mt extract was prepared either by sequential SP Sepharose and Q Sepharose chromatography or by the same method followed by gel filtration on a Superose 6 column, as described (27), starting with mitochondria isolated from T. brucei strain IsTaR 1.7a (15) lysed in 0.7% (vol/vol) Triton X-100 (29). Fractions from the final step of these purification procedures were tested for insertion editing activity, and peak fractions were used for the experiments. Extract was stored in 10 to 15% (vol/vol) glycerol at −70°C between purification and use in editing assays. Both types of extract displayed the same editing activity, though fractions from the three-step purification contained less nonspecific RNase activity (data not shown).
Precleaved and full-round editing assays.
Editing reactions, and the separation and visualization of the edited products, were performed as described by Igo et al. (16). Reactions performed in the absence of UTP were used as background measurements for quantifying U addition products in precleaved insertion reactions. Controls without UTP were also used for measuring background for quantifying full-round insertion editing, rather than without gRNA, because the stable gRNA-mRNA structure caused a small amount of retardation of input RNA during gel electrophoresis. Parallel time course assays were stopped by the addition of 2 μl of stop buffer (16) and incubation on ice prior to phenol-chloroform extraction of RNA. The abundance of products of the editing reaction was initially calculated as the percentage of total input RNA and then normalized to the abundance of the corresponding products in control reactions, except as indicated. Abundance of total U addition products was computed as the sum of ligated and nonligated products with the specified number of added U's. Unless otherwise specified, numerical data are reported as the mean of two independent experiments.
RESULTS
Specification of U insertion by guiding nucleotides.
The role of the guiding nucleotides of gRNA in the U addition and ligation steps of editing was investigated using the precleaved in vitro editing assay (16). A single guiding A in gPCA6-1A resulted in substantial amounts of both total +1 U addition product (48% of input) and edited RNA (20% of input) with one inserted U, but a single guiding G resulted in more +1 U addition product and only about one-fourth as much edited RNA (Fig. 1). Thus, U addition was similar with a guiding A or G, but ligation was lower with the guiding G, perhaps reflecting the less optimal G · U base pair upstream of the site of ligation. A single guiding pyrimidine reduced the production of both U addition products by more than half and of edited products by more than 85%, but increased the generation of ligation products that contain no inserted U's. This ligation occurred with either an A or G immediately upstream of the ES in the 5" mRNA fragment (Fig. 1B and data not shown).
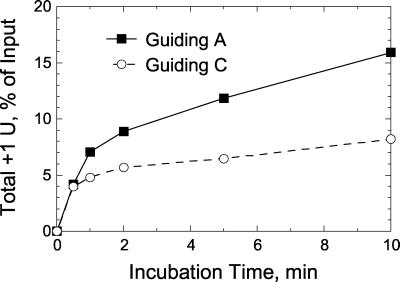
Figure 2 shows that the initial rate of U addition with a single guiding C was similar to that with a guiding A, but that the reduced accumulation of +1 U addition product was reached quickly. This was also the case with a nonligatable 3" fragment which has no 5" phosphate, although the overall rate of addition was lowered, as reported previously (16), resulting in accumulation of +1 U addition product being reduced by about 35-fold (data not shown). These data suggest that the guiding nucleotide does not affect U addition but stabilizes the added U, perhaps by blocking U removal by the 3" exoUase that is present in the editing complex, as has been shown for purified exoUase (2).
FIG. 2.
Initial rates of U addition to 5"CL18 with one guiding A or C. Abundance of total +1 U addition product (ligated plus nonligated, expressed as a percentage of total input RNA) during the first 10 min of precleaved editing reactions is plotted as a function of reaction time. U addition was guided by gPCA6-1A (solid squares, solid line) and by a similar gRNA in which the guiding A was replaced by a C (open circles, dashed line).
Two guiding A's in gPCA6-2A resulted in substantial amounts of both +2 U addition product and edited RNA with two inserted U's (Fig. 3). Replacement of either or both A's with a G resulted in a small reduction of the total +2 U addition product (although nonligated +2 addition product was more abundant) and a small increase in +1 U addition product when a G was at position 1. However, these replacements resulted in a ≥70% reduction in edited product, again indicating an effect on ligation. Replacement of one guiding A with a C or U reduced the amount of +2 U addition product, increased the amount of +1 U addition product, and generated some +3 U addition product. These replacements also substantially reduced the amounts of accurately edited RNA (i.e., containing two inserted U's) and produced prominent ligation products with one or no inserted U's.
