Abstract
Selenocysteine (Sec), the 21st amino acid in protein, is encoded by UGA. The Sec insertion sequence (SECIS) element, which is the stem-loop structure present in 3" untranslated regions (UTRs) of eukaryotic selenoprotein-encoding genes, is essential for recognition of UGA as a codon for Sec rather than as a stop signal. We now report the identification of a new eukaryotic selenoprotein, designated selenoprotein M (SelM). The 3-kb human SelM-encoding gene has five exons and is located on chromosome 22 but has not been correctly identified by either Celera or the public Human Genome Project. We characterized human and mouse SelM cDNA sequences and expressed the selenoprotein in various mammalian cell lines. The 3" UTR of the human, mouse, and rat SelM-encoding genes lacks a canonical SECIS element. Instead, Sec is incorporated in response to a conserved mRNA structure, in which cytidines are present in place of the adenosines previously considered invariant. Substitution of adenosines for cytidines did not alter Sec incorporation; however, other mutant structures did not support selenoprotein synthesis, demonstrating that this new form of SECIS element is functional. SelM is expressed in a variety of tissues, with increased levels in the brain. It is localized to the perinuclear structures, and its N-terminal signal peptide is necessary for protein translocation.
In addition to the 20 common amino acids found in proteins of all living organisms, an unusual cotranslationally incorporated amino acid, selenocysteine (Sec), is present in several proteins in representatives of the three major domains of life, bacteria, archaea, and eukaryotes. Sec is inserted into polypeptide chains in response to the codon UGA, which requires a Sec-specific tRNA and a specific elongation factor in ribosome-based protein synthesis (1).
Despite having its own codon and specific factors for its biosynthesis and insertion into protein, Sec is a rare amino acid. Only 17 proteins in mammals have been reported to contain Sec. These proteins do not have a common amino acid sequence motif, biological function, tissue expression, or intracellular localization. Hence, sequence analysis is generally not sufficient to determine the presence of Sec in protein. Thus far, the only common feature in eukaryotic selenoprotein-encoding genes (besides Sec-encoding UGA) is the presence of a stem-loop structure, called the Sec insertion sequence (SECIS) element, in the 3" untranslated region (UTR) (19). This structure is essential for insertion of Sec in response to UGA.
The functional part of a eukaryotic SECIS element is composed of a helix containing non-Watson-Crick base pairs UGAN. . . . .NGAN (designated the quartet or core), an unpaired A preceding the quartet, and an unpaired AA motif in the apical loop or bulge that is separated from the quartet by 11 to 12 nucleotides (3, 24). While having low sequence conservation, the secondary structure and free energy of eukaryotic SECIS elements are strictly conserved and can help in the identification of these stem-loop structures in nucleotide sequence databases (14, 18). It has been established that the quartet is involved in the interaction with SECIS-binding protein 2 (SBP2) (5), which is, in turn, essential for the formation of a complex with the Sec-specific elongation factor and Sec tRNA (7, 22). This complex functions by inserting Sec in response to in-frame UGA codons and preventing termination of translation at this site (1). The role of the unpaired AA motif in this process has not been established, although it is invariantly present in every eukaryotic selenoprotein mRNA identified thus far and its mutation results in a dramatic decrease in Sec incorporation into nascent polypeptide chains (2). In this work, we identified a new eukaryotic selenoprotein, designated selenoprotein M (SelM). This periplasmic protein was not detected by major public and private genome projects. We found that Sec is inserted into this protein in response to a new form of SECIS element that lacks the invariant adenosines.
MATERIALS AND METHODS
Sequence determination and homology analyses.
Human and mouse expressed sequence tag (EST) clones were obtained from Research Genetics. Their nucleotide sequences were determined in the University of Nebraska—Lincoln DNA sequencing core facility. BLAST programs and GenBank nonredundant and EST databases were used to analyze the sequences and to determine exon/intron structures within the human SelM-encoding gene. Initial homology analyses of the SelM sequence against the nonredundant database revealed no homologs. However, adaptation of SelM and nonredundant selenoprotein sequences to use Cys in place of Sec in advanced BLAST (PSI-BLAST/PHI-BLAST) searches revealed homology between human SelM and human 15-kDa selenoprotein sequences. SECIS elements were analyzed with the SECISearch program (14; G. V. Kryukov, A. V. Lobanov, and V. N. Gladyshev, unpublished data). This program analyzes candidate SECIS elements for the presence of primary-sequence consensus, secondary structure, and the free-energy parameters characteristic of known eukaryotic SECIS elements. Secondary structures of proteins were predicted with an Sspro program.
Constructs with GFP.
