Abstract
The human genome is littered by endogenous retrovirus sequences (HERVs), which constitute up to 8% of the total genomic sequence. The sequencing of the human (Homo sapiens) and chimpanzee (Pan troglodytes) genomes has facilitated the evolutionary study of ERVs and related sequences. We screened both the human genome (version hg16) and the chimpanzee genome (version PanTro1) for ERVs and conducted a phylogenetic analysis of recent integrations. We found a number of recent integrations within both genomes. They segregated into four groups. Two larger gammaretrovirus-like groups (PtG1 and PtG2) occurred in chimpanzees but not in humans. The PtG sequences were most similar to two baboon ERVs and a macaque sequence but neither to other chimpanzee ERVs nor to any human gammaretrovirus-like ERVs. The pattern was consistent with cross-species transfer via predation. This appears to be an example of horizontal transfer of retroviruses with occasional fixation in the germ line.
Human endogenous retrovirus sequences (HERVs), which have become trapped as Mendelian genes and fixed in the germ line (9), constitute 7 to 8% of the human genome (8, 26). The sequencing of the human (Homo sapiens) genome (26) and the draft sequence of the chimpanzee (Pan troglodytes) genome (16) have facilitated the evolutionary studies of ERVs and related sequences. The Homo-Pan speciation has been estimated to have occurred between 4.6 and 6.2 million years ago (15). The genomic nonidentity is approximately 1.2% (15, 17) up to 5% (14), depending on analysis methods. We used a newly developed bioinformatic tool, RetroTector (G. O. Sperber, T. Airola, P. Jern, A. Castell, and J. Blomberg, unpublished data), to screen both the human genome (version hg16) and the chimpanzee genome (version PanTro1) for ERVs. We have successfully used RetroTector-derived data in several papers; the program validation can be traced in these papers and their respective supplemental materials (1, 27-29, 42). The differences in ERV content serve as indicators of dynamic genomes. The proviral long terminal repeats (LTRs) are identical at the integration moment (50). We analyzed recent provirus integrations with less than 2% LTR difference. With a typical value of 0.2% substitutions per million years (34), this LTR difference served as an approximation to 5 million years and the Homo-Pan sp. split. In this paper, we refer to these proviruses as “recent,” knowing that postintegrational mutations can accumulate at different rates depending on the genetic environment (25, 33, 60). The potential effects of retroviral integrations have been studied in several papers. The effects include addition of promoters and enhancers (43) and introduction of alternative splice patterns (18). Recently, it was also shown that the human L1 retrotransposon generates a somatic variation which influences both gene expression and cell differentiation (41). Generation of somatic variation may also apply to retroviruses (57).
In this paper, we demonstrate unique sequences in the human and chimpanzee genomes. We found a difference in recent activity between the beta-like and gamma-like retroviruses. This difference applied to both genomes inversely, with one group expanding in each genome. It indicates the importance of environmental factors and random reactivation events in preexisting elements for determining the retroviral genetic setup. Several cross-species transfers of nonhuman, nonchimpanzee primate gamma-like retroviruses to chimpanzee have occurred since the Homo-Pan sp. split.
MATERIALS AND METHODS
Bioinformatics.
Alignments were performed using ClustalX 1.83 (51) with default settings. The neighbor joining (NJ) dendrogram (Fig. 1) was constructed in the MEGA software package (32) using pairwise deletions, p-distance, and 500 bootstraps. The bootstrap values are, due to limited space in Fig. 1, presented in an NJ cladogram rooted on midpoint in Fig. S1 in the supplemental material. Sequences retrieved by RetroTector (Sperber et al., unpublished data) were verified by BLAT search (http://genome.ucsc.edu/cgi-bin/hgBlat) against the respective genomes. tBLASTn searches (http://www.ncbi.nlm.nih.gov/BLAST/) using putative Pol (“puteins”) were conducted with the following search criteria for PanTro1: Primates NOT Pan Troglodytes [ORGN] alt. for hg16: Primates NOT Homo Sapiens [ORGN]. The rooted NJ cladogram (see Fig. S2 in the supplemental material) was constructed using pairwise deletions, p-distance, and 1,000 bootstraps. The similarity matrix (Fig. S2 in the supplemental material) was calculated from the cladogram data using pairwise deletions and PAM250 scoring. The similarity score of the shortest sequence versus itself was set to 100%. Sequences included into respective groups were at least 80% similar and applied to the clades in Fig. 1. The gammaretroviral groups were confirmed against a maximum parsimony tree constructed in PAUP4.0 (see Fig. S3 in the supplemental material).
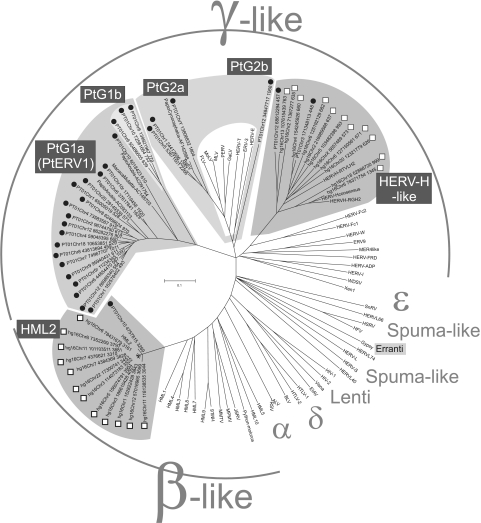
FIG. 1.
Pol NJ phylogeny of recently integrated hg16 proviruses (indicated by white boxes), PanTro1 proviruses (indicated by black circles), and reference retroviral sequences using the entire proteins or “puteins.” Shadows indicate the different groups discussed in the text. Abbreviations are spelled out in the last paragraph of Materials and Methods. Bootstrap supports for the corresponding NJ cladogram rooted on midpoint are presented in Fig. S1 in the supplemental material.
The program RetroTector was used to screen both the human (hg16) and chimpanzee (PanTro1) genomes. Briefly, RetroTector recognizes consensus motifs and constructs putative ERV proteins (“puteins”) from the different reading frames in the gene candidates. The program uses codon statistics, frequency of stop codons, and alignment to known retrovirus proteins to approximate an original open reading frame (ORF). The Pol includes a number of conserved motifs in both reverse transcriptase (RT) and integrase (IN) that facilitates the RetroTector computations. The RetroTector RT1-5 motifs correspond to the conserved Pol RT motif numbers 1 and 3 to 6, presented by Xiong and Eickbush (59). This RetroTector multifactorial sequence analysis is more robust to poor sequence quality than traditional BLAST alignments (Sperber et al., unpublished data), which are based on similarities to predefined query sequences. Validation of the RetroTector-derived data can be found in the supplemental data of several papers (1, 27-29, 42).
Sequences and accession numbers.
The genome sequences for human (hg16) and chimpanzee (PanTro1) were retrieved from the UCSC Genome Browser (http://genome.ucsc.edu/).
