Abstract
An intimate relationship between hepatitis C virus (HCV) replication and the physiological state of the host liver cells has been reported. In particular, a highly reproducible and reversible inhibitory effect of high cell density on HCV replication was observed: high levels of HCV RNA and protein can be detected in actively growing cells but decline sharply when the replicon cells reach confluence. Arrested cell growth of confluent cells has been proposed to be responsible for the inhibitory effect. Indeed, other means of arresting cell growth have also been shown to inhibit HCV replication. Here, we report a detailed study of the effect of cell growth and confluence on HCV replication using a flow cytometry-based assay that is not biased against cytostasis and reduced cell number. Although we readily reproduced the inhibitory effect of cell confluence on HCV replication, we found no evidence of inhibition by serum starvation, which arrested cell growth as expected. In addition, we observed no inhibitory effect by agents that perturb the cell cycle. Instead, our results suggest that the reduced intracellular pools of nucleosides account for the suppression of HCV expression in confluent cells, possibly through the shutoff of the de novo nucleoside biosynthetic pathway when cells become confluent. Adding exogenous uridine and cytidine to the culture medium restored HCV replication and expression in confluent cells. These results suggest that cell growth arrest is not sufficient for HCV replicon inhibition and reveal a mechanism for HCV RNA inhibition by cell confluence.
Hepatitis C virus (HCV) is a positive-strand RNA virus that infects more than 170 million people worldwide. Its infection mainly affects the liver and leads to both acute and chronic liver diseases including cirrhosis and hepatocellular carcinoma. HCV replication has proved to be a good model system for studies of virus-host cell interactions, as the virus often establishes a chronic infection that typifies the intricate relationship between a pathogen and its host.
Research on HCV infection in vitro has been hampered by lack of an efficient cell culture-based production and infection system. The very recent reports of production of infectious HCV particles in tissue culture will hopefully break through this barrier and usher in a new era of HCV research (15, 26, 32). Studies of HCV RNA replication, however, received a great boost with the development of the HCV subgenomic replicon system a number of years ago (1, 16). Many important questions about the RNA replication of the virus were answered through use of the replicon cells. Adaptive mutations that permit efficient RNA replication in cultured hepatoma cells as well as nonhepatic cell lines have been defined (1, 13, 33); critical components of replication have been mapped and the data greatly complemented the limited in vivo data available previously (5, 12, 31). A novel mechanism by which HCV suppresses the innate antiviral responses to establish persistent replication has been uncovered (4). The replicon system is also being used extensively to evaluate antiviral agents such as small-molecular drugs (10) as well as small interfering RNAs (23). The validity of the replicon system for this purpose has been highlighted by the in vivo efficacy in humans of a small-molecule compound that was tested solely in the replicon without any animal efficacy study (14). Even though the future of the compound is uncertain because of in vivo toxicity issues, the excellent correlation between the replicon inhibition in vitro and the in vivo antiviral effect provided good proof of principle for both the class of the drug and the screening assay.
Various derivatives of the HCV replicon that harbor different reporter genes have been constructed to facilitate the measurement of replication by means of surrogate markers (13, 20, 30). Recently, a replicon cell line has been developed in which the green fluorescent protein (GFP) gene is inserted into the coding region of the NS5A gene without abolishing the ability of the RNA to replicate in Huh-7 cells (18). This line allowed direct visualization of the fusion protein, NS5A-GFP, with fluorescent microscopy for study of the dynamics of the HCV replication complex in living cells, as NS5A is one of the nonstructural proteins believed to be in the replication complex. Co-localization of NS5A and newly synthesized viral RNA was observed.
A tight coupling between HCV replication and the physiological state of the host liver cells has been observed. In particular, a highly reproducible and reversible inhibitory effect of cell confluence on HCV replication was reported: high levels of HCV RNA and protein that can be detected in actively growing cells decrease dramatically when the replicon cells reach confluence. When the confluent cells are split into a lower density, replication resumes and HCV expression recovers (22, 24).
Here, we report the development and validation of a new replicon assay that allows flow cytometry-based measurement of HCV expression. This assay measures NS5A-GFP expression in individual cells and is therefore not biased against reduced cell number in samples with cytostasis. Using this assay, combined with traditional viral RNA and protein analysis, we found that cell growth arrest is not the mechanism for cell confluence-based replicon inhibition. Instead, reduced nucleoside level in confluent cells can partly account for the inhibition.
MATERIALS AND METHODS
Compounds.
Alpha interferon (IFN-α), aphidicolin, uridine, and cytidine were purchased from Sigma. Cyclosporine (Cs) was purchased from Alexis Corporation (San Diego, CA). 2′-C-methyl-adenosine (2CMA) was a gift from Steve Carroll (Merck Inc.).
Cell lines.
Clone B and Huh.8 cells were obtained from Apath LLC through the National Institutes of Health (NIH) AIDS reagents program. The I/5A-GFP cell line was provided by Charles Rice. Replicon cells were routinely maintained in Dulbecco modified medium supplemented with antibiotics, 10% fetal bovine serum, and 500 μg/ml G418. Huh-7 cells were maintained in a similar medium but without the G418. When replicon cells were treated with various HCV inhibitors, the G418 was also omitted from the medium.
Confluent cells and exogenously added nucleotides.
