Abstract
The most intensively studied rotavirus strains initially attach to cells when the “heads” of their protruding spikes bind cell surface sialic acid. Rotavirus strains that cause disease in humans do not bind this ligand. The structure of the sialic acid binding head (the VP8* core) from the simian rotavirus strain RRV has been reported, and neutralization epitopes have been mapped onto its surface. We report here a 1.6-Å resolution crystal structure of the equivalent domain from the sialic acid-independent rotavirus strain DS-1, which causes gastroenteritis in humans. Although the RRV and DS-1 VP8* cores differ functionally, they share the same galectin-like fold. Differences between the RRV and DS-1 VP8* cores in the region that corresponds to the RRV sialic acid binding site make it unlikely that DS-1 VP8* binds an alternative carbohydrate ligand in this location. In the crystals, a surface cleft on each DS-1 VP8* core binds N-terminal residues from a neighboring molecule. This cleft may function as a ligand binding site during rotavirus replication. We also report an escape mutant analysis, which allows the mapping of heterotypic neutralizing epitopes recognized by human monoclonal antibodies onto the surface of the VP8* core. The distribution of escape mutations on the DS-1 VP8* core indicates that neutralizing antibodies that recognize VP8* of human rotavirus strains may bind a conformation of the spike that differs from those observed to date.
Rotavirus is the most important cause of severely dehydrating childhood gastroenteritis worldwide (37). To prime the nonenveloped virion for host membrane penetration, intestinal trypsin cleaves the rotavirus spike protein, VP4, into two fragments, VP8* and VP5* (15). In some strains, VP8* mediates initial attachment to target cells by binding cell surface sialic acid (SA) (6, 16). In electron cryomicroscopy image reconstructions of trypsin-primed, SA-dependent virions, the protruding part of the spikes has approximate twofold symmetry (Fig. 1). It is tipped by paired heads, separated by a small gap (41, 44). The heads are formed by a globular domain of VP8* (14). We refer to this globular domain as the “VP8* core,” because it remains intact following limit protease digestion of recombinant VP4 (12). VP5* forms more virion-proximal parts of the spikes (Fig. 1) and has a hydrophobic apex (13), which has been implicated as a potential membrane penetration region (32). VP8* masks the hydrophobic apex of VP5* on primed spikes (13). During cell entry, VP8* probably separates from VP5*, exposing the hydrophobic apex and allowing a fold-back rearrangement of VP5*. Some antibodies that bind VP8* appear to neutralize virus by triggering uncoating—the shedding of VP4 or its fragments and the coat protein VP7 (45). This mechanism of neutralization suggests that conformational changes involving VP8* could trigger subsequent entry events, such as VP5* rearrangement and outer-layer disassembly. VP8* may also function intracellularly, binding intracellular tumor necrosis factor receptor-associated factors to activate cellular signaling pathways (28).
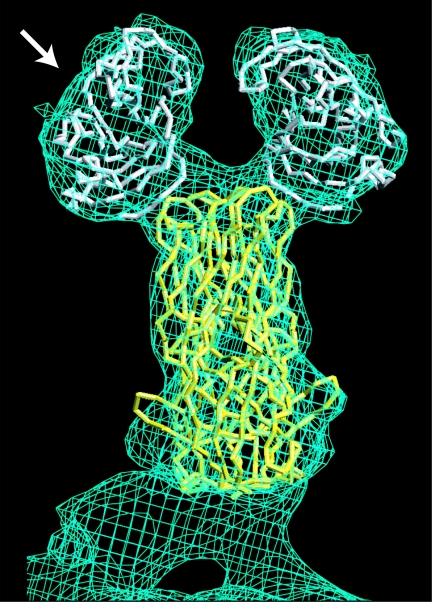
FIG. 1.
The rotavirus VP4 spike. The Cα traces of the VP8* core (white) and a globular domain of VP5* (yellow) from RRV are fitted to the molecular envelope of the VP4 spike in an approximately 12-Å-resolution electron cryomicroscopy image reconstruction of a primed SA11-4F rotavirus virion. The arrow indicates the perspective of the depictions in Fig. 3 and 4B.
Although SA was the first rotavirus receptor identified (2), most rotavirus strains do not, in fact, bind this receptor during entry (6). None of the strains that are known to be virulent in humans bind SA (6). Differences between SA-dependent and SA-independent strains extend beyond the ability or inability of their spike proteins to bind SA: SA-independent strains are generally more fastidious in cell culture than SA-dependent strains (40, 42), and although SA-independent strains infect polarized epithelial cells from either the apical or basolateral membrane, SA-dependent strains enter only at the apical surface (4).
The VP8* core is an important target of neutralizing antibodies against rotavirus (reviewed in reference 14). Some neutralizing monoclonal antibodies (MAbs) that recognize this domain on SA-dependent strains protect mouse pups from rotavirus diarrhea when present in the gut lumen (33). The VP8* core is the major determinant of P serotype (which correlates reasonably well with P genotype) for both SA-dependent and SA-independent strains (22). Therefore, this domain contains key neutralization determinants for both functional variants. While most VP4-specific MAbs that neutralize SA-dependent rotavirus virions map to the VP8* fragment, most VP4-specific MAbs that neutralize SA-independent virions map to the VP5* fragment (reviewed in reference 25). It is not known whether this difference reflects biological differences between SA-dependent and SA-independent strains or the use of different strategies to screen hybridomas.
Although understanding the structure and function of SA-independent strains is particularly important for understanding and preventing rotavirus gastroenteritis in humans, SA-independent strains are less studied than SA-dependent strains. X-ray crystal and nuclear magnetic resonance (NMR) solution structures of the VP8* core of an SA-dependent simian rotavirus strain (RRV) have been obtained (14), but neither electron cryomicroscopy-based image reconstructions nor high-resolution structures are available for any SA-independent strain. Therefore, the structural basis for the differences in VP8* function is not known. Here, we present the X-ray crystal structure of the VP8* core from rotavirus strain DS-1, an SA-independent P genotype 4 strain that causes gastroenteritis in humans. We also report neutralization escape mutations of VP8* selected by two human monoclonal antibodies derived from cDNA of B lymphocytes from naturally infected humans. Mapping neutralization escape mutations onto the DS-1 VP8* core structure suggests that VP8*-specific antibodies that neutralize SA-dependent and SA-independent rotavirus strains recognize different conformations of the spike protein.
MATERIALS AND METHODS
Subcloning of the DS-1 and KU VP8* cores.
Genes encoding the VP8* cores of rotavirus strains DS-1 and KU were subcloned from recombinant baculoviruses that contain the full-length DS-1 and KU VP4 coding sequences (18). Primers to amplify the KU VP8* coding sequence were designed based on a previously banked KU VP4 sequence (GenBank accession code M21014). The sequence of the DS-1 VP8* core was determined from DNA amplified by PCR from the recombinant baculovirus using primers complementary to sequences in the BlueBac2 vector on either side of the VP4 coding sequence. The sequences encoding KU and DS-1 VP4 residues 60 to 223 (the VP8* cores) were isolated as PCR products with XhoI and NotI ends and with a potential trypsin cleavage site just N terminal to residue 60. Each fragment was subcloned into the multiple cloning site of a pGex 4T-1 vector (Amersham Biosciences) that had been digested with SalI and NotI.
Expression and purification of recombinant VP8* cores.
