Abstract
Rhesus monkey rhadinovirus (RRV), a simian gamma-2 herpesvirus closely related to the Kaposi sarcoma-associated herpesvirus, replicates lytically in cultured rhesus monkey fibroblasts and establishes persistence in B cells. Overlapping cosmid clones were generated that encompass the entire 130-kilobase-pair genome of RRV strain 26-95, including the terminal repeat regions required for its replication. Cloned RRV that was produced by cotransfection of overlapping cosmids spanning the entire RRV26-95 genome replicated with growth kinetics and to titers similar to those of the parental, uncloned, wild-type RRV26-95. Expression cassettes for secreted-engineered alkaline phosphatase (SEAP) and green fluorescent protein (GFP) were inserted upstream of the R1 gene, and the cosmid-based system for RRV genome reconstitution was used to generate replication-competent, recombinant RRV that expressed either the SEAP or GFP reporter gene. Using the SEAP and GFP recombinant RRVs, assays were developed to monitor RRV infection, neutralization, and replication. Heat-inactivated sera from rhesus monkeys that were naturally or experimentally infected with RRV were assayed for their ability to neutralize RRV-SEAP and RRV-GFP infectivity using rhesus monkey fibroblasts. Sera from RRV-positive monkeys, but not RRV-negative monkeys, were consistently able to neutralize RRV infectivity when assayed by the production of SEAP activity or by the ability to express GFP. The neutralizing activity was present in the immunoglobulin fraction. Of the 17 rhesus monkeys tested, sera from rhesus monkey 26-95, i.e., the monkey that yielded the RRV 26-95 isolate, had the highest titer of neutralizing activity against RRV26-95. This cosmid-based genetic system and the reporter virus neutralization assay will facilitate study of the contribution of individual RRV glycoproteins to entry into different cell types, particularly fibroblasts and B cells.
Kaposi's sarcoma-associated herpesvirus (KSHV; also called human herpesvirus 8) is the causative agent for Kaposi's sarcoma and is associated with the lymphoproliferative disorders primary effusion lymphoma and multicentric Castleman's disease (5, 14). A distinct simian herpesvirus related to KSHV was isolated at the New England Primate Research Center (NEPRC) after rhesus monkey sera were found to react positively by enzyme-linked immunosorbent assay (ELISA) with herpesvirus saimiri. Coculturing of peripheral blood mononuclear cells (PBMCs) from rhesus monkeys with rhesus monkey fibroblasts resulted in cytopathology characteristic of lytic viral replication and plaque formation within the cultures. Electron microscopy revealed the presence of large numbers of nuclear nonenveloped, cytoplasmic enveloped, and extracellular herpesviruses in these cultures (8). Initial sequencing of a 10.6-kbp DNA fragment isolated from these rhesus monkey herpesvirus particles showed significant homology to genes of KSHV, a gamma-2 herpesvirus (rhadinovirus). Subsequently, the complete primary sequence of the newly isolated rhesus monkey rhadinovirus (RRV) isolate 26-95 was determined. The RRV26-95 genome is 130,733 bp and has the potential of encoding at least 84 distinct polypeptide products (1). In agreement with the previously sequenced 10.6-kbp genome fragment, the overall organization of the RRV26-95 genome is very similar to that of KSHV. Furthermore, RRV26-95 coding sequences share a greater degree of similarity to those of KSHV than other herpesviruses. The majority of RRV26-95 genes are in corresponding genomic locations and in the same polarity as their KSHV homologues. RRV has also been isolated and sequenced by researchers at the Oregon National Primate Research Center (23).
Examination of sera by ELISA revealed a high prevalence of antibodies to RRV in rhesus monkey colonies at multiple facilities for at least 10 years (3, 8, 22). After experimental infection of rhesus monkeys with RRV, animals that were previously seronegative for RRV developed persisting antibody responses to the virus, which could be consistently isolated from peripheral blood. By PCR, RRV was detected in the lymph nodes, oral mucosa, skin, and PBMCs in inoculated animals. PCR analysis of sorted PBMCs revealed a preferential persistence of RRV in CD20-positive B lymphocytes (18, 25). While experimentally infected rhesus monkeys developed lymphadenopathy as evidenced by paracortical expansion and follicular hyperplasia, these pathologies were transient and subsided by 12 weeks postinfection. Coinoculation of rhesus monkeys with RRV and simian immunodeficiency virus (SIV) resulted in an attenuated antibody response and a shorter mean survival time compared to animals infected with SIV alone. Immunocompromised SIV-positive rhesus monkeys infected with RRV displayed weaker and delayed antibody responses to RRV. Furthermore, a study performed at the Oregon National Primate Research Center observed a lymphoproliferative disorder similar to multicentric Castleman's disease in rhesus monkeys experimentally infected with both RRV and SIV (25).
RRV's ability to replicate permissively in standard rhesus monkey fibroblast cultures provides the potential for facile genetic manipulation. When coupled with the ready availability of rhesus monkeys for experimental infection, a genetic system would provide attractive opportunities to study the contributions of individual genes to biological properties relevant to KSHV in the setting of the whole organism. In this report, we describe the generation of overlapping cosmid clones for reconstitution of the RRV26-95 genome and their use in producing recombinant RRV by cotransfection. We inserted genes for green fluorescent protein (GFP) and secreted engineered alkaline phosphatase (SEAP) into an RRV cosmid and subsequently generated recombinant RRV that expressed GFP or SEAP and which displayed no altered growth phenotype compared to uncloned virus. A convenient assay for quantitating antibody-mediated RRV neutralization was developed using these recombinant strains. With a convenient genetic system now available, the contributions of individual RRV genes to infection, immune evasion, replication, and persistence can now be examined in rhesus monkeys.
MATERIALS AND METHODS
Cell culture.
Human embryonic kidney cells (293T) were maintained on Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum, 2 mM l-glutamine, penicillin-streptomycin (50 IU and 50 μg/ml, respectively) at 37°C in a humidified incubator with 5% CO2. Rhesus macaque skin fibroblasts (RF) were maintained in DH20 medium (DMEM supplemented with 20% fetal calf serum, 2 mM l-glutamine, penicillin-streptomycin [50 IU and 50 μg/ml, respectively] and 10 mM HEPES) at 37°C in a humidified incubator with 5% CO2.
Library construction.
The cosmid vector, pSCos/ICeuI, used to make libraries was derived from pSuperCos 1 (Stratagene, La Jolla, CA). Complementary oligonucleotides, 5′-GATCTTAACTATAACGGTCCTAAGGTAGCGAGCGCGCGGATCCGCGCGCTAACTATAACGGTCCTAAGGTAGCGAA-3′ and 5′-GATCTTCGCTACCTTAGGACCGTTATAGTTAGCGCGCGGATCCGCGCGCTCGCTACCTTAGGACCGTTATAGTTAA-3′, were annealed at 55°C and phosphorylated using T4 polynucleotide kinase, forming an adaptomer. The adaptomer featured a cut BglII site at each end flanking ICeuI-BamHI-ICeuI sites. The pSuperCos 1 plasmid was linearized with BamHI and dephosphorylated using calf intestinal phosphatase (CIP). Subsequently, the linearized pSuperCos 1 plasmid was ligated to the BglII-ICeuI-BamHI-ICeuI-BglII adaptomer, yielding pSCos/ICeuI, which was confirmed by sequence analysis.
Cosmid libraries were constructed from purified RRV DNA according to the instructions of the SuperCos 1 Cosmid Vector Kit (Stratagene). Briefly, pSCos/ICeuI was digested with XbaI and dephosphorylated with CIP. Following phenol extraction, the linearized pSCos/ICeuI plasmid was digested with BamHI. Virion DNA was partially digested with Sau3AI to an average length of 38 to 42 kbp and dephosphorylated. Sau3AI-digested RRV DNA and a molar excess of pSCos/ICeuI arms were ligated with T4 DNA ligase for 16 h at 16°C. The ligation products were packaged into bacteriophage lambda particles using the Gigapack III XL packaging extract (Stratagene) according to the manufacturer's instructions. XL-1-Blue MR (recA1 endA1) supercompetent cells (Stratagene) were transduced with the packaged libraries, followed by plating and growth on Luria-Bertani agar media plates containing 25 μg/ml of ampicillin per ml. Cosmid DNA from randomly picked colonies was digested with ICeuI to identify large DNA inserts, which were subsequently sequenced to identify the boundaries of the selected clones. A series of cosmids encompassing the entire RRV genome, including the terminal repeat regions, was selected with this procedure.
Cosmid subclones.
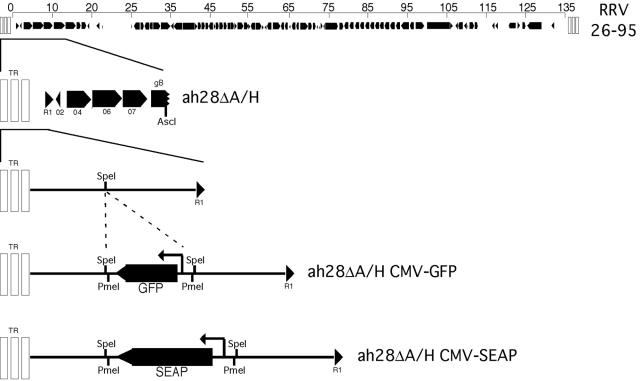
To facilitate reporter gene insertion, the ah28 cosmid (Table 1 and Fig. 1) was digested with AscI and HindIII to remove excess overlapping genomic RRV sequences. The remaining fragment was digested with Klenow fragment to produce blunt ends and ligated, yielding ah28ΔA/H. Complementary oligonucleotides, 5′-CTAGTTGTTTAAACGGGGCGCCGGA-3′ and 5′-CTAGTCCGGCGCCCCGTTTAAACAA-3′, were annealed at 55°C and phosphorylated using T4 polynucleotide kinase, forming an adaptomer. The adaptomer featured a cut SpeI site at each end flanking a central PmeI site. The ah28ΔA/H cosmid was linearized with SpeI and dephosphorylated using CIP. Subsequently, the linearized ah28ΔA/H cosmid was ligated to the SpeI-PmeI-SpeI adaptomer, yielding ah28ΔA/H-PmeI.