To generate confluence samples, we routinely seed four times more cells for confluent samples than the nonconfluent samples. This generated a 100% confluent monolayer of confluent cells the next day. Cells cultured at 39°C or treated with various compound were kept under nonconfluent conditions. The cell numbers for the various experiments involving confluence are as follows. (i) For experiments carried out with six-well plates, 1.5 × 105 cells/well were seeded for nonconfluent samples and 6 ×105 cells/well for confluent samples. These include results shown in Fig. 2A and B and 5B. Note that experiments with fluorescent microscopy were performed with cells grown on glass coverslips placed in six-well plates. (ii) For experiments carried out with 12-well plates, 0.75 ×105 cells/well were seeded for nonconfluent samples and 3 ×105 cells/well for confluent samples. These include results shown in Fig. 5A and C, 6, and 7. (iii) For experiments carried out with T-25 flasks, 3 ×105 cells/flask were seeded for nonconfluent samples and 1.2 ×106 cells/flask for confluent samples. These include results shown in Fig. 2C and D and 5D. For experiments with exogenously added nucleotides, the nucleotides were added at day 0, which is the day when the cells first became confluent (1 day after cell seeding).
FIG. 2.
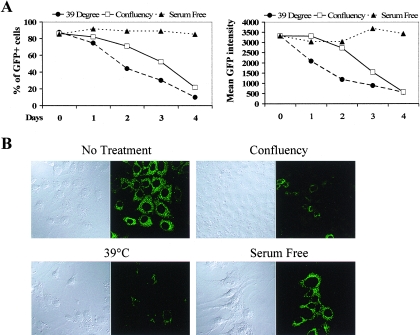
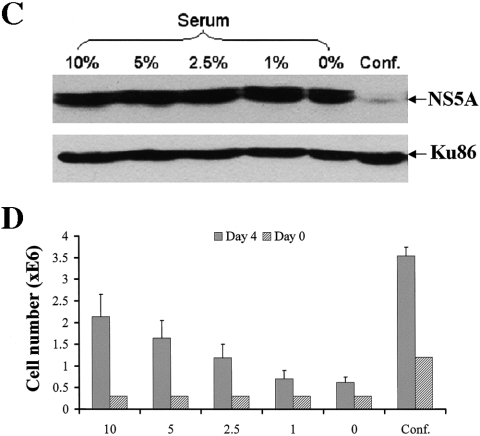
Cell confluence and serum starvation have drastically different effects on HCV replication. (A) Comparison of the effects on HCV expression by three nonchemical treatments: 39°C, confluence, and serum starvation. The cells were cultured under the indicated conditions for 4 days, and samples were taken each day for FACS analysis. Four times more cells were seeded in the confluence sample which resulted in a 100% confluent monolayer of cells in the culture vessels. (B) Microscopic images (DIC and fluorescence) of the day 4 sample. (C) Serum starvation failed to inhibit HCV expression in a replicon cell line without GFP. 1B replicon cells were cultured under the indicated conditions for 4 days, and total proteins were extracted and then subjected to immunoblotting with an anti-NS5A antibody. After detection, the same membrane was stripped of the NS5A signal and probed again with an anti-Ku86 antibody as an internal control for protein loading. (D) Serum starvation effectively arrested cell growth as expected. The cell numbers at the beginning (day 0) and the conclusion of the experiment (day 4) were quantified with a trypan blue exclusion assay. In general, four times more cells were seeded for the confluence sample which resulted in a 100% confluent monolayer of cells in the culture plates or slide chambers (see materials and methods for cell numbers).
FIG. 5.
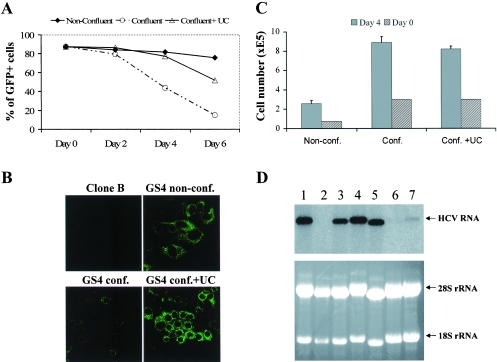
Exogenous added nucleosides counteract the inhibitory effect of cell confluence. (A) GS4 cells were cultured under either nonconfluent or confluent conditions for 6 days. One group of confluent cells was supplemented with uridine and cytidine (200 μM each) for the duration of the experiment. Samples were taken for FACS analysis every 2 days. (B) The microscopic images (DIC and fluorescence) of the day 4 sample. (C) UC addition did not increase cell growth of the confluent cells. The cell numbers at the beginning (day 0) and the conclusion of the experiment (day 4) were quantified with a trypan blue exclusion assay. (D) RNA analysis confirms the results obtained with NS5A-GFP expression. GS4 cells were cultured under various conditions for 3 days before the total RNAs were extracted and subjected to northern analysis as described in the legend to Fig. 1D. A single band of 9.6 kb representing the viral genome was detected. A photograph of the RNA electrophoresis gel before northern transfer shows that equivalent amounts of RNA were loaded for each sample. Lanes: 1, GS4 untreated, nonconfluent cells cultured in 10% serum; 2, 1 μM 2CMA; 3, 10 μg/ml aphidicolin; 4, 0% serum; 5, same as lane 1; 6, GS4 cells cultured under confluence; 7, GS4 cells cultured under confluence but with uridine and cytidine (200 μM each) added to the culture medium. The conditions for generating confluent cells and addition of UC are detailed in the Material and Methods.
FIG. 6.