The RRV, DS-1, and KU VP8* cores were expressed in Escherichia coli strain BL21 and purified as described previously (14). Overnight cultures were diluted 1:100 in LB containing 0.1 mg/ml ampicillin and incubated at 37°C. When the cultures reached an A600 of about 0.6, the temperature was reduced to 25°C. After 30 to 60 min of incubation at 25°C, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to 1 mM. The cultures were then incubated for an additional 4 h at 25°C before being harvested. Pelleted bacteria were lysed by sonication. The lysates were clarified by centrifugation in a Ti45 rotor (Beckman) at 40,000 rpm for 2 h at 4°C. The clarified lysates were passed over a glutathione-Sepharose column (Amersham Biosciences), and the VP8* cores were cleaved from the bound glutathione S-transferase (GST) tags by incubation on the column with 5 μg/ml tosylsulfonyl phenylalanyl chloromethyl ketone-treated trypsin (Worthington Biochemical) in TNC (20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM CaCl2) for 2 h at room temperature. The trypsin was removed from the released VP8* by passage over a benzamidine-Sepharose column (Amersham Biosciences), and any residual tryptic activity was eliminated by the addition of 1 mM phenylmethylsulfonyl fluoride and 2.5 mM benzamidine. The VP8* cores were further purified by size exclusion chromatography on a Superdex 200 column (Amersham Biosciences) in TNE (20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA) at 4°C using a fast-protein liquid chromatography system (Amersham Biosciences).
Crystallization.
The DS-1 VP8* core crystallized in space group P1 with the following unit cell parameters: a = 42.42 Å, b = 84.12 Å, c = 90.77 Å, α = 90.03°, β = 90.02°, and γ = 75.54°. The crystals have a solvent content of 38.5%, and each asymmetric unit contains eight VP8* cores. Crystals formed within 1 week at 20°C in hanging drops, with a sample solution containing 5 to 10 mg/ml protein in TNE-0.02% sodium azide-0.1 mM benzamidine, mixed 1:1 with a well solution containing 20% polyethylene glycol 4000, 500 mM sodium chloride, 100 mM sodium citrate, pH 5.6, and 3% ethanol or ethylene glycol. Harvested crystals were soaked in a cryoprotectant solution containing 22% polyethylene glycol 4000, 550 mM sodium chloride, 100 mM sodium citrate, pH 5.6, 3% ethanol or ethylene glycol, 15% glycerol and then frozen by rapid immersion in liquid nitrogen.
X-ray diffraction data collection and processing.
X-ray diffraction data were collected at 100 K and a wavelength of 0.916 Å using beam line F1 at the Cornell High Energy Synchrotron Source. Diffracted X rays were detected by an ADSC Quantum 4 charge-coupled device. Diffraction data were indexed, integrated, and scaled using HKL2000 (HKL Research, Inc.). (For scaling statistics, see Table 2.)
TABLE 2.
Crystallographic data collection and refinement statistics
| Statistic | Value |
|---|---|
| Data collection | |
| Resolution limit (Å) | 1.6 |
| No. of unique reflections | 153,466 |
| Redundancya | 1.95 (1.72) |
| Completenessa (%) | 96.8 (94.7) |
| I/σa | 17.2 (3.4) |
| Rsyma,b (%) | 5.0 (37.6) |
| Refinement | |
| No. of polypeptide chains | 8 |
| No. of protein atoms | 10,762 |
| No. of water molecules | 1,724 |
| No. of glycerol molecules | 6 |
| No. of ethanol molecules | 4 |
| No. of amino acids with alternative conformations | 43 |
| Residues in allowed regions of Ramachandran plot (%) | 100 |
| Residues in most favored regions of Ramachandran plot (%) | 91.8 |
| RMSD bond lengths (Å) | 0.011 |
| RMSD bond angles (°) | 1.31 |
| Mean B value (Å2) | 24.91 |
| RMSD main chain B (Å2) | 1.019 |
| Resolution range (Å) | 14.9-1.6 |
| R factorc | 0.158 |
| Free R factorc | 0.192 |
Value for the highest-resolution shell is given in parentheses.
Rsym = Σ(I − 〈I〉)/ΣI. 〈I〉 is the average intensity over symmetry-equivalent reflections.
R factor = (Σ‖Fobs| − |Fcalc‖)/Σ|Fobs|, where the summations is over the working set of reflections. For the free R factor, the summation is over the test set of reflections (5% of the total reflections).
Structure determination and refinement.
The DS-1 VP8* core structure was determined by molecular replacement, using an initial phasing model based on the previously determined structure of the SA-bound RRV VP8* core (14). Rotation and translation solutions were determined using CNS (3). The structure was refined by multiple cycles of simulated annealing, energy minimization, and individual B-factor refinement using CNS (3). The model was rebuilt in O (24) using simulated annealing omit maps calculated in CNS (3) to eliminate phasing bias from the molecular replacement model. The geometry of the structures was analyzed using PROCHECK (29). (For refinement statistics, see Table 2.)
NMR spectroscopy.
Purified protein for NMR was prepared as described above, except that size exclusion chromatography was carried out in 20 mM sodium phosphate, pH 7.0-100 mM NaCl, and the resulting protein sample was dialyzed into 20 mM sodium phosphate, pH 7.0-10 mM NaCl. The assayed peptide was 15N labeled on valine and had the sequence TVEPVS, corresponding to DS-1 VP4 residues 60 to 64 plus a C-terminal serine. To avoid competition between the peptide and residues of the authentic N-terminal leader of the unlabeled DS-1 VP8* core, residues 60 to 64 of the core were mutated to SGSGG using PCR. Samples were made 10% in D2O for field locking. Spectra were obtained at 25°C using a 500-MHz Bruker spectrometer equipped with a cryoprobe. Two-dimensional 15N-1H heteronuclear single quantum coherence spectra showed no change in the chemical shifts of the 15N valines of the peptide when 0.29 mM DS-1 VP8* core was mixed with 0.1 mM peptide. NMR data were processed using PROSA (20), and spectra were analyzed using XEASY (1).
Structural analysis and figure production.
Structure alignments were calculated using LSQKAB in CCP4 (7). Amino acid variability was calculated by AMAS (30). To determine the accessibility of VP8* core surfaces for antibody binding on trypsin-primed virions, the DS-1 VP8* core structure was aligned with an RRV VP8* core structure that had been fitted to an approximately 12-Å electron cryomicroscopy-based envelope of trypsin-primed SA11-4F rotavirus particles, contoured at 0.5 σ, as previously described (13). The envelope and fitted crystal structure were probed with the crystal structure of an Fab of a human immunoglobulin G1(κ) in complex with Lewis Y nonoate methyl ester (Protein Data Bank accession code 1CLY) using O (23, 24). Residues that could be brought into contact with any portion of the antigen-combining site were scored as accessible. The figures were made using GRASP (34), MOLSCRIPT (27), Illustrator (Adobe Systems, Inc.), and Photoshop (Adobe Systems, Inc.).
Selection of neutralization escape mutants.
Rotavirus strain KU (30 μl; 3.6 × 105 PFU) was pretreated with trypsin (10 μg/ml) and mixed with 70 μl of purified 1-2H (200 μg/ml) or 2-3E (50 μg/ml) Fab in Eagle's minimum essential medium (MEM). After 1 h of incubation at 37°C, the virus-antibody mixtures were inoculated onto MA104 cell monolayers in 1 ml of Eagle's MEM in roller tubes (15 mm by 15 cm). After adsorption for 1 h, the monolayers were washed with phosphate-buffered saline and then incubated in a rotator (RT-550; Taitec Inc.) with 1 ml of Eagle's MEM containing purified 1-2H (100 μg/ml) or 2-3E (25 μg/ml) antibody and trypsin (5 μg/ml). Cultures were harvested 7 days after infection. After three rounds of propagation in the presence of neutralizing antibody, the selected viruses were plaque purified on CV-1 cells.
