Abstract
Mutations in the human cytomegalovirus DNA polymerase (UL54) can not only decrease but also increase susceptibility to the pyrophosphate (PPi) analogue foscarnet. The proximity of L802M, which confers resistance, and K805Q, which confers hypersusceptibility, suggests a possible unifying mechanism that affects drug susceptibility in one direction or the other. We found that the polymerase activities of L802M- and K805Q-containing mutant enzymes were literally indistinguishable from that of wild-type UL54; however, susceptibility to foscarnet was decreased or increased, respectively. A comparison with the crystal structure model of the related RB69 polymerase suggests that L802 and K805 are located in the conserved α-helix P that is implicated in nucleotide binding. Although L802 and K805 do not appear to make direct contacts with the incoming nucleotide, it is conceivable that changes at these residues could exert their effects through the adjacent, highly conserved amino acids Q807 and/or K811. Our data show that a K811A substitution in UL54 causes reductions in rates of nucleotide incorporation. The activity of the Q807A mutant is only marginally affected, while this enzyme shows relatively high levels of resistance to foscarnet. Based on these data, we suggest that L802M exerts its effects through subtle structural changes in α-helix P that affect the precise positioning of Q807 and, in turn, its presumptive involvement in binding of foscarnet. In contrast, the removal of a positive charge associated with the K805Q change may facilitate access or increase affinity to the adjacent Q807.
The human cytomegalovirus (HCMV), which is a DNA virus that belongs to the Herpesviridae family, is an important human pathogen (16, 23). Immunocompromised patients, for instance, transplant recipients or AIDS patients, are especially vulnerable to such infection, although the clinical use of antiretroviral drugs against human immunodeficiency virus type 1 (HIV-1) has dramatically decreased the incidence of HCMV-related disease for patients who respond to HIV-specific treatment (33). Antiviral drugs that are used to treat HCMV infection target the viral DNA polymerase (9). The nucleoside analogue ganciclovir (GCV) and the nucleotide analogue cidofovir (CDV) are approved anti-HCMV drugs. Both drugs were shown to significantly reduce the viral burden; however, antiviral therapy over protracted periods of time can lead to the emergence of resistance-conferring mutations, which is a major factor associated with treatment failure (18). Resistance to GCV has been associated with mutations in the viral UL97 and UL54 genes (4, 11, 12, 18). The former encodes a kinase that phosphorylates the drug to its monophosphate form, and the latter encodes the viral DNA polymerase that accepts the triphosphate form of this compound. CDV is a phosphonate that requires activation by cellular enzymes to its diphosphate form, which explains why changes in the UL97 gene do not affect susceptibility to this compound (27). However, many of the mutations found in the DNA polymerase confer cross-resistance to both drugs. The pyrophosphate analogue foscarnet (PFA; phosphonoformic acid) is the third approved anti-HCMV compound that is frequently administered when first-line agents have failed.
The triphosphate form of GCV and the diphosphate form of CDV compete with their natural counterparts dGTP and dCTP, respectively, for binding to the DNA polymerase and incorporation into the viral DNA (39). Once incorporated, these compounds were shown to reduce subsequent polymerization steps (38). Biochemical studies with the UL54 enzyme and the related herpes simplex virus (HSV) DNA polymerase (UL30) suggested that resistance-conferring mutations appear to diminish drug binding and/or incorporation (5, 22). The mechanisms of drug action and resistance associated with foscarnet are less well characterized. This compound exhibits a broad spectrum of antiviral activities and has been shown to block replication of related herpesviruses, including HCMV, HSV type 1 (HSV-1), and HSV-2, as well as retroviruses, including HIV-1 (8, 28). Foscarnet acts noncompetitively with respect to the deoxynucleoside triphosphate (dNTP) substrate (13). It has been suggested that the pyrophosphate analogue binds to a site in close proximity to the active center that may overlap at least in part with the pyrophosphate binding site (10). This notion is based on biochemical studies showing that foscarnet blocks pyrophosphate exchange reactions and pyrophosphorolysis, i.e., the reverse reaction, in competitive fashion. However, the precise binding site of foscarnet and the detailed mechanism of drug action remain elusive. Mutations that confer resistance to foscarnet map to different conserved regions in the UL54 gene and do not appear to cluster around a specific site (17). At the same time, it should be mentioned that crystal structure models of UL54 and UL30 are not available, which makes it difficult to define the active center and the putative binding site for foscarnet.
Clinical data have shown that resistance to foscarnet is associated with mutations T700A, M715V, E756Q, V781I, V787L, L802M, A809V, V812L, and T821I (3, 18, 37). Of note, it has been reported that prolonged treatment with GCV and PFA can lead to the emergence of a dually resistant phenotype. One particular isolate harbored mutations T821I and K805Q among others (32). Drug susceptibility measurements with recombinant viruses that contained single point mutations revealed that T821I caused a 21-fold decrease in susceptibility to foscarnet, while K805Q conferred a 5- to 6-fold increase in susceptibility to foscarnet (5). Drug hypersusceptibility is of potential clinical benefit, which merits further investigation. K805 is also located in the vicinity of other residues that were shown to reduce susceptibility to foscarnet, e.g., L802M. It is therefore conceivable that this region plays an important role in drug binding and enzymatic activity.
Subtle structural changes may either decrease or increase the affinity to foscarnet, which may translate into resistant or hypersusceptible phenotypes, respectively. Here we generated wild-type (WT) UL54 as well as the K805Q- and L802M-containing mutant enzymes in order to study the biochemical mechanisms associated with these complex phenotypes. We found that the enzymatic activities are not significantly changed in the absence or in the presence of either of the two mutations. However, K805Q increased the inhibitory effects of foscarnet, while L802M decreased its efficacy in cell-free assays, which is in good agreement with published drug susceptibility data (5). We provide evidence to show that both residues are most likely located within a helical region in close proximity but not in contact distance to the bound dNTP. Mutagenesis studies, based on the structure of the related DNA polymerase (gp43) of the bacteriophage RB69, suggest that both mutations could exert their effects indirectly through the adjacent, highly conserved residue Q807.
MATERIALS AND METHODS
Plasmid constructs.
Wild-type and mutant enzymes were derived from recombinant viruses generated by overlapping cosmids as described previously (5). The UL54 coding sequence was cloned into pCITE4b (Novagen) by use of the EcoRI and HindIII sites to generate pCITE4b/UL54-WT, -L802M, -Q805A, -Q807A, and -K811A. Amino acid substitutions were introduced with Pfu DNA polymerase (Fermentas) according to the manufacturer's recommendations by amplification of the pCITE4b/UL54-WT plasmid with primer pairs that contained the proper changes. All plasmids were sequenced to confirm the presence of the introduced mutation and the absence of undesired changes.
Protein expression.
All enzymes were expressed in rabbit reticulocyte lysate by use of a coupled in vitro transcription-translation system (Promega). Reactions were conducted essentially as previously described (6). In order to assess and compare the yields of the different enzymes, we included 40 mCi [35S]methionine (Amersham Biosciences) in the reaction mixture and analyzed the products through sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The gels were quantified by phosphorimager analysis (Storm 840; Amersham Biosciences). We found that 2.5 to 3% of [35S]methionine was incorporated into the proteins, which corresponds to a protein concentration of approximately 1.5 ng/ml. This amount was similar for wild-type and mutant proteins.
