Abstract
Most viral glycoproteins mediating membrane fusion adopt a metastable native conformation and undergo major conformational changes during fusion. We previously described a panel of compounds that specifically prevent fusion induced by measles virus (MV), most likely by interfering with conformational rearrangements of the MV fusion (F) protein. To further elucidate the basis of inhibition and better understand the mechanism of MV glycoprotein-mediated fusion, we generated and characterized resistant MV variants. Spontaneous mutations conferring drug resistance were confirmed in transient assays and in the context of recombinant virions and were in all cases located in the fusion protein. Several mutations emerged independently at F position 462, which is located in the C-terminal heptad repeat (HR-B) domain. In peptide competition assays, all HR-B mutants at residue 462 revealed reduced affinity for binding to the HR-A core complex compared to unmodified HR-B. Combining mutations at residue 462 with mutations in the distal F head region, which we had previously identified as mediating drug resistance, causes intracellular retention of the mutant proteins. The transport competence and activity of the mutants can be restored, however, by incubation at reduced temperature or in the presence of the inhibitory compounds, indicating that the F escape mutants have a reduced conformational stability and that the inhibitors stabilize a transport-competent conformation of the F trimer. The data support the conclusion that residues located in the head domain of the F trimer and the HR-B region contribute jointly to controlling F conformational stability.
Enveloped viruses, such as retroviruses, paramyxoviruses, orthomyxoviruses, and filoviruses, infect cells through fusion of their lipid envelope with the plasma membrane or intracellular membranes of the target cell (17, 30). For members of these viral families, membrane merger is mediated by homotrimeric type I fusogenic membrane glycoproteins (FMGs), integral membrane proteins displayed on the surfaces of the viral particles (17, 54). All type I FMGs contain an internal hydrophobic domain of approximately 25 amino acids, generally termed the fusion peptide. Proteolytic cleavage at a specific site yields a metastable native FMG that consists of a transmembrane and a membrane-distal subunit. Subsequent activation of the FMG results in insertion of the fusion peptide, which is located in the transmembrane subunit, into the target membrane (21). Depending on the origin of the FMG, activation can be realized at neutral pH, as postulated, for example, for lentiviruses (4, 27) and most paramyxoviruses, including measles virus (MV) (19), or at low-pH conditions in an endosomal compartment of the target cell, as exemplified by influenza virus (54). Insertion of the fusion peptide into the target membrane is then followed by conformational rearrangements of the FMG trimer that bring the fusion peptide and the transmembrane domain, and hence the target and donor membranes, into close proximity (1, 2, 37, 40, 50, 61), ultimately resulting in the formation of a fusion pore.
Instrumental in this process are two highly conserved 4-3 heptad repeat (HR) sequences, one of which is located adjacent to the fusion peptide and near the N terminus of the protein (hence termed the HR-N or HR-A domain), while the other is adjacent to the transmembrane domain and near the C terminus (HR-C or HR-B) (17, 29). Activation of the native FMG and insertion of the fusion peptide into the target membrane are thought to be followed by refolding into a transient hairpin intermediate and the formation of a stable six-helix bundle (6-HB) fusion core structure (17, 54). Analysis of this core structure of lentivirus (7) and paramyxovirus (1, 61) FMGs has revealed a central homotrimeric coiled coil formed by HR-A domains that is surrounded by three HR-B helices in an antiparallel fashion (17, 54). In this model, the process of protein refolding and 6-HB formation is thus coupled to membrane fusion (15, 37, 50). The conformational changes may in fact liberate the free energy required for the membrane fusion event. Indeed, a small-molecule inhibitor of respiratory syncytial virus (RSV) that is postulated to bind to a groove in the HR-A coiled coil (11) and synthetic peptides derived from the HR-B domains of some FMGs are potent inhibitors of viral entry, presumably by competing with the endogenous HR-B sequences for binding to the central HR-A trimer (31, 47, 58, 59).
For paramyxoviruses, the fusion (F) protein precursor F0 is cleaved into a larger transmembrane F1 and a smaller extracellular F2 subunit. In addition to the crystal structures of the RSV and simian virus type 5 (SV5) fusion cores, medium- and high-resolution structural information for paramyxovirus F proteins comes from a three-dimensional cryoelectron microscopy reconstruction of the Sendai virus F protein (36) and X-ray structures of the Newcastle disease virus (NDV) and human parainfluenzavirus type 3 (hPIV3) F ectodomains (9, 60). All of the structures adopt similar overall spatial organizations of the F trimer, with a distal head, a widening neck, and a stalk region proximal to the viral membrane composed of a central triple-helix coiled coil. Since the fusion peptide and adjacent parts of the HR-A domain and the whole HR-B domain could not be localized in the NDV F X-ray structure (9, 12), and since the 6-HB fusion core was already present in the hPIV3 structure, contacts of HR residues in the native or fusion intermediate conformation could not be determined. It is remarkable, though, that the structure of the uncleaved hPIV3 F ectodomain assumes a conformation largely similar to the postfusion state (60). Membrane fusion, however, requires proteolytic maturation, and it has been demonstrated for several F proteins that maturation triggers a conformational rearrangement of the F trimer, resulting in a metastable fusion-competent structure (16, 22, 28, 57). Further refolding must be tightly regulated to prevent 6-HB formation in the absence of a target membrane.
The paramyxovirus attachment protein hemagglutinin (H), hemagglutinin-neuraminidase (HN), or glycoprotein (G), depending on the genus of the virus, is thought to provide fusion support for the F trimer upon receptor binding, thus coupling F refolding to the presence of a target membrane. This is believed to involve specific interactions of H with F, since most paramyxoviruses require coexpression of their homotypic glycoproteins for efficient fusion and hence F activation (23, 55). In this model, receptor binding by H induces changes in the interaction with F that result in lowering of the energy barrier controlling the metastable native F conformation.
For several paramyxoviruses, however, mutant F variants have been described that induce membrane fusion in the absence of fusion support provided by their receptor binding HN protein (38, 52). The anomalous HN independence of the SV5 F protein derived from strain W3A (38) has been linked to the presence of a proline residue at F position 22 (39, 56), while an L289A mutation confers HN independence on NDV F in certain cell lines (32, 52) and mutations in the cytosolic tail of SER virus F protein eliminate the requirement for HN (53). These mutations are considered to lower the energy barrier required for activation of native F, thus increasing the likelihood of spontaneous structural rearrangements. In contrast, H independence has not been demonstrated for F proteins of viruses from the paramyxovirus genus Morbillivirus, such as MV, canine distemper virus, or rinderpest virus.
Through characterization of MV strains with different cytopathicities, we previously identified a cavity-like microdomain located at the intersection of the neck and head regions of the F trimer that is critical for F fusion activity (41, 45). A functional analysis of this microdomain points to a role for the cavity in an early phase of the fusion process (45). Small-molecule compounds postulated to have high affinity for this microdomain proved to be effective and highly specific inhibitors of MV entry, and point mutations in the postulated target area resulted in resistance to inhibition (43).
In addition to primary resistance mutants located in a compound target site, resistance to viral entry inhibitors can also be modulated by domain changes distant from the postulated binding site, as demonstrated for the human immunodeficiency virus (HIV) entry inhibitor T-20 (14, 20). Conceivably, secondary-site resistance mutants can control FMG activation or the stability of fusion-intermediate conformations. Generation and characterization of spontaneous resistance mutants to the MV inhibitors may therefore enable us to identify residues that regulate conformational rearrangements of the MV F protein and hence to better understand the mechanism of MV F fusion activation.
In the current study, we have tested this hypothesis by using an adaptation approach combined with testing of wild-type MV isolates of different genotypes to obtain a panel of resistant MV variants. Having identified a residue in the MV F HR-B domain that prominently contributes to resistance to inhibition, we have generated a panel of double mutants harboring changes in this domain and the previously identified cavity in the F neck region and have explored whether residues in both domains contribute to controlling F fusogenicity. Our findings suggest that the residue in the HR-B domain has a dual function in MV F-mediated fusion. First, it jointly regulates the stability of a prefusion conformation of the F trimer, together with residues in the F neck cavity. Second, at the completion of fusion, residue 462 contributes to formation of the final 6-HB fusion core structure.
MATERIALS AND METHODS
Cell culture, transfection, mutagenesis, and production of MV stocks.