When the guiding A's were replaced with two C's, addition products with up to four U's were generated (Fig. 3). No edited RNA was produced in this case, but ligation products with no inserted U's were formed. The ligation products may reflect base pairing of the terminal G of the 5" fragment with the guiding C's (see Discussion). Replacement of the G with an A along with a compensatory change in the gRNA to retain base pairing still resulted in preferential production of the ligated product with no inserted U's, although a small amount of ligated RNA with one or two inserted U's was generated. Overall, these results indicate that U addition does not require a complementary nucleotide in the gRNA and thus that it is not templated, although base pairing appears to stabilize the added U, since the addition products continue to accumulate quickly after a short period of time only in the presence of a guiding purine (Fig. 2). In addition, base pairing of the added U's with the gRNA substantially enhances ligation.
The effects of pyrimidine guiding nucleotides were also pronounced when insertion editing was examined by the in vitro assay that requires cleavage of the substrate (16) (Fig. 4). Replacement of one of the three guiding A's in gA6[14]USD-3A (Fig. 4A) with a C resulted in cleavage at the same site as with the wild-type gRNA (ES2) except when the C was adjacent to the anchor duplex (Fig. 4B). In the latter case, cleavage occurred one or two nucleotides upstream of ES2, probably due to base pairing of the guiding nucleotides (lowercase in Fig. 4B) with the G or the G and U upstream of ES2 in A6AC. The resultant edited RNA reflected the number of guiding A's adjacent to the anchor duplex, and no edited RNA was produced when the replacing C shifted the cleavage site. Similar results were obtained using A6AC precleaved at ES2 (data not shown). Replacement of one of the three guiding A's with U rather than C resulted in some cleavage at ES2 but also some cleavages further upstream (Fig. 4C). In addition, a similar but low abundance of edited RNA with one, two, or three inserted U's was generated. These multiple cleavage sites and production of multiple edited RNAs may reflect the potential base pairing between the substituted U and the G or A that is on either side of ES2 in the mRNA.
Nucleotides and base pairing adjacent to the ES.
The effects on in vitro editing of various nucleotides adjacent to the ES were examined because there is a nucleotide bias adjacent to the insertion ESs in pre-mRNAs, especially against C's immediately upstream of the ES (6). The efficiency and accuracy of insertion editing were tested for all possible Watson-Crick and G · U base pairs 5" to the ES using the precleaved system (Fig. 5). Conversion of the upstream G · C base pair to G · U (Xm = G in mRNA and Xg = U in gRNA, Fig. 5A) did not substantially affect the amount of resultant +2 U addition product or +2 edited RNA but did increase the amount of +1 addition product (Fig. 5B). Substitution of A · U for G · C resulted in a 70% increase in +2 U addition product and +2 edited RNA. In contrast, editing was significantly reduced when Xm was a pyrimidine. This effect was most pronounced with C in position Xm, where both the +2 addition product and edited RNA were reduced to less than 2% of the wild-type levels. In addition, the 3" terminal U was removed from a small fraction (1 to 2%) of the input RNA, probably by the U-specific exonuclease in the extract, suggesting that exoUase is more efficient than TUTase in this context (Igo et al., submitted for publication).
Precleaved substrate with the upstream U · A underwent insertion editing with about half the efficiency of the wild type, but only small amounts of U addition and edited products were observed with an upstream U · G base pair. The G in gRNA prevented U removal as well as did A. Formation of ligation product with no added U's was greater than the +2 edited product with an upstream pyrimidine (Xm in Fig. 5A). This may again reflect the tendency of a purine guiding nucleotide adjacent to the anchor duplex to bridge with the upstream nucleotide and enhance ligation. In contrast, a pyrimidine downstream of the ES (Zm in Fig. 5A) had little effect on precleaved editing, although a U at this position promoted ligation without U addition. These results may, in part, account for the observed bias against C's upstream of insertion ESs, but not the bias against pyrimidines downstream of the ES (6).
The effects on U addition and editing of gRNA/pre-mRNA base pairing adjacent to the ES were examined (Fig. 6). A single G · G mismatch upstream of the ES substantially reduced the production of +2 U addition product and edited RNA but retained the accuracy of precleaved insertion editing (Fig. 6B). Production of accurately edited RNA was reduced by over 90% with two or three G · G mismatches upstream and substantial +1 U addition product, and inaccurately edited RNA (with one U inserted) was generated with three mismatches. The pattern of U addition with three upstream G · G mismatches resembled that obtained in the absence of gRNA. One, two, or three mismatches downstream of the ES progressively diminished the production of edited RNA but, unlike upstream mismatches, did not greatly affect the accuracy of editing (Fig. 6B).