Sequencing of rotavirus variants.
Full-length cDNA encoding VP4 and VP7 was prepared by reverse transcription-PCR. Nucleotide sequences were determined directly from the PCR products using ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kits (PE Biosystems) with an automated sequencer, the ABI Prism 310 Genetic Analyzer (PE Applied Biosystems). The VP4-encoding genes of the two selected viruses each differed from the parental gene by a single nucleotide substitution. The VP7-encoding genes contained no mutations.
Protein structure and nucleotide sequence accession numbers.
The atomic coordinates and structure factor amplitudes for the DS-1 VP8* core have been deposited in the Protein Data Bank under accession code 2AEN. Nucleotide sequences have been deposited in GenBank with the following accession codes: DS-1 VP8* core, DQ141310; KU VP4 clone 1, AB222784; M-KU-1-2H, AB222785; M-KU-2-3E, AB222786.
RESULTS AND DISCUSSION
Expression, purification, and stability of rotavirus VP8* cores.
The VP8* core is a compact domain that presents conformational neutralizing epitopes. Our previously reported data demonstrate that the RRV VP8* core (P genotype 3) can be expressed in E. coli as a soluble GST fusion protein (14). After glutathione affinity chromatography, the domain can be separated from its GST tag by trypsin cleavage and purified by size exclusion chromatography with high yield as a homogeneous, protease-resistant, and very soluble protein (14).
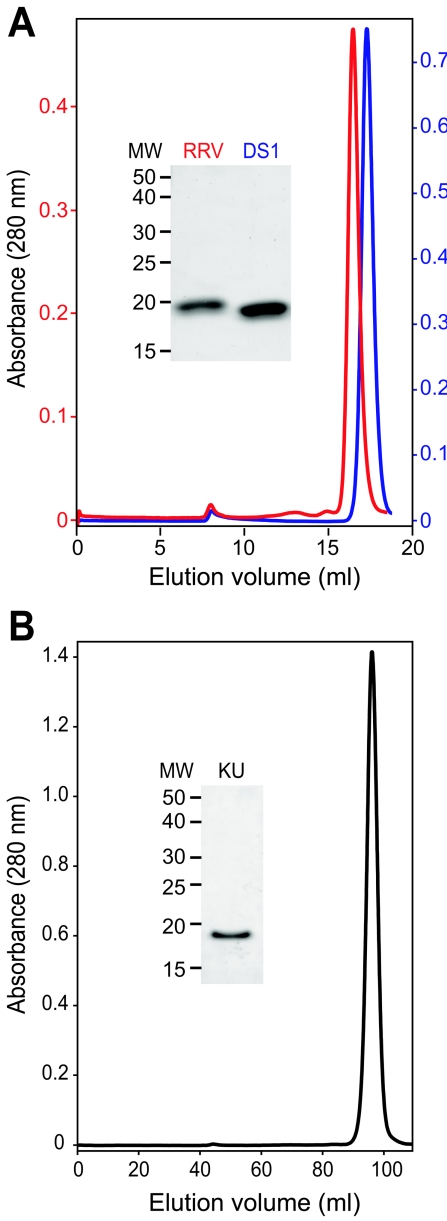
We tested whether the VP8* core of an SA-independent strain that causes gastroenteritis in humans has similarly favorable characteristics. When expressed and purified using the protocol developed for the RRV VP8* core, the DS-1 VP8* core (P genotype 4) was obtained in good yield as a homogeneous, protease-resistant, soluble, pure protein (Fig. 2A and Table 1). As assayed by size exclusion chromatography, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), matrix-assisted laser desorption ionization mass spectrometry, and N-terminal sequencing, the RRV and DS-1 VP8* cores remain intact, homogeneous, and soluble after storage at 4°C for approximately 2 years (Fig. 2A, Table 1, and data not shown).
FIG. 2.
Size exclusion chromatography and SDS-PAGE of purified VP8* cores. (A) Chromatograms of RRV (red) and DS-1 (blue) VP8* cores, which were separated on a Superdex 200 10/30 column at 4°C in TNE after storage for 2 years at 4°C in TNE with 0.02% sodium azide and 0.1 mM benzamidine. The inset image of a Coomassie blue-stained SDS-PAGE gel shows stored samples prior to chromatography. (B) Chromatogram of a freshly prepared sample of the KU VP8* core, which was separated on a Hi-Load Superdex 200 16/60 column at 4°C in TNE. The inset image of a Coomassie-blue stained SDS-PAGE gel shows protein from the pooled peak fractions. Apparent molecular masses that correspond to peak elution volumes are listed in Table 1.
TABLE 1.
Biochemical characteristics of the RRV, DS-1, and KU VP8* cores
| Characteristic | Value
|
||
|---|---|---|---|
| RRV | DS-1 | KU | |
| No. of rotavirus VP4 residues in construct | 60-224 | 60-223 | 60-223 |
| Predicted MW | 18570.6 | 18758.3 | 18882.4 |
| MW fresha | 18592.8 | 18826.8 | 19104.4 |
| MW after storagea | 18568.2 | 18734.3 | Multiple fragments |
| Length of storage (days) | 798 | 718 | 767 |
| Apparent mass (kDa)b | 23.6 (after storage) | 17.4 (after storage) | 7.9 (fresh)c |
| Yield (mg/liter of bacterial culture)d | 16 | 8.6 | 2.6 |
| Solubility (mg/ml) | ≥88 | ≥39.1 | ≥22.7 |
Determined by matrix-assisted laser desorption ionization mass spectrometry.
Based on elution volume by gel filtration chromatography (Fig. 2).
This anomalously low apparent molecular mass probably results from affinity of the KU VP8* core for the chromatography medium.
Yield refers to the final purified VP8* core.
This purification procedure is also effective at producing a homogeneous VP8* core from a P genotype 8, SA-independent rotavirus strain, KU (Fig. 2B and Table 1). Together, P genotypes 4 and 8 cause more than 90% of human cases of rotavirus gastroenteritis in temperate climates (11). Although the KU VP8* core is also very soluble, it is produced in relatively low yield and undergoes substantially more degradation and aggregation with storage than do the RRV and DS-1 VP8* cores (Fig. 2B, Table 1, and data not shown). Thus, the biochemical characteristics of the RRV and DS-1 VP8* cores make them promising potential immunogens for use in recombinant rotavirus vaccines. The less favorable characteristics of the KU VP8* core indicate that developing an antigenic cocktail of VP8* cores to completely cover the P types causing human disease will require screening of additional strains and possibly protein engineering.
Structural comparison of the DS-1 and RRV VP8* cores.
Using the X-ray crystal structure of the RRV VP8* core as an initial phasing model for molecular replacement, we determined the X-ray crystal structure of the DS-1 VP8* core at 1.6-Å resolution (see Materials and Methods) (Table 2). The DS-1 VP8* core, like the RRV VP8* core, resembles the galectins, a family of animal lectins, in its fold. It is built around a central β-sandwich, with a β-hairpin (strands E and F) packed against a six-stranded β-sheet and a C-terminal α-helix packed against a five-stranded β-sheet (Fig. 3A). Each asymmetric unit contains eight molecules of the DS-1 VP8* core. There are no major conformational differences between the eight molecules, which can be superimposed on each other with an average root mean square deviation (RMSD) between Cα atoms of 0.26 Å (not shown).
