Abstract
The flavivirus nonstructural protein NS1 is a highly conserved secreted glycoprotein that does not package with the virion. Immunization with NS1 elicits a protective immune response against yellow fever, dengue, and tick-borne encephalitis flaviviruses through poorly defined mechanisms. In this study, we purified a recombinant, secreted form of West Nile virus (WNV) NS1 glycoprotein from baculovirus-infected insect cells and generated 22 new NS1-specific monoclonal antibodies (MAbs). By performing competitive binding assays and expressing truncated NS1 proteins on the surface of yeast (Saccharomyces cerevisiae) and in bacteria, we mapped 21 of the newly generated MAbs to three NS1 fragments. Prophylaxis of C57BL/6 mice with any of four MAbs (10NS1, 14NS1, 16NS1, and 17NS1) strongly protected against lethal WNV infection (75 to 95% survival, respectively) compared to saline-treated controls (17% survival). In contrast, other anti-NS1 MAbs of the same isotype provided no significant protection. Notably, 14NS1 and 16NS1 also demonstrated marked efficacy as postexposure therapy, even when administered as a single dose 4 days after infection. Virologic analysis showed that 17NS1 protects at an early stage in infection through a C1q-independent and Fc γ receptor-dependent pathway. Interestingly, 14NS1, which maps to a distinct region on NS1, protected through a C1q- and Fc γ receptor-independent mechanism. Overall, our data suggest that distinct regions of NS1 can elicit protective humoral immunity against WNV through different mechanisms.
West Nile virus (WNV) is a single-stranded, positive-sense-enveloped RNA virus that is maintained in nature through a mosquito-bird-mosquito transmission cycle. It is endemic in parts of Africa, Europe, the Middle East, and Asia, and outbreaks now occur annually in North America. Humans, which are dead-end hosts, can develop a febrile illness that progresses to a meningitis or encephalitis syndrome (32). At present, treatment is supportive, and no vaccine exists for human use.
A member of the Flaviviridae family, WNV is closely related to other major human pathogens such as yellow fever (YF), dengue (DEN), tick-borne encephalitis (TBE), Japanese encephalitis (JEV), and Murray Valley encephalitis (MVE) viruses. The 10.7-kilobase genome is translated as a single polyprotein, which is then cleaved into three structural proteins (C, prM/M, and E) and seven nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) by both virus- and host-encoded proteases (5). The NS proteins include an RNA-dependent RNA polymerase (NS5), a helicase/protease (NS3), and other proteins that form part of the viral replication complex (36, 37).
NS1 is a highly conserved 48-kDa glycoprotein with 12 invariant cysteine residues. Although the disulfide linkage arrangement of MVE and DEN NS1 has been described (4, 66), structural analysis is currently lacking. NS1 is inserted into the lumen of the endoplasmic reticulum via a signal peptide that is cleaved cotranslationally by a cellular signalase to generate the mature N terminus of the protein (5). Within infected cells, NS1 is believed to function as a cofactor in viral RNA replication. NS1 colocalizes with the double-stranded RNA replicative form (48), and viral RNA accumulation is attenuated by specific amino acid substitutions in the NS1 gene (36, 46, 51). Unlike the other nonstructural proteins, NS1 is secreted (47, 49, 68, 69), and high levels (up to 50 μg/ml) are detected in the serum of DEN-infected patients (1, 40, 74) and correlate with the development of severe disease. Additionally, NS1 becomes associated with cell surface membranes through an as yet undetermined mechanism (68, 69). The function of secreted and cell-associated NS1 in the pathogenesis of flavivirus infection remains uncertain, although it has been hypothesized to participate in immune complex formation (74), the generation of autoantibodies that react with platelets and matrix proteins (7, 20), and endothelial cell damage (20, 41-43).
Although NS1 is absent from the virion, antibodies against it can protect against infection in vivo. Immunization with purified NS1 or passive administration of some anti-YF and anti-DEN NS1 monoclonal antibodies (MAbs) protects mice against lethal virus challenge (12, 22, 27, 30, 33, 34, 56, 57, 59). However, no significant virologic analysis was performed in these studies to address the mechanism of protection. To gain insight into the mechanisms of NS1-mediated protection, we generated and purified recombinant, glycosylated, soluble WNV NS1 from insect cells and produced and characterized 22 new anti-NS1 MAbs. Using yeast (Saccharomyces cerevisiae) surface display and bacterial expression of truncated NS1 fragments, we mapped the vast majority of the MAbs to one of three regions. Prophylaxis studies demonstrated that several distinct anti-NS1 MAbs could protect against lethal WNV infection in C57BL/6 mice. Furthermore, two MAbs had marked efficiency as postexposure therapy in mice. By performing protection studies in congenic C1q- or Fc γ receptor-deficient mice, we demonstrate that distinct regions of NS1 can elicit protective humoral immunity through independent mechanisms.
MATERIALS AND METHODS
Plasmid construction.
To generate a recombinant baculovirus for WNV NS1 expression in serum-free-adapted SF9 cells, an initiator methionine, the last 72 nucleotides of WNV E (endogenous signal sequence), and the entire NS1 gene from the New York 1999 strain were amplified from an infectious cDNA clone (gift of R. Kinney, Fort Collins, CO) by high-fidelity PCR. A six-histidine tag was added at the C terminus immediately upstream of a stop codon. The following oligonucleotides were used as forward- and reverse-strand DNA primers, with the added restriction enzyme sites underscored: forward oligonucleotide, 5′-ACAGGGATCCACCATGGATAGGTCCATAGCTCTCACGTTT-3′; reverse oligonucleotide, 5′-CTAGTCTAGATCAATGATGATGATGATGATGAGCATTCACTTGTGACTGCACG-3′. The PCR product was digested and inserted into the BamHI and XbaI sites of a baculovirus shuttle vector (pFastBac1; Invitrogen, Carlsbad, CA) and sequenced.
For expression of NS1 on the surface of yeast, plasmids containing different portions of the WNV NS1 gene were constructed by inserting DNA encoding-intact, N- or C-terminal fragments of NS1 in frame downstream of the Aga2 protein in the BamHI-NotI site of the pYD1 plasmid (Invitrogen). For pYD-NS1 (amino acids [aa] 1 to 352), the following primers were used for amplification from the infectious cDNA clone, with the added restriction enzyme sites underscored: forward, 5′-ACAGGGATCCGAGGAGGAGGACACTGGGTGTGCCATAGACATCAGC-3′; reverse, 5′-CATTGACTGCGGCCGCTAAGCATTCACTTGTGACTGCAC-3′. For pYD-NS1 (amino acids 1 to 157): forward (NS1-1F), 5′-CGCGGATCCGGTGGTGGTGACACTGGGTGTGCCATAGACACT-3′; reverse, 5′-CATTGACTGCGGCCGCTAATCCTCCACTTCTAAGCTATTC-3′. For pYD-NS1 (amino acids 158 to 352) and pYD-NS1 (amino acids 236 to 352): forward (NS1-158F), 5′-CGCGGATCCGGTGGTGGTTTTGGATTTGGTCTCACCAGCAC-3′; forward (NS1-236F), 5′-CGCGGATCCGGTGGTGGTATCCTTGAGAGTGACTTGATAATAC-3′; and common reverse, 5′-CATTGACTGCGGCCGCTAAGCATTCACTTGTGACTGCAC-3′.
To express NS1 in Escherichia coli, PCR fragments with the N or C terminus of NS1 were inserted in the NdeI-XhoI or BamHI-XhoI sites of a modified pET21 vector (Novagen, San Diego, CA). For pET-NS1 (amino acids 1 to 157), NS1-1F 5′-ACAGGGATCCGACACTGGGTGTGCCATAGAC-3′ and 5′-GGACCGCTCGAGTCAATCCTCCACTTCTAAGCTATTC-3′ were used as forward- and reverse-strand primers, respectively. The PCR product was inserted into the BamHI-XhoI site of the modified pET21. For pET-NS1 (amino acids 125 to 352) and pET-NS1 (amino acids 236 to 352), the following primers were used: forward (NS1-125F), 5′-ATGGAATTCCATATGGCACCAGAACTCGCCAACAAC-3′; forward (NS1-236F), 5′-ATGGAATTCCATATGATCCTTGAGAGTGACTTGATA-3′; and common reverse, 5′-GGACCGCTCGAGTCAAGCATTCACTTGTGACTGCAC-3′. The PCR products were inserted into the NdeI-XhoI site of pET21 and sequenced.
Cells and viruses.
BHK21-15 and C6/36 Aedes albopictus cells were cultured as previously described (13). The majority of experiments were performed with the WNV strain (3000.0259 isolate, passage 2) that was isolated in New York in 2000 (18). Some experiments were also performed with a lineage II WNV strain (67, 72) or DEN virus-2 (16681) strain. Viruses were diluted and injected into mice as described previously (15). SF9 cells that were adapted to serum-free conditions were obtained commercially and grown according to the manufacturer's instructions in SF-900 II SFM medium (Invitrogen, Carlsbad, CA).
Expression and purification of WNV NS1 from insect cells and E. coli.
For expression of WNV NS1 from insect cells, soluble NS1 protein was generated using a commercially available baculovirus expression system (Bac-to-Bac system; Invitrogen). Three days after the baculovirus infection of serum-free-adapted SF9 insect cells at a multiplicity of infection of 0.25, supernatants were harvested, filtered, buffer exchanged into 10% glycerol, 300 mM NaCl, 25 mM Tris-HCl (pH 7.5), and then concentrated and purified by nickel affinity chromatography according to the manufacturer's instructions (Invitrogen). Subsequently, NS1 was purified by size exclusion chromatography on a Hi-load 16/60 Superdex 200 column (Amersham Biosciences, Piscataway, NJ) in 20 mM HEPES (pH 7.5), 150 mM NaCl, and 0.01% NaN3. The elution peak was then collected, buffer exchanged into 20 mM HEPES (pH 7.5) and 0.01% NaN3, and further purified by Mono Q fast protein liquid chromatography (Amersham Biosciences, NJ). WNV NS1 was eluted to homogeneity using a 0 to 500 mM NaCl gradient over 30 column volumes in 20 mM HEPES (pH 7.5) and 0.01% (wt/vol) NaN3.
