Abstract
Borna disease virus (BDV) is a nonsegmented, negative-strand RNA virus that replicates and transcribes in the nucleus of infected cells. Recently, we have demonstrated that BDV phosphoprotein (P) can modulate its subcellular localization through binding to the protein X, which is encoded in the overlapping open reading frame (T. Kobayashi et al., J. Virol. 77:8099-8107, 2003). This observation suggested a unique strategy of intracellular trafficking of a viral protein that is essential for the formation of a functional BDV ribonucleoprotein (RNP). However, neither the mechanism nor the consequences of the cytoplasmic retention or nuclear export of BDV X-P complex have been elucidated. In this study, we show that BDV P contains a bona fide nuclear export signal (NES) and can actively shuttle between the nucleus and cytoplasm. A transient transfection analysis of cDNA clones that mimic the BDV bicistronic X/P mRNA revealed that the methionine-rich (MetR) domain of P is responsible for the X-dependent cytoplasmic localization of the protein complex. Mutational and functional analysis revealed that the methionine residues within the MetR domain are critical for the activity of the NES of P. Furthermore, leptomycin B or small interfering RNA for inhibition of CRM1 strongly suggested that a CRM1-dependent pathway mediates nuclear export of P. Fluorescence loss in photobleaching analysis confirmed the nucleocytoplasmic shuttling of P. Moreover, we revealed that the nuclear export of P is not involved in the inhibition of the polymerase activity by X in the BDV minireplicon system. Our results may provide a unique strategy for the nucleocytoplasmic transport of viral RNP, which could be critical for the formation of not only infectious virions in the cytoplasm but also a persistent viral state in the nucleus.
Riboviruses, whose RNA biosynthetic processes, replication, and transcription take place in the nucleus of the infected cell, need to control the directional transport of their genomes through nuclear pore complexes. Recent studies have revealed that viral proteins play central roles in the regulation of nucleocytoplasmic trafficking of viral nucleic acids (2, 24). For instance, the nucleocapsid (NP) and nonstructural (NS2/NEP) proteins of influenza A virus are required for nuclear import and export, respectively, of the viral ribonucleoprotein (RNP) complex (16-18). Likewise, the Rev protein of human immunodeficiency virus type 1 mediates the nuclear transport of unspliced or single-spliced viral transcripts (21). The nucleocytoplasmic transport of viral components is an active and energy-dependent process mediated by specific nuclear localization and export signals termed NLS and NES, respectively, which are present within cargo molecules (2, 7). The interaction between NLS or NES with nuclear transport receptors known as importin and exportin, respectively, regulates the direction in which cargo molecules are transported (7, 23). The signal-dependent translocation of cargo molecules most likely contributes to the nucleocytoplasmic transport of viral components in infected cells, but the detailed mechanisms by which viruses control the directional transport of their genome-protein complexes remain poorly understood.
Borna disease virus (BDV) is an enveloped virus with a nonsegmented, negative-strand RNA genome that has a gene organization characteristic of mononegaviruses (MNV). However, based on its unique genetic and biological features, BDV is considered to be the prototypic member of a new virus family, Bornaviridae, within the order Mononegavirales (4, 5, 30). BDV is highly neurotropic and noncytopathic, and it appears to be exquisitely adapted to establish persistent infections (5, 30). BDV has the property, unique among known animal MNV, of a nuclear site for the replication and transcription of its genome. Therefore, the nucleocytoplasmic transport of BDV macromolecules plays a key role in the virus life cycle.
Recent studies have revealed that several BDV proteins including the nucleoprotein (N), phosphoprotein (P), and protein X (also called p10) contribute to the nucleocytoplasmic transport of BDV (8, 9, 28, 31, 34). Two isoforms of the BDV N (p40N and p38N) are found in BDV-infected cells. Whether p40N and p38N are encoded by two different mRNA species (22) or the usage of a second in-frame initiation AUG codon located 13 amino acids (aa) downstream in the BDV N open reading frame remains unsolved. Basic amino acid- and leucine-rich motifs present within BDV N have been shown to function as NLS and NES, respectively (8, 9). The function of the NES of N is mediated through the chromosome region maintenance protein 1 (CRM1)-dependent pathway (9). The NLS of N has been mapped to the 13 N-terminal amino acid residues, and therefore the p38N isoform lacks an NLS signal (8). The significance of the NLS-lacking p38N has not been determined, but our previous findings suggest that p38N might facilitate the nuclear export of the viral RNPs by increasing the relative number of NESs in the N multimer (9). Moreover, we showed that P could counteract the nuclear export activity of p38N (9), which resulted in the nuclear retention of p40N. These findings support the involvement of p38N and P in the regulation of the trafficking of the viral RNPs in BDV-infected cells (2, 9).
We have reported that a P-X interaction might also contribute to the regulation of the nucleocytoplasmic transport of BDV components (10). BDV P contains a bipartite NLS (28), and P can directly bind to all other components of the viral RNP (26, 27). The intracellular localization of P is drastically influenced by its interaction with X (10). Synthesis of protein X starts within the same mRNA transcription unit as P, but 49 nucleotides upstream, and it overlaps, in a different frame, with the 71 N-terminal amino acids of P (3). Notably, P was efficiently retained in the cytoplasm of BDV-infected cells only when expression of X could be detected in the same cell. Conversely, expression of X was below detection levels in BDV-infected cells in which P exhibited predominantly a nuclear location. Recently, studies using a BDV minireplicon system have shown that X has a strong inhibitory effect on RNA synthesis mediated by the BDV polymerase (19, 25). This X inhibitory effect has been proposed to operate via X interaction with P (20). These results suggest that the control of the nucleocytoplasmic transport of P may play a key role in not only the regulation of BDV RNP trafficking but also the activity of the virus polymerase complex. Therefore, the elucidation of the mechanisms underlying the regulation of the nucleocytoplasmic transport of P can provide important information for a better understanding of the biology of BDV.
