Abstract
The expression of ubiquitin-like modifier ISG15 and its conjugation to target proteins are highly induced by interferon (IFN) stimulation and during viral and bacterial infections. However, the biological significance of this modification has not been clearly understood. To investigate the function of protein modification by ISG15, we generated a mouse model deficient in UBE1L, an ISG15-activating enzyme. Ube1L−/− mice did not produce ISG15 conjugates but expressed free ISG15 normally. ISGylation has been implicated in the reproduction and innate immunity. However, Ube1L−/− mice were fertile and exhibited normal antiviral responses against vesicular stomatitis virus and lymphocytic choriomeningitis virus infection. Our results indicate that UBE1L and protein ISGylation are not critical for IFN-α/β signaling via JAK/STAT activation. Moreover, using Ube1L/Ubp43 double-deficient mice, we showed that lack of UBP43, but not the increase of protein ISGylation, is related to the increased IFN signaling in Ubp43-deficient mice.
Interferon (IFN)-stimulated gene 15 (ISG15) encodes a 17-kDa protein whose expression is highly induced upon IFN stimulation (1, 5, 15, 17, 34). ISG15 is composed of two domains, each of which bears high sequence and structural similarity to ubiquitin (30). As such, ISG15 was originally found to react with certain ubiquitin antibodies and has thus also been named ubiquitin cross-reactive protein (7). Like ubiquitin and other members of the ubiquitin-like modifiers, ISG15 can exist as a free protein or as a covalent conjugate to target proteins (25). Paralleling the protein ubiquitination system, ISG15 conjugation and deconjugation processes are controlled by a canonical set of enzymes, including the ISG15-activating enzyme UBE1L (41), ISG15-conjugating enzyme Ubc8 (13, 43), the E3 ligases, and ISG15-deconjugating enzyme UBP43 (27). Unlike components of the ubiquitination system, though, the expression of ISG15 and most of the known ISG15 modification enzymes is IFN inducible (1). Therefore, protein ISG15 modification (ISGylation) is strongly induced upon IFN treatment or viral and bacterial infections. Whether protein ISGylation is involved in the diverse downstream responses of IFN signaling triggered by microbes or other stress remains to be determined.
We have documented that UBP43 (USP18) is a 43-kDa member of the ubiquitin-specific protease family (10, 23, 37, 39). Several other groups also cloned this gene independently during the analysis of cellular responses to IFN treatment or viral infections (12, 22, 42). Studies about the substrate specificity of UBP43 indicated that this enzyme preferentially removes ISG15 from its conjugates, compared to ubiquitin, Sumo, and Nedd8 conjugates (27). Compared to the wild-type control, higher levels of ISGylated proteins, but not ubiquitinated proteins, are detected in Ubp43-deficient cells (36). Ubp43−/− cells are hypersensitive to IFN-α/β treatment, have enhanced and prolonged STAT1 phosphorylation, and show increased expression of IFN-stimulated genes, including ISG15 (28). Furthermore, Ubp43 knockout mice exhibited enhanced resistance to certain viral and bacterial infections (14, 35). These findings suggested a link between protein ISGylation and IFN-α/β responses. However, we could not rule out the possibility that these results were unrelated to protein ISGylation but, rather, were related to the lack of UBP43 expression. To further investigate the functional roles of protein ISGylation, we generated mice deficient in ISG15 conjugation. Free ISG15 has been reported to be a cytokine that enhances IFN-γ production and nature killer cell proliferation (2). Therefore, we generated Ube1L knockout mice lacking ISG15 conjugation but not free ISG15. Homozygous Ube1L knockout mice were healthy and fertile. Furthermore, Ube1L-deficient cells did not show any abnormal responses to IFN treatment, and Ube1L+/+ and Ube1L−/− cells exhibited similar susceptibility to vesicular stomatitis virus (VSV) and lymphocytic choriomeningitis virus (LCMV) infection, indicating that Ube1L and protein ISGylation are not essential for IFN signaling. Using Ube1L/Ubp43 double-deficient mice, we demonstrated that lack of UBP43, not the increase of protein ISGylation, is related to the increased IFN-α/β signaling of Ubp43-deficient mice.
MATERIALS AND METHODS
Generation and genotyping of Ube1L knockout mice.
To generate the targeting construct, the 5′ arm, 3′ arm, and target regions as indicated in Fig. 1A were amplified by PCR using mouse genomic DNA prepared from 129Sv embryonic stem (ES) cells as a template. PCR products were sequenced to check for errors and subsequently cloned into pBluescript (pBS; Stratagene). The target region was first subcloned into pBS with EcoR I/SpeI digestion. The LoxP site between exons 13 and 14 was generated by adding the LoxP sequence to the PCR primer for the target region, and the PCR product was ligated to pBS to create pBS_Target-LoxP. A 4.2-kb fragment from the 3′-arm region digested with Hind III/SalI was ligated to pBS_Target-LoxP digested with the same enzymes, resulting in pBS_Target-LoxP_3′-arm. To add Frt-PGKneo-Frt-LoxP to the construct, a fragment containing the Frt-PGKneo-Frt-LoxP sequence was cleaved from pK11 FRT-PGKneo-FRT-loxP pBSSK (kindly provided by Gail Martin, University of California, San Francisco [29])with SacI/KpnI digestion and blunted. pBS_Target-LoxP_3′-arm was digested with SacI/KpnI, blunted, and ligated with the Frt-PGKneo-Frt-LoxP fragment (pBS_Target-LoxP_3′-arm_Frt-PGKneo-Frt-LoxP). Finally, a NotI fragment of 3.8 kb from the 5′-arm region was ligated to pBS_Target-LoxP_3′-arm_Frt-PGKneo-Frt-LoxP digested with the same enzyme, resulting in the final construct (pBS_Target-LoxP_3′-arm_Frt-PGKneo-Frt-LoxP_5′-arm). The final plasmid was linearized using PvuI and used for electroporation. The electroporation, expansion of ES (from 129 mice) cell clones, and blastocyte injection were performed by inGenious Targeting Laboratory, Inc. (New York, NY).
