Abstract
Marek's disease virus (MDV) is an avian herpesvirus that causes T-cell lymphomas and immune suppression in susceptible chickens. At least one gene product, MDV Eco Q-encoded protein (Meq), is essential for the oncogenicity of MDV. Alternative splicing permits the meq gene to give rise to two major transcripts encoding proteins designated Meq and Meq/vIL8. Meq is a basic leucine zipper protein capable of modulating transcription. The Meq/vIL8 protein retains a modified leucine zipper, along with the mature receptor-binding portion of vIL8, but lacks the domain of Meq responsible for transcriptional modulation. In this report, we describe studies using fusions between either Meq or Meq/vIL8 and fluorescent proteins to characterize the distribution and properties of these products in chicken embryo fibroblasts (CEFs). Meq and Meq/vIL8 both localized to the nucleoplasm, nucleoli, and Cajal bodies of transfected cells. Similar distributions were found for fluorescent fusion proteins and native Meq or Meq/vIL8. Fluorescence recovery after photobleaching and photoactivatable green fluorescent protein revealed that Meq exhibited mobility properties similar to those of other transcription factors, while Meq/vIL8 was far less mobile. In addition, fluorescence resonance energy transfer studies indicated the formation of Meq/vIL8 homodimers in CEFs. Time lapse studies revealed the coordinated elimination of a portion of Meq and Meq/vIL8 from the nucleus. Our data provide new insight regarding the dynamic cellular properties of two forms of a herpesvirus-encoded oncoprotein and suggest that these forms may have fundamentally different functions in MDV-infected cells.
Marek's disease is a lymphoproliferative disease of chickens caused by the alphaherpesvirus Marek's disease virus (MDV). The disease is characterized by mononuclear infiltration, demyelization of peripheral nerves, and the onset of T-cell lymphomas within 6 weeks postinfection (43). The disease spreads through flocks via shedding of infectious virus from the feather follicle epithelia of infected birds. MDV describes a family of antigenically related viruses. MDV serotype 1 (MDV-1) causes disease in chickens; however, two other MDV serotypes, MDV-2 and herpesvirus of turkeys, do not. Unique to MDV-1 serotypes is a 13.4-kilobase block of genes present in the internal repeat region that contains 11 open reading frames including the meq gene, which apparently functions as an oncogene (22, 31, 40).
The meq gene has been found consistently in MDV-induced tumors as well as MDV-transformed cell lines (21). Furthermore, inhibition of meq expression by antisense strategies has shown that maintenance of the transformed phenotype is meq dependent in the MDV-transformed cell line MSB1 (44). Recently, deletion of the meq gene was found to ablate oncogenicity while not attenuating the virus for replication in vivo (31). Overexpression of meq in Rat-2 cells, a rat fibroblast line, resulted in transformation based on the appearance of morphological changes, along with anchorage- and serum-independent growth (29). Taken together, these data directly implicate meq in either the induction or the maintenance of the oncogenic state in transformed T lymphocytes.
The meq gene gives rise to two alternatively spliced mRNAs. One of the mRNAs encodes the Meq protein, which contains several domains that are characteristic of transcription-regulatory proteins. It possesses a DNA binding domain, nuclear and nucleolar localization signals, and a basic leucine zipper (bZIP) that is similar to members of the Jun/Fos family of transcriptional activators (21). Via the leucine zipper, Meq forms homodimers and also heterodimerizes with cellular proteins including JunB, c-Jun, c-Fos, and SNF in vitro (4); it was recently shown to dimerize with c-Jun in vivo (23). These dimers can bind DNA and regulate transcription of target genes (24). Additionally, Meq contains a proline-rich region within its C terminus that has transcriptional repression activity and an adjacent 33-amino-acid domain at the extreme C terminus that activates transcription when fused to the Gal-4 DNA binding domain (44). Furthermore, Meq interacts with non-bZIP proteins such as p53 and CDK2 (4) and may bind RNA through a putative RNP1/arginine fork domain (28). Thus, Meq appears to be a multifunctional protein containing cell cycle-regulatory, transcriptional modulation, and apoptosis-suppressing functions (26, 44).
The second mRNA product of the meq gene is generated using a splice donor site within the region encoding the leucine zipper and the splice acceptor site of the second exon of v-IL-8 (34, 38). This transcript encodes a protein, termed Meq/vIL8, containing the DNA binding domain of Meq, a modified leucine zipper, and the cytokine (receptor-binding) portion of vIL8 but lacking the C-terminal transcriptional regulatory domains of Meq and the amino-terminal secretory signal of vIL8. In vitro, Meq/vIL8 will dimerize with either Meq or c-Jun and binds to the AP-1 site as a heterodimer with c-Jun (38, 39). However, Meq/vIL8 does not function as a transcriptional activator in a Gal4-dependent transcription assay system (V. Arumugaswami et al., unpublished data). The role of Meq/vIL8 in the lytic, latent, or transforming activity of MDV is uncertain, although the absence of transcriptional activation ability has led to the hypothesis that Meq/vIL8 may function as a competitive inhibitor of Meq action (38).
Insight into a protein's function can be gained through study of its localization, its mobility in vivo, and the structures and proteins with which it interacts. A variety of techniques have been devised to examine the subcellular localization of proteins as they are expressed in cells. One of the most powerful uses fusions between the protein of interest and fluorescent reporters such as green, yellow, and cyan fluorescent proteins (GFP, YFP, and CFP, respectively). When expressed as nonfusion proteins, the fluorescent proteins are typically distributed throughout the cell cytoplasm and nucleoplasm. However, when expressed as fusion proteins, they are frequently targeted to the site of action of the protein, with the fluorescent reporter marking its location (3, 6, 8-10, 20, 33, 41, 51). Additional techniques can be used to further characterize the physical properties of the fusion protein. For example, fluorescence recovery after photobleaching (FRAP) (5, 8, 9, 17, 25, 49, 50) and photoactivatable GFP (PA-GFP) constructs (36) have been used to characterize the mobility of proteins, and fluorescence resonance energy transfer (FRET) (7, 46, 53) can be used to examine protein-protein interactions.