TABLE 1.
RRV26-95 genome coordinates of infectious cosmids
| Cosmida | RRV26-95 genome coordinates (nt) |
|---|---|
| 3A | TRb-3985 |
| ah39 | TR-12467 |
| ah12 | TR-30153 |
| ah43 | 130547-38784 |
| ah28 | TR-31122 |
| ah28ΔA/Hc | TR-13284 |
| 48 | 8233-54900 |
| 15A | 11365-50111 |
| 1 | 38781-80580 |
| 12 | 51570-95619 |
| 45 | 58699-97944 |
| 37 | 62132-101561 |
| 14 | 74307-111062 |
| 43 | 78793-121310 |
| ah53 | 61132-101561 |
| 6A | 87320-124261 |
| ah47 | 99098-TR |
| ah34 | 95023-TR |
Cosmid clones were maintained in pSuperCos 1.
Terminal repeat.
Modified cosmid clone not selected in the original cosmid library.
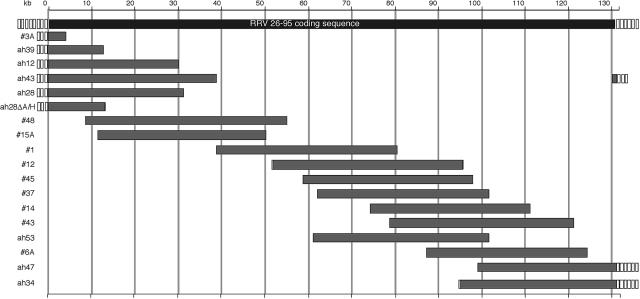
FIG. 1.
Schematic representation of the cosmids derived from RRV26-95. Open boxes represent the terminal repeat regions of various lengths ranging from <1 kb to approximately 28 kb. The gray bars indicate the extent and location of each cosmid insert within the parental RRV26-95 genome (top black bar). Each cosmid insert was cloned into the pSuperCos 1 vector following the addition of an ICeuI homing endonuclease linker. The cosmid insert designated ah28ΔA/H was derived from the removal of the AscI-HindIII fragment from the ah28 cosmid within the pSuperCos 1 vector.
To generate the ah28ΔA/H-CMV-GFP cosmid (Fig. 2), ah28ΔA/H-PmeI was digested with PmeI and dephosphorylated with CIP. The cytomegalovirus (CMV)-GFP cassette was obtained by PCR amplification of pEGFP-C1 (where EGFP is enhanced GFP; BD Biosciences Clontech, Palo Alto, CA) using the primers 5′-AGCTTTGTTTAAACGGGCCATGCATTAGTTATTAATAG-3′ and 5′-CCGGCGCCCCGTTTAAACCAAACCACAACTAGAATGCA-3′. The amplified product contained the CMV-GFP cassette flanked by PmeI restriction sites at its ends. The PCR fragment was digested with PmeI and ligated to the linearized ah28ΔA/H-PmeI cosmid, yielding ah28ΔA/H-CMV-GFP. The pCMV/SEAP (Tropix, Inc., Bedford, MA) expression plasmid was modified to contain PmeI restriction sites flanking the CMV-directed transgene. Complementary oligonucleotides, 5′-GATCTAGCTTTGTTTAAACGGGGCGA-3′ and 5′-GATCTCGCCCCGTTTAAACAAAGCTA-3′, were annealed at 55°C and phosphorylated using T4 polynucleotide kinase, forming an adaptomer. The adaptomer featured a cut BglII site at each end flanking a central PmeI site. The pCMV-SEAP plasmid was linearized with BglII and dephosphorylated with CIP. Subsequently, the linearized pCMV-SEAP plasmid was ligated to the BglII-PmeI-BglII adaptomer, yielding pCMV-SEAP BP. A KpnI-PmeI-KpnI adaptomer, from complementary oligonucleotides 5′-CAGCTTTGTTTAAACGGGGCGGTAC-3′ and 5′-CGCCCCGTTTAAACAAAGCTGGTAC-3′, was annealed at 55°C and phosphorylated using T4 polynucleotide kinase. This adaptomer featured a cut KpnI site at each end flanking a central PmeI site. The pCMV-SEAP BP plasmid was linearized with KpnI and dephosphorylated with CIP. Subsequently, the linearized pCMV-SEAP BP plasmid was ligated to the KpnI-PmeI-KpnI adaptomer, yielding pCMV-SEAP PmeIx2. The pCMV-SEAP PmeIx2 plasmid was digested with PmeI and ligated to the linearized ah28ΔA/H-PmeI cosmid, yielding ah28ΔA/H-CMV-SEAP (Fig. 2).
FIG. 2.
Insertion of GFP and SEAP transgene cassettes into the RRV ah28ΔA/H cosmid. An oligonucleotide linker containing the PmeI restriction endonuclease recognition site was inserted into the SpeI site of the ah28ΔA/H cosmid. A cassette containing either CMV-GFP or CMV-SEAP and flanked by PmeI restriction endonuclease sites was cloned into the PmeI site of ah28ΔA/H to generate ah28ΔA/H CMV-GFP or ah28ΔA/H CMV-SEAP. The size of the CMV-GFP and CMV-SEAP transgene cassettes are 1,636 and 3,882 bp, respectively.
DNA sequencing.
Cosmid and plasmid constructs were sequenced with a CEQ 8000 Genetic Analysis System using a dye terminator cycle sequencing kit as specified by the manufacturer (Beckman Coulter, Fullerton, CA).
Cotransfection and virus preparation.
Prior to transfection, the cosmids were digested overnight with the ICeuI homing endonuclease, removing the RRV26-95 sequence from the pSuperCos 1 backbone vector. The cosmid DNA was precipitated by adding 3 volumes of 5% 3 M sodium acetate-95% ethanol and incubating for >1 h at −20°C. The DNA was then pelleted by centrifugation for 10 min at maximum speed in a microcentrifuge. The pellets were washed in 70% ethanol, dried, and rehydrated in distilled water. One day postseeding, 293T cells (4.5 × 105 cells/well in six-well plates) were transfected with different combinations of digested overlapping cosmids (0.4 μg of each cosmid) using Transfectin reagent (Bio-Rad Laboratories, Hercules, CA) following a scaled-down procedure. As a positive control, 0.25 μg of whole viral RRV DNA isolated from column-purified RRV26-95 was transfected in the same manner. At 5 days posttransfection, cell-free culture supernatant was collected and stored at 4°C. To amplify recombinant stocks generated in 293T cells, fresh RF cultures were inoculated with 1 ml of the supernatant collected from the 293T transfections. Inoculated RF cultures were passaged until the emergence of viral plaques was observed in the cultures, and then cultures were maintained without splitting until complete lysis of the RF monolayer. High-titer recombinant RRV stocks were subsequently generated in fresh RF cultures.
Isolation and analysis of RRV DNA.
For each RRV virus, supernatant collected following complete lysis of RRV-infected RFs was subjected to low-speed centrifugation to remove cellular debris. The supernatant was then filtered through a 0.45-μm-pore-size filter to remove any additional debris. The filtered supernatant was then centrifuged for 3 h at 45,000 × g in a Sorvall type 19 rotor to pellet the virus. The crude virus was resuspended in Tris-EDTA buffer and lysed by adding 0.1 vol. 1% N-lauroylsarcosine and proteinase K and incubating at 60°C for 1 h. The mixture was extracted twice with phenol-chloroform, followed by four chloroform washes. The DNA was recovered by precipitation with 2.5 volumes of 5% 3 M sodium acetate-95% ethanol, rinsed in 80% ethanol, and resuspended in Tris-EDTA buffer. Viral DNA was digested with restriction endonucleases, separated on a 0.5% agarose electrophoretic gel, and stained with ethidium bromide.
Plaque assay.
The titers of parental RRV26-95 and recombinant RRV stocks were determined as previously described (10). Briefly, cell-free culture supernatant was collected following complete lysis of RRV-infected RFs. Fresh RFs were seeded into 12-well plates at 2 × 105 cells/well. The following day, 10-fold serial dilutions of the virus-containing supernatant were made in DH20 medium. The medium was removed from the RF cultures and replaced with 200 μl of diluted virus/well. Cultures were then incubated for 1 h at 37°C with gentle rocking every 15 min. After 1 h, 2 ml of Hank's buffered saline solution (HBSS) was added to each well and subsequently aspirated. Two milliliters of overlay medium (1:1 ratio of 2× DMEM and 1.5% methyl-cellulose [Sigma, St. Louis, MO] supplemented with 2% fetal calf serum) was then applied, and the cultures were incubated at 37°C and 5% CO2 for 1 week. Overlay medium was then aspirated, and a staining solution (0.8% crystal violet in 50% ethanol) was applied for 10 min. Each well was then washed five times with distilled water, and the number of plaques at each dilution of inoculum was determined.
Quantitative real-time PCR.