For expression of WNV NS1 fragments in E. coli, plasmids were transformed into BL21 Codon Plus E. coli cells (Stratagene). Bacteria were grown in LB, induced with 0.5 to 1.0 mM isopropyl thiogalactoside, pelleted, lysed after the addition of lysozyme, and sonicated, and NS1 fragments were recovered as insoluble aggregate from the inclusion bodies. Fragments were solubilized in the presence of 6 M guanidine hydrochloride and 20 mM β-mercaptoethanol and refolded by rapidly diluting out the solubilizing reagents in the presence of 0.4 M l-arginine, 2 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride, 5 mM reduced glutathione, and 0.5 mM oxidized glutathione. Refolded NS1 fragments were separated from aggregates on Superdex 75 or 200 16/60 size exclusion columns (Amersham Biosciences) and concentrated using a Centricon 10 spin column in 20 mM HEPES (pH 7.4).
Glycosidase treatment.
The deglycosylation experiments were performed using N-glycosidase F or PNGase F (New England BioLabs, Beverley, MA). Purified WNV NS1 protein (1.6 μg at a concentration of 100 μg/ml) was heated to 95°C for 10 minutes in the presence of 0.5% sodium dodecyl sulfate (SDS) and 1% β-mercaptoethanol. Immediately prior to adding 15,000 units of PNGase F (500 units/μl), 4 μl of reaction buffer (0.5 M sodium phosphate [pH 7.5], 1% NP-40) was added. After a 2-hour incubation at 37°C, samples were subjected to reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and detected by Coomassie blue staining.
MAb generation.
BALB/c mice were primed and boosted at 3-week intervals with insect cell-generated, purified, recombinant WNV NS1 protein that was complexed with adjuvant (RIBI Immunochemical, Hamilton, MT). Approximately 1 month after the last boost, serum was harvested and tested for immunoreactivity against a lysate prepared from BHK21 cells infected with WNV. Mice with the highest titers (∼1/100,000) were boosted intravenously with purified NS1 protein (5 μg). To generate hybridomas, splenocytes were harvested 3 days after the intravenous boost and fused to P3X63Ag8.653 myeloma cells using polyethylene glycol according to published procedures (29, 52).
Characterization of MAbs.
MAbs were purified from hybridoma culture supernatants or mouse ascitic fluid by protein A or G chromatography according to the manufacturer's instructions (Pharmacia, Piscataway, NJ). An enzyme-linked immunosorbent assay (ELISA) was performed to determine the immunoglobulin G (IgG) subclass. Plates were coated with a polyclonal rabbit antibody against individual mouse IgG isotypes, and bound mouse IgG was detected with horseradish peroxidase (HRP)-labeled goat anti-mouse IgG (Zymed, California).
Competition binding studies.
Purified MAbs were conjugated to biotin-XX-succinimidyl ester according to the manufacturer's instructions (Molecular Probes, Oregon). Purified WNV NS1 protein was adsorbed onto individual wells of a 96-well polystyrene microtiter plate (Nalge Nunc International, New York). Unlabeled MAbs were added to individual wells in duplicate and incubated for 1 hour at room temperature. Without removing the unlabeled antibodies from the wells, biotinylated MAbs were added and incubated for an additional hour at room temperature. The plates were washed extensively, and bound biotinylated antibody was recognized by streptavidin-conjugated HRP (Invitrogen). Enzyme activity was detected using the substrate 3,3′,5, 5′-tetramethyl-benzidine (DakoCytomation, Carpinteria, CA), and 1 M H2SO4 was added to stop the enzyme reaction. Inhibition was defined as absorbance values at 450 nm that were <50% of those obtained with negative control antibody (anti-DEN-3).
ELISA.
WNV NS1 proteins that were generated from E. coli were adsorbed overnight at 4°C to Maxi-Sorp microtiter plates (Nalge Nunc International, Rochester, NY). Nonspecific binding was blocked after incubation with blocking buffer (phosphate-buffered saline [PBS], 0.05% Tween 20, 1% bovine serum albumin, and 3% horse serum) for 1 h at 37°C. Plates were then incubated with MAbs against WNV NS1 (∼50 μg/ml) for 1 h at 4°C. After extensive washing, plates were incubated serially with biotin-conjugated goat anti-mouse IgG (Southern Biotech, Birmingham, AL) and horseradish peroxidase-conjugated streptavidin (Sigma Chemical) at 4°C and developed after addition of 3,3′,5,5′-tetramethyl-benzidine substrate as described above.
Western blotting.
Protein samples were boiled in reducing SDS sample buffer and separated by 10% bis-Tris-glycine polyacrylamide gel electrophoresis (Invitrogen). Following transfer, membranes were blocked with 5% (wt/vol) nonfat dry milk in TBST (20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.5% Tween 20), washed in TBST, and incubated in a 1/1,000 dilution of cross-reactive anti-DEN NS1 MAb (7E11) or in a 1/4,000 dilution of anti-WNV NS1 polyclonal antibody in TBST. After washing, membranes were subsequently incubated with a 1/3,000 dilution of secondary antibody conjugated with HRP. Signal was detected chemiluminescently with ECL Western blotting reagent (Amersham Biosciences, Piscataway, NJ).
Flow cytometry.
After WNV or DEN-2 infection of BHK21 cells, intracellular NS1 levels were measured by flow cytometry of permeabilized cells as described previously (13). Two or three days after infection at a multiplicity of infection of 0.1, infected BHK cells were washed three times in PBS, fixed in PBS with 4% paraformaldehyde for 10 min at room temperature, washed twice in PBS, and permeabilized in Hanks ' balanced salt solution (Sigma Chemical Co., Missouri) containing 10 mM HEPES (pH 7.3), 0.1% saponin (Aldrich Chemical, Missouri), and 0.02% NaN3 (HHSN). For indirect immunofluorescence experiments, cells were resuspended in HHSN and MAb, incubated for 1 h at 4°C, washed three times in HHSN (4°C), resuspended in a 1/250 dilution of fluorescein isothiocyanate-labeled goat anti-mouse IgG (50 μl), and incubated for 1 h on ice in the dark. Cells were subsequently washed three times in HHSN (4°C), fixed in 1% paraformaldehyde in PBS, and stored in the dark prior to analysis on a Becton Dickinson FACSCalibur flow cytometer.
Mouse experiments.
All wild-type C57BL/6 mice were purchased from a commercial source (Jackson Laboratories, Bar Harbor, Maine). Congenic C1q-deficient mice were obtained from M. Botto and G. Stahl (Imperial College, London, United Kingdom, and Beth Israel Deaconess Medical Center, Boston, MA), and congenic Fc γ receptor I- and III-deficient mice were purchased commercially (Taconic, Germantown, NY). Age-matched mice were inoculated subcutaneously with 102 PFU of WNV by footpad injection after anesthetization with xylazine and ketamine. Mouse experiments were approved and performed according to the guidelines of the Washington University School of Medicine Animal Safety Committee.
For antibody transfer experiments, mice were administered a single dose of purified MAb or mouse ascites by intraperitoneal injection at a given time point before or after infection.
Quantitation of viral burden in mice.
For analysis of virus in tissues of infected mice, organs were recovered after cardiac perfusion with PBS, cooled on ice, weighed, homogenized using a Bead-Beater apparatus, and titrated for virus by a plaque assay on BHK21-15 cells as described previously (15). Serum samples were obtained from whole blood by phlebotomy of the axillary vein immediately prior to sacrifice. Viral RNA was harvested from thawed aliquots (50 μl) of sera by using a QIAamp viral RNA mini kit (QIAGEN, Palo Alto, CA). Viral RNA was quantitated by real-time fluorogenic reverse transcriptase PCR (RT-PCR) using an ABI 7000 sequence detection system (Applied Biosystems, Foster City, CA) according to a previously published protocol (38).
Yeast cell culture and transformation.
Yeast cells were grown on yeast extract-peptone-dextrose medium (1% yeast extract, 2% peptone, 2% dextrose, and 1.5% agar [for plate]) or on synthetic minimal medium (0.67% yeast nitrogen base, the appropriate supplements, and 1.5% agar [for plates]) containing 2% dextrose (SD) or 2% galactose (SGAL). Yeast cells were transformed with appropriate plasmids according to the manufacturer's instructions (Invitrogen), and the transformants were selected on the appropriate synthetic minimal medium.
Expression of WNV NS1 protein on yeast.
The intact WNV NS1 or NS1 fragments were fused in frame with the yeast Aga2 protein and expressed on the surface of yeast using a modification of a previously described protocol (52). Briefly, individual NS1 plasmids were introduced into the S. cerevisiae yeast strain EBY100 and cultivated on tryptophan-deficient SD plates at 30°C. Subsequently, individual yeast colonies from each transformant were grown to log phase in tryptophan-deficient media at 30°C, harvested in log phase, and then cultivated in tryptophan-deficient SGAL media at 20°C for 2 days to induce NS1 protein expression on the yeast surface. Yeast cells were harvested, washed with PBS supplemented with 1 mg/ml bovine serum albumin, and stained with individual MAbs against WNV NS1 protein. After 30 min, yeast cells were washed three times and incubated with a goat anti-mouse secondary antibody conjugated to Alexa Fluor 647 (Molecular Probes, Oregon). Subsequently, the yeast cells were analyzed for surface antigen expression by flow cytometry.
Statistical analysis.
All data were analyzed with Prism software (GraphPad Software, Inc., San Diego, CA). For survival analysis, Kaplan-Meier survival curves were analyzed by the log rank and Mantel-Haenszel tests. For viral burden experiments, statistical significance was determined using the nonparametric Mann-Whitney test.
RESULTS
Studies by other groups have shown that passive transfer of MAbs against YF and DEN NS1 prior to infection can protect mice (27, 30, 57, 59). To determine whether anti-NS1 MAbs had therapeutic potential against WNV and to gain insight into the mechanisms of NS1-mediated protection, we produced a soluble recombinant WNV NS1 for immunization to create a large panel of novel anti-WNV NS1 MAbs.
Purification of WNV NS1 and MAb generation.