Green fluorescent protein (GFP)-SelM (wild-type SelM), GFP-SelM(Sec TGA>TGT) (the Sec codon, TGA, was mutated to a cysteine codon, TGT), GFP-SelM(quartet TGA>ACT) (the TGA triplet in the SECIS element was changed to ACT to prevent Sec incorporation), GFP-SelM(CCdel) (the CC at positions 567 and 568 was deleted), GFP-SelM(mouse loop CC>AA) (the CC at positions 567 and 568 was changed to AA), GFP-SelM(mouse loop CC>GG) (the CC at positions 567 and 568 was changed to GG), GFP-SelM(mouse loop CC>TT) (the CC at positions 567 and 568 was changed to TT), and GFP-SelM(human loop CC>AA) (the CC at positions 567 and 568 was changed to AA, and in addition, the apical loop of the mouse SECIS element was replaced with a human sequence) constructs were developed by using expression vector pEGFP-C3 (Clontech). The mouse SelM cDNA and mutant plasmids were amplified with primers Sel-15-2M (5"-CGCAACGTCGACATGAGCATCCTACTGTCG-3") and T3 and cloned into the XhoI/Bsp120I sites of pEGFP-C3. GFP-SelM(CC>AA), GFP-SelM(CCdel), GFP-SelM(CC>TT), GFP-SelM(CC>GG), and GFP-SelM(human loop CC>AA) were obtained by the QuickChange site-directed mutagenesis method (Stratagene) using primers SelM(AA>CC) (5"-GAATGAAGCGCTCAGTATAACGGGAGCATCTCCCTTG-3" and 5"-CAAGGGAGATGCTCCCGTTATACTGAGCGCTTCATTC-3"), SelM(human loop AA>CC) (5"-GCGCTCAGCATAACGGGAATACTTCTCTTGCTGAGGGCCGA-3" and 5"-TCGGCCCTCAGCAAGAGAAGTATTCCCGTTATGCTGAGCGC-3"), SelM(CCdel) (5"-GAATGAAGCGCTCAGTATCGGGAGCATCTCC-3" and 5"-GGAGATGCTCCCGATACTGAGCGCTTCATTC-3"), SelM(CC>TT) (5"-GAATGAAGCGCTCAGTATTTCGGGAGCATCTCC-3" and 5"-GGAGATGCTCCCGAAATACTGAGCGCTTCATTC-3"), and SelM(CC>GG) (5"-GAATGAAGCGCTCAGTATGGCGGGAGCATCTCC-3" and 5"-GGAGATGCTCCCGCCATACTGAGCGCTTCATTC-3"), respectively. The GFP-SelMh construct was obtained by amplification of the mutant (U48C) human SelM cDNA with primers Sal-15-2H (5"-CGCATCGTCGACATGAGCCTCCTGTTGCCTCCGCTGG-3") and T3 and cloning of the product into the XhoI/Bsp120I sites of pEGFP-C3. The SelMh-GFP construct was obtained by amplification of a mutant (U48C) human SelM cDNA with primers T7 and Xho-15-2H (5"-GCCACTCGAGGTCAGCGTGGTCCGAAG-3") and cloning of the product into the XhoI site of pEGFP-N1 (Clontech). The fragment encoding N-terminal sequences of SelM was obtained by amplification of human cDNA with primers T7-Nhe (5"-CGATGCTAGCTAATACGACTCACTATAGGG-3") and Age-15-2H (5"-CGAGACCGGTAGGCCGCTCAGACGGTTCCAGTC-3"). The N-GFP-SelMh and N-GFP constructs were made by cloning this fragment into the NheI/AgeI sites of GFP-SelMh and pEGFP-N1, respectively. The N-GFP-SelMhΔ construct, which codes for a SelM form lacking four C-terminal residues, was obtained by mutagenesis of N-GFP-SelMh with 15-2h-142stopF (5"-CCAGAGGAAACTTCGGACTAGGCTGACCTGTAGGTCCG-3") and 15-2h-142stopR (5"-CGGACCTACAGGTCAGCCTAGTCCGAAGTTTCCTCTGG-3"). All plasmids were transformed into Escherichia coli strain NovaBlue (Novagen), and the plasmids were isolated with a Plasmid Maxi Kit (Qiagen).
Cell growth, transfection, and metabolic labeling with 75Se.
Transfection of CV-1, NIH 3T3, and human embryonic kidney (HEK) 293 cells and metabolic labeling of cells with 75Se were carried out as previously described (14). For CV-1 cells, 5 μg of DNA and 30 μl of Lipofectamine (Gibco BRL) were used for transfection of each 60-mm-diameter plate. For NIH 3T3 cells, transfection was carried out with 4 μg of DNA, 20 μl of Lipofectamine, and 12 μl of PLUS Reagent (Gibco BRL). HEK 293 cells were transfected by the calcium phosphate method (21) using 8.8 μg of DNA. The samples were analyzed on sodium dodecyl sulfate (SDS)-10% NuPAGE gels (Novex). 75Se-labeled proteins were visualized with a Storm PhosphorImager system (Molecular Dynamics).
Dual fluorescence imaging confocal microscopy.
CV-1 cells cultured in 60-mm-diameter culture dishes were transfected with the appropriate constructs in the presence of Lipofectamine (Gibco BRL) and incubated for 12 h in a CO2 incubator. A fluorescent ceramide was used as a reference marker for perinuclear structures (endoplasmic reticulum [ER] and Golgi). This reagent has been shown to accumulate in the ER and Golgi and has been previously used to study protein trafficking (11, 12). The transfected cells were rinsed with serum-free Dulbecco modified Eagle medium-10 mM HEPES and then incubated for 25 min at room temperature in the same medium containing 2 μM BODIPY TR ceramide (Molecular Probes). The cells were washed twice in serum-free Dulbecco modified Eagle medium-10 mM HEPES and immediately used for image collection. Double-labeled images of live cells were collected with a water immersion lens using a dual excitation/emission and dual-channel mode on a Bio-Rad MRC1024ES laser scanning microscope.
Northern blot analysis.
A Mouse Adult Tissue Blot (Seegene) was probed with a 0.7-kb 32P-labeled XhoI-BamHI fragment of mouse SelM. Northern Territory-Human Tumor Panel Blots IV and V (Invitrogen) were probed individually with a labeled 0.7-kb human SelM cDNA. Probes were generated by a Rediprime II random prime labeling system (Amersham Pharmacia Biotech) in accordance with the manufacturer's protocol. To analyze mRNA expression of GFP-SelM constructs, total RNA was isolated from transfected CV-1 cells (∼106) with an RNAqueous Kit (Ambion). RNA was loaded onto a denaturing agarose gel and transferred to a Zeta-Probe Blotting Membrane (Bio-Rad). The membrane was probed with human SelM cDNA as a probe and, after stripping, with a 32P-labeled DECAtemplate-β-actin-mouse template (Ambion) as an internal control.
Nucleotide sequence accession numbers.
The sequence data obtained in this study were submitted to the DDBJ/EMBL/GenBank databases under accession numbers AY043487 (human SelM) and AY043488 (mouse SelM).
RESULTS
SelM sequence characterization.
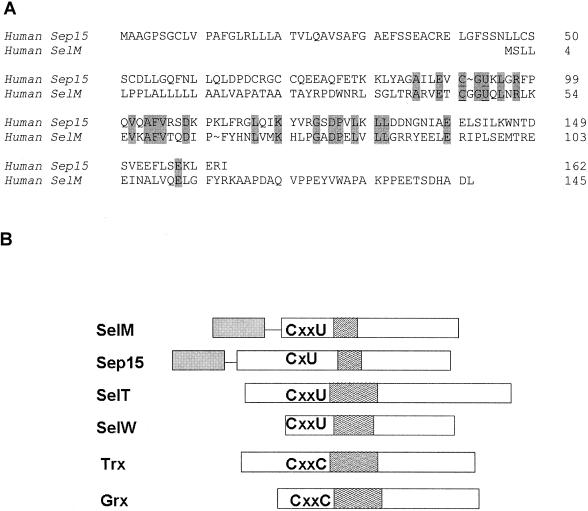
A 0.7-kb cDNA sequence identified in mammalian EST databases encodes an open reading frame of a new protein designated SelM. Sequences of human (Fig. 1A) and mouse SelM cDNA clones were determined. The open reading frame of 145 amino acids begins with an ATG codon in a favorable Kozak context and contains an in-frame TGA as the 48th codon. This codon is predicted to encode Sec. Homologous proteins were found in rats, zebra fish, and other vertebrates (Fig. 1B). Sec was conserved in these homologs and flanked by sequences that exhibited >50% overall sequence identity. A lower eukaryotic (silkworm [Bombyx mori]) homolog of SelM (47% identity to a mammalian SelM) was also identified that contained Cys in place of Sec (Fig. 1B).