Bioinformatics and structural analyses.
Sequence alignments were generated with MacVector software (Accelrys). The structural information of the RB69-associated polymerase gp43 is based on the structure of the ternary complex (PDB no. 1IG9). Figure 1C was generated with a Deep View-Swiss PDB viewer.
FIG.1.
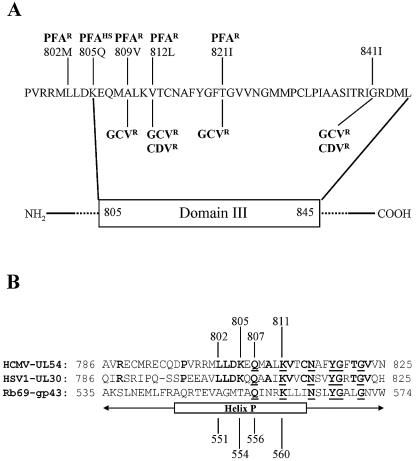
Location of resistance-conferring mutations in helix P. (A) Amino acid sequence of conserved domain III of the HCMV DNA polymerase UL54. Highlighted are residues that have been associated with changes in drug susceptibility: cidofovir (CDV), ganciclovir (GCV), and foscarnet (PFA). Superscript R and HS indicate resistant and hypersusceptible phenotypes, respectively. (B) Amino acid sequence of helix P of the RB69-associated DNA polymerase gp43 aligned against corresponding sequences of UL54 (HCMV) and UL30 (HSV1). Bold letters indicate amino acid residues that are conserved in both UL54 and UL30. Underlined letters indicate amino acid residues conserved among all three related DNA polymerases. Highly conserved residues are found between positions 807 and 822. (C) Interaction between helix P and the bound nucleotide, based on the structure of the ternary complex of gp43 (14). Divalent metal ions are shown as gray spheres. The bound nucleotide is shown in green, the position of the 3′ end of the primer is shown in yellow, and the complementary template positions i + 1 and i are shown in white. Helix P (A535 to G571) is shown in blue, and residues that are relevant to this work are highlighted: A551 (red; changes at this position in UL54 are associated with resistance to foscarnet), T554 (green; changes at this position in UL54 are associated with hypersusceptibility to foscarnet), Q556 (yellow), and K560 (magenta). The last two amino acids are highly conserved among α-DNA polymerases. Amino acids in parentheses indicate the structural equivalents in UL54.
Filter-based DNA polymerase assay.
DNA polymerase activity was tested in the presence of activated calf thymus DNA. The reaction was performed with 25 mM Tris-HCl (pH 8), 90 mM NaCl, 0.5 mM dithiothreitol, 0.2 mg/ml bovine serum albumin, and 5% glycerol (designated buffer P). dATP, dCTP, and dTTP (10 μM each), 100 μg/ml activated calf thymus DNA (Amersham Biosciences), 1 μM [3H]dTTP, and 0.5 μl of the enzyme-containing transcription-translation mixture (∼0.75 ng enzyme) were used in a typical setup. Reactions were carried out in a total volume of 25 μl at 37°C, following the addition of MgCl2 (10 mM). DNA synthesis was stopped after 30 min by addition of 600 μl of cold 10% trichloroacetic acid-1% NaPPi, followed by nucleic acid precipitation on ice for 1 h. The samples were filtered and washed with 10% trichloroacetic acid-1% NaPPi to measure the labeled DNA by scintillation counting.
Gel-based DNA polymerase assay.
Unless otherwise indicated, we used the following synthetic heteropolymeric oligonucleotides (Invitrogen) in gel-based assays to measure the polymerase activity of UL54: 5′-GAGTGGTATAGTGGAGTGAA-3′ (P20) and 5′-CCAATATTCACCATCAAGGCTTGACGTCACTTCACTCCACTATACCACTC-3′ (T50). The underlined sequence represents the region of the template that is complementary to the primer (P20). The synthetic oligonucleotides were purified on 12% polyacrylamide-7 M urea gels containing 50 mM Tris-borate, pH 8.0, and 1 mM EDTA. 5′-end labeling of the primer was performed with [γ-32P]ATP and T4 polynucleotide kinase. The labeled products were again electrophoretically purified to obtain homogeneously labeled nucleic acids. Primer/template sequences were heat annealed prior to the start of the reaction. In a typical reaction, DNA synthesis was conducted with buffer P in the presence of 10 μM of each of the four nucleoside triphosphates, 1.5 pmol of the primer/template substrate, and 1 μl (∼0.0125 pmol UL54) of the enzyme preparation. We used a relatively high ratio of nucleic acid/enzyme to ensure that DNA synthesis was monitored in the linear range. Reactions were initiated by the addition of MgCl2 (10 mM) in a total volume of 20 μl and stopped by addition of 20 μl formamide and bromophenol blue/xylene cyanole dyes. The products were heat denatured for 5 min at 95°C, resolved on 8% polyacrylamide-7 M urea gels, and analyzed by phosphorimager analysis.
Measurements of IC50 values.
Polymerase activity was monitored in the presence of increasing concentrations of foscarnet or pyrophosphate in order to determine the concentrations of inhibitor that were required to block 50% of full-length DNA synthesis (50% inhibitory concentrations [IC50]). The ranges of concentrations used are indicated in the figure legends. The data were analyzed with Prism software (GraphPad Prism version 4.0).
Steady-state kinetics.
Steady-state kinetic parameters Km and Vmax for single-nucleotide incorporation events were determined through gel-based assays, as described previously (2). We used four different primer/template combinations, referred to as P20/T33-A, -C, -G, and -T: 5′-GAGTGGTATAGTGGAGTGAA-3′ (P20), 5′-AGAGAGAGAGAGTTTCACTCCACTATACCACTC-3′ (T33-A), 5′-ACACACACACACGTTCACTCCACTATACCACTC-3′ (T33-C), 5′-GTGTGTGTGGTGCTTCACTCCACTATACCACTC-3′ (T33-G), and 5′-TCTCTCTCTTCTATTCACTCCACTATACCACTC-3′ (T33-T). Each underlined sequence represents the region of the template that is complementary to the primer (P20).