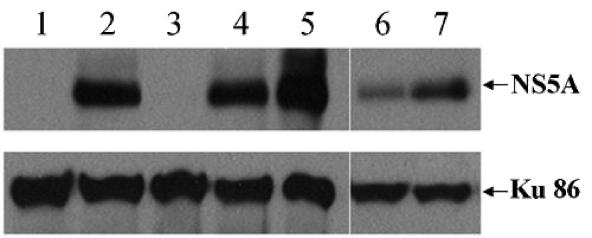
Results obtained with a replicon containing wild-type HCV sequence are consistent with those of cell culture-adapted replicons. Huh.8 cells were cultured under various conditions for 4 days before the total cell lysates were subjected to immunoblotting with anti-NS5A and anti-Ku86 antibodies. Lanes: 1, Huh-7; 2, Huh.8 untreated, nonconfluent cells cultured in 10% serum; 3, 1 μM 2CMA; 4, 10 μg/ml aphidicolin; 5, 0% serum; 6, Huh.8 cells cultured under confluence; 7, Huh.8 cells cultured under confluence but with uridine and cytidine (200 μM each) added to the culture medium.
FIG. 7.
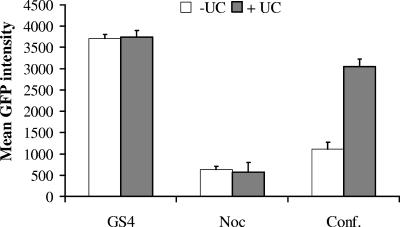
Exogenous nucleotides did not rescue HCV inhibition mediated by nocodazole. GS4 cells were treated with 25 μM nocodazole (Noc) or cultured under confluence (Conf.) conditions in the presence or absence of 200 μM UC for 3 days before being analyzed by FACS. The mean GFP intensity is shown.
Flow cytometry and cell cycle analysis.
Cells were routinely fixed in 2% paraformaldehyde before flow cytometry analysis, which was performed with a FACSCanto flow cytometer (BD Biosciences). Live cell sorting was performed under sterile conditions with a FACSAria flow cytometer (BD Biosciences). The approximately 5% of cells (2 × 107 cells total) with the strongest GFP signal were sorted back for expansion at each round of live-cell sorting. For cell cycle analysis, 1 × 106 cells were stained with 50 μg/ml propidium iodide in the presence of 200 μg/ml RNase A and 0.1% Triton X-100 for 30 min before being subjected to fluorescence-activated cell sorter (FACS) analysis.
Microscopy.
For confocal microscopy, the cells were seeded onto glass coverslips deposited in six-well plates. After treatment, cells were fixed in 4% paraformaldehyde on the coverslip, which was then mounted on a glass slide for microscopic observations. Fluorescence and differential interference contrast (DIC) images of the cells were examined with a Zeiss LSM 510 laser scanning confocal microscope equipped with multi-photon laser and recorded with the accompanying LSMIB-3.2 software.
Antibodies and Western blots.
An anti-NS5A monoclonal antibody was purchased from Virogen (Boston, MA) and an anti-Ku86 monoclonal antibody from Sigma. A monoclonal antibody against GFP was obtained from Clontech/BD Biosciences. To detect membrane-associated HCV proteins, we directly lysed the replicon cells in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis sample buffer. We used 2 × 105 cells per 160 μl of SDS-polyacrylamide gel electrophoresis sample buffer for all our Western blotting experiments. Blotting and detection were all performed according to standard procedures. For detection of the Ku86 signal, the membrane was first stripped of the NS5A signal by incubation in stripping buffer (100 mM 2-mercaptoethanol, 2% SDS, 62.5 mM Tris-HCl, pH 6.7) for 30 min at 50°C.
RNA extraction and Northern blots.
Total RNA was extracted with TRIzol reagent (Invitrogen, San Diego, CA). Equal amounts of RNA (10 μg per sample) were loaded onto formaldehyde-containing agarose gels for electrophoresis. The transfer of RNA onto a nitrocellulose membrane, random labeling of a radioactive probe, and hybridization were all performed according to standard molecular biology protocols. The probe for detecting HCV RNA is a cDNA fragment that corresponds to nucleotides 8024 to 9563 of HCV-1b. The exposure and detection of the radioactive signal was done with a Storm 860 phosphorimaging system (Amersham/GE Healthcare).
RESULTS
A novel GFP-based flow cytometry assay for HCV replication.