At the indicated time postinfection (p.i.), viral DNA was isolated from 200 μl of cell-free culture supernatant from each sample using the QiaAmp DNA Blood Mini Kit (QIAGEN, Valencia, CA) according to the manufacturer's protocol. Tenfold serial dilutions (ranging from 1 to 106 plasmid copies/reaction) of pcDNA3.1/RRV-pol were used in each assay to generate a standard curve for genome copy number. The pcDNA3.1/RRV-pol plasmid was constructed by PCR amplification of RRV polymerase (Pol) from the ah28 cosmid using the primers 5′-CCCAAGCTTATGGATTTCTTTAACCCGTACC-3′ and 5′-CGCGGATCCTCACGAGAACAGCTTATACGGGAC-3′. The amplified product contained the RRV Pol gene flanked by an upstream HindIII site and a BamHI site downstream. The resulting PCR product and the pcDNA3.1 plasmid were digested with HindIII and BamHI, gel purified, and ligated together to generate pcDNA3.1/RRV-pol. Quantitative PCRs were performed using the iQ Supermix kit and the MyiQ Single Color Real-Time PCR Detection System (Bio-Rad). The 94-bp amplicon internal to the RRV pol sequence was amplified using the primers 5′-CCGCTTTCTGTGACGATCTG-3′ and 5′-AGCAGACACTTGAACGTCTT-3′ and the probe 5′-6FAM-CCAGGATCACTGCGGACCTGTTCC-TAMRA-3′. Amplification was performed using the following conditions: 95°C for 3 min, followed by 50 cycles of 95°C for 30 s and 60°C for 30 s. Reactions were performed in triplicate and no-template controls were included in the analysis. The number of RRV genome copies/reaction was calculated from the equation for the standard curve using the MyiQ real-time PCR detection system software.
Reporter gene expression.
SEAP expression was quantitated using the Phosphalight kit (Applied Biosystems, Foster City, CA). GFP expression was observed by fluorescence microscopy, and emission intensity was quantitated using the Victor3V 1420 Multilabel Counter with 480-nm excitation and 510-nm emission filters (PerkinElmer, Wellesley, MA).
RRV ELISA.
RRV26-95 was pelleted and column purified as previously described (8). Purified virus was lysed in 0.1 volume of 10% Triton X-100, and protein concentration was determined using a BCA (bicinchoninic acid) Protein Assay Kit (Pierce Biotechnology, Rockford, IL). ELISA plates were coated with 2 μg/ml RRV26-95 lysate for 1 h at room temperature. ELISAs were then performed as previously described (17).
Viral neutralization.
RF cells were seeded into 24-well plates at 1 × 105 cells/well. One day postseeding, RRV-SEAP (0.006 PFU/cell) or RRV-GFP (0.04 PFU/cell) was incubated with various dilutions of heat-inactivated rhesus monkey serum or concentrations of purified rhesus monkey immunoglobulin G (IgG) in a total volume of 200 μl for 3 h at 37°C with constant gentle rocking. The heat-inactivated serum and purified IgG were diluted in DH20 medium. RRV-SEAP or RRV-GFP was also diluted in DH20 medium without antibody to serve as a no-antibody control for virus neutralization. After the preincubation period, RF cultures were inoculated with medium alone, virus alone, or the virus-serum or virus-IgG mixture. At 16 to 20 h p.i., cultures were rinsed five times with HBSS and refed with DH20 medium. At the indicated day p.i., cultures were examined for either SEAP or GFP expression.
Purification and depletion of serum IgG.
Rhesus monkey serum was diluted 1:5 in HBSS and centrifuged at 640 × g for 5 min at room temperature to remove debris. For large-scale IgG purification, the clarified serum was decanted into a column containing 300 μl of protein A-Sepharose (Amersham Biosciences, Piscataway, NJ). The diluted serum was allowed to flow through the column. The column was then washed with 50 volumes of phosphate-buffered saline (PBS), and IgG was eluted with ImmunoPure IgG Elution Buffer (Pierce) into 0.1 volumes of 10× PBS to neutralize the elution buffer. IgG was concentrated using a Viva Spin concentrator (50 kMWCO PES; Vivascience, Hannover, Germany) and dialyzed overnight in PBS at 4°C. The IgG protein concentration was determined using the BCA Protein Assay Kit (Pierce) according to the manufacturer's instructions. Furthermore, the IgG concentration in serum was approximated by batch immunoprecipitation. Based on the IgG concentration determined by BCA analysis of the purified antibody fraction, known amounts of IgG and serum were diluted to a final volume of 200 μl in PBS. Fifty microliters of a protein A and protein G (protein A/G)-Sepharose mixture (1:5) was added, and the samples were mixed overnight at 4°C. Afterwards, the protein A/G-Sepharose was pelleted in a microcentrifuge, and the IgG-depleted supernatant was removed. The protein A/G-Sepharose pellets were rinsed four times with PBS, resuspended in Laemmli buffer, and boiled for 5 min. The Sepharose was pelleted in a microcentrifuge, and the supernatant was electrophoresed through a 12% polyacrylamide-sodium dodecyl sulfate gel. The gel was then stained with Coomassie blue for 30 min and destained in methanol-acetic acid.
RESULTS
Identification of infectious sets of RRV cosmid clones.
Cosmid libraries of RRV26-95 were constructed using a pSuperCos 1 vector containing a restriction endonuclease adaptomer. The adaptomer modified the BamHI cloning site by the inclusion of flanking ICeuI sites. Since ICeuI sites are not present in the RRV26-95 genome, the inclusion of flanking ICeuI sites allows ready removal of inserted RRV sequences from the cosmid vector. The modified pScos/ICeuI vector was approximately 8 kb and was capable of accepting cloned inserts of up to 47 kb. The RRV26-95 genome was partially digested with Sau3AI, and DNA fragments were inserted into the pScos/ICeuI vector. Random colonies were selected and the sizes of inserted RRV DNA segments were evaluated following digestion with ICeuI. Clones containing large inserts were partially sequenced to determine the boundaries of RRV genome insertion. Subsequently, clones were digested with a variety of restriction endonucleases to confirm the appropriate digestion pattern for the RRV genomic insertion (data not shown). Seventeen overlapping cosmid clones encompassing the entire RRV26-95 genome, including the terminal repeat regions, were selected in this manner (Table 1 and Fig. 1).
To determine which sets of cosmids yielded productive RRV infections, 14 combinations of overlapping cosmids were transfected into 293T cells (Table 2, B to O). As a positive control, 0.25 μg of genomic DNA isolated from column-purified RRV26-95 virions was transfected into 293T cells in a separate six-well plate (Table 2, A). Combination O, containing all the cosmids, was included in the transfection repertoire to reduce the possibility of a null result due to one or more defective cosmids. While no apparent cytopathology was observed in the 293T cells 5 days posttransfection, clarified culture supernatant was collected and used to infect fresh RFs. RFs were passaged until the appearance of viral plaques was observed, and then cultures were maintained without splitting until complete virolysis of the cultures. Of the 14 combinations of overlapping cosmids, only combination N failed to produce infectious RRV, suggesting that cosmid ah43 may be unable to yield infectious virus upon cotransfection. Whereas transfection of whole genomic viral DNA directly into RFs routinely produced infectious virus, transfection of the cosmid combinations into RFs repeatedly failed to yield infectious virus. This inability to produce recombinant RRV by transfection into RFs may be due, at least in part, to a low transfection efficiency for RFs, compounded by the high molecular weight of the cosmid clones. Transfection of a low-molecular-weight EGFP plasmid using numerous transfection methods and reagents into RFs resulted in a maximum transfection efficiency of approximately 1% (data not shown). Higher transfection efficiencies with the EGFP plasmid were obtained using nucleofector technology (Amaxa, GmbH, Cologne, Germany); however, this procedure also failed to produce infectious virus directly from RFs using overlapping cosmid clones (data not shown). While the nucleofector technology is highly efficient for low-molecular-weight plasmids, nucleofection of high-molecular-weight DNA species is inefficient.
TABLE 2.
RRV cosmid combinations transfected into 293T cells
| Cosmid combinations for recombinant RRVs
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | ||||||||||||||
| RRV DNA | ah12 | ah12 | ah12 | ah28 | ah28 | ah28 | ah39 | ah39 | ah39 | ah39 | ah39 | ah39 | ah43 | All cosmids | ||||||||||||||
| 48 | 15A | 15A | 48 | 15A | 15A | 15A | 15A | 15A | 48 | 15A | 48 | 15A | ||||||||||||||||
| 12 | 1 | 1 | 12 | 1 | 1 | 1 | 1 | 1 | 12 | 1 | 12 | 1 | ||||||||||||||||
| 6A | 37 | 37 | 6A | 37 | 37 | 14 | 43 | 37 | ah34 | 37 | 6A | 43 | ||||||||||||||||
| ah47 | ah47 | ah34 | ah47 | ah47 | ah34 | ah47 | ah47 | ah34 | ah47 | ah47 | ah34 | |||||||||||||||||
| Production of RRV infection
|
||||||||||||||||||||||||||||
| + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | ||||||||||||||
Construction of recombinant RRVs containing reporter genes.
