Abstract
Human wild-type (wt) hepatitis A virus (HAV), the causative agent of acute hepatitis, barely grows in cell culture and in the process accumulates attenuating and cell culture-adapting mutations. This genetic instability of wt HAV in cell culture is a major roadblock to studying HAV pathogenesis and producing live vaccines that are not overly attenuated for humans. To develop a robust cell culture system capable of supporting the efficient growth of wt HAV, we transfected different cell lines with in vitro RNA transcripts of wt HAV containing the blasticidin resistance gene. Blasticidin-resistant colonies grew only in transfected Huh7 cells and produced infectious virus. HAV was genetically stable in Huh7 cells for at least nine serial passages and did not accumulate attenuating or cell culture-adapting mutations. Treatment with alpha interferon A/D cured the blasticidin-resistant Huh7 cells of the HAV infection. The cured cells, termed Huh7-A-I cells, did not contain virus or HAV antigens and were sensitive to blasticidin. Huh7-A-I cells were more permissive than parental cells for wt HAV infection, including a natural isolate from a human stool sample, and produced 10-fold-more infectious particles. This is the first report of a cell line that allows the genetically stable growth of human wt HAV. The viral vectors and cells described here should allow better insight into the pathogenesis of HAV and the development of attenuated vaccines. The cell lines susceptible to wt HAV growth may also be used to detect and isolate infectious virus from patient and environmental samples.
Hepatitis A virus (HAV) is a member of the Picornaviridae that causes acute hepatitis in humans, a worldwide preventable infectious disease. In the United States, approximately 25,000 cases of HAV are reported each year, and an estimated average of 263,000 HAV cases occur annually when corrected for underreporting and asymptomatic infections (17). HAV is a nonenveloped virus that contains a 7.5-kb single-stranded positive-sense genomic RNA encapsidated in an icosahedral 27- to 32-nm particle. The viral RNA has a 5′-end nontranslated region of approximately 750 bases containing an internal ribosome entry site (IRES) (39), and a 3′-end short nontranslated region followed by a poly(A) tail. The viral protease 3Cpro cleaves the HAV polyprotein into smaller structural (VP0, VP3, and VP1-2A) and nonstructural (2B, 2C, 3A, 3B, 3C, and 3D) proteins (references 25, 27, and 31 and references therein), and a cellular protease cleaves the VP1-2A precursor (24).
Human wild-type (wt) HAV rarely grows in cell culture, is genetically unstable, and requires several weeks to months in culture before it can be detected (4, 7, 14, 19). HAV has been adapted to a variety of primate (3, 9, 18, 34, 38) and nonprimate (10, 16) cell lines, but this process results in the accumulation of cell culture-adapting and attenuating mutations (4, 5, 9, 11, 20, 34). During adaptation to cell culture, two main hot-spot mutation areas in the HAV genome were identified, one located in the IRES that is required for viral translation and another located within the nonstructural 2B and 2C genes that is required for RNA replication. The efficient growth of HAV in cell culture depends on both hot-spot areas (26), but mutations in the 2B and 2C genes are essential for growth in cell culture (12) whereas mutations in the IRES are growth enhancers and do not function autonomously (37). Interestingly, there is a strong pressure in most cell culture systems that favors the selection of mutants containing a nucleotide change at position 3889 of wt HAV (14, 23), which results in an Ala-to-Val change at amino acid 216 of the 2B protein (2B/A216V) and allows the virus to replicate marginally better in FRhK-4 cells (11) without affecting its virulence in marmosets and chimpanzees (13). However, mutants containing the 2B/A216V change still grow poorly, are genetically unstable, and tend to accumulate additional cell culture-adapting and attenuating mutations.
Studies to understand the restricted growth of wt HAV in cell culture are cumbersome because the virus grows poorly without causing cytopathic effect and there are no markers to select infected cells. Although HAV expression vectors have been developed (2, 41), strains of HAV carrying antibiotic-resistant genes that could allow the rapid selection of infected cells have not been described. In this paper, we show that a blasticidin resistance gene cloned into the 2A-2B junction of HAV can be used to identify and select cells capable of supporting the stable growth of human wt HAV in cell culture. Although additional validation will be required, it is possible that the cell lines described in this paper or other cell lines selected using the vectors described in this work could be used to monitor the presence of infectious wt HAV in patient, food, and water samples. Moreover, our findings could lead to the development of HAV strains that are not overly attenuated for humans and could be used as live vaccines.
MATERIALS AND METHODS
Cells and viruses.
All cell lines were grown in growth medium supplemented with 10% fetal bovine serum, except for FRhK-4 cells, which were grown in the presence of 10% equine serum. Human hepatoma Huh7 cells, obtained from D. Taylor, FDA, Bethesda, MD, were grown in Dulbecco's modified Eagle's medium. The continuous clone GL37 of African green monkey kidney cells (15) was grown in Eagle's minimal essential medium. Human HeLa cells and African green monkey kidney Vero cells, obtained from the ATCC, and rhesus monkey kidney FRhK-4 cells, obtained from S. Emerson, NIH, Bethesda, MD, were grown in Dulbecco's modified Eagle's medium. Chinese hamster ovary (CHO) cells deficient in the enzyme dihydrofolate reductase, obtained from the ATCC, were grown in Iscove's medium supplemented with 100 μM hypoxanthine and 16 μM thymidine (Sigma Chemical Co.). MMH-D3 mouse liver cells (1), derived from transgenic mice carrying a truncated cytoplasmic form of the human Met gene, were grown in RPMI 1640 medium supplemented with 10 μg/ml insulin, 50 ng/ml epidermal growth factor, and 30 ng/ml insulin-like growth factor 2 growth factors on collagen-coated flasks. All cell lines were grown in a 5% CO2 incubator at 37°C.
The stool-derived human wt HM-175 strain of HAV was obtained from S. Feinstone, FDA, Bethesda, MD. A mutant of the wt HM-175 strain of HAV containing only an Ala-to-Val change at position 216 of protein 2B (2B/A216V), termed HAV8Y, was rescued from Huh7 cells transfected with in vitro SP6 polymerase transcripts from pHAV8Y (11, 13, 22). Attenuated and cell culture-adapted HAV/7 was rescued from FRhK-4 cells transfected with in vitro SP6 polymerase transcripts from pHAV/7 (5-7).
Plasmids and constructs.
Plasmids were constructed using PCR and standard molecular biology methods (36). DNA fragments were amplified by PCR using Pfu Turbo Hotstart polymerase (Stratagene) as recommended by the manufacturer in 25 cycles of 95°C for 30 s, 50°C for 1 min, and 72°C for 2 min. For overlap PCR, fragments were denatured at 94°C and annealed at 45°C in 1× PCR buffer for 2 min. Escherichia coli strain DH5α was transformed with the constructs, and plasmids were purified by chromatography (QIAGEN) as suggested by the manufacturer. Constructs were verified by automatic nucleotide sequence analysis. The following plasmids were constructed.
pHAV8Y-MCS.