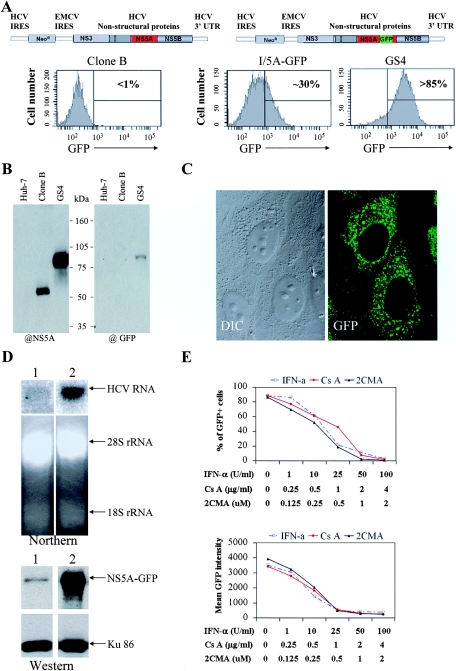
Recently, a modified HCV replicon with the enhanced GFP gene inserted into the coding region of NS5A has been generated for study of the subcellular localization of the HCV replication complex (18). In this replicon cell line (I/5A-GFP), the NS5A-GFP fusion protein exhibits properties of both the fluorescent protein and the HCV NS5A as it supports HCV replication and renders the cells harboring the hybrid replicon fluorescent under a fluorescent microscope. When we subjected I/5A-GFP cells to flow cytometry analysis, we found the GFP signal of the majority (∼70%) of the cells to be indistinguishable from that of Clone B, a replicon cell line without GFP but containing the same S2204I mutation in the NS5A gene (1) (Fig. 1A). To obtain a cell line with minimal FACS profile overlap with the GFP-negative replicon cells, we then performed consecutive rounds of cell sorting on I/5A-GFP-6 cells in which the cells with the strongest GFP were recovered. A population with no greater than 15% GFP-negative cells was obtained after four rounds of sorting and was used for the present study. We designated these cells GS4 (Fig. 1A). Immunoblotting analysis using both anti-NS5A and anti-GFP antibodies revealed only one species of NS5A-GFP hybrid protein in these cells, indicating that no green fluorescence was contributed by unfused or truncated GFP proteins (Fig. 1B). When examined under a fluorescent microscope, the green fluorescence of the GS4 cells exhibited the characteristic punctuate subcellular localization of HCV nonstructural proteins (Fig. 1C). To examine if the increased GFP expression in GS4 cells was the result of increased RNA level, we compared the HCV RNA and protein levels in GS4 and I/5A-GFP cells. As shown in Fig. 1D, higher level of NS5A-GFP expression correlated with increased level of HCV RNA in GS4 cells.
FIG. 1.
A new flow cytometry-based reporter cell line for HCV replication in vitro. (A) Schematics drawings of various replicons and their FACS profiles. Clone B cells were used as negative control for gating purposes to determine the percentage of GFP-positive cells in I/5A-GFP and GS4 samples. IRES, internal ribosom entry site; UTR, untranslated region. (B) Western blotting with anti-NS5A and anti-GFP revealed the correct fusion protein was expressed in GS4 cells. (C) Fluorescent microscopic image of GS4 cells showing the typical punctuate cytoplasmic staining of HCV nonstructural proteins. (D) GS4 cells harbor increased level of HCV replicon RNA and protein. Total RNAs were extracted from I/5A-GFP and GS4 cells and then subjected to northern analysis with a radioactively labeled DNA probe that is complementary to the 3′ end of the NS5B and the 3′ untranslated region of HCV-1b (top). A similar comparison of the NS5A-GFP expression was done (bottom). Lane 1, I/5A-GFP; lane 2, GS4. (E) Dosage-dependent inhibition of NS5A-GFP expression by known HCV inhibitors, IFN, 2CMA, and Cs. The duration of treatment was 4 days.
The GFP signal of these cells was then analyzed by flow cytometry 4 days after treatment with increasing amounts of inhibitors, including IFN-α, 2CMA, and Cs. The inhibitors effectively reduced the percentage of GFP-positive cells as well as the mean GFP intensity of the treated population. Results for both parameters showed good dosage-dependent inhibition by the inhibitors (Fig. 1E). The 50% inhibitory concentration (0.25 μM for 2CMA, 0.5 μg/ml for Cs, and 10 to 25 U/ml for IFN-α) obtained with this assay closely resembles results from previous studies where the HCV RNA level was measured (3, 7, 21, 28). Furthermore, the GFP signal of GS4 cells was inhibited by a small interfering RNA expressed as a hairpin from a lentiviral vector whose direct target is the HCV genome (data not shown), as reported previously (27, 29). Taken together, these results show that the GS4 cell line is a suitable indicator cell line for HCV replication and that the flow cytometry-based assay can be used to study replicon inhibition in vitro.
Serum starvation does not inhibit HCV expression as cell confluence does.
We applied this assay to test a number of non-drug-based treatments that had been reported to inhibit HCV replication in vitro. These included cell confluence, incubation at 39°C, and serum starvation. GS4 cells were treated for 4 days before FACS analysis. To our surprise, even though cell confluence and treatment at 39°C efficiently inhibited replicon as previously reported (8, 22, 24, 33), serum starvation had no effect even when cells were maintained in culture medium with 0% serum for 96 h, after which the replicon cells started to die off (Fig. 2A). Lack of inhibition of NS5A-GFP expression by serum starvation was also confirmed by fluorescent microscopy (Fig. 2B). To rule out the possibility that the failure of serum starvation to suppress replicon expression was a result of our multiple rounds of cell sorting and specific to this GFP-containing replicon, we repeated the experiment using a different replicon cell line that did not contain GFP and had not been subjected to cell sorting. Cell line 1B is a replicon clone derived from an HCV-N-based replicon (8, 11). We subjected these cells to serum starvation for 4 days and then detected NS5A expression using Western blot analysis. Decreasing amount of serum had no effect on the NS5A level, whereas confluence had a dramatic inhibitory effect (Fig. 2C). On the other hand, serum starvation had a significant effect on cell growth (Fig. 2D). Note that 1% or no serum had a greater effect on cell growth than did confluence and yet had no effect on NS5A expression.
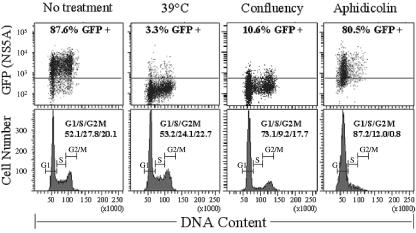
Cell cycle perturbation failed to suppress HCV replication and expression.