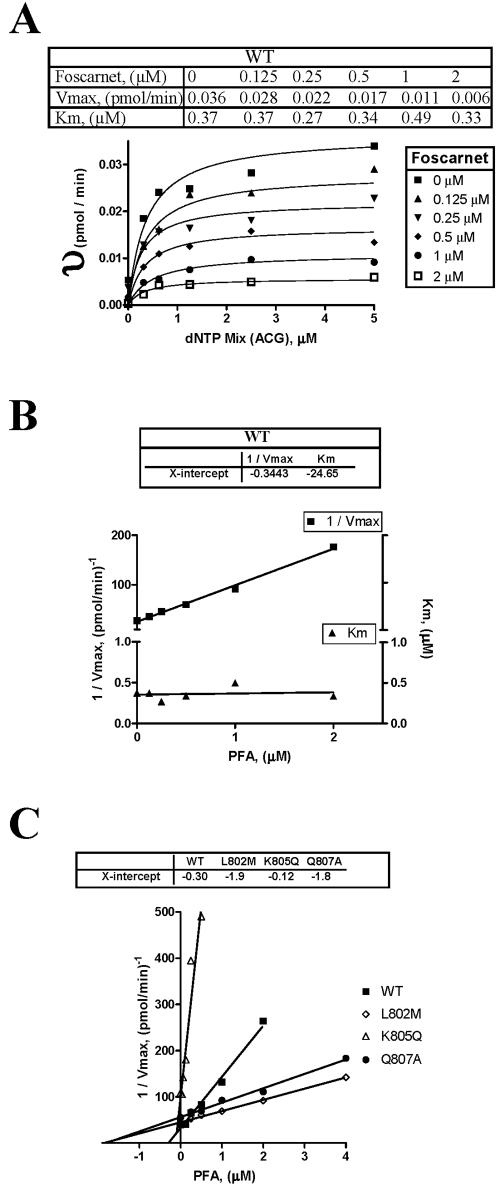
The incorporation of the nucleotide was monitored at a single time point in the presence of increasing concentrations of dATP, dCTP, dGTP, or dTTP. Reactions were allowed to proceed for 10 min at 37°C. Misincorporations have not been observed under these conditions. The data were fitted to Michaelis-Menten equations by use of GraphPad Prism (version 4.0). Ki values were determined on the basis of measurements of multiple incorporation events by using calf thymus DNA as described above. The assay was conducted in the presence of 1 μM [3H]dTTP and increasing concentrations (0, 0.08, 0.16, 0.32, 0.64, 1.3, and 2.5 μM) of dATP, dCTP, and dGTP. This experiment was performed with five different concentrations of foscarnet (0.13, 0.25, 0.5, 1, and 2 μM for WT-UL54; 0.25, 0.5, 1, 2, and 4 μM for the L802M and Q807A mutants; and 0.031, 0.061, 0.13, 0.25, and 0.5 μM for Q805K). The data were fitted to Michaelis-Menten equations to determine Km and Vmax values. Ki values and type of inhibition were determined by plotting 1/Vmax and Km as a function of foscarnet concentration.
RESULTS
Experimental design.
Figure 1A shows the distribution of known foscarnet resistance mutations in domain III of HCMV UL54. This region contains numerous residues that are highly conserved among different members of the α-DNA polymerase family, to which the HSV-, HCMV-, and RB69-associated enzymes belong (17). While structural models of the HSV and HCMV polymerases are not available, the crystal structure of the RB69 enzyme (gp43) has been determined in various forms (15, 20, 36), including the structure of a complex with bound primer/template and nucleotide substrates (14). This model shows that the highly conserved residues of domain III constitute part of helix P, which interacts with the template strand and the nucleoside triphosphate (Fig. 1B and C). K560 in gp43 has been identified as an important residue that contacts the bound nucleotide at its α- and γ-phosphates (40). Pre-steady-state kinetic analyses have shown that a K560A change in gp43 caused a severe reduction in rates of nucleotide incorporation (40). The mutant had much a higher Kd value than the wild-type enzyme. The UL54 counterpart of K560 is located at position 811, which is in close proximity to the mutations that are implicated in resistance (L802M) and hypersusceptibility (K805Q) to foscarnet. However, the gp43 structure suggests that the equivalent residues, A551 and T554, respectively, reside at a distance of approximately 12 Å away from γ-phosphate (Fig. 1C). Thus, one could consider two different explanations to reconcile these observations. The structures of UL54 and gp43 could differ to such an extent that L802 and K805 are actually located closer to the bound nucleotide, where the two mutations may directly affect binding of foscarnet. Alternatively, the structures are indeed very similar and L802M and K805Q may exert their effects indirectly over a longer distance. An HSV-1 recombinant that contains the K805Q change shows the same hypersusceptible phenotype as described for HCMV, which points to a high degree of structural homology in this region, at least among UL54 and UL30 (1).
To better assess to which degree the structures of the RB69 and UL54 enzymes resemble one another, in particular around the dNTP/PPi binding site, we generated two UL54 mutant enzymes that contain alanine residues at positions K811 and Q807. The glutamine is also conserved among various members of the α-like polymerases; however, Q556 in gp43 still resides at a distance of approximately 6 Å away from the γ-phosphate. This may not suffice for direct contacts, although Q556 is located on the same side of the helix as the phosphate. Thus, one would predict that the effects of changes at Q556 (gp43) and Q807 (UL54) on the rates of polymerization are likely to be less pronounced than changes at K560 and K811, respectively. In order to shed light on the molecular mechanisms involved in resistance and hypersusceptibility to foscarnet, we characterized the two UL54-associated mutants K811A and Q807A together with enzymes that contain mutations L802M and K805Q.
Steady-state kinetics.
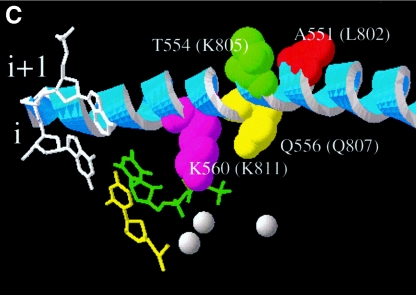
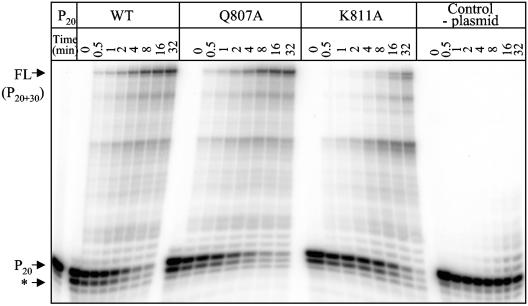
Throughout this study, we used a coupled in vitro transcription-translation system to generate UL54 and its mutant variants (see Materials and Methods). This expression system has proven successful for the determination of steady-state kinetic parameters (7, 41). In order to compare the efficiency of DNA synthesis of wild-type UL54 and that of the four mutant enzymes, we initially looked at multiple nucleotide incorporation events by using a defined primer/template system in gel-based assays (Fig. 2). The activities of wild-type UL54 and the Q807A mutant appeared almost identical in this experiment; however, DNA synthesis with K811A was compromised. The two mutants L802M and K805Q displayed wild-type-like activities as well (data not shown). To translate these observations into quantitative terms, we next determined Km and Vmax values with regards to the incorporation of a single nucleotide. For this purpose, we incrementally increased the concentrations of the first nucleotide and omitted the three other dNTPs from the reaction mixture (Fig. 3). Overall, we found that the efficiency of the reaction (Vmax/Km) was similar for wild-type UL54 and the three mutants Q807A, L802M, and K805Q, while K811A was again compromised (Table 1). We measured the following reductions in the efficiency of a single-nucleotide incorporation event when K811A was compared with the wild-type enzyme: 15-fold (dATP), 12-fold (dCTP), 20-fold (dGTP), and 30-fold (dTTP). In contrast, Q807A caused only relatively minor changes: 2.3-fold (dATP), 2-fold (dCTP), 1.7-fold (dGTP), and 3-fold (dTTP). We determined similar relatively small changes with the other two mutants, L802M and K805Q (Table 1).