The goal of this study was to determine the relative differences in the nuclear properties of the major meq gene products, Meq and Meq/vIL8. Previous antibody staining experiments showed Meq localized to the nucleus, particularly in the nucleolus and Cajal (coiled) bodies (27), in Rat-2 cells. We now extend these findings using fluorescent protein fusions of Meq and Meq/vIL8 expressed in primary chicken embryo fibroblasts (CEF). The use of these fusion proteins, in conjunction with laser scanning confocal microscopy, has allowed us to assess the subcellular localization, interactions, and mobility of Meq and Meq/vIL8 proteins in vivo.
MATERIALS AND METHODS
Plasmids.
The full-length meq gene was amplified by PCR from RB1B-infected CEF DNA using primers SpeI/NheI-T7-Meq For and Meq fusion-Rev (Table 1; also see Fig. 1). The 1,089-bp amplicon was cloned into pCR2.1TOPO, and the DNA sequence was determined. This amplicon was subcloned as a 1,062-bp NheI-HindIII fragment into NheI-HindIII-digested pEYFP-N1 and pECFP-N1. The T7 epitope tag (MASMTGGQQMGR) is identical to that used in the studies by Liu et al. (27) in which immunofluorescence was used to examine the localization of Meq. The Meq/vIL-8 construct was amplified from a clone of this gene product (obtained by rapid amplification of 3′ cDNA ends) (Table 1) using primers SpeI/NheI-T7-Meq For and Meq-vIL-8 fusion-Rev. This 705-bp amplicon was cloned into pCR2.1TOPO, the DNA sequence was determined, and it was subcloned into pEYFP-N1 and pECFP-N1, via NheI-HindIII sites, as a 679-bp fragment, as above. Escherichia coli strain DH10B was used to produce cell culture-grade DNA for transfections. Bacteria containing the plasmids of interest were grown in 500-ml cultures of Luria-Bertani medium to late-log phase, and plasmid DNA was extracted using QIAGEN endotoxin-free MAXI kits.
TABLE 1.
Primers used for the amplification of Meq and Meq/vIL8
| Primer | Sequencea | Template accession no.b |
|---|---|---|
| P1: SpeI/NheI-T7-Meq-For | 5′-GGACTAGTATGGCTAGCATGACTGGTGGACAGCAAATGGGTCGGA TGTCTCAGGAGCCAG-3′ | AY243332 (RB1B Meq) |
| P2: Meq fusion-Rev (HindIII-XbaI) | 5′-GCTCTAGAGCAAGCTTGCGGAATCCTCCGGGTCTCC-3′ | AY243332 (RB1B Meq) |
| P3: Meq/vIL8 fusion-Rev (HindIII-XbaI) | 5′-GCTCTAGAGCAAGCTTGCGGAATCCAAGACAGATATG-3′ | AY243340 (Meq/vIL8) |
Underlined nucleotides are restriction endonuclease sites introduced to facilitate cloning. Italicized nucleotides are encoded by the virus genome.
Accession numbers are from the NCBI nucleotide sequence database.
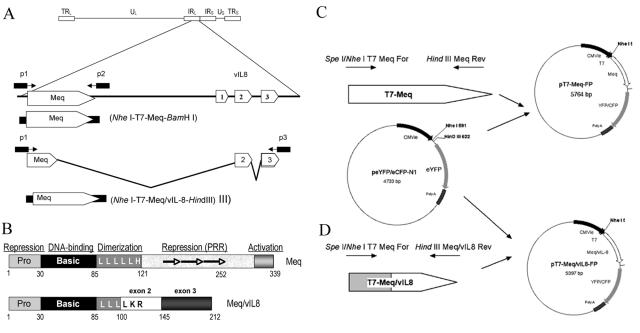
FIG. 1.
(A) Genomic context of Meq and Meq/vIL8. The Meq gene is located in the repeat of the long region (IRL) of Marek's disease virus. Meq is derived from an unspliced transcript, while Meq/vIL8 utilizes a cryptic splice donor site proximal to the region encoding the leucine zipper of Meq and the splice acceptor site of vIL8 exon 2. Primers p1-p2 and p1-p3 were used to amplify Meq and Meq/vIL8, respectively (see Table 1 for primer sequences). (B) Representation of domains present in Meq and Meq/vIL8. Note that while Meq/vIL8 retains a leucine zipper, the splicing event alters the last two amino acids compared with the Meq leucine zipper. (C and D) Construction of Meq or Meq/vIL8 fluorescent fusion protein expression vector. Amplicons were synthesized using primers listed in Table 1, digested with the indicated restriction enzymes, and cloned into either pEYFP or pECFP, in frame with the fluorescent reporter protein. The T7 epitope was included at the amino termini of these constructs (MASMTGGQQMGR). (C) Construction of vectors encoding the T7-Meq fluorescent fusion protein. (D) Construction of vectors encoding the T7-Meq/vIL8 fluorescent fusion protein.
Preparation of cells and transfection.
Primary CEF were prepared from 10-day-old specific-pathogen-free eggs (Spafas, Inc. Charles River, CT) and placed in an incubator at 37°C and 5% CO2 (33, 47). One day later, secondary CEF were plated in a 2-well LabTek Nalgene Nunc chambered coverglass system (Nunc catalog number 155361; Nalgene Nunc International, Naperville, Ill.) at a density of 2 × 105 cells per chamber. The following day, cells were transfected by calcium phosphate precipitation (6 μg of DNA per chamber) followed by a glycerol shock.
Immunofluorescence.
The polyclonal antibody to Meq (23) was obtained from Lucy Lee (USDA-ADOL, East Lansing, Mich.). Cells were fixed in 2% paraformaldehyde 24 to 48 h following transfection in phosphate-buffered saline (PBS) for 30 min at room temperature. Cells were then washed three times with PBS and then incubated for 2 h with blocking buffer (1× PBS, pH 7.4, 3% goat serum, 1% bovine serum albumin, 0.1% Triton X-100, 0.1% saponin). After removal of the blocking buffer, the Meq antibody was added (1:500 dilution in blocking buffer) and incubated overnight at 4°C. Subsequently, slides were washed three times with PBS plus detergent buffer (PD) (1× PBS, pH 7.4, 0.1% Triton X-100, 0.1% saponin, 0.1% NaN3) and then incubated with a goat anti-rabbit immunoglobulin G (IgG) antibody conjugated with Alexa 546 (Molecular Probes, Eugene, OR) (1:200 dilution in blocking buffer) for 2 h. Slides were then washed in PD buffer, with a final wash with 1× PBS.