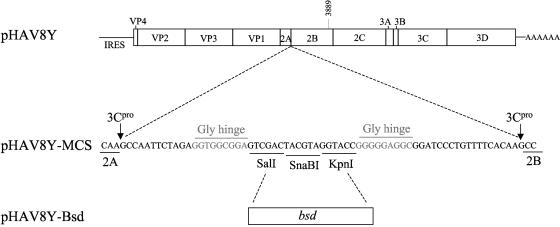
A polylinker containing unique restriction sites SalI, SnaBI, and KpnI flanked by three-residue Gly hinges designed to facilitate processing of the two adjacent 3Cpro cleavage sites and cleavage sites for the HAV protease 3Cpro (2, 41) was introduced into the 2A-2B junction of the HAV infectious cDNA in pHAV8Y using overlap PCR (Fig. 1). Forward PCR primer A (5′-GTTTTATTTTCCCAGAGCTCCATTGAACTCAA-3′), corresponding to nucleotides (nt) 2975 to 3006 of HAV coding for the C terminus of protein VP1 and containing a naturally occurring SacI restriction site, and reverse PCR primer B (5′-GGTACCTACGTAGTCGACTCCGCCACCTCTAGAATTGGCTTGTGAAAACAGTCCCTTCTTCATTTTCCTAGG-3′), coding for nt 3213 to 3242 of HAV corresponding to the C terminus of 2A, a synthetic 3Cpro cleavage site, and the polylinker described above plus a three-residue Gly hinge were used to amplify fragment I using pHAV8Y as template. An additional PCR fragment II was amplified using pHAV8Y as template and oligonucleotides C (5′-GACTACGTAGGTACCGGGGGAGGCGGATCCCTGTTTTCACAAGCCAATATTTCTCTTTTTTATACTGAGGAG-3′) and D (5′-ATTTTTCCACATCTTGGATTTGCAAAATGCAAAATT-3′) as PCR primers. It should be pointed out that the 5′-end 15 nucleotides of forward PCR primer C are complementary to the polylinker of oligonucleotide B followed by three codons of the Gly hinge, a 3Cpro cleavage site, and nt 3243 to 3272 of HAV. Reverse PCR primer D codes for nt 4183 to 4217 of HAV and contains a naturally occurring PflMI restriction site. PCR fragments I and II were annealed and used as template for the amplification of a larger fragment using the forward A and reverse D PCR primers. The resulting PCR fragment was gel purified, digested with SacI and PflMI enzymes, and cloned into pHAV8Y cut with the same enzymes. The resulting construct was termed pHAV8Y-MCS.
FIG. 1.
Schematic representation of HAV vector constructs. A polylinker coding for SalI, SnaBI, and KpnI restriction sites flanked by HAV protease 3Cpro cleavage sites and Gly hinges was cloned at the 2A-2B junction of the HAV cDNA in pHAV8Y and termed pHAV8Y-MCS. The blasticidin resistance bsd gene without translation and initiation codons was cloned into the SalI and KpnI sites of pHAV8Y-MCS. The resultant construct, termed pHAV8Y-Bsd, carried the bsd gene in frame with the polyprotein, and its gene product Bsd should be released from the polyprotein by 3Cpro cleavage.
pHAV8Y-Bsd.
The blasticidin resistance gene, bsd, of 396 bp coding for a protein of 132 amino acids was cloned into the polylinker of pHAV8Y-MCS. A DNA fragment was amplified from pTracer-CMV/Bsd (Invitrogen) using synthetic oligonucleotide primers 5′-GTCGACGTCGACCAGGCCAAGCCTTTGTCTCAAGAA-3′ and 5′-CGGTTAGGTACCGCCCTCCCACACATAACCAGAGGG-3′, which introduced SalI and KpnI restriction sites at the 5′ and 3′ ends of the gene, respectively, and eliminated the translation initiation and termination codons of bsd. The resulting PCR fragment was gel purified, digested with SalI and KpnI, and cloned into pHAV8Y-MCS digested with the same restriction enzymes. The resulting construct was termed pHAV8Y-Bsd and coded for the Bsd resistance protein inserted between the 2A and 2B genes in frame with the rest of the HAV polyprotein.
pHAV.WT-Bsd.
The 2B/A216V residue in HAV8Y was back-mutated to the Ala residue found in natural isolates of wt HAV. Overlap PCR was done using forward primer A1 (5′-GAGTCATGAATTATGCAGATA-3′) coding for nt 3874 to 3894 of wt HAV and reverse primer A2 (5′-AACCAATATCTGCATAATTCA-3′) coding for nt 3900 to 3880 of wt HAV, both coding for an Ala codon (underlined) at position 216 of 2B. Two overlapping PCR DNA fragments amplified from pHAV8Y-Bsd using primers A and A2 or A1 and D were denatured, annealed, and used as templates for the amplification of a longer PCR fragment using primers A and D that was digested with SalI and PflMI, gel purified, and cloned into the SalI and PflMI sites of pHAV8Y-Bsd. The resulting construct was termed pHAV.WT-Bsd.
Immunofluorescence (IF) analysis.
Mock- and HAV-infected cells grown in eight-well Permanox chamber slides (Nunc, Inc.) at 35°C were fixed with cold acetone for 30 min, air dried, blocked with 2% fetal bovine serum in phosphate-buffered saline, and stained with murine anti-HAV neutralizing monoclonal antibodies (MAbs) K2-4F2 and K3-4C8 (29) and fluorescein isothiocyanate-conjugated goat anti-mouse antibody (KPL Inc.). Fluorescent micrographs were taken with a Zeiss Axioscope microscope at a magnification of 400× with an oil-immersion objective.
RNA transfections and HAV infections.
Full-length HAV RNA transcripts were synthesized in vitro using SP6 RNA polymerase (Amersham Pharmacia) and plasmid templates linearized at the HaeII site downstream of the poly(A) of the HAV cDNA (5, 41), and cells were transfected with RNA transcripts using DEAE-dextran as facilitator (33). One day after transfection, cells were split 1:6 and grown in selection medium containing 1 to 3 μg/ml blasticidin (Invitrogen). Cells were split 1:5 weekly into 25-cm2 flasks and eight-well chamber slides for IF analysis. To prepare viral stocks, monolayers with at least 80% of the cells expressing HAV antigens as assessed by IF analysis were subjected to three freeze-and-thaw cycles, cell debris was pelleted by low-speed centrifugation, and supernatants containing the virus were stored at −70°C.
