Abstract
Two lymphoid cell-specific proteins, RAG-1 and RAG-2, initiate V(D)J recombination by introducing DNA breaks at recombination signal sequences (RSSs). Although the RAG proteins themselves bind and cleave DNA substrates containing either a 12-RSS or a 23-RSS, DNA-bending proteins HMG-1 and HMG-2 are known to promote these processes, particularly with 23-RSS substrates. Using in-gel cleavage assays and DNA footprinting techniques, I analyzed the catalytic activity and protein-DNA contacts in discrete 12-RSS and 23-RSS complexes containing the RAG proteins and either HMG-1 or HMG-2. I found that both the cleavage activity and the pattern of protein-DNA contacts in RAG-HMG complexes assembled on 12-RSS substrates closely resembled those obtained from analogous 12-RSS complexes lacking HMG protein. In contrast, 23-RSS complexes containing both RAG proteins and either HMG-1 or HMG-2 exhibited enhanced cleavage activity and displayed an altered distribution of cleavage products compared to 23-RSS complexes containing only RAG-1 and RAG-2. Moreover, HMG-dependent heptamer contacts in 23-RSS complexes were observed. The protein-DNA contacts in RAG-RSS-HMG complexes assembled on 12-RSS or 23-RSS substrates were strikingly similar at comparable positions, suggesting that the RAG proteins mediate HMG-dependent heptamer contacts in 23-RSS complexes. Results of ethylation interference experiments suggest that the HMG protein is positioned 5" of the nonamer in 23-RSS complexes, interacting largely with the side of the duplex opposite the one contacting the RAG proteins. Thus, HMG protein plays the dual role of bringing critical elements of the 23-RSS heptamer into the same phase as the 12-RSS to promote RAG binding and assisting in the catalysis of 23-RSS cleavage.
The generation of immune repertoire diversity in most vertebrate organisms depends on a process called V(D)J recombination (11). In this process, exons encoding the antigen binding domains of immunoglobulin and T-cell receptors are assembled from arrays of component variable (V), diversity (D), and joining (J) segments by a series of site-specific DNA rearrangements. Recombination signal sequences (RSSs) that abut the coding segments direct the site of rearrangement; each RSS is composed of a conserved heptamer (CACAGTG) and nonamer (ACAAAAACC) element separated by 12 or 23 bp of nominally conserved intervening DNA (12-RSS and 23-RSS, respectively). Recombination normally occurs between two gene segments presenting a 12-RSS/23-RSS pair, a restriction termed the 12/23 rule. Two lymphoid cell-specific proteins, RAG-1 and RAG-2 (17, 24), initiate V(D)J recombination by catalyzing a DNA double-strand break at each RSS (introduced at the heptamer-coding-segment junction). Two distinct types of DNA ends are generated by this process: signal ends that are blunt and 5" phosphorylated and coding ends that are terminated in covalently sealed DNA hairpin structures (21, 22, 25). The origin of these products can be traced to a two-step, divalent metal ion-assisted cleavage reaction whose first step involves the introduction of a nick at the 5" end of each heptamer. The resulting 3"-OH at the terminus of each coding segment is then covalently linked to the phosphodiester of the antiparallel DNA strand via direct transesterification (13, 33). Signal ends are generally ligated precisely to form signal joints. However, hairpinned coding ends must be resolved before they are joined. While the RAG proteins can accomplish this task in vitro (3, 26), other candidates capable of resolving these hairpin structures have been reported (18, 19).
While the RAG proteins are capable of binding and cleaving DNA substrates containing isolated RSSs in the absence of other protein factors, certain DNA-bending proteins (e.g., HMG-1 and HMG-2) assist in these processes, particularly with regard to 23-RSS substrates. In addition, 12/23 paired complex formation and cleavage in vitro generally requires the presence of HMG proteins before it can be visualized (for reviews, see references 9 and 23 and references therein). However, the relationship between the RAG-RSS-HMG complexes observed by electrophoretic mobility shift assays (EMSA) and their enzymatic activity remains unclear because these complexes have not been tested directly to determine whether they support the cleavage activity that is attributed to them indirectly from in vitro assays that assess cleavage product formation in samples containing heterogeneous mixtures of protein-DNA complexes.
Using an in-gel cleavage assay that enabled me to assess and compare the catalytic activity of multiple, discrete RAG-RSS complexes fractionated on the same native polyacrylamide gel, I recently examined the active-site organization of V(D)J initiation complexes assembled on single RSS substrates (28). In this study, I used the in-gel cleavage assay and DNA footprinting techniques to relate DNA binding and cleavage activity in discrete RAG-RSS complexes formed in the absence and presence of HMG-1 or HMG-2. By EMSA, I found that HMG-1 and HMG-2 similarly supershifted RAG-DNA complexes assembled on either 12- or 23-RSS substrates. The stoichiometry of RAG-1 in these complexes was not changed by the presence of HMG protein.
Incorporation of HMG-1 or HMG-2 into a 12-RSS complex containing both RAG proteins does not augment the cleavage activity of the complex or significantly alter the pattern of protein-DNA contacts relative to a RAG-RSS complex formed in the absence of HMG protein. In contrast, a 23-RSS complex containing both RAG proteins and either HMG-1 or HMG-2 exhibits enhanced cleavage activity compared to its counterpart lacking HMG protein, particularly with respect to transesterification (hairpin formation). Moreover, additional HMG-dependent protein-DNA contacts are observed at the heptamer element which were not previously seen in 23-RSS complexes formed in the absence of HMG protein (30). Interestingly, the pattern of heptamer contacts observed in 23-RSS complexes formed in the presence of HMG protein resembles those seen in 12-RSS complexes formed in the absence of HMG protein, suggesting that the RAG proteins largely mediate heptamer contact in 23-RSS complexes containing HMG protein. Stimulation of RAG-mediated 23-RSS cleavage by the HMG proteins does not require synapsis of two recombination signals by a single RAG-1 dimer, since these activities are not altered in 23-RSS complexes formed using RAG-1 heterodimers in which one RAG-1 subunit contains a mutant DNA binding domain. Taken together, these data suggest that the HMG proteins assist the RAG proteins in the binding and cleavage of 23-RSS substrates by stabilizing RAG interactions with the heptamer motif.
MATERIALS AND METHODS
Plasmid constructs.
Eukaryotic expression constructs encoding core fragments of RAG-1 and RAG-2 fused at the amino terminus to one or two copies of the maltose binding protein (MBP) and possessing or lacking a carboxyl-terminal myc epitope and/or a polyhistidine tag have been described elsewhere and are depicted in Fig. 1 (28-30). Expression constructs pET11d-hHMG1 (a gift of R. Roeder via M. Schlissel) and pMT-PKA-HMG-2 (a gift of T. Wirth via D. Schatz) have been described previously (10, 36). The former encodes full-length human HMG-1 (residues 1 to 217), and the latter encodes mouse HMG-2 lacking the acidic carboxy-terminal tail (residues 1 to 185). Both expressed HMG proteins contain an amino-terminal hexahistidine tag to facilitate purification.
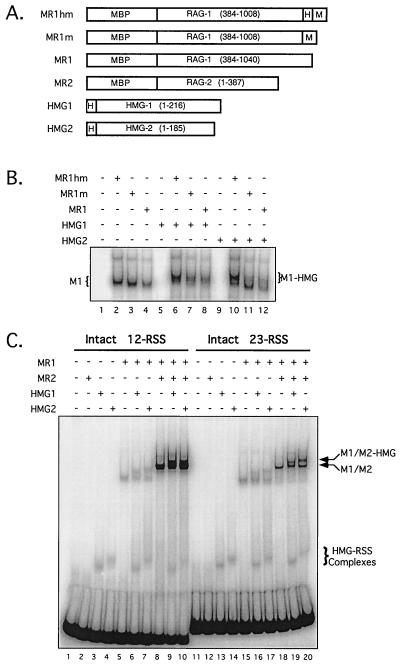
FIG. 1.