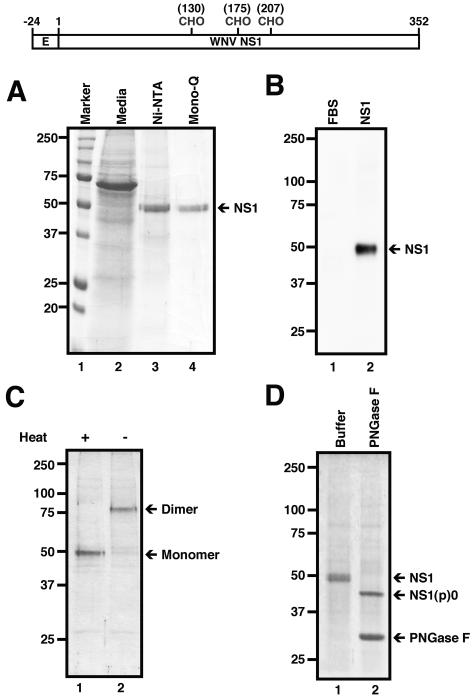
To generate recombinant WNV NS1, we fused the last 72 nucleotides of E (endogenous signal sequence) and the complete NS1 gene from the New York 1999 WNV strain immediately upstream of a histidine repeat in a baculovirus shuttle vector. Soluble NS1 was harvested from baculovirus-infected SF9 insect cell supernatants and purified to homogeneity after sequential nickel affinity, size exclusion, and Mono Q ion-exchange chromatography. A single band (∼48 kDa) was detected after SDS-PAGE and Coomassie staining (Fig. 1A). The identity of this protein was confirmed as NS1 by Western blotting with a cross-reactive anti-DEN NS1 MAb (Fig. 1B). NS1 elutes from size exclusion chromatography as an oligomer with an apparent mass of ∼300 kDa (data not shown); these results agree with prior studies in which DEN NS1 appeared to form a hexamer in solution (25). Under conditions of SDS-PAGE, WNV NS1 migrated as a dimer, with an apparent molecular mass of ∼80 kDa; boiling of samples prior to electrophoresis converted NS1 from a dimer to a monomer (Fig. 1C), as has been observed for DEN, JEV, and TBE NS1 (10, 23, 25, 53). Finally, insect cell-soluble WNV NS1 was glycosylated, as treatment with PNGase F reduced the apparent molecular weight of recombinant NS1 to ∼40 kDa (Fig. 1D).
FIG. 1.
Biochemical characterization of WNV NS1 protein. (Top) Diagram of construct generated for WNV NS1 protein from insect cells. The construct contains the signal sequence from the last 24 C-terminal amino acids of the WNV E protein and the full-length WNV NS1. Numbers and “CHO” indicate the amino acid position of the three N-glycosylated sites. CHO, N-linked carbohydrate. (A) Purification of WNV NS1. Three days after baculovirus infection of SF9 insect cells, supernatants were harvested (lane 2), and NS1 was purified to homogeneity after sequential nickel affinity (lane 3) and size exclusion and Mono Q (lane 4) chromatography. Protein size markers are indicated with respective Mr (lane 1). Samples were separated by 12% SDS-PAGE and stained with Coomassie blue. Ni-NTA, nickel-nitrilotriacetic acid. (B) Western blot analysis. Fetal bovine serum (FBS) (lane 1) and nickel affinity-purified NS1 protein (lane 2) were subjected to Western blot analysis using the cross-reactive anti-NS1 MAb (7E11). Samples were separated by 10% SDS-PAGE. (C) Dimerization of WNV NS1. The protein was incubated at 95°C (lane 1) or room temperature (lane 2) for 3 minutes prior to electrophoresis. Monomeric and dimeric NS1 are indicated by arrows. (D) Analysis of glycosylated WNV NS1. Purified NS1 was mock treated (Buffer; lane 1) or treated for 2 h at 37°C with PNGase F (lane 2). NS1(p)0 denotes the deglycosylated form of NS1. The positions of marker proteins are indicated to the left of the gel.
To generate MAbs against WNV NS1 protein, BALB/c mice were immunized three times with purified WNV NS1 and splenocytes were fused to a nonsecreting myeloma cell line. After screening more than 2,000 hybridomas, 22 new MAbs that recognized soluble and intracellular WNV NS1 protein were generated (Table 1). To define the specificity of the MAbs, each antibody was tested by flow cytometry for immunoreactivity with BHK21-15 cells infected with a lineage I (New York 1999) or II (956) WNV strain (Table 1). In the two WNV strains that we examined, there was 97% similarity and 92% identity between lineage I and II NS1 proteins. Although all MAbs recognized NS1 from the lineage I strain, four (1NS1, 6NS1, 18NS1, and 19NS1) failed to recognize NS1 from the lineage II strain. Nonetheless, some MAbs (2NS1, 3NS1, 7NS1, 14NS1, 15NS1, 17NS1, and 23NS1) showed a decreased avidity for the lineage II NS1 as judged by a reduced mean fluorescence intensity at a given antibody concentration (data not shown). Interestingly, although alignment of NS1 protein sequences between lineage I WNV and DEN-2 NS1 revealed 74% similarity and 55% identity, only one MAb from our panel, 9NS1, cross-reacted strongly with DEN-2 NS1 (Table 1).
TABLE 1.
Monoclonal antibodies against WNV NS1a
| Antibody | Isotype | Lineage I WNV | Lineage II WNV | DEN virus-2 |
|---|---|---|---|---|
| 1NS1 | IgG1 | + | − | − |
| 2NS1 | IgG1 | + | + | − |
| 3NS1 | IgG2b | + | + | − |
| 4NS1 | IgG2b | + | + | − |
| 5NS1 | IgG1 | + | + | − |
| 6NS1 | IgG1 | + | − | − |
| 7NS1 | IgG1 | + | + | − |
| 8NS1 | IgG2a | + | + | − |
| 9NS1 | IgG1 | + | + | + |
| 10NS1 | IgG2a | + | + | − |
| 11NS1 | IgG2b | + | + | − |
| 12NS1 | IgG2a | + | + | − |
| 13NS1 | IgG1 | + | + | − |
| 14NS1 | IgG2a | + | + | − |
| 15NS1 | IgG2a | + | + | − |
| 16NS1 | IgG2a | + | + | − |
| 17NS1 | IgG2a | + | + | − |
| 18NS1 | IgG2b | + | − | − |
| 19NS1 | IgG1 | + | − | − |
| 21NS1 | IgG1 | + | + | − |
| 22NS1 | IgG2a | + | + | − |
| 23NS1 | IgG1 | + | + | − |
Isotype was determined by heavy-chain-specific ELISA. Immunoreactivity of each MAb was defined by flow cytometry with BHK21-15 cells infected with a lineage I WNV (New York strain 3000.0259), lineage II WNV (956), or dengue virus-2 (16681). +, immunoreactivity (>10% of positive cells); −, no immunoreactivity.
Mapping of anti-NS1 MAbs.
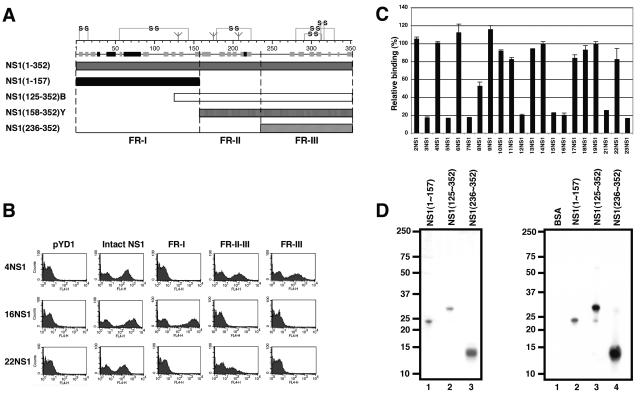
Previous studies that have localized binding regions for individual MAbs against flavivirus NS1 had done so using overlapping linear peptides (21) or MAb competition binding assays (28, 31, 73). These strategies are limited, as they require linear epitopes or define only the relative spatial arrangements of MAb binding sites. To determine where our newly generated MAbs mapped on WNV NS1, we applied a yeast surface display strategy (8, 70) that we recently used to map anti-WNV E MAbs (52). In that study, yeast cells that displayed either domains I and II or domain III of E protein were generated to facilitate rapid domain assignment. As individual functional or structural domains of NS1 have not yet been identified, we initially set out to map the individual anti-NS1 MAbs using error-prone PCR mutagenesis of the NS1 gene and expression on yeast. During the initial screening, we consistently observed the introduction of clusters of frameshift or nonsense mutations at residues 147 to 162 and 200 to 240 that resulted in truncated variants of NS1 that nonetheless were expressed on the yeast surface and reacted with subsets of anti-NS1 MAbs (data not shown). When we analyzed these sites in the context of secondary structure predictions (11) and putative cysteine disulfide bonding patterns (4, 66), three NS1 fragments emerged (Fig. 2A): FR-I, amino acids 1 to 157; FR-II, amino acids 158 to 235; and FR-III, amino acids 236 to 352. Truncated forms of NS1 corresponding to these regions were expressed on the surface of yeast and tested for MAb reactivity (Fig. 2B). Importantly, negative control MAbs against WNV or DEN E proteins did not recognize any version of WNV NS1 on yeast. In contrast, 20 of 22 MAbs against WNV NS1 recognized yeast that displayed the entire NS1 protein (Table 2). Fifty-five percent (11 of 20) MAbs recognized yeast that displayed FR-I of NS1 alone, 40% (8 of 20) detected FR-III of NS1 alone, and 5% (1 of 20, 22NS1) reacted with FR-II/-III of NS1 but not FR-III alone. Interestingly, two MAbs (10NS1 and 17NS1) appeared to have recognition sites in more than one region. For example, at low MAb concentrations (0.4 μg/ml), 10NS1 and 17NS1 recognized a single fragment alone, whereas at high MAb concentrations (20 μg/ml), each MAb recognized an additional fragment (Table 2). Our mapping results also were supported by MAb competition studies (Fig. 2C). 3NS1, a MAb that reacted with FR-I by yeast display, partially competed binding of 8NS1 and substantially reduced binding of 5NS1, 7NS1, 12NS1, 15NS1, 16NS1, 21NS1, and 23NS1. In contrast, 3NS1 did not block binding of any of the anti-NS1 MAbs that reacted with FR-II/-III or FR-III alone. Interestingly, 3NS1 did not compete with three MAbs (10NS1, 11NS1, and 17NS1) that recognized FR-I. Although more detailed mapping experiments are required, it is possible that these MAbs bind FR-I in spatially distinct regions.
FIG. 2.