For use as tools in a variety of studies, we selected the GFP and SEAP reporter genes for insertion into the RRV genome. Plasmids containing either the GFP or SEAP reporter gene under control of the CMV immediate-early promoter were modified to contain PmeI enzyme sites flanking the CMV-GFP or CMV-SEAP cassettes. After considering a number of intergenic sites and nonessential genes for insertion, we elected to insert the reporter genes between the left terminal repeat and the first open reading frame (Fig. 2, R1). Numerous attempts to insert the reporter gene cassettes into the original RRV cosmids were unsuccessful. As the size of the cosmids exceeded 46 kb on average prior to reporter gene insertion, we set out to examine if the size of the relaxed-circular ligation product may have exceeded the limits of bacterial transformation and electroporation. The ah28 cosmid was digested with AscI and HindIII to remove RRV26-95 bp 13285 to 31122 and a segment from the pSuperCos 1 backbone. The remainder of the plasmid was digested with Klenow fragment to generate blunt ends and ligated to produce the ah28ΔA/H cosmid (Table 1 and Fig. 2). An SpeI site was selected for reporter gene insertion within the approximately 26-kb ah28ΔA/H cosmid. The SpeI site, bp 206 in the sequence of Alexander et al. (1), is located upstream of the R1 open reading frame and in a region that shares no similarity with the terminal repeats of RRV. The location of the R1 promoter is currently unknown; however, subsequent real-time reverse transcription-PCR analyses of cells infected with recombinant RRV revealed no decrease in R1 transcript expression when a reporter gene cassette is inserted at this site (data not shown). After insertion of a PmeI adaptomer into ah28ΔA/H, either the PmeI-flanked CMV-GFP or CMV-SEAP reporter gene cassette was inserted into the truncated cosmid, the product of which was confirmed by restriction endonuclease digestion and DNA sequencing (Fig. 2). The ah28ΔA/H CMV-GFP or ah28ΔA/H CMV-SEAP cosmids were cotransfected with the cosmids 15A, 1, 37, and ah34 into 293T cells and amplified in RFs as described above.
Characterization of recombinant RRVs in vitro.
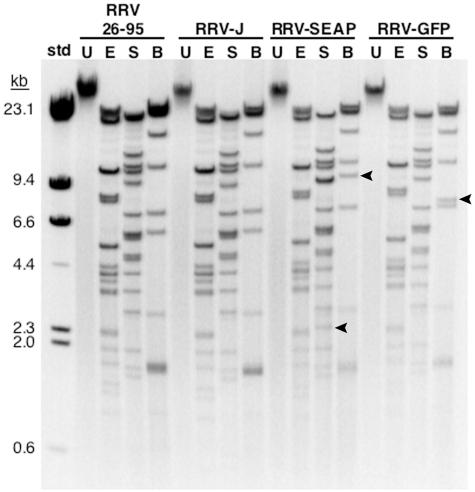
To determine whether recombinant RRV obtained from cosmid transfection accurately resembles wild-type RRV26-95, we compared the restriction endonuclease fragmentation profiles of the cosmid-derived RRV genomic DNA to those of RRV26-95. Genomic DNA was isolated from RRV26-95, recombinant RRV containing no reporter gene (Table 2, RRV-J), or recombinant RRV containing either the CMV-GFP or CMV-SEAP reporter gene cassettes (RRV-GFP or RRV-SEAP). Virion DNA was digested with EcoRV, SphI, or the BbvCIA and BbvCIB nicking endonucleases and separated on an agarose electrophoretic gel. As shown in Fig. 3, the restriction fragmentation profile of the cosmid-derived RRV-J was identical to the parental RRV26-95. The restriction fragmentation profiles of the RRV-GFP and RRV-SEAP genomes also mirrored that of RRV26-95. The fragments with altered mobility in the SphI and/or BbvCIA/B digests of the RRV-GFP and RRV-SEAP genomes resulted from the insertion of the CMV-SEAP or CMV-GFP reporter gene cassettes into the RRV genome.
FIG. 3.
Restriction digests of RRV genomic DNA. DNA from purified RRV26-95 or cosmid-derived recombinant RRV-J, RRV-SEAP, and RRV-GFP was digested with either EcoRV (E), SphI (S), or the nicking endonucleases BbvCIA and BbvCIB (B). Restriction endonuclease-digested and undigested (U) virion DNA were separated on a 0.5% agarose electrophoretic gel and stained with ethidium bromide. Bands marked with an arrow represent altered digestion products resulting from insertion of the CMV-SEAP or CMV-GFP reporter gene cassettes. The molecular size standards (std) are HindIII-digested λ DNA.
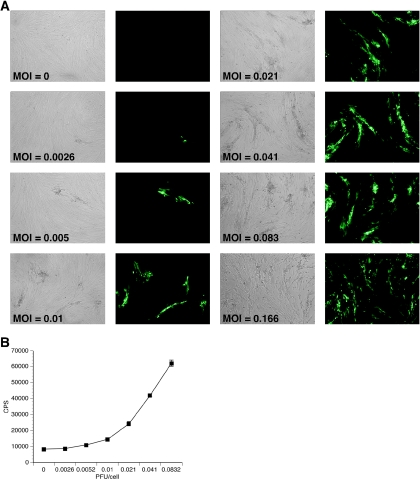
RFs were infected with RRV-GFP at increasing multiplicities of infection (MOIs) ranging from 0.0026 to 0.166 PFU/cell. Infected cultures were examined daily by fluorescence microscopy until a significant GFP signal was detected for the majority of the MOIs tested (day 4). As displayed in Fig. 4A, GFP expression at day 4 in RFs was MOI dependent. To quantitate these results, the GFP emission intensity of each well was determined using a multilabel plate reader (Fig. 4B). The emission intensity measured in this manner also increased with increasing PFU/cell. While GFP expression at MOIs lower than 0.0026 was undetectable, infection at MOIs higher than 0.0832 PFU/cell resulted in a near complete lysis of the cultures by day 4 p.i. (data not shown). Additionally, when cultures were infected at MOIs ranging from 0.0026 to 0.0832 PFU/cell and examined beyond day 4 p.i., the linear relationship between MOI and GFP expression was distorted due to virus spread in the cultures (data not shown).
FIG. 4.
GFP expression in rhesus fibroblasts after infection with RRV-GFP is MOI dependent. At 24 h postseeding, rhesus fibroblasts were infected with increasing MOIs of RRV-GFP. At day 4 p.i., cultures were examined for GFP expression. (A) Fluorescence microscopy of rhesus fibroblasts after infection with increasing MOIs of RRV-GFP. (B) GFP expression by RRV-GFP as measured by emission intensity. Rhesus fibroblasts were seeded into 96-well plates and infected with increasing MOIs of RRV-GFP at 24 h postseeding. At day 4 p.i., cultures were examined for GFP expression using a multiwell plate reader (n = 8). The results show the GFP counts/s (CPS) with standard deviation at the indicated PFU/cell. Where no error bar is shown, the error falls within the size of the symbol.
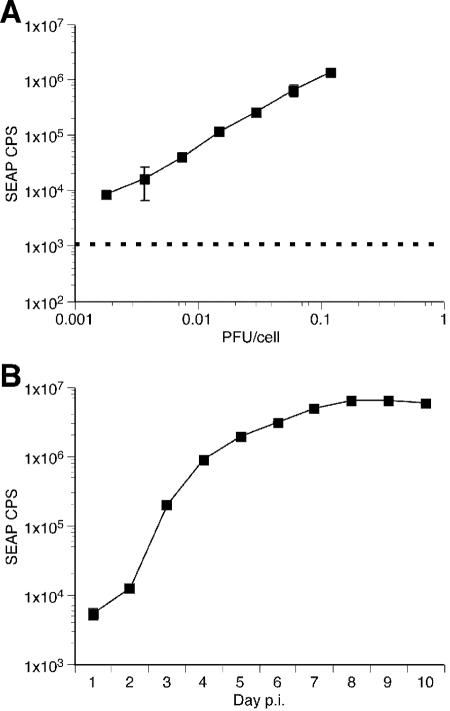
Similarly, RFs were infected with increasing MOIs of RRV-SEAP ranging from 0.0018 to 0.12 PFU/cell. Because SEAP is secreted into the medium along with RRV-SEAP virus during amplification, the viral inoculum contains carryover SEAP activity. Therefore, cultures were washed five times with HBSS and fed DH20 medium at 16 h p.i. A fraction of the culture supernatant was removed from each culture 4 days p.i and examined for SEAP activity. SEAP expression increased in a MOI-dependent manner over approximately two orders of magnitude (Fig. 5A). Using measurement of SEAP expression levels, the time course of RRV-SEAP expansion throughout RF cultures was measured. One day after seeding, RFs were infected with RRV-SEAP at 0.002 PFU/cell. At 16 h p.i., cultures were washed five times with HBSS and refed DH20 medium. At this time point and each day afterwards, an aliquot of cell-free medium was removed from each flask and replaced with DH20 medium until complete lysis of the culture was observed (day 10). SEAP expression in RF cultures infected with RRV-SEAP was determined in triplicate for each day (Fig. 5B). SEAP expression increased steadily until day 7 when lytic replication was observed throughout the culture. Days 7 to 10 represent the time frame during which the remaining cells lysed and detached from the tissue culture flask.
FIG. 5.
SEAP expression in rhesus fibroblasts after infection with RRV-SEAP. (A) SEAP expression in rhesus fibroblasts after infection with RRV-SEAP is MOI dependent. At 24 h postseeding, triplicate cultures of rhesus fibroblasts were infected with increasing MOIs of RRV-SEAP. At day 4 p.i., cultures were examined for SEAP expression (r2 = 0.9973). The results show the SEAP counts/s (CPS) with standard deviation for each PFU/cell indicated. The background SEAP CPS is represented by the dashed line. (B) Time course of SEAP expression after infection of rhesus fibroblasts with RRV-SEAP. At 24 h postseeding, triplicate cultures of rhesus fibroblasts were infected with 0.002 PFU/cell of RRV-SEAP. At day 1 p.i., cultures were rinsed five times with HBSS and refed with DH20 medium. One milliliter of cell-free supernatant was removed each day and replaced with 1 ml of DH20. The SEAP counts/s (CPS), with standard deviation, are shown for each day p.i. Where no error bar is shown, the error falls within the size of the symbol.