HMG-1 and HMG-2 supershift RSS complexes containing RAG-1 alone or both RAG-1 and RAG-2. (A) Schematic diagrams of RAG-1 and HMG fusion proteins (encoded residues in parentheses); the proteins are designated on the left. MBP, myc (M), and polyhistidine (H) sequences are also indicated. (B) EMSA of RAG-1 complexes bound to a 23-RSS in the absence and presence of HMG protein. A 32P-end-labeled 23-RSS substrate was incubated either alone (lane 1) or with (+) or without (−) various RAG-1 and HMG fusion proteins, as indicated at the top of the gel and defined in panel A. The positions of protein-DNA complexes fractionated by EMSA that contained RAG-1 alone (M1) or both RAG-1 and HMG (M1-HMG) are indicated on the left and right, respectively. (C) Formation of RAG-HMG complexes on 12- and 23-RSS substrates. A 32P-end-labeled 12-RSS (lanes 1 to 10) or 23-RSS (lanes 11 to 20) substrate was incubated either alone (lanes 1 and 11, respectively) or with (+) or without (−) various RAG-1, RAG-2, and HMG fusion proteins, as indicated at the top of the gel and defined in panel A. The positions of RSS complexes formed in the presence of HMG only (RSS-HMG) and those containing RAG-1 and RAG-2 formed in the absence (M1/M2) and presence (M1/M2-HMG) of HMG are indicated on the right. Complexes containing only RAG-1 are indicated on the left as described for panel B. Note that at lower exposure, distinct 12-RSS M1/M2 and M1/M2-HMG species can be clearly resolved in lanes 9 and 10; however, at that exposure, 23-RSS M1/M2 and M1/M2-HMG complexes, as well as HMG-RSS, M1, and M1-HMG complexes, are difficult to visualize.
Protein purification.
Single or double MBP-RAG fusion proteins (wild type or mutant) were expressed individually or coexpressed (where noted) in 293 cells and purified by amylose affinity chromatography as previously described (29, 30). Heterodimeric single and double MBP-RAG-1 fusion proteins were prepared by a previously published procedure (28). The protein preparations were judged to be >95% pure by silver staining. The Escherichia coli strain BL21(DE3)pLysS, transformed with pET11d-hHMG1 or pMT-PKA-HMG2, was induced to express HMG protein by adding 400 μl of 100 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to 100 ml of a log-phase culture (optical density at 600 nm, 0.6) and maintaining it at 30°C for 4 h. Thawed cell pellets were resuspended in binding buffer (40 mM Tris [pH 8.0], 0.5 M NaCl, 50 mM imidazole) and sonicated on ice (10 10-s pulses at 10-s intervals), and the lysates were clarified by centrifugation at 22,000 rpm (SW55Ti rotor) for 30 min at 4°C. HMG protein was purified from the supernatant by Ni2+-nitrilotriacetic acid affinity chromatography, dialyzed into buffer C (25 mM Tris [pH 8.0], 150 mM KCl, 2 mM dithiothreitol, 10% glycerol), dispensed into aliquots, snap frozen on liquid nitrogen, and stored at −70°C until use. Commercially available rabbit polyclonal anti-HMG-1 and anti-HMG-2 antibodies were used for immunoblotting and supershift experiments (BD PharMingen).
Oligonucleotide binding and cleavage assays.
The assembly and purification of intact and prenicked radiolabeled 12- and 23-RSS substrates used in binding and cleavage assays have been described previously (12, 28). Binding-reaction mixtures (10 μl) were assembled in the presence of Ca2+ as previously described (30) and contained purified RAG-1 (∼20 ng) and RAG-2 (∼20 ng, where indicated) fusion proteins, purified polyhistidine-tagged HMG-1 or HMG-2 (5 μg/ml final concentration, unless noted), and either an intact or prenicked RSS substrate (0.02 pmol). For in-gel cleavage experiments, the binding-reaction mixtures were scaled up fivefold. Protein-DNA complexes were fractionated on 4% native polyacrylamide gels in 0.5× Tris-borate-EDTA buffer. For DNA binding experiments, the gels were dried and protein-DNA complexes were visualized by autoradiography using a PhosphorImager (Storm 860; Molecular Dynamics). For in-gel cleavage experiments, the polyacrylamide gel was soaked in cleavage buffer (25 mM morpholinepropanesulfonic acid [MOPS]-KOH [pH 7.0], 60 mM potassium glutamate, 5 mM MgCl2) and incubated at 37°C for 1 h to initiate cleavage in fractionated protein-DNA complexes. Reaction products were recovered from the gel and fractionated by denaturing polyacrylamide gel electrophoresis (PAGE) using a previously published procedure (30). PhosphorImager scans of these gels were quantified using the ImageQuaNT software.
Modification interference assays.
Oligonucleotides used in DNA footprinting assays were synthesized and purified by denaturing PAGE at the UNMC Eppley Molecular Biology Core facility. Their composition, modification, and assembly into 12- and 23-RSS substrates have been previously described (30). Unmodified RSS substrates or those chemically modified with ethylnitrosourea (ENU), dimethyl sulfate (DMS), or potassium permanganate (KMnO4) were incubated with purified RAG-1 (MR1), RAG-2 (MR2), and either HMG-1 or HMG-2 in preparative binding reactions as described above and fractionated by EMSA. DNA was recovered from discrete RAG complexes as described previously (30). Chemically modified DNA was then treated with 10% piperidine (DMS or KMnO4) or 150 mM NaOH (ENU) to cleave the modified residues. Reaction products were fractionated by denaturing PAGE and analyzed using a PhosphorImager.
RESULTS
Incorporation of HMG protein into a RAG-RSS complex shifts its electrophoretic mobility.
van Gent et al. first demonstrated that supplementing standard RAG in vitro DNA substrate cleavage reaction mixtures with DNA-bending proteins HMG-1 or HMG-2 stimulated the cleavage of 23-RSS substrates but not of 12-RSS substrates (32). They and others have shown that supplementing standard RAG in vitro DNA binding reaction mixtures with HMG-1 or HMG-2 results in the appearance of supershifted RAG-RSS complexes in an EMSA (1, 15, 32). I wished to further biochemically characterize these supershifted complexes and directly assess their catalytic activity. Toward this end, hexahistidine-tagged forms of human HMG-1 and mouse HMG-2 (Fig. 1A) were overexpressed in bacteria and purified from clarified lysates by affinity chromatography using a Ni2+-chelating resin. These proteins were judged to be at least 90% pure by silver staining and were immunoreactive toward appropriate commercially available antibodies against HMG-1 and HMG-2 (data not shown). Catalytically active core portions of RAG-1 and RAG-2, fused at the amino terminus to MBP and containing or lacking carboxy-terminal myc epitope and polyhistidine tags, were also purified (Fig. 1A).
When purified RAG-1 is incubated with a single RSS substrate, the predominant protein-DNA complex observed by EMSA (M1) contains a RAG-1 dimer (29). A supershifted protein-DNA complex (M1-HMG) is observed by EMSA when standard in vitro binding-reaction mixtures containing RAG-1 alone are supplemented with either purified HMG-1 or HMG-2 (Fig. 1B, compare lanes 2-4 to lanes 6-8 [HMG-1] and lanes 10-12 [HMG-2]). The presence of a carboxy-terminal polyhistidine tag on RAG-1 appears to stimulate the formation of the M1-HMG complex (especially with HMG-2), since formation of this complex is less efficient when the polyhistidine tag on RAG-1 is absent (Fig. 1B, compare lane 6 with lanes 7 and 8, and compare lane 10 with lanes 11 and 12). The basis for this phenomenon is unknown, but it may be due to interactions between the polyhistidine tags on the two proteins. The same effect is observed to a lesser extent in complexes containing both RAG proteins (data not shown). Thus, to avoid potential RAG-HMG binding artifacts, forms of RAG-1 and RAG-2 lacking carboxy-terminal tags were used wherever possible for the remaining studies (Fig. 1A, MR1 and MR2, respectively).