The prominent U addition product contained two U's, as specified by the gRNA. A small fraction of RNA with more than two added U's was produced, especially with three mismatches, perhaps reflecting base pairing of various added U's with the two A's in the gRNA. Overall, base pairing immediately upstream of the ES affects precleaved insertion editing more than similar changes downstream, perhaps reflecting a focus of the catalytic activity on the 3" end of the 5" fragment.
The effects of specific nucleotides and base pairing 5" to the ES on full-round insertion editing (including cleavage) at ES2 of A6 RNA were examined (Fig. 7). Editing was examined using A6-eES1/gA6[14] (18) (Fig. 7A) and A6AC/gA6[14]USD (16) (Fig. 7B) pre-mRNA/gRNA pairs in which the pre-mRNA G upstream of ES2 and the corresponding C in the gRNA were replaced with each of the other nucleotides and tested in all possible combinations. With the A6-eES1/gA6 pair, insertion editing was only detected in the presence of the wild-type G · C upstream base pair. However, cleavage and chimera formation occurred with a purine upstream of the ES regardless of the corresponding nucleotide in the gRNA, and no cleavage was observed with a C upstream of the ES (data not shown). A6-eES1 pre-mRNA with U upstream of the ES was not tested, since the U could base pair with the guiding A in gRNA. Accurate editing occurred when the wild-type G · C pair in ACA6/gA6[14]USD was replaced with an A · U base pair but was reduced by about 50% (Fig. 7B).
Editing also occurred, but at a lower level, when G · C was replaced with other base pairs, including G · U, and the +1 edited product was increased relative to the wild type. The amount of edited RNA was correlated with predicted base pair stability. Cleavage occurred at ES2 in each of these cases, but only in the presence of gRNA. Thus, cleavage, U addition, and ligation can all proceed without an upstream base pair, but this base pair greatly enhances accurate editing. However, no editing or cleavage at ES2 or nearby was observed when the upstream nucleotide was a pyrimidine (data not shown), perhaps because the U or C in the mRNA may base pair with the guiding A in gRNA and block substrate recognition by the endonuclease. Hence, the number of inserted U's was efficiently specified by the guiding A's only when a purine was present immediately upstream of the ES that could form a G · C or A · U base pair with the nucleotide 3" to the guiding A's.
A similar but more limited analysis was performed using CYb pre-mRNA. This analysis used gCYb[558]USD gRNA, modified from the wild-type gRNA (30), which does not support editing in vitro. gCYb[558]USD can form a duplex upstream of ES1, creating a 6-nt bulge in pre-mRNA (Fig. 8A). The major edited product contained two inserted U's, although considerable +1 U and some +3 edited RNA was also produced (Fig. 8B). Use of pre-mRNA with A substituted for the upstream G and gRNA with C substituted for the corresponding U to provide G · C, A · U, and A · C base pairs resulted in reduced editing and cleavage (Fig. 8B). In particular, there was very little cleavage or editing with the G · C base pair. However, precleaved editing of this substrate-gRNA pair was efficient (data not shown), and thus the presence of this G · C upstream base pair appears to specifically affect cleavage. Thus, wild-type A6 and CYb pre-mRNAs were most efficiently edited with wild-type gRNA, and base pairing upstream of the ES appears to be necessary but not sufficient for efficient, specific editing.
Stability and position of an upstream duplex.
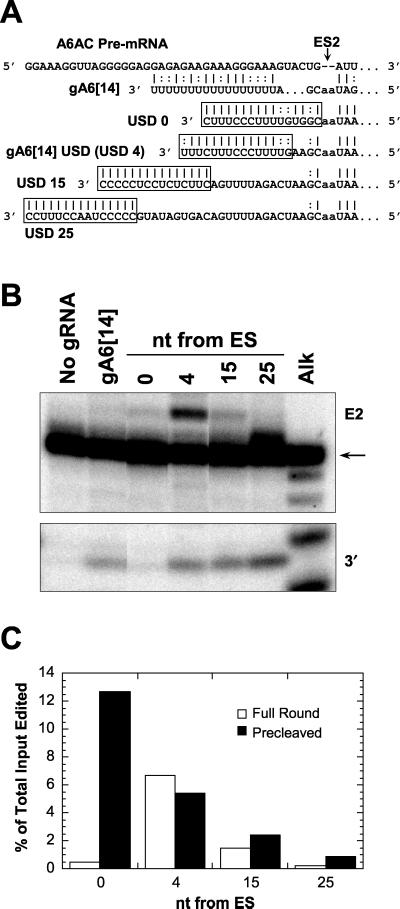
The effect of the location of an upstream duplex on insertion editing was examined, since a strong gRNA/pre-mRNA upstream duplex is known to increase the efficiency of in vitro editing (5, 11, 16, 19) (Fig. 9). Little cleavage or editing of A6AC pre-mRNA was observed with a 15-bp duplex immediately adjacent to ES2 (Fig. 9B). Otherwise, the efficiency of full-round insertion editing correlated with the proximity of the stable duplex to the ES, with duplexes nearer the ES promoting more editing, although production of the 3" cleavage product was similar. The most efficient editing was achieved using gA6[14]USD, which leaves 4 nt of single-stranded RNA upstream of ES2 upon binding to A6AC.