Assays for epitope mapping of NS1 MAbs. (A) Schematic diagram of NS1 fragments. (Top) A secondary structure prediction model, putative disulfide bonding patterns, and N-linked glycosylated sites are depicted. The secondary structure model was generated by JPRED (11) and is a consensus prediction based on NS1 from several different flaviviruses. The disulfide bonding was modeled after previously published experiments (4, 66). (Bottom) Diagram of constructs generated for yeast surface display and bacterial expression of NS1 fragments. NS1(125-352)B and NS1(158-352)Y indicate E. coli- and yeast-expressed FR-II/-III, respectively. (B) Flow cytometry profiles for immunoreactivity against NS1 fragments on the yeast surface by three distinct MAbs (4NS1, 16NS1, and 22NS1). pYD1 represents the vector alone, which expresses the Aga2 protein in the absence of NS1. One representative histogram of several is shown. (C) Competition binding studies with 3NS1. Competition of binding between biotinylated 3NS1 and individual anti-NS1 MAbs for purified NS1 by ELISA. Competition for MAb binding was considered positive if absorbance values at 450 nm were <50% of those obtained with a nonbinding control antibody (anti-E protein of DEN virus-3). (D) Purification of bacterially expressed fragments of WNV NS1 protein. (Left) The fragments were expressed in E. coli, refolded, and purified by gel filtration chromatography. After purification, samples were separated by gel electrophoresis and stained with Coomassie blue. Lane 1, E. coli FR-I, amino acids 1 to 157; lane 2, E. coli FR-II/-III, amino acids 125 to 352; lane 3, E. coli FR-III, amino acids 236 to 352. (Right) Western blot analysis. Bovine serum albumin (lane 1), E. coli FR-I (lane 2), E. coli FR-II/-III (lane 3), and E. coli FR-III (lane 4) were subjected to Western blot analysis using the polyclonal anti-WNV NS1 polyclonal antibody. The migration of marker proteins is indicated to the left of the gel. BSA, bovine serum albumin.
TABLE 2.
Epitope mapping of anti-NS1 MAbs
| Antibody | Binding activity
|
|||||
|---|---|---|---|---|---|---|
| Yeasta
|
E. colib
|
|||||
| Full-length NS1 | FR-I | FR-II/-III | FR-III | FR-II/-III | FR-III | |
| 1NS1 | − | − | − | − | − | − |
| 2NS1 | − | − | − | − | ++ | ++ |
| 3NS1 | ++ | ++ | − | − | − | − |
| 4NS1 | ++ | − | ++ | ++ | ++ | ++ |
| 5NS1 | ++ | ++ | − | − | − | − |
| 6NS1 | ++ | − | + | + | ++ | ++ |
| 7NS1 | ++ | ++ | − | − | − | − |
| 8NS1 | ++ | ++ | − | − | ++ | − |
| 9NS1 | ++ | − | ++ | + | ++ | ++ |
| 10NS1 | ++ | ++ | ++ | − | ++ | − |
| 11NS1 | ++ | ++ | − | − | − | − |
| 12NS1 | ++ | ++ | − | − | − | − |
| 13NS1 | ++ | − | ++ | ++ | ++ | ++ |
| 14NS1 | ++ | − | ++ | + | ++ | ++ |
| 15NS1 | ++ | ++ | − | − | − | − |
| 16NS1 | ++ | ++ | − | − | − | − |
| 17NS1 | ++ | + | ++ | ++ | ++ | ++ |
| 18NS1 | + | − | + | + | ++ | ++ |
| 19NS1 | ++ | − | + | + | ++ | ++ |
| 21NS1 | ++ | − | − | − | − | − |
| 22NS1 | ++ | − | ++ | − | ++ | − |
| 23NS1 | ++ | ++ | − | − | − | − |
Yeast fragment binding was determined by flow cytometric analysis of binding to yeast expressing intact NS1 (NS1 [aa 1 to 352]), yeast FR-I (NS1 [aa 1 to 157]), yeast FR-II/-III (NS1 [aa 158 to 352]) or yeast FR-III (NS1 [aa 236 to 352]). Binding activity was scored as follows: −, <10%; +, 10 to 20%; ++, >20% positive compared to the negative control yeast transformed with the pYD1 vector.
Binding to E. coli-expressed NS1 fragments was determined by ELISA using E. coli FR-II/-III (NS1 [aa 125 to 352]) and E. coli FR-III (NS1 [aa 236 to 352]). Binding activity was scored as follows: −, <0.09; ++, >0.5 optical density at 450 nm.
To further confirm our mapping results, we expressed, refolded, and purified three NS1 fragments from bacteria (Fig. 2D, left): E. coli FR-I, amino acids 1 to 157; E. coli FR-II/-III, amino acids 125 to 352; and E. coli FR-III, amino acids 236 to 352. After purification of each fragment, a single band of the expected Mr (E. coli FR-I, ∼24 kDa; E. coli FR-II/-III, ∼26 kDa; and E. coli FR-III, ∼13 kDa) was detected after SDS-PAGE and Coomassie staining. The identity of these proteins was confirmed by Western blotting with an anti-WNV NS1 polyclonal antibody (Fig. 2D, right). Mapping studies with the bacterially refolded FR-III of NS1 corroborated the yeast display data, as the same eight MAbs localized to this region. In addition, one additional MAb that was not mapped by yeast display, 2NS1, recognized E. coli-derived FR-III strongly by ELISA (Table 2). Mapping studies with bacterial FR-II/-III also showed a reactivity pattern similar to the yeast display results. However, NS1 FR-I generated in E. coli recognized only a subset of the MAbs (8NS1 and 16NS1) that bound to FR-I on the yeast surface, suggesting that it did not adopt a native conformation when adsorbed to ELISA plates (data not shown). Overall, using yeast display, competitive binding, and bacterial expression, we mapped 21 of 22 MAbs to one of three fragments of NS1.
In vivo protection of NS1 MAbs against WNV infection.
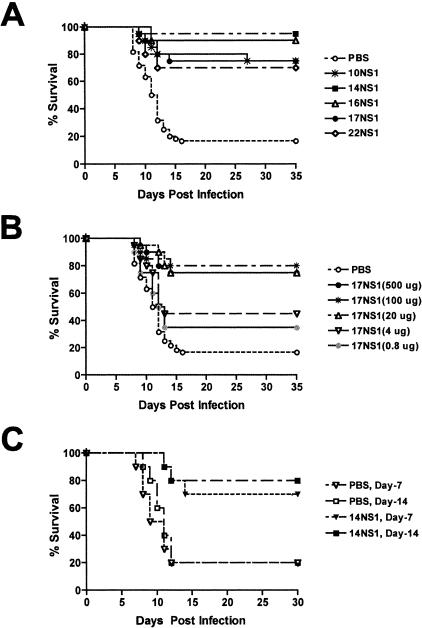
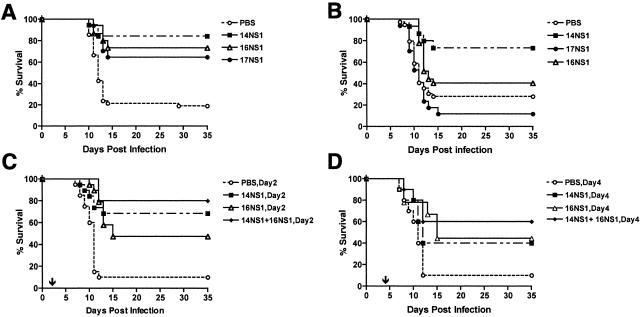
To determine whether anti-NS1 MAbs provided protection against lethal WNV infection in vivo, we evaluated their inhibitory activity in our well-characterized mouse model (15, 16, 19, 52, 60). Prophylaxis studies were performed in 6-week-old C57BL/6 mice which have a baseline survival rate of ∼15 to 20% (Table 3 ); mice were inoculated subcutaneously with 102 PFU of WNV and administered a single dose of purified MAb (500 μg) or ascites fluid (200 μl) at the same time through an intraperitoneal route. Notably, a single dose of 14NS1 protected 95% of mice from lethal infection (n = 40, P < 0.0001). Similarly, a single dose of 10NS1, 16NS1, 17NS1, or 22NS1 improved the survival rate to 70 to 90% (Table 3 and Fig. 3A). As expected, the inhibitory effect of anti-NS1 MAbs was dose dependent, as smaller amounts provided less protection; administration of 4 μg of 17NS1 resulted in a 45% survival rate (Fig. 3B, P = 0.02). The protective effect was not dependent on isotype or antibody preparation (purified versus ascites); passive transfer of 7NS1, 12NS1, 15NS1, 18NS1, and 23NS1 provided no significant benefit in vivo against WNV infection (Table 3), and administration of 10NS1 and 14NS1 as either purified MAb or ascites fluid provided equivalent protection (data not shown). As another measure of the prophylactic effect, we performed duration of protection experiments with 14NS1. Four-week-old mice were administered a single 1-mg dose of 14NS1 or PBS and then infected with WNV 7, 14, or 21 days later (Fig. 3C). The half-life of mouse MAbs has been shown by others to range from 4 to 8 days, depending on the isotype (55, 64). Notably, a single dose of 14NS1 administered either 7 or 14 days before infection provided significant protection (70 to 80% survival, P < 0.007). As expected, a greater interval between prophylaxis and infection was associated with decreased benefit, as no survival advantage was observed if 14NS1 was given 21 days prior to WNV infection (data not shown).
TABLE 3.
Protective activity of anti-NS1 MAbsa
| Antibody or control | % Surviving mice | No. of mice | P value | Antibody source |
|---|---|---|---|---|
| PBS | 17 | 60 | ||
| 1NS1 | 25 | 20 | 0.29 | PUR |
| 2NS1 | 40 | 10 | 0.09 | ASC |
| 3NS1 | 50 | 10 | 0.02* | PUR |
| 4NS1 | 50 | 10 | 0.02* | PUR |
| 5NS1 | 40 | 10 | 0.10 | ASC |
| 6NS1 | 20 | 10 | 0.93 | ASC |
| 7NS1 | 0 | 10 | 0.66 | ASC |
| 8NS1 | 20 | 10 | 0.50 | ASC |
| 9NS1 | 40 | 10 | 0.15 | ASC |
| 10NS1 | 75 | 20 | 0.0001* | ASC/PUR |
| 11NS1 | 50 | 10 | 0.04* | ASC |
| 12NS1 | 10 | 10 | 0.95 | ASC |
| 13NS1 | 30 | 10 | 0.30 | ASC |
| 14NS1 | 95 | 40 | <0.0001* | ASC/PUR |
| 15NS1 | 0 | 10 | 0.74 | ASC |
| 16NS1 | 90 | 10 | 0.0001* | ASC |
| 17NS1 | 75 | 20 | <0.0001* | PUR |
| 18NS1 | 10 | 10 | 0.14 | ASC |
| 19NS1 | 20 | 10 | 0.36 | ASC |
| 21NS1 | 40 | 10 | 0.27 | ASC |
| 22NS1 | 70 | 10 | 0.004* | ASC |
| 23NS1 | 0 | 10 | 0.21 | ASC |
For in vivo protection studies with MAbs, 6-week-old C57BL/6 mice were inoculated with 102 PFU of WNV via the footpad and administered a single dose of purified MAbs (500 μg) or ascitic fluid (200 μl) at the same time through an intraperitoneal route. The mice were followed for 35 days. The data were obtained and P values were determined after comparison to the PBS-treated control group. Asterisks indicate statistical significance by the log rank test. ASC, ascites fluid; PUR, purified antibody; ASC/PUR, same result obtained when antibody was administered as either ascites or in a purified form in independent experiments.