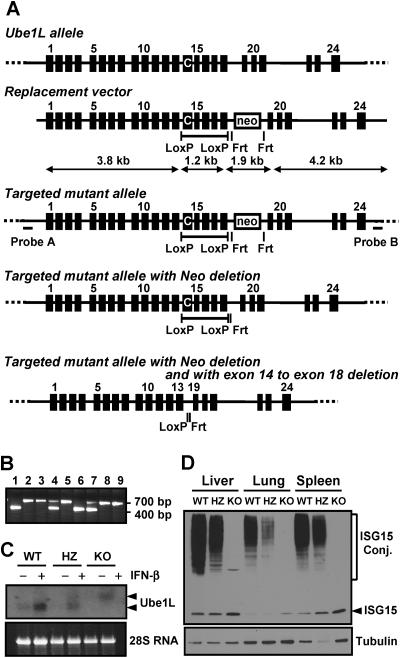
FIG. 1.
Generation of Ube1L knockout mice. (A) Schematics of Ube1L knockout strategy. C on exon 14 indicates active site cysteine of UBE1L. (B) PCR genotyping of mouse tail DNA. (C) Northern blot analysis of Ube1L mRNA. A mixture of cells from spleen and thymus from mice of each genotype was cultured in vitro in the absence or presence of 100 U/ml of IFN-β for 12 h. The mRNA for the Ube1L gene was detected by Northern blotting. (D) Western blot analysis of protein ISGylation upon LPS challenge in mice. Eighteen hours after LPS injection, livers, lungs, and spleens were harvested, and protein ISGylation was detected by Western blotting using anti-mouse ISG15 polyclonal antibody. WT, wild type; HZ, heterozygous; KO, knockout.
Gene targeting in ES clones was determined by Southern blot analysis (Fig. 1A, probes A and B, with SacI [wild type, 16 kb; knockout, 8.4 kb] and EcoRV [wild type, 7.9 kb; knockout, 6.0 kb] digestion, respectively [data not shown]). Germ line-transmitted chimeric mice were produced using standard techniques. Mice homozygous for disruption of the Ube1L allele were then obtained by intercrossing F1 heterozygotes (129 × C57BL/6 mixed). Genotyping of resultant and successive progeny was performed by PCR using one forward primer on exon 18 (Ube1LGT-FW, ACCTCTTCTCTGCTGAGCATGG) and two reverse primers, one on the intron between exon 18 and exon 19 (Ube1LGT-RV, CTACTGAGGGACTAGAGGATGG) and the other on the neo gene (NEO-RV, CTTCCTCGTGCTTACGGTATCG). The PCR generates two bands; one is ∼400 bp, indicating the wild-type allele from the Ube1LGT-FW and Ube1LGT-RV primer pair; the other is ∼700 bp, showing the targeted allele from the Ube1LGT-FW and NEO-RV pair. All animals used in the studies were handled in accordance with guidelines of The Scripps Research Institute, and procedures were approved by the Institutional Animal Care and Use Committee of the institute. All mice used for this work were in a mixed background of C57BL/6 and 129Sv.
Generation of Ube1L/Ubp43 double-knockout mice.
Ubp43 and Ube1L double-heterozygous mice (Ubp43+/−/Ube1L+/−) were generated from crossing Ube1L−/− and Ubp43+/− mice (36). Ubp43+/−/Ube1L+/− mice were intercrossed to generate double-knockout mice and their littermates.
LPS and poly(I · C) injection and blood cell counting.
Lipopolysaccharide (LPS) (Escherichia coli serotype O127:B8; Sigma, St. Louis, MO) was dissolved in phosphate-buffered saline (PBS) and intraperitoneally injected to mice at 15 mg/kg of body weight. Poly(I · C) (Sigma) was dissolved in PBS and injected intraperitoneally into mice at 10 mg/kg of body weight. The total white blood cell count and blood parameters were measured by using the HEMAVET multispecies hematology analyzer (Drew Scientific, Oxford, CT) following the manufacturer's instructions. Peripheral blood smear was stained with ACCUSTAIN Wright stain and ACCUSTAIN Giemsa stain solutions (Sigma) following the two-step staining instruction from the manufacturer. Differential counts of blood cells were obtained by counting 200 nucleated cells for each sample.
Antibodies and Western blot analyses.
Rabbit anti-mouse ISG15 polyclonal antibody has been described previously (26). Phosphospecific antibody for STAT1 (Tyr701) was purchased from Cell Signaling (Beverly, MA). Antibodies for ubiquitin and tubulin-α were from Sigma. Antibodies for STAT1 and inducible nitric oxide synthase (iNOS) were from Santa Cruz Biotechnology (Santa Cruz, CA) and from Cayman Chemicals (Ann Arbor, MI), respectively. Western blotting was performed as described previously (27).
Northern blotting.
Total RNA from bone marrow-derived macrophages was isolated using RNA Bee reagent according to the manufacturer's instructions (Tel-Test Inc., Friendswood, TX). Ten micrograms of total RNA from each time point was separated on an agarose-formaldehyde gel (0.22 M), blotted on Hybond N+ membrane (GE Healthcare, Piscataway, NJ), and probed with 32P-labeled cDNAs. UV detection of 28S rRNA was used as an equal loading control.
In vitro bone marrow cell culture and apoptosis assay.
Liquid culture of bone marrow cells was performed in RPMI 1640 medium with 10% fetal bovine serum, 10 ng/ml interleukin-3 (PeproTech, Rocky Hill, NJ), 10 ng/ml interleukin-6 (PeproTech), and 100 ng/ml stem cell factor (PeproTech) in the absence or presence of the indicated amount of IFN-β (Calbiochem, San Diego, CA). An annexin V-PE/7-AAD (7-amino-actinomycin D) apoptosis assay was performed using an apoptosis detection kit according to the manufacturer's instructions (BD PharMingen, San Diego, CA).