GenBank accession numbers or chromosomal positions in hg16 for reference (putein) sequences used in the analysis were as follows: avian leukosis virus (ALV) (NC001408), Rous sarcoma virus (RSV) (NC001407), mouse mammary tumor virus (MMTV)/(MPMV) (NC001503), Mason-Pfizer monkey virus (MMTV)/(MPMV) (NC001550), Jaagsiekte sheep retrovirus (JSRV) (M80216), HML1 (ch19-21849393), HML2 (chr11-101600013), HML3 (chr1-48344461), HML4 (chr8-75679221), HML5 (AC004536), HML6 (consensus), HML7 (chr6-121300220), HML8 (chr3-131452286), HML9 (chr9-62700428), HML10 (chr6-32017925), HERV-H (consensus), HERV-H/RGH2 (D11078), HERV-H/RTVLH2 (M18048), HERV-Fc1 (AL354685), HERV-Fc2 (AC019088), HERV-W (chr7-9105739), ERV9 (AC073410), ERV3 (Chr7-63865366), HERV-E (M10976), murine leukemia virus (MLV) (NC001501), Moloney murine leukemia virus (MoMLV) (AF033811), baboon endogenous retrovirus (BaEV) (D10032), gibbon ape leukemia virus (GaLV) (M26927), HERV-ADP (AC005741), HERV-FRD (AC004022), HERV-I (chr16-72821350), HERV-T (chr14-104635791), HERV-S (AC004385), feline leukemia virus (FLV) (NC001940), porcine endogenous retrovirus (PERV) (AJ293656), walleye dermal sarcoma virus (WDSV) (NC001867), Xenopus laevis endogenous retrovirus (Xen1) (AJ506107), snakehead fish retrovirus (SnRV) (NC001724), bovine leukemia virus (BLV) (NC001414), human T-cell leukemia virus 1 (HTLV-1) (NC001436), HTLV-2 (NC001488), Gypsy (AJ000387), HERV-L (RepBase), human spumaretrovirus (HSRV) (AF033816), human foamy virus (HFV) (NC001736), MER4like (chr13-54208300), HERVL66 (RepBase), HERVL74 (RepBase), HERVL40 (RepBase), and Python molurus endogenous retrovirus (AAN77283).
RESULTS AND DISCUSSION
Genomic differences in recent ERV integrations.
In a screening for ERVs in the human (Homo) genome (version hg16) and the chimpanzee (Pan) genome (version PanTro1), using the RetroTector bioinformatic tool, we found 24 integrations in Homo and 36 integrations in Pan with less than 2% LTR divergence (Table 1). As mentioned, this LTR difference corresponds to 4 to 5 million years of selection-neutral evolution, approximating the Homo-Pan sp. split, based on a typical value of 0.2% substitutions per million years (34).
TABLE 1.
Properties of recent ERV integrations in the human (hg16) and chimpanzee (PanTro1) genomes
| Element detected | No. in hg16 (24)a
|
No. in PanTro1 (36)a
|
||
|---|---|---|---|---|
| Beta (B) | Gamma (C) | Beta (B) | Gamma (C) | |
| ERVs detected | 12 | 12 | 1 | 35 |
| Detected gag | 12 | 10 | 1 | 18 |
| Detected pro | 12 | 4 | 1 | 9 |
| Detected pol | 12 | 11 | 1 | 27 |
| Detected env | 12 | 2 | 1 | 22 |
| Detected “LTR-gag-pro-pol-env-LTR” | 12 | 1 | 1 | 1 |
Numbers in parentheses indicate the number of ERVs with <2% LTR nonidentity. “Beta” and “Gamma” indicate beta- and gamma-like genera assigned by RetroTector.
The 51 pol-containing proviruses with <2% LTR divergence were analyzed further. The whole RetroTector-derived hg16 and PanTro1 ERV nucleotide sequences (Table 2) were used in BLAT searches against both the Homo and Pan sp. genomes to verify the positions and uniqueness (i.e., occurring in only one or the other species [underlined in Table 2]). Before a Homo sp. ERV was labeled as unique, we analyzed the occurrence and matching of orthologous flanking sequences of the proviruses in the Pan sp. Proviral integrations with doubtful chromosomal flanking sequences in the opposite species were not considered unique. False conclusions due to poorly sequenced or missing chromosomal regions were thus minimized. Using these rather stringent criteria, 31 of the 51 pol-containing proviruses were assessed to be unique to the respective genomes and to have integrated after the Homo-Pan speciation (underlined sequences in Table 2). These are in this paper operationally referred to as “recent” integrations. Further, the uniqueness of 10 integrations was uncertain (Table 2). Their BLAT results are presented in Fig. S6 in the supplemental material. In an attempt to test the <2% LTR divergence limit, an extended analysis of the human genome (hg16), using <3% LTR divergence, resulted in 4 additional beta-like and 19 additional gamma-like Pol-containing proviral integrations. Among those integrations, only two additional uncertain unique gamma-like proviruses (HERV-H like) at chromosomes 3p12 and 6p22 were detected (data not shown), and two probably unique beta-like (HML2) proviruses were found at chromosomes 19p13 and 3q21 (data not shown). This indicates that the LTR divergences are not exact measures of integration times. They are probably influenced by recombination events or gene conversions (24, 30) but useful in screenings. The extended analysis, in this case by an additional 1% LTR divergence, thus resulted in an increasing proportion of uncertain proviruses (Fig. S6 in the supplemental material) and very little unique integration.
TABLE 2.
Grouping and characterization of recent Pol-containing ERV integrations in the Homo (hg16) and Pan (PanTro1) genomes