All cell lines were maintained at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin, and streptomycin. Vero-CD150w cells stably expressing human CD150w were incubated in the additional presence of G-418 at a concentration of 100 μg/ml. Lipofectamine 2000 (Invitrogen) was used for transient-transfection experiments. For some experiments, transfected cells were photographed 24 h posttransfection at a magnification of ×200. Site-directed mutagenesis was carried out using appropriate primers and the Quick Change mutagenesis system (Stratagene) according to the manufacturer's instructions. To prepare virus stocks, cells were infected at a multiplicity of infection (MOI) of 0.01 PFU/cell and incubated at 37°C. The cells were scraped into OPTIMEM (Invitrogen), and virus was released by two freeze-thaw cycles. MV strain Edmonston (MV-Edm) stocks were grown and titered by 50% tissue culture infective dose (TCID50) determination on Vero cells according to the Spearman-Karber method as previously described (44), while primary MV strains were grown and titered on Vero-CD150w cells.
Generation of spontaneous resistance mutants.
For directed evolution, Vero or Vero-CD150w cells were infected with MV strain Edmonston or MVi/Kansas.USA/43.00 (MV-KS), respectively, at an MOI of 0.1 PFU/ml and incubated in the presence of 60 μM 5-amino-2-benzylbenzoxazole (OX-1) (43) or 15 μM 4-nitro-2-phenylacetylamino-benzamide (AS-48) (42), as specified. To prepare inhibitor stocks, all compounds were dissolved at 300 mM in dimethyl sulfoxide (DMSO). In experiments, final DMSO concentrations never exceeded 0.1%, at which no adverse effect on cell viability or viral growth could be detected in control samples. At 48 h postinfection, cell-associated viral particles were released by two freeze-thaw cycles, diluted 10-fold, and used for infection of fresh cell monolayers in the presence of compound at the same concentration. Spontaneous mutants were subjected to two consecutive rounds of plaque purification when extensive cytopathicity was detected in the presence of compound, usually after four to seven passages, and purified clones were reassessed for resistance, followed by reverse transcription-PCR (RT-PCR) and DNA sequencing.
RT-PCR and subcloning of envelope glycoproteins from MV isolates B3-2 and B1.
Total RNA was prepared from Vero-CD150w cells infected with the respective wild-type isolate using the RNeasy Mini Kit (QIAGEN) and subjected to reverse transcription using Superscript II Reverse Transcriptase (Invitrogen) and random hexamer primers. Genome fragments containing the F gene were then further amplified using TaqHiFi DNA polymerase (Invitrogen), followed by transfer into TOPO 2.1 vectors (Invitrogen) and further cloning into pCG expression plasmids containing the constitutive cytomegalovirus promoter.
Recovery of recombinant viruses.
Recombinant MVs were generated essentially as described previously (46). Briefly, the helper cell line 293-3-46 stably expressing MV N, MV P, and T7 polymerase was transfected by calcium phosphate precipitation using the ProFection kit (Promega) with a cDNA copy of the relevant MV genome and MV polymerase L. Helper cells were overlaid on Vero cells 76 h posttransfection, and the resulting infectious centers were passaged on Vero cells. The integrity of recombinant MV particles was confirmed by RT-PCR and DNA sequencing of the modified genes.
Transient-inhibition assays.
To determine the abilities of compounds to inhibit cell-to-cell fusion, 6 × 105 cells per well were transfected with 4 μg plasmid DNA each encoding MV H and F genes, respectively, and the cells were transferred 4 h posttransfection to 96-well plates containing twofold compound dilutions ranging from 75 μM to 4.6875 μM in four replicates each for AS-48 and from 300 μM to 18.75 μM for OX-1. Fusion activity was assessed microscopically 48 h posttransfection, and the extent of cytotoxicity as a consequence of extensive syncytium formation was quantified, using a nonradioactive cytotoxicity assay (Promega) according to the manufacturer's instructions, by the following formula: percent cytotoxicity = ((experimental − background / (maximum − background)) × 100.
Dose-response inhibition curves of virus replication.
To test the sensitivity of recombinant virions to compound-mediated inhibition, Vero cells were infected in four replicates per compound concentration in a 96-well plate format with the respective recombinant MV-Edm (rMV-Edm) variant at an MOI of 0.1 PFU/cell in the presence of OX-1 ranging from 300 μM to 18.75 μM or AS-48 ranging from 75 μM to 4.6875 μM in twofold dilutions. At 96 h postinfection, virus-induced cytopathicity was quantified using a nonradioactive proliferation assay (Promega), and the results were calculated according to the following formula: percent relative cytopathic effect = 100 − ((experimental − background)/ (maximum − background)) × 100. For peptide competition experiments, Vero cells were infected with rMV-Edm variants as indicated at an MOI of 0.1 PFU/cell in the presence of peptide in twofold dilutions starting at 600 μg/ml or 300 μg/ml as specified and incubated in the presence of peptide at 37°C for 36 h. Yields of cell-associated virus were then determined by TCID50 titration.
Molecular modeling of the MV 6-HB core.
Sequence alignment was performed with ClustalW (10), using a gap open penalty of 10.0, a gap extension penalty of 0.1, and the Gonnet matrix. When the MV fusion protein sequence was aligned with the sequence of the SV5 fusion protein, 35% sequence identity, 22% strongly similar, and 13% weakly similar results were found for the portion of the protein to be modeled. Homology modeling was accomplished with Prime (Schrödinger), using the SV5 fusion protein fragment structure (1) as a template (PDB code 1sv5). This fragment (residues 132 to 195 and 450 to 487 by MV numbering) encompasses the 6-HB (both N1 and C1 heptad repeats). The MV F sequence was modeled onto this structure, and the other two subunits of the trimer were generated by transposing the structure to the symmetrical positions of the other subunits. Sections absent in the crystal structure were not modeled. The resulting structure was refined using Prime's structure refinement module. The lowest-energy rotamers for all nonconserved side chains were predicted and incorporated into the model. The final structure (see Fig. 2) was evaluated using the WHAT IF Ramachandran-based z score and the WHAT IF z score for the local amino acid environments (49). The Ramachandran z score was 4.73, similar to the z score of the crystal structure (4.64). The z score for the local amino acid environments was slightly better for the model (2.22) than for the crystal structure (2.66). Models of the 6-HB mutants (N462S, N462D, and N462K) were generated by mutating the residue within the model and using Prime's side chain prediction feature to determine the lowest-energy rotamers for the mutant residue. The Lovell rotamer library (34), derived from Protein Data Bank side chain populations, was used to select the lowest-energy rotamer.
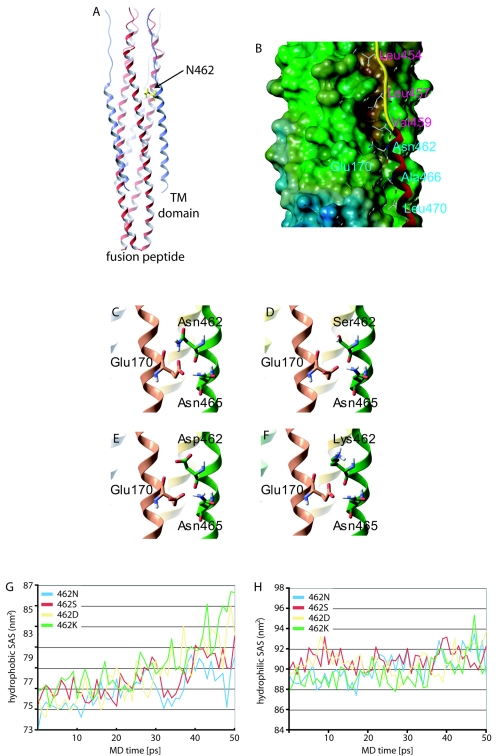
FIG. 2.
Localization of residue 462 in the final MV F core structure. (A) Ribbon model of the MV 6-HB; N termini of HR-B domains are facing up. For clarity, residue 462 is highlighted in only one of the three HR-B domains. TM, transmembrane. (B) Surface model of the MV 6-HB, shown with only one HR-B ribbon for clarity. Residues V459, L457, and L454 are predicted to interact with a hydrophobic groove in the HR-A trimer. HR-A residue E170 is predicted to engage in hydrogen bonding with residues N462 and N465. (C to F) Enlarged ribbon models of HR-A and HR-B highlighting the interaction described above (C). Dynamic structural modeling predicts disruption of the hydrogen bonding, destabilizing the interaction for either of the resistant mutants based on distance (D), charge (E), or steric hindrance (F). (G and H) MD simulations predict greater destabilization of the mutant 6-HBs compared to the wild type. The mutants increase the peptides' hydrophobic exposure to water through the course of the simulation, indicating greater dissociation (G). Hydrophobic solvent-accessible surface (SAS) areas are shown for 50-ps MD simulation. Hydrophilic exposures to water were similar for the four structures, with average hydrophilic SAS areas over the last 10 picoseconds of simulation between 91.1 nm2 and 92.0 nm2 (H).