Multiphoton and confocal microscopy.
Expression of fusion proteins was allowed to proceed for 24 to 48 h. In some cases, cells were stained with 10 μg/ml of Hoechst 33342 (vital DNA probe) 1 h prior to imaging. Imaging was carried out using a Zeiss LSM 510 NLO confocal microscope (Carl Zeiss, Inc., Oberkoken, Germany) with a Plan-Neofluar 40× oil immersion objective (numerical aperture, 1.3). The 458-nm and 514-nm laser lines of a 15-mW argon laser (Coherent Enterprises, Santa Clara, Calif.) were used to excite enhanced CFP (ECFP) and EYFP, respectively. Hoechst 33342 fluorescence was excited at 745 nm using a 5-W pumped MIRA-900 titanium:sapphire laser (Coherent Enterprises, Santa Clara, Calif.) with a 400- to 460-nm band-pass emission filter. Alexa 546 was excited using a 543-nm helium neon laser with a 560 long-pass emission filter. All multichannel imaging was performed in fastline switch mode to eliminate cross talk.
FRAP.
Transfected cells were observed as described above. Localized areas were targeted with brief high-intensity laser light to bleach the fluorescent molecules in the defined area, followed by continuous image collection at appropriate time intervals until fluorescence had recovered to equilibrium (2). Zeiss LSM 510 AIM Software (release 3.2) coupled with an Excel spreadsheet was used to plot fluorescence recovery (pixel intensity) over time in specified regions. Cells were maintained at 37°C and 5% CO2 during all time course experiments by using a microscope-adapted environmental chamber (PECON Instruments, Inc.).
Photoactivation of GFP.
Cells were transfected with plasmids encoding PA-GFP (35, 36) fused in frame with either Meq or Meq/vIL8. At 24 h posttransfection, PA-GFP-expressing cells were activated in specific regions of transfected cells by brief illumination at 830 nm with the titanium:sapphire laser, followed by continuous image collection at appropriate time intervals until the activated region equilibrated with its surroundings. Cells were maintained and analyzed as described for FRAP experiments.
FRET.
Cells cotransfected with ECFP and EYFP fusion constructs were examined as described above. If both fluorescent proteins were observed within the same cell, protein interaction was assessed for FRET by bleaching the acceptor, EYFP, so that resonant energy transfer could no longer occur. This resulted in a concomitant increase in the intensity of ECFP (the resonance energy donor) if the fusion proteins were close enough for FRET. Signal intensity was plotted before and after photobleaching in selected areas, and FRET values were measured and reported as the percent increase in fluorescence of the donor following bleaching of the acceptor (52) using the Zeiss LSM 510 FRET Macro (version 1.5).
RESULTS
Subcellular localization of Meq and Meq/vIL8.
Genes were constructed to express Meq or Meq/vIL8 fused in frame with either ECFP or EYFP (Fig. 1). Initially, the distributions of Meq and Meq/vIL8 fusion constructs were compared with those of native ECFP and EYFP. Both ECFP and EYFP were evenly distributed throughout the cytoplasm and nucleus, except that they were excluded from the nucleolus and cytoplasmic vacuoles (not shown). In contrast, both the Meq and Meq/vIL8 fusion proteins with ECFP or EYFP were largely concentrated within distinct bodies in the nucleus (Fig. 2). Cells expressing either Meq-EYFP or Meq-ECFP exhibited fluorescence concentrated within the nucleolus and weaker fluorescence throughout the nucleus and cytoplasm (Fig. 2A; cytoplasmic fluorescence not visible). Meq/vIL8-ECFP or -EYFP also localized to the nucleus and was typically concentrated within the nucleolus, with less fluorescence detected in the nucleoplasm and cytoplasm (Fig. 2B; cytoplasmic fluorescence not visible). Meq and Meq/vIL8, not fused to a fluorescent reporter, were also expressed in CEF and visualized with an anti-Meq polyclonal antibody that recognizes an epitope shared by Meq and Meq/vIL8 (23). These proteins exhibited localization patterns similar to those seen with the Meq (27) and Meq/vIL8 fluorescent fusion proteins (Fig. 2I to O).
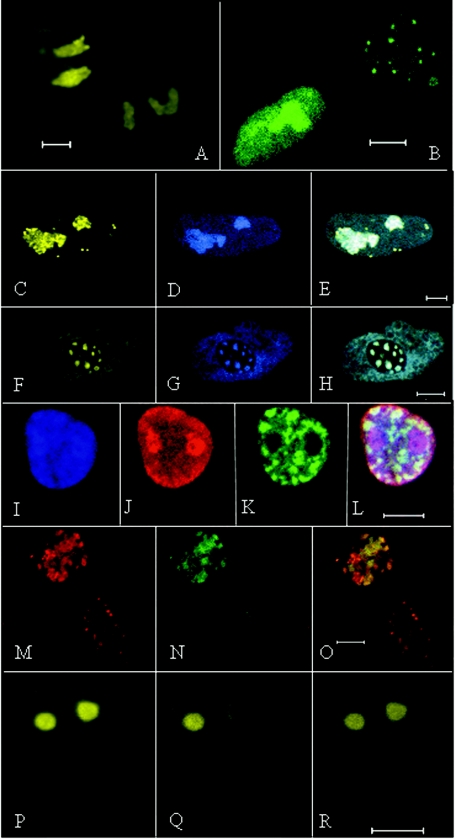
FIG. 2.