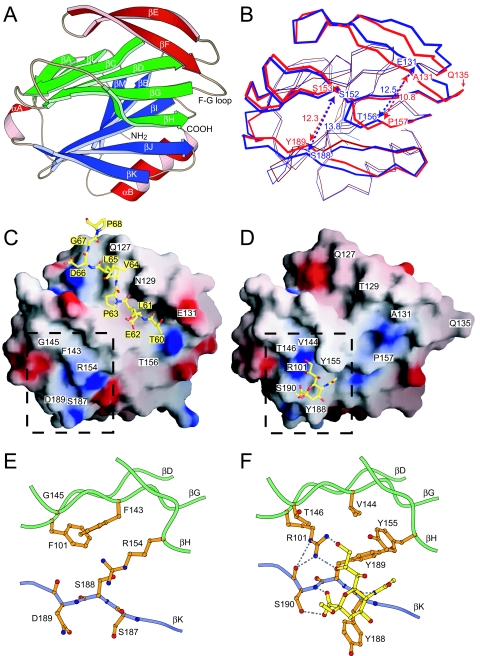
FIG. 3.
Comparison of DS-1 and RRV VP8* cores. (A) Ribbon diagram of the DS-1 VP8* core. Labeling of secondary-structure elements is as previously described for the RRV VP8* core (14), except that strand βH, which splits into strands βH and βH′ in RRV, is continuous in DS-1. The EF β-hairpin is red, the six-stranded β-sheet is green, and the five-stranded β-sheet is blue. (B) Superimposed Cα traces of the DS-1 VP8* core (blue) and the RRV VP8* core (red). Residue Q135 of RRV, which lacks a structural equivalent in DS-1, is indicated. The blue and red arrows indicate the widths of surface clefts, measured in Å between Cα atoms of the labeled residues, for the DS-1 and RRV VP8* cores, respectively. (C) Surface representation of the DS-1 VP8* core colored by electrostatic potential. Blue is positive; red is negative. The bound leader of an adjacent molecule in the crystal is depicted with a ball-and-stick model. Residues in the space-filling model are labeled in white text boxes. Residues in the ball-and-stick model are labeled in yellow text boxes. (D) Surface representation of the RRV VP8* core colored by electrostatic potential. The bound sialoside is depicted with a ball-and-stick model. (E) Molecular details of the site in DS-1 VP8* that corresponds to the RRV SA binding pocket. The depicted area is indicated by the dashed outline in panel C. (F) Molecular details of the SA binding pocket of RRV VP8*. The depicted area is indicated by a dashed outline in panel D. Selected hydrogen bonds are indicated by dashed gray lines. In panels E and F, residues on strand βK are depicted with backbone and side chain atoms; only Cα and side chain atoms of other residues are shown. The perspective of all the panels is indicated by the arrow in Fig. 1.
Although the DS-1 and RRV VP8* cores have only 45% amino acid identity (for residues 65 to 224 of RRV and 65 to 223 of DS-1), they can be superimposed on each other with an RMSD of 1.04 Å for 159 equivalent Cα atoms (Fig. 3B). The broad surface that is formed by the EF β-hairpin, strands H and G of the six-stranded β-sheet, and strands J and K of the five-stranded β-sheet is furrowed by two clefts (Fig. 3A to D). Both clefts are wider in the DS-1 VP8* core than in the RRV VP8* core (Fig. 3B). In the DS-1 VP8* core, the architecture of the cleft corresponding to the RRV SA binding site, which lies between the five-stranded and six-stranded β-sheets, is extensively reworked (Fig. 3C to F). In the RRV VP8* core, the R101 side chain amide makes key contacts with SA. It projects from strand D to the floor of the binding site to form a positively charged surface patch and makes bidentate hydrogen bonds to the glycerol group of the bound carbohydrate (Fig. 3D and F). In the DS-1 VP8* core, phenylalanine replaces arginine at this site and has a very different structural role. F101 of DS-1 makes no contribution to the molecular surface (Fig. 3C); instead, its aromatic ring forms part of a hydrophobic core in the interface between the β-sheets (Fig. 3E). In the RRV VP8* core, the aromatic rings of Y155 and Y188 project out into solvent to form walls on either side of the SA binding pocket (Fig. 3D and F). In the DS-1 VP8* core, replacement of these residues by R154 and S187 removes these walls. In fact, the R154 side chain replaces the floor of the SA binding pocket with a low ridge, as it stretches across the gap between the six- and five-stranded β-sheets (Fig. 3C and E). Although the structural data do not exclude the possibility that an alternative carbohydrate ligand binds in place of SA in DS-1, the surface of the DS-1 VP8* core that corresponds to the RRV SA binding site is not an obvious binding pocket.
DS-1 VP8* binds a polypeptide chain in a second surface cleft.
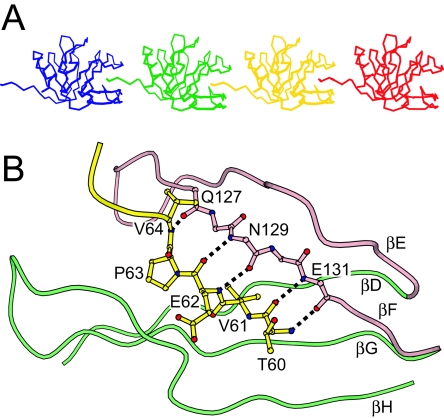
Does the DS-1 VP8* core structure suggest an alternative ligand to SA? The packing of the EF β-hairpin against the six-stranded β-sheet creates a cleft, which is adjacent to the cleft between the β-sheets that forms the SA binding pocket in RRV (Fig. 3A, C, and D). In the DS-1 VP8* core crystal, this cleft is occupied by the five N-terminal residues (residues 60 to 64) of an adjacent molecule, so that each DS-1 VP8* core “bites its neighbor's tail,” linking the cores into chains (Fig. 4A). The bound N-terminal residues are not part of the tightly folded structure of the core, but instead form an extended N-terminal “leader.” The equivalent residues are disordered in the RRV VP8* core crystal and solution structures (14). In the DS-1 VP8* core crystal, this leader is held in alignment by five backbone amide-to-carbonyl hydrogen bonds that form between residues 60, 62, and 64 of the leader and residues 131, 129, and 127 of strand F in the β-hairpin, thus making a new intermolecular three-stranded β-sheet with strands E and F (Fig. 4B). Leader binding is also stabilized by insertion of the aliphatic V61 side chain into a pocket lined by hydrophobic residues at the base of the cleft (Fig. 5B).
FIG. 4.
Leader binding in the DS-1 VP8* core crystal. (A) Cα traces of representative molecules in the crystal. The cleft between the β-hairpin and the six-stranded β-sheet of each molecule binds the leader of an adjacent molecule. (B) Molecular details of the cleft-to-leader interaction. The leader is depicted in yellow. Hydrogen bonds are depicted as dotted black lines. Residues on strand βF are depicted without side chains. The perspective and coloring match Fig. 3A.
FIG. 5.