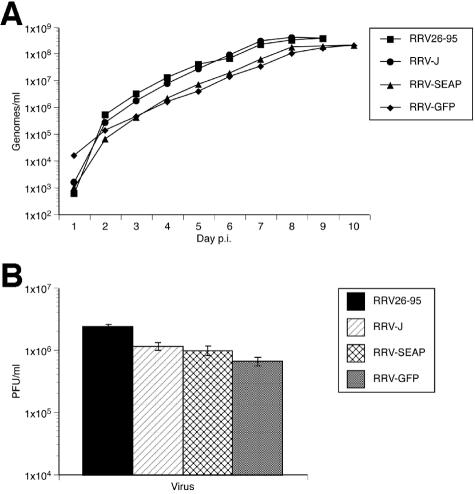
We next determined if the replication kinetics and/or yield titers differed between the parental RRV26-95 and the recombinant RRVs generated by cotransfection of overlapping cosmids, including RRV-GFP and RRV-SEAP. One day postseeding, triplicate cultures of RFs were infected at 0.002 PFU/cell with either RRV26-95, RRV-J (Table 2; contains no reporter gene), RRV-SEAP, or RRV-GFP. As before, cultures were washed five times with HBSS and refed DH20 medium at 16 h p.i. An aliquot of cell-free medium was removed from each flask daily and replaced with DH20 medium until complete lysis of the cultures. Viral DNA was isolated from a fraction of each sample and analyzed for RRV genome copies by real-time PCR. Measurement of viral genome copy number by this method was previously shown to correlate strongly with infectious titer (10). Complete lysis and cell detachment occurred on day 10 p.i. for the RRV-SEAP and RRV-GFP viruses and on day 9 p.i. with RRV26-95 and RRV-J. In addition, the genome copy number at each time point was not significantly different between the parental and recombinant viruses (Fig. 6A). The growth rate was also similar among the parental and recombinant RRV strains. Additionally, titers of samples of the cell-free supernatant collected at the completion of lytic replication (day 9 for RRV26-95 and RRV-J or day 10 for RRV-SEAP and RRV-GFP) were determined in triplicate on fresh cultures of RFs. End-point titers were similar between the parental and recombinant RRV strains (Fig. 6B).
FIG. 6.
Growth kinetics of parental and recombinant RRV in rhesus fibroblasts. (A) Recombinant RRVs replicate with similar kinetics to the parental RRV26-95 as measured by real-time PCR. At 24 h postseeding, triplicate cultures of rhesus fibroblasts were infected at 0.002 PFU/cell with RRV26-95, RRV-J, RRV-SEAP, or RRV-GFP. At day 1 p.i., cultures were rinsed five times with HBSS and refed with DH20. One milliliter of cell-free supernatant was removed each day and replaced with 1 ml of DH20 medium. Triplicate samples of viral DNA isolated at each day were examined by real-time PCR. Results show the average genomes/ml, with standard deviation, at the indicated day p.i. (B) End-point titers of recombinant RRVs are similar to RRV26-95. Cell-free supernatant isolated at either day 9 (RRV26-95 and RRV-J) or 10 (RRV-SEAP and RRV-GFP) p.i. was diluted in DH20 medium and used to determine the end-point titer for each virus at the completion of lytic replication in rhesus fibroblasts. Each bar represents the average PFU/ml, with standard deviation, for the indicated virus.
Neutralization of RRV by sera from rhesus monkeys naturally infected with RRV.
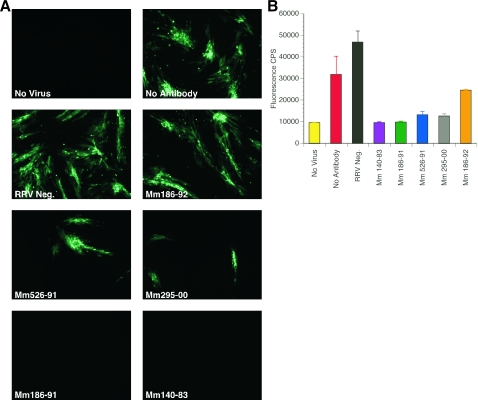
Previous studies showed that detection of the presence or absence of anti-RRV antibodies by whole-virus ELISA was a reliable means of identifying RRV-positive and RRV-negative monkeys (8). Sera from 14 rhesus monkeys from the NEPRC colony were used to validate our assays for RRV neutralization. RRV-negative samples included sera from two monkeys (Mm 288-94 and 320-98) that were delivered by cesarean section, hand-reared separately from other rhesus monkeys, and subsequently kept only with other such super-clean monkeys free of eight common infectious agents of rhesus monkeys, including RRV. Heat-inactivated sera from one RRV-negative and five RRV-positive monkeys were used initially to assay neutralization of RRV-GFP. RRV-GFP (0.04 PFU/cell) was preincubated with the sera at a 1:10 dilution for 3 h and then inoculated onto RFs in 24-well plates. After 16 h p.i., cultures were washed five times with HBSS and refed DH20 medium. Cultures were examined at day 4 p.i. for GFP expression by fluorescence microscopy (Fig. 7). As expected, RRV-GFP preincubated with medium alone (Fig. 7, No Antibody) or with sera from an RRV-negative animal (Fig. 7, RRV Neg.) was not neutralized. The ability of sera from RRV-positive monkeys to neutralize RRV-GFP varied among the animals tested. While sera from Mm 186-91 and 140-83 appeared to completely neutralize RRV-GFP at a 1:10 dilution, sera from Mm 526-91 and 295-00 only partially inhibited infection of RFs (Fig. 7). Serum from Mm 186-92 did not neutralize RRV-GFP (Fig. 7) at a detectable level. No correlation was observed between the ability of a serum sample to neutralize RRV-GFP and the level of RRV ELISA positivity.
FIG. 7.
Neutralization of RRV-GFP by sera from rhesus monkeys naturally infected with RRV. (A) Fluorescence microscopy of RRV-GFP neutralization. RRV-GFP (MOI of 0.04 PFU/cell) was incubated with either DH20 medium (No Antibody), sera from a RRV-negative rhesus monkey (RRV Neg.) or sera from rhesus monkeys naturally infected with RRV (Mm 186-92, 526-91, 295-00, 186-91, and 140-83). After a 3-h incubation at 37°C, rhesus fibroblasts were inoculated with either DH20 medium (No Virus) or the virus-sera mixture. At 24 h p.i., cultures were rinsed five times with HBSS and refed with DH20. At day 4 p.i., cultures were examined by fluorescence microscopy for GFP expression. (B) Detection of RRV-GFP neutralization by GFP emission intensity. Each bar represents the average GFP counts/s (CPS), with standard deviation, for each rhesus monkey sera tested.
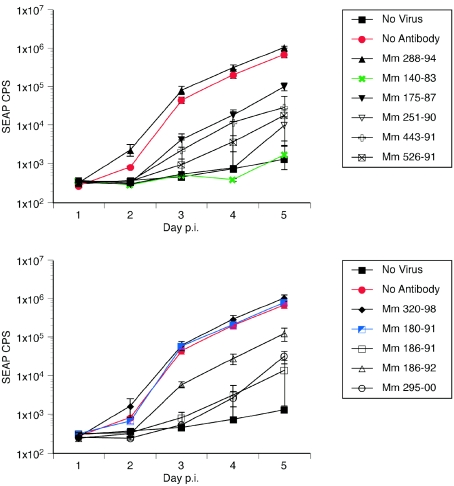
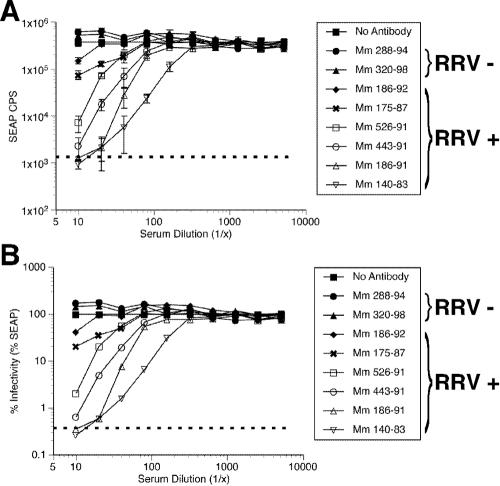
We next investigated use of the SEAP reporter RRV for measurement of neutralization. The ease of measurement of SEAP enzymatic activity, greater sensitivity, and greater dynamic range of measurement predict distinct potential advantages to the use of the RRV-SEAP reporter virus for assay of neutralization. Heat-inactivated sera at a 1:10 dilution from two RRV-negative and nine RRV-positive monkeys were used to assay neutralization of RRV-SEAP. RRV-SEAP (0.006 PFU/cell) was preincubated with the diluted sera for 3 h and then inoculated onto RFs in 24-well plates. At 16 h p.i., cultures were washed five times with HBSS and refed with DH20. An aliquot of cell-free medium was removed from each well daily for 5 days and replaced with DH20 medium. The ability of the sera to inhibit RRV-SEAP infection of RFs was then examined (Fig. 8). As was observed in RRV-GFP neutralization studies, the sera from RRV-positive monkeys neutralized RRV-SEAP with various rates of effectiveness. With the exception of Mm 180-91, sera from RRV-positive monkeys inhibited the infectivity of RRV-SEAP (Fig. 8). Sera from the two RRV-negative monkeys did not neutralize the RRV-SEAP virus. To determine the 50% neutralization titer, serial twofold dilutions of sera from naturally infected RRV-positive monkeys were preincubated with 0.006 PFU of RRV-SEAP/cell. SEAP counts measured at day 4 p.i. further demonstrate the precision of the assay and illustrate the wide range of neutralization abilities among the sera from RRV positive animals (Fig. 9A). At dilutions of 1:10, sera from Mm 186-92 and 175-87 decreased RRV-SEAP infectivity by approximately 50 to 80%, while sera from Mm 526-91, 443-91, 186-91, and 140-91 neutralized RRV-SEAP by >98% (Fig. 9B). The 50% neutralization titers calculated from these data ranged from approximately 1:14 for Mm 186-92 to 1:220.5 for Mm 140-83 (Table 3). Sera from younger RRV-positive rhesus monkeys emulated the 50% neutralization titers determined for the aged animals (Table 3).