Confocal images of nuclei of transfected cells. Bars, 5 μm. (A) Meq-EYFP distributed throughout nuclei and nucleoplasm of two cells. (B) Meq/vIL8-EYFP associated with Cajal bodies (punctate) and nucleus in cell at upper right and with the nucleoplasm and nucleolus in the cell at bottom left. (C to E) Cell cotransfected with Meq-ECFP and Meq/vIL8-EYFP. (C) Meq/vIL8-ECFP exhibiting nucleoplasmic, nucleolar, and Cajal body association. (D) Meq-ECFP showing association with the nucleoplasm and nucleolus. (E) Combined image of panels C and D. (F to H) Nuclear expression of p80-coilin and Meq/vIL8. (F) p80 coilin-EYFP. (G) Same cell as in panel F, displaying expressed Meq/vIL8-ECFP. (H) Merged image showing colocalization of Meq/vIL8-ECFP with p80-coilin-EYFP. (I to L) CEF expressing p80-coilin-EYFP (green) and Meq-ECFP. (I) Meq-ECFP found in the nucleolus, nucleoplasm, and Cajal bodies. (J) Same cell as in panel I, visualized with the anti-Meq polyclonal antibody followed by an Alexa 546-conjugated goat anti-rabbit IgG. (K) Same cell as in panel I, showing the localization of p80-coilin-EYFP (green). (L) Merged image of panels I through K. (M to O) Cells expressing Meq/vIL8 (red) and p80-coilin-EYFP (green). (M) Meq/vIL8 expressed in the nuclei and Cajal bodies of two separate cells detected with an anti-Meq polyclonal antibody followed by an Alexa 546-conjugated goat anti-rabbit IgG. (N) The same cells visualized for p80-coilin. In this case the nucleus of the cell on the lower right is difficult to discern. (O) Merged image of panels M and N. (P) CEF expressing Meq-YFP prior to photobleaching of the nucleolus on the right. (Q) The same cell as in panel P after photobleaching of the nucleolus on the right. (R) The same cell 20 s after photobleaching.
Previous studies have shown colocalization of Meq with Cajal bodies when Meq was expressed in Rat-2 cells (26). To determine if Meq or Meq/vIL8 fused to fluorescent reporters would also localize to Cajal bodies, cells were cotransfected with p80-coilin fused to YFP and either Meq or Meq/vIL8 fused to CFP. p80-coilin localizes to, and nucleates the formation of, Cajal bodies (1, 45) (Fig. 2F and K), and both Meq (Fig. 2I to L) and Meq/vIL8 (Fig. 2M to O) exhibited colocalization with p80-coilin. In many of the p80-coilin-transfected cells, the p80-coilin protein was distributed in a diffuse pattern throughout the nucleus, and either Meq or Meq/vIL8 colocalized with this p80-coilin (Fig. 2I to O). However, in approximately 20% of the transfected cells, Meq or Meq/vIL8 fluorescent protein was distributed in a punctate nuclear pattern, and these punctate bodies also accumulated p80-coilin-YFP. To test if Meq/vIL8 and Meq would colocalize to these punctate bodies, cells were cotransfected with both constructs, one fused to ECFP and one to EYFP. Cells with punctate Meq/vIL8 bodies were identified, but none of these bodies were found associated with Meq (Fig. 2C to E). Taken together, these studies reveal that Meq fused to either YFP or CFP exhibits the localization properties of native Meq. Meq/vIL8, like Meq, also localizes to the nucleus, nucleolus, and Cajal bodies. The observation that Meq and Meq/vIL8 do not colocalize to the punctate Cajal bodies suggest that they may not form Meq-Meq/vIL8 heterodimers at these sites.
FRAP.
In vivo protein dynamics of the various forms of Meq were explored using FRAP, which provides insight into the mobility of a protein in its natural environment. We expressed Meq and Meq/vIL8 as fusion proteins with fluorescent tags and observed their subcellular localizations. Relevant areas of fluorescence within the nucleoplasm or nucleolus were bleached, and by using time lapse confocal microscopy, the time required for bleached fluorescence to recover was measured (Fig. 2P to R). Meq showed rapid recovery, with a half-life (t1/2) to recovery of 15 (±7) s (Table 2) in both the nucleolus and the nucleoplasm. This observation was consistent with the recovery dynamics observed with other transcription factors (12, 25, 32, 41, 51). In contrast, the t1/2 of Meq/vIL8 to recovery was 15 (±3) min in both the nucleolus and the nucleoplasm, or approximately 60-fold longer than that of Meq (Table 2).
TABLE 2.
FRAP and photoactivation t1/2 valuesa
| Process and protein | Nuclear t1/2 | SD |
|---|---|---|
| FRAP-Meq | 15 s | 7 s |
| PA-Meq | 35 s | 17 s |
| FRAP-Meq/vIL8 | 15 min | 3 min |
| PA-Meq/vIL8 | 20 min | 1 min |
t1/2 values were determined by normalizing the fluorescence intensity and taking the time point at 50% recovery (FRAP) or 50% reduction (photoactivation).
Photoactivatable constructs.
To complement the FRAP analysis, we also examined the motion of Meq and Meq/vIL8 fused to PA-GFP (35). This form of GFP exhibits very low fluorescence until activated by UV light or the appropriate two-photon excitation wavelength (830 nm). By activating PA-GFP fluorescence in discrete regions of the nucleoplasm or nucleolus, we monitored the redistribution of fusion proteins in the presence of very low background. Time lapse confocal microscopy showed that activated Meq-PA-GFP had a t1/2 to equilibrium of 35 (±17) s within either the nucleolus or the nucleoplasm (Table 2). The t1/2 for equilibration of Meq/vIL8-PA-GFP following activation in either the nucleolus or the nucleoplasm was 20 (±1) min (Table 2). Hence, the Meq- and Meq/vIL8-PA-GFP data were consistent with the results obtained by FRAP analysis, with Meq exhibiting much greater mobility in the nucleoplasm and nucleolus than Meq/vIL8.
FRET.
Because Meq has been shown to form homodimers in vitro, we tested for the ability of appropriate pairs of fluorescent reporters to exhibit FRET. Efficient resonance energy transfer requires the close proximity of fluorochromes, which, for ECFP and EYFP, is on the order of 1 to 10 nm (13). Because these distances are typical of the dimensions of many proteins, detection of FRET implies a direct interaction between the fluorescent fusion proteins. However, the pairing of unfused ECFP with Meq-EYFP resulted in FRET, although the difference in efficiency was clearly significant compared to that of the Meq homodimers (not shown). Because of this, we were unable to conclude that Meq-ECFP and Meq-EYFP were capable of forming close complexes detectable by FRET. In contrast, the Meq/vIL8 control experiments did not exhibit FRET (Table 3). Cotransfection of Meq/vIL8-ECFP and Meq/vIL8-EYFP yielded positive FRET results with the nucleolus, nucleoplasm, and Cajal bodies, exhibiting FRET efficiencies of 14.1% (±2%), 11.4% (±3.3%), and 7.5% (±1.5%), respectively. These data suggested that FRET did occur with the formation of Meq/vIL8 homodimers in both these nuclear regions.
TABLE 3.