| Sequence from Pol putein tree (Score > 300, LTR nonidentity < 2%) (species “unique”)a | Genomic positions (PanTro1/hg16)b | Nucleotide frequency (%)
|
Chain (genus) | PBS | Stops and frameshifts (genus)
|
LTR nonidentity (%) | BLAT 5′ and 3′ LTRs >98% of max. score | Best Pol hits in tBLASTn (against nr database) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | T | G | C | gag | pro | pol | env | |||||||
| PtG1a | 26.5 | 21.7 | 23.7 | 28.2 | Using 75% Pol consensus | |||||||||
| pt01Chr1_162576402_493 | chr1: 162576402/(−) | 28.4 | 23.2 | 21.6 | 26.8 | gamma (C) | 12 (C) | 6 (C) | 1.58 | 4 | AC148703Macaca mulatta | |||
| pt01Chr1_65005019_528 | chr1: 65005019/(−?) | 26.1 | 22 | 23.4 | 28.5 | gamma (C) | 9 (C) | 4 (C) | 0.48 | 4 | AC093133Papio anubis | |||
| pt01Chr10_6018423_610 | chr10: 6018423/(−) | 27.3 | 22.5 | 22.4 | 27.8 | gamma (C) | P | 11 (C) | 4 (C) | 0.53 | 9 | AF142988Papio cynocephalus | ||
| pt01Chr10r_17119458_1920 | chr10: 17111172/(−) | 26.9 | 21.5 | 23.6 | 28 | gamma (C) | P | 2 (C) | 0 (C) | 7 (C) | 3 (C) | 1.99 | 2 | |
| pt01Chr12_86282276_907 | chr12: 86282276/(−) | 26.2 | 21.5 | 24 | 28.3 | gamma (C) | P | 4 (C) | 8 (C) | 2 (C) | 0.98 | 19 | ||
| pt01Chr12_86988340_584 | chr12: 86981358/(−) | 26.0 | 21.8 | 23.8 | 28.4 | gamma (C) | 10 (C) | 1 (C) | 0.27 | 19 | ||||
| pt01Chr18_10653851_529 | chr18: 10646172/(−) | 25.6 | 21.6 | 24.5 | 28.3 | gamma (C) | 8 (C) | 4 (C) | 0.97 | 20 | ||||
| pt01Chr2_96744750_613 | chr2: 96737061/(−) | 24.3 | 22.1 | 25.1 | 28.4 | gamma (C) | 0 (C) | 7 (C) | 3 (C) | 0.5 | 2 | |||
| pt01Chr20_29149338_571 | chr20: 29142410/(−) | 27.1 | 21.6 | 23.3 | 28 | gamma (C) | 3 (C) | 11 (C) | 1.22 | 4 | ||||
| pt01Chr4_59045395_660 | chr4: 59038147/(−) | 26.1 | 21.6 | 24.1 | 28.1 | gamma (C) | 5 (C) | 4 (C) | 1.23 | 8 | ||||
| pt01Chr4_73583587_915 | chr4: 73584507/(−) | 26.7 | 21.4 | 23.6 | 28.3 | gamma (C) | 4 (C) | 6 (C) | 8 (C) | 0.52 | 13 | |||
| pt01Chr5r_11234345_1097 | chr5r: 11234345/(−) | 25.9 | 21.5 | 24.2 | 28.4 | gamma (C) | P | 3 (C) | 3 (C) | 2 (C) | 0.76 | 17 | ||
| pt01Chr6_37617641_1629 | chr6: 37617641/(−) | 26.7 | 21.5 | 23.5 | 28.3 | gamma (C) | 4 (C) | 0 (C) | 8 (C) | 5 (C) | 0.79 | 4 | ||
| pt01Chr6_44854470_567 | chr6: 44854470/(−) | 26.8 | 21.7 | 23.2 | 28.3 | gamma (C) | 4 (C) | 7 (C) | 7 (C) | 1.06 | 2 | |||
| pt01Chr7_74967707_1011 | chr7: 74960368/(−) | 27.2 | 21.4 | 23.4 | 28.1 | gamma (C) | P | 2 (C) | 4 (C) | 2 (C) | 0.79 | 8 | ||
| pt01Chr8_43813694_459 | chr8: 43806427/(−) | 27.7 | 21.4 | 23 | 27.9 | gamma (C) | 11 (C) | 5 (C) | 0.95 | 3 | ||||
| pt01Chr8_62466624_970 | chr8: 62466624/(−) | 25.2 | 21.2 | 24.9 | 28.7 | gamma (C) | P | 5 (C) | 1 (C) | 0.73 | 25 | |||
| pt01Chr9_95940431_611 | chr9: 95940431/(−) | 26.2 | 21.6 | 24.1 | 28.2 | gamma (C) | 7 (C) | 9 (C) | 0.98 | 8 | ||||
| PtG1b | 25.4 | 22.4 | 24.2 | 28.0 | Using pt01Chr6_139921967_779 | |||||||||
| pt01Chr10_72581664_733 | chr10: 72573964/(−) | 25.6 | 22 | 24.2 | 28.2 | gamma (C) | 4 (C) | 7 (C) | 8 (C) | 0 | 4 | AC148703Macaca mulatta | ||
| pt01Chr6_139921967_779 | chr6: 139915850/(−) | 24.9 | 22.5 | 24.6 | 28 | gamma (C) | 2 (C) | 7 (C) | 2 (C) | 0.74 | 11 | AC091754Papio anubis | ||
| pt01Chr6_50486605_639 | chr6:50486605/(−) | 25.7 | 22.6 | 23.9 | 27.8 | gamma (C) | 7 (C) | 3 (C) | 0.49 | 5 | AF142988Papio cynocephalus | |||
| PtG2a | 25.9 | 21.4 | 23.9 | 28.9 | Using pt01Chr5_53871501_226 | |||||||||
| pt01ChrY_13806832_1804 | chrY: 13806832/(−) | 25.7 | 21.1 | 23.9 | 29.3 | gamma (C) | P | 10 (C) | 0 (C) | 3 (C) | 0 | 2 | AF142988Papio cynocephalus | |
| pt01Chr1_24412696_1251 | chr1: 24412696/(−) | 26.3 | 20.9 | 24.1 | 28.7 | gamma (C) | P | 3 (C) | 1 (C) | 7 (C) | 4 (C) | 0.29 | 4 | AC148703Macaca mulatta |
| pt01Chr5_53871501_2266 | chr5: 53871501/(−) | 25.7 | 22.1 | 23.6 | 28.6 | gamma (C) | P | 6 (C) | 0 (C) | 9 (C) | 1 (C) | 1.67 | 4 | AC091754Papio anubis |
| PtG2b | Using pt01Chr12_34847717_1068 | |||||||||||||
| pt01Chr12_34847717_1068 | chr12: 34840755/(−) | 25.9 | 22 | 23.6 | 28.5 | gamma (C) | P | 1 (C) | 1 (C) | 8 (C) | 0.79 | 4 | AC148703Macaca mulatta | |
| AC091754 Papio anubis | ||||||||||||||
| AF142988 Papio cynocephalus | ||||||||||||||
| HML2 | 32.2 | 26.1 | 21.3 | 20.4 | Using pt01Chr10_4757815_326 | |||||||||
| pt01Chr10_4757815_3269 | chr10: 4757815/(−) | 32.1 | 26 | 21.2 | 20.6 | beta (B) | K | 6 (B) | 3 (B) | 6 (B) | 10 (B) | 0.63 | 4 | AC016577Homo sapiens |
| hg16Chr1_152822428_3843 | (−?)/1q22 | 32.2 | 26.2 | 21.3 | 20.3 | beta (B) | K | 3 (B) | 0 (B) | 1 (B) | 0 (B) | 0.21 | 28 | |
| hg16Chr11_101103511_3881 | (−)/11q22 | 32.2 | 26 | 21.3 | 20.5 | beta (B) | K | 3 (B) | 0 (B) | 1 (B) | 1 (B) | 0.41 | 61 | |
| hg16Chr11_118138535_3044 | (−?)/11q22 | 32.1 | 26.2 | 21.4 | 20.3 | beta (B) | K | 3 (B) | 0 (B) | 7 (B) | 2 (B) | 0.41 | 4 | |
| hg16Chr12_57016965_3699 | (−)/12q14 | 32.3 | 26 | 21.2 | 20.5 | beta (B) | K | 2 (B) | 0 (B) | 4 (B) | 0 (B) | 0.41 | 58 | |
| hg16Chr22_17300741_3451 | (−?)/22q11 | 32.2 | 26.1 | 21.3 | 20.4 | beta (B) | 2 (B) | 1 (B) | 3 (B) | 0 (B) | 0.41 | 21 | ||