FIG. 2.
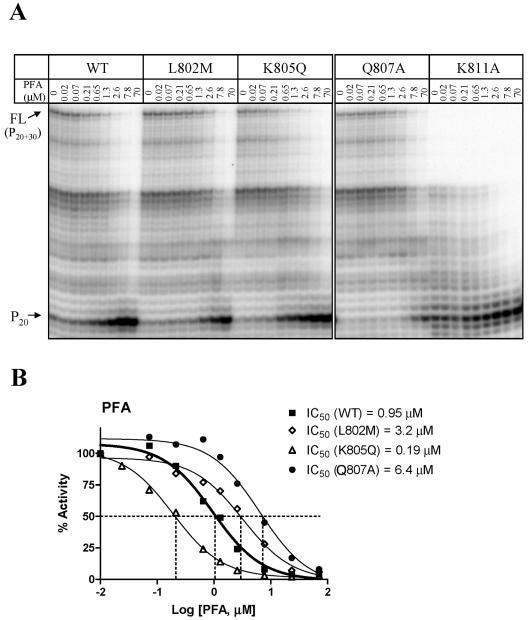
Efficiency of DNA synthesis with WT and mutant enzymes. The levels of DNA synthesis were measured with the heteropolymeric primer/template substrate P20/T50 DNA as a function of time. FL denotes the full-length product, and P20 denotes the unextended primer. The asterisk points to the exonuclease activity of UL54 enzymes. This band is less pronounced in the control without enzyme (left, primer only) and in reaction mixtures that contain plasmid pCITE4b without the coding sequence for UL54 (right).
FIG. 3.
Steady-state kinetic analyses. (A) Graphic representation of the experimental setup used to monitor single-nucleotide incorporations. The substrate P20/T33-T DNA allows the incorporation of dTTP, which extends primer P20 by a single nucleotide to yield P20 + 1. Complete sequences are shown in Materials and Methods. (B) Incorporation of dTMP by WT UL54. efficiency of single-nucleotide incorporations was monitored in the presence of increasing concentrations of dTTP, while all other deoxyribonucleotides were omitted from the reaction. The reaction was allowed to proceed for 10 min. Under these conditions, no significant extensions in the control reactions that contain plasmid pCITE4b without the coding sequence for UL54 were seen. This approach was used to determine the kinetic parameters that are listed in Table 1.
TABLE 1.
Kinetic parameters for DNA polymerizationa
| Enzyme | dATP
|
dCTP
|
dGTP
|
dTTP
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Km (μM) | Vmax (pmol/min) | Km/Vmax | Km (μM) | Vmax (pmol/min) | Km/ Vmax | Km (μM) | Vmax (pmol/min) | Km/Vmax | Km (μM) | Vmax (pmol/min) | Km/Vmax | |
| WT | 0.79 ± 0.03 | 0.07 ± 0.003 | 0.09 | 0.10 ± 0.02 | 0.12 ± 0.08 | 1.2 | 0.11 ± 0.01 | 0.11 ± 0.002 | 1.0 | 0.31 ± 0.04 | 0.10 ± 0.005 | 0.3 |
| L802M | 1.01 ± 0.11 | 0.07 ± 0.002 | 0.07 | 0.10 ± 0.03 | 0.07 ± 0.005 | 0.7 | 0.22 ± 0.03 | 0.06 ± 0.004 | 0.3 | 0.24 ± 0.02 | 0.06 ± 0.002 | 0.3 |
| K805Q | 0.57 ± 0.05 | 0.07 ± 0.004 | 0.13 | 0.16 ± 0.02 | 0.05 ± 0.001 | 0.3 | 0.21 ± 0.02 | 0.04 ± 0.006 | 0.2 | 0.20 ± 0.01 | 0.06 ± 0.003 | 0.3 |
| Q807A | 1.16 ± 0.09 | 0.04 ± 0.020 | 0.04 | 0.19 ± 0.05 | 0.12 ± 0.08 | 0.6 | 0.17 ± 0.01 | 0.11 ± 0.005 | 0.6 | 0.76 ± 0.45 | 0.10 ± 0.008 | 0.1 |
| K811A | 1.65 ± 0.66 | 0.01 ± 0.002 | 0.006 | 0.65 ± 0.07 | 0.08 ± 0.03 | 0.1 | 2.09 ± 0.38 | 0.10 ± 0.01 | 0.05 | 3.55 ± 0.6 | 0.05 ± 0.03 | 0.01 |
DNA synthesis reactions of one nucleotide incorporation were performed using P20/T33 DNA substrates as described in Materials and Methods. Data were fitted to the Michaelis-Menten equation in order to determine Km and Vmax constants. Values are means of at least three determinations ± standard deviations.
These data provide strong evidence to suggest that the two conserved residues in helix P of gp43 and UL54 exhibit similar functions with regards to dNTP binding and catalysis. With the exception of dATP, the reduced efficiency of nucleotide incorporation seen with the K811A mutant was largely attributable to higher Km values. For yet-unknown reasons, the incorporation of dAMP, relative to that of the three other nucleotides, was generally associated with elevated Km values for each of the enzymes studied. The relatively small effects on single-nucleotide incorporation events seen with the Q807A mutant suggest that the distance between the side chain of the conserved glutamine and the γ-phosphate of the incoming nucleotide might be too long to allow direct contacts. This notion is supported by the fact that mutations L802M and K805Q showed similar minor changes with respect to the efficiency of incorporation of a single nucleotide.
Inhibitory effects of foscarnet and pyrophosphate.
We utilized the same primer/template substrate as shown in Fig. 2 to analyze the effects of L802M- and K805Q-containing mutant enzymes on DNA synthesis in the presence of increasing concentrations of foscarnet. In this context, we measured the concentration of foscarnet that is required to inhibit 50% of the production of full-length DNA (Fig. 4). The L802M mutant showed a threefold-increased IC50 value, while the K805Q mutant showed a fivefold-decreased IC50 value. These biochemical data are remarkably consistent with drug susceptibility measurements based on plaque reduction assays (5). It would be of interest to conduct the kinetic measurements also in the presence of UL44, which acts as a processivity factor (24); however, our data suggest that resistance- or hypersusceptibility-conferring mutations affect intrinsic properties of the polymerase UL54. It is unlikely that L802M and K805Q may directly affect binding of foscarnet. Our data with mutants K811A and Q807A suggest that positions 802 and 805 are too far away from the presumptive binding site of the drug. This site may partially overlap with the PPi binding site, considering that foscarnet inhibits pyrophosphorolysis in a competitive fashion. Crystal structures of complexes of T7 RNA polymerase with and without PPi or dNTP substrates show that PPi binds to the same location that was formerly occupied by the β- and γ-phosphates of the bound nucleotide (42).