FIG. 1.
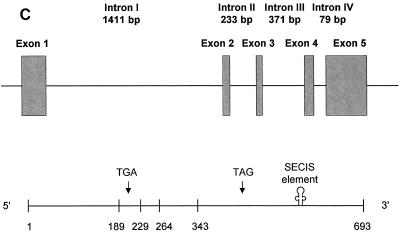
SelM sequences. (A) Human SelM cDNA and protein sequences. The Sec-encoding TGA codon and the TAG stop signal are in bold, and TGA is underlined. U represents Sec. The numbers on the right indicate amino acid residues in the SelM sequence. The SECIS element in the 3" UTR is underlined and in italics. The predicted ER signal peptide is also in italics. (B) Alignment of multiple eukaryotic SelM sequences. Human and mouse sequences were determined in the present study. The GenBank accession numbers for the rat, zebra fish, and B. mori ESTs are AW433967, BE606173, and AU005812, respectively. U indicates Sec. This amino acid residue and the two conserved Cys amino acids upstream of Sec are underlined. In addition, a star above the sequences indicates the position of Sec. Residues conserved in all sequences are highlighted. The N-terminal signal peptide sequences are in italics. The numbers on the right indicate amino acid residues within corresponding sequences. The alignment was generated with the ClustalW program. (C) Organization of the human SelM-encoding gene. The upper portion of the panel shows the locations of exons (boxes) and introns (horizontal lines) in the human SelM-encoding gene. The lower portion of the panel shows the locations of exon-exon junctions and other functional features in the human SelM cDNA. The numbers under the boxes indicate nucleotide numbers within the human SelM cDNA that correspond to exon-exon junctions. The locations of the Sec-encoding TGA codon, the TAG stop codon, and the SECIS element in the 3" UTR are indicated.
The human SelM cDNA sequence matched a region on chromosome 22 (accession numbers AC005005 and ref|NT__011520.2). Analyses of these genomic sequences revealed that the human SelM-encoding gene spanned 3 kb and had five exons and four introns with the splice junctions following the GT…AG rule for the major class introns (Table 1 and Fig. 1C). The coding sequence was present in all five exons with the Sec-encoding UGA located in the second exon and the 3" UTR located in the fifth exon. Chromosome 22 is the first human chromosome whose complete nucleotide sequence was determined by the Human Genome Project (6). It accounts for ∼1.5% of the human genome and is rich in protein-encoding genes (545 genes have been identified) and pseudogenes (134 pseudogenes have been identified). However, the SelM-encoding gene was not correctly annotated by either the public Human Genome Project (17) or the Celera private project (23).
TABLE 1.
Organization of the human SelM-encoding gene
| Exon (size [bp]) | Coding information | Intron (size [bp]) | Splice donora | Splice acceptora |
|---|---|---|---|---|
| 1 (189) | 5" UTR + coding region | I (1411) | TAGAGgtgag | tccagACCTG |
| 2 (40) | Coding region (TGA) | II (233) | GGTGAgtttg | cccagGTGAA |
| 3 (35) | Coding region | III (371) | TTCTAgtatc | tccagTCACA |
| 4 (79) | Coding region | IV (79) | TAGAGgtgag | ctcagCGCAT |
| 5 (350) | Coding region + 3" UTR |
Exon sequences are in uppercase, and intron sequences are in lowercase.
SelM is a selenoprotein.
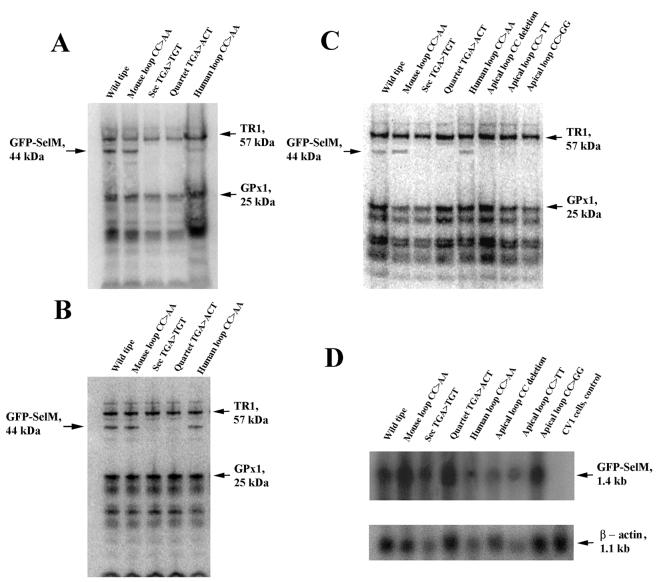
We expressed SelM in mouse NIH 3T3 (Fig. 2A), human HEK 293 (Fig. 2B), and monkey CV-1 (Fig. 2C) cells as a fusion protein with the GFP. The construct encoded GFP, followed by SelM sequences, and also contained the 3" UTR of the SelM-encoding gene, as 3" UTRs of eukaryotic selenoprotein-encoding genes are necessary for Sec insertion (19). Transient expression of the fusion protein was predicted to result in an ∼44-kDa selenoprotein. The fusion selenoprotein was designed such that its mobility on SDS-polyacrylamide gel electrophoresis would be different from those of major naturally occurring selenoproteins expressed in mammalian cells. Indeed, metabolic labeling of cells with 75Se revealed a 44-kDa selenoprotein band (wild-type lanes in Fig. 2A to C). This band was not present in cells transfected with a construct in which the Sec-encoding TGA codon was mutated to a cysteine codon (Sec TGA>TGT in Fig. 2A to C).