Viral infection.
A VSV antiviral assay was performed as reported previously (40). Ube1L+/+ and Ube1L−/− mice were inoculated intracerebrally with 30 μl of serum-free Dulbecco's modified Eagle's medium containing 1,000 PFU of Armstrong strain of LCMV.
Generation of MEFs.
Embryos (day 12.5) were aseptically taken out from the mother and washed once with PBS. The head and liver were removed from the body, and the rest was soaked in 2 ml of trypsin, crushed by passing through a needle, and then incubated at 37°C for 30 min. Cells were harvested, resuspended in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) with 10% fetal bovine serum (HyClone, Logan, UT) and 2 mM l-glutamine (Invitrogen, Carlsbad, CA), and then plated in one 10-cm dish per embryo. All mouse embryonic fibroblast (MEF) cells used in this report are from breeding of Ubp43 and Ube1L double-heterozygous mice (Ubp43+/− Ube1L+/−).
RESULTS
Generation of Ube1L-deficient mice.
To investigate the role of protein ISGylation without compromising any possible functions of free ISG15 itself, we generated the Ube1L knockout mice that lack the only known E1 enzyme for the ISGylation system. For the conditional disruption of the Ube1L gene (Fig. 1A) we added two loxP sites to the introns encompassing exons 14 to 18, where the active-site cysteine on exon 14 is located (13). This construct also contains the bacterial neomycin gene as a selection marker in an opposite direction to the Ube1L gene. Chimeric mice were produced by injection of two independent clones of heterozygous ES cells into C57BL/6 blastocyst stage embryos, and germ line transmission was determined by breeding with wild-type C57BL/6 mice.
Defective protein ISGylation but normal ubiquitination in Ube1L knockout mice.
Mice were genotyped by PCR (Fig. 1B), and expression levels of Ube1L mRNA were compared among wild-type, heterozygous, and Ube1L knockout mice. A mixture of cells from the spleen and thymus of each genotype was cultured in vitro in the absence or presence of 100 U/ml of IFN-β for 12 h. The mRNA for the Ube1L gene was detected by Northern blotting (Fig. 1C). Wild-type cells showed a single band of mRNA; heterozygous cells contained two bands of Ube1L mRNA, one the same size as that of the wild type and another slower-migrating band, indicating a larger mRNA resulting from the insertion of the Neo gene. Ube1L knockout cells showed a single band of mRNA, which matched the larger band of heterozygous cells. To determine whether lack of Ube1L mRNA correlated with decreased ISGylation in vivo, we injected wild-type, heterozygous, and knockout mice with LPS, which should result in IFN-α/β production and subsequent increased ISGylation (14). Approximately 18 h later, livers, lungs, and spleens were harvested, and protein ISGylation was detected by Western blotting (Fig. 1D). Protein ISGylation was readily detected in tissues from wild-type but not from Ube1L knockout mice. Tissues from heterozygote mice showed a decreased amount of ISG15 conjugates, indicating that the lack of expression of Ube1L from one allele caused reduced conjugation of ISG15 to target proteins. Ube1L-deficient cells showed an increased amount of free ISG15 as a result of the lack of conjugation. We obtained similar results with mice derived from two independent ES clones. These results indicate that Ube1L knockout mice are defective in protein ISGylation but not in free ISG15 expression.
Ube1L was identified as a gene encoding a protein homologous to the ubiquitin-activating enzyme (UBE1) (16). UBE1L shares significant homology with UBE1 including similar domain structure and high conservation in their amino acid sequences (46% identical between human UBE1 and UBE1L) (Fig. 2A). Furthermore, ISG15 E2 Ubc8 is a known ubiquitin E2 (18). These findings led us to consider whether UBE1L contributes to protein ubiquitination. For this we compared protein ISGylation and ubiquitination in UBE1L-deficient macrophages treated with LPS (Fig. 2B) and MEFs treated with IFN (see Fig. 6). ISGylated proteins were not detected in macrophages or MEFs derived from Ube1L−/− mice. Protein ubiquitination was normal. Taken together, these results strongly support the view that UBE1L is the only strictly required E1 for the ISGylation pathway, but UBE1L is not involved in the cellular ubiquitination process.
FIG. 2.
Analysis of protein ISGylation and ubiquitination in Ube1L knockout cells. (A) Schematics of human ubiquitin-activating enzyme (UBE1) and ISG15-activating enzyme (UBE1L) indicating similar domain structures. ThiF, a repeated domain in ubiquitin-activating enzyme E1 and members of the bacterial ThiF/MoeB/HesA family; UBACT, a repeat in ubiquitin-activating enzyme E1. The numbers on the right side indicate the number of amino acids in each protein. (B) Bone marrow-derived macrophages from wild-type and Ube1L knockout mice were treated with 100 ng/ml of LPS for the time periods indicated in the figure. Cells were harvested and subjected to immunoblotting against ISG15 and ubiquitin.
FIG. 6.
Detection of ISGylation, STAT phosphorylation, iNOS expression and ubiquitination in wild-type (WT), Ube1L−/−, Ubp43−/−, and Ube1L/Ubp43 double-knockout (DKO) cells. MEFs prepared from littermates of wild-type, Ube1L−/−, Ubp43−/−, and Ube1L/Ubp43 double-knockout mice were treated with 1,000 U/ml of IFN-β for the indicated time periods. Protein ISGylation, phosphorylation of STAT1, STAT1 itself, iNOS expression, and protein ubiquitination were analyzed by Western blotting.
Normal responses of Ube1L-deficient cells and mice to IFN-α/β.