FIG. 8.
SEAP expression by RRV-SEAP following neutralization by sera from rhesus monkeys naturally infected with RRV. RRV-SEAP (MOI of 0.006 PFU/cell) was incubated with either DH20 medium (No Antibody), sera from RRV-negative rhesus monkeys (Mm 288-94 and 320-98) or sera from rhesus monkeys naturally infected with RRV (Mm 140-83, 175-87, 180-91, 186-91, 186-92, 251-90, 295-00, 443-91, and 526-91). After a 3-h incubation at 37°C, rhesus fibroblasts were inoculated with either DH20 medium (No Virus) or the virus-sera mixture. At 24 h p.i., cultures were rinsed five times with HBSS and refed with DH20. At days 1 to 5 p.i., an aliquot of medium was removed to determine the level of SEAP expression and replaced with DH20 medium. The results show the average SEAP counts/s (CPS), with standard deviation, for each day p.i.
FIG. 9.
Neutralization of RRV-SEAP by sera from rhesus monkeys naturally infected with RRV. RRV-SEAP (MOI of 0.006 PFU/cell) was incubated with either DH20 medium (No Antibody), serial dilutions of sera from RRV-negative rhesus monkeys (Mm 288-94 and 320-98) or serial dilutions of sera from rhesus monkeys naturally infected with RRV (Mm 186-92, 175-87, 526-91, 443-91, 186-91, and 140-83). After a 3-h incubation at 37°C, rhesus fibroblasts were inoculated with either DH20 medium (dotted line) or the virus-sera mixture. At 24 h p.i., cultures were rinsed five times with HBSS and refed with DH20. At day 4 p.i., an aliquot of medium was removed to determine the level of SEAP expression. The results show the average SEAP counts/s (CPS) (A), with standard deviation, and the percent infectivity (B), for each serum dilution.
TABLE 3.
Fifty percent neutralization antibody titers in sera from uninfected and naturally infected RRV+ rhesus macaques
| Animal | 50% RRV neutralization titera | RRV statusb |
|---|---|---|
| 288-94 | <10 | − |
| 320-98 | <10 | − |
| 232-03 | <10 | − |
| 248-03 | <10 | + |
| 186-92 | 13.99 | + |
| 288-03 | 17.7 | + |
| 526-91 | 36.25 | + |
| 175-87 | 40.26 | + |
| 443-91 | 65.48 | + |
| 488-03 | 60.87 | + |
| 269-03 | 61.27 | + |
| 186-91 | 77.3 | + |
| 140-83 | 220.5 | + |
| 541-03 | 230.5 | + |
Reciprocal dilution using RRV-SEAP.
Determined by whole-virus ELISA (8).
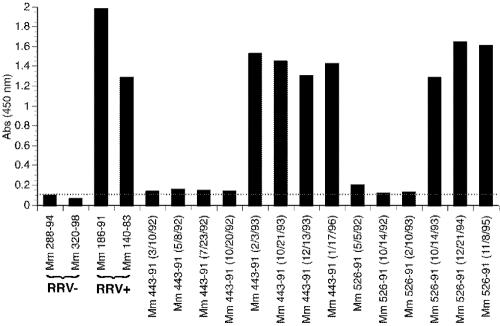
Since we observed a wide variation in neutralizing activity in the sera of RRV-positive monkeys regardless of age, we next analyzed sequential serum samples from individual monkeys to assess the consistency of neutralization titers over time. By ELISA, seroconversion occurred between serum sampling dates of 20 October 1992 and 3 February 1993 for Mm 443-91 and 10 February 1993 and 14 October 1993 for Mm 526-91 (Fig. 10 and Table 4). Detection of neutralizing activity lagged somewhat behind seroconversion measured by whole-virus ELISA (Table 4). Nonetheless, the time of seroconversion correlated well with the appearance of neutralizing activity. Positive sera obtained from Mm 443-91 and 526-91 over an 8- to 9-year period consistently neutralized RRV relatively weakly (Table 4). Positive sera obtained from monkey 26-95, the donor animal of RRV26-95, consistently neutralized RRV relatively strongly over time (Table 4).
FIG. 10.
ELISA for RRV antibodies in sera from naturally infected rhesus monkeys. Maxi-sorb plates coated with RRV were incubated with sera from Mm 443-91 and 526-91 isolated at different dates after birth. After exposure, RRV positivity was determined for these sera by comparison to sera from RRV-negative (Mm 288-94 and 320-98) and RRV-positive (Mm 186-91 and 140-83) monkeys.
TABLE 4.
Fifty percent neutralization titers over time of rhesus monkeys naturally infected with RRV
| Animal | RRV statusa | Date (mo/day/yr) | 50% RRV neutralization titerb |
|---|---|---|---|
| 26-95c | − | 3/7/96 | <10 |
| 26-95 | + | 7/16/96 | <10 |
| 26-95 | + | 4/10/97 | 224 |
| 26-95 | + | 2/25/98 | 363.5 |
| 443-91 | − | 10/20/92 | <10 |
| 443-91 | + | 2/3/93 | <10 |
| 443-91 | + | 10/21/93 | <10 |
| 443-91 | + | 12/13/94 | <10 |
| 443-91 | + | 1/17/96 | 22.1 |
| 443-91 | + | 11/16/04 | 65.48 |
| 526-91 | − | 2/10/93 | <10 |
| 526-91 | + | 10/14/93 | <10 |
| 526-91 | + | 12/21/94 | 19.32 |
| 526-91 | + | 11/8/95 | 15 |
| 526-91 | + | 11/16/04 | 36.25 |
Determined by whole-virus ELISA (8).
Reciprocal dilution using RRV-SEAP.
Mm 26-95 was born in 1995, and the other monkeys were born in 1991 within the NEPRC colony.
Neutralization of RRV-SEAP by serum is antibody mediated.
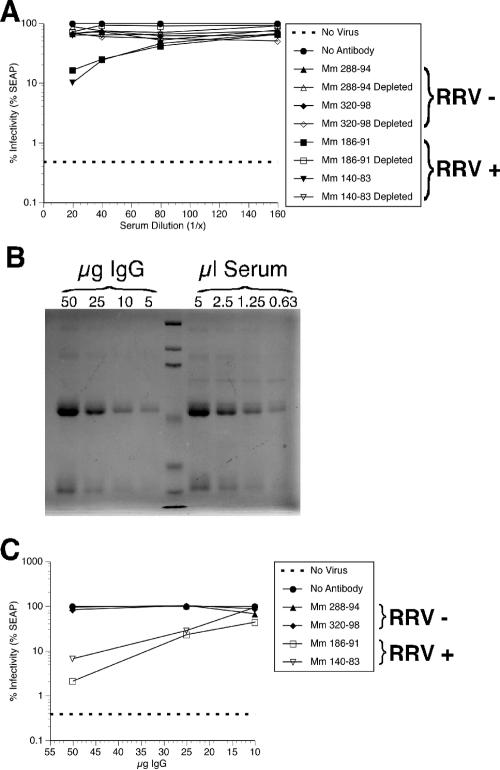
Control experiments were performed to determine if the neutralization of RRV-SEAP by heat-inactivated sera from RRV-positive rhesus monkeys was antibody mediated. Initially, IgG was depleted from the serum of RRV-negative and -positive rhesus monkeys by incubation with a mixture of protein A/G-Sepharose overnight at 4°C. RRV-SEAP (0.006 PFU/cell) was preincubated with dilutions of the IgG-depleted sera and undepleted sera and inoculated onto fresh RF cultures. At day 4 p.i., SEAP activity was measured. RRV-negative and IgG-depleted sera failed to neutralize RRV-SEAP, whereas the undepleted sera isolated from RRV-positive monkeys neutralized RRV-SEAP (Fig. 11A). To further demonstrate that the neutralization of RRV by sera was antibody mediated, IgG was purified from the sera of RRV-negative and -positive monkeys by passage through a protein A column. The relative levels of IgG in the monkey sera were determined by immunoprecipitation of known micrograms of the purified IgG and various volumes of sera. The samples were then electrophoresed through polyacrylamide gels and examined by Coomassie staining. Comparison of the staining densities between the purified IgG and serum samples (50 μg is approximately 5 μl, e.g.) revealed that the IgG concentration in the sera was approximately 10 mg/ml (Fig. 11B; results from Mm 186-91 are shown), which agrees well with known averages for rhesus monkeys. To relate the IgG concentration to neutralization efficiencies, 0.006 PFU of RRV-SEAP/cell was preincubated with increasing concentrations of purified IgG isolated from the sera of RRV-negative and -positive rhesus monkeys. Fresh cultures of RFs were then inoculated with the IgG and virus mixture, and SEAP activity was measured at day 4 p.i. Purified IgG isolated from RRV-positive monkey sera neutralized RRV-SEAP; however, IgG isolated from the sera of RRV-negative monkeys did not neutralize RRV-SEAP (Fig. 11C). Specifically, the infectivity of RRV-SEAP after preincubation with 25 μg of IgG from RRV-positive monkeys was approximately 20%. As shown in Fig. 11A, 20% RRV-SEAP infectivity corresponds to an RRV-positive serum dilution of between 1:20 and 1:40, or 2.5 μl and 1.25 μl of serum, respectively. Therefore, based on the approximated IgG concentration of sera shown in Fig. 11B, the dilutions required to reduce infectivity of 0.006 PFU of RRV-SEAP/cell to 20% correspond to between 12.5 and 25 μg. This result agrees with the concentration of IgG required to reduce RRV-SEAP infectivity to 20% shown in Fig. 11C.