Here, we show that BDV P has nuclear export activity that is mediated by an NES contained within a methionine-rich (MetR) domain spanning amino acid residues 145 to 158 of P, which has been previously shown to be required also for P oligomerization. We document that P proteins with a mutated MetR domain accumulate in the nucleus in the presence of X. Moreover, we present evidence that the methionine (M) residues within the MetR domain are critical for the nuclear export activity of P, which operates via the CRM1 pathway. We also demonstrate that BDV P shuttles between the nucleus and cytoplasm and that X interaction with P favors the activity of the NES over the NLS present in P, which leads to a predominant cytoplasmic distribution of the X-P complex. Finally, we provide data supporting the view that BDV X may modulate the BDV polymerase activity via direct interaction with the polymerase complex rather than by altering of the subcellular distribution of P. Our findings illustrate the complexity of interactions underlying the regulation of the intracellular trafficking of the X-P complex, which may be critical for the regulation of both the viral polymerase activity and nucleocytoplasmic trafficking of BDV RNP.
MATERIALS AND METHODS
Cells.
The OL cell line, derived from a human oligodendroglioma (1), was cultured in Dulbecco's modified Eagle's medium-high glucose (4.5%) supplemented with 10% fetal bovine serum, 100 U of penicillin G per ml, 100 μg of streptomycin per ml, and 4 mM glutamine. The 293T, HeLa, and NIH 3T3 cell lines were cultured in Dulbecco's modified Eagle's medium-low glucose (1.0%) supplemented with 10% fetal bovine serum.
Plasmid construction.
Expression vectors encoding X/P-GFP (pgX/P; GFP is green fluorescent protein), P (pgP) and FLAG-tagged P (pcPF) have been previously described (10). Expression vectors pcPHA and pcXHA encoding hemagglutinin (HA)-tagged P and X BDV proteins, respectively, were generated by subcloning the P and X open reading frames, respectively, into the EcoRI and XhoI sites of plasmid pcDNA3 (Invitrogen, San Diego, CA). Plasmids expressing mutant forms of X/P-GFP and P-FLAG were generated from pgX/P and pcPF, respectively, using appropriate PCR procedures. To generate pgX/PNLS, the NLS located between aa 18 and 41 of P was PCR amplified and subcloned into the NotI and XhoI sites of pcDNA3 to yield pcNLS. Finally, a fragment containing X/P-GFP was subcloned into the EcoR I and NotI sites in pcNLS. Plasmids pCFN-βGal and pCFNrev-βGal were kindly provided by M. Dobbelstein (Philipps-Universität, Germany). Detailed information about the primers and PCR procedures used to generate these plasmids is available from the authors. Nucleotide sequences of the recombinant plasmids were confirmed by DNA sequencing.
Cell transfection and gene expression assays.
Cells were seeded in 60-mm tissue culture plates or eight-well chamber slides (Lab-Tek Nunc Inc., Naperville, Ill.). After an overnight culture, cells were transfected with Lipofectamine 2000 (Invitrogen). Gene expression in transfected cells was examined 24 to 48 h later using one, or several, of the following procedures: (i) indirect immunofluorescence, (ii) GFP fluorescence, (iii) immunoprecipitation, and (iv) Western blotting.
Protein pull-down assay.
Transfected cells were lysed in NP-40 lysis buffer as described previously (9). Proteins in the soluble fraction were immunoreacted for 2 h with an anti-FLAG antibody (Sigma-Aldrich, St. Louis, Mo.) at 4°C, and the precipitates were then recovered by incubation with protein G agarose beads (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.) for 24 h at 4°C. After a thorough washing, proteins bound to the agarose beads were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by Western blotting with an anti-HA antibody (Sigma-Aldrich). The specific reactions were detected by an ECL Western blotting kit (Amersham Pharmacia Biotech, Uppsala, Sweden).
LMB treatment assay.
Leptomycin B (LMB) was kindly provided by M. Yoshida (The University of Tokyo). At 48 h posttransfection, the medium was replaced with fresh medium containing LMB (20 ng/ml). The cells were incubated for 3 h in the presence of LMB. After the LMB treatment, the cells were fixed, and then the subcellular localization of GFP fusion proteins was visualized by epifluorescence.
Heterokaryon assay.
Nucleocytoplasmic shuttling of P was examined using a heterokaryon assay. Transfected HeLa cells were seeded on eight-well chamber slides (Lab-Tek Nunc Inc.) together with an equal number of NIH 3T3 cells. Protein synthesis was blocked with 50 μg of cycloheximide per ml for 1 h prior to the fusion. The cells were washed in phosphate-buffered saline and fused by the addition of 50% (wt/wt) polyethylene glycol. After 2 min, the cells were washed in phosphate-buffered saline and were incubated in medium containing cycloheximide (50 μg/ml) for 3 h. After fusion, the cells were fixed and stained with Hoechst 33258 (Sigma-Aldrich) and anti-β-galactosidase (β-Gal) (GIBCO/BRL, Rockville, Md.).
RNA interference.
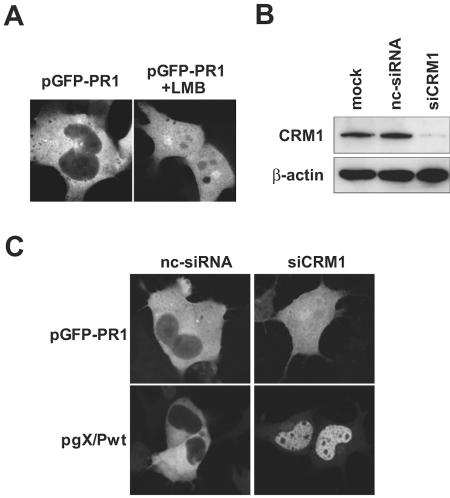
Sequences of small interfering RNA (siRNA) for depletion of human CRM1 (siCRM1) were described previously (13, 15). Transfection of OL cells with siCRM1, nonsilencing negative control siRNA (QIAGEN K.K., Tokyo, Japan), or mock transfection (transfection reagents only) was performed in 12-well plates using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Twenty-four hour after siCRM1 transfection, pGFP-PR1 or pgX/Pwt was transfected into the cells, and the cells were harvested at 48 h posttransfection and lysed. Then, Western blot analysis was performed using a rabbit polyclonal anti-CRM1 antibody (Santa Cruz Biotechnology) or a mouse monoclonal anti-β-actin antibody (Sigma-Aldrich).
FLIP analysis.