FIG. 4.
Measurements of IC50 values for inhibition of DNA synthesis in the presence of foscarnet. (A) The amount of full-length (FL) DNA synthesis was measured in the presence of increasing concentrations of foscarnet as indicated. Reactions were allowed to proceed for 30 min. (B) Dose-response curves and measurements of IC50 values for inhibition of DNA synthesis by WT and mutant enzymes. Changes in the ratios of the full-length product and the unextended primer were quantified to generate dose-response curves. Dotted lines point to the concentration of the inhibitor that reduces the activity of the enzyme by 50% (IC50).
L802M and K805Q may exert their effects indirectly through structural changes of other residues that are located in close proximity to the nucleotide binding site. Q807 might be a good candidate in this regard. Thus, we included the Q807A mutant in this set of experiments and found that this change caused a sixfold-increased IC50 value, which is twice as high as the change we measured with the naturally occurring resistance mutation L802M. These data support the notion that L802M may exert its resistance-conferring effects through structural perturbations at or around position 807. However, these data do not explain the hypersusceptible phenotype associated with K805Q.
If the binding site for foscarnet indeed overlaps with the binding site for PPi, one can predict that the three mutations L802M, K805Q, and Q807A affect the inhibition of DNA synthesis in the presence of PPi in a fashion similar to that described for foscarnet. To address this problem, we utilized a polymerase assay that is based on measurements of DNA synthesis with activated calf thymus DNA (see Materials and Methods). This assay differs from our gel-based setup, as it involves longer heteropolymeric DNA templates, which potentially provides higher sensitivity since the inhibitory effects of the drug can theoretically affect each of the multiple nucleotide incorporation events. We found that the resistance values (n-fold) for foscarnet are similar for both assays (Table 2): L802M (3.5-fold), K805Q (0.3-fold), and Q807M (5.3-fold). The measurements with PPi do not reveal any significant differences between wild-type UL54 and enzymes that contain mutations L802M and Q807A, which increase the IC50 for foscarnet. Although we observed subtle increases in IC50 values, these increases are by far not as pronounced as those seen with foscarnet.
TABLE 2.
Inhibition of DNA synthesis in the presence of foscarnet and pyrophosphate
| Enzyme | Foscarnet
|
Pyrophosphate
|
||
|---|---|---|---|---|
| IC50 (μM)a | Fold resistanceb | IC50 (mM)a | Fold resistanceb | |
| WT | 0.47 ± 0.17 | 1 | 1.03 ± 0.22 | 1 |
| L802M | 1.68 ± 0.15 | 3.5 | 1.11 ± 0.02 | 1.1 |
| K805Q | 0.16 ± 0.05 | 0.3 | 0.62 ± 0.04 | 0.6 |
| Q807A | 2.98 ± 0.73 | 5.3 | 1.18 ± 0.23 | 1.1 |
IC50 values were determined in the presence of activated calf thymus DNA as described in Materials and Methods. Values are means of at least three independent determinations ± standard deviations.
Values of less than 1 represent hypersusceptibility.
These data suggest that the binding sites for foscarnet and PPi may not be identical. It appears that Q807A can affect binding of foscarnet but not binding of PPi. These data are consistent with the crystal structure of gp43, which shows the functional equivalent of Q807 at a distance that does not allow contacts with the β- and γ-phosphates of the incoming nucleotide. Unlike mutations L802M and Q807A, K805Q, which decreased the IC50 for foscarnet (0.3-fold), also decreased the IC50 for PPi (0.6-fold). Thus, it appears that K805Q diminishes the inhibitory effects of PPi and the PPi analogue through a similar mechanism. This mechanism does not appear to involve increased drug binding through 805Q, because position 805 is too far away from the β- and γ-phosphates of the bound nucleotide.
Type of inhibition.
Previous biochemical studies have shown that foscarnet is a noncompetitive inhibitor with respect to the nucleotide substrate (13). Thus, we next determined steady-state kinetic parameters (Vmax, Km, and Ki) in the presence of increasing concentrations of foscarnet in order to analyze whether the same or a different type of inhibition is seen with the resistance- and hypersusceptibility-conferring mutants (Fig. 5 and Table 3). With respect to all four enzymes, i.e., wild-type UL54, L802M, K805Q, and Q807A, we found that changes for Km values were insignificant, while Vmax values decreased dramatically with increasing concentrations of the drug. These data show that foscarnet acts as a noncompetitive inhibitor in each case, regardless of whether the phenotype shows resistance or hypersusceptibility. As expected, the extent of the decrease in Vmax values depends on the phenotype. Ki values were determined by plotting 1/Vmax against the different concentrations of foscarnet (Table 3). Ki values higher than those for wild-type UL54 are consistent with a resistant phenotype (L802M and Q807A), while the lower Ki value determined for K805Q is consistent with the hypersusceptible phenotype.
FIG. 5.
Type of inhibition and measurements of Ki values. (A) DNA synthesis was measured at a single time point (30 min) in the presence of increasing concentrations of each the four nucleotides. This experiment was repeated with different concentrations of foscarnet to determine Ki values as described in Materials and Methods. This figure shows changes in Vmax and Km for WT UL54 as an example. υ, velocity of the reaction. The data for WT and mutant enzymes are summarized in Table 3. (B) Replot of 1/Vmax and Km values determined as for panel A, shown here as a function of foscarnet concentration. The x intercept provides the Ki value. Changes in Vmax at constant Km point to a noncompetitive type of inhibition. This is shown for WT UL54 as an example. (C) Values of 1/Vmax determined as for panel A and replotted as for panel B, shown here for WT and mutant enzymes.
TABLE 3.
Ki values for foscarnet-mediated inhibition of DNA synthesisa
| Enzyme | Foscarnet
|
|
|---|---|---|
| Ki (μM)b | Fold resistancec | |
| WT | 0.30 ± 0.08 | 1 |
| L802M | 2.0 ± 0.4 | 6.7 |
| K805Q | 0.14 ± 0.03 | 0.47 |
| Q807A | 1.9 ± 0.7 | 6.3 |
As described in Materials and Methods.
Values are means of at least three independent determinations ± standard deviations.
Values of less than 1 represent hypersusceptibility.
DISCUSSION
The pyrophosphate analogue foscarnet is clinically used to treat infection with HSV, HCMV, and related herpesviruses. As with other antiviral drugs, success in therapy is sometimes severely compromised in the presence of resistance-conferring mutations that eventually emerge over longer periods of treatment. Specific mutations have been identified for various conserved domains of the HCMV DNA polymerase and do not appear to cluster around a single site (18). In this study, we focused on the biochemical characterization of relevant mutations that are found in helix P (A786 to N825). Changes in this region have also been associated with resistance to nucleoside/nucleotide inhibitors (Fig. 1A), and in this context it is interesting to note that a V823A change was also shown to decrease susceptibility to a non-nucleoside analogue inhibitor (34). These combined data suggest that α-helix P plays important roles in enzymatic function.