FIG. 11.
Neutralization of RRV-SEAP is antibody mediated. (A) Depletion of IgG from rhesus monkey serum abrogated neutralization of RRV. To remove IgG from the serum, serum from either RRV-negative rhesus monkeys (Mm 288-94 and 320-98) or rhesus monkeys naturally infected with RRV (Mm 186-91 and 140-83) was incubated with immobilized protein A/G overnight at 4°C. The resulting IgG-depleted serum and nondepleted serum were diluted as indicated and analyzed for their ability to neutralize RRV-SEAP as described previously. At each indicated serum dilution, the percent infectivity or RRV-SEAP is shown. (B) Coomassie staining to reveal relative levels of IgG in serum (results from Mm 186-91 is shown). Known amounts of previously isolated IgG and decreasing volumes of sera from Mm 186-91 were immunoprecipitated with immobilized protein A/G. After polyacrylamide gel electrophoresis, gels were stained with Coomassie blue to reveal the resulting proteins. (C) Purified IgG from the sera of rhesus monkeys naturally infected with RRV neutralizes RRV-SEAP. RRV-SEAP (MOI of 0.006 PFU/cell) was incubated with either DH20 medium (No Antibody) or known amounts of IgG isolated from either RRV-negative rhesus monkeys (Mm 288-94 and 320-98) or rhesus monkeys naturally infected with RRV (Mm 186-91 and 140-83) diluted in DH20 medium. After a 3-h incubation at 37°C, rhesus fibroblasts were inoculated with either DH20 medium (No Virus) or the virus-IgG mixture. At 24 h p.i., cultures were rinsed five times with HBSS and refed with DH20. At day 4 p.i., an aliquot of medium was removed to determine the level of SEAP expression. The results show the percent infectivity for each IgG concentration.
Neutralization of RRV by sera from rhesus monkeys experimentally infected with RRV26-95.
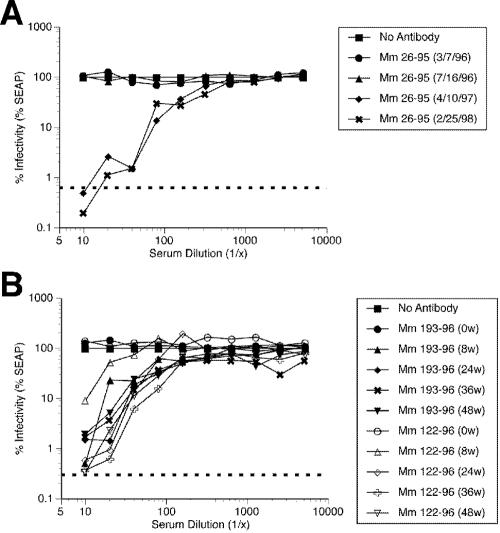
We next investigated the kinetics of appearance and strength of neutralization of RRV26-95 (RRV-SEAP) using sequential sera from monkeys infected naturally or experimentally with RRV26-95. Mm 26-95 is the naturally infected monkey that was the source of the RRV26-95 isolate used to generate RRV-GFP and RRV-SEAP. Similar to results from Mm 443-91 and 526-91, the ability to detect neutralizing activity in serum from Mm 26-95 lagged somewhat in time relative to the first detection of anti-RRV antibodies by ELISA. Sera from Mm 26-95 yielded the highest neutralizing titers (1:224 and 1:363) against RRV26-95 (RRV-SEAP) that we have observed to date (Fig. 12A and Table 4). The rhesus monkeys infected experimentally with RRV26-95 that were the source of the sera for neutralization assays have been described previously (18). The neutralization titers in sera from rhesus monkeys experimentally infected with RRV26-95 were also relatively high (Fig. 12B and Table 5). Serum samples were obtained at 8, 24, 36, and 48 weeks after intravenous injection of RRV26-95. Sera obtained prior to RRV26-95 infection failed to neutralize RRV-SEAP; however, sera obtained after infection neutralized RRV-SEAP with increasing efficacy over time after infection. The neutralization titers plateaued by week 36, where they remained until the conclusion of the experiment (week 48).
FIG. 12.
Neutralization of RRV-SEAP by sera from Mm 26-95 and rhesus monkeys experimentally infected with RRV26-95. (A) Neutralization of RRV-SEAP by Mm 26-95 sera. RRV-SEAP (MOI of 0.006 PFU/cell) was incubated with either DH20 medium (No Antibody) or various dilutions of serum from Mm 26-95 isolated at different dates. After a 3-h incubation at 37°C, rhesus fibroblasts were inoculated with either DH20 medium (dotted line) or the virus-sera mixture. At 24 h p.i., cultures were rinsed five times with HBSS and refed with DH20. At day 4 p.i., an aliquot of medium was removed to determine the level of SEAP expression. The results show the percent infectivity for each serum dilution at the serum isolation dates indicated. (B) Neutralization of RRV-SEAP by sera from rhesus monkeys experimentally infected with RRV26-95. At 8, 24, 36, and 48 weeks after inoculation of RRV-naïve rhesus monkeys with RRV26-95, sera was collected and used to determine its efficacy in neutralizing RRV-SEAP as previously described. The results show the percent infectivity for each serum dilution at the indicated week postinoculation.
TABLE 5.
Fifty percent RRV-SEAP neutralization titers of sera from rhesus monkeys experimentally infected with RRV26-95
| Animal | RRV statusa | Inoculum | Week(s) p.i. | 50% RRV neutralization titerb |
|---|---|---|---|---|
| 193-96 | − | RRV26-95 | 0 | <10 |
| 193-96 | + | RRV26-95 | 8 | 67.59 |
| 193-96 | + | RRV26-95 | 24 | 127.2 |
| 193-96 | + | RRV26-95 | 36 | 141.3 |
| 193-96 | + | RRV26-95 | 48 | 156 |
| 122-96 | − | RRV26-95 | 0 | <10 |
| 122-96 | + | RRV26-95 | 8 | 19.7 |
| 122-96 | + | RRV26-95 | 24 | 71 |
| 122-96 | + | RRV26-95 | 36 | 145.1 |
| 122-96 | + | RRV26-95 | 48 | 126.4 |
Determined by whole-virus ELISA (8).
Reciprocal dilution using RRV-SEAP.
DISCUSSION
Since its initial description in 1997 (8), RRV has served as a useful model for the study of KSHV infection in cultured cells and in vivo (3, 18, 25). The availability of a lytic cell culture system has enabled studies of RRV viral structure (20, 26), transcriptional profiling (9), and replication (10). However, studies focusing on the contribution of individual RRV genes thus far have been limited by the lack of an efficient genetic system with which to manipulate the RRV genome. We report here the development of a genetic system for the generation of mutant and recombinant RRV. The genomes of numerous herpesviruses, including herpes simplex virus (7, 21), Epstein-Barr virus (24), varicella-zoster virus (6, 19), herpesvirus saimiri (12), and human and mouse cytomegaloviruses (11, 16), have been reconstituted by homologous recombination of overlapping cosmids, yielding replication-competent virus in cell culture. In an analogous manner, we set out to define an infectious set of overlapping cosmid clones that span the entire RRV genome including the terminal repeats. Of the 14 combinations of overlapping cosmids that were examined, only one combination failed to yield replication-competent RRV. Replication-competent RRV was produced only when cosmid clones were first transfected into 293T cells, presumably due in part to the high transfection efficiency of these cells. Because 293T cells release only low levels of RRV, amplification to high titer was performed in the permissive Rf388-93 rhesus monkey cell line.
Both intergenic regions and open reading frames deemed nonessential in other herpesviruses were considered for reporter gene insertion. Previous studies have demonstrated a natural tendency for retroviruses (reticuloendotheliosis virus and avian leukosis virus) to insert in regions close to terminal or internal repeat regions of herpesviruses, including herpesvirus of turkeys and Marek's disease virus (13, 15). Furthermore, these insertions were genetically stable over multiple viral passages. As the unique long region of the RRV genome is flanked by terminal repeats, we selected a convenient restriction site upstream from the R1 open reading frame for insertion of GFP and SEAP reporter gene cassettes. In agreement with other herpesvirus cosmid systems (4, 11), the RRV cosmids were too large (>46 kbp including terminal repeat regions) for routine genetic manipulation. A sub-cosmid plasmid clone was generated by truncation of the existing ah28 cosmid. Having made this alteration, the CMV immediate-early promoter-directed GFP or SEAP cassettes were cloned into the truncated sub-cosmid, which was subsequently cotransfected with the remaining overlapping cosmids that span the RRV genome. Characterization of the resultant RRV-GFP and RRV-SEAP viruses in rhesus fibroblasts demonstrated that these recombinant viruses were replication competent, that reporter gene expression was MOI dependent, and that reporter gene expression could be used as a measure of virus replication. Recombinant RRV generated by cotransfection of overlapping cosmids replicated in rhesus fibroblasts with growth kinetics and to titers that were similar to those of the uncloned parental RRV. Furthermore, with the exception of the inserted reporter gene cassettes, cosmid-derived recombinant RRVs were genetically indistinguishable from wild-type RRV26-95 on the basis of restriction endonuclease fragmentation patterns.