The four models generated were subjected to molecular-dynamics (MD) simulation using the GROMACS package (3, 33). The structures were simulated using the OPLS-AA force field (26) and solvated with the TIP4P water model (25). A hydrated rectangular box (5 by 5 by 12 nm; volume, ∼327 nm3) containing ∼9,300 water molecules was built around each structure. Sodium cations were randomly placed throughout the water to bring the total charge in each system to zero. Each hydrated complex was energy minimized to remove any high-energy contacts and subjected to 50 ps of position-restrained MD at 300 K to allow water to soak into the structure. Each system was then subjected to 50 ps of MD at 300 K, followed by 50 ps of simulated annealing MD with temperature increases of 5 K/ps (300 to 550 K). Time steps of 0.002 ps were used in all simulations. HR-A was constrained using position restraints of 1,000 kJ · mol−1 · nm−2 in the simulations to account for the absence of the remainder of the fusion protein in this model. Solvent-accessible surface areas over the last 50 ps were calculated using the g_sas function within the GROMACS package.
Membrane permeability predictions for OX-1 and AS-48 were made with the QikProp package (24). For both compounds, the predicted apparent Caco-2 and MDCK cell permeabilities exceed 25 nm/s, indicating membrane permeability based on a range of 95% of known drugs.
Expression and purification of MV F-derived peptides.
For expression of HR domain-derived peptides, previously identified suitable target sequences (31) were amplified by PCR and transferred in frame into pET-30a expression vectors (Novagen), followed by transformation into BL21(DE3) expression-capable Escherichia coli (Novagen). The bacteria were cultured in terrific broth (Invitrogen) to an optical density at 600 nm of 0.6 and then induced with IPTG (isopropyl-β-d-thiogalactopyranoside; 1 mM) for 3 hours. Samples were harvested by centrifugation and treated with lysozyme (Sigma) at a concentration of 200 kU/g of wet biomass. Lysates were digested with RNase A (10 μg/ml) and DNase I (20 kU/ml) overnight and then purified using NiNTA Superflow resin (1 ml resin/10 ml lysate). The bound product was eluted in imidazole buffer (250 mM imidazole, 50 mM NaH2PO4, 300 mM NaCl, pH 8.0) and then dialyzed against phosphate-buffered saline (PBS). The final peptide concentration was estimated by aromatic UV (λ = 280 nm) absorbance, using extinction coefficients calculated by the following formula: Ex = a × Etyrosine + b × Etryptophan + c × Ecysteine, where Ex is the unknown extinction coefficient; a, b, and c are the numbers of tyrosine, tryptophan, and cysteine amino acids per peptide; and Etyrosine, Etryptophan, and Ecysteine are the respective extinction coefficients for each residue. Flag-tagged HR-B peptide for coprecipitation assays was synthesized at the microchemical core facility of Emory University.
Coimmunoprecipitation.
Vero cells were transfected with 4 μg each of plasmid DNA encoding MV H and MV F or 4 μg of plasmid DNA encoding MV F and incubated in the presence of 200 μM fusion inhibitory peptide (Bachem). At 24 h posttransfection, the cells were washed five times with cold PBS, overlaid with PBS containing 100 μg Flag-tagged HR-B peptide (41 μM) and 100 μM AS-48 to achieve a molar excess of the compound, and incubated at 37°C for 20 min. Samples were then washed five times with cold PBS, and the cells were lysed in immunoprecipitation buffer (10 mM Tris, pH 7.4, 150 mM NaCl, 1% deoxycholate, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS], 1 mM phenylmethylsulfonyl fluoride [PMSF], protease inhibitors). The cleared lysates (20,000 × g; 60 min; 4°C) were incubated with monoclonal anti-Flag M2 antibodies (Sigma) at 4°C, followed by precipitation with immobilized protein G (Pierce) overnight. The precipitates were washed five times with immunoprecipitation buffer, resuspended in urea buffer, fractionated on 12% SDS-polyacrylamide gels, blotted onto polyvinylidene difluoride membranes (Millipore), and subjected to enhanced-chemiluminescence detection (Amersham Pharmacia Biotech) using antisera directed against the MV F tail.
Surface biotinylation.
Cells were transfected with 4 μg of plasmid DNA encoding MV F variants as indicated using Lipofectamine 2000. After being washed in cold PBS, the cells were incubated in PBS with 0.5 mg/ml sulfosuccinimidyl-2-(biotinamido)ethyl-1,3-dithiopropionate (Pierce) for 20 min at 4°C, followed by washing and quenching for 10 min at 4°C in Dulbecco's modified Eagle's medium. Samples were scraped into immunoprecipitation buffer (10 mM HEPES, pH 7.4, 50 mM sodium pyrophosphate, 50 mM sodium fluoride, 50 mM sodium chloride, 5 mM EDTA, 5 mM EGTA, 1% Triton X-100) containing protease inhibitors (Roche complete mix) and 1 mM PMSF, and the lysates were cleared by centrifugation for 20 min at 20,000 × g and 4°C. The biotinylated proteins were adsorbed to Sepharose-coupled streptavidin (Amersham Pharmacia Biotech) for 90 min at 4°C, washed in buffer 1 (100 mM Tris, pH 7.6, 500 mM lithium chloride, 0.1% Triton X-100) and then buffer 2 (20 mM HEPES, pH 7.2, 2 mM EGTA, 10 mM magnesium chloride, 0.1% Triton X-100), and incubated in urea buffer (200 mM Tris, pH 6.8, 8 M urea, 5% SDS, 0.1 mM EDTA, 0.03% bromphenol blue, 1.5% dithiothreitol) for 25 min at 50°C. Samples were then subjected to Western analysis as described above. For statistical analysis, Western blots were developed using a VersaDoc imaging system (Bio-Rad) and arbitrary densitometric units determined using the QuantityOne software package.
EndoH treatment.
Cells transfected with 2.5 μg plasmid DNA encoding MV F variants were lysed 24 h posttransfection at 4°C in lysis buffer (50 mM Tris, pH 8.0, 62.5 mM EDTA, 0.4% deoxycholate, 1% Igepal [Sigma], 1 mM PMSF, protease inhibitors [Roche]), and the cleared lysates were incubated in denaturing buffer (final concentration, 0.5% SDS, 1% β-mercaptoethanol) for 25 min at 50°C. Deglycosylation of denatured samples was achieved in deglycosylation buffer (final concentration, 50 mM sodium citrate, pH 5.5) for 2 h at 37°C, using 2 units of endoglycosidase H (EndoH) (New England Biochemicals). Urea buffer was then added, and the samples were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotting using antisera directed against the MV F tail.
RESULTS
To identify residues that confer resistance to inhibition by the MV inhibitor OX-1 (43) or AS-48 (42), we analyzed a panel of primary MV isolates of different genotypes and employed a directed-evolution strategy by repeated passaging of laboratory-adapted MV-Edm or the wild-type strain MV-KS in the presence of the compounds.
Spontaneous escape mutants cluster at F residue 462.