To infect cells, 50 to 80% confluent cell monolayers were inoculated with a multiplicity of infection of 1 to 2 50% tissue culture infectious doses (TCID50)/cell in 25-cm2 flasks. After 4 h of adsorption, cells were washed three times and grown at 35°C in a CO2 incubator. At 24 h postinfection, blasticidin (2 μg/ml) was added to the medium to select for cells infected with HAV constructs coding for Bsd. For one-step growth curve analysis, six-well plates were infected using the same conditions described above. However, infected cells were grown in the absence of blasticidin. At different times postinfection, plates were frozen at −70°C and, after the last time point sample was collected, plates were thawed and viral stocks were prepared as indicated above.
HAV titer determination.
HAV titers were determined by an enzyme-linked immunosorbent assay (ELISA) endpoint dilution assay in 96-well plates containing 20 to 50% confluent cell monolayers as described previously (16). Briefly, eight replicate wells were inoculated with 100 μl of 10-fold dilutions of HAV, plates were incubated at 35°C in a CO2 incubator, and viral titers were determined 2 weeks after infection by ELISA staining with a 1:4,000 dilution of anti-HAV MAb K2-4F2, a 1:5,000 dilution of peroxidase-labeled goat anti-mouse antibodies (KPL, Inc.), and TMB one-component substrate (KPL, Inc.) as suggested by the manufacturer. Wells that developed at least two times the absorbance of the uninfected control wells were considered positive. Alternatively, HAV8Y-Bsd and HAV.WT-Bsd titers were determined by a blasticidin resistance endpoint dilution assay in 96-well plates containing 20 to 50% confluent monolayers of Huh7 cells. Blasticidin (2 μg/ml) was added to the cell culture medium of the 96-well plates 24 h after infection, and cells were incubated at 35°C under CO2. Five to seven days after infection, the 96-well plates were inspected under the microscope and wells containing monolayers or live cells forming colonies were considered positive. Viral titers of the ELISA and blasticidin resistance endpoint titrations were determined using the Reed and Muench method (35).
Cure of HAV-infected cells with interferon.
Huh7 cells transfected with HAV8Y-Bsd synthetic transcripts and selected with 1 μg/ml blasticidin were treated with human leukocyte-derived alpha interferon A/D (IFN-αA/D) (Sigma Chemical Co.) to eliminate the virus from the cells. Prior to IFN-αA/D treatment, cells were split twice in growth medium lacking blasticidin. Huh7 cells infected with HAV8Y-Bsd were grown in 12-well plates in the presence of 250 U/ml IFN-αA/D in the absence of blasticidin and split weekly in medium containing IFN-αA/D. Cells treated with IFN-αA/D were assessed for the presence of HAV antigens by IF analysis, sensitivity to 1 to 10 μg/ml blasticidin, and production of infectious HAV by titration in Huh7 cells. Cells that did not immunofluoresce, were sensitive to Bsd treatment, and did not produce infectious HAV were considered cured, termed Huh7-A-I, and stored in liquid N2.
Nucleotide sequence analysis.
HAV RNA was extracted from viral stocks using Trizol (Invitrogen). The nucleotide sequence of the full-length HAV genome was obtained by reverse transcription-PCR (RT-PCR). cDNA was synthesized using primers complementary to HAV nt 7494 to 7446 or 4219 to 4190 and the Superscript II enzyme kit (Invitrogen). PCR fragments were amplified using primers corresponding to HAV nt 1 to 21 and 4219 to 4190 or 3800 to 3829 and 7429 to 7408 with Expand High-Fidelity PCR enzyme (Roche). Automatic sequencing of the gel-purified PCR fragments was done using the ABI Prism BigDye terminator cycle sequencing ready reaction kit (Applied Biosystems) and HAV-specific forward primers 6961-6992, 5900-5926, 4760-4785, 3000-3029, 3800-3829, 1802-1822, 900-929, and 1-21 or reverse primers 7429-7408, 7020-6995, 5996-5971, 4900-4871, 4219-4190, 3721-3696, 3209-3189, 3000-2978, 2081-2046, 1206-1171, and 600-577. The cDNAs corresponding to the 5′ and 3′ ends of the HAV genome were amplified by rapid amplification of cDNA ends (RACE) using the 5′ (Roche) and 3′ (Clontech) RACE kits. The cDNA for the 5′ RACE reaction was synthesized using an HAV-specific oligonucleotide corresponding to nt 800 to 771. The cDNA for the HAV 3′-end RACE was synthesized using the oligo(dT)-anchored primer from the kit. cDNA fragments from the 5′ and 3′ ends were amplified by PCR using the RACE anchor primers and HAV synthetic oligonucleotides corresponding to nt 450 to 421 and 6961 to 6982 of HAV, respectively. Automatic nucleotide sequence of the 5′- and 3′-end HAV PCR fragments was obtained as indicated above using the 450-421 and 6961-6982 HAV primers.
RESULTS
Rescue of wt HAV from cells transfected with in vitro transcripts.