FIG. 3.
Survival of mice with anti-NS1 MAbs. (A) Six-week-old C57BL/6 mice were inoculated with 102 PFU of WNV subcutaneously and administered a single dose of purified MAbs (500 μg) or ascites fluid (200 μl) at the same time by an intraperitoneal route. The survival curves were constructed using data from one to three independent experiments with 10 animals each. As indicated in Table 3, 10NS1, 14NS1, 16NS1, 17NS1, and 22NS1 all provided statistically significant protection compared to treatment with saline alone. (B) Dose-response analysis of MAb 17NS1. Six-week-old mice were inoculated with 102 PFU of WNV via footpad and administered a single dose of purified 17NS1 MAb (0.8, 4, 20, 100, or 500 μg) at the same time by an intraperitoneal route. The survival curves were constructed using data from two independent experiments with 10 animals, resulting in 20 animals tested at each dose. Statistical differences compared to the PBS control were as follows: 0.8 μg, P = 0.13; 4 μg, P = 0.02; 20 μg, 100 μg, and 500 μg, P < 0.0001. (C) Four-week-old C57BL/6 mice were administered a single dose of purified MAb 14NS1 (1 mg) through an intraperitoneal route and inoculated subcutaneously with 102 PFU of WNV 7 or 14 days later. Statistical differences compared to the PBS control were as follows: prophylaxis at day −7, P = 0.007; prophylaxis at day −14, P = 0.005.
Effect of 17NS1 on viral spread and burden.
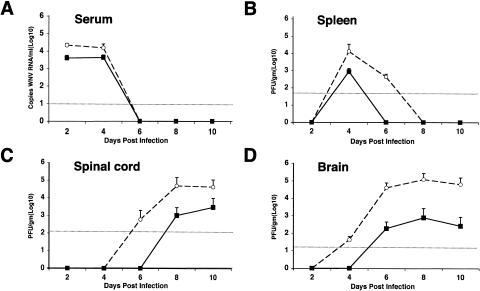
To better define the mechanism of inhibition by MAbs against WNV NS1, we evaluated the effects of one inhibitory MAb, 17NS1, on the viral burden in the periphery and in central nervous system (CNS) tissues. Six-week-old wild-type mice were infected with 102 PFU and administered 17NS1 or PBS, and the viral load was measured at days 2, 4, 6, 8, and 10 in the serum, spleen, spinal cord, and brain (Fig. 4).
FIG. 4.
Effect of MAb 17NS1 on viral burden over time. (A) Levels of viral RNA in serum. Six-week-old mice were treated immediately after WNV infection with MAb 17NS1 (500 μg) or saline (PBS). Serum was harvested at the indicated days, and viral RNA levels were determined by a real-time fluorogenic RT-PCR assay. Data are expressed as genomic equivalents of WNV RNA per milliliter of serum and reflect the average of at least four independent mice per time point. The dashed line represents the limit of sensitivity of the assay. (B to D) Infectious virus levels in tissues. Virus levels were measured from the spleen (B), spinal cord (C), and brain (D) by a viral plaque assay in BHK21-15 cells after tissues were harvested at the indicated days. Data are shown as the average PFU per gram of tissue at four or eight mice per point. The dotted line represents the limit of sensitivity of the assay. Open circles and filled squares indicate PBS and 17NS1-treated mice, respectively.
(i) Serum and spleen.
In both PBS- and 17NS1-treated mice, viremia was undetectable by viral plaque assay throughout the time course; these results were consistent with our previous publications (16, 60). When viral RNA in serum was measured by a more sensitive quantitative RT-PCR assay (38), viremia was detected at day 2 after subcutaneous infection but rapidly decreased to a level below detection by day 6. However, in mice that received 17NS1, approximately four- to fivefold- lower levels of WNV RNA were detected at days 2 and 4 (P < 0.04) (Fig. 4A). A similar pattern was observed in the spleen. Although the level of infectious WNV peaked in both groups at day 4 after infection, there was an approximately 15-fold-lower level of infectious virus in mice that received 17NS1 (Fig. 4B, P = 0.01). Consistent with this, infectious WNV was cleared more rapidly in 17NS1-treated mice. Thus, 17NS1 appeared to decrease viral spread and/or enhance viral clearance in the serum and spleen at early stages in WNV infection.
(ii) Spinal cord and brain.
WNV was detected earlier and at higher levels in the spinal cord of PBS-treated mice (Fig. 4C). At day 6 after infection, 25% (2 of 8) of PBS-treated but no (0of 4) 17NS1-treated mice had detectable levels of infectious virus. By day 8, PBS-treated mice had viral burdens (∼105 PFU/g) that were ∼100-fold higher (n = 8, P = 0.02) than that observed in 17NS1-treated mice. Similar results were observed in the brain (Fig. 4D). At day 4, infectious virus was detected in 62% (5 of 8) of PBS-treated but no (0 of 4) 17NS1-treated mice. As the time course progressed, the gap in viral burden in the brain widened such that by days 8 and 10, there were ∼250-fold-lower levels (n = 8, P < 0.005) of infectious virus in the mice treated with 17NS1. Collectively, the virologic analyses suggest that treatment of mice with 17NS1 enhances the control of WNV infection in the periphery, which limits virus spread into the CNS.
Mechanism of protection by anti-NS1 MAbs.
A previous study with MAbs against YF NS1 suggested that protection in vivo was dependent on the Fc region of the antibody (59). Although direct experiments were not performed, it was hypothesized that antibody-dependent complement activation, phagocytosis, or cell-mediated cytotoxicity could explain the effect. To test whether the effector functions of antibody were linked to the protective effect of three (14NS1, 16NS1, and 17NS1) of our inhibitory MAbs, survival studies were performed with 8- to 10-week-old C1q- or Fc γ receptor I- and III-deficient congenic C57BL/6 mice. At baseline, these mice all show increased susceptibility to lethal WNV infection compared to age-matched wild-type control mice (Fig. 5) (52). In C1q-deficient mice, which cannot activate complement by the antibody-dependent classical pathway, 14NS1, 16NS1, and 17NS1 maintained most of their protective effects (Fig. 5A). In contrast, in Fc γ receptor I- and III-deficient mice, which are severely impaired in antibody-dependent phagocytic and cellular cytotoxic responses, much of the protective effect of 16NS1 and 17NS1 was lost (Fig. 5B), suggesting that the Fc regions determined the potency of 16NS1 and 17NS1 by virtue of their ability to bind to Fc γ receptors I or III. Surprisingly, only a small loss of efficacy was observed with 14NS1, as treated wild-type and Fc γ receptor I- and III-deficient mice had 95% and 73% survival rates, respectively (Table 3 and Fig. 5B). Taken together, our data suggest that different anti-NS1 MAbs protect in vivo by distinct mechanisms.
FIG. 5.
Efficacy of anti-NS1 MAbs in wild-type, C1q-deficient, or Fc γ receptor I- and III-deficient mice. (A) C1q-deficient or (B) Fc γ receptor I- and III-deficient mice were inoculated subcutaneously with 102 PFU of WNV at day zero. At the same time, mice were administered PBS or a single dose of 14NS1, 16NS1, or 17NS1 via an intraperitoneal route. The number of animals for each antibody dose ranged from 15 to 27. The difference in survival curves was statistically significant for all WNV NS1-specific monoclonal antibodies shown in the C1q-deficient mice (P ≤ 0.0004). The difference in survival curves for the Fc γ receptor I- and III-deficient mice was significant for MAb 14NS1 (P = 0.003) and 16NS1 (P = 0.04) but not for 17NS1 (P = 0.3). (C and D) Therapeutic activity of MAbs 14NS1 and 16NS1. At day 2 (C) or 4 (D) after WNV infection, 5-week-old C57BL/6 mice were passively transferred saline or a single intraperitoneal inoculation of 14NS1, 16NS1, or 14NS1 plus 16NS1 as ascites fluid. The survival curves were constructed with a total of 10 to 20 mice for each treatment group. The statistical difference in survival curves compared to saline was as follows: day 2, mice treated with 14NS1 (P = 0.0003), 16NS1 (P < 0.0001), or 14NS1 plus 16NS1 (P = 0.0001); day 4, mice treated with 14NS1 (P > 0.1), 16NS1 (P = 0.03), or 14NS1 plus 16NS1 (P = 0.02). An arrow shows the timing of MAb administration.
Therapeutic studies in mice.
Although other studies have shown that prophylaxis of mice with MAbs against NS1 can protect against lethal YF and DEN infections (27, 30, 57, 59), the therapeutic potential of anti-NS1 MAbs has not been explored. To evaluate the potential of 14NS1 and 16NS1 as possible immunotherapeutics against WNV infection, postexposure treatment experiments were performed in 5-week-old C57BL/6 mice (Fig. 5C and D). Mice were inoculated with 102 PFU of WNV at day zero and then administered a single dose of MAb or PBS at day 2 or 4 after infection. As observed previously, treatment of mice with PBS or an isotype control MAb resulted in a baseline survival rate of ∼10% (19, 52). A single dose of 14NS1 or 16NS1 at day 2 after infection resulted in 68% (P = 0.0003) and 47% (P < 0.0001) survival rates, respectively. When 5-week-old mice were given a combination of anti-NS1 MAbs (14NS1 and 16NS1) that mapped to distinct fragments, an enhanced survival rate was observed (80% survival, P = 0.0001). As WNV spreads to the CNS in C57BL/6 mice within 4 days of infection (Fig. 4D) (19, 52), we evaluated the therapeutic efficacy of anti-NS1 at a later and perhaps more clinically meaningful time point. Notably, a single dose of 14NS1 and 16NS1 at day 4 after infection resulted in significant protection (60% survival, P = 0.02) at a rate that was superior to that observed with a single dose of either 14NS1 (40% survival, P = 0.15) or 16NS1 (44% survival, P = 0.03).