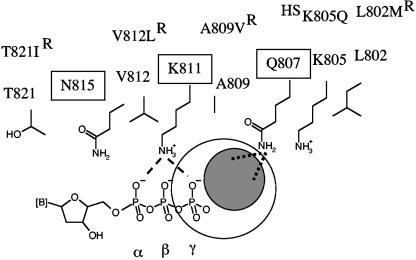
The structure of the related RB69-associated polymerase gp43 shows that N564, K560, and Q556 of helix P reside in the vicinity of the incoming dNTP (40). The functional equivalents in UL54 are N815, K811, and Q807. Several mutations that confer either reduced (L802M, A809V, V812L, and T821I) or increased (K805Q) susceptibility to foscarnet are located in close proximity to the conserved residues that are implicated in nucleotide binding. Based on our biochemical studies, we propose a model that helps to explain the role of helix P in mechanisms involved in foscarnet drug action and drug resistance. We suggest that the presumptive binding site for foscarnet is located in close proximity to the γ-phosphate of the bound nucleotide and that Q807 provides important contacts (Fig. 6). The development of this model is discussed next.
FIG. 6.
Possible interactions between helix P and foscarnet. This simplified model shows side chains of important residues in helix P that are either highly conserved (boxed) among α-DNA polymerases or associated with changes in susceptibility to the drug (R, resistant; HS, hypersusceptible). Helix P of UL54 is shown here in an orientation to the bound nucleotide similar to that shown for helix P of gp43 in Fig. 1C. The structural data suggest that positions 805 and 802 are likely to be too far away to provide direct contacts with the drug. Our data suggest that foscarnet may bind in close proximity to Q807. The gray circle indicates the possible location of the bound drug relative to the nucleotide. The orientation of the bound foscarnet cannot be predicted on the basis of the data presented in this study. Dotted lines illustrate possible interactions between the negatively charged foscarnet and the side chain of Q807. There might be a partial overlap with the position of the γ-phosphate (large open circle), which would help to explain previous data that suggested a competitive mode of inhibition of pyrophosphorolysis. Dashed lines illustrate contacts between the structural equivalent of K811 (K560) in gp43 and the α- and γ-phosphates of the bound dNTP. Possible interaction between K811 and foscarnet cannot be confirmed, because mutations at this position were shown to affect dNTP binding at the same time.
Nucleotide binding characteristics.
Previous biochemical and crystallographic data for gp43 suggested that K560 is involved in nucleotide binding (40). Here we propose a similar function for K811 of UL54. The notion that this residue is involved in nucleotide binding is supported by steady-state kinetic analyses showing that DNA synthesis with the K811A mutant is compromised. This effect is largely attributable to increases in Km values, which is reminiscent of the higher Kd values obtained with the functionally equivalent RB69 mutant. The Km and Kd values are not necessarily identical (29); however, given the established position of K560 in the RB69 enzyme (40), it is reasonable to assume that K811 is likewise involved in nucleotide binding. In contrast, the function of the conserved residue Q807 is less well defined, because the model of gp43 shows the equivalent amino acid at a distance of approximately 6 Å away from the γ-phosphate, which appears to be too far to permit direct contacts with the bound nucleotide. In fact, the polymerase activity of the Q807A mutant of UL54 was not significantly altered. These data provide strong evidence to suggest that the structures and functions of helix P of enzymes gp43 and UL54 are very similar. It remains to be elucidated why the glutamine at position 807 is actually conserved among the various members of the α-polymerase family. Given the proximity to the phosphates of the bound nucleotide, this residue might be involved in the release of PPi, which is difficult to study under steady-state conditions. Pre-steady-state kinetic analyses along with cell culture-based fitness measurements may help to shed light on this problem.
Binding of foscarnet.
Here we have shown that the Q807A mutant confers sixfold resistance to foscarnet, which suggests that Q807 might be directly involved in binding of foscarnet. Previous studies of related polymerases have shown that foscarnet acts as a competitive inhibitor with respect to the reverse reaction, i.e., pyrophosphorolysis (10). These findings indicate that the binding sites for PPi and foscarnet may overlap to a certain degree. Such overlap is unlikely to be extensive, since foscarnet acts as a noncompetitive inhibitor with respect to the nucleotide substrate (13). Moreover, high levels of resistance to foscarnet are usually not associated with high levels of resistance to PPi (Table 2) (10), which is what one would predict were the binding sites identical. Together, these data provide evidence to suggest that the binding site for foscarnet is located in close proximity to the position of the γ-phosphate and that Q807 provides important contacts (Fig. 6). Both the carboxyl and phosphonate moieties may interact either with Q807 or with residues that are normally in contact with the γ-phosphate. Of note, residue K65 in HIV-1 reverse transcriptase (RT) contacts the γ-phosphate of the bound nucleotide and a K65R change confers resistance to foscarnet (31). In UL54, K811 could be seen as a functional equivalent in this regard; however, the activity of the K811A mutant is impaired per se, which makes it difficult to accurately measure changes in IC50 values. Drug binding in the proposed mode may prevent simultaneous binding of PPi, particularly if one considers the high IC50 values compared to those for foscarnet (Table 2). In contrast, the noncompetitive mode of inhibition of DNA synthesis suggests that the incoming dNTP may still bind to its designated binding site; thus, simultaneous binding of foscarnet could affect the precise positioning of the dNTP substrate rather than its affinity to the enzyme. Binding of the dNTP substrate and foscarnet may also involve different complexes, as previously suggested for HIV-1 RT (26). Site-specific footprinting experiments revealed that the nucleotide substrate can bind only to a posttranslocation complex, in which the dNTP binding site is freely accessible (25). In contrast, foscarnet may (also) bind to the pretranslocation complex, in which the newly incorporated nucleoside monophosphate occupies the dNTP binding site.
Other residues that are not necessarily part of helix P may likewise provide contacts to the bound foscarnet. For instance, the gp43 structure suggests that basic amino acids R785 and R789 in UL54 are found within 6 Å of the γ-phosphate (14). Of note, changes at the adjacent amino acid V781 have been associated with resistance to foscarnet, and changes at positions D780 and L782 of the related herpes simplex virus DNA polymerase (UL30) reduce susceptibility to foscarnet and to acyclovir and adefovir (17). All of these residues belong to helix N, which was also shown to provide important contacts to the bound dNTP binding (14, 40). As for helix P, many mutations that affect susceptibility to foscarnet and nucleoside/nucleotide analogue inhibitors are found in close proximity to highly conserved residues. It is thus likely that some of these mutations exert their effects indirectly, possibly through subtle rearrangements of the side chains of adjacent highly conserved residues.
Resistance and hypersusceptibility.
In this study, we have focused on mutations L802M and K805Q. Our model predicts that the distance between L802M and the bound foscarnet might be too long to allow contacts. Drug binding in close proximity to the highly conserved residues K811 and Q807, as shown in Fig. 6, literally excludes direct interactions with L802. The L802M substitution may affect the precise positioning of Q807, which in turn affects binding of foscarnet. In this model, subtle rearrangements of the side chain do not necessarily affect the affinity to PPi. Based on crystal structures of other polymerase complexes, it is reasonable to assume that PPi occupies the positions of the β- and γ-phosphates of a nucleoside triphosphate substrate (42). This region is not within contact distance of Q807, which is supported by our data that show that Q807A- and L802M-associated mutations increase IC50 values for foscarnet, while no discernible differences were measured with PPi.