Consistent with previous studies, both HMG-1 and HMG-2 are able to bind 12-RSS and 23-RSS substrates in the absence of the RAG proteins (HMG-RSS; Fig. 1C, lanes 3 and 4 [12-RSS] and lanes 13 and 14 [23-RSS]). Under these conditions, a single protein-DNA complex predominates. The HMG1-RSS complex migrates slightly faster than the HMG2-RSS complex, probably due to the presence of an acidic tail on HMG-1 that is absent from the form of HMG-2 used in these experiments. The HMG-RSS complexes are also observed in samples containing the RAG proteins (Fig. 1C, lanes 6, 7, 9, and 10 [12-RSS] and lanes 16, 17, 19, and 20 [23-RSS]). In contrast to a previous report (14), however, no apparent stimulation of HMG-RSS formation was observed in the presence of the RAG proteins.
When assayed with 12-RSS and 23-RSS substrates on the same gel, similar levels of M1-HMG complex formation were observed in reaction mixtures containing RAG-1 alone (Fig. 1C, compare lanes 6 and 7 to lanes 15 and 16). Supershifted RAG-RSS complexes were also formed to similar levels when binding-reaction mixtures containing both RAG proteins were supplemented with HMG-1 or HMG-2 (M1/M2-HMG; Fig. 1C, compare lane 8 with lanes 9 and 10, and compare lane 18 with lanes 19 and 20). However, both M1/M2 and M1/M2-HMG complexes assembled on 12-RSS substrates were routinely three- to fivefold more abundant than those assembled on 23-RSS substrates when equivalent amounts of DNA substrate were used in the binding reaction (Fig. 1C, compare lanes 9 and 10 with lanes 19 and 20; see also Fig. 3B). In this experiment, the M1/M2 complex was not fully supershifted by the addition of HMG to 1 μg/ml in the binding reaction. However, the intent of this experiment was to clearly demonstrate the difference in electrophoretic mobility between the M1/M2 and M1/M2-HMG complexes. Increasing the amount of HMG to 5 μg/ml in the binding-reaction mixtures resulted in a complete supershift of the M1/M2 complex (data not shown, but see Fig. 3B for an example).
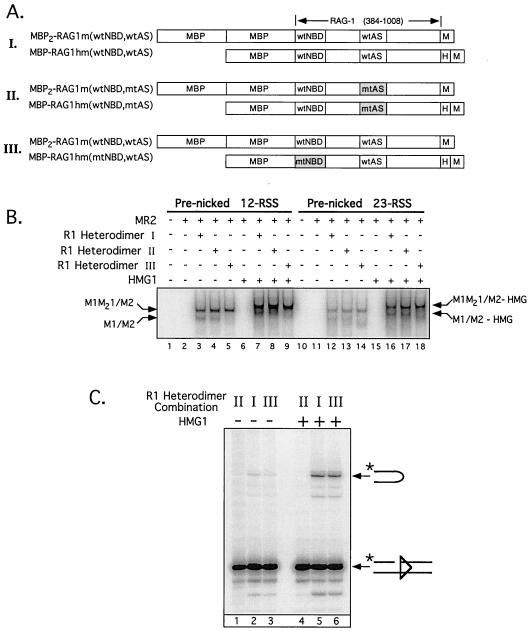
FIG. 3.
HMG-dependent stimulation of RAG-mediated 23-RSS cleavage does not involve synapsis of two 23-RSS substrates to a single RAG-1 dimer. (A) RAG-1 heterodimers (rows I to III) containing wild-type or mutant forms of single and double MBP-RAG-1 (as diagrammed and designated at left) were prepared for a previous study (see reference 28 for details of the composition, purification, and analysis of these proteins). The positions of the NBD (wild type or mutant; wtNBD or mtNBD, respectively) and active site (wild type or mutant; wtAS or mtAS, respectively) are shown. The mtNBD carries alanine substitutions at residues 384 to 393; the mtAS carries the D708A mutation. Note that homodimers of the single but not the double MBP-RAG-1 fusion protein are copurified in this procedure. (B) A 32P-end-labeled, prenicked 12-RSS (lanes 1 to 9) or 23-RSS (lanes 10 to 18) substrate was incubated either alone (lanes 1 and 10, respectively) or with RAG-2 and different RAG-1 heterodimer preparations (I to III) in the absence and presence of HMG-1, as indicated at the top of the gel (with [+] or without [−]) and defined in panel A. Positions of protein-DNA complexes, fractionated by EMSA, containing RAG-2 and only single MBP-RAG-1 subunits (M1/M2) or both single- and double MBP-RAG-1 subunits (M1M21/M2) or their counterparts incorporating HMG-1 (M1/M2-HMG and M1M21/M2-HMG, respectively) are designated. (C) Preparative binding-reaction mixtures were prepared for the samples indicated in panel B, lanes 12 to 14 and 16 to 18 (containing a prenicked 23-RSS substrate), and the protein-DNA complexes were fractionated by EMSA (on the same native gel). Reaction products were recovered after an in-gel cleavage assay from M1M21/M2 (lanes 1 to 3) and M1M21/M2-HMG (lanes 4 to 6) complexes containing the RAG-1 heterodimers indicated above the gel. The positions of nicked and hairpin species are indicated on the right.
Anti-myc antibody supershift experiments using pairwise combinations of myc epitope-tagged and untagged forms of RAG-1 and RAG-2, described previously (30), demonstrate that the HMG-M1/M2 complex contains both RAG proteins (data not shown). The presence of HMG protein in the M1/M2-HMG complex, although inferred from the distinct electrophoretic mobilities of complexes containing HMG-1 or HMG-2, was formally demonstrated for HMG-1 by antibody supershift experiments using a commercially available polyclonal antibody against HMG-1 (data not shown). Finally, RAG-1 was found to retain its dimer configuration in the presence of HMG protein, since EMSA showed that binding-reaction mixtures containing a mixture of single- and double-MBP RAG-1 dimers (29) and HMG-1 or HMG-2 yielded supershifted M1-HMG and M1/M2-HMG complexes resembling those obtained from reaction mixtures lacking HMG in their abundance and distribution (i.e., no species consistent with the formation of a dimer of RAG-1 dimers was observed) (data not shown).
HMG-1 and HMG-2 similarly stimulate RAG-mediated cleavage activity in 23-RSS but not 12-RSS preinitiation complexes.
Others have used mutant DNA substrates to determine how HMG alters RAG binding to RSS substrates (1, 14, 20); therefore, this line of investigation was not pursued. Rather, I chose to examine how HMG alters the intrinsic enzymatic activity of the complex and then attempt to relate these changes to differences in protein-DNA recognition as assessed by DNA footprinting. To address the first issue, I used an in-gel cleavage assay to directly compare enzymatic catalysis in M1/M2 and M1/M2-HMG complexes (28). In this assay, RSS substrate cleavage in native gel-fractionated precleavage RAG-RSS complexes (assembled in the presence of Ca2+) is initiated by soaking the whole gel (after electrophoresis) in cleavage buffer containing Mg2+ for 1 h at 37°C. Cleavage products are subsequently recovered from discrete protein-DNA complexes and analyzed by denaturing PAGE.
In the first experiment, the catalytic activities of M1/M2 and M1/M2-HMG complexes assembled on either an intact 12- or 23-RSS substrate were directly compared (Fig. 2A). As expected, an M1/M2 complex obtained with a RAG-1 active-site mutant (D600A) was essentially inactive on either RSS substrate (Fig. 2A, lanes 1 and 5). A wild-type M1/M2 complex catalyzed both nicking and hairpin formation (the latter to a small extent) on a 12-RSS substrate (lane 2). Significant nicking was also observed in an M1/M2 complex formed with a 23-RSS substrate, although the amount of nicked product obtained was about fivefold less than that observed from the M1/M2 complex assembled on a 12-RSS substrate (compare lanes 2 and 6). The hairpin species obtained with the 12-RSS substrate was virtually undetectable when the 23-RSS substrate was used, although two other cleavage products whose composition has not been determined were observed in this vicinity.