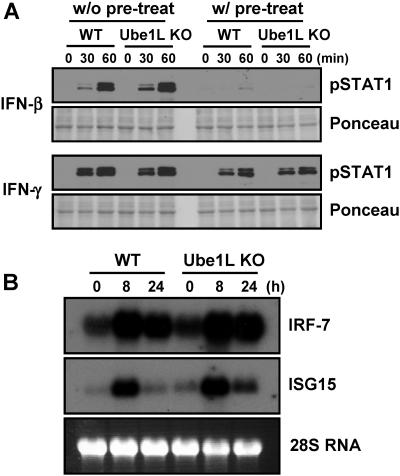
Offspring of heterozygous mice (Ube1L+/−) breeding were born at the expected Mendelian ratio of 1:2:1 of Ube1L+/+, Ube1L+/−, and Ube1L−/− (Ube1L+/+, 19; Ube1L+/−, 42; and Ube1L−/−, 18). This indicates that Ube1L and protein ISGylation are not essential for embryonic development and agrees with previous observations on the ISG15 knockout mice (31). Ube1L knockout mice appeared normal, healthy, and fertile. Since the ISGylation system is induced upon IFN challenge and UBP43-deficient cells showed hypersensitivity to IFN-α/β (28), we examined the IFN-response of Ube1L-deficient cells. Macrophages derived from bone marrow cells of wild-type or Ube1L knockout mice were cultured in vitro and treated with 100 U/ml of IFN-β or IFN-γ, and STAT1 phosphorylation was detected as an indication of the activation of the signaling pathway. Increased phosphorylation of STAT1 upon IFN treatment was observed. However, there was no difference in STAT1 phosphorylation between wild-type and Ube1L-deficient cells (Fig. 3A, w/o pretreat). We then analyzed the effect of IFN-pretreatment, which induces protein ISGylation in wild-type cells but not in knockout cells. IFN-β or IFN-γ (100 U/ml) was added to the cells for 24 h, and then the same amounts of IFNs were added again. Pretreatment desensitizes cells to additional stimulation by IFN, and as shown in Fig. 3A (w/ pretreatment), we found significantly less phospho-STAT1 but no difference between wild-type and knockout cells. We also compared the expression of ISGs (ISG15 and IRF-7) in macrophages from wild-type and Ube1L knockout mice upon LPS treatment. LPS activates the expression of these genes via the IFN signaling pathway (14). Wild-type and Ube1L-deficient cells showed quite similar expression patterns for both genes upon LPS treatment (Fig. 3B).
FIG. 3.
Analysis of IFN-response in Ube1L knockout cells. (A) Bone marrow-derived macrophages from wild-type (WT) and Ube1L knockout (KO) mice were cultured in the absence or presence of 100 U/ml of IFN-β or IFN-γ for 24 h and then treated with the same amount of IFN-β or IFN-γ for the indicated time periods. Cells were harvested and subjected to immunoblotting against pSTAT1 and STAT1. (B) Macrophages from wild-type (WT) and Ube1L knockout (KO) mice were treated with 100 ng/ml of LPS and harvested at the indicated time points, and the induction of genes for ISG15 and IRF-7 was detected by Northern blotting.
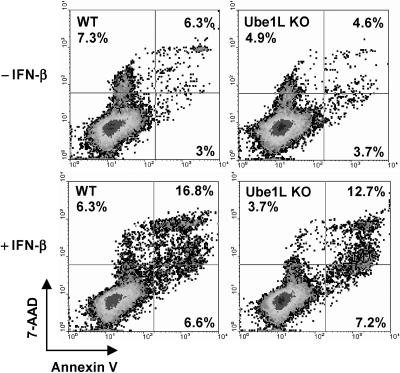
We have previously reported that bone marrow cells from Ubp43-deficient cells are hypersensitive to apoptosis caused by IFN-α/β (28). Therefore, we examined the Ube1L knockout mouse model to see whether a defect in the ISGylation system plays a role in IFN-induced cell death. A low dose of IFN-β (100 U/ml) was first added to the bone marrow cells to induce ISGylation for 24 h; then the cells were treated with a high IFN-β dose (1,000 U/ml) for 48 h to induce apoptosis. Apoptosis of bone marrow cells was measured by annexin V/7-AAD staining. As shown in Fig. 4, there was no significant difference between wild-type and Ube1L knockout cells in their sensitivity to IFN-induced cell death. Furthermore, no differences in peripheral blood cell count and other blood parameters were observed between wild-type and Ube1L knockout mice either without any treatment or after poly(I · C) stimulation (Table 1). In summary, Ube1L-deficient cells do not show any detectable differences from wild-type cells in their responses to IFN treatment.
FIG. 4.
Ube1L-deficient cells are not hypersensitive to IFN-α/β-induced cell death. Bone marrow cells isolated from wild-type (WT) and Ube1L knockout (Ube1L KO) mice were cultured in vitro and treated with 100 U/ml IFN-β for 24 h and then treated with 1,000 U/ml IFN-β for an additional 48 h. An apoptosis assay was performed by staining cells with annexin V/7-AAD, followed by fluorescence-activated cell sorting analysis.
TABLE 1.