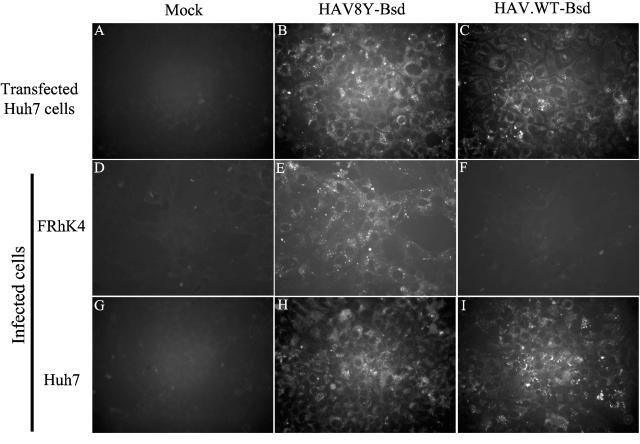
We hypothesized that cells susceptible to wt HAV infection could be selected using a recombinant virus coding for a selectable marker. To do so, we constructed an HAV vector by inserting a polylinker with unique SalI, SnaBI, and KpnI restriction sites flanked by Gly hinges and 3Cpro protease cleavage sites at the 2A-2B junction of HAV in pHAV8Y and termed the construct pHAV8Y-MCS (Fig. 1). We chose the HAV8Y background because this virus contains the cell culture-adapting 2B/A216V mutation that marginally enhances growth in cell culture (14) but does not affect wt HAV virulence (13). The coding region of the blasticidin resistance gene bsd lacking translation initiation and termination codons was inserted into the SalI and KpnI sites of pHAV8Y-MCS (Fig. 1) in frame with the HAV polyprotein, and the construct was termed pHAV8Y-Bsd. Processing of the encoded HAV8Y-Bsd polyprotein by 3Cpro should release the bsd gene product (Bsd). To rescue virus, SP6 in vitro transcripts of pHAV8Y-Bsd were transfected into Huh7, FRhK-4, GL37, HeLa, Vero, CHO, and MMH-D3 cells. One day after transfection, cells were split 1:6 and grown in medium containing 1 to 3 μg/ml blasticidin. After 14 days of selection, approximately 10 blasticidin-resistant colonies grew in Huh7 cells transfected with HAV8Y-Bsd RNA and selected with 1 μg/ml blasticidin. None of the other mock-transfected or HAV-transfected cells survived selection with blasticidin. IF analysis (Fig. 2) showed that the blasticidin-resistant Huh7 cells had the characteristic cytoplasmic granular fluorescence of HAV-infected cells (Fig. 2B), which was not observed in mock-transfected cells (Fig. 2A). To assess the role of the 2B/A216V mutation, we back-mutated nucleotide 3889 of HAV8Y-Bsd from T to C to restored the sequence observed in natural isolates of the virus and termed the construct pHAV.WT-Bsd. Huh7 cells transfected with in vitro RNA transcripts of pHAV.WT-Bsd and treated with 1 μg/ml blasticidin for 14 days also resulted in the selection of approximately 10 resistant colonies that contained HAV antigens (Fig. 2C). We estimated that approximately 1:30,000 cells gained resistance to blasticidin after transfection with in vitro-synthesized RNA from HAV constructs carrying the bsd gene. To study whether viruses rescued from transfected Huh7 cells could grow in monkey kidney and human liver cells, we prepared viral stocks and infected FRhK-4 and Huh7 cells. IF analysis showed that FRhK-4 cells became infected with HAV8Y-Bsd (Fig. 2E) but not HAV.WT-Bsd (Fig. 2F) whereas Huh7 cells were infected with both viruses (Fig. 2H and I). As expected, HAV antigens were not detected in mock-infected FRhK-4 (Fig. 2D) and Huh7 (Fig. 2G) cells. These findings are consistent with previous data showing that wt HAV does not grow in FRhK-4 cells and that the 2B/A216V change is an important cell culture-adapting mutation that allows viral growth in infected cells (11, 13).
FIG. 2.
IF analysis of wt HAV rescued from transfected Huh7 cells. Huh7 cells were mock transfected (A) or transfected with HAV8Y-Bsd (B) or HAV.WT-Bsd (C) in vitro-synthesized RNA, selected with 1 μg/ml blasticidin for 2 weeks, and grown in eight-well slides. FRhK-4 cells were mock infected (D) or infected with HAV8Y-Bsd (E) or HAV.WT-Bsd (F). Huh7 cells were mock infected (G) or infected with HAV8Y-Bsd (H) or HAV.WT-Bsd (I). Infected cells were incubated for 2 weeks without selection and then grown in eight-well slides in the absence of blasticidin. Cells were fixed with cold acetone and stained with anti-HAV MAbs K2-4F2 and K3-4C8 and fluorescein isothiocyanate-conjugated goat anti-mouse antibodies. Immunofluorescent micrographs were taken with a Zeiss Axioscope microscope at 400× magnification using an oil-immersion objective.
To analyze whether the rescued viruses could transmit resistance to blasticidin, Huh7 cells were infected with viral stocks prepared from transfected Huh7 cells containing HAV antigens shown by IF analysis. Approximately 50% of the Huh7 cells infected with HAV8Y-Bsd or HAV.WT-Bsd survived treatment with 2 μg/ml blasticidin and had HAV antigens (Fig. 3A) whereas mock-infected Huh7 cells did not survive the same treatment, which indicated that the functional bsd gene cloned into the HAV genome was packaged into infectious particles.
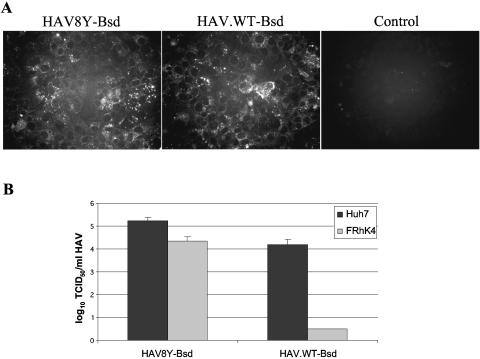
FIG. 3.
Stability of wt HAV in Huh7 cells. (A) IF analysis of Huh7 cells infected with HAV8Y-Bsd or HAV.WT-Bsd and selected with 2 μg/ml blasticidin for 2 weeks. Mock-infected Huh7 cells grown in the absence of blasticidin were used as a negative control (Control). Cells were grown in eight-well slides and stained with anti-HAV antibodies as indicated in Fig. 2. (B) Titers of HAV8Y-Bsd and HAV.WT-Bsd grown in Huh7 cells were assessed in Huh7 and FRhK-4 cells using the blasticidin resistance endpoint titration assay in the presence of 2 μg/ml blasticidin. Plates were observed under the microscope 7 days after infection, and wells containing surviving cells were counted as positive for the titer determination. The titration was repeated at least three times with similar results, and the values represent one titration. Values are log10 of the HAV titers determined by the Reed and Muench method (35), and the standard deviations are shown as lines.
Stable growth of wt HAV in Huh7 cells.
To study whether the strong selective pressure for the accumulation of cell culture-adapting and attenuating mutations was also present in Huh7 cells, nine serial passages of HAV8Y-Bsd and HAV.WT-Bsd were done in Huh7 cells in the presence of 1 μg/ml blasticidin. RT-PCR amplification and nucleotide sequence analysis of HAV RNA extracted from the nine serial passages revealed that the bsd gene was stable. The full-length nucleotide sequences of passage 9 HAV8Y-Bsd and HAV.WT-Bsd were identical to those of their respective parental cDNAs, demonstrating that these viruses did not accumulate cell culture-adapting mutations. To further assess viral stability, passage 9 HAV8Y-Bsd and HAV.WT-Bsd were titrated in parallel in Huh7 and FRhK-4 cells using the blasticidin resistance endpoint dilution assay in 96-well plates (Fig. 3B). HAV8Y-Bsd produced approximately 104 TCID50/ml and 105 TCID50/ml in Huh7 and FRhK-4 cells, respectively. HAV.WT-Bsd produced approximately 104 TCID50/ml in Huh7 cells but did not grow in FRhK-4 cells, which further confirmed that this virus did not accumulate cell culture-adapting mutations during the nine serial passages in Huh7 cells.
IFN-αA/D-cured HAV8Y-Bsd-infected Huh7 cells are susceptible to wt HAV infection.