In contrast to the data obtained with L802M, the K805Q mutant that confers hypersusceptibility to foscarnet also reduces the IC50 value for PPi. It is unlikely that the K805Q change can directly affect binding of foscarnet or PPi, because the distance to the presumptive binding sites would be too long, as outlined above. However, the natural K805 could affect the precise orientation of foscarnet and/or its interaction with Q807. The net loss of positive charge in K805Q may counteract such an effect, which would provide an explanation for the hypersusceptible phenotype. Mechanisms that are not based on affinity changes cannot be excluded at this point. Previous biochemical and crystallographic data have shown that the L103N mutation in HIV-1 RT acts as a “gate keeper” and restricts the access of certain non-nucleoside analogue RT inhibitors to their designated target site (21). Conversely, it is conceivable that hypersusceptibility-conferring mutations, such as the K805Q change, may facilitate drug access.
The mechanisms by which mutations V812L, A809V, and T821I confer resistance to the drug can also differ from the mechanisms discussed in this study. V812L and A809V are located in close proximity to the incoming nucleotide and may thus directly affect binding of foscarnet. In contrast, T821I, which confers relatively high levels of resistance to foscarnet, is too far away from the bound nucleotide. However, the structural equivalent in RB69 contacts the template, which may provide an alternative mechanism of drug resistance. Resistance-conferring mutations in HIV-1 RT were also found to map to various regions (26), which likewise points to different mechanisms. One of these mutations, e.g., E89K, may affect the interaction with the template, as suggested for T821I in HCMV UL54, but many other changes do not make direct contacts with the active site, the bound dNTP, or the nucleic acid substrate, which is again reminiscent of the mutational patterns described for HSV and HCMV. Several studies have shown that mutations in the conserved region II (around positions 696 and 724) of the related HSV-1, HSV-2, and bacteriophage T4 DNA polymerases can increase or decrease susceptibility to foscarnet or to its derivative phosphonoacetic acid (1, 19, 30, 35). Given that this region is likewise implicated in nucleotide binding, it would be interesting to study the effects of corresponding mutations in this region of HCMV UL54.
Taken together, the data presented in this study provide strong evidence to suggest that the structures and functions of helix P of RB69 gp43 and HCMV UL54 are highly related, at least in the vicinity of the nucleotide binding site. The characterization of enzymes with mutations at highly conserved residues sheds light on the possible binding site for the pyrophosphate analogue foscarnet. We propose a model that places foscarnet in close proximity to Q807 and the position of the γ-phosphate of the bound dNTP. In this model, mutations L802M and K805Q exert their effects indirectly through structural changes that affect the interaction between foscarnet and Q807.
Acknowledgments
E.T. performed this work in partial fulfillment of the requirements for a Ph.D. degree, Faculty of Graduate Studies and Research, McGill University, Montreal, Quebec, Canada. We thank Thomas Cihlar for providing some of the HCMV recombinant mutants.
This research was supported by grants to M.G. and G.B. from the Canadian Institutes of Health Research (CIHR). M.G. is recipient of a career award from the CIHR.
REFERENCES
- 1.Bestman-Smith, J., and G. Boivin. 2003. Drug resistance patterns of recombinant herpes simplex virus DNA polymerase mutants generated with a set of overlapping cosmids and plasmids. J. Virol. 77:7820-7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boosalis, M. S., J. Petruska, and M. F. Goodman. 1987. DNA polymerase insertion fidelity. Gel assay for site-specific kinetics. J. Biol. Chem. 262:14689-14696. [PubMed] [Google Scholar]
- 3.Chou, S., N. S. Lurain, K. D. Thompson, R. C. Miner, and W. L. Drew. 2003. Viral DNA polymerase mutations associated with drug resistance in human cytomegalovirus. J. Infect. Dis. 188:32-39. [DOI] [PubMed] [Google Scholar]
- 4.Chou, S. W. 2001. Cytomegalovirus drug resistance and clinical implications. Transpl. Infect. Dis. 3(Suppl. 2):20-24. [DOI] [PubMed] [Google Scholar]
- 5.Cihlar, T., M. D. Fuller, and J. M. Cherrington. 1998. Characterization of drug resistance-associated mutations in the human cytomegalovirus DNA polymerase gene by using recombinant mutant viruses generated from overlapping DNA fragments. J. Virol. 72:5927-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cihlar, T., M. D. Fuller, and J. M. Cherrington. 1997. Expression of the catalytic subunit (UL54) and the accessory protein (UL44) of human cytomegalovirus DNA polymerase in a coupled in vitro transcription/translation system. Protein Expr. Purif. 11:209-218. [DOI] [PubMed] [Google Scholar]
- 7.Cihlar, T., M. D. Fuller, A. S. Mulato, and J. M. Cherrington. 1998. A point mutation in the human cytomegalovirus DNA polymerase gene selected in vitro by cidofovir confers a slow replication phenotype in cell culture. Virology 248:382-393. [DOI] [PubMed] [Google Scholar]
- 8.Crumpacker, C. S. 1992. Mechanism of action of foscarnet against viral polymerases. Am. J. Med. 92:3S-7S. [DOI] [PubMed] [Google Scholar]
- 9.De Clercq, E. 2004. Antiviral drugs in current clinical use. J. Clin. Virol. 30:115-133. [DOI] [PubMed] [Google Scholar]
- 10.Derse, D., K. F. Bastow, and Y. Cheng. 1982. Characterization of the DNA polymerases induced by a group of herpes simplex virus type I variants selected for growth in the presence of phosphonoformic acid. J. Biol. Chem. 257:10251-10260. [PubMed] [Google Scholar]
- 11.Drew, W. L., C. V. Paya, and V. Emery. 2001. Cytomegalovirus (CMV) resistance to antivirals. Am. J. Transplant. 1:307-312. [PubMed] [Google Scholar]
- 12.Erice, A. 1999. Resistance of human cytomegalovirus to antiviral drugs. Clin. Microbiol. Rev. 12:286-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eriksson, B., B. Oberg, and B. Wahren. 1982. Pyrophosphate analogues as inhibitors of DNA polymerases of cytomegalovirus, herpes simplex virus and cellular origin. Biochim. Biophys. Acta 696:115-123. [DOI] [PubMed] [Google Scholar]
- 14.Franklin, M. C., J. Wang, and T. A. Steitz. 2001. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell 105:657- 667. [DOI] [PubMed] [Google Scholar]