DISCUSSION
WNV infects the CNS, leading to encephalitis, paralysis, and death in humans, birds, and other animals. Although its precise role in flavivirus infection and pathogenesis has not been clearly defined, NS1 functions as a cofactor in replication (37, 45, 46, 48), elicits protective antibodies (56, 58), and accumulates to high levels in the serum of patients with severe DEN disease (1, 40). Based on analogy to DNA viruses, flavivirus NS1 may have an immune evasive function as it is secreted, glycosylated, and not packaged in the virion (9, 14). In this study, we purified to homogeneity full-length glycosylated WNV NS1 from insect cells and used it to generate 22 new NS1-specific MAbs. Using competitive binding studies, yeast surface display, and bacterial expression systems, we mapped 21 of 22 MAbs to one of three regions on NS1. Prophylaxis studies in mice demonstrated that 36% of the anti-NS1 antibodies had significant protective activity in vivo against WNV, yet there was no strict correlation with domain localization. Virologic studies with one inhibitory MAb, 17NS1, indicated that viral infection was impaired at a relatively early stage of infection. Studies with immunodeficient mice suggest that distinct regions of NS1 can elicit humoral protective immunity through complement- and/or Fc γ receptor-independent mechanisms. Finally, in therapeutic postexposure trials in mice, a single dose of either of two anti-NS1 MAbs, 14NS1 or 16NS1, protected mice against WNV-induced mortality.
Our study is the first to express and purify full-length, recombinant, glycosylated NS1 to homogeneity from a pathogenic strain of WNV. Using a similar insect cell expression system, NS1s from Kunjin (35), DEN (6, 39), and JEV (24) viruses also have been purified. Size exclusion chromatography experiments suggested that secreted WNV NS1 had an apparent mass of ∼300 kDa and thus behaved as an oligomer, which agrees with prior studies that purified DEN or TBE NS1 from infected mammalian cells (10, 25). We also observed similar results after purifying WNV NS1 from replicon-expressing mammalian cells by affinity and size exclusion chromatography (K. M. Chung and M. Diamond, unpublished observations). Similar to the results of others (10, 23, 25, 53), by SDS-PAGE analysis, only the NS1 dimer was observed, and this was converted to a monomer by heat denaturation. Our biochemical results are consistent with a model (25) in which secreted NS1 is organized as a hexamer composed of three dimers.
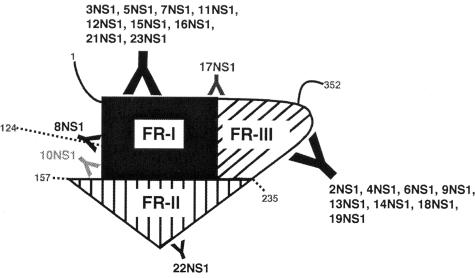
Previous studies that have localized MAb epitopes on DEN or MVE NS1 have used linear peptides (21), Western blotting of bacterially expressed fragments (54), and competition binding (28, 31, 73) assays. We combined yeast surface display and bacterial expression and oxidative refolding of NS1 fragments with competition binding to map the vast majority of our anti-NS1 MAbs. Yeast surface display requires proper folding and secretion of proteins (70) and has been recently adapted to map MAb epitopes in a high-throughput manner (8, 52). Based on antibody reactivity and putative disulfide bonding patterns, we generated three distinct fragments of WNV NS1, I (1 to 157), II (158 to 235), and III (236 to 352), with 11, 1, and 8 of our MAbs, respectively, binding to them. However, two MAbs (10NS1 and 17NS1) had recognition sites in more than one fragment, suggesting a physical proximity between different parts of the protein. Using these results, we developed a model for the topography of WNV NS1 and its antibody epitopes (Fig. 6). Although informative, we acknowledge that domain assignments and their quaternary assembly can only be schematic in the absence of NS1 atomic-resolution structural results. Such studies are currently under way in our laboratory.
FIG. 6.
A model of the topography of WNV NS1 and antibody binding sites. Nine, one (8NS1), one (22NS1), and eight of our MAbs recognized amino acids 1 to 124, amino acids 1 to 157, amino acids 158 to 235, and amino acids 236 to 352, respectively. 10NS1 recognized both FR-I and FR-II. 17NS1 recognized both FR-I and FR-III. FR-I, FR-II, and FR-III indicate amino acids 1 to 157, amino acids 158 to 235, and amino acids 236 to 352, respectively. Numbers indicate the amino acid position on WNV NS1.
Although all of our MAbs recognized NS1 in WNV-infected cells by flow cytometry, three (1NS1, 2NS1, and 21NS1) were not mapped by yeast display, possibly because of differences in carbohydrate modification between yeast and mammalian cells or epitope interference associated with the Aga2 fusion. Nonetheless, 21NS1 likely localizes to or near FR-I, as it was competed by 3NS1, a FR-I MAb, and 2NS1 maps to FR-III, as it strongly bound the bacterially expressed fragment by ELISA. Future studies using error-prone PCR mutagenesis and yeast surface display are planned to identify the amino acid contact residues for many of the individual MAbs against WNV NS1.
Because some antibodies against flavivirus NS1 protect against lethal infection in vivo, immunization with purified NS1 has been proposed as an alternative vaccination strategy against flaviviruses (26, 33, 34, 44, 62, 63, 71, 75). Indeed, immunization with a single NS1 peptide (amino acids 37 to 55) of TBE elicited antibodies that protected mice against lethal challenge (65). Subunit-based vaccines containing NS1 could prove relevant if live-attenuated vaccines facilitate the development of enhancing antibodies against the viral envelope proteins. In our experiments, several antibodies that mapped to either FR-I (10NS1 and 16NS1), FR-II (22NS1), or FR-III (14NS1 and 17NS1) of WNV NS1 strongly protected against infection in mice when they were administered at the time of infection. Moreover, therapeutic administration of two different MAbs (14NS1 and 16NS1) that strongly reacted with FR-III and FR-I, respectively, provided significantly increased survival even when given as a single dose 2 days after WNV infection. A combination of 14NS1 and 16NS1 increased the survival rate beyond that observed with either MAb alone, even when administrated 4 days after infection. A synergistic interaction of anti-NS1 MAbs also was observed in a preexposure prophylaxis model of DEN-2 infection (30). Given this, studies are planned with specific combinations of anti-E and anti-NS1 MAbs to improve the efficiency of antibody-based therapeutics against WNV (52).
Although previous experiments demonstrated that passive transfer of anti-NS1 MAbs prior to infection protects against flaviviruses (27, 57, 59), virologic studies were never performed to define the stage in pathogenesis at which the MAbs were acting. Protective anti-NS1 MAbs could improve survival by limiting hematogenous spread, facilitating immune system effector function, or blocking a specific immunomodulatory or virologic function of NS1. To our knowledge, our experiments with 17NS1 are novel, as they define how an antibody against a nonstructural protein alters the course of pathogenesis at a virologic level. Prophylaxis with 17NS1 reduced the WNV burden in serum and lymphoid compartments early during the course of infection, resulting in decreased spread to the brain and spinal cord and enhanced survival.
Our studies also investigated the mechanism of protection of three of our inhibitory anti-NS1 MAbs. Previous studies with isotype switch variants and F(ab′)2 fragments of anti-YF NS1 MAbs suggested that the Fc region of anti-NS1 MAbs was required for protection (59). Because protection against YF infection was not altered in mice treated with cobra venom factor, an agent that depletes complement component C3, these authors suggested that protection was complement independent. In support of this, vaccination of C5-deficient mice with TBE NS1 was effective against lethal challenge (34). Our experiments in C1q-deficient mice support this, as 14NS1, 16NS1, and 17NS1 retained protective activity without triggering the classical pathway of complement activation. It should be noted that in other animals, the inhibitory effect of NS1 MAbs could be magnified by complement. Mouse complement C4 has an inherently low classical pathway C5 convertase subunit activity in vitro and in vivo (3, 17, 61), resulting in depressed complement-dependent antibody neutralization of WNV (50). Because anti-NS1 MAbs prevented YF infection in a complement-independent yet Fc-dependent manner, it was speculated that protection was dependent on Fc γ receptors (59). Protective anti-NS1 MAbs could enhance survival in an Fc γ receptor-dependent manner by triggering viral clearance by phagocytic cells and/or by promoting antibody-dependent cytotoxicity. For 17NS1, which completely lost protective activity in Fc γ receptor I- and III-deficient mice, we confirmed this mechanism. Ongoing studies in NK cell-depleted mice should define the precise mechanism by which Fc γ receptors engage 17NS1 to mediate protection. In contrast, 14NS1 still protected mice that were deficient in Fc γ receptors I and III through an as yet undefined mechanism, possibly by directly inhibiting a virologic or immunomodulatory function of NS1. Indeed, a recent paper suggested that treatment of cells with soluble NS1 could enhance DEN viral infection (2); however, addition of 14NS1 in the presence or absence of soluble NS1 in cell culture had no apparent effect on WNV infection (K. M. Chung and M. S. Diamond, unpublished observations). It also remains possible that 14NS1 inhibits WNV infection through a combination of complement- and Fc γ receptor-dependent mechanisms, and elimination of one pathway is not sufficient to abolish the protection.
The lack of available therapy and the expanding WNV epidemic necessitates the evaluation of agents that inhibit infection and can be rapidly transferred into the clinical setting. Here, we demonstrate that prophylaxis or therapy with anti-NS1 MAbs can efficiently inhibit lethal WNV infection through at least two independent mechanisms. The generation of a selected panel of protective MAbs against different WNV proteins may facilitate a combination-based therapeutic strategy that increases potency, prevents the emergence of resistance, and provides superior clinical protection.
Acknowledgments
We thank A. Pekosz, K. Blight, D. Leib, L. Morrison, R. Klein, P. Olivo, and T. Pierson and their laboratories for experimental advice and E. Mehlhop, T. Oliphant, and S. Schaecher for help with some of the experiments. We also thank G. Stahl and M. Botto for complement-deficient mice, R. Levis for the anti-DEN NS1 MAb, and S. Burke and MacroGenics, Inc., for growing and purifying some of the anti-NS1 MAbs.
The work was supported by grants from NIH (U01 AI061373 [to M.S.D.] and U54 AI057160 [to M.S.D. and D.H.F.] to the Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research) and the Pediatric Dengue Vaccine Initiative (to D.H.F. and M.S.D.).