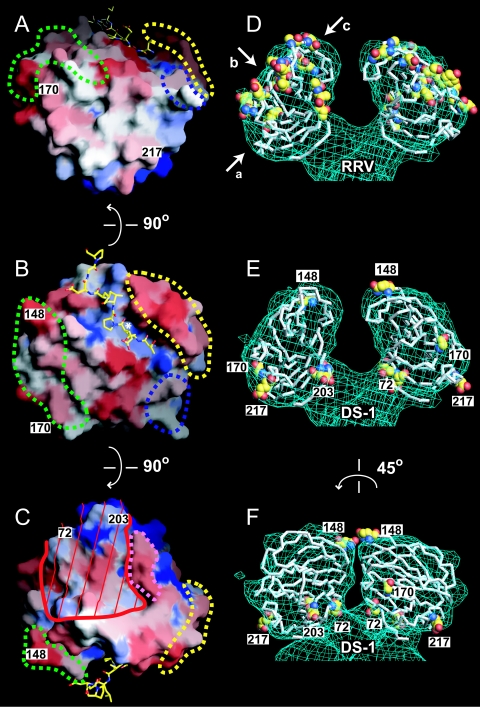
Neutralization surfaces of the RRV and DS-1 VP8* cores. Panels A to C show three views of a surface representation of the DS-1 VP8* core, colored by conservation among a set of VP8* sequences that contains one representative from each of 14 SA-independent P genotypes (Table 3). Blue is conserved; red is variable. Residue numbers mark neutralization escape mutations selected in SA-independent human rotavirus strains. Dashed outlines mark the previously described (14) neutralization epitopes of SA-dependent strains: green, 8-1; blue, 8-2; yellow, 8-3; pink, 8-4. The white asterisk in panel B indicates a hydrophobic pocket at the base of the peptide binding cleft. Red lines in panel C mark a surface that is inaccessible to antibody binding when the DS-1 VP8* core is fitted to the head of the SA11-4F VP4 spike. The bound leader is depicted with a ball-and-stick model. The perspectives of panels A, B, and C are indicated by arrows a, b, and c, respectively, in panel D. An observer looking down arrow c would be “head down” to view the perspective in panel C. In panels D to F, Cα traces of VP8* cores are fitted to the molecular envelope of the head from an electron cryomicroscopy image reconstruction of the SA11-4F spike on primed virions. Residues selected in neutralization escape mutants are indicated with space-filling models. (D) The RRV VP8* core with escape mutations selected in SA-dependent animal rotavirus strains. (E) The DS-1 VP8* core with escape mutations selected in SA-independent human rotavirus strains. Exposed escape mutations are labeled by residue number. (F) The model from panel E rotated 45° to show more clearly the inaccessible escape mutations in the cleft between the heads.
We used NMR spectroscopy to assay the binding in solution of a peptide based on the DS-1 VP8* core leader sequence to this potential peptide binding cleft (see Materials and Methods). The free peptide did not bind in the cleft with measurable affinity (data not shown). Close inspection of the DS-1 VP8* core crystal structure indicates that the bound leader does not fit the cleft optimally: the cleft continues beyond the N terminus of the bound leader (Fig. 3C), the hydrophobic pocket that holds the V61 side chain could accommodate a bulkier moiety (not shown), the P63 side chain prevents formation of potential hydrogen bonds between the leader and residues in strand H and the GH loop (Fig. 4B), and the leader is forced out of the cleft C terminal to P63 by steric hindrance from the tightly folded region of its own VP8* core (Fig. 4A).
The peptide binding cleft of the DS-1 core is one of the only exposed VP8* core surfaces that is conserved among SA-independent strains (Fig. 5B), suggesting a conserved function. A similar, but narrower, cleft is also present on the surface of the RRV VP8* core (Fig. 3D). Similar residues line this cleft in SA-dependent and SA-independent strains (see Table S1 in the supplemental material). Fitting to electron cryomicroscopy image reconstructions of virions from SA-dependent strains shows that this cleft is exposed at the tips of the VP4 spikes in a position favoring interaction with host cell proteins (Fig. 5B and D). As described previously, the VP8* β-hairpin appears to be an elaboration of a much shorter loop in the galectins, and it blocks the galectin carbohydrate binding site (14). The DS-1 VP8* core crystal structure suggests that this elaboration of the β-hairpin may also have created a new ligand binding site at the tip of the primed VP4 spike.
Structural polymorphism.
Many rotavirus strains, such as DS-1, have a deletion in the FG loop (Table 3) so that they lack a residue that is structurally equivalent to RRV residue Q135 (Fig. 3B). The FG loop links the EF β-hairpin to the six-stranded β-sheet (Fig. 3A). Near the deletion, the potential peptide binding cleft between the EF β-hairpin and the six-stranded β-sheet is wider in the DS-1 VP8* core than in the RRV VP8* core (Fig. 3B), possibly because the shorter loop does not permit as close an approximation of the proximal portion of the β-hairpin to the six-stranded β-sheet.
TABLE 3.
FG loop length correlations
| Strain | Host | P typea | RRV Q135 equivalentb | SA dependencec | Residue at position 101 | Genbank accession no. |
|---|---|---|---|---|---|---|
| SA11-4f | Simian | 6[1] | Present | + | R | X57319 |
| NCDV-Lin | Bovine | 6[1] | Present | +h | R | M63267 |
| RF | Bovine | 6[1] | Present | + | R | U65924 |
| BRV033 | Bovine | 6[1] | Present | + | R | U62155 |
| SA11 cl3 | Simian | 5B[2] | Present | +h | R | M23188 |
| RRV | Simian | 5B[3] | Present | +h | R | AY033150 |
| CU-1 | Canine | 5A[3] | Present | + | R | D13401 |
| HCR3 | Human | 5A[3] | Present | + | R | L19712 |
| K9 | Canine | 5A[3] | Present | + | R | D14725 |
| Cat97 | Feline | 5A[3] | Present | + | R | D13402 |
| DS-1 | Human | 1B[4] | Absent | −h | F | DQ141310 |
| L26 | Human | 1B[4] | Absent | − | F | M58292 |
| 4S | Porcine | 7[5] | Present | − | R | L10358 |
| UK | Bovine | 7[5] | Present | −d,h | R | M22306 |
| WC3 | Bovine | 7[5] | Present | − | R | AY050271 |
| B461 | Bovine | 7[5] | Present | − | R | M63267 |
| 678 | Bovine | 7[5] | Present | − | R | D32054 |
| M37 | Human | 2A[6] | Absent | − | I | Nonee |
| 1076 | Human | 2A[6] | Absent | −h | I | Nonee |
| McN13 | Human | 2A[6] | Absent | − | I | Nonee |
| ST3 | Human | 2A[6] | Absent | − | I | L33895 |
| Gott | Porcine | 2B[6] | Absent | − | V | M33516 |
| CRW-8 | Porcine | 9[7] | Present | + | R | L07888 |
| OSU | Porcine | 9[7] | Present | +h | R | X13190 |
| H1 | Equine | 9[7] | Present | + | R | D16341 |
| TFR-41 | Porcine | 9[7] | Present | + | R | L07889 |
| YM | Porcine | 9[7] | Present | + | R | M63231 |
| Wa | Human | 1A[8] | Absent | − | F | M96825 |
| KU | Human | 1A[8] | Absent | −h | F | M21014 |
| MO | Human | 1A[8] | Absent | − | F | AB008278 |
| YO | Human | 1A[8] | Absent | − | F | AB008279 |
| Ito | Human | 1A[8] | Absent | − | F | AB008280 |
| VA70 | Human | 1A[8] | Absent | − | F | AJ540229 |
| Hochi | Human | 1A[8] | Absent | − | F | AB039943 |
| K8 | Human | 3[9] | Present | −h | R | D90260 |
| O264 | Human | 3[9] | Present | − | R | AB008665 |
| Cat2 | Feline | 3[9] | Present | − | R | D13403 |
| 69M | Human | 4[10] | Present | −h | R | M60600 |
| B233 | Bovine | 8[11] | Absent | −h | F | D13394 |
| I321 | Human | 8[11] | Absent | − | F | L07657 |
| H-2 | Equine | 4[12] | Present | −h | R | L04638 |
| FI-14 | Equine | 4[12] | Present | − | R | D13398 |
| FI23 | Equine | 4[12] | Present | − | R | D16342 |
| A46 | Porcine | [13] | Present | −h | M | AY070274 |
| Ala | Lapine | 11[14] | Present | −h | R | U62149 |
| C-11 | Lapine | 11[14] | Present | − | R | U62150 |
| BAP-2 | Lapine | 11[14] | Present | − | R | U62151 |
| R-2 | Lapine | 11[14] | Present | − | R | U62152 |
| PA169 | Lapine | 11[14] | Present | − | R | D14724 |
| HAL1166 | Human | 11[14] | Present | − | R | L20885 |
| Mc35 | Human | 11[14] | Present | − | R | D14032 |
| Lp14 | Ovine | [15] | Present | −h | R | L11599 |
| EW | Murine | 10[16] | Present | − | R | U08429 |
| EC | Murine | 10[16] | Present | +/−f | R | U08421 |
| EB | Murine | 10[16] | Present | −h | R | U08419 |
| EDIM | Murine | 10[16] | Present | − | R | AF039219 |
| Ty-1 | Avian | [17] | Present | −h | R | L41493 |
| L338 | Equine | 12[18] | Present | +/−f | R | D13399 |
| 4F | Porcine | [19] | Absent | −h | V | L10359 |
| EHP | Murine | [20] | Present | +/−g | R | U08424 |
P serotype[P genotype]. Some strains have not been serotyped. Many of the strain classifications were obtained from reference 6.