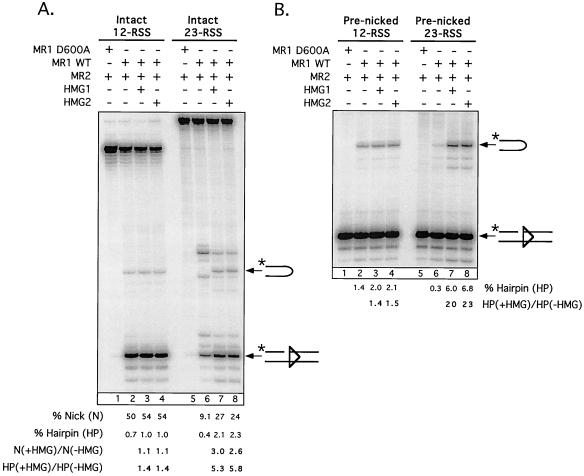
FIG. 2.
HMG proteins stimulate RAG-mediated cleavage of 23-RSS but not 12-RSS substrates in M1/M2-HMG complexes. Preparative binding-reaction mixtures containing an intact (A) or prenicked (B) 12-RSS (left) or 23-RSS (right) substrate, wild-type or catalysis-deficient (D600A) RAG-1, RAG-2, and either HMG-1 or HMG-2 in the combinations indicated above the gel (with [+] or without [−]) were assembled, and protein-DNA complexes were fractionated by EMSA. Catalysis was initiated in the gel by addition of Mg2+ (see Materials and Methods). Cleavage products were recovered from M1/M2 (lanes 1, 2, 5, and 6) or M1/M2-HMG (lanes 3, 4, 7, and 8) complexes and fractionated by denaturing gel electrophoresis. The positions of nicked and hairpin species are indicated on the right. Quantitative analysis of substrate cleavage is shown below the gel and is representative of several experiments. Note that all eight samples represented in panels A and B were recovered from a single native gel subjected to the in-gel cleavage reaction.
The M1/M2-HMG complexes assembled on a 12-RSS substrate did not exhibit significantly (< twofold) different cleavage activity from their M1/M2 counterpart (Fig. 2A, compare lane 2 with lanes 3 and 4). However, the M1/M2-HMG complexes assembled on a 23-RSS showed marked stimulation of both nicking and transesterification activity (∼threefold and >five fold, respectively, but see below) relative to their M1/M2 counterpart (compare lane 6 with lanes 7 and 8). The presence of HMG clearly altered the pattern and distribution of the cleavage products obtained, with formation of the predicted nicked and hairpin products being favored over other species. Similar patterns of cleavage products were obtained when binding-reaction mixtures prepared for EMSA (Fig. 2A) were supplemented with fivefold excess Mg2+ and incubated at 37°C for 1 h without prior gel fractionation of the protein-DNA complexes (data not shown), suggesting that the cleavage specificity intrinsic to a given RAG-RSS-(HMG) complex is similar whether the complex is in solution or embedded in a native polyacrylamide gel.
The presence of a reaction product derived from the M1/M2 complex whose mobility is slightly higher than that of the hairpin species made accurate quantification of this band difficult for comparative purposes. Therefore, a second experiment was performed that was identical to the first, except that the RSS substrates used in the assay were prenicked rather than intact, which enabled me to specifically analyze hairpin formation (Fig. 2B). The results were consistent with data obtained using intact RSS substrates, but they more effectively demonstrated that HMG-1 and HMG-2 promote hairpin formation on 23-RSS, but not 12-RSS, substrates, stimulating this activity ∼20-fold. It is important to note that in all of these experiments, HMG-1 and HMG-2 stimulated RAG-mediated cleavage activity to essentially the same degree.
HMG-dependent stimulation of RAG-mediated 23-RSS cleavage does not involve synapsis of two 23-RSS substrates by a single RAG-1 dimer.
Previous studies have demonstrated that detectable hairpin product formation is observed when an isolated 23-RSS substrate is incubated in a standard RAG in vitro cleavage reaction mixture (containing Mg2+) that is supplemented with HMG protein (32). Similar results were found in this study. The basis for this observation is unclear, but a recent study suggests that synapsis of two identical 23-RSS substrates in a “paired complex” might enable the complex to override constraints that normally prevent hairpin formation from occurring when individual RSSs are physically prevented from forming a paired complex (35). Alternatively, or in addition, van Gent et al. have speculated that the HMG proteins may distort the DNA in the RAG-RSS complex, thereby facilitating the catalysis of hairpin formation (32).
If a single RAG-1 dimer can support synapsis of RSS pairs through RAG-RSS interactions mediated by each of the RAG-1 subunits, as one model has suggested (28, 29), the former possibility could be addressed by analyzing the cleavage activity of M1/M2-HMG complexes that cannot effectively support synapsis due to mutation in a requisite DNA binding domain of one of the RAG-1 subunits. To explore this possibility, RAG-1 heterodimers were prepared in which one RAG-1 subunit possesses a mutant nonamer binding domain (NBD) that impairs RSS binding by disrupting RAG-1-nonamer interactions. This approach, described previously (28), involves cotransfecting single or double MBP-RAG-1 expression constructs into 293 cells in pairwise combinations and purifying the protein by amylose affinity chromatography followed by affinity chromatography over a Ni2+-chelating resin. RAG-1 heterodimers and single MBP-RAG-1 homodimers were recovered by this two-step purification process (double MBP-RAG-1 homodimers were not retained), since only the single MBP-RAG-1 fusion protein contains a carboxyl-terminal polyhistidine tag (Fig. 3A). Fully wild-type and active-site mutant (D708A) RAG-1 heterodimers were prepared similarly (29). The composition of each RAG-1 heterodimer used for these experiments is shown in Fig. 3A (rows I to III).
The ability of the RAG-1 dimers to bind a 32P-labeled prenicked 12- or 23-RSS substrate in the presence of RAG-2 with or without the addition of HMG-1 was assessed by an EMSA (Fig. 3B). As expected from previous results (28), both the RAG-1 homodimers and the RAG-1 heterodimers obtained from transfections I and II assembled RSS complexes containing RAG-2 (M1/M2 and M1M21/M2, respectively) (Fig. 3B, lane 3 and 4 [12-RSS] and 12 and 13 [23-RSS]), since both RAG-1 dimers contain at least one RAG-1 subunit with an intact NBD. No significant M1/M2 complex was observed from samples obtained from transfection III (Fig. 3B, lane 5 [12-RSS] and lane 14 [23-RSS]), since the copurified RAG-1 homodimer bears mutant NBDs on both subunits, thereby precluding stable protein-DNA complex formation. Importantly, all three RAG-1 heterodimers supported the formation of a supershifted complex in the presence of HMG-1 (M1M21/M2-HMG; Fig. 3B, lanes 7 to 9 [12-RSS] and 16 to 18 [23-RSS]), suggesting that HMG-1 can interact with a RSS complex containing both RAG proteins, even if one RAG-1 subunit cannot bind DNA.
To determine whether HMG-dependent stimulation of RAG-mediated transesterification activity on 23-RSS substrates requires an intact NBD on both RAG-1 subunits, M1M21/M2 and M1M21/M2-HMG complexes assembled on a prenicked 23-RSS substrate using RAG-1 heterodimers obtained from transfections I to III were directly compared using the in-gel cleavage assay (Fig. 3C). Consistent with previous results, M1M21/M2 and M1M21/M2-HMG complexes assembled using a RAG-1 heterodimer bearing an active-site mutation on both subunits failed to support substrate cleavage (Fig. 3C, lanes 1 and 4, respectively). Importantly, M1M21/M2-HMG complexes assembled using wild-type RAG-1 heterodimers or heterodimers bearing a mutant NBD on a single RAG-1 subunit exhibited similar levels of enhanced cleavage activity to those of their M1M21/M2 counterparts (Fig. 3C, compare lanes 2 and 3 to lanes 5 and 6). This result suggests that the ability of HMG to stimulate cleavage may be an intrinsic property of HMG when incorporated into a complex containing both RAG proteins bound to a single 23-RSS.
HMG-1 and HMG-2 similarly promote heptamer occupancy on 23-RSS substrates.