| hg16Chr3_114073183_3522 | (−)/3q13 | 32.3 | 26.1 | 21.3 | 20.4 | beta (B) | K | 2 (B) | 2 (B) | 2 (B) | 1 (B) | 0 | 106 | |
| hg16Chr3_186610428_4007 | (−)/3q27 | 32.2 | 26.2 | 21.1 | 20.4 | beta (B) | K | 2 (B) | 1 (B) | 1 (B) | 1 (B) | 0.31 | 11 | |
| hg16Chr5_156074791_3951 | (−)/5q33 | 32.2 | 26.2 | 21.3 | 20.3 | beta (B) | K | 2 (B) | 0 (B) | 1 (B) | 1 (B) | 0.21 | 24 | |
| hg16Chr6_78431679_3161 | (−)/6q14 | 32.2 | 25.9 | 21.4 | 20.5 | beta (B) | K | 2 (B) | 0 (B) | 3 (B) | 0 (B) | 0.31 | 33 | |
| hg16Chr7_4376621_3310 | (−?)/7q22 | 32.1 | 26.1 | 21.4 | 20.5 | beta (B) | 3 (B) | 0 (B) | 1 (B) | 0 (B) | 0.86 | 4 | ||
| hg16Chr7_4384369_3828 | (−?)/7q22 | 32.4 | 25.8 | 21.3 | 20.5 | beta (B) | K | 3 (B) | 1 (B) | 1 (B) | 0 (B) | 0.21 | 4 | |
| hg16Chr8_7352269_3797 | (−?)/8p23 | 32.3 | 25.9 | 21.3 | 20.5 | beta (B) | 2 (B) | 1 (B) | 1 (B) | 0 (B) | 1.36 | 16 | ||
| HERV-H like | 24.2 | 28.1 | 15.3 | 32.4 | Using pt01Chr12_68612294_457 | |||||||||
| pt01Chr12_68612294_457 | chr12: 68612294/2p14 | 25.5 | 29.6 | 16.2 | 28.7 | gamma (C) | 2 (C) | 13 (C) | 1.55 | 15 | AC003009Homo sapiens | |||
| pt01Chr3_171348113_448 | chr3: 171342431/4q31.3 | 24.4 | 28.4 | 14.6 | 32.6 | gamma (C) | H | 15 (C) | 1.26 | 2 | ||||
| hg16Chr13_107615439_763 | chr14: 92137066/13q33 | 23.5 | 28.5 | 14.7 | 33.2 | gamma (C) | 0 (C) | 18 (C) | 1.16 | 2 | ||||
| hg16Chr18_52888720_999 | chr17: 49759585/18q21 | 24.1 | 28.6 | 14.9 | 32.5 | gamma (C) | H | 8 (C) | 4 (C) | 16 (C) | 3 (C) | 1.33 | 2 | |
| hg16Chr2_216989568_637 | (−?)/2q35 | 24.5 | 27.5 | 14.7 | 33.3 | gamma (C) | H | 6 (C) | 19 (C) | 1.98 | 2 | |||
| hg16Chr2_71367277_635 | (−?)/2p13 | 23.2 | 27.3 | 17.8 | 31.7 | gamma (C) | H | 6 (C) | 16 (C) | 1.39 | 2 | |||
| hg16Chr20_12321779_626 | chr21: 12124144/20p12 | 24.4 | 28.1 | 14.7 | 32.8 | gamma (C) | H | 7 (C) | 33 (C) | 0.6 | 3 | |||
| hg16Chr4_154245926_660 | chr4: 171342440/4q31 | 24.3 | 28 | 14.8 | 32.9 | gamma (C) | H | 9 (C) | 13 (C) | 1.36 | 2 | |||
| hg16Chr5_103482398_993 | chr4: 109273559/5q21 | 24.8 | 27.7 | 15 | 32.5 | gamma (C) | H | 11 (C) | 12 (C) | 1.33 | 2 | |||
| hg16Chr6_125192129_682 | (−?)/6q22 | 23.7 | 26.5 | 17.3 | 32.5 | gamma (C) | 7 (C) | 2 (C) | 22 (C) | 1.62 | 2 | |||
| hg16Chr6_127150561_671 | chr5: 129741796/6q22 | 24.8 | 28.6 | 14.5 | 32.1 | gamma (C) | H | 8 (C) | 17 (C) | 1.39 | 2 | |||
| hg16Chr6_78371754_1345 | chr5: 79852317/6q14 | 24.4 | 28 | 14.7 | 32.9 | gamma (C) | H | 11 (C) | 5 (C) | 1 (C) | 1.8 | 2 | ||
| hg16ChrX_5657489_573 | chrX: 6014174/Xp22 | 23.5 | 28 | 15.1 | 33.4 | gamma (C) | H | 8 (C) | 2 (C) | 17 (C) | 1.71 | 2 | ||
Sequences are referred according to their chromosomal start positions and RetroTector scores. Species-unique integrations are underlined. BLAT results for uncertain species-unique integrations are presented in Fig. S6 in the supplemental material.
(−) indicates no detected proviral integration. (−?) indicates uncertain proviral integration or possible deletion.
Among the recent integrations, with <2% LTR divergence, we found mainly beta-like proviruses in hg16, whereas the recent PanTro1 integrations were dominated by gammaretrovirus-like integrations (Fig. 1). Given that the International Committee on Taxonomy of Viruses (ICTV) does not classify endogenous retroviruses, the beta-like and gamma-like proviral integrations described here will, with support from their positions in the phylogenetic analyses (Fig. 1 and Fig. S1 to S3 in the supplemental material) and their genomic structures (see Fig. S4 in the supplemental material), henceforth in this paper be referred to as betaretroviruses and gammaretroviruses, respectively. Only a few of the recently integrated gammaretroviral sequences in hg16 had full retroviral gene structures (Tables 1 and 2). In contrast, the recent betaretroviruses in hg16 (Tables 1 and 2) were all full-length, with one or several ORF genes per provirus. We used the entire reconstructed Pol protein (putein) sequences as computed by RetroTector (23 for hg16 and 28 for PanTro1) together with reference sequences and constructed an unrooted NJ dendrogram (Fig. 1). Bootstrap supports are presented in a corresponding NJ cladogram (Fig. S1 in the supplemental material). The gammaretroviral NJ clades were similar to those of a maximum-parsimony analysis (Fig. S3 in the supplemental material).
In the Homo sp., one betaretroviral and one gammaretroviral group were detected (Fig. 1). The groups were selected using an 80% Pol similarity criterion. All recent human betaretroviruses were members of the HERV-K(HML2) group (2), hereafter called “HML2.” They were 97.5 to 99.9% similar to each other in Pol (see the similarity matrix in Fig. S2 in the supplemental material) and 98.7 to 99.8% similar to an HML2 consensus sequence (V. Blikstad, G. O. Sperber, and J. Blomberg, unpublished).This homogeneous subgroup of HML2 corresponds to what was earlier named “human-specific HERV-K” (4, 38, 39). All recent human gammaretroviruses grouped within the HERV-H-like group (28).