Indicates the presence or absence of a residue that is structurally equivalent to residue 135 of strain RRV.
SA dependence is based on published data that define SA independence as a preserved infectious titer on cultured cells or enterocytes digested with Arthrobacter ureafaciens neuraminidase (5, 6, 10, 31).
Although UK entry is not SA dependent, it binds glycosphingolipids with a specificity dependent upon the SA moiety in the oligosaccharide chain (10).
Sequences from reference 17.
Weak inhibition by neuraminidase digestion of MA104 cells (6).
Cell-type-specific neuraminidase sensitivity (31).
Strain included in the SA-dependent and SA-independent sets used to calculate variability for Table S1 in the supplemental material and Fig. 5A to C.
Most human rotavirus strains have this deletion in the FG loop (Table 3). The infrequently isolated human rotavirus strains without the deletion have P types (such as 5A[3], 3[9], 4[10], and 11[14]), that also include animal rotavirus strains. These strains may have been introduced into human populations relatively recently. The deletion is consistently associated with SA independence (as verified by the presence of a hydrophobic residue at position 101), but it is not required for SA independence (Table 3). The phylogeny of rotavirus VP4 (6) does not clearly demonstrate whether this common structural feature is an adaptation to replication and spread in humans or simply a consequence of common ancestry: the clustering of VP4 molecules with the deletion in a single clade (containing P genotypes 4, 6, 8, and 19) is not absolute, and some bovine strains (in the outlying P[11] group) also have the deletion. Potential ligand binding in the cleft between the β-hairpin and the six-stranded β-sheet suggests that the structural polymorphism may reflect differences in ligand specificity.
Escape mutations selected by human neutralizing MAbs against rotavirus.
Three human neutralizing MAbs against rotavirus have been derived from a phage display library of B-lymphocyte cDNA from naturally infected humans (21). The phage antibodies were selected for binding to rotavirus strain KU virions, tested for neutralization of strain KU, and converted to immunoglobulin G1 MAbs through recombinant DNA manipulation. One of the MAbs, 2-11G, binds VP7. The other two bind VP4 and neutralize heterotypically (i.e., neutralize more than one P type): 1-2H neutralizes P[4] and P[8]; 2-3E neutralizes P[6] and P[8].
We have mapped the residues recognized by 1-2H and 2-3E in strain KU, using neutralization escape mutant analysis (Table 4). MAb 1-2H selects a unique G-to-D mutation at VP4 residue 170 (virus strain m-KU-1-2H), and MAb 2-3E selects a unique E-to-K mutation at VP4 residue 203 (virus strain m-KU-2-3E). Both mutations are in the VP8* fragment of VP4. Three other VP8*-specific antibodies that neutralize human strains of rotavirus have been described (26, 35, 36). Two of these MAbs, HS11 and RV5:2, neutralize homotypically, but the other MAb, HS6, neutralizes P[6] and some P[8] viruses (Table 4). Thus, three of five neutralizing monoclonal antibodies that recognize VP8* of human rotavirus strains are heterotypic in their neutralization specificities. A more limited degree of heterotypic neutralization has been observed among the 20 mapped MAbs that bind VP8* and neutralize animal rotavirus strains (see Table S2 in the supplemental material). Heterotypic neutralization by monoclonal antibodies derived from naturally infected humans may reflect selection for heterotypic antibodies by repeated rotavirus infection. This result correlates well with the increasingly broad serum neutralizing response against rotavirus elicited by reinfection with rotaviruses of the same or different rotavirus serotypes (19, 43).
TABLE 4.
Neutralization escape mutations selected by MAbs that recognize VP8* of human rotavirus strains
| MAb | Escape mutation (strain)a | Immunization regimen | P genotypes neutralized (not neutralized)b | Immunized species | Initial screen | Reference |
|---|---|---|---|---|---|---|
| 1-2H | G170D (KU) | Natural infection | P[4, 8] (P[5, 6, 9, 10]) | Human | Binding | 21 |
| 2-3E | E203K (KU) | Natural infection | P[6, 8] (P[4, 5, 9, 10]) | Human | Binding | 21 |
| HS6 | T721 (ST3) | IPc with ST3 | P[6, +/−8]d (P[4]) | Mouse | Neutralization | 35 |
| HS11 | E217K (ST3) | IP with ST3 | P[6] (P[4, 8]) | Mouse | Neutralization | 35 |
| RV-5:2 | Q148R (RV-5) | IP and IVe with RV-5 | P[4] (P[2, 3, 5, 6, 8, 10]) | Mouse | Neutralization | 8 |
P genotypes of rotavirus strains: KU, P[8]; ST3, P[6]; RV-5, P[4].
Only listed P genotypes were tested.
IP, intraperitoneal hyperimmunization.
Some, but not all, P[8] strains are neutralized.
IV, intravenous hyperimmunization.
Although VP5* is more conserved among strains than is VP8*, the presence of heterotypic neutralization epitopes on VP8* of human strains suggests that immunization with recombinant VP8* of human strains could induce a heterotypic neutralizing antibody response. Such a response has been demonstrated against VP8* from an animal rotavirus strain, as primary immunization of laboratory animals with recombinant VP8* of the simian strain RRV does produce heterotypically neutralizing antibodies (16).
Mapping of neutralization escape mutations on the DS-1 VP8* core structure.
As previously described, the 20 neutralization escape mutations mapped to VP8* of SA-dependent animal rotavirus strains cluster in four epitopes (14). The five neutralization escape mutations now mapped to VP8* of SA-independent human rotavirus strains do not cluster in these epitopes or in any easily identifiable new epitopes (Fig. 5A to C, labeled residues). Only the mutation at residue 148 of human strain RV-5 lies within one of the previously described epitopes (designated 8-1). Because the DS-1 and RRV VP8* cores have a common fold, gross structural differences do not explain the distinct distributions of escape mutations.