Blood analysisa
| Mice and treatment | WBCs (103/μl) | RBCs (103/μl) | Hb (g/dl) | PLT (103/μl) | % in blood WBCs
|
|||
|---|---|---|---|---|---|---|---|---|
| Lymphocytes | Neutrophils | Eosinophils | Monocytes | |||||
| Before poly(I · C) | ||||||||
| WT (n = 4) | 9.5 ± 2.9 | 9.8 ± 0.3 | 12.6 ± 2.4 | 995 ± 119 | 81.1 ± 2.6 | 13.5 ± 2.5 | 1.9 ± 1.8 | 3.5 ± 0.6 |
| Ube1L−/− (n = 4) | 9.5 ± 1.3 | 8.9 ± 1.3 | 13.2 ± 2.3 | 930 ± 102 | 80.9 ± 1.9 | 12.8 ± 3.4 | 2.9 ± 1.3 | 3.6 ± 1.4 |
| After poly (I · C) | ||||||||
| WT (n = 4) | 7.3 ± 1.4 | 8.2 ± 1.5 | 12.6 ± 3.4 | 954 ± 224 | 67.4 ± 2.9 | 26.1 ± 4.3 | 2.9 ± 1.7 | 3.6 ± 1.4 |
| Ube1L−/− (n = 4) | 9.5 ± 1.3 | 8.5 ± 1.0 | 12.5 ± 1.5 | 928 ± 50 | 67.0 ± 2.4 | 26.6 ± 3.8 | 2.6 ± 1.3 | 3.6 ± 0.8 |
Samples were collected via retro-orbital bleeding from wild-type (WT) and Ube1L knockout (Ube1L−/−) mice before and after daily poly(I·C) injections for 2 days. WBCs, white blood cells; RBCs, red blood cells; Hb, hemoglobin; PLT, platelets. Values are means ± standard deviations.
Similar antiviral responses between wild-type and Ube1L knockout cells against VSV and LCMV.
In addition to enhanced protein ISGylation and hypersensitivity to IFN stimulation, Ubp43−/− mice also exhibited increased resistance to viral infection (35). Therefore, we examined the consequences of an absence of protein ISGylation on the antiviral effects of IFN in MEFs (Fig. 5A). Treatment with increasing concentrations of IFN-β correlated positively with the antiviral stage of both wild-type and Ube1L knockout MEFs upon infection with VSV, with no detectable differences in the response between the two genotypes. To further investigate the antiviral potential of protein ISGylation, we determined mouse survival after intracranial LCMV infection. Five wild-type and five knockout mice were intracranially injected with 1,000 PFU of LCMV and examined for mouse lethality caused by virus-induced LCM. All of the mice died between days 6 and 7 (Fig. 5B), indicating that there is no significant difference between wild-type and Ube1L knockout mice for LCMV-caused death. These two viral infection experiments suggest that protein ISGylation is not significantly involved in the antiviral response against VSV and LCMV.
FIG. 5.
VSV protection assay. (A) Wild-type and Ube1L knockout MEFs were left untreated or treated with 100 or 1,000 U/ml of IFN-β for 24 h, followed by VSV infection ranging from a multiplicity of infection of 0 to 104 per well for an additional 24 h. Cell viability was assessed by crystal violet staining. (B) Ube1L+/+ (n = 5) and Ube1L−/− (n = 5) mice were infected intracerebrally with 1,000 PFU of LCMV and monitored daily for development of clinical symptoms and survival.
Hypersensitivity of Ubp43-deficient cells to IFN-α/β is not rescued in a Ube1L/Ubp43 double knockout.
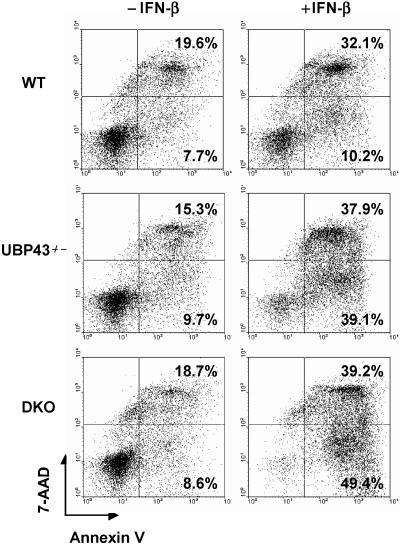
Ubp43-deficient cells have enhanced protein ISGylation and are hypersensitive to IFN-α/β treatment, which led to the originally proposed role of ISGylation in regulating IFN signaling. However, as the data indicate, Ube1L-deficient cells lack protein ISGylation but are normal in inducing IFN signaling. To further verify these observations, we generated Ubp43/Ube1L double-deficient mice by intercrossing Ubp43 and Ube1L double-heterozygous mice (Ubp43+/−/Ube1L+/−). We first checked IFN-induced ISGylation and STAT1 phosphorylation in primary MEFs derived from wild-type, Ube1L−/−, Ubp43−/−, and Ube1L/Ubp43 double-deficient mice. As expected, elevated protein ISGylation was detected in Ubp43−/− cells compared to wild-type cells, and no ISGylation was detected in either Ube1L−/− or Ube1L/Ubp43 double-deficient cells (Fig. 6). However, the prolonged phosphorylation of STAT1 in Ubp43−/− cells was not affected in Ube1L/Ubp43 double-deficient cells even though these cells did not show protein ISGylation, indicating that the prolonged phosphorylation of STAT1 in Ubp43−/− cells is unaffected by protein ISGylation levels. Next, we examined IFN-induced apoptosis in bone marrow cells prepared from wild-type, Ubp43−/−, and Ube1L/Ubp43 double-knockout mice. As shown in Fig. 7, there is no significant difference in IFN-induced cell death between Ubp43−/− and Ube1L/Ubp43 double-knockout cells. These results demonstrate that the hypersensitivity to IFN-α/β in UBP43-deficient cells is not related to protein ISGylation.
FIG. 7.
Ube1L/Ubp43 double-knockout cells remained hypersensitive to IFN-induced cell death. Bone marrow cells from wild-type (WT), Ubp43 knockout (Ubp43−/−), and Ubp43/Ube1L double-knockout (DKO) mice were treated with 50 U/ml of IFN-β for 48 h. The apoptosis analysis was performed as described in the legend of Fig. 4.