OL cells transfected with P containing either a wild-type (Pwt) or mutated NES were used in fluorescence loss in photobleaching (FLIP) experiments with a Digital Eclipse Spectral Imaging Confocal Laser Microscope C1si (Nikon Co., Japan) using the 488-nm laser line of an Ar laser, 2-mW optical fiber output, and detection at 500 to 530 nm. Cells were bleached in a spot with a radius of 3 μm with the 408-nm laser line (100% laser power) and 17-mW optical fiber output and were imaged at interval of 1.0 s.
Minireplicon assay.
293T cells in 12-well plates were transfected with Pol I-MG (polymerase I-driven BDV minigenome [MG] construct), pc-N, pc-P, or its mutant form pc-PM1,2,3A, and pc-L as previously described (19), together with the indicated amounts of plasmid pc-XF that expresses a FLAG-tagged X protein. Forty-eight hours later, the cells were lysed, and cell lysates were prepared for chloramphenicol acetyltransferase (CAT) assays. CAT activity was quantified with a CAT enzyme-linked immunosorbent assay (Roche Molecular Diagnostics, Pleasanton, CA) according to the manufacturer's recommendations.
RESULTS
Identification of the P region required for the protein X-dependent cytoplasmic localization of P.
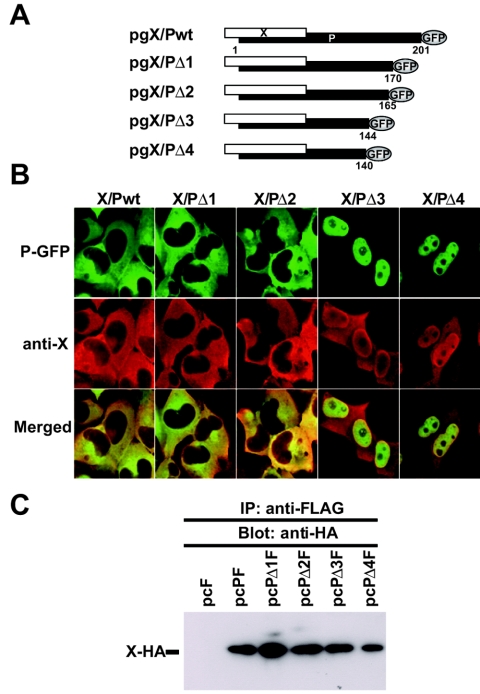
We have documented that the intracellular localization of BDV P is influenced by its interaction with X (10), but the mechanisms underlying the X-dependent subcellular distribution of P remained unsolved. To address this question, we constructed a series of plasmids expressing X and a GFP-tagged Pwt or a GFP-tagged variety of C-terminal deletion mutants of P (Fig. 1A). Both X and P-GFP were expressed from the same bicistronic mRNA that recreated the P transcription unit responsible for X and P expression in BDV-infected cells. This approach allowed us to simultaneously examine the subcellular distribution of X and P by using an anti-X-specific serum and GFP fluorescence, respectively. Both X and P were predominantly found in the cytoplasm of cells transfected with pgX/Pwt, or pgX/PΔ1 and pgX/PΔ2, which lacked, respectively, the 31 and 36 C-terminal amino acids of P (Fig. 1B). In contrast, cells transfected with pgX/PΔ3 and pgX/PΔ4, which lacked, respectively, the 57 and 61C-terminal amino acids of P, showed a nuclear accumulation of X and P (Fig. 1B). We next examined whether the C-terminal deletion mutants of P retained their ability to interact with X. We conducted coimmunoprecipitation (CoIP) assays using lysates from OL cells cotransfected with the corresponding FLAG-tagged versions of the P constructs shown in Fig. 1A and pcXHA that expresses an HA-tagged X protein. Pwt and all the C-terminal P deletion mutants immunoprecipitated X-HA with similar efficiency (Fig. 1C). These results suggested that the formation of an X-P complex does not suffice to determine the subcellular localization of P and that the region between aa 145 to 164 of P was required for the cytoplasmic accumulation of P in the presence of X.
FIG. 1.
Intracellular localization of deletion mutants of BDV phosphoprotein. (A) Schematic of plasmids expressing BDV X and GFP-tagged P proteins from a single mRNA that mimics the BDV bicistronic X/P mRNA. (B) Subcellular localization of BDV X and P in transfected cells. Cells were transfected with the indicated expression plasmids. P expression was monitored by GFP fluorescence, whereas X expression was examined by indirect immunofluorescence using a serum specific to X and a Cy3-labeled secondary antibody. The overlap in the distribution of X and P is revealed in the merged image. (C) CoIP of BDV P and X. OL cells were cotransfected with a plasmid expressing an HA-tagged wild-type X (Xwt) and plasmids expressing FLAG-tagged versions of the P mutant shown in panel A. Cell lysates were prepared and subjected to immunoprecipitation with an anti-FLAG antibody. Immunoprecipitated proteins were analyzed by Western blotting using an anti-HA antibody. Prior immunoprecipitation, the expression of each FLAG-tagged P and the HA-Xwt proteins was verified by Western blotting.
Role of the MetR domain of P in the cytoplasmic accumulation of the X-P complex.