FRETa
| Donor | Acceptor | nc | % FRET efficiencyb
|
||
|---|---|---|---|---|---|
| Nucleolus | Nucleoplasm | Cajal bodies | |||
| ECFP | EYFP | 3 | None present | 1.6 ± 2.8 | None present |
| ECFP | Meq/vIL8-EYFP | 3 | −2.2 ± 0.2 | −3.0 ± 1.3 | None present |
| Meq/vIL8-ECFP | EYFP | 5 | 0.7 ± 2.0 | −2.1 ± 3.8 | None present |
| Meq/vIL8-ECFP | Meq/vIL8-EYFP | 3 | 14.1 ± 2.0 | 11.4 ± 3.3 | 7.5 ± 1.5 |
Resonance energy transfer was measured by the acceptor photobleaching method. Initial fluorescence intensities were recorded from ECFP and EYFP constructs, and then EYFP fluorescence was photobleached using 514-nm excitation light.
The percentage of FRET efficiency (E) was determined by the equation E = (IA − IB)/IA, where IA and IB are the ECFP fluorescence intensities after and before EYFP photobleaching, respectively. None present, these combinations of donor and acceptor were not found. A negative value for FRET efficiency arises from slight drift during data collection.
n, number of measurements.
Time lapse imaging.
CEF were cotransfected with Meq-ECFP and Meq/vIL8-EYFP and imaged using a modified tissue culture incubator attached to the confocal microscope stage. This permitted observation of the subcellular distribution of these fluorescent proteins for up to several days. During time course experiments, the levels of nuclear Meq and Meq/vIL8 proteins typically remained high, with very little cytoplasmic fluorescence. In approximately 20 to 30% of the cells observed, we noted a decrease in the nuclear Meq and Meq/vIL8 fluorescence with a concomitant increase in cytoplasmic fluorescence (Fig. 3). This phenomenon was repeated several times over a 24-hour observation period.
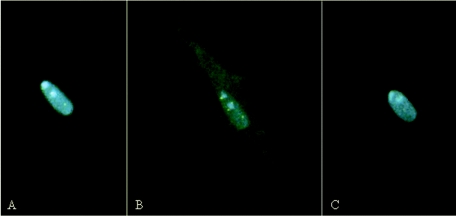
FIG. 3.
Expulsion of nuclear Meq and Meq/vIL8. CEFs were transfected with Meq-ECFP and Meq/vIL8-EYFP and subjected to time lapse acquisition. (A, B and C) Three time points, separated by 20 min, for an individual cell that appeared to expel Meq and Meq/vIL8 from the nucleus (A→B) and then reaccumulate the two proteins in the nucleus (B→C).
DISCUSSION
The meq gene of MDV encodes at least two major sense gene products that are generated by alternative splicing. The Meq gene product is a bZIP transcription modulator that contains multiple domains (Fig. 1B) including nuclear and nucleolar localization domains, an arginine fork RNA binding motif, a bZIP leucine zipper, and transcriptional activation and repression domains (26). The second product, Meq/vIL8, arises by a splicing event (Fig. 1A) that eliminates the transcription modulation domains and fuses the region encoding the leucine zipper in frame with the coding region of the second exon of vIL8 (38). Meq/vIL8 retains a leucine zipper, although the sequence differs from that found in Meq. Using fusions to fluorescent reporter proteins, we have examined the subcellular locations of Meq and Meq/vIL8 in chicken embryonic fibroblasts (Fig. 2). Both Meq and Meq/vIL8 were typically found localized to the nucleolus and nucleoplasm. Meq or Meq/vIL8 was also found associated with p80-coilin, which nucleates the formation of Cajal bodies (1). In addition, antibody-staining experiments indicate that Meq and Meq/vIL8 (22) (Fig. 2I to O) have localization properties similar to those of the fluorescent fusion proteins used in these studies. This suggests that fusion with the fluorescent reporter proteins did not significantly alter the localization properties of either Meq or Meq/vIL8.
The FRAP and PA-GFP mobility studies indicate that Meq and Meq/vIL8 had distinct intracellular properties (Fig. 2P to R; Table 2). FRAP analysis indicated that Meq had a recovery time of approximately 15 s in the nucleolus and nucleoplasm, which is similar to that seen for many other nuclear proteins (11, 15, 18, 19, 25, 41, 42, 50). This recovery time was much slower than that of GFP alone, which had a t1/2 of <0.3 s (not shown) within nuclei, suggesting that the mobility of Meq relative to that of GFP was reduced, perhaps due to specific and nonspecific interactions with nuclear components such as chromatin. Meq/vIL8 was far less mobile than Meq in the nucleolus and nucleoplasm, exhibiting kinetic properties similar to those of proteins that bind tightly to their targets, such as the TATA binding protein, which interacts with chromatin (8), or p80-coilin, which tightly binds Cajal bodies (14, 17, 48).
The mobility studies also suggested that nucleolar Meq and Meq/vIL8 do not have a direct role in ribosomal biogenesis. Nucleolar proteins involved in ribosome biogenesis typically have slower mobility inside the nucleolus than outside (9). This difference is apparently due to the nucleolar proteins forming larger, less mobile complexes involved in ribosomal biogenesis while the nucleoplasmic forms are in much smaller complexes, with correspondingly higher mobilities. In contrast, both Meq and Meq/vIL8 had equivalent t1/2 values in the nucleolus and the nucleoplasm. Accumulating evidence suggests that the nucleolus functions not only in ribosome biogenesis but also in a variety of regulatory processes including modulation of p53 stability, sequestration of Rb protein, and control of mRNA export (55). Given these observations, it is possible that the nucleolar forms of Meq and Meq/vIL8 are involved in regulating cellular functions other than ribosomal biogenesis.
The Cajal bodies have been implicated in a variety of functions, but the most consistent finding is their involvement in RNA maturation directed by small nuclear RNAs affecting splicing or rRNAs. Possibly, Meq or Meq/vIL8 either controls the identity of specific RNAs found in the Cajal bodies or affects their modification pathways. In addition, the RNA component of telomerase associates with Cajal bodies (37, 54). MDV also encodes a telomerase RNA (16), and the association of Meq or Meq/vIL8 with Cajal bodies or p80-coilin could play a role in the function of either the MDV or cellular telomerase RNAs.