DISCUSSION
Ubiquitin and the family of ubiquitin-like modifiers are important players in regulating multiple cellular events ranging from protein stability to signal transduction (9, 11, 32). In contrast to ubiquitin and other ubiquitin-like modifiers, though, much less is known about the function of ISG15 and protein ISGylation. ISG15 was discovered during the studies of proteins and genes differentially expressed upon IFN treatment (5). Further studies indicated that LPS, retinoic acid, and other chemicals and stresses can also induce ISG15 expression and protein ISGylation (21, 24, 33). However, IFN-α/β signaling via ISGF3 complex formation and activation of IRF3 via its phosphorylation remain as major mechanisms for the induction of ISG15 expression and conjugation. Since IFN and LPS signaling are closely linked to viral and bacterial infections, protein ISGylation may play a role in innate immunity to microbes (1).
Cells derived from ISG15-deconjugating enzyme UBP43 knockout mice (36) showed markedly higher levels of protein ISGylation and also displayed a hypersensitivity to IFN-α/β treatment (28), suggesting that increased protein ISGylation causes enhanced IFN signaling. This, however, does not conclusively demonstrate whether increased ISGylation was the cause or a consequence of the IFN hypersensitivity. To address this issue we generated knockout mice for Ube1L, the E1 for ISG15 conjugation.
To study protein ISGylation in different cell types and at different developmental stages, we decided to generate a conditional Ube1L knockout model. Interestingly, due to a change in exon-intron splicing with the insertion of an Frt-flanked pGK-Neo DNA fragment (Fig. 1C), the homozygous mice carrying the initial targeting construct via homologous recombination had already lost UBE1L activity and protein ISGylation. No significant change in total protein ubiquitination was detected in Ube1L-deficient cells, indicating that UBE1L is not involved directly in the ubiquitination process, despite its high homology to UBE1. The absence of ISGylation also plays no general role in altering cellular ubiquitination levels.
Ube1L knockout mice were born at the expected Mendelian ratio, healthy and fertile, suggesting no significant role of ISGylation system in normal embryonic development, life, and reproduction. ISGylation is induced by IFN-α/β, which led us to examine a possible role of ISGylation in IFN-α/β signaling and in viral infection. Ube1L+/+ and Ube1L−/− mice injected with poly(I · C) or LPS showed similar responses in terms of lethality and white blood cell counts. Furthermore, no obvious difference was detected regarding antiviral responses against VSV and LCMV, and the response to IFN in Ube1L-deficient cells was normal. These results clearly demonstrate that Ube1L and protein ISGylation are not essential for IFN signaling via JAK/STAT activation.
Protein ISGylation is a tightly regulated energy-consuming (ATP-dependent) process. ISG15 and all of the known enzymes associated with protein ISGylation are encoded by IFN-inducible genes. It is therefore reasonable to suspect that ISGylation plays important roles during innate immune responses. The lack of any noticeable phenotype associated with Ube1L−/− mice may be due to compensatory pathways among the existing several hundreds of IFN-inducible genes (3, 4, 19). A detailed characterization of Ube1L−/− mice may uncover new aspects about the function of the ISGylation system. In addition, crossing of Ube1L-deficient mice with other transgenic or knockout mice, especially for genes with demonstrated functions in innate immune responses could provide new insights about the role of ISGylation in innate immunity. Likewise, it would be important to elucidate the biological consequences of Serpin 2A, Stat1, Jak1, PLCγ1, and Erk1/2 ISGylation (8, 26). Most recently, ISG15 modification via the ɛ-amino group of a lysine residue has been confirmed via mass spectrometry analysis of ISGylated Ubc13 (45), and a large number of additional potential ISG15 target proteins have been identified from immunoaffinity purification and mass spectrometry analysis (6, 38, 44, 45). The study of these new target proteins may reveal a role of protein ISGylation in additional biological pathways. Recent studies with Sindbis virus have provided further support for a contribution of protein ISGylation in the host antiviral response (20). Chimeric Sindbis virus expressing wild-type ISG15, but not a nonfunctional mutant of ISG15, exhibited attenuation upon infection of IFN receptor gene ifnar1 knockout mice.
As predicted Ubp43 and Ube1L double-knockout mice lack detectable protein ISGylation. However, their hypersensitivity to IFN—as judged by enhanced Stat1 phosphorylation and the increased number of apoptotic cells upon IFN treatment (Fig. 6 and 7)—remained unchanged compared to the levels observed in single-knockout mice. These results indicate that lack of Ubp43, not an increase of protein ISGylation, is responsible for the IFN-hypersensitive responses in Ubp43-deficient cells. These new findings disprove our original hypothesis stating that increased protein ISGylation was directly linked to enhanced IFN-α/β signaling (28). Additional studies have demonstrated that UBP43 is a negative regulator of IFN-α/β signaling independent of its ISG15 isopeptidase activity (O. Malakhova et al., unpublished data). Further investigation is necessary to elucidate the complete range of UBP43 biological activities and underlying molecular mechanisms.
Acknowledgments
We thank members of the laboratory of D.-E. Zhang for valuable discussions.
This work was supported by National Institutes of Health grants CA079849 and GM066955 (D.-E.Z.). J.-K.L. was partially supported by the Leukemia Research Foundation. W.Z. is a Leukemia and Lymphoma Society fellow. The Stein Endowment Fund has partially supported the Department of Molecular and Experimental Medicine departmental molecular biology service laboratory for DNA sequencing and oligonucleotide synthesis.
This report is paper 17626-MEM from The Scripps Research Institute.