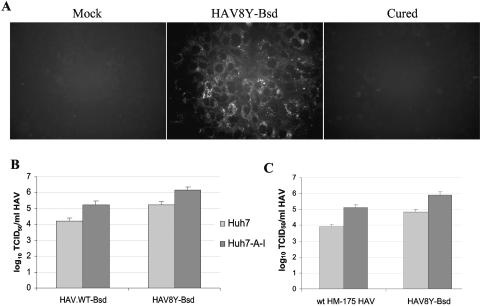
Our data showed that a rather small proportion of the Huh7 cells transfected with HAV8Y-Bsd or HAV.WT-Bsd RNA survived selection with blasticidin, which suggested that a subset of these cells were more permissive for wt HAV growth than the general population of cells and/or the transfection efficiency was low. To test this hypothesis, we cured the blasticidin-resistant Huh7 cells of the HAV infection by treatment with interferon (8). Since HAV8Y-Bsd grew better than HAV.WT-Bsd, we reasoned that HAV8Y-Bsd-cured Huh7 cells could support a higher viral yield and, therefore, cured these cells of the HAV infection. To do so, blasticidin-resistant Huh7 cells transfected with HAV8Y-Bsd were grown for several passages in the presence of 250 IU/ml IFN-αA/D in medium lacking blasticidin. IF analysis (Fig. 4A) showed that cells treated with IFN-αA/D for seven passages lost HAV antigens (Cured) and looked similar to mock-infected cells (Mock) while untreated control cells (HAV8Y-Bsd) had the characteristic cytoplasmic fluorescence of HAV-infected cells. To determine whether the cured cells gained sensitivity to blasticidin, cells were grown in the presence of 1 to 10 μg/ml blasticidin for 10 days and analyzed under the microscope. Control HAV8Y-Bsd-infected cells grew in the presence of blasticidin at up to 8 μg/ml whereas the cured cells, which were termed Huh7-A-I cells, and control Huh7 cells did not survive the blasticidin treatment. In addition, RT-PCR analysis failed to detect HAV RNA in Huh7-A-I cells (data not shown). Huh7-A-I cells grown in the absence of IFN-αA/D for two passages retained sensitivity to blasticidin and did not produce HAV antigens as assessed by IF analysis. These data clearly indicated that Huh7 cells infected with HAV8Y-Bsd were cured of the HAV infection by treatment with IFN-αA/D in the absence of blasticidin. Although we did not cure HAV.WT-Bsd-infected Huh7 cells, it is possible that these cured cells may support similar or higher levels of wt HAV.
FIG. 4.
Huh7 cells cured of HAV8Y-Bsd infection. (A) IF analysis. Huh7 cells mock infected (Mock), HAV8Y-Bsd infected (HAV8Y-Bsd), or HAV8Y-Bsd infected and cured by treatment with 250 U/ml IFN-αA/D (Cured) were grown in eight-well slides and stained with anti-HAV antibodies for IF analysis as described for Fig. 2. (B) Susceptibility of Huh7 and interferon-cured Huh7-A-I cells to infection with HAV8Y-Bsd and HAV.WT-Bsd. Titers were assessed in both cell lines using the blasticidin resistance endpoint titration assay as described for Fig. 3B. (C) Susceptibility of Huh7 and interferon-cured Huh7-A-I cells to infection with wt HM-175 HAV derived from human stools. Cell lines were infected with HAV8Y-Bsd as a positive control. Titers were assessed by the ELISA endpoint titration assay. The titration was repeated at least three times with similar results, and the values represent one titration. Values are log10 of the HAV titers determined by the Reed and Muench method (35), and the standard deviations are shown as lines.
To compare the susceptibility of Huh7 and Huh7-A-I to wt HAV infection, we titrated HAV8Y-Bsd and HAV.WT-Bsd in parallel in both cell lines using the blasticidin resistance endpoint dilution assay (Fig. 4B). Titers of both viruses were 10-fold higher in Huh7-A-I cells than in parental Huh7 cells, which confirmed that the Huh7-A-I subline supported higher levels of HAV infection than the parental cell line. It should be pointed out that the Huh7-A-I cells maintained in culture for more than 40 passages in the absence of selective pressure did not lose their ability to support 10-fold-higher titers than parental Huh7 cells. We also analyzed the susceptibility of both cell lines to infection with a natural isolate of wt HAV obtained from human stools (wt HM-175 HAV). Interestingly, Huh7-A-I cells also supported 10-fold-more wt HM-175 HAV particles than parental Huh7 cells as assessed by an ELISA endpoint dilution assay in 96-well plates (Fig. 4C). As an internal control of the ELISA endpoint titration assay, we included the same stock of HAV8Y-Bsd titrated in the blasticidin resistance endpoint titration assay. The HAV8Y-Bsd titers were similar in the two assays (compare Fig. 4B and C), indicating that the colorimetric and cell survival assays are comparable and that the ELISA did not underestimate viral titers due to lower levels of viral antigens present in the Huh7 cells. We concluded that the increased susceptibility of Huh7-A-I cells to wt HAV infection was independent of the cell culture-adapting mutation at position 3889 as well as the presence of the Bsd selectable marker.
Stable and more efficient growth of wt HAV in the cured Huh7-A-I cells.
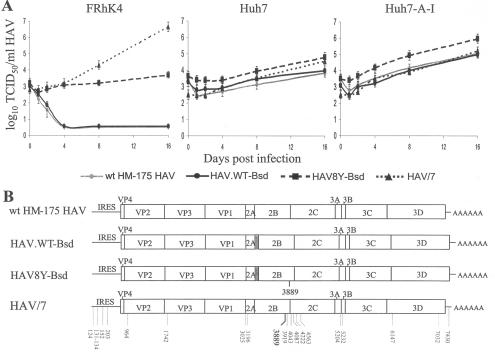
It was of interest to confirm that wt HAV could grow to higher levels in Huh7-A-I cells than in parental Huh7 cells and that the virus will remain genetically stable. A one-step growth curve analysis of wt HM-175 HAV, HAV8Y-Bsd, and HAV.WT-Bsd grown for nine passages in Huh7 cells and cell culture-adapted HAV/7 grown in FRhK-4 was performed in Huh7, Huh7-A-I, and control FRhK-4 cells. Time points were titrated by the ELISA endpoint dilution assay in 96-well plates containing Huh7-A-I cells (Fig. 5A). In FRhK-4 cells, attenuated HAV/7 grew to approximately 106 to 107 TCID50/ml and HAV8Y-Bsd grew marginally, whereas wt viruses wt HM-175 HAV and HAV.WT-Bsd did not grow. All viruses grew in Huh7 cells to titers of approximately 104 to 105 TCID50/ml and in Huh7-A-I cells to titers of approximately 105 to 106 TCID50/ml. Interestingly, attenuated HAV/7 grew similarly in Huh7 and Huh7-A-I cells but viruses with a wt background, wt HM175 HAV and HAV.WT-Bsd, grew approximately 1 log better in Huh7-A-I cells than in Huh7 parental cells. It should be pointed out that HAV8Y-Bsd, which was used to select the Huh7-A-I cell line, grew in Huh7-A-I cells better than did any of the other viruses, reaching a titer of 106 TCID50/ml.