In the Pan sp., the recent integrations were dominated by 27 gammaretroviral sequences, but there was also one betaretroviral sequence (Fig. 1). The single recent betaretroviral Pan sp. sequence was assigned to the HML2 group, based on 98.8% Pol similarity to the HML2 consensus Pol and the dendrogram position (Fig. 1). Among the recent gammaretroviral Pan sp. integrations, two were similar to the HERV-H-like group (28). We defined two major sequence groups, PtG1 (18 PtG1a elements and 3 PtG1b elements) and PtG2 (3 PtG2a elements and 1 PtG2b element), which had no similarity to other chimpanzee or human proviruses (Fig. 1). The names were derived from Pan troglodytes (Pt) and gammaretrovirus-like (G). The numerals refer to the sequences joined by the ≥80% Pol similarity criterion (distance matrix in Fig. S2 of the supplemental material). Subgroups (a, b, etc.) derive from seemingly monophyletic branches within the groups (Fig. 1 and Fig. S1 to S3 in the supplemental material). We favor the use of this Pol similarity limit together with data from phylogenetic analyses in grouping ERVs over the use of phylogenetic analyses alone. The similarity criterion is unambiguously related to evolutionary distance, regardless of exogenous (with higher evolutionary rate) or endogenous (with lower evolutionary rate) retroviral states. Classification based on phylogenetic branching is relative and can split closely related retroviruses into separate clades, depending on the selection of included sequences. The rapidly growing number of retroviral sequences will facilitate classification (29). The PtG nomenclature may have to be revised then. The recently described PtERV1 (61), whose sequence kindly was provided by the group of Evan Eichler, clustered together with our PtG1a subgroup, with >80% (median, 89%) Pol similarity (Fig. S2 in the supplemental material). The average Pol similarities within the respective subgroups were 92% (PtG1a), 92% (PtG1b), 93% (PtG2a), and 82% in the HERV-H-like group (Fig. S2 in the supplemental material). The branch orders were essentially the same for the gammaretroviral sequences in an NJ cladogram and a maximum-parsimony (MP)-derived cladogram (Fig. S3 in the supplemental material). The minor inconsistencies within and at the borderlines of the subgroups of PtG1 and PtG2 are reflected in the somewhat lower MP bootstrap values. Similar to Fig. 1, the monophyletic PtG2a is paraphyletic to PtG2b in the additional NJ and MP cladograms (Fig. S3 in the supplemental material). In an attempt to increase the resolution for the gammaretroviruses and the PtG2 subgroups, we included additional nonmammalian gamma-like RT sequences (22) in the analysis. Using these shorter sequences, the early gammaretrovirus-like branch topology in the RT tree was consistent with the Pol tree in Fig. 1 (data not shown), although with less confidence. The recent PtG2b provirus came out in an ancestral position to HERV-E. Currently, no additional Pol sequences are available to further “pin down” the relationships of the PtG2b element to other gamma-like retroviruses. PtG1a and 1b (Fig. 1) could be treated as a group according to the similarity matrix (Fig. S2 in the supplemental material). However, an exception from the otherwise robust PtG groups is the Papio anubis (clone AC091754) provirus, which groups inconsistently in the different analyses (see Fig. S1 to S3 in the supplemental material). This may theoretically be caused by convergent evolution, or recombination, within the two PtG groups and AC091754. The gammaretrovirus-like PtG1 group is distinct from the ICTV-defined gammaretroviruses (including MLV and BaEV), despite sequence similarity to another baboon ERV. There are thus distinct gammaretrovirus-like baboon ERVs as well (see below).
Horizontal transfers.
There were signs of cross-species transfer involving the PtG retroviruses. We performed tBLASTn searches against the whole nonredundant database at GenBank with a consensus of the aligned PtG1a Pol sequences and with original Pol sequences to represent PtG1b, PtG2a, and PtG2b (Table 2). A few primate nonchimpanzee sequences, yielding high tBLASTn scores (data not shown), proved to be closely similar to the Pol of each PtG group (Table 2). They grouped as novel gammaretroviruses in Fig. 1. Also, they had typical gammaretroviral gene structures, with gag, pro, and pol in one reading frame separated from env and no obvious accessory genes (see Fig. S4 in the supplemental material). It was further noted that the human retroviral sequences had primer binding sites (PBSs) complementary to either Lys-tRNA in HML2 or His-tRNA in the HERV-H-like group, while detected PBSs in the recent Pan sp. ERV groups (PtG1 and PtG2) were complementary to Pro-tRNA(Table 2). Other primate gammaretroviral sequences, like HUERSP3 and a number of ERV3-like sequences (1), also have a Pro-PBS but were only 50 to 76% similar in Pol to PtG1 (data not shown).
The nonchimpanzee sequences were also included in the Pol similarity matrix (Fig. S2 in the supplemental material). Both PtG groups were >80% similar in Pol to both murine and feline leukemia viruses (Fig. 1). The PtG1 elements were highly (84 to 96%) similar in Pol to two previously not described ERVs: a baboon (Papio anubis) sequence, clone AC093133, and a macaque (Macaca mulatta) sequence, clone AC148703 (Fig. S2 in the supplemental material and Table 2). The PtG2 elements were similar to the baboon endogenous retrovirus (BaEV) and to a Papio cynocephalus sequence, clone AF142988 (Fig. S2 in the supplemental material). The three PtG2a elements were approximately 96% similar to each other (Fig. S2 in the supplemental material). They were more than 92% similar to BaEV Pol (56) and to the Papio cynocephalus ERV Pol (Fig. 1 and Fig. S2 in the supplemental material). Further, they were 85% similar to MLV Pol. The single PtG2b sequence was more separate in the retroviral tree (Fig. 1) but was 93% similar to a Papio anubis ERV in clone AC091754 and 88% similar to MLV. A supplementary comparison of the PtG groups with a range of previously described BaEV-related Pol (RT, 108 amino acids) sequences (55), obtained through the courtesy of Antoinette van der Kuyl, showed that the PtG1 group also was similar to Papio and Colobus ERVs other than those described in Table 2 (see Fig. S5 in the supplemental material). The PtG2a elements grouped within the BaEV (55) clade. The PtG2b element grouped together with the Papio anubis sequence, however inconsistently (see above) (Fig. S1 to S3 in the supplemental material). Although a recent integration, it came out in an ancestral position relative to much older gamma-like HERVs, relatively close to HERV-E (Fig. 1 and Fig. S1 to S3 in the supplemental material). This could mean that more-or-less close relatives of gammaretrovirus-like HERVs may still be spreading among primates.
Subsequent LTR analysis (Fig. S5 in the supplemental material) showed that the PtG1 elements were highly similar (average, 88% nucleotide identities using pairwise deletion) to the chimpanzee LTR homologues of the colobus CPC-1 proviruses described by Bonner et al. (10). PtG1 LTRs were also similar to the macaque MAC-1 LTR (average, 80%) but less similar to the colobus CPC-1 LTR (average, 58%). However, the reference LTR sequences were not full-length and therefore merely indicate kinship between the ERVs. Due to separate ERV data sets and genes, a strict comparison between the Pol and LTR phylogenies (Fig. S5 in the supplemental material) is not possible. What is clear is that there are several novel sequences in the chimpanzee. They were probably transmitted to chimpanzees several times, in a complex way, from other primates in the recent past. We here demonstrate close similarities of the PtG groups to at least three different primate ERV groups, including BaEV.
The PtG1a, the PtERV1 (61), and the CPC-1 elements may derive from the same virus. CPC-1 was earlier described to occur in chimpanzee and gorilla but not in human, gibbon, and orangutan. Further, it was supposed to have transmitted to chimpanzees from colobus (10). Although mostly based on hybridization data derived from the presequencing era, that information on type C retroviral sequences in chimpanzees (6, 12) supports our findings. It was here corroborated and extended by a bioinformatic approach. The transmission hypothesis gained support from the (however limited) Pol and LTR phylogenies (Fig. S5 in the supplemental material), in addition to the pairwise LTR similarities from that alignment (see above).