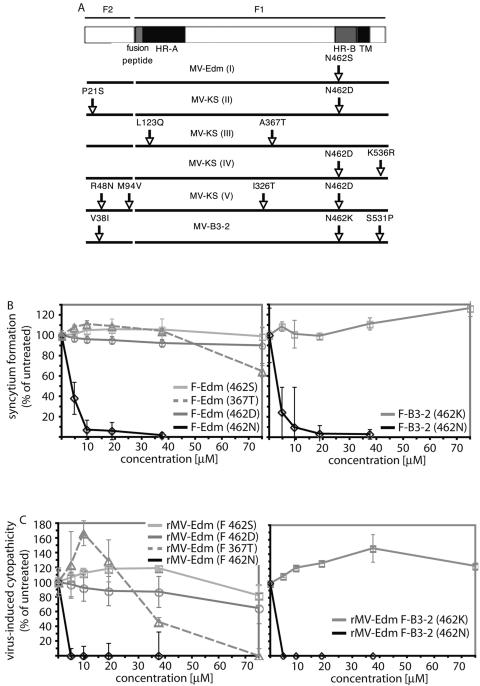
Escape mutants that induced extensive syncytium formation in the presence of compound were isolated by plaque purification. The F-encoding genes were then analyzed by reverse transcription-PCR, followed by DNA sequencing and transfer into expression vectors. The biological activity of all expression constructs was verified by monitoring their abilities to induce cell-to-cell fusion upon cotransfection of MV H and F expression plasmids in the presence and absence of compound (data not shown). For all escape mutants analyzed, resistance to inhibition was achieved by changes in the F protein. Figure 1A shows all mutations found in the F proteins of five independent escape mutants based on MV-Edm or MV-KS. In addition, of 25 primary strains analyzed (mostly described in reference 42), a single sub-Saharan strain of genotype B3-2, MVi/Ibadan.NIE/97/1 (MV-B3-2), likewise revealed resistance to inhibition. The glycoprotein genes of this strain and the closely related wild-type strain MVi/Y22 of genotype B1 (MV-B1), which is readily inhibited by the compounds, were sequenced and transferred into expression plasmids. Changes in MV-B3-2 F are shown in comparison to MV-B1 F protein (Fig. 1A).
FIG. 1.
Escape mutants resistant to inhibition cluster predominantly at F residue 462. (A) Mutations found in the F proteins of adapted MV-Edm (I) or MV-KS (II to V) variants compared to the parental strains. Sequence changes in the F protein of a resistant primary isolate (MV-B3-2) are shown in comparison to the related sensitive isolate MV-B1. TM, transmembrane. (B) Transient dose-response assay to determine the contributions of selected mutations to resistance. Individual mutations were rebuilt by site-directed mutagenesis in the F-Edm or F-B3-2 background, respectively, and all F expression plasmids were cotransfected with equal amounts of MV H encoding plasmid of the corresponding genotype (MV H-Edm [left] or MV H-B3-2 [right]). Syncytium formation in the presence of different AS-48 concentrations was quantified and values were normalized for cells treated with solvent (DMSO) only. The means of four replicates are shown. The error bars indicate standard deviations. (C) Quantitative cytopathicity assay to assess resistance in the context of viral infection. Mutants confirmed to confer resistance in transient assays were rebuilt in the MV-Edm genome, recombinant virions were recovered, and virus-induced cytopathicities in the presence of different AS-48 concentrations were determined. For comparison, unmodified rMV-Edm (left) and rMV-Edm harboring F B3-2 (462N) (right) are shown. The values were normalized as described above and represent the means of four replicates.
To assess the contributions of individual mutations to the resistant phenotype, all changes were rebuilt individually in the F-Edm or, in the case of the B3-2 primary isolate, the F-B1 background and tested for the ability to mediate cell-to-cell fusion in the presence of compound in a quantitative assay. In five out of six resistant strains, resistance was mediated by a mutation of F residue N462 (Fig. 1B). In the case of mutant V, an accompanying M94V exchange in the F cavity domain was found, which we had previously observed to decrease sensitivity to inhibition (43). In one case (mutant III), resistance was based on an F A367T exchange (Fig. 1B). To test the relevance of these findings in the context of viral infection, recombinant virions harboring these point mutations were recovered and tested for sensitivity to inhibition by AS-48 (Fig. 1C) and OX-1 (not shown). All strains with a mutation of residue 462 showed >25-fold resistance to inhibition, while the recombinant harboring the A367T mutation was found to be approximately 15-fold less sensitive to inhibition (Table 1). These observations thus confirm the results of the plasmid-based fusion assay.
TABLE 1.
Identifications of mutations conferring resistance to compound-mediated inhibition
| Identifier | Straina | Compoundb | Passagec | Resistanced | Molecular basise |
|---|---|---|---|---|---|
| I | MV-Edm | OX-1 | 4 | >25 | F-N462S |
| II | MV-KS | AS-48 | 6 | >25 | F-N462D |
| III | MV-KS | AS-48 | 6 | 15 | F-A367T |
| IV | MV-KS | AS-48 | 7 | >25 | F-N462D |
| V | MV-KS | AS-48 | 7 | >25 | F-V94M |
| F-N462D | |||||
| VIf | MV-B3-2 | >25 | F-462K |
Input strain used for each independent adaptation procedure.
Compound used for adaptation (OX-1, 60 μM; AS-48, 15 μM [final concentration]).
Passage number at which extensive cytopathic effect was observed in the presence of compound.
Resistance (n-fold) to AS-48 of recombinant virions harboring the identified point mutation compared to that of the input strain or an rMV-Edm F-B3-2 (462N) variant.
Mutation conferring resistance as determined by transient-inhibition assay and resistance of recombinant virions.
Naturally resistant wild-type isolate MVi/lbadan.NIE/97/1 (MV-B3-2; genotype B3-2), assessed for compounds OX-1 and AS-48.
Residue 462 is part of the MV 6-HB fusion core structure.
In this study, we concentrated on residue 462 for further analysis, considering its high frequency of mutation. This residue is located in the N-terminal half of the HR-B domain of the F protein. Since the HR-B helix is part of the paramyxovirus 6-HB fusion core, for which structural information is available (1, 61), we generated a structural model of the MV F 6-HB on the basis of the coordinates reported for SV5 F (1) (Fig. 2A). In this model, N462 is localized in the vicinity of the contacting hydrophobic faces of HR-B and the HR-A trimer (Fig. 2B) and is postulated to engage in an electrostatic interaction with E170 in HR-A (Fig. 2C).
Synthetic HR-B-derived peptides have been shown to inhibit membrane fusion and hence viral entry for many viruses, including MV (31, 47, 58, 59), most likely by competing with endogenous HR-B for binding to HR-A. Indeed, a correlation between inhibitory potency and the stability of the corresponding helix bundle complexes has been demonstrated for peptidic, C-helix-derived inhibitors of HIV membrane fusion (6). In light of these findings, the compounds could directly interfere with 6-HB formation, and the microdomain around residue 462 in the MV F 6-HB structure could be part of an additional or alternative target site for OX-1 and AS-48.
AS-48 does not directly impair HR-B interaction with HR-A.
To test whether AS-48 directly interferes with interaction of the HR-B domain with the HR-A core trimer, we employed multiple strategies. First, we introduced the various mutations of residue 462 identified in the screen into the MV 6-HB structural model (Fig. 2D to F). If the compounds compete with HR-B for binding to HR-A, then mutations of residue 462 that confer resistance would increase the affinity of HR-B for HR-A. In all cases, however, molecular modeling predicted disruption of the hydrogen bonding and hence decreased rather than increased 6-HB stability due to lack of contact with 170E (mutation 462S) (Fig. 2D), charge incompatibility (mutation 462D) (Fig. 2E), or steric constraints (mutation 462K) (Fig. 2F). These predictions were quantitated by molecular-dynamics simulations. The simulations indicated that when subjected to molecular motion, the mutants dissociate to a greater extent than the wild type, allowing more of the mutant hydrophobic surfaces to be exposed to solvent, while the hydrophilic surfaces remain largely unaffected (Fig. 2G and H). Over the last 10 picoseconds of MD, wild-type HR-B exhibits an average hydrophobic solvent-accessible area of 78.5 nm2, compared to 80.2 nm2, 81.9 nm2, and 83.6 nm2 for 462S, 462D, and 462K, respectively.
Second, we employed a peptide competition assay to experimentally assess these modeling predictions. His-tagged variants of MV HR-B-derived peptides harboring either wild-type asparagine (N) or mutant serine (S) at the equivalent of position 462 were expressed, purified, and tested for the ability to inhibit viral entry and hence compete with endogenous HR-B for docking to HR-A. The peptide containing the N462S exchange was unable to inhibit MV-Edm even at very high concentrations of 600 μg/ml. A recombinant MV-Edm with an endogenous F N462S mutation, although more sensitive than the parent virus, likewise showed only partial inhibition by this peptide (Fig. 3A, left). In contrast, the unmodified peptide (462N) reduced yields of MV-Edm by >99% at 300 μg/ml, and recombinant virions containing point mutations at F residue 462 conferring resistance to the compounds revealed even greater sensitivity to this peptide (Fig. 3A, right). These findings thus corroborate the molecular-dynamics predictions and indicate a reduced affinity of all mutant HR-B variants for the HR-A trimer.