FIG. 2.
Characterization of 75Se-labeled GFP-SelM fusion proteins. Transfected cells were grown in the presence of 75Se-selenite, and 75Se-labeled proteins were resolved by SDS-polyacrylamide gel electrophoresis and visualized with a PhosphorImager. The locations and molecular masses of the major selenoproteins TR1 and glutathione peroxidase 1 (GPx1) are indicated on the right. The location of the GFP-SelM fusion selenoprotein is on the left. (A) GFP-SelM expressed in mouse NIH 3T3 cells. NIH 3T3 cells were transfected with the plasmids encoding GFP-SelM (Wild type), GFP-SelM(CC>AA) (Mouse loop CC>AA), GFP-SelM(U48C) (Sec TGA>TGT), GFP-SelM(TGA−) (Quartet TGA>ACT), and GFP-SelM(mAL>hAL) (Human loop CC>AA). (B) GFP-SelM expressed in human HEK 293 cells. HEK 293 cells were transfected with the same plasmids as in panel A. (C) GFP-SelM expressed in monkey CV-1 cells. CV-1 cells were transfected with the plasmids encoding GFP-SelM (Wild type), GFP-SelM(CC>AA) (Mouse loop CC>AA), GFP-SelM(U48C) (Sec TGA>TGT), GFP-SelM(TGA−) (Quartet TGA>ACT), GFP-SelM(mAL>hAL) (Human loop CC>AA), GFP-SelM(CCdel) (Apical loop CC deletion), GFP-SelM(CC>TT) (Apical loop CC>TT), and GFP-SelM(CC>GG) (Apical loop CC>GG). (D) Northern blot analyses of GFP-SelM mRNAs. Cells were transfected with the constructs shown in panel C, and mRNAs were isolated and probed in Northern blot assays with the probe corresponding to SelM cDNA (top). The blot was reprobed to determine actin mRNA levels (bottom).
SelM has a new form of SECIS element that lacks invariant adenosines.
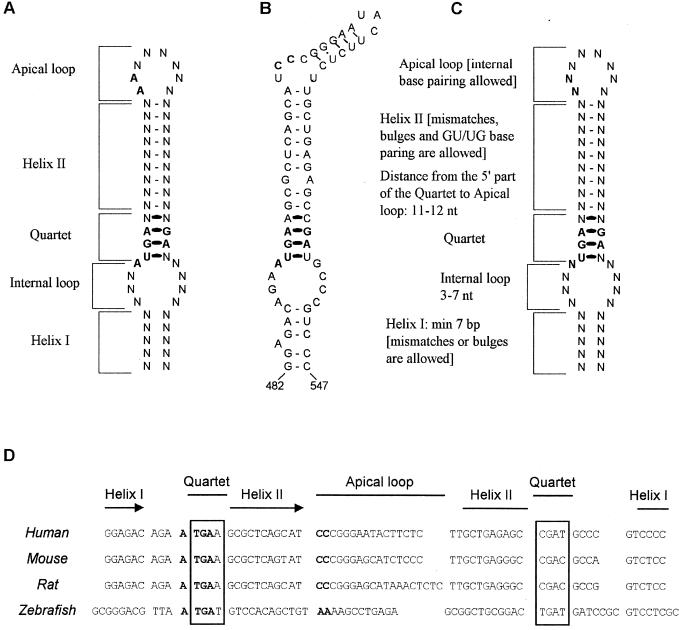
The presence of 75Se in the GFP-SelM fusion protein implied that its 3" UTR contains a functional SECIS element. All previously characterized eukaryotic selenoprotein genes contain a conserved SECIS element in the 3" UTR (Fig. 3A to C). SECIS elements could be found in these genes with SECISearch, a program that identified SECIS elements by searching for primary-sequence consensus, secondary structure, and free-energy parameters of stem-loop structures (14). However, no SECIS elements were initially identified by SECISearch in either human, mouse, or rat SelM mRNAs. Subsequent in silico structural and homology analyses of mammalian SelM mRNAs identified an mRNA structure that conserved the secondary structure and the free-energy criteria of eukaryotic SECIS elements but had two cytidines in place of the adenosines in the apical bulge (Fig. 3B). The predicted stem-loop structure and the CC motif were conserved among rats, mice, and humans (Fig. 3D). A modified version of SECISearch that accepted a CC sequence in place of AA recognized the new structure as a SECIS element. The predicted new form of the eukaryotic SECIS element was most similar to the type 2 SECIS element, which differs from the type I SECIS element in that it contains an additional minihelix in the apical loop. Similarly to both previously proposed SECIS element forms (Fig. 3A and C), the SelM SECIS contained the UGA. . . . .GA motif that forms the non-Watson-Crick base-paired quartet and is involved in SBP2 binding (Fig. 3B).
FIG. 3.
SECIS elements in vertebrate selenoprotein mRNAs. (A) Previously proposed eukaryotic SECIS element structure. The locations of structural features in the stem-loop (helix I, internal loop, quartet, helix II, and apical loop, or bulge) are indicated. N indicates any base. The quartet has non-Watson-Crick interactions. Previously invariant sequences in the SECIS element are in bold. (B) SECIS element in human SelM mRNA. Conserved sequences in the apical bulge (CC replaces the AA motif) and the quartet are in bold. The numbers under the structure indicate the locations of this SECIS element in human SelM mRNA. (C) Eukaryotic SECIS element consensus structure proposed in the present study. Characteristics of different features in the consensus SECIS element are indicated. See the text for further details. (D) Alignment of the SECIS elements in the human, mouse, rat, and zebra fish SelM-encoding genes. Locations of structural features in SECIS elements are indicated. The quartet is boxed. The CC motif in the apical loop of mammalian SelM SECIS elements and the AA motif in the zebra fish structure are in bold. nt, nucleotide.
To determine if the predicted structure was a functional SECIS element, the UGA sequence in the quartet was replaced with ACU. Transient transfection of the resulting construct (Fig. 2A to C, lanes quartet TGA>ACT) into CV-1, NIH 3T3, and HEK 293 cells did not result in expression of the Sec-containing (75Se-labeled) polypeptide. Thus, this 3" UTR mutation disrupted Sec insertion into SelM in human, monkey, and mouse cells and the predicted stem-loop structure is therefore a new form of SECIS element. While its actual structure is not known, comparison of the SelM SECIS element and those SECIS elements for which secondary structures have been demonstrated (24) revealed similar structural features in the SelM SECIS element and previously characterized SECIS elements.