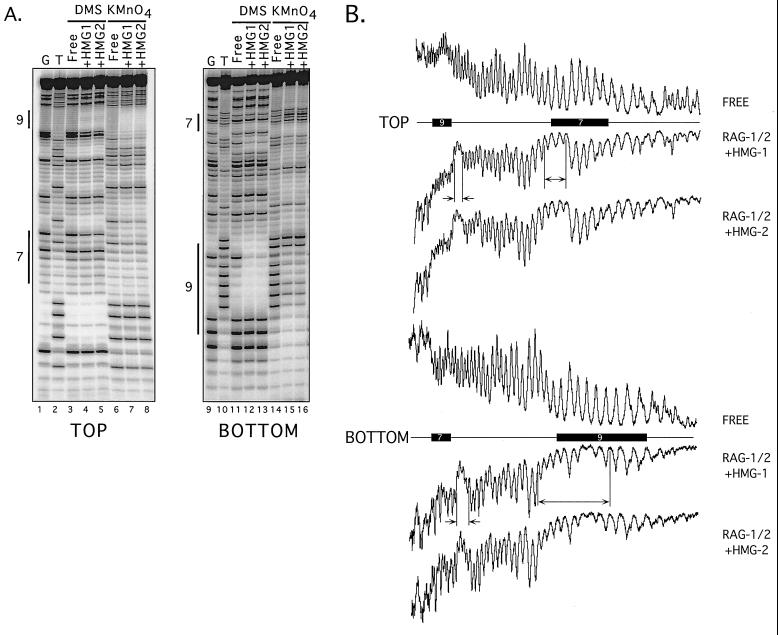
I next wished to determine whether DNA substrate recognition is altered when HMG-1 or HMG-2 is incorporated into RSS complexes containing both RAG proteins. Therefore, DNA contacts in M1/M2-HMG complexes assembled on intact 12- or 23-RSS substrates were mapped by DNA footprinting, an approach I had previously used to define protein-DNA contacts in discrete RSS complexes containing RAG-1 alone or both RAG-1 and RAG-2 (30). For modification interference experiments, 12- or 23-RSS substrates were prepared whose top or bottom strand (top strand is nicked by the RAGs) has been modified with ENU (which ethylates phosphates), DMS (which reacts with guanine to form N-7 methylguanine), or potassium permanganate (KMnO4) (which reacts with thymine to form a thymine glycol). Modified RSS substrates were incubated with the RAG proteins in binding-reaction mixtures (containing Ca2+) supplemented with HMG-1 or HMG-2, and protein-DNA complexes were separated by an EMSA. DNA was recovered from M1/M2-HMG complexes and chemically treated to break the DNA at modified residues. Cleavage products derived from bound and free DNA were compared on a denaturing polyacrylamide gel. Underrepresentation or overrepresentation of a cleavage product in bound DNA, relative to free DNA, indicates that the modified residue inhibits or promotes, respectively, the formation of the M1/M2-HMG complex.
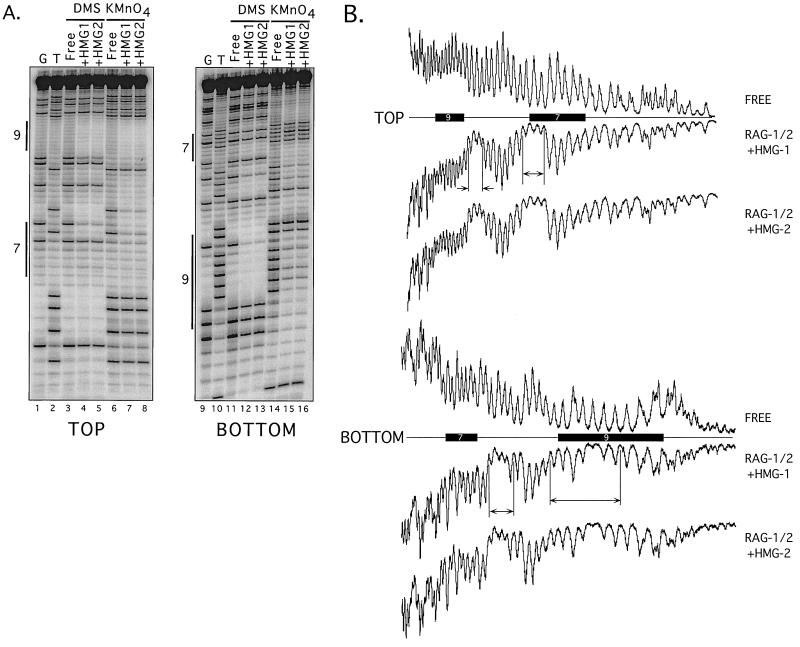
The data obtained from modification interference footprinting of M1/M2-HMG-1 and M1/M2-HMG-2 complexes assembled on 12-RSS substrates reveal that the patterns of protein-DNA contacts were qualitatively and quantitatively similar to each other (Fig. 4; also Fig. 6 and data not shown). Phosphate and base-specific contacts extend from the 3" end of the heptamer through the spacer region and include all but the last two or three residues of the nonamer. On both the top and bottom strands, two regions of ethylation interference are observed. On the top strand, the two regions each span three residues; one region encompasses the two residues at the 3" end of the heptamer and the first residue in the spacer and the other region includes three residues in the spacer arm located 5" of the nonamer. On the bottom strand, four residues in the spacer arm proximal to the heptamer exhibit ethylation interference, as do seven residues spanning the 5" end of the nonamer and adjacent spacer arm. Consistent with earlier results (30), these phosphate contacts are biased toward one side of the DNA helix (see Fig. 6) (data not shown).
FIG. 4.
Modification interference footprinting of 12-RSS complexes containing RAG-1, RAG-2, and either HMG-1 or HMG-2. (A) DMS and KMnO4 modification interference. Piperidine-generated cleavage products from free DNA or DNA recovered from M1/M2-HMG complexes containing HMG-1 (+HMG1) or HMG-2 (+HMG2) were fractionated by denaturing gel electrophoresis and visualized using a PhosphorImager. Reaction products in which the DNA was radiolabeled and modified with DMS or KMnO4 on the top (left) or bottom (right) strand are shown, as well as guanine- (G) and thymine- (T) specific sequencing tracts. Heptamer (shown as 7 on the left) and nonamer (shown as 9) sequences are indicated by vertical bars. Quantitative analysis of the distribution of cleavage products is found in Fig. 6. (B) Ethylation interference. PhosphorImager traces of alkali-generated cleavage products obtained from free DNA or DNA bound in an M1/M2-HMG complex are shown from samples in which the RSS substrate was radiolabeled on the top (upper) or bottom (lower) strand. Horizontal bars delineate the positions of heptamer (bar labeled 7) and nonamer (bar labeled 9) sequences, determined by comparison to a G-specific sequencing tract. Regions of interference are bounded by vertical bars and marked by arrows.