FIG. 3.
Resistance mutations at position 462 reduce the efficiency of 6-HB formation. (A) Peptide competition assay determining the efficiencies of virus inhibition by synthetic HR-B-derived peptides. (Left) Cells were infected with unmodified rMV-Edm or mutant rMV-Edm F (462S) in the presence of increasing concentrations of a modified peptide resembling one of the variants identified (462S; sequence shown above graph), and cell-associated viral titers were determined by TCID50 titration. (Right) Peptide competition assay of all rMV F variants described, using an unmodified synthetic peptide (462N; sequence shown above the graph). Shaded letters in the peptide sequences indicate the position of residue 462. To facilitate comparison of different strains, the values were normalized (norm.) for growth in the absence of compound and represent the means of two experiments. The error bars indicate standard deviations. (B) Quantitative cytopathicity assay to assess resistance of rMV-Edm F (E170A) virions to AS-48. Virus-induced cytopathicities were determined in the presence of different AS-48 concentrations. For comparison, unmodified rMV-Edm is shown. The values were normalized for cells infected in the presence of equal amounts of solvent (DMSO) only and represent the means of four replicates. (C) Coimmunoprecipitation of MV F with HR-B peptide in the presence (+) or absence (−) of AS-48. Cells expressing MV H and F (left) were incubated with 41 μM Flag-tagged peptide (pep) and 100 μM AS-48 or an equal amount of solvent (DMSO) and subjected to immunoprecipitation (IP) using anti-Flag antibodies, and precipitates were analyzed by Western blotting using anti-F tail antibodies. For control, immunoprecipitation was carried out in the absence of peptide. Cells expressing MV F only (right) were otherwise treated and analyzed in the same way.
Third, we addressed whether mutation of the HR-A binding partner of residue 462 in the MV 6-HB structure results in resistance to compound-mediated inhibition. This would again suggest for the compounds a mechanism of activity comparable to that of the peptidic HIV entry inhibitor T-20, for which escape mutants in HR-A have been reported (48). MV residue E170, which is predicted to engage with N462 in hydrogen bonding (Fig. 2C), was thus changed to alanine, and the corresponding recombinant virions were recovered. This rather drastic change should result in increased resistance of these recombinants to compound-mediated inhibition if the drugs physically engage this microdomain in HR-A. Dose-response curves generated for OX-1 (data not shown) and AS-48, however, showed no change in sensitivity to inhibition of the rMV-Edm F (E170A) virus compared to unmodified rMV-Edm (Fig. 3B), underlining the findings of the peptide competition assays.
Fourth, we compared the coimmunoprecipitation efficiencies of MV F with a Flag-tagged variant of HR-B-derived peptide in the presence or absence of a molar excess of AS-48 compound (Fig. 3C, left). Consistent with the previous assays, the presence of the compound did not prevent HR-B peptide interaction with MV F but rather resulted in increased coprecipitation efficiency, potentially due to arrest of the F trimer in a conformation that facilitates docking of the peptide. In the absence of coexpressed MV H, the coimmunoprecipitation efficiency of MV F with HR-B-derived peptides was overall very low and could not be increased by addition of AS-48 (Fig. 3C, right).
Taken together, these experiments indicate that while residue 462 contributes to 6-HB formation, the compounds do not directly interfere with this process.
Residues in the F cavity and at position 462 jointly control F activity.
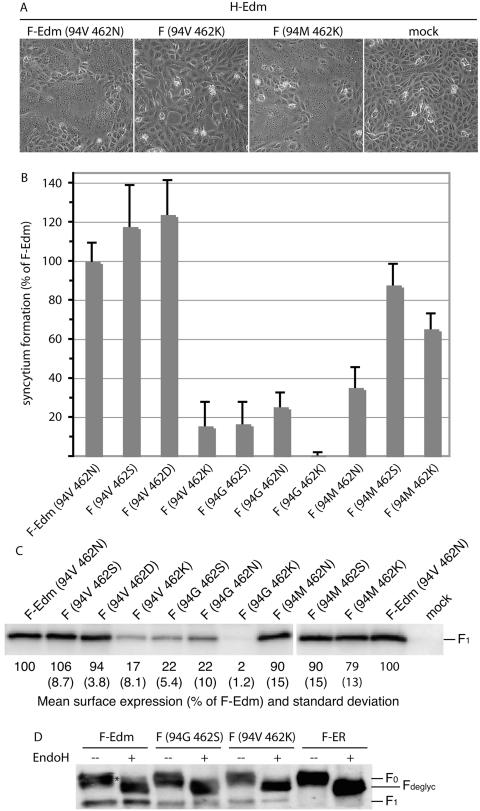
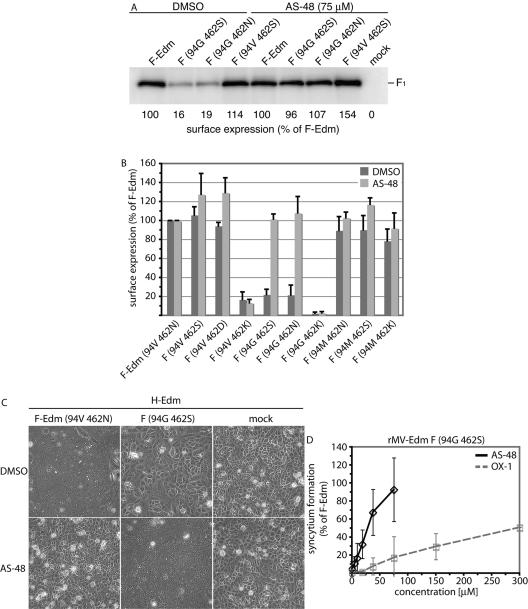
The inhibitors OX-1 and AS-48 were originally designed to dock in a defined microdomain located at the intersection of the neck and head domains of the F trimer that we previously found to be involved in the initiation of fusion (45). Mutations in this domain also cause resistance to inhibition by the compounds (43). To test whether this cavity-like domain and residue 462, although predicted to be located at different ends of the F structure, functionally interact in regulating F activity, we generated a series of F variants harboring mutations in both domains. When these constructs were coexpressed with MV H and their abilities to induce syncytium formation were assessed and quantified, several variants indeed demonstrated interdependent effects on fusion activity (exemplified in Fig. 4A and summarized in Fig. 4B). In particular, the 94V 462K and 94G 462S combinations resulted in an approximate 90% loss of activity, and the 94G 462K construct failed to induce syncytium formation. Loss of activity of these F variants mostly coincided with a reduction in their intracellular transport competence (see below). By contrast, the 94M 462K variant reached about 65% of the activity of F-Edm, and 94V 462S was fully active (Fig. 4B).
FIG. 4.
The nature of residue 94 in the F cavity determines the effects of changes of residue 462. (A) Syncytium formation after cotransfection of cells with plasmid DNA encoding MV H-Edm and MV F-Edm or F-Edm variants as indicated. Mock-transfected cells (mock) received MV H encoding plasmid only; the plates were photographed 20 h posttransfection at a magnification of ×200 (shown at ×186). (B) Fusogenicity of F-Edm variants cotransfected with H-Edm. Cytotoxicity as an indicator for the ability to induce syncytium formation was quantified as described for Fig. 1B. The values were normalized for unmodified F-Edm and represent the means of three independent experiments. The error bars indicate standard deviations. (C) Surface biotinylation of cells expressing different F-Edm variants to determine F plasma membrane steady-state levels. Biotinylated proteins were precipitated and separated by SDS-PAGE, and F-antigenic material was detected with specific antisera directed against the cytosolic domain of F. The values are based on densitometric quantification using a VersaDoc system and indicate the average percentage of surface material relative to unmodified F-Edm calculated from three to six independent experiments. Standard deviations are shown in parentheses. Mock-transfected cells received vector DNA only. (D) EndoH of F-antigenic material. The F0 fractions of both mutants analyzed were sensitive to EndoH treatment (Fdeglyc material), indicating an ER-type carbohydrate chain conformation. The asterisk marks the small Golgi fraction of EndoH-resistant F-Edm prior to proteolytic maturation. For control, an F variant (F-ER) carrying a KKXX ER retention signal was included. All samples were analyzed by SDS-PAGE and immunoblotting subsequent to EndoH treatment.