When the RRV VP8* core crystal structure is fitted to the spike envelope of a 12-Å-resolution electron cryomicroscopy image reconstruction of trypsin-primed, SA-dependent rotavirus particles, all of the escape mutations on VP8* of SA-dependent strains are accessible for antibody binding, forming “caps” at the tips of the spikes' heads (Fig. 5D). Although the DS-1 VP8* core fits the same molecular envelope, the escape mutations of SA-independent human strains are not in the most accessible locations, and three (at residues 72, 203, and 217) ring the base of the head (Fig. 5E and F). In fact, the escape mutation at residue 72 contributes to the point of attachment of head to body and to the surface of the gap between the paired heads, where it is not accessible for antibody binding (Fig. 5C, E, and F). The escape mutation at residue 203 also contributes to the surface of the gap between head and body and is on the boundary between accessible and inaccessible surfaces.
Thus, the distribution of escape mutations suggests that the state of the VP4 spike recognized by some VP8*-specific antibodies that neutralize SA-independent human rotavirus strains differs from the state recognized by VP8*-specific antibodies that neutralize SA-dependent animal rotavirus strains. Alternatively, the selected residues could mediate escape from neutralization by indirect steric effects, rather than by direct disruption of an epitope. No electron cryomicroscopy image reconstructions of SA-independent rotavirus virions are currently available to test the hypothesis that the conformations of trypsin-primed spikes on SA-independent and SA-dependent strains expose different molecular surfaces of the VP8* core for potential antibody binding.
VP4 spikes have multiple conformations during rotavirus entry. Prior to trypsin priming, the spikes are flexible and therefore not visible in averaged icosahedral image reconstructions (9). Trypsin priming rigidifies pairs of VP4 molecules to produce spikes. Threefold symmetry of the portion of VP4 buried under the VP7 shell (44), the trimeric appearance of altered VP4 spikes on virions that have been treated with alkali (39), and the stable trimer formed by a rearranged VP5* fragment (13) suggest that each VP4 cluster on the virion surface may contain three molecules, one of which remains flexible after trypsin priming. In addition, electron cryomicroscopy image reconstructions demonstrate subtle conformational differences in spike morphology among SA-dependent rotavirus strains (38). The 22- to 23-Å resolution limit of these reconstructions does not permit a precise determination of the boundaries of the accessible surfaces on the variants. Thus, either strain differences in spike morphology or the multiple conformational states of VP4 could explain the distribution of neutralization escape mutations on VP8* of human rotavirus strains.
In summary, although the VP8* cores of an SA-dependent strain (RRV) and of an SA-independent strain (DS-1) are substantially similar, there are significant structural differences between the two phenotypic variants. The biochemical characteristics of both variants, including ease of expression and purification, high solubility, and chemical stability make them promising components for a potential second-generation recombinant rotavirus vaccine. In this regard, heterotypic neutralization by MAbs recognizing VP8* of human strains is a particularly promising finding. Differences between the RRV and DS-1 VP8* cores in the region that corresponds to the RRV SA binding site make it unlikely that DS-1 VP8* binds an alternative carbohydrate ligand in this location. A widened cleft between the EF β-hairpin and the six-stranded β-sheet in the DS-1 VP8* core and the binding of a peptide chain in this cleft suggest that VP8* may bind a protein ligand. The very different neutralization surfaces of SA-dependent and SA-independent viruses suggest different mechanisms of neutralization and, possibly, differences in spike morphology. Further structural studies of SA-independent rotavirus strains could reveal differences in the VP4 spike that are directly relevant to the pathogenesis of rotavirus gastroenteritis in children.
Supplementary Material
Acknowledgments
We thank Marina Babyonyshev for technical assistance, Mary Estes and Mario Gorziglia for recombinant baculoviruses containing cloned KU and DS-1 genes, Eric Vogan for assistance in analyzing X-ray diffraction data, Gerhard Wagner for use of NMR spectrometers, Stephen C. Harrison for scientific advice and support, the MACCHESS staff at the Cornell High Energy Synchrotron Source for assistance with data collection, the Harvard Medical School Biopolymers Facility for DNA sequencing and oligonucleotide synthesis, and Michael Berne and the Tufts Protein Chemistry Facility for mass spectrometry, N-terminal sequencing, and peptide synthesis. We acknowledge the use of electron cryomicroscopy facilities at the National Center for Macromolecular Imaging, funded by NIH, at Baylor College of Medicine.
This work was supported by NIH grant R37 AI 36040 to B.V.V.P. and by NIH grant R01 AI 053174 and an Ellison Medical Foundation New Scholars in Global Infectious Disease Award to P.R.D.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org.
REFERENCES
- 1.Bartels, C., T.-H. Xia, M. Billeter, P. Guntert, and K. Wuthrich. 1995. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR 5:1-10. [DOI] [PubMed] [Google Scholar]
- 2.Bastardo, J. W., and I. H. Holmes. 1980. Attachment of SA-11 rotavirus to erythrocyte receptors. Infect. Immun. 29:1134-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 4.Ciarlet, M., S. E. Crawford, and M. K. Estes. 2001. Differential infection of polarized epithelial cell lines by sialic acid-dependent and sialic acid-independent rotavirus strains. J. Virol. 75:11834-11850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciarlet, M., and M. K. Estes. 1999. Human and most animal rotavirus strains do not require the presence of sialic acid on the cell surface for efficient infectivity. J. Gen. Virol. 80:943-948. [DOI] [PubMed] [Google Scholar]
- 6.Ciarlet, M., J. E. Ludert, M. Iturriza-Gomara, F. Liprandi, J. J. Gray, U. Desselberger, and M. K. Estes. 2002. The initial interaction of rotavirus strains with N-acetyl-neuraminic (sialic) acid residues on the cell surface correlates with VP4 genotype, not species of origin. J. Virol. 76:4087-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collaborative Computational Project. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 8.Coulson, B. S., J. M. Tursi, W. J. McAdam, and R. F. Bishop. 1986. Derivation of neutralizing monoclonal antibodies to human rotaviruses and evidence that an immunodominant neutralization site is shared between serotypes 1 and 3. Virology 154:302-312. [DOI] [PubMed] [Google Scholar]
- 9.Crawford, S. E., S. K. Mukherjee, M. K. Estes, J. A. Lawton, A. L. Shaw, R. F. Ramig, and B. V. Prasad. 2001. Trypsin cleavage stabilizes the rotavirus VP4 spike. J. Virol. 75:6052-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delorme, C., H. Brussow, J. Sidoti, N. Roche, K. A. Karlsson, J. R. Neeser, and S. Teneberg. 2001. Glycosphingolipid binding specificities of rotavirus: identification of a sialic acid-binding epitope. J. Virol. 75:2276-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desselberger, U., M. Iturriza-Gomara, and J. J. Gray. 2001. Rotavirus epidemiology and surveillance. Novartis Found. Symp. 238:125-152. [DOI] [PubMed] [Google Scholar]