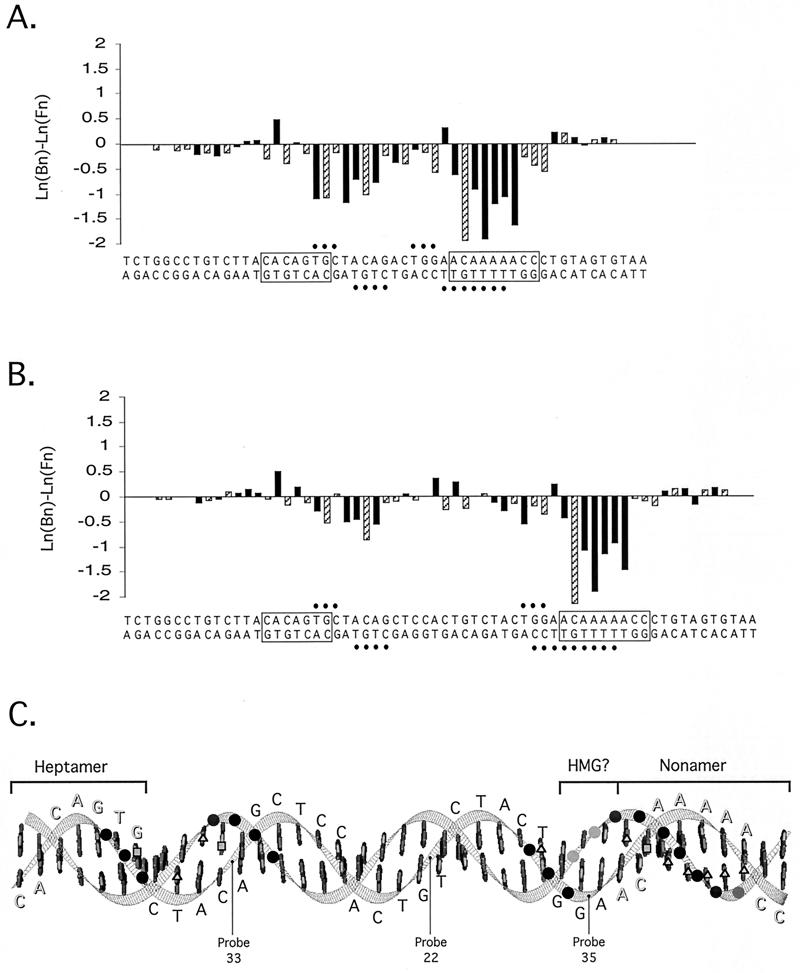
FIG. 6.
Summary of protein-DNA contacts in 12- and 23-RSS complexes containing RAG-1, RAG-2, and HMG-1. (A and B) Patterns of modification interference in M1/M2-HMG-1 complexes on 12-RSS (A) and 23-RSS (B) substrates. The magnitude of DMS (hatched bars) and KMnO4 (solid bars) interference was quantified from data shown in Fig. 4A and 5A by subtracting the natural logarithm of the fractional abundance of the cleaved product at each position (n) in free DNA [ln (Fn)] from its counterpart in bound DNA [ln (Bn)]. The resulting values for ln (Bn) - ln (Fn) are presented in bar graph format above the corresponding positions in the RSS. Negative or positive values reflect interference or enhanced binding, respectively. Sites of ethylation interference (solid circles) are summarized from data shown in Fig. 4B and 5B. Quantitation of data obtained from RAG-RSS complexes containing HMG-2 yielded essentially identical results (data not shown). (C) Arrangement of protein-DNA contacts on a 23-RSS substrate. A ribbon diagram of the minimal 23-RSS (as linear, B-form DNA) was generated on a Silicon Graphics workstation running the MidasPlus software package (8) (Computer Graphics Laboratory, University of California, San Francisco). The sequence of the top strand is shown; residues in the heptamer and nonamer are shadowed, and the two motifs are overlined. Phosphodiesters exhibiting interference when modified with ENU are shown as shaded circles: those arrayed on the front face of the DNA are darkly shaded, while those on the back face are lightly shaded. Significant base-specific contacts [ln (Bn) - ln (Fn) < −0.5] defined in this study by DMS (squares) or KMnO4 (triangles) interference footprinting are highlighted on the appropriate residue. The positions of selected phosphodiesters derivatized in another photo-cross-linking study (14) are also shown. A putative site of HMG contact is overlined. Note that this model is not meant to imply the actual positions of the protein-DNA contacts in the M1/M2-HMG complex, since DNA bending induced by the RAG and HMG proteins is expected to alter their relative orientation (1).
The patterns of protein-DNA contacts obtained from 23-RSS M1/M2-HMG-1 and M1/M2-HMG-2 complexes were also very similar to one another (Fig. 5; also Fig. 6 and data not shown). In contrast to the 23-RSS M1/M2 complex examined previously (30), both base-specific and phosphate contacts are clearly detected on the top and bottom strands near the 5" end of the spacer and the abutting heptamer element of the 23-RSS. The phosphate contacts in and near the heptamer encompass the same positions as those observed in the 12-RSS M1/M2-HMG complex, as do the phosphate contacts located 5" of the nonamer on the top strand. Interestingly, on the bottom strand, the region of ethylation interference previously observed at the nonamer in the 23-RSS M1/M2 complex (30) is expanded in the M1/M2-HMG complexes to include three additional residues at the nonamer-proximal end of the spacer.
FIG. 5.
Modification interference footprinting of 23-RSS complexes containing RAG-1, RAG-2, and either HMG-1 or HMG-2. Patterns of DMS and KMnO4 interference (A) and ethylation interference (B) are displayed as described the legend to in Fig. 4.
The pattern of modification interference in the 23-RSS M1/M2-HMG complex qualitatively resembled that found in the 12-RSS M1/M2-HMG complex at the corresponding positions, but the degree of base-specific interference was quantitatively not as severe in and near the heptamer element (Fig. 6). Modified thymine residues at two comparable positions, one in the heptamer proximal to the coding end and one in the spacer arm abutting the nonamer (both on the bottom strand), were overrepresented to the same degree in both 12-RSS and 23-RSS M1/M2-HMG complexes (Fig. 6), suggesting that structural perturbations induced by KMnO4 at these positions enhance complex formation to a similar extent. As seen in the 12-RSS M1/M2-HMG complex, the phosphate contacts observed in the 23-RSS M1/M2-HMG complex were arrayed largely on one face of the DNA; the exceptions are the phosphate contacts located at the 5" end of the nonamer, which extend toward the opposite side of the duplex (Fig. 6C).
DISCUSSION
HMG-1 and HMG-2 similarly promote 23-RSS substrate recognition and cleavage by the RAG proteins.
Studies from several laboratories have established that the DNA-bending proteins HMG-1 and HMG-2 stimulate the binding and cleavage of isolated RSS substrates (especially those containing a 23-RSS) by the RAG proteins (1, 14, 20, 32). In reviewing these studies, however, one finds that the RAG-RSS-HMG complexes detected by EMSA have not been directly tested for cleavage activity. Hence, it is still unclear whether HMG-dependent stimulation of RAG-mediated cleavage occurs in the context of a discrete RAG-RSS-HMG complex detectable by EMSA or, rather, occurs in some higher-order protein-DNA complex that is unstable toward electrophoresis conditions. Moreover, while RAG-RSS contacts have been extensively mapped using DNA footprinting and photo-cross-linking techniques (2, 6, 15, 16, 29, 30), less is known about how incorporation of HMG into a RAG-RSS complex alters substrate DNA recognition at the molecular level.
To address these issues, I examined the DNA substrate cleavage activity and protein-DNA contacts in discrete RAG-RSS-HMG complexes using a combination of in-gel cleavage assays and DNA footprinting techniques. The results presented here extend previous studies by demonstrating that (i) HMG-1 and HMG-2 similarly stimulate cleavage (and preferentially promote transesterification) of 23-RSS substrates, but not 12-RSS substrates, in a defined RAG-RSS-HMG complex detectable by EMSA; (ii) HMG-dependent stimulation of RAG-mediated hairpin formation on 23-RSS substrates does not proceed via synapsis of two RSS substrates by a single RAG-1 dimer; (iii) enhanced RAG-mediated cleavage of 23-RSS substrates in discrete RAG-RSS-HMG complexes is correlated with the acquisition of stable protein-DNA contacts in and near the heptamer element that are otherwise absent in 23-RSS complexes lacking HMG protein; and (iv) the patterns of protein-DNA contacts in 12- and 23-RSS complexes containing the RAG proteins and either HMG-1 or HMG-2 are strikingly similar to each other at comparable positions within the RSS.
Comparison of protein-DNA contacts in RAG-RSS complexes lacking or containing HMG-1 or HMG-2.