The fact that introduction of either 94M or 462K alone in the F-Edm background drastically reduces activity while the combination of 94M and 462K restores the ability to induce syncytium formation points to a functional interaction of both microdomains rather than an independent synergistic effect of phenotypes associated with the individual exchanges.
Loss of activity is linked to reduced intracellular transport of F mutants.
To determine whether such changes in activity are due to a loss of F functionality or to a lack of intracellular-transport competence, plasma membrane steady-state levels were assessed (Fig. 4C). Surface expression of the F variants 94V 462K and 94G 462S was greatly reduced, and mutant 94G 462K was almost completely retained, while insertion of a methionine residue in the F cavity (construct 94M 462K) greatly restored transport competence in the 462K background. This suggests intracellular retention as a predominant mechanism for the reduction or complete loss of fusion activity observed for certain double-mutant constructs.
Lack of transport competence could be based on misfolding or decreased conformational stability of the mutant F trimers, resulting in exposure of hydrophobic domains on the surfaces of the molecules that are normally oriented toward the inside of the native structure and hence shielded from recognition by host chaperones. In the case of the F protein, this may also extend to the hydrophobic fusion peptide domain, which is thought to be masked within the properly folded F trimer to facilitate transport competence (9, 12, 36). When selected mutants were subjected to endoglycosidase H treatment, we indeed observed complete sensitivity of the intracellular F material to deglycosylation by this enzyme, indicating retention in the endoplasmic reticulum (ER) of the host cell (Fig. 4D). A small fraction of endoglycosidase H-resistant uncleaved F0 material reflecting the F protein steady-state level in the Golgi is visible in the wild-type sample (Fig. 4D, lane 2). For comparison, an F variant (F-ER) carrying a KKXX ER retention motif was included, which we previously characterized as fully retained in the ER (44).
These observations indicate that the majority of the mutant F antigenic material is retained in the ER of the host cell rather than in downstream compartments of the secretory system.
Residues 94 in the cavity and 462 in HR-B are involved in controlling the conformational stability of the F trimer.
If mutations in the cavity domain and at position 462 reduce the conformational stability of the F trimer, rendering it intracellularly retained, incubation at reduced temperature should restore both transport competence and fusion activity, since it reduces the likelihood of intracellular F misfolding. Indeed, when we assessed transport competence upon incubation at 30°C or 37°C, we observed a fully restored plasma membrane steady-state level at lower temperature for several mutants (Fig. 5A). Furthermore, we quantified syncytium formation upon coexpression of these F mutants with MV H and incubation at 30°C or 37°C (Fig. 5B). In contrast to the unmodified F-Edm, all mutant constructs revealed a bona fide temperature-sensitive phenotype characterized by increased fusion activity at 30°C. Recovery of recombinant virions harboring one of these constructs (F 94G 462S) further confirmed this observation. While the final titers reached by MV-Edm were not significantly influenced by the incubation temperature, the mutant variant, when incubated at 37°C, reached only 10% of the maximal yield obtained at 30°C (data not shown).
FIG. 5.
F mutants 94G 462S and 94G 462N are temperature sensitive. (A) Surface biotinylation and detection of F-antigenic material, as previously described, after incubation of cells at 30°C or 37°C. The values represent densitometric quantification of surface-expressed material and are expressed as the percentage of unmodified F-Edm found at 30°C or 37°C, respectively. (B) Quantification of syncytium formation of selected F-Edm variants after cotransfection of cells with equal amounts of plasmid DNA encoding MV H and F and incubation at 30°C or 37°C as indicated. The values represent the means of four experiments and are expressed as the percentage of syncytium formation activity observed for unmodified F-Edm after incubation at 30°C or 37°C, respectively. For each F variant, the percent syncytium formation at 30°C compared to the same variant at 37°C is given above the graph. The error bars indicate standard deviations.
Compounds stabilize a transport-competent prefusion conformation.
Our previous characterization of the inhibitory compounds showed that the drugs prevent lipid mixing and hence an early phase of the fusion process (42, 43), possibly by stabilizing a native conformation of the F trimer. The compounds may thus be able to restore surface expression of the mutant F trimers, if a reduced conformational stability indeed constitutes the basis for intracellular retention of these constructs. Based on permeability predictions for OX-1 and AS-48 using the QikProp package (24), we expect the compounds to be membrane permeable and thus capable of docking to the F protein in the secretory system of the host cell. Strikingly, expression of temperature-sensitive F variants at 37°C in the presence of 75 μM AS-48 resulted in complete restoration of transport competence (Fig. 6A). While the double mutants 94G 462S and 94G 492N demonstrated the greatest increase in surface expression levels in the presence of compound, this finding extended to a lesser degree to several of the mutant F variants tested (Fig. 6B).
FIG. 6.
Compounds OX-1 and AS-48 stabilize a transport-competent conformation of mutant F trimers. (A) Surface expression of F mutants 94G 462S and 94G 462N subsequent to incubation of cells in the presence of 75 μM AS-48 or equal amounts of solvent (DMSO) only. The values represent densitometric quantification of surface material as described previously. (B) Quantification of surface expression of all F double mutants generated subsequent to incubation in the presence or absence of compound AS-48 as detailed in the legend to panel A. The values represent the means of three to six independent experiments, and standard deviations are shown. (C) Syncytium formation after cotransfection of cells with plasmid DNA encoding unmodified F-Edm or the 94G 462S F-Edm variant and MV H-Edm and incubation in the presence of 75 μM AS-48 or equal amounts of solvent (DMSO) only. Mock-transfected cells received H-Edm encoding plasmid only, and the plates were photographed 24 h posttransfection at a magnification of ×200 (shown at ×168). (D) Dose-dependent increase of fusion activity after cotransfection of cells with F variant 94G 462S and H-Edm and incubation in the presence of different concentrations of compound OX-1 or AS-48 as indicated. The values represent three independent experiments and are expressed as percentages of fusion activity induced by unmodified F-Edm in the presence of solvent (DMSO) only. The error bars indicate standard deviations.
Moreover, cotransfection of several double mutants, in particular the 94G 462S variant, with MV H in the presence of OX-1 or AS-48 resulted in a dose-dependent increase of fusion activity, achieving an F-Edm-like degree of activity at approximately 75 μM for AS-48 (shown for AS-48 in Fig. 6C and quantified for both drugs in Fig. 6D). This observation confirms that the compounds not only restore transport competence, but that, when incubated at 37°C in the presence of compound, the mutant F trimers reach the surface in a native, prefusion conformation. While high enough to restore intracellular transport, compound affinity for the mutant F variants once surface exposed is most likely too low to further prevent membrane fusion in the presence of fusion support provided by the H protein upon receptor binding. Although less likely on the basis of the available data, we cannot completely exclude the possibility that compound-facilitated F folding and fusion inhibition alternatively constitute mechanistically distinct events.
Taken together, these findings support a role for residue 462, together with residues in the cavity domain, in controlling the stability of a prefusion conformation of the F trimer in addition to its function in 6-HB formation at completion of the fusion process.
DISCUSSION
Taking advantage of a series of novel MV entry inhibitors developed in our laboratories, we report a role of residues in two distinct microdomains in the MV F protein in folding of the F trimer into a transport-competent conformation. While postulated to be physically distant in a structural model that is based on the coordinates reported for NDV F, residues in both domains contribute jointly to controlling the stability of a prefusion conformation of the F trimer, in addition to the role of a residue in one of these domains in 6-HB formation.
Screening of a panel of viral isolates representing MV strains of all genotypes currently endemic worldwide revealed effective inhibition of most isolates by the lead compound, AS-48 (42). A sub-Saharan strain, however, showed strong resistance to inhibition, mediated by a mutation of residue 462 in the F protein. Mutation of the same residue was also predominantly responsible for the resistance of four out of five spontaneous escape mutants that were generated by viral adaptation. In all cases, resistance was based on mutations in the F protein, corroborating the findings of the previous characterization of the activities of these inhibitors (42, 43). Furthermore, mutation of residue 462 was induced by adaptation in the presence of either the first-generation compound OX-1 or the second-generation analog AS-48, and all escape mutants analyzed were resistant to both compounds, suggesting that they share the same mechanism of fusion inhibition and recognize the same physical target site.