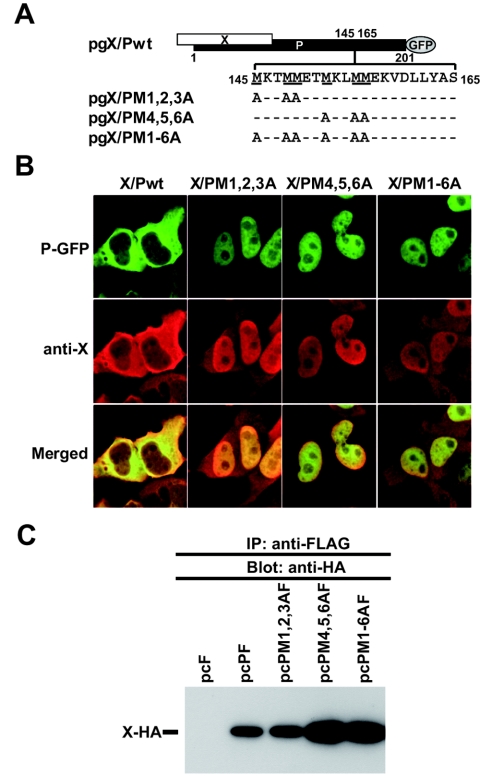
The region spanning aa 145 to 164 of P contains a unique methionine-rich sequence, 145MKTMMET-MKLMMEK158 (methionine residues in boldface), termed the MetR, which has been suggested to stabilize the tertiary structure of P as well as participate in P interactions with other viral and cellular proteins (27, 29). To investigate the role of the MetR domain in the X-dependent subcellular distribution of P, we generated constructs pgX/PM1,2,3A, pgX/PM4,5,6A, and pgX/PM1-6A (Fig. 2A). Cells transfected with plasmids pgX/PM1,2,3A (where the first three M residues are replaced by A), pgX/PM4,5,6A, and pgX/PM1-6A exhibited a nuclear localization of P (Fig. 2B). The substitution of A for M residues might have altered the ability of P to interact with X, which could explain the nuclear, rather than cytoplasmic, accumulation of P with substitutions of A for M. To address this issue, we conducted CoIP assays using cell lysates from cells expressing an HA-tagged X and FLAG-tagged versions of Pwt or of each of the P mutants with substitutions of A for M. Our CoIP results indicated that the three P mutants with substitutions of A for M were not impaired in their ability to bind X (Fig. 2C).
FIG. 2.
Mutants with substitution in the methionine-rich region fail to localize in the cytoplasm of BDV P. (A) Schematic diagram of the MetR mutants of BDV P. Alanine (A) substitutions within the MetR domain were introduced into the plasmid pgX/Pwt. (B) Subcellular localization of BDV X and P in transfected cells. Cells were transfected with the indicated plasmids. The expression of P and X was detected by GFP-fluorescence and indirect immunofluorescence, respectively, as described in the legend of Fig. 1B. The overlap in the distribution of X and P is revealed in the merged image. (C) CoIP of BDV P and X. OL cells were cotransfected with a plasmid expressing an HA-tagged wild-type X (Xwt) and FLAG-tagged versions of P-expressing plasmids shown in panel A. Cell lysates were immunoprecipitated with an anti-FLAG antibody, and precipitants were examined by Western blotting using an anti-HA antibody. Prior to immunoprecipitation, the expression level of each recombinant protein was confirmed by Western blotting.
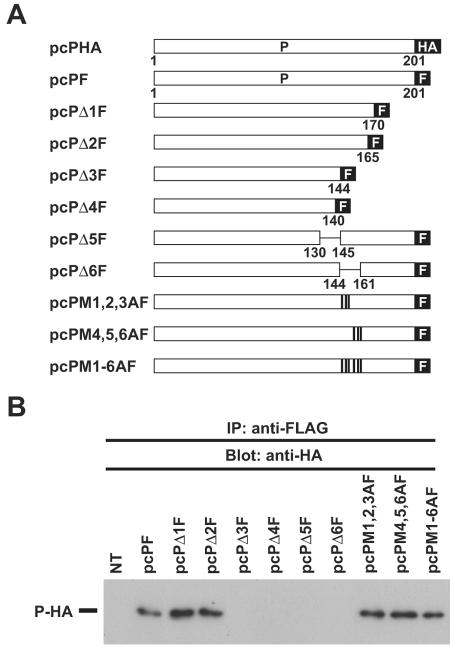
As with the P of other MNV, the oligomerization of BDV P is likely required for its functions (26). The P oligomerization domain has been mapped to amino acid residues 141 to 165 of P (26, 27), a region that contains the MetR domain. We therefore assessed whether the MetR mutants we generated (Fig. 2A) were competent for oligomerization. We conducted CoIP assays using cell lysates from OL cells expressing an HA-tagged Pwt and FLAG-tagged versions of either Pwt or a series of P mutants (Fig. 3A). Proteins immunoprecipitated with the anti-FLAG antibody were analyzed by immunoblotting using an antibody to HA. Consistent with previous findings, P-HA was immunoprecipitated with FLAG-tagged Pwt and P mutants with C-terminal deletions of 31 (Δ1F) and 36 (Δ2F) aa, but not with P mutants lacking the 57 (Δ3F) or 61 (Δ4F) C-terminal amino acids (Fig. 3B). Notably, we found that all the P mutants within the MetR domain examined (M1,2,3AF, M4,5,6AF, and M1-6AF) could immunoprecipitate P-HA. These results indicated that the M residues within the MetR domain are not necessary for P oligomerization or for the X-P intermolecular interaction, but rather these M residues appear to play a key role in the X-dependent subcellular localization of P.
FIG. 3.
Oligomerization of BDV P mutants. (A) Schematic representation of HA- and FLAG (F)-tagged Pwt and FLAG-tagged P mutants. (B) Oligomerization of BDV P mutants. OL cells were cotransfected with pcPHA expressing an HA-tagged Pwt and the indicated plasmids expressing FLAG-tagged P mutant proteins. Cell lysates were immunoprecipitated with anti-FLAG antibody, and the precipitants were detected by Western blotting with anti-HA antibody.
The MetR of P contains a CRM1-dependent NES.
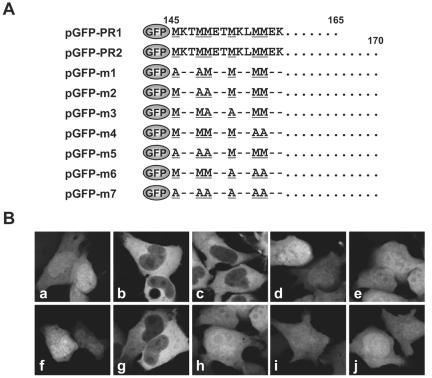
We next examined whether the MetR region of P may actively contribute to the nuclear export of P. We fused GFP to aa 145 to 165 (pGFP-PR1) and 145 to 170 (pGFP-PR2) of P (Fig. 4A). As expected, GFP alone was found in both the nucleus and cytoplasm (Fig. 4B, frame a), whereas in cells transfected with pGFP-PR plasmids, GFP expression was restricted to the cytoplasm (Fig. 4B, frames b and c), suggesting that the MetR region contains the nuclear export activity of P. To assess the contribution of the M residues within the MetR domain to the nuclear export activity of the GFP fusion peptides, we generated several mutants with substitutions of A for M residues based on plasmid pGFP-PR2 (Fig. 4A). GFP-fusion proteins were diffusely distributed in both the nucleus and cytoplasm of the cells transfected with pGFP-m1, -m2, -m3, -m5, -m6, and -m7, as well as with pGFP alone (Fig. 4B, frames d to f and h to j). In contrast, GFP expression was mainly restricted to the cytoplasm in cells transfected with pGFP-m4 (Fig. 4B, frame g). These results suggested that the MetR domain is the NES of P and that the M residues at positions 145, 148, 149, and 152 but not those at 155 and 156 are essential for the activity of this novel NES.