FRET results clearly detected energy transfer between Meq/vIL8-ECFP and Meq/vIL8-EYFP in the nucleoli, nucleoplasm, and Cajal bodies, suggesting that this protein formed homodimers (Table 3). Due to an apparent interaction between ECFP and Meq-EYFP, we are unable to use FRET to evaluate the formation of Meq-Meq homodimers or Meq-Meq/vIL8 heterodimers. However, in vitro and recent in vivo evidence indicates that Meq forms homodimers (4, 44), and chromosomal immunoprecipitation studies demonstrated the binding of Meq homodimers to the MDV origin of replication in MSB cells (24).
In vitro studies using reticulocyte-translated proteins also suggested that Meq can form heterodimers with Meq/vIL8 (38). However, our localization and mobility studies suggested that Meq and Meq/vIL8 may not form a large population of heterodimers when coexpressed in CEF. In cotransfected cells expressing both Meq and Meq/vIL8 fusion proteins, Meq/vIL8 was seen associated with Cajal bodies, but no Meq colocalized with the Meq/vIL8 bound to these bodies. In fact, when punctate Cajal bodies were observed in cells cotransfected with Meq plus Meq/vIL8, those bodies were exclusively associated with Meq/vIL8 (Fig. 2C to E). If Meq and Meq/vIL8 formed stable heterodimers, we might expect colocalization of the two proteins at the Cajal bodies, unless heterodimers were unable to bind Cajal bodies. Furthermore, given the disparity between the mobilities of Meq and Meq/vIL8, a reasonable prediction would be that Meq-Meq/vIL8 heterodimers would form a new population of molecules with intermediate mobility between Meq and Meq/vIL8 homodimers. Either Meq (in the context of heterodimers) would shift toward a less mobile form or we would detect a more mobile form of Meq/vIL8. Neither was observed. Taken together, these data suggested that heterodimers of Meq and Meq/vIL8 may not form in abundance inCEF.
Time lapse studies demonstrated a periodic increase in cytoplasmic Meq and Meq/vIL8 in some transfected cells (Fig. 3). This could result from the expulsion of nuclear Meq and Meq/vIL8 into the cytoplasm. At the time of increased cytoplasmic fluorescence, decreased fluorescence was detected in the nucleus, suggesting that Meq and Meq/vIL8 were expelled from the nucleus. Alternatively, newly synthesized Meq and Meq/vIL8 might be excluded from the nucleus yet maintained in a stable cytoplasmic form, while nuclear Meq and Meq/vIL8 are being actively degraded. These models are not mutually exclusive, and both expulsion and degradation may play roles in controlling the levels of nuclear Meq isoforms.
The coordinated regulation of Meq and Meq/vIL8 nuclear localization suggests that a mechanism exists to target them for nuclear expulsion or exclusion. Liu et al. (30) reported elevated cytoplasmic Meq levels in Rat-2 cells during S phase and demonstrated that this change in localization could be regulated by CDK2-mediated phosphorylation. It is not clear what role this phenomenon may play in the function of Meq or Meq/vIL8, but the CDK2 phosphorylation site is in the common amino terminus of each protein. This is possibly a general regulatory mechanism, in which excess transcription factors are cleared from the nucleus during some critical phase of the cell cycle. Alternatively, Meq and Meq/vIL8 may mediate nuclear export of other proteins or RNAs (e.g., through Meq binding of RNA via its arginine fork motif) at this time.
These studies have shown that Meq and Meq/vIL8 occupy similar subcellular nuclear compartments, but their distinct mobilities suggest that they may exist and function as distinct populations. Given the identical amino-terminal regions of Meq and Meq/vIL8, it is reasonable to hypothesize that the two may regulate MDV lytic, latency, or transformation pathways by competitive mechanisms. The absence of the transcription regulatory domain from Meq/vIL8, combined with its mobility characteristics, suggested models for such mechanisms. For example, Meq/vIL8 might form heterodimers with Meq or Meq targets, such as c-Jun, thereby reducing the activity of Meq. The mobility properties of Meq are similar to those of transcription factors, while the mobility of Meq/vIL8 is similar to those of structural components. Conceivably, Meq/vIL8 may also function by anchoring factors, such as Meq or c-Jun, and reduce their dynamic ability to form the transcription complexes necessary for cellular transformation or latency. Future studies examining the dynamic properties of Meq, Meq/vIL8, and proteins such as c-Jun should help distinguish among these possibilities.
Acknowledgments
We thank Judith Sleeman for providing the p80-coilin construct, George Patterson for providing the PA-GFP construct, and Lucy Lee for the anti-Meq antibody.
REFERENCES
- 1.Andrade, L. E., E. K. Chan, I. Raska, C. L. Peebles, G. Roos, and E. M. Tan. 1991. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J. Exp. Med. 173:1407-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Axelrod, D., D. E. Koppel, J. Schlessinger, E. Elson, and W. W. Webb. 1976. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys. J. 16:1055-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birbach, A., S. T. Bailey, S. Ghosh, and J. A. Schmid. 2004. Cytosolic, nuclear and nucleolar localization signals determine subcellular distribution and activity of the NF-κB inducing kinase NIK. J. Cell Sci. 117:3615-3624. [DOI] [PubMed] [Google Scholar]
- 4.Brunovskis, P., Z. Qian, D. Li, L. F. Lee, and J.-J. Jung. 1996. Functional analysis of the MDV basic-leucine zipper product—MEQ, p. 265-270. In R. F. Silva, H. H. Cheng, P. M. Coussens, L. F. Lee, and L. F. Velicer (ed.), Current research on Marek's disease. American Association of Avian Pathologists, Kennett Square, Pa.