FIG. 9.
Effects of a stable upstream duplex on insertion editing of full-round and precleaved A6AC mRNA. (A) Alignments of gRNAs with A6AC mRNA, showing the positions of potential upstream duplexes (boxed). Most of the gA6[14] informational sequence is not shown, and its oligo(U) tail is arbitrarily aligned with a portion of the purine-rich region. The distance in nucleotides (i.e., predicted single-stranded region) between ES2 and the upstream duplex is indicated in the names of the modified gRNAs. (B) Products of cleavage (bottom panel) and editing (top panel) of A6AC mRNA reactions with the gRNAs in A. The distance of the upstream duplex from the ES, the alkaline hydrolysis ladder of A6AC RNA (Alk), and no-gRNA lanes are indicated. Other labeling is as in Fig. 6. (C) Comparison of upstream duplex distance from the ES effect on precleaved and full-round insertion editing. Quantification of accurately edited RNA is expressed as a percentage of total input.
The efficiency of insertion editing of the A6AC substrate increased 30-fold over the native gA6 (14) using the partially purified mt extract (see Materials and Methods) (Fig. 9B and data not shown). Placement of the duplex 15 or 25 nt upstream of the ES progressively diminished the amount of edited RNA that was produced. However, efficiency of precleaved editing of the same substrate was maximal with the upstream duplex adjacent to the ES (Fig. 9C), indicating that the reduction in full-round editing occurs at the cleavage step. In addition, the accuracy of +2 U precleaved insertion was optimized when no single-stranded RNA adjoined the ES (data not shown). Thus, the efficiency and accuracy of the sum of U addition and ligation improved with a 5" mRNA/gRNA duplex upstream of and near the ES. Cleavage was largely unaffected by the distance between the ES and upstream duplex, except that it was sensitive to duplex RNA directly adjacent to the ES, and thus required some single-stranded RNA upstream of the ES. The latter observation agrees with the results of Cruz-Reyes et al. (11) for deletion editing of A6 pre-mRNA at ES1.
DISCUSSION
This study shows that guiding nucleotide sequences, the nucleotides immediately flanking the ES and their potential for mRNA/gRNA base pairing, and the position of an mRNA/gRNA duplex upstream of the ES all impact the efficiency and accuracy of insertion editing. In general, the guiding nucleotides of gRNA do not affect U addition. Rather, base pairing of the added U's with the gRNA stabilizes their addition and consequently enhances ligation (3) and thus accurate editing. Base pairing with gRNA immediately downstream of the ES (i.e., at the 5" end of the anchor) has little or no effect on editing after cleavage, while such base pairing immediately upstream of the ES strongly affects insertion editing, but neither is absolutely necessary or sufficient. The interactions upstream of the ES affect the cleavage, U addition, and ligation steps of editing. Overall, the region upstream of the ES appears to have key impacts on editing.
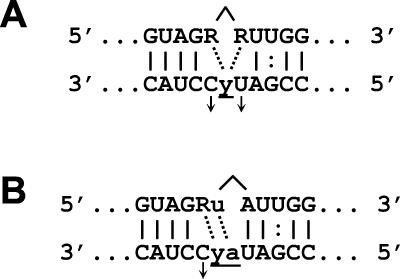
Increased formation of aberrant editing products when gRNA contains guiding pyrimidines (Fig. 1 and 3) may result from base pairing of the guiding pyrimidine with the substrate purine that is next to the ES in either the 5" or 3" mRNA fragment, as illustrated in Fig. 10. Such base pairing would act to bulge out a nucleotide in the gRNA and would enhance ligation by bringing the mRNA termini together. These bulged structures are less energetically favorable than structures in which the helices both 5" and 3" of the ES are continuous, but thermodynamic considerations (35) suggest that they occur as a significant proportion of all editing intermediates. For example, if the guiding A in gPCA6-1A is replaced with a C (Fig. 1A and 1B, lane C), the 3"-terminal G of 5"CL18 can pair with the guiding C, bulging out a C in gRNA, as shown in Fig. 10A (left arrow). The G°37 of this alternate structure is estimated to be 4.2 kcal/mol, less favorable than the unbulged structure (35), but some of this difference is expected to be compensated for by the stacking interaction between the 3"-terminal G and the 5"-terminal A of the 3" fragment.