FIG. 5.
Growth of wt HAV in different cell lines. (A) One-step growth curve analysis of wt HAV recombinant viruses containing the Bsd selectable marker (HAV8Y-Bsd and HAV.WT-Bsd), wt HAV isolated from human stools (wt HM-175 HAV), and cell culture-adapted HAV (HAV/7) was done in monkey kidney (FRhK-4) cells, human hepatoma (Huh7) cells, and interferon-cured (Huh7-A-I) cells. Viral titers were assessed in Huh7-A-I cells using the ELISA endpoint titration assay as indicated in Fig. 4C. (B) Schematic representation of nucleotide sequences. Full-length nucleotide sequences of HAV8Y-Bsd, HAV.WT-Bsd, wt HM-175 HAV, and HAV/7 RNA extracted from the last time point sample of the growth curve in Huh7-A-I cells were obtained by RT-PCR. Sequences that differ from the wt HM-175 HAV are indicated with the nucleotide position under the viral genomes. The main cell culture-adapting mutation at nt 3889 is indicated in boldface. Viral genes, the IRES, and the 3′-end poly(A) tail are indicated. The bsd gene clone in HAV8Y-Bsd and HAV.WT-Bsd is shown as a gray box.
To verify that the viruses produced in the different cell lines resembled the input virus, we performed nucleotide sequence analysis of RT-PCR fragments generated from viral RNA extracted at the 16-day time point of Huh7-A-I cells infected with wt HM-175 HAV, HAV8Y-Bsd, and HAV.WT-Bsd (Fig. 5A). Nucleotide sequence analysis of the complete viral genomes confirmed that wt HM-175 HAV and HAV.WT-Bsd did not contain cell culture-adapting mutations, HAV8Y-Bsd contained the expected cell culture-adapting mutation at nucleotide 3889, and HAV/7 had the expected 23 cell culture-adapting mutations (Fig. 5B). Therefore, the genotypes of these viruses correlated with their phenotypes in FRhK-4 cells. Since this analysis showed that wt HAV was genetically stable in Huh7 cells, it was of interest to determine whether wt HAV was also genetically stable in the cured Huh7-A-I cells. To do so, we performed nucleotide sequence analysis of the whole genome of wt HM-175 HAV that was passaged 10 times in Huh7-A-I cells, which revealed that the virus was genetically stable and did not accumulate a cell culture-adapting or any other mutation.
DISCUSSION
Hepatitis A is an age-dependent disease that in general has a subclinical manifestation in young children but can be devastating in older children and adults, resulting in rare cases of fulminant hepatitis. The pathogenesis of HAV is poorly understood, and the basis for the age dependency and diverse outcomes of hepatitis A infections is unknown. Extrahepatic manifestations of the disease include acute kidney failure (reference 30 and references therein) and hematopoietic dysregulation (reference 40 and references therein). The lack of a cell culture system to grow human wt HAV has been a major roadblock to studying pathogenesis of HAV. The data presented in this paper addressed this major roadblock and resulted in the selection of the Huh7-A-I cells that are more permissive to wild-type HAV growth than parental Huh7 cells (Fig. 4 and 5).
In general, wt HAV grows marginally in cell culture and tends to accumulate cell culture-adapting mutations that result in its attenuation. For instance, the prototype wt HM-175 strain of HAV required months to grow in African green monkey kidney cell primary cultures (9) and accumulated 23 mutations that attenuated the virus in marmosets and chimpanzees. In this paper, we showed that Huh7 cells are permissive for wt HAV growth and that HAV antigens can be detected by IF analysis a few days after infection (Fig. 2). We also showed that Huh7-A-I cells, a selected subline of Huh7 cells cured of the HAV infection by IFN treatment, are more permissive to wt HAV growth than parental Huh7 cells (Fig. 5A). A similar strategy was used to select Huh7 cells that allowed the efficient growth of hepatitis C virus replicons containing the neomycin (Neo) resistance gene (28). Interestingly, wt HAV grew approximately 10-fold better in Huh7-A-I cells than in parental Huh7 cells. Although this may seem a small increase in viral yield, it represents a very significant increment for a virus that grows poorly in cell culture, which most likely will facilitate further experimentation and vaccine development. It is possible that the low yield of wt HAV in parental Huh7 cells was due to a small proportion of Huh7-A-I-like cells present in the population. However, IF analysis showed that most Huh7 cells contain HAV antigens 2 weeks postinfection, which indicated that the majority of the parental Huh7 cells were susceptible to wt HAV infection but produced lower viral yields.
wt HAV was stable in Huh7 and Huh7-A-I cells and did not accumulate cell culture-adapting mutations as assessed by the lack of growth in FRhK-4 cells (Fig. 5A) and nucleotide sequence analysis (Fig. 5B). The genetic stability of wt HAV indicated that Huh7-A-I cells did not impose the strong pressure for the selection of cell culture-adapting mutations found in most other cell lines. Indeed, Huh7-A-I cells most likely contain cellular factors similar to those found in human liver cells that allow the growth of wt HAV.
Rescue of wt HAV from full-length cDNA was difficult because in vitro transcripts are marginally infectious in cell culture. For instance, FRhK-4 cells transfected with in vitro RNA transcripts derived from the wt HM-175 full-length cDNA required 132 days to show HAV-specific IF in only 5% of the cells (14). This inefficient growth of wt HAV most likely favors the selection of mutants that replicate more efficiently in cell culture. To circumvent this problem, wt HAV was rescued from inoculated marmoset livers with a mixture containing cDNA and full-length genomic RNA transcripts (13). This direct transfection of marmoset livers is a complicated and expensive technique that contrasts with the simplicity of deriving wt HAV from transfected Huh7 or Huh7-A-I cells as described in this paper. Molecular clones of wt and attenuated HAV have been available for approximately two decades (5-7). However, the lack of a robust cell culture system that could allow the rescue and efficient growth of wt viruses limited the use of reverse genetics to understand the pathogenesis of HAV and develop cost-effective attenuated vaccines. This is the first report showing that wt HAV can be efficiently rescued from Huh7 cells transfected with in vitro-synthesized transcripts containing a selectable marker irrespective of the presence of the main cell culture-adapting mutation at position 3889. The availability of the permissive Huh7-A-I cells for wt HAV growth will most likely allow the unraveling of the full potential of reverse genetics applied to the study of the pathogenesis and live vaccine development of HAV.