REFERENCES
- 1.Dao, C. T., and D. E. Zhang. 2005. ISG15: a ubiquitin-like enigma. Front. Biosci. 10:2701-2722. [DOI] [PubMed] [Google Scholar]
- 2.D'Cunha, J., E. Knight, Jr., A. L. Haas, R. L. Truitt, and E. C. Borden. 1996. Immunoregulatory properties of ISG15, an interferon-induced cytokine. Proc. Natl. Acad. Sci. USA. 93:211-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Veer, M. J., M. Holko, M. Frevel, E. Walker, S. Der, J. M. Paranjape, R. H. Silverman, and B. R. Williams. 2001. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 69:912-920. [PubMed] [Google Scholar]
- 5.Farrell, P. J., R. J. Broeze, and P. Lengyel. 1979. Accumulation of an mRNA and protein in interferon-treated Ehrlich ascites tumour cells. Nature 279:523-525. [DOI] [PubMed] [Google Scholar]
- 6.Giannakopoulos, N. V., J. K. Luo, V. Papov, W. Zou, D. J. Lenschow, B. S. Jacobs, E. C. Borden, J. Li, H. W. Virgin, and D. E. Zhang. 2005. Proteomic identification of ISGylated proteins in mouse and human cells. Biochem. Biophys. Res. Commun. 336:496-506. [DOI] [PubMed] [Google Scholar]
- 7.Haas, A. L., P. Ahrens, P. M. Bright, and H. Ankel. 1987. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J. Biol. Chem. 262:11315-11323. [PubMed] [Google Scholar]
- 8.Hamerman, J. A., F. Hayashi, L. A. Schroeder, S. P. Gygi, A. L. Haas, L. Hampson, P. Coughlin, R. Aebersold, and A. Aderem. 2002. Serpin 2a is induced in activated macrophages and conjugates to a ubiquitin homolog. J. Immunol. 168:2415-2423. [DOI] [PubMed] [Google Scholar]
- 9.Hicke, L. 2001. A new ticket for entry into budding vesicles-ubiquitin. Cell 106:527-530. [DOI] [PubMed] [Google Scholar]
- 10.Hochstrasser, M. 1996. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30:405-439. [DOI] [PubMed] [Google Scholar]
- 11.Hochstrasser, M. 2000. Evolution and function of ubiquitin-like protein-conjugation systems. Nat. Cell Biol. 2:E153-E157. [DOI] [PubMed] [Google Scholar]
- 12.Kang, D., H. Jiang, Q. Wu, S. Pestka, and P. B. Fisher. 2001. Cloning and characterization of human ubiquitin-processing protease-43 from terminally differentiated human melanoma cells using a rapid subtraction hybridization protocol RaSH. Gene 267:233-242. [DOI] [PubMed] [Google Scholar]
- 13.Kim, K. I., N. V. Giannakopoulos, H. W. Virgin, and D. E. Zhang. 2004. Interferon-inducible ubiquitin E2, Ubc8, is a conjugating enzyme for protein ISGylation. Mol. Cell. Biol. 24:9592-9600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, K. I., O. A. Malakhova, K. Hoebe, M. Yan, B. Beutler, and D. E. Zhang. 2005. Enhanced antibacterial potential in UBP43-deficient mice against Salmonella typhimurium infection by up-regulating type I IFN signaling. J. Immunol. 175:847-854. [DOI] [PubMed] [Google Scholar]
- 15.Kim, K. I., and D. E. Zhang. 2003. ISG15, not just another ubiquitin-like protein. Biochem. Biophys. Res. Commun. 307:431-434. [DOI] [PubMed] [Google Scholar]
- 16.Kok, K., R. Hofstra, A. Pilz, A. van den Berg, P. Terpstra, C. H. Buys, and B. Carritt. 1993. A gene in the chromosomal region 3p21 with greatly reduced expression in lung cancer is similar to the gene for ubiquitin-activating enzyme. Proc. Natl. Acad. Sci. USA 90:6071-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korant, B. D., D. C. Blomstrom, G. J. Jonak, and E. Knight, Jr. 1984. Interferon-induced proteins. Purification and characterization of a 15,000-dalton protein from human and bovine cells induced by interferon. J. Biol. Chem. 259:14835-14839. [PubMed] [Google Scholar]
- 18.Kumar, S., W. H. Kao, and P. M. Howley. 1997. Physical interaction between specific E2 and Hect E3 enzymes determines functional cooperativity. J. Biol. Chem. 272:13548-13554. [DOI] [PubMed] [Google Scholar]
- 19.Leaman, D. W., M. Chawla-Sarkar, B. Jacobs, K. Vyas, Y. Sun, A. Ozdemir, T. Yi, B. R. Williams, and E. C. Borden. 2003. Novel growth and death related interferon-stimulated genes (ISGs) in melanoma: greater potency of IFN-beta compared with IFN-alpha2. J. Interferon Cytokine Res. 23:745-756. [DOI] [PubMed] [Google Scholar]
- 20.Lenschow, D. J., N. V. Giannakopoulos, L. J. Gunn, C. Johnston, A. K. O'Guin, R. E. Schmidt, B. Levine, and H. W. Virgin. 2005. Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J. Virol. 79:13974-13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, J., G. W. Peet, D. Balzarano, X. Li, P. Massa, R. W. Barton, and K. B. Marcu. 2001. Novel NEMO/IkappaB kinase and NF-kappa B target genes at the pre-B to immature B cell transition. J. Biol. Chem. 276:18579-18590. [DOI] [PubMed] [Google Scholar]
- 22.Li, X. L., J. A. Blackford, C. S. Judge, M. Liu, W. Xiao, D. V. Kalvakolanu, and B. A. Hassel. 2000. RNase-L-dependent destabilization of interferon-induced mRNAs. A role for the 2-5A system in attenuation of the interferon response. J. Biol. Chem. 275:8880-8888. [DOI] [PubMed] [Google Scholar]