FIG. 10.
Possible alternate RNA base pairings in editing intermediates with guiding pyrimidines. Alternate base pairings adjacent to the ES are indicated by dotted lines, and the ligations promoted by these pairings are indicated by carets. Potential bulging nucleotides in gRNA are represented by arrows. (A) Base pairing of terminal purines in the 5" and 3" cleavage fragments with a single guiding pyrimidine. The two pairings shown are not expected to occur simultaneously. (B) Base pairing of a 3"-terminal purine with its added U to the guiding A and adjacent pyrimidine.
The structure in Fig. 10B, in which a 3"-terminal purine and added U pair with two guiding nucleotides (Fig. 3), has a similar difference in G°37. A free-energy difference of 4.2 kcal/mol corresponds to a relative abundance of the alternate structure of about 1 in 930 at 37°C. Thus, the alternate structure comprises, at the very least, 1 in 930 of the total RNA intermediates in these cases. Taking into consideration the stacking interaction across the ligation site (for which the free energy is uncertain), the actual proportion of the alternate structure is likely much greater. Thus, these alternate structures, in which the ligatable termini are closely apposed, may promote ligation of 5" fragments with fewer added U's than specified by gRNA.
U addition during precleaved insertion occurred opposite guiding C's (Fig. 3), and neither precleaved nor full-round insertion editing strictly required a potential base pair to secure the 3" end of the 5" fragment (Fig. 6 and 7). That this was not observed previously (18) is most likely due to the greater sensitivity of the editing assays used here. This U addition to the 5" fragment in the absence of base pairing is indicative of the terminal transferase rather than polymerase activity for the addition of U's during editing. The specificity of this activity for U's (N. Ernst, R. P. Igo, Jr., B. Panicucci, A. K. Panigrahi, and K. Stuart, unpublished data) contributes to accuracy of the editing. The gene for the editing 3" TUTase(s) has not yet been identified. The guiding purines appear to stabilize the retention of the correct number of U's that are added to the 5" fragment. This stabilization may entail protecting the added U's from removal by exoUase (7, 16, 23; R. P. Igo, Jr., et al., unpublished data) and/or enhancing ligation by bringing the 3"-terminal hydroxyl of the 5" mRNA fragment in close proximity to the 5"-terminal phosphate of the 3" mRNA fragment (3, 16). The slight reduction in abundance of U addition products with two guiding G's rather than A's (Fig. 3) suggests that less-stable base pairing renders the added U's more susceptible to exoUase activity and less well tethered for ligation.
The presence of base pairs and certain single nucleotides immediately upstream of the ES has a more profound effect on insertion editing than do specific nucleotides and base pairing downstream of the ES. Cleavage, U addition, and ligation all occur in the absence of an upstream base pair, but only in the presence of this base pair do all three activities act together to produce gRNA-specified sequence, as has been observed by others (11). Upstream mismatches greatly reduce precleaved editing to the extent that U addition with three mismatches resembles that obtained without gRNA (Fig. 6). Accurately edited RNA is the most prominent edited product in full-round in vitro editing only when there is an upstream base pair (Fig. 7 and 8) and the upstream base pair closest to the anchor duplex influences the number of inserted U's (Fig. 4), as can G · C, G · U, and even A · C base pairs, even with a strong potential upstream duplex (Fig. 1 and 2).
Ligation seems to be efficient only when the terminal nucleotide is paired with gRNA, and the efficiency is greater with more stable base pairing (Fig. 1 and 2). An upstream base pair is required but nevertheless is not sufficient for accurate insertion editing (Fig. 8). The anchor duplex is required for editing (10, 34; N. Ernst, unpublished results), and base pairing immediately downstream of the ES affects the site of cleavage and hence ES selection but not the number of inserted U's, although the efficiency of ligation is reduced (Fig. 6). These data suggest that the editing complex binds the anchor duplex but also interacts with RNA upstream of the ES.
The results reported here suggest that additional factors contribute to the rarity of C's immediately 5" to insertion ESs (6) beyond the possible selection against C's as a result of their potential base pairing with guiding G's or even A's. Such upstream C's impair both cleavage (data not shown) and U addition (Fig. 5), even in the absence of gRNA (N. Ernst et al., unpublished data). These data suggest that characteristics of the editing machinery itself contribute to the sequence bias. The lack of cleavage downstream of insertion site C's is surprising, given the bias for C's upstream of deletion sites (5). This may be due to mRNA/gRNA structure differences between insertion and deletion substrates. In addition, this may reflect the specificity of the editing endonuclease, or the editing complex may contain two (or more) endonucleases, each with a substrate specificity biased for insertion or deletion substrates.