Huh7 cells are derived from a hepatoma cell line and do not represent an ideal substrate for vaccine production. However, this limitation may be circumvented by the fact that HAV is a nonenveloped virus highly resistant to detergent, protease, and nuclease treatment which could be purified to homogeneity. Alternatively, the Huh7 cell system could be used to develop attenuated vaccine strains that could be adapted to grow in cell substrates that are more adequate for vaccine production. Viral titers of attenuated viruses capable of inducing a protective immune response vary and depend on the particular pathogen and route of administration. For instance, licensed oral poliovirus vaccines contain approximately 105 to 106 TCID50 per vaccine dose whereas injectable measles, mumps, rubella, and varicella attenuated vaccines require only approximately 103 to 104 TCID50 per dose. Although the dose of a not-overly attenuated HAV required to induce a protective response in humans is an unknown, it is possible that the yield of HAV in Huh7-A-I cells (105 to 106 TCID50/ml) will be sufficient for the development of an attenuated vaccine. Alternatively, it is possible that vaccine strains could be adapted to grow in Huh7 cells or other cell substrates to higher titers without becoming overly attenuated.
Current diagnostic tests of HAV infection are based on the detection of anti-HAV antibodies and HAV genomes by RT-PCR, which does not distinguish between infectious and noninfectious viruses. Detection of infectious HAV before antibody production (window period) could prevent transmission of HAV in outbreak settings since this virus has a long incubation period of 2 to 7 weeks with peak infectivity 2 weeks before symptoms (17). Although RT-PCR can be used to detect HAV in the window period, an infectivity-based assay could increase detection sensitivity and also be used as a confirmatory test. It has been recently reported that HAV RNA was detected by RT-PCR in patients more than a year after infection (32). A cell culture infectivity assay could be used to determine whether the detected HAV RNA is associated with infectious particles. Moreover, relapsing HAV infection is well documented (21), indicating that HAV can reemerge even in the presence of anti-HAV antibodies and cause jaundice. Consequently, a cell culture assay to detect HAV infectivity can have an important impact on diagnosis of HAV and patient management.
PCR-based assays can be used to detect HAV RNA in environmental samples but do not provide information about infectivity. Cell culture infectivity assays could be used to determine whether the detected HAV genomes are associated with infectious particles. In addition, cell culture assays could also be used as confirmatory assays for HAV contamination of the food and water supply.
In this paper, we showed that a wt HAV construct containing the blasticidin selectable marker can be used to screen cell lines that support wt virus replication. A similar construct reported previously containing the bleomycin selectable marker (2) was not suitable for our experiments because of the slow nature of the bleomycin selection. Indeed, cells transfected with an HAV construct containing the bleomycin selectable marker instead of the blasticidin marker did not survive selection with zeocin initiated 24 h after transfection (K. Konduru and G. G. Kaplan, unpublished data). The HAV constructs coding for Bsd are an excellent genetic tool that could allow the identification of genes required for the growth of HAV, development of rapid titration and neutralization tests for research and diagnosis, selection of HAV vaccine strains that could be grown more rapidly and/or to higher titers by increasing the blasticidin concentration, and isolation of cells that support higher levels of HAV growth. Further experimentation will be required to validate the use of the HAV vectors described in this paper for the above-mentioned applications. The HAV vectors coding for effective selectable markers and the identification of cell lines capable of supporting the genetically stable growth of wt HAV should provide a significant insight into how HAV replicates and causes disease and also contribute to the development of cost-effective attenuated vaccines.
Acknowledgments
We thank Susan Zullo, Robert Duncan, and Barry Falgout for critical review of the manuscript.
REFERENCES
- 1.Amicone, L., F. M. Spagnoli, G. Spath, S. Giordano, C. Tommasini, S. Bernardini, V. De Luca, C. Della Rocca, M. C. Weiss, P. M. Comoglio, and M. Tripodi. 1997. Transgenic expression in the liver of truncated Met blocks apoptosis and permits immortalization of hepatocytes. EMBO J. 16:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beard, M. R., L. Cohen, S. M. Lemon, and A. Martin. 2001. Characterization of recombinant hepatitis A virus genomes containing exogenous sequences at the 2A/2B junction. J. Virol. 75:1414-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binn, L. N., S. M. Lemon, R. H. Marchwicki, R. R. Redfield, N. L. Gates, and W. H. Bancroft. 1984. Primary isolation and serial passage of hepatitis A virus strains in primate cell cultures. J. Clin. Microbiol. 20:28-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen, J. I., B. Rosenblum, S. M. Feinstone, J. Ticehurst, and R. H. Purcell. 1989. Attenuation and cell culture adaptation of hepatitis A virus (HAV): a genetic analysis with HAV cDNA. J. Virol. 63:5364-5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen, J. I., B. Rosenblum, J. R. Ticehurst, R. J. Daemer, S. M. Feinstone, and R. H. Purcell. 1987. Complete nucleotide sequence of an attenuated hepatitis A virus: comparison with wild-type virus. Proc. Natl. Acad. Sci. USA 84:2497-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, J. I., J. R. Ticehurst, S. M. Feinstone, B. Rosenblum, and R. H. Purcell. 1987. Hepatitis A virus cDNA and its RNA transcripts are infectious in cell culture. J. Virol. 61:3035-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, J. I., J. R. Ticehurst, R. H. Purcell, A. Buckler-White, and B. M. Baroudy. 1987. Complete nucleotide sequence of wild-type hepatitis A virus: comparison with different strains of hepatitis A virus and other picornaviruses. J. Virol. 61:50-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crance, J. M., F. Leveque, S. Chousterman, A. Jouan, C. Trepo, and R. Deloince. 1995. Antiviral activity of recombinant interferon-alpha on hepatitis A virus replication in human liver cells. Antivir. Res. 28:69-80. [DOI] [PubMed] [Google Scholar]