FIG. 4.
BDV P nuclear export activity is mediated by its MetR domain. (A) Schematic diagram of the GFP-tagged P MetR region. Methionine (M) residues at positions 145, 148, 149, 152, 155, and 156 were replaced with alanine (A) (underlined) residues. (B) Subcellular localization of GFP fusion proteins in OL cells. Frame a, pGFP (control); frame b, pGFP-PR1; frame c, pGFP-PR2; frame d, pGFP-m1; frame e, pGFP-m2; frame f, pGFP-m3; frame g, pGFP-m4; frame h, pGFP-m5; frame i, pGFP-m6; frame j, pGFP-m7.
The nuclear export of a variety of proteins is mediated by the interaction of their respective NES and the cellular transporter receptor CRM1. The CRM1-dependent nuclear export pathway is specifically inhibited by LMB (11). Treatment with LMB of OL cells transfected with pGFP-PR1 resulted in nuclear accumulation of GFP expression (Fig. 5A), suggesting that the MetR domain mediates nuclear export of P in a CRM1-dependent manner. For further confirmation of CRM1-dependent nuclear export of BDV P, we performed RNA interference targeting CRM1 (siCRM1) as described in previous reports (13, 15). At 24 h after transfection of siCRM1 into the OL cells, the cells were further transfected with pGFP-PR1 or pgX/Pwt. Forty-eight hours posttransfection, the expression level of endogenous CRM1 was analyzed by Western blotting. As shown in Fig. 5B, this treatment specifically reduced the expression level of CRM1. The effect of the down-regulation of CRM1 on the subcellular localization of the GFP constructs containing MetR domain or full-length P was analyzed by GFP fluorescence. As shown in Fig. 5C, GFP-PR1 was found to accumulate in the nucleus of GFP expression, which was consistent with the results obtained by using LMB, whereas the down-regulation of CRM1 resulted in the predominant nuclear localization of P-GFP in the pgX/Pwt-transfected cells (Fig. 5C). This result strongly confirmed the CRM1-dependent nuclear export of BDV P.
FIG. 5.
The nuclear export of BDV P uses a CRM1-dependent pathway. pGFP-PR1-transfected OL cells untreated (left panel) or treated with LMB (20 ng/ml for 2 h) (right panel) were examined for GFP fluorescence after fixation. (B) OL cells were mock transfected or transfected with negative control siRNA (nc-siRNA) or siRNA for human CRM1 (siCRM1). At 48 h after the GFP construct transfection, the cells were harvested, and Western blot analysis was performed using an anti-CRM1 antibody or an anti-β-actin antibody. (C) The pGFP-PR1 or pgX/Pwt construct was transfected into the OL cells expressing negative control siRNA or siCRM1. At 48 h after transfection, GFP fusion proteins were visualized by fluorescence microscopy.
BDV P shuttles between the nucleus and cytoplasm in the absence of X.
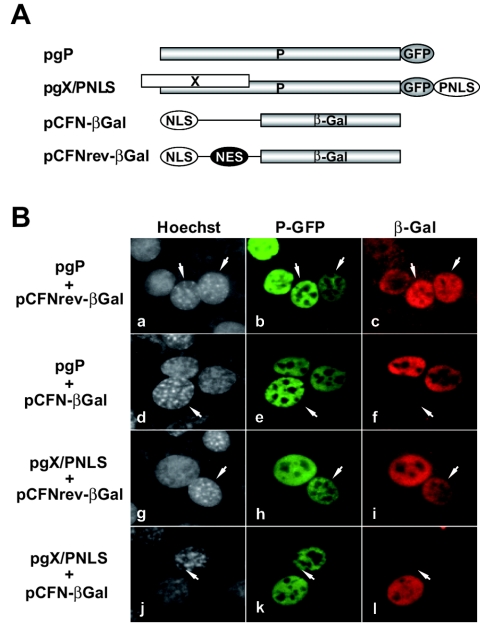
Our finding that BDV P contains both NLS and NES suggested that P could shuttle between the nucleus and cytoplasm. To examine this possibility, we performed interspecies heterokaryon assays. Human HeLa cells transfected with a P-GFP expression plasmid (Fig. 6A, pgP) were fused to mouse NIH 3T3 cells as described above (see Materials and Methods). We monitored the nucleocytoplasmic shuttling of P by examining GFP expression in NIH 3T3 nuclei. Human and mouse cells were distinguished based on their distinct staining pattern with Hoechst 33258. As an internal control, we cotransfected HeLa cells, together with pgP, with plasmids expressing β-Gal containing the simian virus 40 T antigen NLS either alone (pCFN-βGal) or together with the human immunodeficiency virus type 1 Rev NES (pCFNrev-βGal) (Fig. 6A). P-GFP was detected in both human and mouse nuclei (Fig. 6B, frames a, b, d, and e). As predicted, β-Gal derived from pCFNrev-βGal was also detected in both human and mouse nuclei (Fig. 6B, frames a and c), whereas β-Gal derived from pCFN-βGal lacking the Rev NES was detected only in human nuclei (Fig. 6B, frames d and f). To investigate the possible influence of X on this process, we subjected pgX/PNLS-transfected HeLa cells (Fig. 6A) to the same heterokaryon assay. GFP derived from pgX/PNLS was detected in human and mouse nuclei (Fig. 6B, frames g, h, j, and k). Likewise, as predicted, β-Gal derived from pCFNrev-βGal, but not from pCFN-βGal, was also detected in both human and mouse nuclei (Fig. 6B, frames g, i, j, and l). These results indicated that P can exhibit nucleocytoplasmic shuttling activity in the absence of X.