- 5.Carrero, G., D. McDonald, E. Crawford, G. de Vries, and M. J. Hendzel. 2003. Using FRAP and mathematical modeling to determine the in vivo kinetics of nuclear proteins. Methods 29:14-28. [DOI] [PubMed] [Google Scholar]
- 6.Catez, F., H. Yang, K. J. Tracey, R. Reeves, T. Misteli, and M. Bustin. 2004. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol. Cell. Biol. 24:4321-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centonze, V. E., B. A. Firulli, and A. B. Firulli. 2004. Fluorescence resonance energy transfer (FRET) as a method to calculate the dimerization strength of basic helix-loop-helix (bHLH) proteins. Biol. Proced. Online 6:78-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, D., C. S. Hinkley, R. W. Henry, and S. Huang. 2002. TBP dynamics in living human cells: constitutive association of TBP with mitotic chromosomes. Mol. Biol. Cell 13:276-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, D., and S. Huang. 2001. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J. Cell Biol. 153:169-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Contreras, A., T. K. Hale, D. L. Stenoien, J. M. Rosen, M. A. Mancini, and R. E. Herrera. 2003. The dynamic mobility of histone H1 is regulated by cyclin/CDK phosphorylation. Mol. Cell. Biol. 23:8626-8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cushman, I., D. Stenoien, and M. S. Moore. 2004. The dynamic association of RCC1 with chromatin is modulated by Ran-dependent nuclear transport. Mol. Biol. Cell 15:245-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Day, R. N., T. C. Voss, J. F. Enwright III, C. F. Booker, A. Periasamy, and F. Schaufele. 2003. Imaging the localized protein interactions between Pit-1 and the CCAAT/enhancer binding protein alpha in the living pituitary cell nucleus. Mol. Endocrinol. 17:333-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.dos Remedios, C. G., and P. D. Moens. 1995. Fluorescence resonance energy transfer spectroscopy is a reliable “ruler” for measuring structural changes in proteins. Dispelling the problem of the unknown orientation factor. J. Struct. Biol. 115:175-185. [DOI] [PubMed] [Google Scholar]
- 14.Dundr, M., M. D. Hebert, T. S. Karpova, D. Stanek, H. Xu, K. B. Shpargel, U. T. Meier, K. M. Neugebauer, A. G. Matera, and T. Misteli. 2004. In vivo kinetics of Cajal body components. J. Cell Biol. 164:831-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elbi, C., D. A. Walker, G. Romero, W. P. Sullivan, D. O. Toft, G. L. Hager, and D. B. DeFranco. 2004. Molecular chaperones function as steroid receptor nuclear mobility factors. Proc. Natl. Acad. Sci. USA 101:2876-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fragnet, L., M. A. Blasco, W. Klapper, and D. Rasschaert. 2003. The RNA subunit of telomerase is encoded by Marek's disease virus. J. Virol. 77:5985-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Handwerger, K. E., C. Murphy, and J. G. Gall. 2003. Steady-state dynamics of Cajal body components in the Xenopus germinal vesicle. J. Cell Biol. 160:495-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrer, M., H. Luhrs, M. Bustin, U. Scheer, and R. Hock. 2004. Dynamic interaction of HMGA1a proteins with chromatin. J. Cell Sci. 117:3459-3471. [DOI] [PubMed] [Google Scholar]
- 19.Harrington, K. S., A. Javed, H. Drissi, S. McNeil, J. B. Lian, J. L. Stein, A. J. Van Wijnen, Y. L. Wang, and G. S. Stein. 2002. Transcription factors RUNX1/AML1 and RUNX2/Cbfa1 dynamically associate with stationary subnuclear domains. J. Cell Sci. 115:4167-4176. [DOI] [PubMed] [Google Scholar]
- 20.Howell, B. J., B. Moree, E. M. Farrar, S. Stewart, G. Fang, and E. D. Salmon. 2004. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 14:953-964. [DOI] [PubMed] [Google Scholar]
- 21.Jones, D., L. Lee, J. L. Liu, H. J. Kung, and J. K. Tillotson. 1992. Marek disease virus encodes a basic-leucine zipper gene resembling the fos/jun oncogenes that is highly expressed in lymphoblastoid tumors. Proc. Natl. Acad. Sci. USA 89:4042-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kingham, B. F., V. Zelnik, J. Kopacek, V. Majerciak, E. Ney, and C. J. Schmidt. 2001. The genome of herpesvirus of turkeys: comparative analysis with Marek's disease viruses. J. Gen. Virol. 82:1123-1135. [DOI] [PubMed] [Google Scholar]
- 23.Lee, L. F., J. L. Liu, X. P. Cui, and H. J. Kung. 2003. Marek's disease virus latent protein MEQ: delineation of an epitope in the BR1 domain involved in nuclear localization. Virus Genes 27:211-218. [DOI] [PubMed] [Google Scholar]
- 24.Levy, A. M., Y. Izumiya, P. Brunovskis, L. Xia, M. S. Parcells, S. M. Reddy, L. Lee, H. W. Chen, and H. J. Kung. 2003. Characterization of the chromosomal binding sites and dimerization partners of the viral oncoprotein Meq in Marek's disease virus-transformed T cells. J. Virol. 77:12841-12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lillemeier, B. F., M. Koster, and I. M. Kerr. 2001. STAT1 from the cell membrane to the DNA. EMBO J. 20:2508-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, J. L., and H. J. Kung. 2000. Marek's disease herpesvirus transforming protein MEQ: a c-Jun analogue with an alternative life style. Virus Genes 21:51-64. [PubMed] [Google Scholar]
- 27.Liu, J. L., L. F. Lee, Y. Ye, Z. Qian, and H. J. Kung. 1997. Nucleolar and nuclear localization properties of a herpesvirus bZIP oncoprotein, MEQ. J. Virol. 71:3188-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, J. L., S. F. Lin, L. Xia, P. Brunovskis, D. Li, I. Davidson, L. F. Lee, and H. J. Kung. 1999. MEQ and V-IL8: cellular genes in disguise? Acta Virol. 43:94-101. [PubMed] [Google Scholar]