Failure of growth arrest to inhibit replicon expression prompted us to examine the effect of cell cycle perturbation on replicon expression. Propidium iodide staining of the cells permitted acquisition of dual fluorescence FACS profiles showing the cell cycle distribution of GFP-positive and -negative cells. We found no correlation between positive GFP signal and any of the cell cycle stages in the cycling cells (Fig. 3, no treatment). Incubation at 39°C inhibited NS5A-GFP expression without any effect on cell cycle profile. More importantly, although cell confluence inhibited HCV expression, aphidicolin failed to do so even though it effectively arrested cell cycle at the G1/S transition as expected (Fig. 3). Even though confluence also affected cell cycle to a certain degree, its effect on cell cycle progress was far less dramatic than that of aphidicolin.
FIG. 3.
The effect of aphidicolin on cell cycle and HCV expression. A total of 1 × 106 GS4 cells were cultured under the indicated conditions for 4 days before being stained with propidium iodide for DNA analysis. The simultaneous cell cycle and NS5A-GFP analysis was carried out on a FACSCanto flow cytometer with the accompanying FACSDiva software (BD Biosciences). The concentrations of aphidicolin tested ranged from 5 to 20 μg/ml, and all failed to affect NS5A-GFP expression significantly. The result with 10 μg/ml aphidicolin is shown.
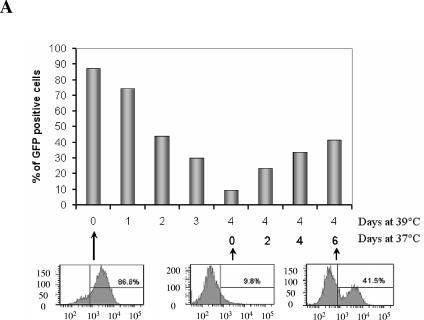
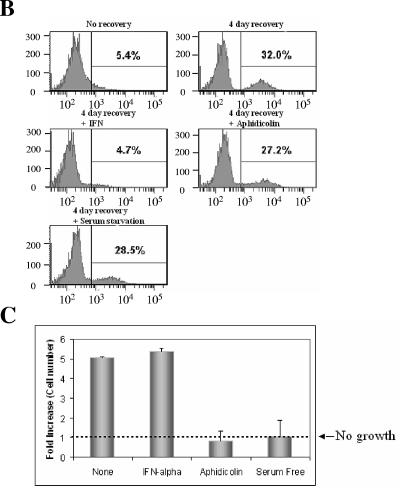
To eliminate the possibility that these growth arrest treatments somehow stabilize the GFP and maintain it throughout the treatment, we took advantage of our finding that 39°C treatment results in reversible inhibition of HCV replication in our assay. We first eliminated the GFP signal from GS4 cells by incubating them at 39°C for 4 days and then transferred the cells back to 37°C so that HCV replication could recover. As shown in Fig. 4A, over a recovery period of 6 days at 37°C, more than 40% of the cells regained the GFP signal, indicating HCV replication and viral protein expression had resumed. We tested aphidicolin and serum starvation using this assay along with IFN-α. Although IFN-α efficiently blocked the recovery of the NS5A-GFP expression in this assay, aphidicolin and serum starvation again did not prevent the resumption of HCV replication, which led to the recovery of NS5A-GFP expression (Fig. 4B). Measurement of cell number in these samples during the course of the recovery indicated that aphidicolin and serum starvation effectively blocked cell growth as expected (Fig. 4C). Taken together, these results show that neither serum starvation nor aphidicolin-induced cell cycle arrest can produce the same inhibitory effect exerted by cell confluence.
FIG. 4.
Aphidicolin and serum starvation do not prevent resumption of HCV expression. (A) The inhibition of HCV replication by 39°C treatment is reversible. The GS4 cells were cultured at 39°C for 4 days then at 37°C for another 6 days (cells were split when appropriate to prevent confluence); samples were taken for FACS everyday during the 39°C treatment period and every 2 days during the 37°C recovery period. (B) Neither aphidicolin nor serum starvation could prevent the recovery of HCV replication and expression. After 4 days in culture at 39°C, the GS4 cells were allowed to recover for 4 days at 37°C in the presence of various inhibitors or in serum-free medium. Although 100 U/ml IFN-α efficiently blocked the resumption of HCV expression, 10 μg/ml aphidicolin and serum starvation failed to do so. (C) Aphidicolin and serum starvation inhibited cell growth as expected. The same number of cells (2 × 105) were seeded for recovery for all samples; at the end of the experiment, the cell numbers in each sample were obtained with the trypan blue exclusion assay, and the fold of increase in cell numbers is shown.
Reversal of confluence-mediated replicon inhibition by exogenous nucleosides.
Inhibitors of de novo synthesis of nucleosides can suppress HCV replication (25), suggesting that lowered nucleotide level in confluent cells accounts for the reduction in viral RNA level. We tested this hypothesis by adding exogenous nucleosides to the culture media of confluent GS4 cells and then comparing the NS5A-GFP expression with that of confluent cells without added nucleosides. Different nucleosides were tested, and the combination of uridine and cytidine (UC; 50 to 200 μM each) was found to rescue HCV expression to various degrees. A nearly complete reversal of the inhibitory effect was observed when the cells were kept in the confluence state for up to 4 days in the presence of 200 μM UC, as demonstrated by both flow cytometry and fluorescent microscopy (Fig. 5A and B). The reversal is evident throughout the duration of the experiment (6 days after confluence), although a longer incubation time in the confluent state caused the percentage of GFP-positive cells to decrease even in the cells to which UC had been added. The reversal of HCV expression was not result of release from growth arrest by UC as the addition did not result in more cell growth (Fig. 5C).