FIG. 6.
BDV P shuttles between the nucleus and cytoplasm. (A) Schematic representation of the plasmids used for heterokaryon assays. (B) HeLa cells were transiently transfected with pgP and pCFNrev-βGal (frames a to c), pgP and pCFN-βGal (frames d to f), pgX/PNLS and pCFNrev-βGal (frames g to i), and pgX/PNLS and pCFN-βGal (frames j to l). Transfected cells were subjected to the interspecies heterokaryon assay as described in the text. Fields containing representative interspecies heterokaryons are shown. Frames a, d, g, and j, Hoechst staining images; frames b, e, h, and k, GFP fluorescence; frames c, f, i, and l, β-Gal staining images. Arrows indicate mouse nuclei.
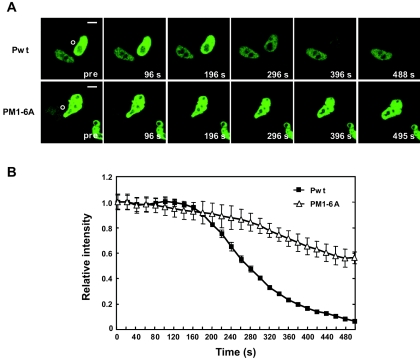
To confirm the nucleocytoplasmic properties of BDV P, we performed a FLIP experiment using OL cells transfected with either pgPwt or pgX/PM1-6A (Fig. 2A). We subjected a defined surface (3-μm radius) of the cytoplasm of transfected cells to repeated photobleaching while monitoring the intensity of nuclear GFP fluorescence (Fig. 7). Cells transfected with pgPwt, but not with pgX/PM1-6A lacking a functional P NES, exhibited a gradual and large (>95%) decrease in nuclear GFP fluorescence over the 500-s observation period (Fig. 7). The modest decrease in GFP nuclear fluorescence observed in pgX/PM1-6A-transfected cells might be due to nuclear import of newly synthesized P. These findings further support that P is endowed with nucleocytoplasmic shuttling properties and that the M residues within the MetR domain are essential for the nuclear export of P.
FIG. 7.
BDV P dynamics in living cells. OL cells were transfected with GFP-tagged either Pwt- or PM1-6A-expressing plasmids and subjected to FLIP analysis. (A) FLIP imaging. The circled area in the cytoplasm (pre panel) was bleached repeatedly, and cells were imaged at interval of 1.0 s. A neighboring cell nucleus was not affected. Scale bars, 5 μm. (B) Quantification of GFP fluorescence in transfected OL cells. The GFP intensity was monitored in the cytoplasm in at least three different areas of transfected cells using the ImageJ program.
Nuclear export activity of P is not critical for X-mediated inhibition of BDV MG expression.
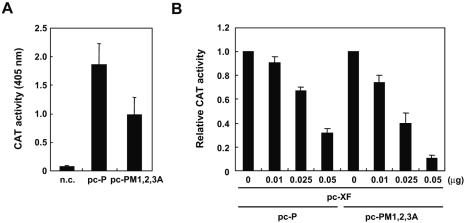
To investigate a possible role of the nuclear export of P in the virus polymerase activity, we examined the activity in the BDV minireplicon system of a P mutant, pc-PM1,2,3A, which lacked a functional NES. Although slightly lower than Pwt, mutant PM1,2,3A supported high levels of BDV RNA analogue (MG) expression (Fig. 8A), suggesting that the M residues within the MetR domain of P are not strictly required for the polymerase cofactor activity of P. BDV X is known to have a powerful inhibitory effect on BDV MG expression (19, 20). It has been suggested that X-mediated redistribution of P from the nucleus to the cytoplasm might contribute to this X inhibitory activity by affecting the assembly of the functional polymerase complex in the nucleus. We therefore investigated whether the nuclear export activity of P influences BDV polymerase activity. We compared the effect of X on BDV MG expression in cells transfected with either Pwt or the mutant PM1,2,3A lacking a functional NES. BDV MG expression was similarly inhibited in both cases (Fig. 8B), suggesting that X-mediated subcellular redistribution of P is not involved in X-mediated inhibition of the activity of the BDV polymerase.
FIG. 8.
The MetR mutant of P cannot escape from the inhibitory effect of X on the BDV minireplicon system. (A) NES mutant of P supports BDV polymerase activity. 293T cells were transfected with Pol I-MG (0.25 μg), pc-N (0.25 μg), pc-L (0.25 μg), and either pc-P (0.025 μg) or pc-PM1,2,3A (0.025 μg), and 48 h later CAT activity in cell lysates was determined with the CAT enzyme-linked immunosorbent assay system. n.c., negative control (mock-transfected cell lysate). (B) BDV X-mediated inhibition of BDV MG expression. 293T cells were transfected with the plasmids expressing the BDV MG RNA and supporting viral trans-acting factors L, N, and P, together with the indicated amounts of X expression plasmid (pc-XF). Relative CAT values from at least three independent experiments are expressed as means plus standard deviations.
DISCUSSION
The completion of the BDV life cycle requires a bidirectional transport of viral RNP across the nuclear envelope. In newly BDV-infected cells, the incoming viral RNP needs to be imported into the nucleus, where viral RNA synthesis takes place. At later stages of the infection, newly synthesized RNP must egress from the nucleus to complete the formation of infectious particles that can propagate the infection to uninfected neighboring cells. The directional control of the BDV RNP trafficking is likely determined by the ratios and interactions between viral proteins containing appropriate NLS or NES or both, as well as by host cell factors including dedicated transporters of cargo molecules carrying appropriate signals.