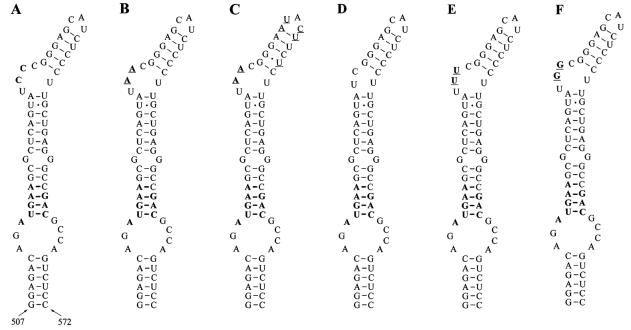
We further tested whether the formation of a typical SECIS element by replacement of CC with AA would change the efficiency of Sec insertion into SelM. For this purpose, we developed a SelM construct containing a mouse SelM SECIS element in which AA was present in place of CC (Fig. 4B). A construct was also developed in which the mouse SECIS element had AA in place of CC, and in addition, its apical loop and minihelix were replaced with the corresponding human sequences (Fig. 4C). Interestingly, these changes had little effect on Sec insertion into mouse SelM expressed in CV-1, NIH 3T3, and HEK 293 cells (Fig. 2). However, other mutations, including deletion of the CC motif (Fig. 4D) and replacement of the CC with UU (Fig. 4E) and GG (Fig. 4F), completely disrupted Sec insertion into SelM (Fig. 2C). Thus, unpaired cytidines are essential for SelM SECIS element function and adenosines, but not other residues, could be tolerated at this position.
FIG. 4.
Mouse SelM SECIS structures. The quartet, the nucleotide preceding the quartet, and the CC motif are in bold. (A) Wild-type SelM SECIS element. (B) Mouse SelM SECIS element in which CC was replaced with AA. Nucleotides that differ from the wild-type structure are underlined. (C) Mouse SelM SECIS element in which CC was replaced with AA and, in addition, the minihelix downstream of this motif and the apical loop were replaced with human sequences. Nucleotides that are different from the wild-type structure are underlined. (D) Mouse SelM SECIS element in which CC was deleted. (E) Mouse SelM SECIS element in which CC was replaced with TT. Nucleotides that differ from the wild-type structure are underlined. (F) Mouse SelM SECIS element in which CC was replaced with GG. Nucleotides that differ from the wild-type structure are underlined.
These effects were not due to decreased stability of SelM mRNA in response to mutations in the SECIS element. We determined GFP-SelM mRNA levels in CV-1 cells that were transfected with various expression constructs. Comparison of fusion protein mRNA levels with those of actin (Fig. 2D) revealed that they did not correlate with Sec insertion into SelM (Fig. 2C).
Further analysis revealed that within SelM-encoding genes, the CC-containing form of the SECIS element was restricted to mammalian genes. Indeed, in contrast to the type 2 SECIS-like CC-containing mammalian structure, the zebra fish SelM-encoding gene contained a typical type 1 SECIS element that exhibited 60% identity to the human SelM SECIS element but had an AA sequence in place of CC and lacked a minihelix (Fig. 3D). This SECIS element was easily recognized when the zebra fish database of ESTs (dbEST) was analyzed with SECISearch. In fact, zebra fish SelM was independently identified as a selenoprotein by applying SECISearch to zebra fish dbEST (Kryukov et al., unpublished). It is possible that, during evolution, SelM SECIS elements not only evolved into either type 1 or type 2 structures but also gave rise to a structure that differs from any other known SECIS element by the presence of cytidines in the loop. These observations and the apparent evolutionary linkage between mammalian and zebra fish SelM SECIS elements provided further support for the conclusion that mammalian SelM has a new form of SECIS element.
SelM is distantly homologous to Sep15, but its Sec-containing motif resembles those of SelW and SelT.
SelM exhibited no homology to any known protein in the nonredundant database when analyzed by default BLAST programs. However, the use of advanced sequence analysis tools revealed a distant homology to the 15-kDa selenoprotein (Sep15) (31% identity in a 73-amino-acid overlap). Moreover, the location of Sec was conserved between these two proteins (Fig. 5A). However, Sep15 had a Cys-Gly-Sec-Lys motif whereas SelM contained Sec in a Cys-Gly-Gly-Sec motif. The latter was similar to sequences found in eukaryotic selenoproteins SelW and SelT. Interestingly, one of the zebra fish SelW forms also had the Cys-Gly-Gly-Sec motif (15).
FIG. 5.
Homology analyses involving SelM. (A) Homology between human SelM and Sep15 selenoproteins. The numbers on the right indicate amino acids in selenoprotein sequences. Conserved residues are highlighted. U is Sec. (B) Comparison of SelM; selenoproteins Sep15, SelT, and SelW; and the thiol/disulfide oxidoreductases thioredoxin (Trx) and glutaredoxin (Grx). Sequences are aligned according to CxxU, CxU, and CxxC motifs. Partially filled boxes downstream of these motifs indicate α-helices. Signal peptides in SelM and Sep15 are shown by filled boxes linked by horizontal lines to other sequences. See the text for other details.
Analysis of predicted secondary structures in SelM and its potential homologs or functional analogs revealed that the CxxU motif in SelM, SelT, and SelW and the CxU motif in Sep15 were followed by an α-helix (Fig. 5B). The presence of CxxU and CxxC motifs upstream of an α-helix is rare in proteins and is often characteristic of a redox center (V. N. Gladyshev, unpublished data). For example, thioredoxins, protein disulfide isomerase, glutaredoxins, and other disulfide oxidoreductases contain the redox CxxC motif that is located upstream of an α-helix and serves as the protein active center (20).
Expression of SelM in mammalian tissues.