Previously, I examined protein-DNA contacts in 12- and 23-RSS complexes containing only RAG-1 and RAG-2 (30). A comparison between the data presented here and those obtained in the earlier study reveals that HMG proteins exert some obvious and some subtle effects on RAG recognition of recombination signals. With respect to 12-RSS substrates, the patterns of DMS and KMO4 interference in 12-RSS M1/M2-HMG complexes are qualitatively and quantitatively similar to those found earlier in 12-RSS M1/M2 complexes. Interestingly, however, subtle differences between the complexes are observed in the ethylation interference patterns. While phosphate contacts in the M1/M2-HMG complex are located in the same region as in the M1/M2 complex, they are more restricted in the former complex. This observation suggests the possibility that HMG alters the conformation or orientation of the RAG proteins in the 12-RSS M1/M2-HMG complex, thereby reducing nonspecific RAG interactions with the phosphate backbone.
The contrast is more striking when 23-RSS M1/M2 and M1/M2-HMG complexes are compared. In the earlier study, modification interference in and near the heptamer was not evident in the 23-RSS M1/M2 complex. In the 23-RSS M1/M2-HMG complex, however, both base-specific and phosphate contacts are clearly observed in this region. The similarity between the heptamer contacts observed in the 23-RSS M1/M2-HMG complex and those seen previously in the 12-RSS M1/M2 complex (lacking HMG-1 or HMG-2) strongly suggests that the HMG-dependent heptamer contacts detected in the former complex are mediated by the RAG proteins themselves rather than by HMG protein. In addition, the close resemblance between the patterns of contacts in 12- and 23-RSS M1/M2-HMG complexes indicates that HMG plays an important role in enabling the RAG proteins to recognize the same elements on both RSSs. It is interesting that, in general, the same modified thymine residues overrepresented in the 12- and 23-RSS M1/M2 complexes are also overrepresented in their M1/M2-HMG counterparts, except for those near the heptamer-proximal end of the spacer in the 23-RSS M1/M2-HMG complex (where interference is now observed). However, the magnitude of the overrepresentation in the M1/M2-HMG complexes is somewhat lower than in their M1/M2 counterparts. One possible explanation for this observation is that DNA bending induced or stabilized by the HMG proteins may offset enhanced binding promoted by alterations in DNA structure imposed by base modification.
Localization and role of HMG proteins in RAG-RSS-HMG complexes.
The results from in-gel cleavage assays presented here confirm the previous observation that HMG proteins selectively stimulate RAG-mediated catalysis of transesterification on 23-RSS substrates in the presence of Mg2+ (32). Two lines of evidence suggest that HMG protein may exert this effect at a step subsequent to RAG-RSS-HMG complex formation. First, RAG binding to 23-RSS substrates is three- to fivefold less efficient than to 12-RSS substrates, regardless of whether HMG is present, yet HMG stimulates RAG-mediated transesterification more that 20-fold in 23-RSS complexes while having minimal effects on 12-RSS substrate cleavage. Thus, RAG binding and cleavage activity are clearly dissociable from one another. Second, the protein-DNA contacts at and near the 23-RSS heptamer in RAG-RSS-HMG complexes, although requiring the presence of HMG protein before they can be clearly visualized, are nevertheless strikingly similar to those found in RAG-RSS complexes assembled on 12-RSS substrates, where HMG-dependent stimulation of substrate cleavage is not observed. Since the binding experiments were performed in the presence of Ca2+, one could explain the distinct cleavage activities on 12- and 23-RSS substrates if the RAG-HMG complex undergoes RSS substrate-dependent conformational changes in the presence of Mg2+, altering the way in which it interacts with the RSS prior to cleavage in ways that differ according to the length of the RSS spacer arm.
Alternatively or in addition, the HMG-dependent stimulation of 23-RSS hairpin activity in Mg2+ could be occurring in the context of a complex containing a pair of 23-RSS signals. However, RAG-1 heterodimers in which the NBD on one of the RAG-1 subunits was mutated to prevent the binding of two RSS substrates to the same RAG-1 dimer were found to support HMG-dependent stimulation of 23-RSS substrate cleavage as well as wild-type heterodimers, suggesting that HMG has an intrinsic ability to promote RAG-mediated cleavage of 23-RSS substrates in the absence of synapsis. The level of hairpin formation is quite low under these conditions, about 6% of bound DNA. These data are consistent with those published in another report demonstrating that RAG-HMG complexes preassembled (in Ca2+) on 23-RSS substrates that are physically isolated from one another to prevent synapsis support low but detectable levels of hairpin formation when cleavage is initiated by addition of Mg2+ (35). Taken together, these data raise the possibility that RAG-HMG complexes may support low levels of single-site cleavage at RSSs in vivo, generating DNA ends that could be a source of chromosomal translocations.
Where is HMG protein localized within the RAG-RSS-HMG complex? The similarity between the pattern of protein-DNA contacts in 12-RSS M1/M2 complexes described previously (30) and M1/M2-HMG complexes analyzed here suggests that recognition of the 12-RSS in the M1/M2-HMG complex is largely mediated by the RAG proteins. In contrast, the expansion of phosphate contacts in the spacer region in 23-RSS M1/M2-HMG complexes compared to 23-RSS M1/M2 complexes may be attributed to HMG binding. Localization of HMG protein to this site is attractive for two reasons. First, this location is adjacent to positions in the nonamer contacted by the NBD of RAG-1 (5, 27), which interacts with HMG protein (1). Second, the phosphate contacts occur on the side opposite those attributed to the RAG proteins (see Fig. 6C). The binding of HMG protein to this site is predicted to promote the flexure of the DNA away from the HMG-RSS interface (for reviews, see references 4 and 31) and thereby facilitate coordinated heptamer and nonamer contact by the RAG proteins. A similar model has been recently proposed to explain the process of enhanceosome assembly (7). In that study, the effect of HMG-1 on the binding of Epstein-Barr virus activator ZEBRA to noncontiguous ZEBRA recognition sites within the BHLF-1 promoter was analyzed. The authors concluded that HMG-1 localizes between two adjacent ZEBRA sites and, by bringing the two sites into direct apposition, promotes cooperative binding of ZEBRA. In contrast to RAG-1, however, specific protein-protein interactions between HMG-1 and ZEBRA have not been detected (7). Thus, HMG-1 and HMG-2 appear to promote RSS binding by the RAG proteins through two mechanisms: by increasing the protein surface area contacting DNA via specific protein-protein interactions with RAG-1, analogous to HMG-facilitated DNA binding of certain transcription factors (4, 31), and by promoting and/or stabilizing a bent DNA structure that brings noncontiguous heptamer and nonamer recognition elements into an appropriate alignment for cooperative RAG binding, comparable to the process of enhanceosome assembly.
Recently, a photo-cross-linking study was reported that used RSS substrates containing photoreactive aryl azide moieties appended to specific phosphodiesters along the DNA backbone to probe protein-DNA contacts in RAG-RSS-HMG complexes (14). The authors concluded that HMG-1 is localized to the 3" end of the heptamer and adjacent residues within the spacer region in 12-RSS RAG-HMG complexes but is localized primarily to the spacer region in analogous 23-RSS complexes. The data presented here, in contrast, do not provide any evidence that HMG protein contacts the 12-RSS at or near the heptamer and do not indicate a significant functional role for HMG-1 or -2 in 12-RSS binding or cleavage (e.g., bending). The lack of heptamer contacts potentially attributable to HMG protein might reflect the choice of DNA-footprinting techniques used here. However, the length of the aryl azide moiety used in the previous cross-linking study (∼11 ÅA) (34) raises the possibility that HMG-1 does not directly interact with the RSS heptamer but, rather, is simply positioned within the radius of the cross-linker as a result of protein-protein interactions between HMG and the RAG-1 NBD (on either or both subunits of the RAG-1 dimer). The previous study did not reveal significant cross-linking near the spacer-proximal end of the nonamer in 23-RSS complexes containing both RAG proteins and HMG-1. However, only one position in this region was derivatized (probe 35 [Fig. 6C]). Closer inspection of its position in the duplex reveals that the probe is located on the DNA face opposite where HMG is putatively localized in this study, potentially limiting its ability to cross-link HMG-1. Interestingly, probes in the spacer region that are positioned on the same DNA face as the phosphate contacts attributed to HMG protein in this study readily cross-link HMG-1 (probes 22 and 33 [Fig. 6C]).