To validate our results at the viral RNA level, we repeated the inhibition and rescue experiments and directly examined HCV RNA by Northern blotting. GS4 cells were treated with various inhibitors/agents for 3 days before the total RNAs were subjected to northern analysis for HCV RNA. 2CMA and cell confluence effectively eliminated HCV RNA signal (Fig. 5D, top panel, lanes 2 and 6), whereas serum starvation had no effect (lane 4). Aphidicolin also only had a minimal effect, not enough to explain the confluence-based inhibition (lane 3). Importantly, exogenously added UC partially rescued RNA replication from confluence-based inhibition (compare lane 7 with lane 6). These confirm that previous results obtained at the GFP expression level reflect HCV RNA replication.
Additional experiments with a replicon cell line containing a wild type HCV sequence.
We carried out additional experiments in a replicon cell line that does not contain the two modifications of GS4 cells: the S2204I mutation and the GFP insertion in NS5A. Huh.8 is replicon cell line that contains the wild-type sequence of NS5A (1). Huh.8 cells were cultured under various conditions for 4 days before being analyzed by Western blot for NS5A expression. As a positive control, 1 μM 2CMA effectively eliminated any NS5A expression; aphidicolin and serum starvation again did not inhibit HCV expression (Fig. 6). Serum starvation actually appeared to have increased NS5A expression, as it did with the GS4 cells (Fig. 2A and data not shown). In contrast, confluence inhibited NS5A expression, and more importantly, the exogenously added UC again counteracted this inhibition (Fig. 6, compare lane 7 with lane 6). These data indicate that the lack of HCV inhibition by serum starvation and aphidicolin and the mechanism of confluence-based viral inhibition are not results of the modifications found in GS4 cells.
Exogenous nucleotides could not rescue viral inhibition by nocodazole.
Other cytostatic agents have been observed to exert an inhibitory effect on viral replication (2, 19, 25). We tested transforming growth factor β (TGF-β; 1 to 4 ng/ml), hydroxyurea (100 to 400 μM), and nocodazole (10 to 50 μM) using the GS4 cells. Of these, only nocodazole produced a significant inhibitory effect on HCV expression comparable to that by cell confluence (Fig. 7; see also Discussion). We thus tested if exogenously added nucleotides could counteract this inhibition. Cells were treated for 75 h with 25 μM nocodazole in the absence or presence of 200 μM UC. Even though nocodazole effectively inhibited NS5A-GFP expression, UC addition did not counteract the inhibition, while it did with cell confluence (Fig. 7). This is consistent with the proposed cytoskeletal requirement for HCV replication in replicon cells (2).
DISCUSSION
We have previously used a GFP-based cell-sorting assay to identify cellular proteins involved in human immunodeficiency virus replication (27) and wished to develop a similar assay for HCV replication in vitro. Taking advantage of a modified HCV replicon with the enhanced GFP gene inserted into the coding region of NS5A (18), we obtained a derivative cell line that we named GS4 by live cell sorting and expansion. The GS4 cells we developed have a much higher level of HCV RNA and protein expression than that of the original I/5A-GFP cells and yet still respond very well to inhibitors. Even though the stronger GFP signal also makes it much easier to visualize the fluorescence under a microscope, the real strength of the assay based on the GS4 cells lies in its amenability to flow cytometry analysis. The assay is simple and yet quantitative. No cell lysis or addition of exogenous substrates is needed. It is also rapid, and the results are visual. The speed of flow cytometry analysis makes the test very fast, and many samples can be processed in parallel. It measures and displays results from individual cells, eliminating the need to normalize the results to cell numbers in different samples. This feature is especially important in situations where antiviral effects must be distinguished from those of cytostasis, for example in the case of cell confluence. Most importantly, the more favorable flow cytometry profile (minimized overlapping between the positive and the negative populations) makes it more suitable for screening projects using a reverse genetics approach, where distinct populations with desired fluorescence intensities can be recovered by cell sorting.
In the study reported here, we applied the assay to examine the effect of cell growth on HCV replication and the mechanism of cell confluence-based inhibition of HCV RNA. The most straightforward mechanism for confluence-mediated inhibition is that exponential cell growth is required for efficient HCV replication. Indeed, other means of arresting cell growth have also been shown to inhibit HCV replication to various degrees (19, 24). In one study, serum starvation reduced the RNA abundance in a replicon cell harboring a selectable full-length HCV RNA based on the genotype 1b of HCV-N, whereas aphidicolin treatment, which arrested the cell cycle as expected, had no effect on the abundance of HCV RNA or protein in these cells (24). In contrast, Murata et al. observed an inhibitory effect by aphidicolin on HCV replicon in their report of TGF-β as an inhibitor of HCV replication in vitro (19). During the course of our study, we found that serum starvation or aphidicolin failed to inhibit HCV RNA replication or protein expression without killing the replicon cells, whereas cell confluence did so very effectively. TGF-β had only a marginal effect in our system (data not shown). The reason for the difference in the sensitivity of different replicons to these different treatments is not clear. Properties of specific replicons and sensitivities of different assays were some of the possibilities. We performed these experiments in three different replicon lines and used various assays, including flow cytometry, fluorescent microscopy, Western blotting, and Northern blotting, and all gave similar results. In any event, because the replicon cells that we used here did respond as expected to cell confluence, we concluded that growth arrest was not the general mechanism of confluence-based viral inhibition. Note also that, because all these treatments significantly inhibit cell growth, the cell numbers at the time of the assay are dramatically lower in the treated group, making accurate normalization, for distinguishing antiviral from cytostatic effects, difficult. Because our flow cytometry-based assay looks at the HCV protein expression in individual cells, it does not need the normalization.