Bona fide NLS have been identified for BDV N (8), P (28), X (34), and L (31) proteins. These signals could contribute to the nuclear import of viral RNP, although the underlying mechanisms remain to be determined. Likewise, the signals and mechanisms by which BDV RNP egresses from the nucleus are poorly understood. We have documented that N contains a bona fide NES that utilizes a CRM1-dependent pathway for traveling through the nuclear pore complex (9). We have also shown that the intracellular distribution of BDV P is influenced by its binding to X, which promotes the cytoplasmic accumulation of P (10). Both BDV X and P contain functional NLS, suggesting that the activity of these signals must be impaired in the context of the X-P complex. The P-binding region in X completely overlaps with the NLS of X (14, 34), whereas the X-binding region in P does not involve residues contributing to any of the two NLS of P (26, 27). The apparent availability of two NLS in the X-P complex raises the question of why it is not retained in the nucleus but, rather, accumulates in the cytoplasm. An X-induced conformational change in P could impair the function of the P NLS. Alternatively, but not mutually exclusively, a functionally active NES might be present in either X or P or both, and this NES could become dominant upon formation of the X-P complex, thus leading to its efficient nuclear export. NES usually consist of four to five hydrophobic residues within a region of around 10 aa (12). The hydrophobic residues in the NES are predominantly leucine, but isoleucine, valine, methionine, and phenylalanine residues may be also tolerated (12). Several leucine- or isoleucine-rich motifs are conserved among known BDV P sequence (residues in boldface): 39LTQPVDQLLKDL50, 73LIKKLVTEL82 and 131IRILGENIKIL141. However, our findings indicate that none of these motifs accounted for the nuclear export activity of P. Instead, we found that a hydrophobic region within the MetR domain spanning P residues 145 to 165 can operate as a bona fide NES to mediate CRM1-dependent nuclear export of P. The MetR domain, 145MKTMMETMKLMMEK-VDLLYAS165, contains six M and three L residues (boldface). Substitutions of A for M dramatically affected MetR-mediated nuclear export, supporting an essential role of these M residues as part of the P NES. This finding also indicates that the L residues within domain MetR may contribute to, but are not sufficient for, this NES activity. To our knowledge, this newly identified P NES represents a unique CRM1-dependent nuclear export signal. It should be noted that a leucine-rich NES-like motif is present in the N terminus of X, but its activity as NES could not be demonstrated (14). Consistent with this, our data also indicate that this NES-like motif does not contribute to the cytoplasmic localization of the X-P complex.
Overexpression of P in infected cells drastically inhibited viral replication and transcription (6). Moreover, studies using a BDV minireplicon system have further illustrated the need of maintaining an exquisitely balanced stoichiometry of N and P for the intracellular reconstitution of a functional polymerase complex (19, 25). These results suggest that to prevent inhibition of the virus polymerase activity, BDV needs to control tightly the N/P ratio levels in the nucleus, and it is plausible that the X-dependent cytoplasmic localization of P might contribute to the regulation of the correct nuclear N/P ratio. However, we found that the BDV MG expression remained susceptible to X-mediated inhibition when the polymerase cofactor P lacked a functional NES (Fig. 8). This result indicates that X-mediated inhibition of the BDV polymerase complex is due not only to the X-dependent cytoplasmic localization of P. Because X does not appear to interfere with the binding of P to N and L (26), it seems plausible that an X-induced conformational change of P may be responsible for its polymerase inhibitory activity.
We have reported changes in the N/P ratio during the different stages of BDV infection (32, 33). Intriguingly, in BDV persistently infected cells, P was found to be present in about an eightfold molar excess over N (32). Based on data from the BDV MG rescue system, such increased P levels would be expected to result in the inhibition of the virus polymerase. A possible way for BDV to avoid this could be by also increasing levels of the X protein to help sequester the excess of P into the cytoplasm compartment or away from the site of formation of the active polymerase complex. This hypothesis would call for a regulatory mechanism, yet unknown, by which BDV could coordinate the synthesis of both P and X proteins from the same P transcriptional unit. Both BDV X and P are found in the BDV-specific nuclear speckles characteristically observed in BDV-infected cells, suggesting that the nuclear export activity of the X-P complex could be modulated by other viral or cellular proteins associated with these structures.
A detailed knowledge of the mechanisms that control the trafficking of BDV RNP would be important for a better understanding of the virus life cycle and its ability to readily establish long-term persistent infections in a variety of cell types, including postmitotic matured neurons. Studies using newly developed reverse genetic approaches will facilitate the task of elucidating the unique life cycle and pathogenesis of BDV.
Acknowledgments
This article is supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan, a Grant-in-Aid from the Zoonosis Control Project of the Ministry of Agriculture, Forestry and Fisheries of Japan, and a Research Grant for Nervous and Mental Disorders from the Ministry of Health, Labor and Welfare of Japan.