Based on gag and env analyses, Yohn et al. recently showed that the PtERV1 elements (similar to our PtG1a group in the Pol analysis) (Fig. S2 in the supplemental material) may possibly have more than one origin (61). Exogenous retroviruses from at least three host groups, including (i) chimpanzee together with gorilla, (ii) baboon, and (iii) macaque, were suggested to have contributed. Their phylogenetic trees differed from generally accepted primate species trees, thus indicating horizontal transfer (61). Our data agree with those results. Although theoretically transmissions could first have reached chimpanzees and then gone from chimpanzees to other primates, the simplest explanation is transmission of nonhuman, nonchimpanzee primate gamma-like retroviruses to chimpanzee since the human-chimpanzee split. The presence of the BaEV-like PtG2 proviruses in Pan sp. but not Homo sp., and the presence of additional PtG2 LTR BLAST hits in PanTro1 but not in hg16, signifies transmissions after the Homo-Pan speciation (Table 2). As noted by van der Kuyl et al., BaEV-like viruses spread among African primates, and probably also to cats, in recent evolutionary time (54-56). In fact, MLV-like gammaretroviruses infected, and occasionally became endogenized in, a number of mammals during this period (11, 37, 49). This is an ongoing process, demonstrated by the widespread polymorphism of MLV integrations among inbred mice (46, 47, 52, 53). The inferred pattern of horizontal spread of the PtG groups is similar to that of simian immunodeficiency virus SIVcpz (3) and human immunodeficiency virus (20), which most likely arose by transfer from smaller primates to chimpanzee and from chimpanzee to human, respectively. Based on the phylogenetic analysis (Fig. S5 in the supplemental material), the PtG2 elements were, like SIVcpz (3), judged to originate from one or several small primates. The relative scarcity of primate sequences prevented the demonstration of a specific transmission route(s) for the PtG1 elements. They were similar to baboon and macaque sequences. The macaque retroviral similarity was unexpected. African macaques are present in only a small North African population, remote from the ancestral chimpanzee habitats in forests and forested savanna. Consequently, they are unlikely to have been in close contact with the chimpanzees. However, the possibility that macaques and chimpanzees have overlapped geographically in the past should not be excluded. The relatedness of PtG1 to macaque proviral sequences may represent similarity to widely distributed non-ape primate retroviruses. Our observations are thus consistent with the existence of a network of relatively frequent horizontal retroviral transmissions, followed by occasional endogenization, among primates. Exposure of wounds to prey blood during predation or eating of retrovirus-rich placentae (3, 20, 54) are possible explanations. Interspecies retroviral transmission via blood-sucking insects may also occur (58). Finally, it cannot be excluded that retrotransposon DNA may be integrated after uptake from the alimentary canal (19). Chimpanzees frequently eat other primates, like baboons, geladas, and colobi (21, 45), which harbor ERVs similar to the PtG retroviral sequences. The probable cases of horizontal transfer of gammaretroviruses to chimpanzees thus agree with the predatory practices of chimpanzees. It is, however, intriguing that the human genome seems to have been spared from the PtG integrations. Human ancestors and chimpanzees may have been differentially exposed through differing hunting practices (13, 40, 45). Differences in ERV fixation due to population size and distribution could also be reasons.
Differences in recent activity between beta- and gamma-like retroviruses.
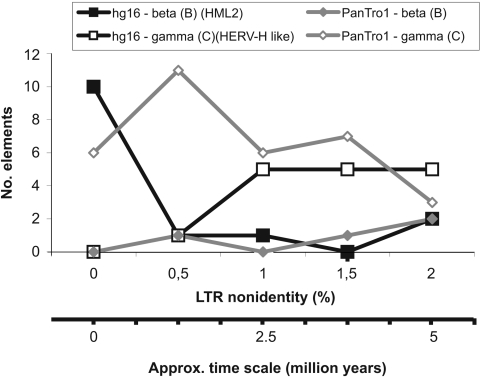
If LTR divergences for all RetroTector-derived ERVs are used as surrogate markers for integration times, an expansion of a limited number of human HML2 (i.e., hg16-beta) integrations appears to have started around 1.5 to 2 million years ago (approximately 0.5% LTR nonidentities) (Fig. 2). The expansion may still be ongoing, because the curve peaks at 0% LTR divergence. However, LTR-LTR homogenization by gene conversion could lead to falsely low LTR divergences, precluding an exact interpretation (24, 30). The betaretrovirus-like HML2 is a relatively large HERV group, of which the majority is common to Homo and Pan spp. (data not shown). A simple explanation for the recent HML2 expansion in humans could be back mutation to replication competence (“breakout”; see below). Even if these recently integrated HML2 elements have been labeled “human specific” (4, 38, 39), the sequence record of primates and other possible contributors of HMLs is incomplete. Cross-species transmissions of HML2 thus cannot be entirely excluded. Figue 2 indicates that the gammaretroviral integrational activity was separate in time from that of betaretroviruses. The recent gammaretroviral integrations of both species differ from the HML2 ones by containing more disrupted genomes yet with low LTR differences (Table 2). Retroviruses may differ substantially in mutation frequency (23, 31, 44). Although the frequency of gene conversion is highly dependent on sequence similarity, the low LTR divergences in otherwise disrupted, and thus probably ancient, gammaretroviral elements can have been caused by LTR homogenization (24, 30).
FIG. 2.
LTR differences in recently integrated beta-like (HML2 group) and gamma-like ERVs in the human (hg16) and chimpanzee (PanTro1) genomes.
Complementation in trans, where particles formed by more complete (“midwife”) proviruses package RNA of less complete ones, as proposed for the HERV-H-like group (28, 36), is another alternative. As expected, the markedly skewed nucleotide distribution of HERV-H elements in general (27) is also present in the recently integrated and disrupted HERV-H elements (Table 2).
Thus, both the human and chimpanzee genomes have been subjected to different kinds of recent retroviral integrations. A BLAT search for LTRs of recent unique ERVs with a stringent criterion (>98% of maximum BLAT score, using either of the 5′ and 3′ LTRs) resulted in numerous hits (Table 2), but only in the cognate genome. This also applies to the otherwise homogeneous HML2 group. Consequently, in the past, there must have been many more integrations of these elements in the chimpanzee and human genomes than the currently residing ones (Table 2). They may have become looped out through homologous recombination between the LTRs, as postulated previously (48). PCR would be a suitable method to address the amount of solitary LTRs but was out of scope for this study. It is also noteworthy that a BLAT search with LTRs of the selected human HERV-H-like elements resulted in fewer hits (Table 2), suggesting that retroviral RNAs with mutated R, U5, and U3 LTR portions resulted in these integrations, as expected from the “midwife” master model (28, 36). HERV-H probably did not reintegrate recently to the same extent as the HML2 elements did (Fig. 2 and Table 1). It is likely that, for more than 30 million years, the HML2 group multiplied mainly through reinfections rather than cis retrotransposition or trans complementation (5). The high Pol similarities among the recent HML2 elements (approximately 98% [Fig. S2 in the supplemental material]), together with the low (<2%) LTR divergence, indicate a common origin after Homo-Pan speciation. LTR homogenizations by gene conversions are unlikely to have occurred simultaneously in all different HML2 loci (Table 2) after the Homo-Pan sp. split, which concurs with the uniqueness of these ERVs in Homo sp. The recent expansion of highly related HML2 integrations (Fig. 2) may have derived either (i) from a random mutational activation of a slightly damaged, preexisting HML2 element or (ii) by reinfection of humans with HML2 from another species. We currently favor the first hypothesis, since there is no known source of infectious HML2 in animals close to humans or human predecessors. According to the “breakout” hypothesis (7), copackaged RNA of partially defective ERV elements occasionally may recombine, thereby rescuing and optimizing retroviral function during reinfections from within. HML2 elements are in general the most complete of all HERVs. Old, relatively intact HML2 elements could have assisted in a stochastic fashion in the recent HML2 activation in Homo sp. and done so to a lesser extent in Pan sp. This possibility should be further investigated.
Conclusion and a caveat.