A second hypothesis, not mutually exclusive with that of growth arrest, is that the reduced intracellular pools of nucleosides account for the decrease of HCV RNA in confluent cells because these cells depend on salvage nucleoside biosynthesis pathways and thus have a lower level of intracellular NTPs (25). We reasoned that, if this were the case, adding back exogenous nucleoside should alleviate the block on HCV replication and rescue viral expression. Our results indicate that this was indeed at least partly the case, as exogenously added uridine and cytidine rescued HCV RNA replication and protein expression in confluent cells of several different replicon lines. These exogenous nucleosides probably rescue HCV replication by increasing the level of building blocks for the synthesis of viral genome RNA. The failure of the nucleosides to reverse completely the inhibitory effect posed by confluence suggests that additional mechanisms exist for confluence-based HCV inhibition.
The finding that serum starvation and cell confluence have drastically different effects on steady-state HCV replicon RNA level is somewhat surprising, as they both induce a cytostatic state of the cells that is believed to be detrimental to HCV replication. There are also other cytostatic compounds shown to inhibit HCV replication that we did not test in this study (25). Our results nevertheless suggest that serum starvation and confluence cause distinct changes in the physiological state of the cells that is relevant for HCV replication. The results with exogenously added nucleosides imply that serum starvation does not result in the reduction of intracellular NTP pools as confluence does, at least not to the extent that it limits HCV replication. It is also interesting to note that confluence does not inhibit HCV replication in HeLa cells (33), suggesting that nucleotide depletion or other unidentified changes caused by confluence could be cell type specific. Alternatively, since the replicon in HeLa cells contains distinct mutations, this difference in response to cell confluence could also be possibly explained by the viral RNA.
It also has been reported that HCV internal ribosome entry site-mediated translation is regulated by cell cycle (9). The activity of the internal ribosome entry site was shown to be highest in actively growing cells and relatively low in resting cells measured by bicistronic reporter constructs. The significance of this observation in the replicon setting is unclear since we and other all failed to detect any correlation of replicon protein level with different cell cycle stages (Fig. 3) (24).
Cell growth/cycle arrest induced by serum starvation or aphidicolin has been reported to increase replication/infection of other viruses (6, 17). The transcription of the Borna disease virus, a negative-stranded RNA virus, was enhanced by serum starvation. Interestingly, the RNA species that was up-regulated the most was the 1.9-kb RNA without the 5′ cap and 3′ poly(A) tail, similar to the HCV genomic RNA (17). The finding that serum starvation can increase HCV expression under certain culture conditions is intriguing.
The dynamics of HCV replication in response to cellular environment may be relevant in vivo. Our results clearly demonstrate that growth arrest does not automatically lead to lower nucleotide pools and inhibition of HCV replication, this may contribute to the ability of HCV to replicate efficiently in liver where most of the cells are spontaneous quiescent.
Acknowledgments
This work was supported by FSU Department of Biological Science start-up funding to H.T.
We thank the following people for providing cell lines/reagents and useful discussions: Charles Rice (Rockefeller), Steve Carroll (Merck), Chen Liu (U. Florida), Christoph Seeger (Fox Chase), and Myra Hurt (FSU). We thank the FSU College of Medicine Flow Cytometry Core and Ruth Didier for assistance with flow cytometry. We also thank Brad Sirak and Chris Green for technical assistance and Anne B. Thistle for proofreading the manuscript.