In contrast to the upstream C, the presence of a pyrimidine 3" to the ES had little effect on precleaved insertion editing (Fig. 5). Addition at low frequency of one more U than specified may reflect breathing at the 5" end of the anchor duplex, which would permit base pairing of the additional U with the Zg purine, perhaps protecting it from removal by 3" exoUase. Thus, the sequence bias and effects of specific nucleotides 5" to the ES on editing activities further indicate the focus of these activities on the 3" end of the 5" cleavage fragment.
A stable upstream duplex that can tether the 5" fragment to gRNA increases insertion editing substantially (5, 11, 16, 19), although this duplex is not present in vivo. We found that the amount and fidelity of editing increased with duplex proximity to the ES, although no full-round editing occurs when the duplex is adjacent to the ES, since no endonucleolytic cleavage occurs (Fig. 9 and data not shown). A larger information region, as in the constructs here, can reduce editing (11). However, placement of an upstream duplex close to the ES in constructs in which the informational region size remains constant increases editing (5), revealing the effect of the upstream duplex.
The large reduction in editing when the upstream duplex abuts the ES reported here for full-round insertion editing and seen in deletion editing (11) is due to the lack of endonucleolytic cleavage. This suggests that the editing endonuclease(s) requires at least one unpaired nucleotides in the substrate. However, cleavage does not always occur immediately 5" to the anchor duplex, even when a bulge is present in pre-mRNA (Fig. 4) (S. D. Lawson, unpublished data). Thus, our understanding of the factors determining the cleavage site is still incomplete. These data suggest that, in vitro, the upstream duplex positions the 3" end of the 5" fragment close to the 3" TUTase and ligase active-site domains. The absence of such upstream duplexes in vivo implies that the 3" oligo(U) tail that is present on all gRNAs could play a comparable role by binding with purine-rich sequences upstream of the ES, and indeed it can bind close to the ES (21). Protein components of the editing complex may also play a role in positioning the 3" end of the 5" fragment, and some of these proteins may bind the oligo(U) tail. The role of the oligo(U) tail and/or editing complex proteins may be especially important for editing the most 5" ESs of an editing block (i.e., the region of editing directed by one gRNA).
Editing activity is focused on the 3" end of the 5" cleavage fragment in both insertion and deletion editing, yet the lack of an upstream base pair dramatically reduces insertion but not deletion editing (11, 20a). This difference may reflect the presentation of insertion versus deletion substrates to the editing catalysts or the use of different catalysis for steps in insertion versus deletion editing (11). The editing complex contains two RNA ligases (24, 27, 32, 33), the significance of which is unclear. However, one is essential for editing (33; A. Schnaufer, unpublished results), and the ligases have been suggested to have differential roles relative to insertion versus deletion editing (32).
Several other proteins of the editing complex have been identified (20, 22, 27a; M. Drożdż, R. Salavati, J. O'Rear, S. S. Palazzo, R. P. Igo, Jr., C. Clayton, and K. Stuart, unpublished data), but only the functions of the RNA ligases and an RNA helicase (26) are evident. However, four related proteins, three of which have zinc finger domains and hence may have RNA and/or protein binding functions, have been identified (27a). This may suggest some functional redundancy in the editing complex. Thus, the enzymatic activities observed here may reflect the composite characteristics of multiple proteins with related activities. Analyses of the catalytic characteristics of the individual enzymes may help to elucidate their roles in editing, although they may be affected by their integration into the editing complex.
Acknowledgments
We thank Nicole Carmean, Brian Panicucci, Jeff O'Rear, and Micaiah Evans for producing the T. brucei mitochondria used in this study; Aswini Panigrahi and Barbara Morach for purification of mitochondrial extract; and Gonzalo Domingo-Villegas, Reza Salavati, and Nancy Lewis Ernst for their comments on the manuscript.
This work was supported by NIH Postdoctoral Fellowship AI10312 to R.P.I. and NIH grant GM42188 and HFSPO grant RG/97 to K.S.