- 23.Liu, L.-Q., R. Ilaria, Jr., P. D. Kingsley, A. Iwama, R. van Etten, J. Palis, and D. E. Zhang. 1999. A novel ubiquitin-specific protease, UBP43, cloned from leukemia fusion protein AML1-ETO-expressing mice, functions in hematopoietic cell differentiation. Mol. Cell. Biol. 19:3029-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, M., B. T. Hummer, X. Li, and B. A. Hassel. 2004. Camptothecin induces the ubiquitin-like protein, ISG15, and enhances ISG15 conjugation in response to interferon. J. Interferon Cytokine Res. 24:647-654. [DOI] [PubMed] [Google Scholar]
- 25.Loeb, K. R., and A. L. Haas. 1992. The interferon-inducible 15-kDa ubiquitin homolog conjugates to intracellular proteins. J. Biol. Chem. 267:7806-7813. [PubMed] [Google Scholar]
- 26.Malakhov, M. P., K. I. Kim, O. A. Malakhova, B. S. Jacobs, E. C. Borden, and D. E. Zhang. 2003. High-throughput immunoblotting. Ubiquitiin-like protein ISG15 modifies key regulators of signal transduction. J. Biol. Chem. 278:16608-16613. [DOI] [PubMed] [Google Scholar]
- 27.Malakhov, M. P., O. A. Malakhova, K. I. Kim, K. J. Ritchie, and D. E. Zhang. 2002. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J. Biol. Chem. 277:9976-9981. [DOI] [PubMed] [Google Scholar]
- 28.Malakhova, O. A., M. Yan, M. P. Malakhov, Y. Yuan, K. J. Ritchie, K. I. Kim, L. F. Peterson, K. Shuai, and D. E. Zhang. 2003. Protein ISGylation modulates the JAK-STAT signaling pathway. Genes Dev. 17:455-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyers, E. N., M. Lewandoski, and G. R. Martin. 1998. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat. Genet. 18:136-141. [DOI] [PubMed] [Google Scholar]
- 30.Narasimhan, J., M. Wang, Z. Fu, J. M. Klein, A. L. Haas, and J. J. Kim. 2005. Crystal structure of the interferon-induced ubiquitin-like protein ISG15. J. Biol. Chem. 280:27356-27365. [DOI] [PubMed] [Google Scholar]
- 31.Osiak, A., O. Utermohlen, S. Niendorf, I. Horak, and K. P. Knobeloch. 2005. ISG15, an interferon-stimulated ubiquitin-like protein, is not essential for STAT1 signaling and responses against vesicular stomatitis and lymphocytic choriomeningitis rirus. Mol. Cell. Biol. 25:6338-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pickart, C. M. 2001. Ubiquitin enters the new millennium. Mol. Cell 8:499-504. [DOI] [PubMed] [Google Scholar]
- 33.Pitha-Rowe, I., B. A. Hassel, and E. Dmitrovsky. 2004. Involvement of UBE1L in ISG15 conjugation during retinoid-induced differentiation of acute promyelocytic leukemia. J. Biol. Chem. 279:18178-18187. [DOI] [PubMed] [Google Scholar]
- 34.Reich, N., B. Evans, D. Levy, D. Fahey, E. Knight, Jr., and J. E. Darnell, Jr. 1987. Interferon-induced transcription of a gene encoding a 15-kDa protein depends on an upstream enhancer element. Proc. Natl. Acad. Sci. USA 84:6394-6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ritchie, K. J., C. S. Hahn, K. I. Kim, M. Yan, D. Rosario, L. Li, J. C. de la Torre, and D. E. Zhang. 2004. Role of ISG15 protease UBP43 (USP18) in innate immunity to viral infection. Nat. Med. 10:1374-1378. [DOI] [PubMed] [Google Scholar]
- 36.Ritchie, K. J., M. P. Malakhov, C. J. Hetherington, L. Zhou, M. T. Little, O. A. Malakhova, J. C. Sipe, S. H. Orkin, and D. E. Zhang. 2002. Dysregulation of protein modification by ISG15 results in brain cell injury. Genes Dev. 16:2207-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwer, H., L. Q. Liu, L. Zhou, M. T. Little, Z. Pan, C. J. Hetherington, and D. E. Zhang. 2000. Cloning and characterization of a novel human ubiquitin-specific protease, a homologue of murine UBP43 (Usp18). Genomics 65:44-52. [DOI] [PubMed] [Google Scholar]
- 38.Takeuchi, T., and H. Yokosawa. 2005. ISG15 modification of Ubc13 suppresses its ubiquitin-conjugating activity. Biochem. Biophys. Res. Commun. 336:9-13. [DOI] [PubMed] [Google Scholar]
- 39.Wilkinson, K. D. 1997. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 11:1245-1256. [DOI] [PubMed] [Google Scholar]
- 40.Wong, A. H., J. E. Durbin, S. Li, T. E. Dever, T. Decker, and A. E. Koromilas. 2001. Enhanced antiviral and antiproliferative properties of a STAT1 mutant unable to interact with the protein kinase PKR. J. Biol. Chem. 276:13727-13737. [DOI] [PubMed] [Google Scholar]
- 41.Yuan, W., and R. M. Krug. 2001. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 20:362-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, X., J. Shin, T. W. Molitor, L. B. Schook, and M. S. Rutherford. 1999. Molecular responses of macrophages to porcine reproductive and respiratory syndrome virus infection. Virology 262:152-162. [DOI] [PubMed] [Google Scholar]
- 43.Zhao, C., S. L. Beaudenon, M. L. Kelley, M. B. Waddell, W. Yuan, B. A. Schulman, J. M. Huibregtse, and R. M. Krug. 2004. The UbcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN-alpha/beta-induced ubiquitin-like protein. Proc. Natl. Acad. Sci. USA 101:7578-7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao, C., C. Denison, J. M. Huibregtse, S. Gygi, and R. M. Krug. 2005. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc. Natl. Acad. Sci. USA 102:10200-10205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zou, W., V. Papov, O. Malakhova, K. I. Kim, C. Dao, J. Li, and D. E. Zhang. 2005. ISG15 modification of ubiquitin E2 Ubc13 disrupts its ability to form thioester bond with ubiquitin. Biochem. Biophys. Res. Commun. 336:61-68. [DOI] [PubMed] [Google Scholar]