Acknowledgments
I thank Garrett Soukup for help in constructing DNA models and for critical reading of the manuscript. I am grateful to Mark Schlissel, David Schatz, Thomas Wirth, and Robert Roeder for providing valuable reagents. I appreciate the expert technical assistance of Lei Wang.
This work was supported by a grant to P.C.S. from the American Cancer Society (RSG-01-020-01-CCE) and by the Health Future Foundation.
REFERENCES
- 1.Aidinis, V., T. Bonaldi, M. Beltrame, S. Santagata, M. E. Bianchi, and E. Spanopoulou. 1999. The RAG1 homeodomain recruits HMG1 and HMG2 to facilitate recombination signal sequence binding and to enhance the intrinsic DNA-bending activity of RAG1-RAG2. Mol. Cell. Biol. 19:6532-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akamatsu, Y., and M. A. Oettinger. 1998. Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences. Mol. Cell. Biol. 18:4670-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besmer, E., J. Mansilla-Soto, S. Cassard, D. J. Sawchuk, G. Brown, M. Sadofsky, S. M. Lewis, M. C. Nussenzweig, and P. Cortes. 1998. Hairpin coding end opening is mediated by RAG1 and RAG2 proteins. Mol. Cell 2:817-828. [DOI] [PubMed] [Google Scholar]
- 4.Bianchi, M. E., and M. Beltrame. 1998. Flexing DNA: HMG-box proteins and their partners. Am. J. Hum. Genet. 63:1573-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Difilippantonio, M. J., C. J. McMahan, Q. M. Eastman, E. Spanopoulou, and D. G. Schatz. 1996. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell 87:253-262. [DOI] [PubMed] [Google Scholar]
- 6.Eastman, Q. M., I. J. Villey, and D. G. Schatz. 1999. Detection of RAG protein-V(D)J recombination signal interactions near the site of DNA cleavage by UV cross-linking. Mol. Cell. Biol. 19:3788-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellwood, K. B., Y. M. Yen, R. C. Johnson, and M. Carey. 2000. Mechanism for specificity by HMG-1 in enhanceosome assembly. Mol. Cell. Biol. 20:4359-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrin, T. E., C. C. Huang, L. E. Jarvis, and R. Langridge. 1988. The MIDAS display system. J. Mol. Graph. 6:13-27. [Google Scholar]
- 9.Fugmann, S. D., A. I. Lee, P. E. Shockett, I. J. Villey, and D. G. Schatz. 2000. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol. 18:495-527. [DOI] [PubMed] [Google Scholar]
- 10.Ge, H., and R. G. Roeder. 1994. The high mobility group protein HMG1 can reversibly inhibit class II gene transcription by interaction with the TATA-binding protein. J. Biol. Chem. 269:17136-17140. [PubMed] [Google Scholar]
- 11.Lewis, S. M. 1994. The mechanism of V(D)J joining: lessons from molecular, immunological, and comparative analyses. Adv. Immunol. 56:27-150. [DOI] [PubMed] [Google Scholar]
- 12.Li, W., P. Swanson, and S. Desiderio. 1997. RAG-1 and RAG-2-dependent assembly of functional complexes with V(D)J recombination substrates in solution. Mol. Cell. Biol. 17:6932-6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McBlane, J. F., D. C. van Gent, D. A. Ramsden, C. Romeo, C. A. Cuomo, M. Gellert, and M. A. Oettinger. 1995. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell 83:387-395. [DOI] [PubMed] [Google Scholar]
- 14.Mo, X., T. Bailin, S. Noggle, and M. J. Sadofsky. 2000. A highly ordered structure in V(D)J recombination cleavage complexes is facilitated by HMG1. Nucleic Acids Res. 28:1228-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mo, X., T. Bailin, and M. J. Sadofsky. 1999. RAG1 and RAG2 cooperate in specific binding to the recombination signal sequence in vitro. J. Biol. Chem. 274:7025-7031. [DOI] [PubMed] [Google Scholar]
- 16.Nagawa, F., K. Ishiguro, A. Tsuboi, T. Yoshida, A. Ishikawa, T. Takemori, A. J. Otsuka, and H. Sakano. 1998. Footprint analysis of the RAG protein recombination signal sequence complex for V(D)J type recombination. Mol. Cell. Biol. 18:655-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oettinger, M. A., D. G. Schatz, C. Gorka, and D. Baltimore. 1990. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science 248:1517-1523. [DOI] [PubMed] [Google Scholar]
- 18.Paull, T. T., and M. Gellert. 1998. The 3" to 5" exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell 1:969-979. [DOI] [PubMed] [Google Scholar]
- 19.Paull, T. T., and M. Gellert. 1999. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 13:1276-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodgers, K. K., I. J. Villey, L. Ptaszek, E. Corbett, D. G. Schatz, and J. E. Coleman. 1999. A dimer of the lymphoid protein RAG1 recognizes the recombination signal sequence and the complex stably incorporates the high mobility group protein HMG2. Nucleic Acids Res. 27:2938-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roth, D. B., J. P. Menetski, P. B. Nakajima, M. J. Bosma, and M. Gellert. 1992. V(D)J recombination: broken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell 70:983-991. [DOI] [PubMed] [Google Scholar]
- 22.Roth, D. B., C. Zhu, and M. Gellert. 1993. Characterization of broken DNA molecules associated with V(D)J recombination. Proc. Natl. Acad. Sci. USA 90:10788-10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sadofsky, M. J. 2001. The RAG proteins in V(D)J recombination: more than just a nuclease. Nucleic Acids Res. 29:1399-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schatz, D. G., M. A. Oettinger, and D. Baltimore. 1989. The V(D)J recombination activating gene, RAG-1. Cell 59:1035-1048. [DOI] [PubMed] [Google Scholar]
- 25.Schlissel, M., A. Constantinescu, T. Morrow, M. Baxter, and A. Peng. 1993. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5"-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev. 7:2520-2532. [DOI] [PubMed] [Google Scholar]
- 26.Shockett, P. E., and D. G. Schatz. 1999. DNA hairpin opening mediated by the RAG1 and RAG2 proteins. Mol. Cell. Biol. 19:4159-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spanopoulou, E., F. Zaitseva, F. H. Wang, S. Santagata, D. Baltimore, and G. Panayotou. 1996. The homeodomain region of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell 87:263-276. [DOI] [PubMed] [Google Scholar]
- 28.Swanson, P. C. 2001. The DDE motif in RAG-1 is contributed in trans to a single active site that catalyzes the nicking and transesterification steps of V(D)J recombination. Mol. Cell. Biol. 21:449-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swanson, P. C., and S. Desiderio. 1999. RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol. Cell. Biol. 19:3674-3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swanson, P. C., and S. Desiderio. 1998. V(D)J recombination signal recognition: distinct, overlapping DNA-protein contacts in complexes containing RAG1 with and without RAG2. Immunity 9:115-125. [DOI] [PubMed] [Google Scholar]
- 31.Thomas, J. O., and A. A. Travers. 2001. HMG1 and 2, and related ‘architectural' DNA-binding proteins. Trends Biochem. Sci. 26:167-174. [DOI] [PubMed] [Google Scholar]
- 32.van Gent, D. C., K. Hiom, T. T. Paull, and M. Gellert. 1997. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 16:2665-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Gent, D. C., K. Mizuuchi, and M. Gellert. 1996. Similarities between initiation of V(D)J recombination and retroviral integration. Science 271:1592-1594. [DOI] [PubMed] [Google Scholar]
- 34.Yang, S. W., and H. A. Nash. 1994. Specific photocrosslinking of DNA-protein complexes: identification of contacts between integration host factor and its target DNA. Proc. Natl. Acad. Sci. USA 91:12183-12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu, K., and M. R. Lieber. 2000. The nicking step in V(D)J recombination is independent of synapsis: implications for the immune repertoire. Mol. Cell. Biol. 20:7914-7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zwilling, S., H. Konig, and T. Wirth. 1995. High mobility group protein 2 functionally interacts with the POU domains of octamer transcription factors. EMBO J. 14:1198-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]