Resistance mediated by residue 462 could arise in several ways. It could be due to an indirect secondary effect, or the microdomain around the residue could be part of an alternative binding site in addition to the microdomain in the F neck region previously described (43, 45). Alternatively, the residues in this cavity-like domain might mediate secondary resistance, while N462 is part of the primary compound-binding site. The observation that all resistance-conferring mutations of residue 462 lower the affinity of HR-B for HR-A and are hence predicted to decrease the stability of the 6-HB, however, argues against direct interference of the compounds with 6-HB formation. This conclusion was corroborated both by coprecipitation of MV F with HR-B-derived peptides in the presence of compound and by the computationally predicted dynamic behavior of the MV 6-HB cores incorporating the mutant HR-B variants.
A hydrophobic pocket in RSV F HR-A has been postulated as the binding site for a small-molecule inhibitor of RSV (11). In MV HR-A, however, the analogous hydrophobic groove is located downstream from residue 462 in the 6-HB, and binding of AS-48 or OX-1 in this domain is hence not immediately compatible with resistance mediated by residue 462. In contrast to escape mutants reported for peptidic entry inhibitors of HIV (48), the absence of mutations in HR-A directly altering this hydrophobic region in response to our adaptation approach further argues against the region as a primary compound target site. This is corroborated by our observation that mutagenesis of the predicted HR-A binding partner of residue 462 in the MV 6-HB fusion core did not result in resistance to the drugs.
A previous analysis of the cavity domain in the MV F neck region has pointed to a role for the microdomain in the initiation phase of the fusion process (45). Certain double-mutant combinations of residues 462 and 94 lose intracellular-transport competence under standard conditions, resulting in a temperature-sensitive phenotype. Since this does not reflect an independent synergistic effect of both microdomains—rather, the physical nature of residues in one domain determines the outcome of mutations in the other regarding F transport competence—both domains appear functionally linked. The F neck region and HR-B are found at opposing ends of the F structural model, and it thus seems unlikely that residues in these domains make direct contact or that the compound physically engages both domains simultaneously, although we cannot exclude the possibility that the inhibitor binds to an intermediate conformation in which these domains are closer together (12).
A dose-dependent increase in intracellular transport and ultimately fusion activity when certain double-mutant F variants are incubated in the presence of compound at 37°C supports the conclusion that the inhibitors stabilize a transport-competent prefusion conformation of the F trimer. Multiple reports have shown that proteolytic maturation of the F0 protein in the late Golgi apparatus results in structural rearrangements of the F trimer (16, 22, 28, 57). Antibodies directed against the HR domains of SV5 recognize only the uncleaved F protein (16), and hydrophobic domains were found to be exposed in the cleaved F trimer of Sendai virus (22). Burying these domains in the F trimer prior to proteolytic cleavage, and hence shielding them from recognition by the quality control machinery of the host cell monitoring proper protein folding, appears to be critical for efficient surface expression.
Several lines of evidence suggest that residues located within or adjacent to HR domains have distinct functions in different stages of the fusion process to conserve the metastable native structure of type I FMGs until a target membrane is present: residues in the HR-A and HR-B domains of HIV gp41 contribute to the stability of the native gp120-gp41 complex (18, 35), several residues located in the influenza hemagglutinin HR-A domain have been shown to change binding partners during the transition of hemagglutinin from the native to the fusogenic conformation (5, 8, 13), and two residues upstream of the HR-B domain of the paramyxovirus SV5 F are postulated to function as a regulatory switch for activation and membrane fusion (51). These SV5 F residues, 447 and 449, are thought both to control the initiation phase of the fusion process and to influence 6-HB stability. Since our experiments indicate that MV F residue 462 is linked to maintaining the F trimer in a transport-competent prefusion conformation, in addition to its function in 6-HB formation at the completion of the fusion process, we suggest a similar dual role for this residue in MV F. Conceivably, a dual regulatory function of residues located near the beginning of HR-B is conserved within paramyxovirus F proteins.
Our findings confirm that the novel MV entry inhibitors, in addition to their therapeutic potential, constitute valuable tools for studying the mechanism of MV F-mediated membrane fusion. In turn, further elucidation of the mechanism of membrane fusion may allow better optimization of the inhibitor compounds and thus increase of their antiviral potentials.
Acknowledgments
We are grateful to P. A. Rota for generously providing MV isolates B3-2 and B1, R. Cattaneo for antibodies directed against the MV F tail, and A. L. Hammond for critical reading of the manuscript.
This work was supported by Public Health Service grant AI057157 to the Southeastern Regional Center of Excellence for Emerging Infections and Biodefense (to R.W.C.) and a pilot grant from the Southeastern Center for Emerging Biological Threats (Grant/Cooperative Agreement number U38/CCU423095 from the CDC), a research grant from the American Lung Association, and Public Health Service grant AI056179 from NIH/NIAID (all to R.K.P.).
The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the CDC.
REFERENCES
- 1.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Efraim, I., Y. Kliger, C. Hermesh, and Y. Shai. 1999. Membrane-induced step in the activation of Sendai virus fusion protein. J. Mol. Biol. 285:609-625. [DOI] [PubMed] [Google Scholar]
- 3.Berendson, H. J. C., D. Vanderspoel, and R. Vandrunen. 1995. Gromacs—a message-passaging parallel molecular-dynamics implementation. Comp. Phys. Commun. 91:43-56. [Google Scholar]
- 4.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 5.Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37-43. [DOI] [PubMed] [Google Scholar]
- 6.Chan, D. C., C. T. Chutkowski, and P. S. Kim. 1998. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc. Natl. Acad. Sci. USA 95:15613-15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 8.Chen, J., J. J. Skehel, and D. C. Wiley. 1999. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc. Natl. Acad. Sci. USA 96:8967-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, L., J. J. Gorman, J. McKimm-Breschkin, L. J. Lawrence, P. A. Tulloch, B. J. Smith, P. M. Colman, and M. C. Lawrence. 2001. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure 9:255-266. [DOI] [PubMed] [Google Scholar]
- 10.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cianci, C., D. R. Langley, D. D. Dischino, Y. Sun, K. L. Yu, A. Stanley, J. Roach, Z. Li, R. Dalterio, R. Colonno, N. A. Meanwell, and M. Krystal. 2004. Targeting a binding pocket within the trimer-of-hairpins: small-molecule inhibition of viral fusion. Proc. Natl. Acad. Sci. USA 101:15046-15051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colman, P. M., and M. C. Lawrence. 2003. The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell Biol. 4:309-319. [DOI] [PubMed] [Google Scholar]
- 13.Daniels, P. S., S. Jeffries, P. Yates, G. C. Schild, G. N. Rogers, J. C. Paulson, S. A. Wharton, A. R. Douglas, J. J. Skehel, and D. C. Wiley. 1987. The receptor-binding and membrane-fusion properties of influenza virus variants selected using anti-haemagglutinin monoclonal antibodies. EMBO J. 6:1459-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, X. Wu, W. A. O'Brien, L. Ratner, J. C. Kappes, G. M. Shaw, and E. Hunter. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 74:8358-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doms, R. W., and J. P. Moore. 2000. HIV-1 membrane fusion: targets of opportunity. J. Cell Biol. 151:F9-F14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dutch, R. E., R. N. Hagglund, M. A. Nagel, R. G. Paterson, and R. A. Lamb. 2001. Paramyxovirus fusion (F) protein: a conformational change on cleavage activation. Virology 281:138-150. [DOI] [PubMed] [Google Scholar]
- 17.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 18.Follis, K. E., S. J. Larson, M. Lu, and J. H. Nunberg. 2002. Genetic evidence that interhelical packing interactions in the gp41 core are critical for transition of the human immunodeficiency virus type 1 envelope glycoprotein to the fusion-active state. J. Virol. 76:7356-7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffin, D. E. 2001. Measles virus, p. 1401-1442. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 20.Heil, M. L., J. M. Decker, J. N. Sfakianos, G. M. Shaw, E. Hunter, and C. A. Derdeyn. 2004. Determinants of human immunodeficiency virus type 1 baseline susceptibility to the fusion inhibitors enfuvirtide and T-649 reside outside the peptide interaction site. J. Virol. 78:7582-7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hernandez, L. D., L. R. Hoffman, T. G. Wolfsberg, and J. M. White. 1996. Virus-cell and cell-cell fusion. Annu. Rev. Cell Dev. Biol. 12:627-661. [DOI] [PubMed] [Google Scholar]