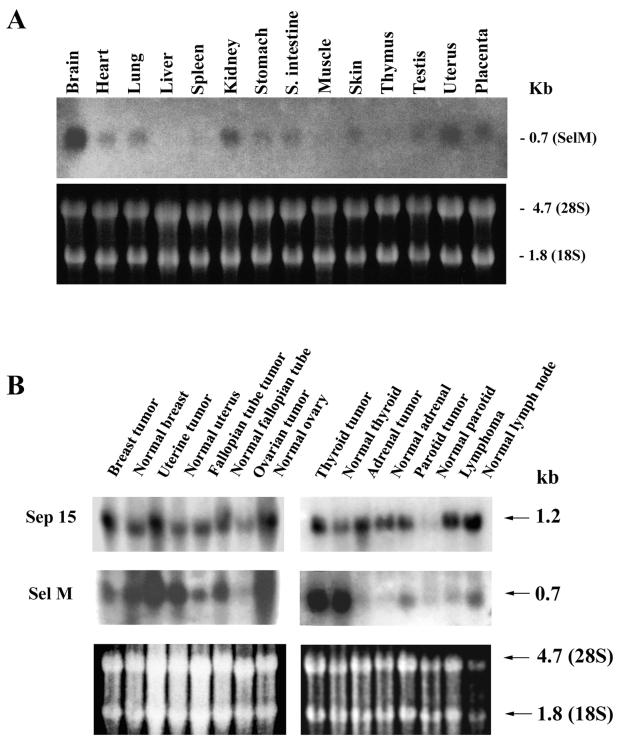
Analyses of the GenBank EST database revealed the presence of 91 full or partial cDNA sequences that matched the human SelM cDNA and more than 50 mouse ESTs. These clones were derived from a variety of different organs and tissues, suggesting a very broad spectrum of moderate expression of SelM mRNAs in many cell types. Direct analyses of SelM mRNA expression in mouse tissues by Northern blot assays also revealed expression of SelM in various tissues. The highest levels of SelM mRNA were observed in the brain (Fig. 6A).
Since SelM is a distant homolog of Sep15, we were interested in comparing the ways in which these two proteins are expressed. Interestingly, Sep15 has been implicated in the role of selenium in cancer prevention and exhibits altered expression in several cancers (10, 16). Thus, we compared matched pairs of tumors and normal samples derived from various human tissues for expression of SelM mRNA and analyzed Sep15 mRNA expression in parallel (Fig. 6B). We found that the expression patterns of the SelM and Sep15 mRNAs were different in human tissues. In addition, although mRNAs for both proteins had altered expression levels in several of the tumors tested (compared to matched control tissues), these changes in mRNA levels did not always correlate between the two proteins.
FIG. 6.
Expression of SelM mRNA. (A) Expression of mouse SelM mRNA. The upper portion shows expression of SelM mRNA in various mouse tissues. The lower portion shows expression of rRNA. (B) Expression of human SelM and Sep15 mRNAs in tumor and matched normal tissues. The upper portion shows expression of Sep15 mRNA. The middle portion shows expression of SelM mRNA. The lower portion shows expression of rRNA. The Northern Territory blot was first probed with a human Sep15 probe, stripped, and then reprobed with a human SelM probe.
Intracellular localization of SelM.
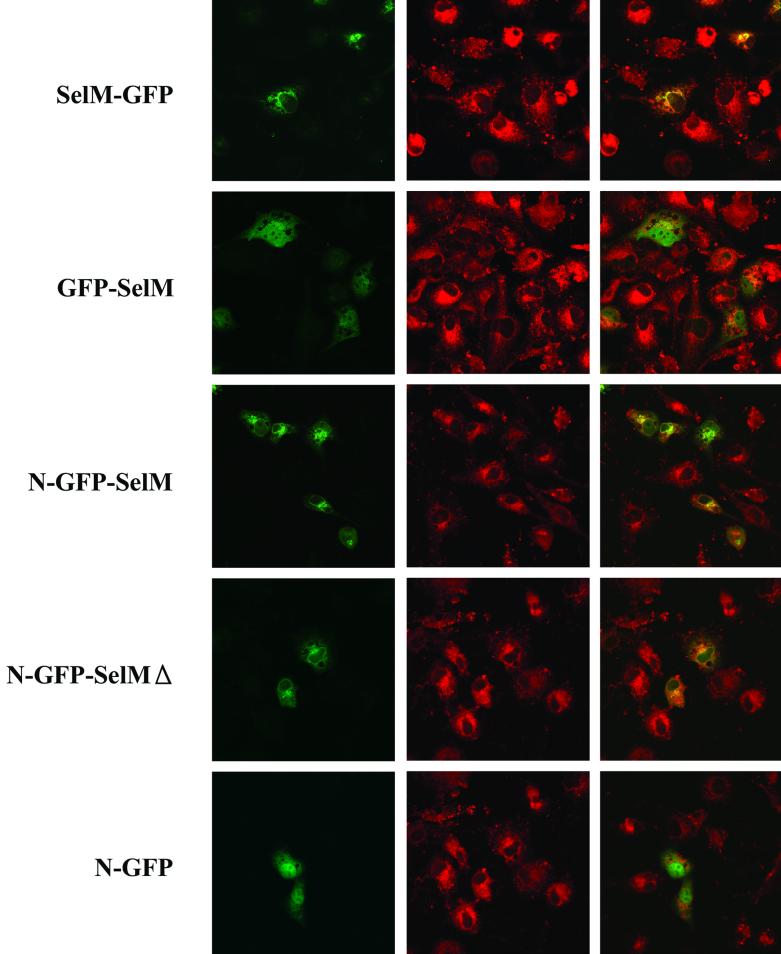
Sequence analyses revealed a putative N-terminal signal peptide within the SelM sequence (Fig. 1B). To determine whether this sequence indeed directs SelM to a particular cellular compartment, we transiently expressed various GFP-fused Cys-for-Sec mutant forms of SelM (Table 2 and Fig. 7). The location of GFP fluorescence was then determined by dual-wavelength confocal microscopy in parallel with the fluorescence of a probe that was targeted to a specific cellular compartment. We found that when a signal peptide of SelM was present as an N-terminal sequence in fusion proteins, the green fluorescence was localized to perinuclear structures (Golgi/ER), whereas no specific localization was seen in the absence of the signal peptide. The insertion of GFP between the N-terminal peptide of SelM and the rest of the SelM sequence also directed the fusion protein to the ER/Golgi structures. ER-resident proteins generally contain a C-terminal tetrapeptide that functions as an ER retention signal. Although the C-terminal sequences of vertebrate SelM proteins exhibited little homology, they terminated with the DL dipeptide (Fig. 1B) that could potentially be a novel retention signal. We tested whether the C-terminal peptide of SelM was essential to the intracellular localization of the protein by developing a construct that encoded a protein lacking the C-terminal tetrapeptide. The fusion protein encoded by this construct localized to the Golgi/ER, suggesting that SelM is maintained in this cellular compartment by a different retention mechanism.