Inevitably, though the screening strategy is likely to detect most young integrations, the genome may contain an undefined portion of older elements where gene conversion caused LTR homogenization. If supplemented with a check for uniqueness (i.e., ERVs occurring in only one or the other species) against the next genome, as conducted here, the approach should correctly detect recent integrations which areunique to humans or chimpanzees. However, a complication is that the assessment of uniqueness for Pan versus Homo sp. elements is stronger than vice versa due to the poorer Pan sp. draft sequence quality. Eventually, this could lead to an erroneous impression of uniqueness for a human sequence. The matching of flanks of the seemingly human-specific HERV-H integrations into the chimpanzee sequence was not convincing. They were therefore not underlined in Table 2. The low number of LTRs with high similarity to the suspected human unique HERV-H proviruses (Table 2) is consistent with the “midwife” master hypothesis (28, 36), because (re)integration-competent proviruses would be more likely to give such single LTRs, while copackaged defective ones would not. Thus, the human unique HERV-H-like sequences (Fig. 1 and Table 2) eventually will need additional experimental analysis. Allelic variation and deletions are additional obstacles. However, precise deletions of proviruses are unlikely to occur (35), especially in the many different loci on different chromosomes presented here. Thus, it is more likely that our observed genomic ERV differences are the results of gain rather than loss. Further, as shown here, LTRs corresponding to the unique proviruses occur frequently but only in the cognate genome. In the BLAT search, they outnumber their proviral counterparts (Table 2). Instead of precise proviral deletions, the higher number of recognized LTRs shows a more likely event of ERV loss through homologous recombination and looped-out proviruses (48). Allelic variation cannot be covered in the single-sequence genome assemblies. A locus-specific PCR, preferably with many individuals, could be used to address this problem but was out of the scope of this study. The false LTR similarity (gene conversion) and false uniqueness problems were addressed by bioinformatic means, as discussed above.
The numerous recent species-unique proviruses, and the larger number of similar species-unique (mainly solitary) LTRs, show that both Homo and Pan sp. genomes have distinct sets of recently active ERVs. The comparison of retroviral sequences in Homo and Pan sp. genomes highlights the importance of (i) habitat, interspecies contact, and predator-prey relations facilitating cross-species retroviral infection from “outside” and/or (ii) probable stochastic reactivation of preexisting ERVs followed by reinfection from “inside” as determinants of the retroviral genetic setup of a species.
Supplementary Material
Acknowledgments
This work was supported by the Swedish Research Council (grant K2004-32X-14252-03A) and Stanley Foundation (grant 03R-584).
We also thank Antoinette van der Kuyl and Evan Eichler for sequence contributions, Tove Airola for assistance in genomic data collection, and Michael Tristem for valuable discussions.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Andersson, A. C., Z. Yun, G. O. Sperber, E. Larsson, and J. Blomberg. 2005. ERV3 and related sequences in humans: structure and RNA expression. J. Virol. 79:9270-9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, M. L., M. Lindeskog, P. Medstrand, B. Westley, F. May, and J. Blomberg. 1999. Diversity of human endogenous retrovirus class II-like sequences. J. Gen. Virol. 80:255-260. [DOI] [PubMed] [Google Scholar]
- 3.Bailes, E., F. Gao, F. Bibollet-Ruche, V. Courgnaud, M. Peeters, P. A. Marx, B. H. Hahn, and P. M. Sharp. 2003. Hybrid origin of SIV in chimpanzees. Science 300:1713. [DOI] [PubMed] [Google Scholar]
- 4.Barbulescu, M., G. Turner, M. I. Seaman, A. S. Deinard, K. K. Kidd, and J. Lenz. 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 9:861-868. [DOI] [PubMed] [Google Scholar]
- 5.Belshaw, R., V. Pereira, A. Katzourakis, G. Talbot, J. Paces, A. Burt, and M. Tristem. 2004. Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl. Acad. Sci. USA 101:4894-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birkenmeier, E. H., T. I. Bonner, K. Reynolds, G. H. Searfoss, and G. J. Todaro. 1982. Colobus type C virus: molecular cloning of unintegrated viral DNA and characterization of the endogenous viral genomes of colobus. J. Virol. 41:842-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blomberg, J., D. Ushameckis, and P. Jern. 2005. Evolutionary aspects of human endogenous retroviral sequences (HERVs) and disease, p. 227-262. In E. D. Sverdlov (ed.), Retroviruses and primate genome evolution. Eurekah.com/Landes Bioscience, Georgetown, Tex.
- 8.Bock, M., and J. P. Stoye. 2000. Endogenous retroviruses and the human germline. Curr. Opin. Genet. Dev. 10:651-655. [DOI] [PubMed] [Google Scholar]
- 9.Boeke, J. D., and J. P. Stoye. 1997. Retrotransposons, endogenous retroviruses, and the evolution of retroelements, p. 343-436. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, New York, N.Y. [PubMed]
- 10.Bonner, T. I., E. H. Birkenmeier, M. A. Gonda, G. E. Mark, G. H. Searfoss, and G. J. Todaro. 1982. Molecular cloning of a family of retroviral sequences found in chimpanzee but not human DNA. J. Virol. 43:914-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonner, T. I., and G. J. Todaro. 1979. Carnivores have sequences in their cellular DNA distantly related to the primate endogenous virus, MAC-1. Virology 94:224-227. [DOI] [PubMed] [Google Scholar]
- 12.Bonner, T. I., and G. J. Todaro. 1980. The evolution of baboon endogenous type C virus: related sequences in the DNA of distant species. Virology 103:217-227. [DOI] [PubMed] [Google Scholar]
- 13.Bramble, D. M., and D. E. Lieberman. 2004. Endurance running and the evolution of Homo. Nature 432:345-352. [DOI] [PubMed] [Google Scholar]
- 14.Britten, R. J. 2002. Divergence between samples of chimpanzee and human DNA sequences is 5%, counting indels. Proc. Natl. Acad. Sci. USA 99:13633-13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, F. C., and W. H. Li. 2001. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 68:444-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chimpanzee Sequencing and Analysis Consortium. 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437:69-87. [DOI] [PubMed] [Google Scholar]
- 17.Ebersberger, I., D. Metzler, C. Schwarz, and S. Paabo. 2002. Genomewide comparison of DNA sequences between humans and chimpanzees. Am. J. Hum. Genet. 70:1490-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feuchter-Murthy, A. E., J. D. Freeman, and D. L. Mager. 1993. Splicing of a human endogenous retrovirus to a novel phospholipase A2 related gene. Nucleic Acids Res. 21:135-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forsman, A., D. Ushameckis, A. Bindra, Z. Yun, and J. Blomberg. 2003. Uptake of amplifiable fragments of retrotransposon DNA from the human alimentary tract. Mol. Genet. Genomics 270:362-368. [DOI] [PubMed] [Google Scholar]
- 20.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 21.Goodall, J. 1971. In the shadow of man. Mariner Books, London, United Kingdom.