- 12.Dormitzer, P. R., H. B. Greenberg, and S. C. Harrison. 2001. Proteolysis of monomeric recombinant rotavirus VP4 yields an oligomeric VP5* core. J. Virol. 75:7339-7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dormitzer, P. R., E. B. Nason, B. V. Prasad, and S. C. Harrison. 2004. Structural rearrangements in the membrane penetration protein of a non-enveloped virus. Nature 430:1053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dormitzer, P. R., Z.-Y. J. Sun, G. Wagner, and S. C. Harrison. 2002. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J. 21:885-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Estes, M. K., D. Y. Graham, and B. B. Mason. 1981. Proteolytic enhancement of rotavirus infectivity: molecular mechanisms. J. Virol. 39:879-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fiore, L., H. B. Greenberg, and E. R. Mackow. 1991. The VP8 fragment of VP4 is the rhesus rotavirus hemagglutinin. Virology 181:553-563. [DOI] [PubMed] [Google Scholar]
- 17.Gorziglia, M., K. Green, K. Nishikawa, K. Taniguchi, R. Jones, A. Z. Kapikian, and R. M. Chanock. 1988. Sequence of the fourth gene of human rotaviruses recovered from asymptomatic or symptomatic infections. J. Virol. 62:2978-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorziglia, M., G. Larralde, A. Z. Kapikian, and R. M. Chanock. 1990. Antigenic relationships among human rotaviruses as determined by outer capsid protein VP4. Proc. Natl. Acad. Sci. USA 87:7155-7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green, K. Y., K. Taniguchi, E. R. Mackow, and A. Z. Kapikian. 1990. Homotypic and heterotypic epitope-specific antibody responses in adult and infant rotavirus vaccinees: implications for vaccine development. J. Infect. Dis. 161:667-679. [DOI] [PubMed] [Google Scholar]
- 20.Guntert, P., V. Dotsch, G. Wider, and K. Wuthrich. 1992. Processing of multi-dimensional NMR data with the new software PROSA. J. Biomol. NMR 2:619-629. [Google Scholar]
- 21.Higo-Moriguchi, K., Y. Akahori, Y. Iba, Y. Kurosawa, and K. Taniguchi. 2004. Isolation of human monoclonal antibodies that neutralize human rotavirus. J. Virol. 78:3325-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoshino, Y., and A. Z. Kapikian. 1996. Classification of rotavirus VP4 and VP7 serotypes. Arch. Virol. Suppl. 12:99-111. [DOI] [PubMed] [Google Scholar]
- 23.Jeffrey, P. D., J. Bajorath, C. Y. Chang, D. Yelton, I. Hellstrom, K. E. Hellstrom, and S. Sheriff. 1995. The X-ray structure of an anti-tumour antibody in complex with antigen. Nat. Struct. Biol. 2:466-471. [DOI] [PubMed] [Google Scholar]
- 24.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 25.Kapikian, A. Z., Y. Hoshino, and R. M. Chanock. 2001. Rotaviruses, p. 1787-1833. In D. M. Knipe, P. M. Howley, D. D. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams and Wilkins, Philadelphia, Pa. [Google Scholar]
- 26.Kirkwood, C. D., R. F. Bishop, and B. S. Coulson. 1996. Human rotavirus VP4 contains strain-specific, serotype-specific and cross-reactive neutralization sites. Arch. Virol. 141:587-600. [DOI] [PubMed] [Google Scholar]
- 27.Kraulis, J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 28.LaMonica, R., S. S. Kocer, J. Nazarova, W. Dowling, E. Geimonen, R. D. Shaw, and E. R. Mackow. 2001. VP4 differentially regulates TRAF2 signaling, disengaging JNK activation while directing NF-kappa B to effect rotavirus-specific cellular responses. J. Biol. Chem. 276:19889-19896. [DOI] [PubMed] [Google Scholar]
- 29.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the sterochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 30.Livingstone, C. D., and G. J. Barton. 1993. Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. Comput. Appl. Biosci. 9:745-756. [DOI] [PubMed] [Google Scholar]
- 31.Ludert, J. E., B. B. Mason, J. Angel, B. Tang, Y. Hoshino, N. Feng, P. T. Vo, E. M. Mackow, F. M. Ruggeri, and H. B. Greenberg. 1998. Identification of mutations in the rotavirus protein VP4 that alter sialic-acid-dependent infection. J. Gen. Virol. 79:725-729. [DOI] [PubMed] [Google Scholar]
- 32.Mackow, E. R., R. D. Shaw, S. M. Matsui, P. T. Vo, M. N. Dang, and H. B. Greenberg. 1988. The rhesus rotavirus gene encoding protein VP3: location of amino acids involved in homologous and heterologous rotavirus neutralization and identification of a putative fusion region. Proc. Natl. Acad. Sci. USA 85:645-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsui, S. M., P. A. Offit, P. T. Vo, E. R. Mackow, D. A. Benfield, R. D. Shaw, L. Padilla-Noriega, and H. B. Greenberg. 1989. Passive protection against rotavirus-induced diarrhea by monoclonal antibodies to the heterotypic neutralization domain of VP7 and the VP8 fragment of VP4. J. Clin. Microbiol. 27:780-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicholls, A., K. A. Sharp, and B. Honig. 1991. Protein folding and association: insights from interfacial and thermodynamic properties of hydrocarbons. Proteins 11:281-296. [DOI] [PubMed] [Google Scholar]
- 35.Padilla-Noriega, L., S. J. Dunn, S. Lopez, H. B. Greenberg, and C. F. Arias. 1995. Identification of two independent neutralization domains on the VP4 trypsin cleavage products VP5* and VP8* of human rotavirus ST3. Virology 206:148-154. [DOI] [PubMed] [Google Scholar]
- 36.Padilla-Noriega, L., R. Werner-Eckert, E. R. Mackow, M. Gorziglia, G. Larralde, K. Taniguchi, and H. B. Greenberg. 1993. Serologic analysis of human rotavirus serotypes P1A and P2 by using monoclonal antibodies. J. Clin. Microbiol. 31:622-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parashar, U. D., E. G. Hummelman, J. S. Bresee, M. A. Miller, and R. I. Glass. 2003. Global illness and deaths caused by rotavirus disease in children. Emerg. Infect. Dis. 9:565-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pesavento, J. B., A. M. Billingsley, E. J. Roberts, R. F. Ramig, and B. V. Prasad. 2003. Structures of rotavirus reassortants demonstrate correlation of altered conformation of the VP4 spike and expression of unexpected VP4-associated phenotypes. J. Virol. 77:3291-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pesavento, J. B., S. E. Crawford, E. Roberts, M. K. Estes, and B. V. Prasad. 2005. pH-induced conformational change of the rotavirus VP4 spike: implications for cell entry and antibody neutralization. J. Virol. 79:8572-8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sato, K., Y. Inaba, T. Shinozaki, R. Fujii, and M. Matumoto. 1981. Isolation of human rotavirus in cell cultures: brief report. Arch. Virol. 69:155-160. [DOI] [PubMed] [Google Scholar]
- 41.Shaw, A. L., R. Rothnagel, D. Chen, R. F. Ramig, W. Chiu, and B. V. Prasad. 1993. Three-dimensional visualization of the rotavirus hemagglutinin structure. Cell 74:693-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ward, R. L., D. R. Knowlton, and M. J. Pierce. 1984. Efficiency of human rotavirus propagation in cell culture. J. Clin. Microbiol. 19:748-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ward, R. L., D. S. Sander, G. M. Schiff, and D. I. Bernstein. 1990. Effect of vaccination on serotype-specific antibody responses in infants administered WC3 bovine rotavirus before or after a natural rotavirus infection. J. Infect. Dis. 162:1298-1303. [DOI] [PubMed] [Google Scholar]
- 44.Yeager, M., J. A. Berriman, T. S. Baker, and A. R. Bellamy. 1994. Three-dimensional structure of the rotavirus haemagglutinin VP4 by cryo-electron microscopy and difference map analysis. EMBO J. 13:1011-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou, Y. J., J. W. Burns, Y. Morita, T. Tanaka, and M. K. Estes. 1994. Localization of rotavirus VP4 neutralization epitopes involved in antibody-induced conformational changes of virus structure. J. Virol. 68:3955-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.