REFERENCES
- 1.Allen, T. E., S. Heidmann, R. Reed, P. J. Myler, H. U. Göringer, and K. D. Stuart. 1998. The association of guide RNA binding protein gBP21 with active RNA editing complexes in Trypanosoma brucei. Mol. Cell. Biol. 18:6014-6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aphasizhev, R., and L. Simpson. 2001. Isolation and characterization of a U-specific 3"-5" exonuclease from mitochondria of Leishmania tarentolae. J. Biol. Chem. 276:21280-21284. [DOI] [PubMed] [Google Scholar]
- 3.Blanc, V., J. D. Alfonzo, R. Aphasizhev, and L. Simpson. 1999. The mitochondrial RNA ligase from Leishmania tarentolae can join RNA molecules bridged by a complementary RNA. J. Biol. Chem. 274:24289-24296. [DOI] [PubMed] [Google Scholar]
- 4.Blum, B., N. Bakalara, and L. Simpson. 1990. A model for RNA editing in kinetoplastid mitochondria: “guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 60:189-198. [DOI] [PubMed] [Google Scholar]
- 5.Burgess, M. L. K., S. Heidmann, and K. Stuart. 1999. Kinetoplastid RNA editing does not require the terminal 3" hydroxyl of guide RNA, but modifications to the guide RNA terminus can inhibit in vitro U insertion. RNA 5:883-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burgess, M. L. K., and K. Stuart. 2000. Sequence bias in edited kinetoplastid RNAs. RNA 6:1492-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byrne, E. M., G. J. Connell, and L. Simpson. 1996. Guide RNA-directed uridine insertion RNA editing in vitro. EMBO J. 15:6758-6765. [PMC free article] [PubMed] [Google Scholar]
- 8.Connell, G. J., and L. Simpson. 1998. Role of RNA structure in RNA editing, p. 641-667. In R. W. Simons and M. Grunberg-Manago (ed.), RNA structure and function. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 9.Corell, R. A., L. K. Read, G. R. Riley, J. K. Nellissery, T. E. Allen, M. L. Kable, M. D. Wachal, S. D. Seiwert, P. J. Myler, and K. D. Stuart. 1996. Complexes from Trypanosoma brucei that exhibit deletion editing and other editing-associated properties. Mol. Cell. Biol. 16:1410-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cruz-Reyes, J., and B. Sollner-Webb. 1996. Trypanosome U-deletional RNA editing involves guide RNA-directed endonuclease cleavage, terminal U exonuclease, and RNA ligase activities. Proc. Natl. Acad. Sci. USA 93:8901-8906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruz-Reyes, J., A. Zhelonkina, L. Rusché, and B. Sollner-Webb. 2001. Trypanosome RNA editing: simple guide RNA features enhance U deletion 100-fold. Mol. Cell. Biol. 21:884-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Decker, C. J., and B. Sollner-Webb. 1990. RNA editing involves indiscriminate U changes throughout precisely defined editing domains. Cell 61:1001-1011. [DOI] [PubMed] [Google Scholar]
- 13.Estévez, A. M., and L. Simpson. 1999. Uridine insertion/deletion RNA editing in trypanosome mitochondria: a review. Gene 240:247-260. [DOI] [PubMed] [Google Scholar]
- 14.Feagin, J. E., D. P. Jasmer, and K. Stuart. 1987. Developmentally regulated addition of nucleotides within apocytochrome b transcripts in Trypanosoma brucei. Cell 49:337-345. [DOI] [PubMed] [Google Scholar]
- 15.Harris, M. E., D. R. Moore, and S. L. Hajduk. 1990. Addition of uridines to edited RNAs in trypanosome mitochondria occurs independently of transcription. J. Biol. Chem. 265:11368-11376. [PubMed] [Google Scholar]
- 16.Igo, R. P., Jr., S. S. Palazzo, M. L. K. Burgess, A. K. Panigrahi, and K. Stuart. 2000. Uridylate addition and RNA ligation contribute to the specificity of kinetoplastid insertion RNA editing. Mol. Cell. Biol. 20:8447-8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Igo, R. P., Jr., D. S. Weston, N. L. Ernst, A. K. Panigrahi, R. Salavati, and K. Stuart. 2002. Role of uridylate-specific exoribonuclease activity in Trypanosoma brucei RNA editing. Euk. Cell 1:112-118. [DOI] [PMC free article] [PubMed]
- 17.Kable, M. L., S. Heidmann, and K. D. Stuart. 1997. RNA editing: getting U into RNA. Trends Biochem. Sci. 22:162-166. [DOI] [PubMed] [Google Scholar]
- 18.Kable, M. L., S. D. Seiwert, S. Heidmann, and K. Stuart. 1996. RNA editing: a mechanism for gRNA-specified uridylate insertion into precursor mRNA. Science 273:1189-1195. [DOI] [PubMed] [Google Scholar]
- 19.Kapushoc, S. T., and L. Simpson. 1999. In vitro uridine insertion RNA editing mediated by cis-acting guide RNAs. RNA 5:656-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Köller, J., U. F. Müller, B. Schmid, A. Missel, V. Kruft, K. Stuart, and H. U. Göringer. 1997. Trypanosoma brucei gBP21: an arginine-rich mitochondrial protein that binds to guide RNA with high affinity. J. Biol. Chem. 272:3749-3757. [DOI] [PubMed] [Google Scholar]