- 22.Hsu, M., A. Scheid, and P. W. Choppin. 1981. Activation of the Sendai virus fusion protein (f) involves a conformational change with exposure of a new hydrophobic region. J. Biol. Chem. 256:3557-3563. [PubMed] [Google Scholar]
- 23.Hu, X. L., R. Ray, and R. W. Compans. 1992. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J. Virol. 66:1528-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jorgensen, W. L. 2004. The many roles of computation in drug discovery. Science 303:1813-1818. [DOI] [PubMed] [Google Scholar]
- 25.Jorgenson, W. L., J. Chandrasekhar, J. D. Madura, R. W. Impey, and M. L. Klein. 1983. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79:926-935. [Google Scholar]
- 26.Jorgenson, W. L., and J. Tirado-Rives. 1988. The Opls potential functions for proteins—energy minimizations for crystals of cyclic-peptides and crambin. J. Am. Chem. Soc. 110:1657-1666. [DOI] [PubMed] [Google Scholar]
- 27.Kielian, M., and S. Jungerwirth. 1990. Mechanisms of enveloped virus entry into cells. Mol. Biol. Med. 7:17-31. [PubMed] [Google Scholar]
- 28.Kohama, T., W. Garten, and H. D. Klenk. 1981. Changes in conformation and charge paralleling proteolytic activation of Newcastle disease virus glycoproteins. Virology 111:364-376. [DOI] [PubMed] [Google Scholar]
- 29.Lamb, R. A. 1993. Paramyxovirus fusion: a hypothesis for changes. Virology 197:1-11. [DOI] [PubMed] [Google Scholar]
- 30.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 31.Lambert, D. M., S. Barney, A. L. Lambert, K. Guthrie, R. Medinas, D. E. Davis, T. Bucy, J. Erickson, G. Merutka, and S. R. Petteway, Jr. 1996. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc. Natl. Acad. Sci. USA 93:2186-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, J., V. R. Melanson, A. M. Mirza, and R. M. Iorio. 2005. Decreased dependence on receptor recognition for the fusion promotion activity of L289A-mutated Newcastle disease virus fusion protein correlates with a monoclonal antibody-detected conformational change. J. Virol. 79:1180-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindahl, E., B. Hess, and D. van der Spoel. 2001. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. 7:306-317. [Google Scholar]
- 34.Lovell, S. C., J. M. Word, J. S. Richardson, and D. C. Richardson. 2000. The penultimate rotamer library. Proteins 40:389-408. [PubMed] [Google Scholar]
- 35.Lu, M., M. O. Stoller, S. Wang, J. Liu, M. B. Fagan, and J. H. Nunberg. 2001. Structural and functional analysis of interhelical interactions in the human immunodeficiency virus type 1 gp41 envelope glycoprotein by alanine-scanning mutagenesis. J. Virol. 75:11146-11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ludwig, K., B. Baljinnyam, A. Herrmann, and C. Bottcher. 2003. The 3D structure of the fusion primed Sendai F-protein determined by electron cryomicroscopy. EMBO J. 22:3761-3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paterson, R. G., S. W. Hiebert, and R. A. Lamb. 1985. Expression at the cell surface of biologically active fusion and hemagglutinin/neuraminidase proteins of the paramyxovirus simian virus 5 from cloned cDNA. Proc. Natl. Acad. Sci. USA 82:7520-7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paterson, R. G., C. J. Russell, and R. A. Lamb. 2000. Fusion protein of the paramyxovirus SV5: destabilizing and stabilizing mutants of fusion activation. Virology 270:17-30. [DOI] [PubMed] [Google Scholar]
- 40.Peisajovich, S. G., O. Samuel, and Y. Shai. 2000. Paramyxovirus F1 protein has two fusion peptides: implications for the mechanism of membrane fusion. J. Mol. Biol. 296:1353-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plemper, R. K., and R. W. Compans. 2003. Mutations in the putative HR-C region of the measles virus F2 glycoprotein modulate syncytium formation. J. Virol. 77:4181-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plemper, R. K., J. Doyle, A. Sun, A. Prussia, L. T. Cheng, P. A. Rota, D. C. Liotta, J. P. Snyder, and R. W. Compans. 2005. Design of a small-molecule entry inhibitor with activity against primary measles virus strains. Antimicrob. Agents Chemother. 49:3755-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plemper, R. K., K. J. Erlandson, A. S. Lakdawala, A. Sun, A. Prussia, J. Boonsombat, E. Aki-Sener, I. Yalcin, I. Yildiz, O. Temiz-Arpaci, B. Tekiner, D. C. Liotta, J. P. Snyder, and R. W. Compans. 2004. A target site for template-based design of measles virus entry inhibitors. Proc. Natl. Acad. Sci. USA 101:5628-5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plemper, R. K., A. L. Hammond, and R. Cattaneo. 2001. Measles virus envelope glycoproteins hetero-oligomerize in the endoplasmic reticulum. J. Biol. Chem. 276:44239-44246. [DOI] [PubMed] [Google Scholar]
- 45.Plemper, R. K., A. S. Lakdawala, K. M. Gernert, J. P. Snyder, and R. W. Compans. 2003. Structural features of paramyxovirus F protein required for fusion initiation. Biochemistry 42:6645-6655. [DOI] [PubMed] [Google Scholar]
- 46.Radecke, F., P. Spielhofer, H. Schneider, K. Kaelin, M. Huber, C. Dotsch, G. Christiansen, and M. A. Billeter. 1995. Rescue of measles viruses from cloned DNA. EMBO J. 14:5773-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rapaport, D., M. Ovadia, and Y. Shai. 1995. A synthetic peptide corresponding to a conserved heptad repeat domain is a potent inhibitor of Sendai virus-cell fusion: an emerging similarity with functional domains of other viruses. EMBO J. 14:5524-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rimsky, L. T., D. C. Shugars, and T. J. Matthews. 1998. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J. Virol. 72:986-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodriguez, R., G. Chinea, N. Lopez, T. Pons, and G. Vriend. 1998. Homology modeling, model and software evaluation: three related resources. Bioinformatics 14:523-528. [DOI] [PubMed] [Google Scholar]
- 50.Russell, C. J., T. S. Jardetzky, and R. A. Lamb. 2001. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 20:4024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Russell, C. J., K. L. Kantor, T. S. Jardetzky, and R. A. Lamb. 2003. A dual-functional paramyxovirus F protein regulatory switch segment: activation and membrane fusion. J. Cell Biol. 163:363-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sergel, T. A., L. W. McGinnes, and T. G. Morrison. 2000. A single amino acid change in the Newcastle disease virus fusion protein alters the requirement for HN protein in fusion. J. Virol. 74:5101-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seth, S., A. Vincent, and R. W. Compans. 2003. Mutations in the cytoplasmic domain of a paramyxovirus fusion glycoprotein rescue syncytium formation and eliminate the hemagglutinin-neuraminidase protein requirement for membrane fusion. J. Virol. 77:167-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 55.Tanabayashi, K., and R. W. Compans. 1996. Functional interaction of paramyxovirus glycoproteins: identification of a domain in Sendai virus HN which promotes cell fusion. J. Virol. 70:6112-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsurudome, M., M. Ito, M. Nishio, M. Kawano, H. Komada, and Y. Ito. 2001. Hemagglutinin-neuraminidase-independent fusion activity of simian virus 5 fusion (F) protein: difference in conformation between fusogenic and nonfusogenic F proteins on the cell surface. J. Virol. 75:8999-9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Umino, Y., T. Kohama, T. A. Sato, A. Sugiura, H. D. Klenk, and R. Rott. 1990. Monoclonal antibodies to three structural proteins of Newcastle disease virus: biological characterization with particular reference to the conformational change of envelope glycoproteins associated with proteolytic cleavage. J. Gen. Virol. 71:1189-1197. [DOI] [PubMed] [Google Scholar]
- 58.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Matthews. 1994. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA 91:9770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yao, Q., and R. W. Compans. 1996. Peptides corresponding to the heptad repeat sequence of human parainfluenza virus fusion protein are potent inhibitors of virus infection. Virology 223:103-112. [DOI] [PubMed] [Google Scholar]
- 60.Yin, H. S., R. G. Paterson, X. Wen, R. A. Lamb, and T. S. Jardetzky. 2005. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. USA 102:9288-9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao, X., M. Singh, V. N. Malashkevich, and P. S. Kim. 2000. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc. Natl. Acad. Sci. USA 97:14172-14177. [DOI] [PMC free article] [PubMed] [Google Scholar]