REFERENCES
- 1.Alcon, S., A. Talarmin, M. Debruyne, A. Falconar, V. Deubel, and M. Flamand. 2002. Enzyme-linked immunosorbent assay specific to Dengue virus type 1 nonstructural protein NS1 reveals circulation of the antigen in the blood during the acute phase of disease in patients experiencing primary or secondary infections. J. Clin. Microbiol. 40:376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcon-LePoder, S., M. T. Drouet, P. Roux, M. P. Frenkiel, M. Arborio, A. M. Durand-Schneider, M. Maurice, I. Le Blanc, J. Gruenberg, and M. Flamand. 2005. The secreted form of dengue virus nonstructural protein NS1 is endocytosed by hepatocytes and accumulates in late endosomes: implications for viral infectivity. J. Virol. 79:11403-11411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkinson, J. P., K. McGinnis, and D. Shreffler. 1980. Development and characterization of a hemolytic assay for mouse C4. J. Immunol. Methods 33:351-368. [DOI] [PubMed] [Google Scholar]
- 4.Blitvich, B. J., D. Scanlon, B. J. Shiell, J. S. Mackenzie, K. Pham, and R. A. Hall. 2001. Determination of the intramolecular disulfide bond arrangement and biochemical identification of the glycosylation sites of the nonstructural protein NS1 of Murray Valley encephalitis virus. J. Gen. Virol. 82:2251-2256. [DOI] [PubMed] [Google Scholar]
- 5.Chambers, T. J., C. S. Hahn, R. Galler, and C. M. Rice. 1990. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44:649-688. [DOI] [PubMed] [Google Scholar]
- 6.Chan, L. C., P. R. Young, C. Bletchly, and S. Reid. 2002. Production of the baculovirus-expressed dengue virus glycoprotein NS1 can be improved dramatically with optimised regimes for fed-batch cultures and the addition of the insect moulting hormone, 20-Hydroxyecdysone. J. Virol. Methods 105:87-98. [DOI] [PubMed] [Google Scholar]
- 7.Chang, H. H., H. F. Shyu, Y. M. Wang, D. S. Sun, R. H. Shyu, S. S. Tang, and Y. S. Huang. 2002. Facilitation of cell adhesion by immobilized dengue viral nonstructural protein 1 (NS1): arginine-glycine-aspartic acid structural mimicry within the dengue viral NS1 antigen. J. Infect. Dis. 186:743-751. [DOI] [PubMed] [Google Scholar]
- 8.Chao, G., J. R. Cochran, and K. D. Wittrup. 2004. Fine epitope mapping of anti-epidermal growth factor receptor antibodies through random mutagenesis and yeast surface display. J. Mol. Biol. 342:539-550. [DOI] [PubMed] [Google Scholar]
- 9.Chua, J. J., R. Bhuvanakantham, V. T. Chow, and M. L. Ng. 2005. Recombinant non-structural 1 (NS1) protein of dengue-2 virus interacts with human STAT3β protein. Virus Res. 112:85-94. [DOI] [PubMed] [Google Scholar]
- 10.Crooks, A. J., J. M. Lee, L. M. Easterbrook, A. V. Timofeev, and J. R. Stephenson. 1994. The NS1 protein of tick-borne encephalitis virus forms multimeric species upon secretion from the host cell. J. Gen. Virol. 75:3453-3460. [DOI] [PubMed] [Google Scholar]
- 11.Cuff, J. A., M. E. Clamp, A. S. Siddiqui, M. Finlay, and G. J. Barton. 1998. JPred: a consensus secondary structure prediction server. Bioinformatics 14:892-893. [DOI] [PubMed] [Google Scholar]
- 12.Despres, P., J. Dietrich, M. Girard, and M. Bouloy. 1991. Recombinant baculoviruses expressing yellow fever virus E and NS1 proteins elicit protective immunity in mice. J. Gen. Virol. 72:2811-2816. [DOI] [PubMed] [Google Scholar]
- 13.Diamond, M. S., T. G. Roberts, D. Edgil, B. Lu, J. Ernst, and E. Harris. 2000. Modulation of dengue virus infection in human cells by alpha, beta, and gamma interferons. J. Virol. 74:4957-4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diamond, M. S. 2003. Evasion of innate and adaptive immunity by flaviviruses. Immunol. Cell Biol. 81:196-206. [DOI] [PubMed] [Google Scholar]
- 15.Diamond, M. S., B. Shrestha, A. Marri, D. Mahan, and M. Engle. 2003. B cells and antibody play critical roles in the immediate defense of disseminated infection by West Nile encephalitis virus. J. Virol. 77:2578-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diamond, M. S., E. Sitati, L. Friend, B. Shrestha, S. Higgs, and M. Engle. 2003. Induced IgM protects against lethal West Nile Virus infection. J. Exp. Med. 198:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ebanks, R. O., and D. E. Isenman. 1996. Mouse complement component C4 is devoid of classical pathway C5 convertase subunit activity. Mol. Immunol. 33:297-309. [DOI] [PubMed] [Google Scholar]
- 18.Ebel, G. D., A. P. Dupuis III, K. Ngo, D. Nicholas, E. Kauffman, S. A. Jones, D. Young, J. Maffei, P. Y. Shi, K. Bernard, and L. Kramer. 2001. Partial genetic characterization of West Nile Virus strains, New York State, 2000. Emerg. Infect. Dis. 7:650-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engle, M. J., and M. S. Diamond. 2003. Antibody prophylaxis and therapy against West Nile virus infection in wild-type and immunodeficient mice. J. Virol. 77:12941-12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falconar, A. K. 1997. The dengue virus nonstructural-1 protein (NS1) generates antibodies to common epitopes on human blood clotting, integrin/adhesin proteins and binds to human endothelial cells: potential implications in haemorrhagic fever pathogenesis. Arch. Virol. 142:897-916. [DOI] [PubMed] [Google Scholar]
- 21.Falconar, A. K., P. R. Young, and M. A. Miles. 1994. Precise location of sequential dengue virus subcomplex and complex B cell epitopes on the nonstructural-1 glycoprotein. Arch. Virol. 137:315-326. [DOI] [PubMed] [Google Scholar]
- 22.Falgout, B., M. Bray, J. J. Schlesinger, and C. J. Lai. 1990. Immunization of mice with recombinant vaccinia virus expressing authentic dengue virus nonstructural protein NS1 protects against lethal dengue virus encephalitis. J. Virol. 64:4356-4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flamand, M., M. Chevalier, E. Henchal, M. Girard, and V. Deubel. 1995. Purification and renaturation of Japanese encephalitis virus nonstructural glycoprotein NS1 overproduced by insect cells. Protein Expr. Purif. 6:519-527. [DOI] [PubMed] [Google Scholar]
- 24.Flamand, M., V. Deubel, and M. Girard. 1992. Expression and secretion of Japanese encephalitis virus nonstructural protein NS1 by insect cells using a recombinant baculovirus. Virology 191:826-836. [DOI] [PubMed] [Google Scholar]
- 25.Flamand, M., F. Megret, M. Mathieu, J. Lepault, F. A. Rey, and V. Deubel. 1999. Dengue virus type 1 nonstructural glycoprotein NS1 is secreted from mammalian cells as a soluble hexamer in a glycosylation-dependent fashion. J. Virol. 73:6104-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibson, C. A., J. J. Schlesinger, and A. D. Barrett. 1988. Prospects for a virus non-structural protein as a subunit vaccine. Vaccine 6:7-9. [DOI] [PubMed] [Google Scholar]
- 27.Gould, E. A., A. Buckley, A. D. Barrett, and N. Cammack. 1986. Neutralizing (54K) and non-neutralizing (54K and 48K) monoclonal antibodies against structural and non-structural yellow fever virus proteins confer immunity in mice. J. Gen. Virol. 67:591-595. [DOI] [PubMed] [Google Scholar]
- 28.Hall, R. A., B. H. Kay, G. W. Burgess, P. Clancy, and I. D. Fanning. 1990. Epitope analysis of the envelope and non-structural glycoproteins of Murray Valley encephalitis virus. J. Gen. Virol. 71:2923-2930. [DOI] [PubMed] [Google Scholar]
- 29.Harlow, E., and D. Lane. 1988. Antibodies, a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Henchal, E. A., L. S. Henchal, and J. J. Schlesinger. 1988. Synergistic interactions of anti-NS1 monoclonal antibodies protect passively immunized mice from lethal challenge with dengue 2 virus. J. Gen. Virol. 69:2101-2107. [DOI] [PubMed] [Google Scholar]
- 31.Henchal, E. A., L. S. Henchal, and B. K. Thaisomboonsuk. 1987. Topological mapping of unique epitopes on the dengue-2 virus NS1 protein using monoclonal antibodies. J. Gen. Virol. 68:845-851. [DOI] [PubMed] [Google Scholar]
- 32.Hubalek, Z., and J. Halouzka. 1999. West Nile fever—a reemerging mosquito-borne viral disease in Europe. Emerg. Infect. Dis. 5:643-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobs, S. C., J. R. Stephenson, and G. W. Wilkinson. 1992. High-level expression of the tick-borne encephalitis virus NS1 protein by using an adenovirus-based vector: protection elicited in a murine model. J. Virol. 66:2086-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobs, S. C., J. R. Stephenson, and G. W. Wilkinson. 1994. Protection elicited by a replication-defective adenovirus vector expressing the tick-borne encephalitis virus non-structural glycoprotein NS1. J. Gen. Virol. 75:2399-2402. [DOI] [PubMed] [Google Scholar]
- 35.Khromykh, A. A., T. J. Harvey, M. Abedinia, and E. G. Westaway. 1996. Expression and purification of the seven nonstructural proteins of the flavivirus Kunjin in the E. coli and the baculovirus expression systems. J. Virol. Methods 61:47-58. [DOI] [PubMed] [Google Scholar]
- 36.Khromykh, A. A., P. L. Sedlak, K. J. Guyatt, R. A. Hall, and E. G. Westaway. 1999. Efficient trans-complementation of the flavivirus Kunjin NS5 protein but not of the NS1 protein requires its coexpression with other components of the viral replicase. J. Virol. 73:10272-10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khromykh, A. A., P. L. Sedlak, and E. G. Westaway. 2000. cis- and trans-acting elements in flavivirus RNA replication. J. Virol. 74:3253-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lanciotti, R. S., A. J. Kerst, R. S. Nasci, M. S. Godsey, C. J. Mitchell, H. M. Savage, N. Komar, N. A. Panella, B. C. Allen, K. E. Volpe, B. S. Davis, and J. T. Roehrig. 2000. Rapid detection of West Nile virus from human clinical specimens, field-collected mosquitoes, and avian samples by a TaqMan reverse transcriptase-PCR assay. J. Clin. Microbiol. 38:4066-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leblois, H., and P. R. Young. 1995. Maturation of the dengue-2 virus NS1 protein in insect cells: effects of downstream NS2A sequences on baculovirus-expressed gene constructs. J. Gen. Virol. 76:979-984. [DOI] [PubMed] [Google Scholar]