Several lines of evidence document the specificity of the neutralizing assays that we describe here. Monkeys negative for RRV antibodies by ELISA consistently failed to neutralize RRV infectivity. RRV-positive monkeys did exhibit neutralizing activity against RRV26-95 as long as sufficient time had elapsed from the time of seroconversion. RRV-negative monkeys that had been inoculated with RRV26-95 developed neutralizing activity by 8 weeks postinoculation. Naturally infected monkeys with low levels of neutralizing activity against RRV26-95 exhibited consistent low titers over time. A naturally infected monkey with high neutralizing activity against RRV26-95 consistently exhibited high titers over time. The neutralizing activity was assigned to the IgG fraction. There was no apparent correlation of strength of RRV26-95 neutralizing activity with anti-RRV antibody titer determined by whole-virus ELISA. The neutralizing assay that employs SEAP reporter activity from RRV-SEAP has a number of distinct advantages over conventional plaque assays for RRV. Use of the SEAP-based neutralization assay is rapid (5 days as opposed to 7 days for plaque assays) and highly sensitive while maintaining a high level of precision. Furthermore, this assay is not technically demanding and is therefore amenable to high throughput analyses.
Our ability to detect neutralizing activity in sera lagged temporally with our ability to detect seroconversion by whole-virus ELISA in naturally infected monkeys. There are several possible explanations or factors that may contribute to this observation. The simplest explanation is that ELISA may be a more sensitive assay for the detection of anti-RRV antibodies. However, it is also possible that neutralizing activity may require a higher affinity interaction that results from antibody maturation. It must also be considered that these naturally infected monkeys were likely infected with strain variants that differ in the sequences of their glycoproteins from those found in RRV26-95 that is the target in the neutralization assay (2). If there is a strain specificity to the strength of the neutralizing antibody response, it may take longer to develop detectable levels of cross-neutralizing activity.
Several lines of evidence suggest that there may indeed be a strain specificity to the strength of the neutralizing antibody response. Of the 12 naturally infected RRV-positive monkeys that we studied, Mm 26-95, the source of the RRV26-95 isolate, had the highest neutralizing activity measured against RRV26-95. The two monkeys infected experimentally with RRV26-95 that were followed for 48 weeks developed neutralizing activities that were only slightly less than those observed in Mm 26-95. The titer of neutralizing activity did not appear to correlate with anti-RRV antibody titer measured by whole-virus ELISA. Monkeys with low titers of neutralizing activity against RRV26-95 consistently exhibited low titers over time. Auerbach et al. (2) have previously documented significant variation in gB sequences among RRV isolates obtained from different rhesus monkeys. More work will be needed to document the influence of glycoprotein sequence variation on susceptibility to neutralization by individual sera and whether indeed there is a strain specificity to the strength of the neutralizing antibody response.
Acknowledgments
We thank Keith Mansfield and Audra Hachey for providing historical rhesus monkey serum samples, Hannah Sanford for technical support, and Jae Jung and Eloisa Yuste for helpful insights and discussions. We also thank Marvin Sommer and Ann Arvin for helpful technical advice.
This work was supported by Public Health Service grants 1R01AI063928 and 1P01DE1438804 to R.C.D., RR00168 to NEPRC, and 5T32AI0724522 to J.P.B.
REFERENCES
- 1.Alexander, L., L. Denekamp, A. Knapp, M. R. Auerbach, B. Damania, and R. C. Desrosiers. 2000. The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J. Virol. 74:3388-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auerbach, M. R., S. C. Czajak, W. E. Johnson, R. C. Desrosiers, and L. Alexander. 2000. Species specificity of macaque rhadinovirus glycoprotein B sequences. J. Virol. 74:584-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergquam, E. P., N. Avery, S. M. Shiigi, M. K. Axthelm, and S. W. Wong. 1999. Rhesus rhadinovirus establishes a latent infection in B lymphocytes in vivo. J. Virol. 73:7874-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bestman-Smith, J., and G. Boivin. 2003. Drug resistance patterns of recombinant herpes simplex virus DNA polymerase mutants generated with a set of overlapping cosmids and plasmids. J. Virol. 77:7820-7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 6.Cohen, J. I., and K. E. Seidel. 1993. Generation of varicella-zoster virus (VZV) and viral mutants from cosmid DNAs: VZV thymidylate synthetase is not essential for replication in vitro. Proc. Natl. Acad. Sci. USA 90:7376-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunningham, C., and A. J. Davison. 1993. A cosmid-based system for constructing mutants of herpes simplex virus type 1. Virology 197:116-124. [DOI] [PubMed] [Google Scholar]
- 8.Desrosiers, R. C., V. G. Sasseville, S. C. Czajak, X. Zhang, K. G. Mansfield, A. Kaur, R. P. Johnson, A. A. Lackner, and J. U. Jung. 1997. A herpesvirus of rhesus monkeys related to the human Kaposi's sarcoma-associated herpesvirus. J. Virol. 71:9764-9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeWire, S. M., M. A. McVoy, and B. Damania. 2002. Kinetics of expression of rhesus monkey rhadinovirus (RRV) and identification and characterization of a polycistronic transcript encoding the RRV Orf50/Rta, RRV R8, and R8.1 genes. J. Virol. 76:9819-9831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeWire, S. M., E. S. Money, S. P. Krall, and B. Damania. 2003. Rhesus monkey rhadinovirus (RRV): construction of a RRV-GFP recombinant virus and development of assays to assess viral replication. Virology 312:122-134. [DOI] [PubMed] [Google Scholar]
- 11.Ehsani, M. E., T. W. Abraha, C. Netherland-Snell, N. Mueller, M. M. Taylor, and B. Holwerda. 2000. Generation of mutant murine cytomegalovirus strains from overlapping cosmid and plasmid clones. J. Virol. 74:8972-8979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ensser, A., A. Pfinder, I. Muller-Fleckenstein, and B. Fleckenstein. 1999. The URNA genes of herpesvirus saimiri (strain C488) are dispensable for transformation of human T cells in vitro. J. Virol. 73:10551-10555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isfort, R. J., Z. Qian, D. Jones, R. F. Silva, R. Witter, and H. J. Kung. 1994. Integration of multiple chicken retroviruses into multiple chicken herpesviruses: herpesviral gD as a common target of integration. Virology 203:125-133. [DOI] [PubMed] [Google Scholar]
- 14.Jenner, R. G., and C. Boshoff. 2002. The molecular pathology of Kaposi's sarcoma-associated herpesvirus. Biochim. Biophys. Acta 1602:1-22. [DOI] [PubMed] [Google Scholar]
- 15.Jones, D., R. Isfort, R. Witter, R. Kost, and H. J. Kung. 1993. Retroviral insertions into a herpesvirus are clustered at the junctions of the short repeat and short unique sequences. Proc. Natl. Acad. Sci. USA 90:3855-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kemble, G., G. Duke, R. Winter, and R. Spaete. 1996. Defined large-scale alterations of the human cytomegalovirus genome constructed by cotransfection of overlapping cosmids. J. Virol. 70:2044-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kodama, T., D. P. Silva, M. D. Daniel, J. E. Phillips-Conroy, C. J. Jolly, J. Rogers, and R. C. Desrosiers. 1989. Prevalence of antibodies to SIV in baboons in their native habitat. AIDS Res. Hum. Retrovir. 5:337-343. [DOI] [PubMed] [Google Scholar]
- 18.Mansfield, K. G., S. V. Westmoreland, C. D. DeBakker, S. Czajak, A. A. Lackner, and R. C. Desrosiers. 1999. Experimental infection of rhesus and pig-tailed macaques with macaque rhadinoviruses. J. Virol. 73:10320-10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niizuma, T., L. Zerboni, M. H. Sommer, H. Ito, S. Hinchliffe, and A. M. Arvin. 2003. Construction of varicella-zoster virus recombinants from parent Oka cosmids and demonstration that ORF65 protein is dispensable for infection of human skin and T cells in the SCID-hu mouse model. J. Virol. 77:6062-6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Connor, C. M., B. Damania, and D. H. Kedes. 2003. De novo infection with rhesus monkey rhadinovirus leads to the accumulation of multiple intranuclear capsid species during lytic replication but favors the release of genome-containing virions. J. Virol. 77:13439-13447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Register, R. B., and J. A. Shafer. 1996. A facile system for construction of HSV-1 variants: site directed mutation of the UL26 protease gene in HSV-1. J. Virol. Methods 57:181-193. [DOI] [PubMed] [Google Scholar]
- 22.Ruff, K., G. B. Baskin, L. Simpson, M. Murphey-Corb, and L. S. Levy. 2003. Rhesus rhadinovirus infection in healthy and SIV-infected macaques at Tulane National Primate Research Center. J. Med. Primatol. 32:1-6. [DOI] [PubMed] [Google Scholar]
- 23.Searles, R. P., E. P. Bergquam, M. K. Axthelm, and S. W. Wong. 1999. Sequence and genomic analysis of a Rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 73:3040-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomkinson, B., E. Robertson, R. Yalamanchili, R. Longnecker, and E. Kieff. 1993. Epstein-Barr virus recombinants from overlapping cosmid fragments. J. Virol. 67:7298-7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong, S. W., E. P. Bergquam, R. M. Swanson, F. W. Lee, S. M. Shiigi, N. A. Avery, J. W. Fanton, and M. K. Axthelm. 1999. Induction of B cell hyperplasia in simian immunodeficiency virus-infected rhesus macaques with the simian homologue of Kaposi's sarcoma-associated herpesvirus. J. Exp. Med. 190:827-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu, X. K., C. M. O'Connor, I. Atanasov, B. Damania, D. H. Kedes, and Z. H. Zhou. 2003. Three-dimensional structures of the A, B, and C capsids of rhesus monkey rhadinovirus: insights into gammaherpesvirus capsid assembly, maturation, and DNA packaging. J. Virol. 77:13182-13193. [DOI] [PMC free article] [PubMed] [Google Scholar]