REFERENCES
- 1.Briese, T., J. C. de la Torre, A. Lewis, H. Ludwig, and W. I. Lipkin. 1992. Borna disease virus, a negative-strand RNA virus, transcribes in the nucleus of infected cells. Proc. Natl. Acad. Sci. USA 89:11486-11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cros, J. F., and P. Palese. 2003. Trafficking of viral genomic RNA into and out of the nucleus: influenza, Thogoto and Borna disease viruses. Virus Res. 95:3-12. [DOI] [PubMed] [Google Scholar]
- 3.Cubitt, B., C. Oldstone, and J. C. de la Torre. 1994. Sequence and genome organization of Borna disease virus. J. Virol. 68:1382-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de la Torre, J. C. 1994. Molecular biology of Borna disease virus: prototype of a new group of animal viruses. J. Virol. 68:7669-7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de la Torre, J. C. 2002. Molecular biology of Borna disease virus and persistence. Front. Biosci. 7:D569-579. [DOI] [PubMed] [Google Scholar]
- 6.Geib, T., C. Sauder, S. Venturelli, C. Hassler, P. Staeheli, and M. Schwemmle. 2003. Selective virus resistance conferred by expression of Borna disease virus nucleocapsid components. J. Virol. 77:4283-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldfarb, D. S., A. H. Corbett, D. A. Mason, M. T. Harreman, and S. A. Adam. 2004. Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell. Biol. 14:505-514. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi, T., Y. Shoya, T. Koda, I. Takashima, P. K. Lai, K. Ikuta, M. Kakinuma, and M. Kishi. 1998. Nuclear targeting activity associated with the amino terminal region of the Borna disease virus nucleoprotein. Virology 243:188-197. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi, T., W. Kamitani, G. Zhang, M. Watanabe, K. Tomonaga, and K. Ikuta. 2001. Borna disease virus nucleoprotein requires both nuclear localization and export activities for viral nucleocytoplasmic shuttling. J. Virol. 75:3404-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi, T., G. Zhang, B. J. Lee, S. Baba, M. Yamashita, W. Kamitani, H. Yanai, K. Tomonaga, and K. Ikuta. 2003. Modulation of Borna disease virus phosphoprotein nuclear localization by the viral protein X encoded in the overlapping open reading frame. J. Virol. 77:8099-8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kudo, N., B. Wolff, T. Sekimoto, E. P. Schreiner, Y. Yoneda, M. Yanagida, S. Horinouchi, and M. Yoshida. 1998. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell. Res. 242:540-547. [DOI] [PubMed] [Google Scholar]
- 12.la Cour, T., R. Gupta, K. Rapacki, K. Skriver, F. M. Poulsen, and S. Brunak. 2003. NESbase version 1.0: a database of nuclear export signals. Nucleic Acids Res. 31:393-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lund, E., S. Guttinger, A. Calado, J. E. Dahlberg, and U. Kutay. 2004. Nuclear export of microRNA precursors. Science 303:95-98. [DOI] [PubMed] [Google Scholar]
- 14.Malik, T. H., M. Kishi, and P. K. Lai. 2000. Characterization of the P protein-binding domain on the 10-kilodalton protein of Borna disease virus. J. Virol. 74:3413-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miki, T., and Y. Yoneda. 2004. Alternative splicing of Staufen2 creates the nuclear export signal for CRM1 (Exportin 1). J. Biol. Chem. 279:47473-47479. [DOI] [PubMed] [Google Scholar]
- 16.Neumann, G., M. R. Castrucci, and Y. Kawaoka. 1997. Nuclear import and export of influenza virus nucleoprotein. J. Virol. 71:9690-9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neumann, G., M. T. Hughes, and Y. Kawaoka. 2000. Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1. EMBO J. 19:6751-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Neill, R. E., J. Talon, and P. Palese. 1998. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 17:288-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez, M., A. Sanchez, B. Cubitt, D. Rosario, and J. C. de la Torre. 2003. A reverse genetics system for Borna disease virus. J. Gen. Virol. 84:3099-3104. [DOI] [PubMed] [Google Scholar]
- 20.Poenisch, M., G. Unterstab, T. Wolff, P. Staeheli, and U. Schneider. 2004. The X protein of Borna disease virus regulates viral polymerase activity through interaction with the P protein. J. Gen. Virol. 85:1895-1898. [DOI] [PubMed] [Google Scholar]
- 21.Pollard, V. W., and M. H. Malim. 1998. The HIV-1 Rev protein. Annu. Rev. Microbiol. 52:491-532. [DOI] [PubMed] [Google Scholar]
- 22.Pyper, J. M., and A. E. Gartner. 1997. Molecular basis for the differential subcellular localization of the 38- and 39-kilodalton structural proteins of Borna disease virus. J. Virol. 71:5133-5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez, M. S., C. Dargemont, and F. Stutz. 2004. Nuclear export of RNA. Biol. Cell. 96:639-655. [DOI] [PubMed] [Google Scholar]
- 24.Sandri-Goldin, R. M. 2004. Viral regulation of mRNA export. J. Virol. 78:4389-4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider, U., M. Naegele, P. Staeheli, and M. Schwemmle. 2003. Active Borna disease virus polymerase complex requires a distinct nucleoprotein-to-phosphoprotein ratio but no viral X protein. J. Virol. 77:11781-11789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider, U., K. Blechschmidt, M. Schwemmle, and P. Staeheli. 2004. Overlap of interaction domains indicates a central role of the P protein in assembly and regulation of the Borna disease virus polymerase complex. J. Biol. Chem. 279:55290-55296. [DOI] [PubMed] [Google Scholar]
- 27.Schwemmle, M., M. Salvatore, L. Shi, J. Richt, C. H. Lee, and W. I. Lipkin. 1998. Interactions of the Borna disease virus P, N, and X proteins and their functional implications. J. Biol. Chem. 273:9007-9012. [DOI] [PubMed] [Google Scholar]
- 28.Shoya, Y., T. Kobayashi, T. Koda, K. Ikuta, M. Kakinuma, and M. Kishi. 1998. Two proline-rich nuclear localization signals in the amino- and carboxyl-terminal regions of the Borna disease virus phosphoprotein. J. Virol. 72:9755-9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thierer, J., H. Riehle, O. Grebenstein, T. Binz, S. Herzog, N. Thiedemann, L. Stitz, R. Rott, F. Lottspeich, and H. Niemann. 1992. The 24K protein of Borna disease virus. J. Gen. Virol. 73:413-416. [DOI] [PubMed] [Google Scholar]
- 30.Tomonaga, K., T. Kobayashi, and K. Ikuta. 2002. Molecular and cellular biology of Borna disease virus infection. Microbes Infect. 4:491-500. [DOI] [PubMed] [Google Scholar]
- 31.Walker, M. P., and W. I. Lipkin. 2002. Characterization of the nuclear localization signal of the Borna disease virus polymerase. J. Virol. 76:8460-8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe, M., Q. Zhong, T. Kobayashi, W. Kamitani, K. Tomonaga, and K. Ikuta. 2000. Molecular ratio between Borna disease viral-p40 and -p24 proteins in infected cells determined by quantitative antigen capture ELISA. Microbiol. Immunol. 44:765-772. [DOI] [PubMed] [Google Scholar]
- 33.Watanabe, M., B. J. Lee, W. Kamitani, T. Kobayashi, H. Taniyama, K. Tomonaga, and K. Ikuta. 2001. Neurological diseases and viral dynamics in the brains of neonatally Borna disease virus-infected gerbils. Virology 282:65-76. [DOI] [PubMed] [Google Scholar]
- 34.Wolff, T., G. Unterstab, G. Heins, J. A. Richt, and M. Kann. 2002. Characterization of an unusual importin alpha binding motif in the Borna disease virus p10 protein that directs nuclear import. J. Biol. Chem. 277:12151-12157. [DOI] [PubMed] [Google Scholar]