TABLE 2.
Characteristics of SelM-GFP fusion proteinsa
| Fusion proteinb | Compositionc | Predicted molecular mass (kDa)d |
|---|---|---|
| GFP-SelM | 239-aa GFP, 5-aa linker, 145-aa mouse SelM | 43.9 |
| GFP-SelMh | 239-aa GFP, 5-aa linker, 145-aa human SelM | 43.7 |
| SelMh-GFP | 145-aa human SelM, 21-aa linker, 239-aa GFP | 45.4 |
| N-GFP-SelMh | N-terminal 37 aa; 4-aa linker, 239-aa GFP, 5-aa linker, 145-aa human SelM | 47.9 |
| N-GFP-SelMhΔ | N-terminal 37 aa, 4-aa linker, 239-aa GFP, 5-aa linker, 141-aa human SelM | 47.4 |
| N-GFP | N-terminal 37 aa, 4-aa linker, 239-aa GFP | 31.2 |
| GFP | 239-aa GFP | 26.9 |
Design of the constructs is described in Materials and Methods.
SelM is a protein without its 28 N-terminal residues. N is the N-terminal signal peptide of SelM. Δ indicates truncation of four C-terminal residues of SelM.
Composition reflects sizes of SelM fragments, linkers, and GFP that are shown in numbers of amino acids (aa).
Molecular masses were calculated for full-length proteins expressed from the constructs.
FIG. 7.
Expression of GFP-SelM fusion proteins. Confocal images of CV-1 cells expressing various GFP-tagged SelM and control proteins are shown. A set of three images is shown for each construct. Each left panel shows green fluorescence corresponding to transiently expressed fusion proteins, each center panel shows fluorescence of the ER/Golgi marker, and each right panel is an image obtained by merging the left and center panels. Bar, 100 μm. The GFP fusion constructs used in this experiment are shown on the left.
DISCUSSION
We identified a new eukaryotic selenoprotein and described some of its expression and localization characteristics. We also provided evidence that a new type of SECIS element occurs in mammalian SelM and functions in Sec insertion. Prior to this study, the AA motif in the apical loop of SECIS elements was considered to be one of two major invariant characteristics of these stem-loop structures and was used to identify and functionally characterize eukaryotic SECIS elements (14, 18, 19). Moreover, mutation of an unpaired AAA motif to CCC in the rat type 1 deiodinase SECIS element decreased Sec incorporation to 7.9% of that of the wild-type form and most other mutations had even more dramatic effects (3). Likewise, we found that type 2 SECIS elements require AA for their function and the AA → CC mutation disrupts Sec insertion (S. V. Novoselov and V. N. Gladyshev, unpublished data).
In contrast to all other known eukaryotic SECIS elements, the SECIS element in mammalian SelM does not have adenosines in the apical loop and, instead, contains a CC motif. Nevertheless, this stem-loop structure was functional, as demonstrated by the incorporation of 75Se into the selenoprotein in response to the wild-type SelM SECIS element, but not in response to the structure containing a mutated quartet sequence. The CC motif in the apical loop and the overall SECIS element sequence were conserved among mammalian SelM mRNAs and resembled the type 2 SECIS element (9). In the zebra fish SelM-encoding gene, however, a typical type 1 SECIS element containing the AA sequence was present. It is possible that compensatory changes were responsible for the observed accommodation of CC in the unpaired apical bulge in mammalian sequences.
Our study suggests that the absolutely conserved primary sequence in SECIS elements is limited to the UGA. . . . .GA motif in the quartet, which serves as a recognition site for SBP2. Besides the quartet, the only other recognition feature of the SECIS element is its actual three-dimensional structure. These two factors of the stem-loop structure appear to be important for SECIS element function. In addition to an unpaired motif in the apical loop or bulge, a nucleoside preceding the quartet was thought to be conserved throughout eukaryotic SECIS elements. However, this previously invariant A is replaced with G in the Caenorhabditis elegans thioredoxin reductase (TR)-Se-encoding gene (4) and several other eukaryotic selenoprotein-encoding genes (8) and with C in the mouse TR2-encoding gene (unpublished observations). In addition, replacement of A with G, U, or C supported Sec insertion at ∼70, 70, and 30% of A, respectively (8). These observations suggest a new consensus structure of the eukaryotic SECIS element, as shown in Fig. 3C.
The identification of SelM is itself of great interest, as only 17 proteins in mammals are known to contain Sec in their polypeptide chains. The actual numbers of selenoproteins in mammalian genomes are not known, but the steady increase in the number of selenoproteins in recent years illustrates the importance of this class of proteins.
SelM appears to be distantly related to Sep15 and has similarities to selenoproteins containing the CxxU motif. The Sec location is conserved in the Sep15 and SelM sequences, but the Sec-flanking sequences are organized differently. In Sep15, Sec is separated from a conserved Cys residue by only a single residue whereas Sec in SelM is separated from a Cys residue by two glycines. The latter tetrapeptide sequence is similar to the Sec centers of SelT and SelW and is, in fact, identical to that of zebra fish SelW (15). In addition, these motifs and secondary structure patterns relate these selenoproteins to thiol/disulfide oxidoreductases. It is thus possible that Sec and Cys in SelM, SelT, and SelW form a reversible selenosulfide bond.
SelM is located in the perinuclear structures, a rare location for selenoproteins. Only its distant homolog Sep15 has been demonstrated to reside in the ER/Golgi structures. Interestingly, Sep15 was found to be associated with UDP-glucose glycoprotein glucosyltransferase, an ER-resident protein that is involved in the quality control of protein folding (13). Sep15 has also been implicated in cancer prevention (10, 16). Analyses of expression patterns of Sep15 and SelM in matched tumor and control samples, described in this report, revealed changes in mRNA expression linked to malignant transformation. However, whether SelM has any role in cancer prevention remains to be established.