REFERENCES
- 1.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 2.Bost, A. G., D. Venable, L. Liu, and B. A. Heinz. 2003. Cytoskeletal requirements for hepatitis C virus (HCV) RNA synthesis in the HCV replicon cell culture system. J. Virol. 77:4401-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carroll, S. S., J. E. Tomassini, M. Bosserman, K. Getty, M. W. Stahlhut, A. B. Eldrup, B. Bhat, D. Hall, A. L. Simcoe, R. LaFemina, C. A. Rutkowski, B. Wolanski, Z. Yang, G. Migliaccio, R. De Francesco, L. C. Kuo, M. MacCoss, and D. B. Olsen. 2003. Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J. Biol. Chem. 278:11979-11984. [DOI] [PubMed] [Google Scholar]
- 4.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 5.Friebe, P., and R. Bartenschlager. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 76:5326-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groschel, B., and F. Bushman. 2005. Cell cycle arrest in G2/M promotes early steps of infection by human immunodeficiency virus. J. Virol. 79:5695-5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo, J. T., J. A. Sohn, Q. Zhu, and C. Seeger. 2004. Mechanism of the interferon alpha response against hepatitis C virus replicons. Virology 325:71-81. [DOI] [PubMed] [Google Scholar]
- 9.Honda, M., S. Kaneko, E. Matsushita, K. Kobayashi, G. A. Abell, and S. M. Lemon. 2000. Cell cycle regulation of hepatitis C virus internal ribosomal entry site-directed translation. Gastroenterology 118:152-162. [DOI] [PubMed] [Google Scholar]
- 10.Horscroft, N., V. C. Lai, W. Cheney, N. Yao, J. Z. Wu, Z. Hong, and W. Zhong. 2005. Replicon cell culture system as a valuable tool in antiviral drug discovery against hepatitis C virus. Antivir. Chem. Chemother. 16:1-12. [DOI] [PubMed] [Google Scholar]
- 11.Ikeda, M., M. Yi, K. Li, and S. M. Lemon. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamarre, D., P. C. Anderson, M. Bailey, P. Beaulieu, G. Bolger, P. Bonneau, M. Bos, D. R. Cameron, M. Cartier, M. G. Cordingley, A. M. Faucher, N. Goudreau, S. H. Kawai, G. Kukolj, L. Lagace, S. R. LaPlante, H. Narjes, M. A. Poupart, J. Rancourt, R. E. Sentjens, R. St. George, B. Simoneau, G. Steinmann, D. Thibeault, Y. S. Tsantrizos, S. M. Weldon, C. L. Yong, and M. Llinas-Brunet. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186-189. [DOI] [PubMed] [Google Scholar]
- 15.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 16.Lohmann, V., F. Korner, J., Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 17.Mizutani, T., H. Inagaki, D. Hayasaka, H. Kariwa, and I. Takashima. 1999. Enhancement of Borna disease virus transcription in persistently infected cells by serum starvation. J. Vet. Med. Sci. 61:831-834. [DOI] [PubMed] [Google Scholar]
- 18.Moradpour, D., M. J. Evans, R. Gosert, Z. Yuan, H. E. Blum, S. P. Goff, B. D. Lindenbach, and C. M. Rice. 2004. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J. Virol. 78:7400-7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murata, T., T. Ohshima, M. Yamaji, M. Hosaka, Y. Miyanari, M. Hijikata, and K. Shimotohno. 2005. Suppression of hepatitis C virus replicon by TGF-beta. Virology 331:407-417. [DOI] [PubMed] [Google Scholar]
- 20.Murray, E. M., J. A. Grobler, E. J. Markel, M. F. Pagnoni, G. Paonessa, A. J. Simon, and O. A. Flores. 2003. Persistent replication of hepatitis C virus replicons expressing the beta-lactamase reporter in subpopulations of highly permissive Huh7 cells. J. Virol. 77:2928-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakagawa, M., N. Sakamoto, N. Enomoto, Y. Tanabe, N. Kanazawa, T. Koyama, M. Kurosaki, S. Maekawa, T. Yamashiro, C. H. Chen, Y. Itsui, S. Kakinuma, and M. Watanabe. 2004. Specific inhibition of hepatitis C virus replication by cyclosporin A. Biochem. Biophys. Res. Commun. 313:42-47. [DOI] [PubMed] [Google Scholar]
- 22.Pietschmann, T., V. Lohmann, G. Rutter, K. Kurpanek, and R. Bartenschlager. 2001. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J. Virol. 75:1252-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Randall, G., and C. M. Rice. 2004. Interfering with hepatitis C virus RNA replication. Virus Res. 102:19-25. [DOI] [PubMed] [Google Scholar]
- 24.Scholle, F., K. Li, F. Bodola, M. Ikeda, B. A. Luxon, and S. M. Lemon. 2004. Virus-host cell interactions during hepatitis C virus RNA replication: impact of polyprotein expression on the cellular transcriptome and cell cycle association with viral RNA synthesis. J. Virol. 78:1513-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stuyver, L. J., T. R. McBrayer, P. M. Tharnish, A. E. Hassan, C. K. Chu, K. W. Pankiewicz, K. A. Watanabe, R. F. Schinazi, and M. J. Otto. 2003. Dynamics of subgenomic hepatitis C virus replicon RNA levels in Huh-7 cells after exposure to nucleoside antimetabolites. J. Virol. 77:10689-10694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Corrigendum: production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waninger, S., K. Kuhen, X. Hu, J. E. Chatterton, F. Wong-Staal, and H. Tang. 2004. Identification of cellular cofactors for human immunodeficiency virus replication via a ribozyme-based genomics approach. J. Virol. 78:12829-12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watashi, K., M. Hijikata, M. Hosaka, M. Yamaji, and K. Shimotohno. 2003. Cyclosporin A suppresses replication of hepatitis C virus genome in cultured hepatocytes. Hepatology 38:1282-1288. [DOI] [PubMed] [Google Scholar]
- 29.Wilson, J. A., S. Jayasena, A. Khvorova, S. Sabatinos, I. G. Rodrigue-Gervais, S. Arya, F. Sarangi, M. Harris-Brandts, S. Beaulieu, and C. D. Richardson. 2003. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc. Natl. Acad. Sci. USA 100:2783-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yi, M., F. Bodola, and S. M. Lemon. 2002. Subgenomic hepatitis C virus replicons inducing expression of a secreted enzymatic reporter protein. Virology 304:197-210. [DOI] [PubMed] [Google Scholar]
- 31.Yi, M., and S. M. Lemon. 2003. Structure-function analysis of the 3′ stem-loop of hepatitis C virus genomic RNA and its role in viral RNA replication. RNA 9:331-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu, Q., J. T. Guo, and C. Seeger. 2003. Replication of hepatitis C virus subgenomes in nonhepatic epithelial and mouse hepatoma cells. J. Virol. 77:9204-9210. [DOI] [PMC free article] [PubMed] [Google Scholar]