- 20a.Lawson, S. D., R. P. Igo, Jr., R. Salavati, and K. Stuart. The specificity of nucleotide removal during RNA editing in Trypanosoma brucei. RNA, in press. [DOI] [PMC free article] [PubMed]
- 21.Leung, S. S., and D. J. Koslowsky. 1999. Mapping contacts between gRNA and mRNA in trypanosome RNA editing. Nucleic Acids Res. 27:778-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madison-Antenucci, S., R. S. Sabatini, V. W. Pollard, and S. L. Hajduk. 1998. Kinetoplastid RNA-editing-associated protein 1 (REAP-1): a novel editing complex protein with repetitive domains. EMBO J. 17:6368-6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McManus, M. T., B. K. Adler, V. W. Pollard, and S. L. Hajduk. 2000. Trypanosoma brucei guide RNA poly(U) tail formation is stabilized by cognate mRNA. Mol. Cell. Biol. 20:883-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McManus, M. T., M. Shimamura, J. Grams, and S. L. Hajduk. 2001. Identification of candidate mitochondrial RNA editing ligases from Trypanosoma brucei. RNA 7:167-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milligan, J. F., D. R. Groebe, G. W. Witherell, and O. C. Uhlenbeck. 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15:8783-8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Missel, A., A. E. Souza, G. Norskau, and H. U. Göringer. 1997. Disruption of a gene encoding a novel mitochondrial DEAD-box protein in Trypanosoma brucei affects edited mRNAs. Mol. Cell. Biol. 17:4895-4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panigrahi, A. K., S. P. Gygi, N. L. Ernst, R. P. Igo, Jr., S. S. Palazzo, A. Schnaufer, D. S. Weston, N. Carmean, R. Salavati, R. Aebersold, and K. D. Stuart. 2001. Identification of two novel proteins, TbMP52 and TbMP48, associated with the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 21:380-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a.Panigrahi, A. K., A. Schnaufer, N. Carmean, R. P. Igo, Jr., S. P. Gygi, N. L. Ernst, S. S. Palazzo, D. S. Weston, R. Aebersold, R. Salavati, and K. D. Stuart. 2001. Four related proteins of the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 21:6833-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piller, K. J., C. J. Decker, L. N. Rusché, and B. Sollner-Webb. 1995. Trypanosoma brucei mitochondrial guide RNA-mRNA chimera-forming activity cofractionates with an editing-domain-specific endonuclease and RNA ligase and is mimicked by heterologous nuclease and RNA ligase. Mol. Cell. Biol. 15:2925-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pollard, V. W., M. E. Harris, and S. L. Hajduk. 1992. Native mRNA editing complexes from Trypanosoma brucei mitochondria. EMBO J. 11:4429-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riley, G. R., R. A. Corell, and K. Stuart. 1994. Multiple guide RNAs for identical editing of Trypanosoma brucei apocytochrome b mRNA have an unusual minicircle location and are developmentally regulated. J. Biol. Chem. 269:6101-6108. [PubMed] [Google Scholar]
- 31.Rusché, L. N., J. Cruz-Reyes, K. J. Piller, and B. Sollner-Webb. 1997. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 16:4069-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rusché, L. N., C. E. Huang, K. J. Piller, M. Hemann, E. Wirtz, and B. Sollner-Webb. 2001. The two RNA ligases of the Trypanosoma brucei RNA editing complex: cloning the essential band IV gene and identifying the band V gene. Mol. Cell. Biol. 21:979-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schnaufer, A., A. K. Panigrahi, B. Panicucci, R. P. Igo, Jr., R. Salavati, and K. Stuart. 2001. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science 291:2159-2162. [DOI] [PubMed] [Google Scholar]
- 34.Seiwert, S. D., S. Heidmann, and K. Stuart. 1996. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell 84:831-841. [DOI] [PubMed] [Google Scholar]
- 35.Serra, M. J., and D. H. Turner. 1995. Predicting thermodynamic properties of RNA. Methods Enzymol. 259:242-261. [DOI] [PubMed] [Google Scholar]
- 36.Simpson, L., S. H. Wang, O. H. Thiemann, J. D. Alfonzo, D. A. Maslov, and H. A. Avila. 1998. U-insertion/deletion edited sequence database. Nucleic Acids Res. 26:170-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stuart, K., T. E. Allen, S. Heidmann, and S. D. Seiwert. 1997. RNA editing in kinetoplastid protozoa. Microbiol. Mol. Biol. Rev. 61:105-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stuart, K., M. L. Kable, T. E. Allen, and S. Lawson. 1998. Investigating the mechanism and machinery of RNA editing. Methods 15:3-14. [DOI] [PubMed] [Google Scholar]