It is also of interest that the human SelM-encoding gene is located on human chromosome 22, which is the first chromosome to be sequenced by the Human Genome Project and is known to contain at least 545 protein-encoding genes. However, the SelM-encoding gene was not previously correctly identified, possibly because currently available gene annotation programs recognize in-frame TGA codons as stop signals. The use of such programs is expected to miss selenoprotein-encoding genes, especially those that, like the SelM-encoding gene, have short coding regions and contain Sec in their N-terminal sequences.
Acknowledgments
K.V.K. and S.V.N. contributed equally to this work.
We thank You Zhou for helping with microscopy and protein localization experiments and Gregory Kryukov for discussions and help with computer analyses.
This work was supported by NIH grants CA80946 and GM61603 (V.N.G.).
REFERENCES
- 1.Atkins, J. F., and R. F. Gesteland. 2000. The twenty-first amino acid. Nature 407:465.. [DOI] [PubMed] [Google Scholar]
- 2.Berry, M. J., L. Banu, J. W. Harney, and P. R. Larsen. 1993. Functional characterization of the eukaryotic SECIS elements which direct selenocysteine insertion at UGA codons. EMBO J. 12:3315-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berry, M. J., G. W. Martin, 3rd, and S. C. Low. 1997. RNA and protein requirements for eukaryotic selenoprotein synthesis. Biomed. Environ. Sci. 10:182-189. [PubMed] [Google Scholar]
- 4.Buettner, C., J. W. Harney, and M. J. Berry. 1999. The Caenorhabditis elegans homologue of thioredoxin reductase contains a selenocysteine insertion sequence (SECIS) element that differs from mammalian SECIS elements but directs selenocysteine incorporation. J. Biol. Chem. 274:21598-21602. [DOI] [PubMed] [Google Scholar]
- 5.Copeland, P. R., J. E. Fletcher, B. A. Carlson, D. L. Hatfield, and D. M. Driscoll. 2000. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 19:306-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunham, I., et al. 1999. The DNA sequence of human chromosome 22. Nature 402:489-495. [DOI] [PubMed] [Google Scholar]
- 7.Fagegaltier, D., N. Hubert, K. Yamada, T. Mizutani, P. Carbon, and A. Krol. 2000. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 19:4796-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fagegaltier, D., A. Lescure, R. Walczak, P. Carbon, and A. Krol. 2000. Structural analysis of new local features in SECIS RNA hairpins. Nucleic Acids Res. 28:2679-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grundner-Culemann, E., G. W. Martin 3rd, J. W. Harney, and M. J. Berry. 1999. Two distinct SECIS structures capable of directing selenocysteine incorporation in eukaryotes. RNA 5:625-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu, Y. J., K. V. Korotkov, R. Mehta, D. L. Hatfield, C. N. Rotimi, A. Luke, T. E. Prewitt, R. S. Cooper, W. Stock, E. E. Vokes, M. E. Dolan, V. N. Gladyshev, and A. M. Diamond. 2001. Distribution and functional consequences of nucleotide polymorphisms in the 3"-untranslated region of the human Sep15 gene. Cancer Res. 61:2307-2310. [PubMed] [Google Scholar]
- 11.Ilgoutz, S. C., K. A. Mullin, B. R. Southwell, and M. J. McConville. 1999. Glycosylphosphatidylinositol biosynthetic enzymes are localized to a stable tubular subcompartment of the endoplasmic reticulum in Leishmania mexicana. EMBO J. 18:3643-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kok, L. W., T. Babia, K. Klappe, G. Egea, and D. Hoekstra. 1998. Ceramide transport from endoplasmic reticulum to Golgi apparatus is not vesicle-mediated. Biochem. J. 333:779-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korotkov, K. V., E. Kumaraswamy, Y. Zhou, D. L. Hatfield, and V. N. Gladyshev. 2001. Association between the 15 kDa selenoprotein and UDP-glucose:glycoprotein glucosyltransferase in the endoplasmic reticulum of mammalian cells. J. Biol. Chem. 276:15330-15336. [DOI] [PubMed] [Google Scholar]
- 14.Kryukov, G. V., V. M. Kryukov, and V. N. Gladyshev. 1999. New mammalian selenocysteine-containing proteins identified with an algorithm that searches for selenocysteine insertion sequence elements. J. Biol. Chem. 274:33888-33897. [DOI] [PubMed] [Google Scholar]
- 15.Kryukov, G. V., and V. N. Gladyshev. 2000. Selenium metabolism in zebra fish: multiplicity of selenoprotein genes and expression of a protein containing 17 selenocysteine residues. Genes Cells 5:1049-1060. [DOI] [PubMed] [Google Scholar]
- 16.Kumaraswamy, E., A. Malykh, K. V. Korotkov, S. Kozyavkin, Y. Hu, S. Y. Kwon, M. E. Moustafa, B. A. Carlson, M. J. Berry, B. J. Lee, D. L. Hatfield, A. M. Diamond, and V. N. Gladyshev. 2000. Structure-expression relationships of the 15-kDa selenoprotein gene: possible role of the protein in cancer etiology. J. Biol. Chem. 275:35540-35547. [DOI] [PubMed] [Google Scholar]
- 17.Lander, E. S., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 18.Lescure, A., D. Gautheret, P. Carbon, and A. Krol. 1999. Novel selenoproteins identified in silico and in vivo by using a conserved RNA structural motif. J. Biol. Chem. 274:38147-38154. [DOI] [PubMed] [Google Scholar]
- 19.Low, S. C., and M. J. Berry. 1996. Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biochem. Sci. 21:203-208. [PubMed] [Google Scholar]
- 20.Martin, J. L. 1995. Thioredoxin-a fold for all reasons. Structure 3:245-250. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., p. 16.32-16.36. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Tujebajeva, R. M., P. R. Copeland, X.-M. Xu, B. A. Carlson, J. W. Harney, D. M. Driscoll, D. L. Hatfield, and M. J. Berry. 2000. Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep. 1:158-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venter, J. C., et al. 2001. The sequence of the human genome. Science 16:1304-1351. [DOI] [PubMed] [Google Scholar]
- 24.Walczak, R., E. Westhof, P. Carbon, and A. Krol. 1996. A novel RNA structural motif in the selenocysteine insertion element of eukaryotic selenoprotein mRNAs. RNA 2:367-379. [PMC free article] [PubMed] [Google Scholar]