- 22.Herniou, E., J. Martin, K. Miller, J. Cook, M. Wilkinson, and M. Tristem. 1998. Retroviral diversity and distribution in vertebrates. J. Virol. 72:5955-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hollsberg, P. 1999. Mechanisms of T-cell activation by human T-cell lymphotropic virus type I. Microbiol. Mol. Biol. Rev. 63:308-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes, J. F., and J. M. Coffin. 2005. Human endogenous retroviral elements as indicators of ectopic recombination events in the primate genome. Genetics 171:1183-1194. (First published 12 September 2005; doi: 10.1534/genetics.105.043976.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hughes, J. F., and J. M. Coffin. 2004. Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: implications for human and viral evolution. Proc. Natl. Acad. Sci. USA 101:1668-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 27.Jern, P., G. O. Sperber, G. Ahlsen, and J. Blomberg. 2005. Sequence variability, gene structure, and expression of full-length human endogenous retrovirus H. J. Virol. 79:6325-6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jern, P., G. O. Sperber, and J. Blomberg. 2004. Definition and variation of human endogenous retrovirus H. Virology 327:93-110. [DOI] [PubMed] [Google Scholar]
- 29.Jern, P., G. O. Sperber, and J. Blomberg. 2005. Use of endogenous retroviral sequences (ERVs) and structural markers for retroviral phylogenetic inference and taxonomy. Retrovirology 2:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson, W. E., and J. M. Coffin. 1999. Constructing primate phylogenies from ancient retrovirus sequences. Proc. Natl. Acad. Sci. USA 96:10254-10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katz, R. A., and A. M. Skalka. 1990. Generation of diversity in retroviruses. Annu. Rev. Genet. 24:409-445. [DOI] [PubMed] [Google Scholar]
- 32.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 33.Lebedev, Y. B., O. S. Belonovitch, N. V. Zybrova, P. P. Khil, S. G. Kurdyukov, T. V. Vinogradova, G. Hunsmann, and E. D. Sverdlov. 2000. Differences in HERV-K LTR insertions in orthologous loci of humans and great apes. Gene 247:265-277. [DOI] [PubMed] [Google Scholar]
- 34.Li, W. H. 1997. Molecular evolution. Sinnauer Associates, Inc., Publishers, Sunderland, Mass.
- 35.Liu, G., S. Zhao, J. A. Bailey, S. C. Sahinalp, C. Alkan, E. Tuzun, E. D. Green, and E. E. Eichler. 2003. Analysis of primate genomic variation reveals a repeat-driven expansion of the human genome. Genome Res. 13:358-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mager, D. L., and J. D. Freeman. 1995. HERV-H endogenous retroviruses: presence in the New World branch but amplification in the Old World primate lineage. Virology 213:395-404. [DOI] [PubMed] [Google Scholar]
- 37.Martin, J., E. Herniou, J. Cook, R. W. O'Neill, and M. Tristem. 1999. Interclass transmission and phyletic host tracking in murine leukemia virus-related retroviruses. J. Virol. 73:2442-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayer, J., M. Sauter, A. Racz, D. Scherer, N. Mueller-Lantzsch, and E. Meese. 1999. An almost-intact human endogenous retrovirus K on human chromosome 7. Nat. Genet. 21:257-258. [DOI] [PubMed] [Google Scholar]
- 39.Medstrand, P., and D. L. Mager. 1998. Human-specific integrations of the HERV-K endogenous retrovirus family. J. Virol. 72:9782-9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morris, K., and J. Goodall. 1977. Competition for meat between chimpanzees and baboons of the Gombe National Park. Folia Primatol. (Basel) 28:109-121. [DOI] [PubMed] [Google Scholar]
- 41.Muotri, A. R., V. T. Chu, M. C. Marchetto, W. Deng, J. V. Moran, and F. H. Gage. 2005. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435:903-910. [DOI] [PubMed] [Google Scholar]
- 42.Oja, M., G. O. Sperber, J. Blomberg, and S. Kaski. 2005. Self-organizing map-based discovery and visualization of human endogenous retroviral sequence groups. Int. J. Neural Syst. 15:163-179. [DOI] [PubMed] [Google Scholar]
- 43.Samuelson, L. C., K. Wiebauer, C. M. Snow, and M. H. Meisler. 1990. Retroviral and pseudogene insertion sites reveal the lineage of human salivary and pancreatic amylase genes from a single gene during primate evolution. Mol. Cell. Biol. 10:2513-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon, V., and D. D. Ho. 2003. HIV-1 dynamics in vivo: implications for therapy. Nat. Rev. Microbiol. 1:181-190. [DOI] [PubMed] [Google Scholar]
- 45.Stanford, C. B., J. Wallis, H. Matama, and J. Goodall. 1994. Patterns of predation by chimpanzees on red colobus monkeys in Gombe National Park, 1982-1991. Am. J. Phys. Anthropol. 94:213-228. [DOI] [PubMed] [Google Scholar]
- 46.Steffen, D. L., S. Bird, and R. A. Weinberg. 1980. Evidence for the Asiatic origin of endogenous AKR-type murine leukemia proviruses. J. Virol. 35:824-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steffen, D. L., B. A. Taylor, and R. A. Weinberg. 1982. Continuing germ line integration of AKV proviruses during the breeding of AKR mice and derivative recombinant inbred strains. J. Virol. 42:165-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stoye, J. P. 2001. Endogenous retroviruses: still active after all these years? Curr. Biol. 11:R914-R916. [DOI] [PubMed] [Google Scholar]
- 49.Stoye, J. P., and J. M. Coffin. 1987. The four classes of endogenous murine leukemia virus: structural relationships and potential for recombination. J. Virol. 61:2659-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Telesnitsky, A., and S. P. Goff. 1997. Reverse transcriptase and generation of retroviral DNA, p. 121-160. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, New York, N.Y. [PubMed]
- 51.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomonaga, K., and J. M. Coffin. 1998. Structure and distribution of endogenous nonecotropic murine leukemia viruses in wild mice. J. Virol. 72:8289-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tomonaga, K., and J. M. Coffin. 1999. Structures of endogenous nonecotropic murine leukemia virus (MLV) long terminal repeats in wild mice: implication for evolution of MLVs. J. Virol. 73:4327-4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Kuyl, A. C., J. T. Dekker, and J. Goudsmit. 1996. Baboon endogenous virus evolution and ecology. Trends Microbiol. 4:455-459. [DOI] [PubMed] [Google Scholar]
- 55.van der Kuyl, A. C., J. T. Dekker, and J. Goudsmit. 1995. Distribution of baboon endogenous virus among species of African monkeys suggests multiple ancient cross-species transmissions in shared habitats. J. Virol. 69:7877-7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van der Kuyl, A. C., J. T. Dekker, and J. Goudsmit. 1995. Full-length proviruses of baboon endogenous virus (BaEV) and dispersed BaEV reverse transcriptase retroelements in the genome of baboon species. J. Virol. 69:5917-5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vartanian, J. P., U. Plikat, M. Henry, R. Mahieux, L. Guillemot, A. Meyerhans, and S. Wain-Hobson. 1997. HIV genetic variation is directed and restricted by DNA precursor availability. J. Mol. Biol. 270:139-151. [DOI] [PubMed] [Google Scholar]
- 58.Vobis, M., J. D'Haese, H. Mehlhorn, and N. Mencke. 2003. Evidence of horizontal transmission of feline leukemia virus by the cat flea (Ctenocephalides felis). Parasitol. Res. 91:467-470. [DOI] [PubMed] [Google Scholar]
- 59.Xiong, Y., and T. H. Eickbush. 1988. Similarity of reverse transcriptase-like sequences of viruses, transposable elements, and mitochondrial introns. Mol. Biol. Evol. 5:675-690. [DOI] [PubMed] [Google Scholar]
- 60.Yin, H. 1999. Human mouse mammary tumour virus like elements and their relation to breast cancer. Ph.D. dissertation. Uppsala University, Uppsala, Sweden.
- 61.Yohn, C. T., Z. Jiang, S. D. McGrath, K. E. Hayden, P. Khaitovich, M. E. Johnson, M. Y. Eichler, J. D. McPherson, S. Zhao, S. Paabo, and E. E. Eichler. 2005. Lineage-specific expansions of retroviral insertions within the genomes of African great apes but not humans and orangutans. PLoS Biol. 3:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.