- 40.Libraty, D. H., P. R. Young, D. Pickering, T. P. Endy, S. Kalayanarooj, S. Green, D. W. Vaughn, A. Nisalak, F. A. Ennis, and A. L. Rothman. 2002. High circulating levels of the dengue virus nonstructural protein NS1 early in dengue illness correlate with the development of dengue hemorrhagic fever. J. Infect. Dis. 186:1165-1168. [DOI] [PubMed] [Google Scholar]
- 41.Lin, C. F., S. C. Chiu, Y. L. Hsiao, S. W. Wan, H. Y. Lei, A. L. Shiau, H. S. Liu, T. M. Yeh, S. H. Chen, C. C. Liu, and Y. S. Lin. 2005. Expression of cytokine, chemokine, and adhesion molecules during endothelial cell activation induced by antibodies against dengue virus nonstructural protein 1. J. Immunol. 174:395-403. [DOI] [PubMed] [Google Scholar]
- 42.Lin, C. F., H. Y. Lei, A. L. Shiau, C. C. Liu, H. S. Liu, T. M. Yeh, S. H. Chen, and Y. S. Lin. 2003. Antibodies from dengue patient sera cross-react with endothelial cells and induce damage. J. Med. Virol. 69:82-90. [DOI] [PubMed] [Google Scholar]
- 43.Lin, C. F., H. Y. Lei, A. L. Shiau, H. S. Liu, T. M. Yeh, S. H. Chen, C. C. Liu, S. C. Chiu, and Y. S. Lin. 2002. Endothelial cell apoptosis induced by antibodies against dengue virus nonstructural protein 1 via production of nitric oxide. J. Immunol. 169:657-664. [DOI] [PubMed] [Google Scholar]
- 44.Lin, Y. L., L. K. Chen, C. L. Liao, C. T. Yeh, S. H. Ma, J. L. Chen, Y. L. Huang, S. S. Chen, and H. Y. Chiang. 1998. DNA immunization with Japanese encephalitis virus nonstructural protein NS1 elicits protective immunity in mice. J. Virol. 72:191-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindenbach, B. D., and C. M. Rice. 1999. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J. Virol. 73:4611-4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lindenbach, B. D., and C. M. Rice. 1997. trans-complementation of yellow fever virus NS1 reveals a role in early RNA replication. J. Virol. 71:9608-9617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macdonald, J., J. Tonry, R. A. Hall, B. Williams, G. Palacios, M. S. Ashok, O. Jabado, D. Clark, R. B. Tesh, T. Briese, and W. I. Lipkin. 2005. NS1 protein secretion during the acute phase of West Nile virus infection. J. Virol. 79:13924-13933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mackenzie, J. M., M. K. Jones, and P. R. Young. 1996. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology 220:232-240. [DOI] [PubMed] [Google Scholar]
- 49.Mason, P. W. 1989. Maturation of Japanese encephalitis virus glycoproteins produced by infected mammalian and mosquito cells. Virology 169:354-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mehlhop, E., K. Whitby, T. Oliphant, A. Marri, M. Engle, and M. S. Diamond. 2005. Complement activation is required for the induction of a protective antibody response against West Nile virus infection. J. Virol. 79:7466-7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muylaert, I. R., T. J. Chambers, R. Galler, and C. M. Rice. 1996. Mutagenesis of the N-linked glycosylation sites of the yellow fever virus NS1 protein: effects on virus replication and mouse neurovirulence. Virology 222:159-168. [DOI] [PubMed] [Google Scholar]
- 52.Oliphant, T., M. Engle, G. Nybakken, C. Doane, S. Johnson, L. Huang, S. Gorlatov, E. Mehlhop, A. Marri, K. M. Chung, G. D. Ebel, L. D. Kramer, D. H. Fremont, and M. S. Diamond. 2005. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat. Med. 11:522-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pryor, M. J., and P. J. Wright. 1993. The effects of site-directed mutagenesis on the dimerization and secretion of the NS1 protein specified by dengue virus. Virology 194:769-780. [DOI] [PubMed] [Google Scholar]
- 54.Putnak, J. R., P. C. Charles, R. Padmanabhan, K. Irie, C. H. Hoke, and D. S. Burke. 1988. Functional and antigenic domains of the dengue-2 virus nonstructural glycoprotein NS-1. Virology 163:93-103. [DOI] [PubMed] [Google Scholar]
- 55.Roopenian, D. C., G. J. Christianson, T. J. Sproule, A. C. Brown, S. Akilesh, N. Jung, S. Petkova, L. Avanessian, E. Y. Choi, D. J. Shaffer, P. A. Eden, and C. L. Anderson. 2003. The MHC class I-like IgG receptor controls perinatal IgG transport, IgG homeostasis, and fate of IgG-Fc-coupled drugs. J. Immunol. 170:3528-3533. [DOI] [PubMed] [Google Scholar]
- 56.Schlesinger, J. J., M. W. Brandriss, C. B. Cropp, and T. P. Monath. 1986. Protection against yellow fever in monkeys by immunization with yellow fever virus nonstructural protein NS1. J. Virol. 60:1153-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schlesinger, J. J., M. W. Brandriss, and E. E. Walsh. 1985. Protection against 17D yellow fever encephalitis in mice by passive transfer of monoclonal antibodies to the nonstructural glycoprotein gp48 and by active immunization with gp48. J. Immunol. 135:2805-2809. [PubMed] [Google Scholar]
- 58.Schlesinger, J. J., M. W. Brandriss, and E. E. Walsh. 1987. Protection of mice against dengue 2 virus encephalitis by immunization with the dengue 2 virus non-structural glycoprotein NS1. J. Gen. Virol. 68:853-857. [DOI] [PubMed] [Google Scholar]
- 59.Schlesinger, J. J., M. Foltzer, and S. Chapman. 1993. The Fc portion of antibody to yellow fever virus NS1 is a determinant of protection against YF encephalitis in mice. Virology 192:132-141. [DOI] [PubMed] [Google Scholar]
- 60.Shrestha, B., and M. S. Diamond. 2004. The role of CD8+ T cells in control of West Nile virus infection. J. Virol. 78:8312-8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Späth, G. F., L. A. Garraway, S. J. Turco, and S. M. Beverley. 2003. The role(s) of lipophosphoglycan (LPG) in the establishment of Leishmania major infections in mammalian hosts. Proc. Natl. Acad. Sci. USA 100:9536-9541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Timofeev, A. V., V. M. Butenko, and J. R. Stephenson. 2004. Genetic vaccination of mice with plasmids encoding the NS1 non-structural protein from tick-borne encephalitis virus and dengue 2 virus. Virus Genes 28:85-97. [DOI] [PubMed] [Google Scholar]
- 63.Timofeev, A. V., S. V. Ozherelkov, A. V. Pronin, A. V. Deeva, G. G. Karganova, L. B. Elbert, and J. R. Stephenson. 1998. Immunological basis for protection in a murine model of tick-borne encephalitis by a recombinant adenovirus carrying the gene encoding the NS1 non-structural protein. J. Gen. Virol. 79:689-695. [DOI] [PubMed] [Google Scholar]
- 64.Vieira, P., and K. Rajewsky. 1988. The half-lives of serum immunoglobulins in adult mice. Eur. J. Immunol. 18:313-316. [DOI] [PubMed] [Google Scholar]
- 65.Volpina, O. M., T. D. Volkova, D. O. Koroev, V. T. Ivanov, S. V. Ozherelkov, M. V. Khoretonenko, M. F. Vorovitch, J. R. Stephenson, and A. V. Timofeev. 2005. A synthetic peptide based on the NS1 non-structural protein of tick-borne encephalitis virus induced a protective immune response against fatal encephalitis in an experimental animal model. Virus Res. 112:95-99. [DOI] [PubMed] [Google Scholar]
- 66.Wallis, T. P., C. Y. Huang, S. B. Nimkar, P. R. Young, and J. J. Gorman. 2004. Determination of the disulfide bond arrangement of dengue virus NS1 protein. J. Biol. Chem. 279:20729-20741. [DOI] [PubMed] [Google Scholar]
- 67.Wengler, G., and H. J. Gross. 1978. Studies on virus-specific nucleic acids synthesized in vertebrate and mosquito cells infected with flaviviruses. Virology 89:423-437. [DOI] [PubMed] [Google Scholar]
- 68.Winkler, G., S. E. Maxwell, C. Ruemmler, and V. Stollar. 1989. Newly synthesized dengue-2 virus nonstructural protein NS1 is a soluble protein but becomes partially hydrophobic and membrane-associated after dimerization. Virology 171:302-305. [DOI] [PubMed] [Google Scholar]
- 69.Winkler, G., V. B. Randolph, G. R. Cleaves, T. E. Ryan, and V. Stollar. 1988. Evidence that the mature form of the flavivirus nonstructural protein NS1 is a dimer. Virology 162:187-196. [DOI] [PubMed] [Google Scholar]
- 70.Wittrup, K. D. 2001. Protein engineering by cell-surface display. Curr. Opin. Biotechnol. 12:395-399. [DOI] [PubMed] [Google Scholar]
- 71.Wu, S. F., C. L. Liao, Y. L. Lin, C. T. Yeh, L. K. Chen, Y. F. Huang, H. Y. Chou, J. L. Huang, M. F. Shaio, and H. K. Sytwu. 2003. Evaluation of protective efficacy and immune mechanisms of using a non-structural protein NS1 in DNA vaccine against dengue 2 virus in mice. Vaccine 21:3919-3929. [DOI] [PubMed] [Google Scholar]
- 72.Yamshchikov, V. F., G. Wengler, A. A. Perelygin, M. A. Brinton, and R. W. Compans. 2001. An infectious clone of the West Nile flavivirus. Virology 281:294-304. [DOI] [PubMed] [Google Scholar]
- 73.Young, P. R. 1990. Antigenic analysis of dengue virus using monoclonal antibodies. Southeast Asian J. Trop. Med. Public Health 21:646-651. [PubMed] [Google Scholar]
- 74.Young, P. R., P. A. Hilditch, C. Bletchly, and W. Halloran. 2000. An antigen capture enzyme-linked immunosorbent assay reveals high levels of the dengue virus protein NS1 in the sera of infected patients. J. Clin. Microbiol. 38:1053-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang, Y.-M., E. P. Hayes, T. C. McCarty, D. R. Dubois, P. L. Summers, K. H. Eckels, R. M. Chanock, and C.-J. Lai. 1988. Immunization of mice with dengue structural proteins and nonstructural protein NS1 expressed by baculovirus recombinant induces resistance to dengue virus encephalitis. J. Virol. 62:3027-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]