- 29.Liu, J. L., Y. Ye, L. F. Lee, and H. J. Kung. 1998. Transforming potential of the herpesvirus oncoprotein MEQ: morphological transformation, serum-independent growth, and inhibition of apoptosis. J. Virol. 72:388-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, J. L., Y. Ye, Z. Qian, Y. Qian, D. J. Templeton, L. F. Lee, and H. J. Kung. 1999. Functional interactions between herpesvirus oncoprotein MEQ and cell cycle regulator CDK2. J. Virol. 73:4208-4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lupiani, B., L. F. Lee, X. Cui, I. Gimeno, A. Anderson, R. W. Morgan, R. F. Silva, R. L. Witter, H. J. Kung, and S. M. Reddy. 2004. Marek's disease virus-encoded Meq gene is involved in transformation of lymphocytes but is dispensable for replication. Proc. Natl. Acad. Sci. USA 101:11815-11820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsushita, K., and Y. Sugiura. 2003. GFP-linked zinc finger protein sp1. fluorescence study and implication for N-terminal zinc finger 1 as hinge finger. Bioorg. Med. Chem. 11:53-58. [DOI] [PubMed] [Google Scholar]
- 33.O'Donnell, L. A., J. A. Clemmer, K. Czymmek, and C. J. Schmidt. 2002. Marek's disease virus VP22: subcellular localization and characterization of carboxyl terminal deletion mutations. Virology 292:235-240. [DOI] [PubMed] [Google Scholar]
- 34.Parcells, M. S., S. F. Lin, R. L. Dienglewicz, V. Majerciak, D. R. Robinson, H. C. Chen, Z. Wu, G. R. Dubyak, P. Brunovskis, H. D. Hunt, L. F. Lee, and H. J. Kung. 2001. Marek's disease virus (MDV) encodes an interleukin-8 homolog (vIL-8): characterization of the vIL-8 protein and a vIL-8 deletion mutant MDV. J. Virol. 75:5159-5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patterson, G. H., and J. Lippincott-Schwartz. 2002. A photoactivatable GFP for selective photolabeling of proteins and cells. Science 297:1873-1877. [DOI] [PubMed] [Google Scholar]
- 36.Patterson, G. H., and J. Lippincott-Schwartz. 2004. Selective photolabeling of proteins using photoactivatable GFP. Methods 32:445-450. [DOI] [PubMed] [Google Scholar]
- 37.Pederson, T. 2004. Can telomerase be put in its place? J. Cell Biol. 164:637-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng, Q., and Y. Shirazi. 1996. Characterization of the protein product encoded by a splicing variant of the Marek's disease virus Eco-Q gene (Meq). Virology 226:77-82. [DOI] [PubMed] [Google Scholar]
- 39.Peng, Q., and Y. Shirazi. 1996. Isolation and characterization of Marek's disease virus (MDV) cDNAs from a MDV-transformed lymphoblastoid cell line: identification of an open reading frame antisense to the MDV Eco-Q protein (Meq). Virology 221:368-374. [DOI] [PubMed] [Google Scholar]
- 40.Petherbridge, L., A. C. Brown, S. J. Baigent, K. Howes, M. A. Sacco, N. Osterrieder, and V. K. Nair. 2004. Oncogenicity of virulent Marek's disease virus cloned as bacterial artificial chromosomes. J. Virol. 78:13376-13380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404:604-609. [DOI] [PubMed] [Google Scholar]
- 42.Phair, R. D., and T. Misteli. 2001. Kinetic modelling approaches to in vivo imaging. Nat. Rev. Mol. Cell Biol. 2:898-907. [DOI] [PubMed] [Google Scholar]
- 43.Powell, P. C., and L. N. Payne. 1993. Marek's disease, p. 37-75. In J. B. McFerran and M. S. McBulty (ed.), Virus infections of birds, vol. 4. Elsevier Science, New York, N.Y. [Google Scholar]
- 44.Qian, Z., P. Brunovskis, F. Rauscher III, L. Lee, and H. J. Kung. 1995. Transactivation activity of Meq, a Marek's disease herpesvirus bZIP protein persistently expressed in latently infected transformed T cells. J. Virol. 69:4037-4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raska, I., R. L. Ochs, L. E. Andrade, E. K. Chan, R. Burlingame, C. Peebles, D. Gruol, and E. M. Tan. 1990. Association between the nucleolus and the coiled body. J. Struct. Biol. 104:120-127. [DOI] [PubMed] [Google Scholar]
- 46.Shimi, T., T. Koujin, M. Segura-Totten, K. L. Wilson, T. Haraguchi, and Y. Hiraoka. 2004. Dynamic interaction between BAF and emerin revealed by FRAP, FLIP, and FRET analyses in living HeLa cells. J. Struct. Biol. 147:31-41. [DOI] [PubMed] [Google Scholar]
- 47.Silim, A., M. A. El Azhary, and R. S. Roy. 1982. A simple technique for preparation of chicken-embryo-skin cell cultures. Avian Dis. 26:182-185. [PubMed] [Google Scholar]
- 48.Sleeman, J. E., L. Trinkle-Mulcahy, A. R. Prescott, S. C. Ogg, and A. I. Lamond. 2003. Cajal body proteins SMN and Coilin show differential dynamic behaviour in vivo. J. Cell Sci. 116:2039-2050. [DOI] [PubMed] [Google Scholar]
- 49.Stavreva, D. A., and J. G. McNally. 2004. Fluorescence recovery after photobleaching (FRAP) methods for visualizing protein dynamics in living mammalian cell nuclei. Methods Enzymol. 375:443-455. [DOI] [PubMed] [Google Scholar]
- 50.Stenoien, D. L., K. Patel, M. G. Mancini, M. Dutertre, C. L. Smith, B. W. O'Malley, and M. A. Mancini. 2001. FRAP reveals that mobility of oestrogen receptor-alpha is ligand- and proteasome-dependent. Nat. Cell Biol. 3:15-23. [DOI] [PubMed] [Google Scholar]
- 51.Tanaka, M., M. Nishi, M. Morimoto, T. Sugimoto, and M. Kawata. 2003. Yellow fluorescent protein-tagged and cyan fluorescent protein-tagged imaging analysis of glucocorticoid receptor and importins in single living cells. Endocrinology 144:4070-4079. [DOI] [PubMed] [Google Scholar]
- 52.Wouters, F. S., P. J. Verveer, and P. I. Bastiaens. 2001. Imaging biochemistry inside cells. Trends Cell Biol. 11:203-211. [DOI] [PubMed] [Google Scholar]
- 53.Zheng, L., K. P. Hoeflich, L. M. Elsby, M. Ghosh, S. G. Roberts, and M. Ikura. 2004. FRET evidence for a conformational change in TFIIB upon TBP-DNA binding. Eur. J. Biochem. 271:792-800. [DOI] [PubMed] [Google Scholar]
- 54.Zhu, Y., R. L. Tomlinson, A. A. Lukowiak, R. M. Terns, and M. P. Terns. 2004. Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol. Biol. Cell 15:81-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zolotukhin, A. S., and B. K. Felber. 1999. Nucleosporins Nup98 and Nup214 participate in nuclear export of human immunodeficiency virus type 1 Rev. J. Virol. 73:120-127. [DOI] [PMC free article] [PubMed] [Google Scholar]