- 15.Freisinger, E., A. P. Grollman, H. Miller, and C. Kisker. 2004. Lesion (in)tolerance reveals insights into DNA replication fidelity. EMBO J. 23:1494-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gandhi, M. K., and R. Khanna. 2004. Human cytomegalovirus: clinical aspects, immune regulation, and emerging treatments. Lancet Infect. Dis. 4:725-738. [DOI] [PubMed] [Google Scholar]
- 17.Gilbert, C., J. Bestman-Smith, and G. Boivin. 2002. Resistance of herpesviruses to antiviral drugs: clinical impacts and molecular mechanisms. Drug Resist. Updates 5:88-114. [DOI] [PubMed] [Google Scholar]
- 18.Gilbert, C., and G. Boivin. 2005. Human cytomegalovirus resistance to antiviral drugs. Antimicrob. Agents Chemother. 49:873-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall, J. D., and S. Woodward. 1989. Aphidicolin resistance in herpes simplex virus type 1 appears to alter substrate specificity in the DNA polymerase. J. Virol. 63:2874-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hogg, M., S. S. Wallace, and S. Doublie. 2004. Crystallographic snapshots of a replicative DNA polymerase encountering an abasic site. EMBO J. 23:1483-1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsiou, Y., J. Ding, K. Das, A. D. Clark, Jr., P. L. Boyer, P. Lewi, P. A. Janssen, J. P. Kleim, M. Rosner, S. H. Hughes, and E. Arnold. 2001. The Lys103Asn mutation of HIV-1 RT: a novel mechanism of drug resistance. J. Mol. Biol. 309:437-445. [DOI] [PubMed] [Google Scholar]
- 22.Huang, L., K. K. Ishii, H. Zuccola, A. M. Gehring, C. B. Hwang, J. Hogle, and D. M. Coen. 1999. The enzymological basis for resistance of herpesvirus DNA polymerase mutants to acyclovir: relationship to the structure of alpha-like DNA polymerases. Proc. Natl. Acad. Sci. USA 96:447-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landolfo, S., M. Gariglio, G. Gribaudo, and D. Lembo. 2003. The human cytomegalovirus. Pharmacol. Ther. 98:269-297. [DOI] [PubMed] [Google Scholar]
- 24.Loregian, A., B. A. Appleton, J. M. Hogle, and D. M. Coen. 2004. Specific residues in the connector loop of the human cytomegalovirus DNA polymerase accessory protein UL44 are crucial for interaction with the UL54 catalytic subunit. J. Virol. 78:9084-9092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchand, B., and M. Gotte. 2003. Site-specific footprinting reveals differences in the translocation status of HIV-1 reverse transcriptase. Implications for polymerase translocation and drug resistance. J. Biol. Chem. 278:35362-35372. [DOI] [PubMed] [Google Scholar]
- 26.Meyer, P. R., S. E. Matsuura, D. Zonarich, R. R. Chopra, E. Pendarvis, H. Z. Bazmi, J. W. Mellors, and W. A. Scott. 2003. Relationship between 3′-azido-3′-deoxythymidine resistance and primer unblocking activity in foscarnet-resistant mutants of human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 77:6127-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morphin, F., and D. Thouvenot. 2003. Herpes simplex virus resistance to antiviral drugs. J. Clin. Virol. 26:29-37. [DOI] [PubMed] [Google Scholar]
- 28.Oberg, B. 1982. Antiviral effects of phosphonoformate (PFA, foscarnet sodium). Pharmacol. Ther. 19:387-415. [DOI] [PubMed] [Google Scholar]
- 29.Reardon, J. E., and W. H. Miller. 1990. Human immunodeficiency virus reverse transcriptase. Substrate and inhibitor kinetics with thymidine 5′-triphosphate and 3′-azido-3′-deoxythymidine 5′-triphosphate. J. Biol. Chem. 265:20302-20307. [PubMed] [Google Scholar]
- 30.Reha-Krantz, L. J., R. L. Nonay, and S. Stocki. 1993. Bacteriophage T4 DNA polymerase mutations that confer sensitivity to the PPi analog phosphonoacetic acid. J. Virol. 67:60-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sluis-Cremer, N., D. Arion, N. Kaushik, H. Lim, and M. A. Parniak. 2000. Mutational analysis of Lys65 of HIV-1 reverse transcriptase. Biochem. J. 348:77-82. [PMC free article] [PubMed] [Google Scholar]
- 32.Smith, I. L., J. M. Cherrington, R. E. Jiles, M. D. Fuller, W. R. Freeman, and S. A. Spector. 1997. High-level resistance of cytomegalovirus to ganciclovir is associated with alterations in both the UL97 and DNA polymerase genes. J. Infect. Dis. 176:69-77. [DOI] [PubMed] [Google Scholar]
- 33.Springer, K. L., and A. Weinberg. 2004. Cytomegalovirus infection in the era of HAART: fewer reactivations and more immunity. J. Antimicrob. Chemother. 54:582-586. [DOI] [PubMed] [Google Scholar]
- 34.Thomsen, D. R., N. L. Oien, T. A. Hopkins, M. L. Knechtel, R. J. Brideau, M. W. Wathen, and F. L. Homa. 2003. Amino acid changes within conserved region III of the herpes simplex virus and human cytomegalovirus DNA polymerases confer resistance to 4-oxo-dihydroquinolines, a novel class of herpesvirus antiviral agents. J. Virol. 77:1868-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsurumi, T., K. Maeno, and Y. Nishiyama. 1987. A single-base change within the DNA polymerase locus of herpes simplex virus type 2 can confer resistance to aphidicolin. J. Virol. 61:388-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, J., A. K. Sattar, C. C. Wang, J. D. Karam, W. H. Konigsberg, and T. A. Steitz. 1997. Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69. Cell 89:1087-1099. [DOI] [PubMed] [Google Scholar]
- 37.Weinberg, A., D. A. Jabs, S. Chou, B. K. Martin, N. S. Lurain, M. S. Forman, and C. Crumpacker. 2003. Mutations conferring foscarnet resistance in a cohort of patients with acquired immunodeficiency syndrome and cytomegalovirus retinitis. J. Infect. Dis. 187:777-784. [DOI] [PubMed] [Google Scholar]
- 38.Xiong, X., J. L. Smith, and M. S. Chen. 1997. Effect of incorporation of cidofovir into DNA by human cytomegalovirus DNA polymerase on DNA elongation. Antimicrob. Agents Chemother. 41:594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiong, X., J. L. Smith, C. Kim, E. S. Huang, and M. S. Chen. 1996. Kinetic analysis of the interaction of cidofovir diphosphate with human cytomegalovirus DNA polymerase. Biochem. Pharmacol. 51:1563-1567. [DOI] [PubMed] [Google Scholar]
- 40.Yang, G., M. Franklin, J. Li, T. C. Lin, and W. Konigsberg. 2002. Correlation of the kinetics of finger domain mutants in RB69 DNA polymerase with its structure. Biochemistry 41:2526-2534. [DOI] [PubMed] [Google Scholar]
- 41.Ye, L. B., and E. S. Huang. 1993. In vitro expression of the human cytomegalovirus DNA polymerase gene: effects of sequence alterations on enzyme activity. J. Virol. 67:6339-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin, Y. W., and T. A. Steitz. 2004. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell 116:393-404. [DOI] [PubMed] [Google Scholar]