Abstract
Herpes simplex virus type 1 (HSV-1) capsids have an icosahedral structure with capsomers formed by the major capsid protein, VP5, linked in groups of three by distinctive structures called triplexes. Triplexes are heterotrimers formed by two proteins in a 1:2 stoichiometry. The single-copy protein is called VP19C, and the dimeric protein is VP23. We have carried out insertional and deletional mutagenesis on VP19C and have examined the effects of the mutations on virus growth and capsid assembly. Insertional mutagenesis showed that the N-terminal ∼100 amino acids of the protein, which correspond to a region that is poorly conserved among herpesviruses, are insensitive to disruption and that insertions into the rest of the protein had various effects on virus growth. Some, but not all, severely disabled mutants were compromised in the ability to bind VP23 or VP5. Analysis of deletion mutants revealed the presence of a nuclear localization signal (NLS) near the N terminus of VP19C, and this was mapped to a 33-amino-acid region by fusion of specific sequences to a green fluorescent protein marker. By replacing the endogenous NLS with that from the simian virus 40 large T antigen, we were able to show that the first 45 amino acids of VP19C were not essential for assembly of functional capsids and infectious virus particles. However, removing the first 63 amino acids resulted in formation of aberrant capsids and prevented virus growth, suggesting that the poorly conserved N-terminal sequences have some as-yet-unidentified function.
Herpesviruses particles have complex structures consisting of four components: envelope, tegument, capsid, and core (7, 24, 41). The core consists of the double-stranded DNA genome, which is packaged into a preformed capsid within the nuclei of infected cells. The capsid is surrounded by a proteinaceous layer of variable thickness called the tegument, and the entire entity is enclosed by a glycoprotein-containing lipid envelope.
The herpes simplex virus type 1 (HSV-1) capsid has been the subject of intense study, and its structure and composition are relatively well characterized compared to the other virion compartments (31). It consists of a protein shell, which has T = 16 icosahedral symmetry. Three distinctive structural elements, designated pentons, hexons, and triplexes, make up the bulk of this shell; 150 hexons form the faces and edges of the icosahedron, while the vertices are formed by the pentons. The triplexes occupy the local threefold positions between hexons and between hexons and pentons. The icosahedral geometry of the capsid largely determines the number of each component, but the precise numbers of pentons and triplexes are uncertain. This is because 1 of the 12 vertices is believed to be the location of a portal complex through which the genome enters and leaves the capsid (16). It seems probable that the portal replaces 1 of the pentons, leaving 11 occupying the remaining icosahedral vertices. Similarly, for the triplex, there are 320 local threefold positions in the capsid, including 5 surrounding each vertex. However, the relationship between the triplexes and the portal is unknown, making it unclear whether triplexes are present at the five positions surrounding the portal.
Assembly of functional capsids is thought to initiate through an interaction between the internal scaffolding protein and the portal (17). Stepwise addition of shell and scaffold complexes then results in the formation of an unstable spherical procapsid (14, 15). This undergoes a maturational reconfiguration (8, 37), during which the scaffolding protein is proteolytically cleaved and removed and the DNA is packaged. The reconfiguration results in altered interactions between capsid proteins and generates a stable polyhedral capsid shell (14, 19, 26).
The HSV-1 capsid shell is primarily composed of four types of protein. Five copies of the major capsid protein, VP5 (149 kDa), form the pentons, and six copies form the bulk of the hexons (18, 46), while six copies of VP26 (12 kDa) occupy the outer surfaces of the hexons (39, 44). The remaining two proteins, VP19C (50 kDa) and VP23 (34 kDa), make up the triplexes. The triplex is a characteristic structural feature of herpesvirus capsids and is a heterotrimer with αβ2 organization, which in HSV-1 is formed by a single molecule of VP19C (α-subunit) and two copies of VP23 (β-subunit) (18, 29). The triplex forms connections with two of the three surrounding capsomers through domains that have been designated the head and tail (11), and all three triplex proteins interact with the capsid floor (14, 43). VP19C is required for efficient transport of VP23 to the nucleus, which is the site of capsid assembly (25). It is also able to transport VP5 into the nucleus, although in HSV-1-infected cells this function is predominantly provided by the scaffolding protein, preVP22a (20), a transient capsid component that is required during assembly.
The structure of the HSV-1 capsid has been determined to ∼8.5 Å by electron cryomicroscopy (43). Detailed examination of this structure, combined with the analysis of particles formed by subsets of the capsid proteins (29, 39, 44, 45), has allowed the boundaries of the component protein molecules to be defined with good precision. However, with the exception of a 60-kDa domain of VP5 that forms the tops of the hexons and pentons (2), the structures of the capsid proteins are unknown at the atomic level, and relatively little is known about the functional organization of the protein sequences. In this paper, we describe mutagenesis of VP19C to analyze the roles of different parts of the protein in virus growth and capsid assembly.