- 9.Daemer, R. J., S. M. Feinstone, I. D. Gust, and R. H. Purcell. 1981. Propagation of human hepatitis A virus in African green monkey kidney cell culture: primary isolation and serial passage. Infect. Immun. 32:388-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dotzauer, A., S. M. Feinstone, and G. Kaplan. 1994. Susceptibility of nonprimate cell lines to hepatitis A virus infection. J. Virol. 68:6064-6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emerson, S. U., Y. K. Huang, C. McRill, M. Lewis, and R. H. Purcell. 1992. Mutations in both the 2B and 2C genes of hepatitis A virus are involved in adaptation to growth in cell culture. J. Virol. 66:650-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emerson, S. U., Y. K. Huang, and R. H. Purcell. 1993. 2B and 2C mutations are essential but mutations throughout the genome of HAV contribute to adaptation to cell culture. Virology 194:475-480. [DOI] [PubMed] [Google Scholar]
- 13.Emerson, S. U., M. Lewis, S. Govindarajan, M. Shapiro, T. Moskal, and R. H. Purcell. 1992. cDNA clone of hepatitis A virus encoding a virulent virus: induction of viral hepatitis by direct nucleic acid transfection of marmosets. J. Virol. 66:6649-6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emerson, S. U., C. McRill, B. Rosenblum, S. Feinstone, and R. H. Purcell. 1991. Mutations responsible for adaptation of hepatitis A virus to efficient growth in cell culture. J. Virol. 65:4882-4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feigelstock, D., P. Thompson, P. Mattoo, Y. Zhang, and G. G. Kaplan. 1998. The human homolog of HAVcr-1 codes for a hepatitis A virus cellular receptor. J. Virol. 72:6621-6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feigelstock, D. A., P. Thompson, and G. G. Kaplan. 2005. Growth of hepatitis A virus in a mouse liver cell line. J. Virol. 79:2950-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiore, A. E. 2004. Hepatitis A transmitted by food. Clin. Infect. Dis. 38:705-715. [DOI] [PubMed] [Google Scholar]
- 18.Flehmig, B., A. Vallbracht, and G. Wurster. 1981. Hepatitis A virus in cell culture. III. Propagation of hepatitis A virus in human embryo kidney cells and human embryo fibroblast strains. Med. Microbiol. Immunol. 170:83-89. [DOI] [PubMed] [Google Scholar]
- 19.Frosner, G. G., F. Deinhardt, R. Scheid, V. Gauss-Muller, N. Holmes, V. Messelberger, G. Siegl, and J. J. Alexander. 1979. Propagation of human hepatitis A virus in a hepatoma cell line. Infection 7:303-305. [DOI] [PubMed] [Google Scholar]
- 20.Funkhouser, A. W., R. H. Purcell, E. D'Hondt, and S. U. Emerson. 1994. Attenuated hepatitis A virus: genetic determinants of adaptation to growth in MRC-5 cells. J. Virol. 68:148-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glikson, M., E. Galun, R. Oren, R. Tur-Kaspa, and D. Shouval. 1992. Relapsing hepatitis A. Review of 14 cases and literature survey. Medicine 71:14-23. [DOI] [PubMed] [Google Scholar]
- 22.Graff, J., and S. U. Emerson. 2003. Importance of amino acid 216 in nonstructural protein 2B for replication of hepatitis A virus in cell culture and in vivo. J. Med. Virol. 71:7-17. [DOI] [PubMed] [Google Scholar]
- 23.Graff, J., A. Normann, S. M. Feinstone, and B. Flehmig. 1994. Nucleotide sequence of wild-type hepatitis A virus GBM in comparison with two cell culture-adapted variants. J. Virol. 68:548-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graff, J., O. C. Richards, K. M. Swiderek, M. T. Davis, F. Rusnak, S. A. Harmon, X. Y. Jia, D. F. Summers, and E. Ehrenfeld. 1999. Hepatitis A virus capsid protein VP1 has a heterogeneous C terminus. J. Virol. 73:6015-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harmon, S. A., W. Updike, X. Y. Jia, D. F. Summers, and E. Ehrenfeld. 1992. Polyprotein processing in cis and in trans by hepatitis A virus 3C protease cloned and expressed in Escherichia coli. J. Virol. 66:5242-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jansen, R. W., J. E. Newbold, and S. M. Lemon. 1988. Complete nucleotide sequence of a cell culture-adapted variant of hepatitis A virus: comparison with wild-type virus with restricted capacity for in vitro replication. Virology 163:299-307. [DOI] [PubMed] [Google Scholar]
- 27.Jia, X. Y., D. F. Summers, and E. Ehrenfeld. 1993. Primary cleavage of the HAV capsid protein precursor in the middle of the proposed 2A coding region. Virology 193:515-519. [DOI] [PubMed] [Google Scholar]
- 28.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 29.MacGregor, A., M. Kornitschuk, J. G. Hurrell, N. I. Lehmann, A. G. Coulepis, S. A. Locarnini, and I. D. Gust. 1983. Monoclonal antibodies against hepatitis A virus. J. Clin. Microbiol. 18:1237-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malbrain, M. L., X. De Meester, A. P. Wilmer, E. Frans, J. Peeters, and F. Nevens. 1997. Another case of acute renal failure (ARF) due to acute tubular necrosis (ATN), proven by renal biopsy in non-fulminant hepatitis A virus (HAV) infection. Nephrol. Dial. Transplant. 12:1543-1544. [DOI] [PubMed] [Google Scholar]
- 31.Martin, A., N. Escriou, S. F. Chao, M. Girard, S. M. Lemon, and C. Wychowski. 1995. Identification and site-directed mutagenesis of the primary (2A/2B) cleavage site of the hepatitis A virus polyprotein: functional impact on the infectivity of HAV RNA transcripts. Virology 213:213-222. [DOI] [PubMed] [Google Scholar]
- 32.Normann, A., C. Jung, A. Vallbracht, and B. Flehmig. 2004. Time course of hepatitis A viremia and viral load in the blood of human hepatitis A patients. J. Med. Virol. 72:10-16. [DOI] [PubMed] [Google Scholar]
- 33.Pagano, J. S., and A. Vaheri. 1965. Enhancement of infectivity of poliovirus RNA with diethylaminoethyl-dextran (DEAE-D). Arch. Gesamte Virus forsch. 17:456-464. [DOI] [PubMed] [Google Scholar]
- 34.Provost, P. J., and M. R. Hilleman. 1979. Propagation of human hepatitis A virus in cell culture in vitro. Proc. Soc. Exp. Biol. Med. 160:213-221. [DOI] [PubMed] [Google Scholar]
- 35.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty per cent end points. Am. J. Hyg. 27:493-497. [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 37.Schultz, D. E., M. Honda, L. E. Whetter, K. L. McKnight, and S. M. Lemon. 1996. Mutations within the 5′ nontranslated RNA of cell culture-adapted hepatitis A virus which enhance cap-independent translation in cultured African green monkey kidney cells. J. Virol. 70:1041-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siegl, G., J. deChastonay, and G. Kronauer. 1984. Propagation and assay of hepatitis A virus in vitro. J. Virol. Methods 9:53-67. [DOI] [PubMed] [Google Scholar]
- 39.Weitz, M., B. M. Baroudy, W. L. Maloy, J. R. Ticehurst, and R. H. Purcell. 1986. Detection of a genome-linked protein (VPg) of hepatitis A virus and its comparison with other picornaviral VPgs. J. Virol. 60:124-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wunschmann, S., B. Becker, and A. Vallbracht. 2002. Hepatitis A virus suppresses monocyte-to-macrophage maturation in vitro. J. Virol. 76:4350-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, Y., and G. G. Kaplan. 1998. Characterization of replication-competent hepatitis A virus constructs containing insertions at the N terminus of the polyprotein. J. Virol. 72:349-357. [DOI] [PMC free article] [PubMed] [Google Scholar]