MATERIALS AND METHODS
Plasmids. (i) VP19C insertional mutants.
pFBpCI was made by inserting a BglII-EcoRI fragment containing the human cytomegalovirus (HCMV) promoter/enhancer from pCI-neo (Promega) into BamHI/EcoRI-digested pFASTBAC HTa (Invitrogen). As a first step, a single NotI site in pFASTBAC HTa was removed by digesting the DNA with NotI, making the ends flush with T4 polymerase, and religation. The UL38 open reading frame (ORF) encoding VP19C was isolated from pBJ382 (33) by digestion with XbaI and HincII and ligated into NheI/StuI-digested pFBpCI to produce pUL38FBpCI, which allows expression of VP19C under the control of the HCMV immediate-early (IE) promoter. Insertional mutants were generated using the transposon-based Mutation Generation System (Finnzymes). Following the in vitro transposition reaction, pUL38FBpCI clones containing the entranceposon (M1-Kanr) sequence inserted at random positions, were isolated by selection on kanamycin (50 μg/ml) agar plates. Individual colonies were grown, and the position of the insertion within the pUL38FBpCI DNA was determined by digestion with PstI, which released a fragment of 2,918 bp if the transposon had inserted into or near the UL38 ORF or a fragment of 1,790 bp if it had inserted elsewhere in the plasmid. Plasmid DNAs containing inserts within the UL38 ORF were digested with NotI and religated. This removed the bulk of the transposon sequence, leaving a 15-bp insertion, which introduced five additional amino acids into the protein. The positions of insertions were determined by DNA sequencing.
(ii) VP19C N-terminal truncations.
Truncated UL38 ORFs were generated by PCR using the forward primers UL38-45F and UL38-63F and the reverse primer UL38R (Table 1). The PCR products were purified and cloned into pGEM-T Easy (Promega). Individual colonies were grown and screened by restriction enzyme digestion. Clones with the inserts in the correct orientation were designated pUL38-45T Easy and pUL38-63T Easy. For expression in insect cells, the UL38 sequences were isolated from pUL38-45T Easy and pUL38-63T Easy by digestion with XbaI and PstI and cloned downstream from the polyhedrin promoter in XbaI/PstI-digested pFASTBAC1 (Invitrogen) to generate pUL38-45FB1 and pUL38-63FB1. For expression in mammalian cells, the UL38 sequences were isolated from pUL38-45T Easy and pUL38-63T Easy by digestion with EcoRI and SpeI and cloned downstream from the HCMV IE1 promoter in EcoRI/XbaI-digested pFBpCI to generate pUL38-45FBpCI and pUL38-63FBpCI.
TABLE 1.
PCR primers used in cloning UL38 mutants
| Primer | Sequencea |
|---|---|
| UL38-45F | GTCTAGAGGATCCATATGGGCCCAGGGCTC CCCAGGGGGTC |
| UL38-63F | GTCTAGAGGATCCATATGTTGCTTGGCCTG GACGGCACAGAC |
| UL38R | GACAAGATCTCACGCGCATGCCCGCCACTC GCCGGGG |
| UL38-45PCR F1 | GAAGAAGAAGCGCAAGGTCGACGGCCCAGG GCTCCCCAGGGGGTCG |
| UL38-63PCR F1 | GAAGAAGAAGCGCAAGGTCGACTTGCTTGG CCTGGACGGCACAGACG |
| 38R-Xba | TCTAGATCACGCGCATGCCCGCCACTCGC |
| UL38NLS-45PCR F3 | CGCAATGGCTCCGAAGAAGAAGCGCAAGGT CGACGGCCCAGGGCTCCCCAGGGGGTCGG |
| UL38ATG | GACAGAATTCGCAATGAAGACCAATCCGC |
| UL38AA46 | ACAGTCGACCCGTCGCGGCGAGAGATCGG |
| UL38AA56 | ACAGTCGACCGGGGTCCCGACCCCCTGGG |
| UL38AA66 | ACAGTCGACCCAAGCAACCACAGCGTGC |
| UL38AA76 | ACAGTCGACGCCCCAGGGGGCGCGTCTG |
| UL38AA83 | ACAGTCGACGTATCGTCGTTGGGGGTCAGC |
| UL38AA118 | ACAGTCGACAGATCCGTCAGGATCACTTGG |
| UL38-24 NTERM | GCAATGACACGCGATACCGCGGGACAG |
| UL38-33 NTERM | GCAATGCTTCGGCGCGTCCTGCGCCCC |
| GFP UPSTREAM | GCTGATTATGATCTAGAGTCGC |
The recognition sequences for BamHI (GGTACC), BglII (AGATCT), SalI (GTCGAC), and XbaI (TCTAGA) used in cloning are singly or doubly underlined.
(iii) VP19C-NLS fusions.
PCRs were carried out using the forward primers UL38-45PCR F1 and UL38-63PCR F1 and the reverse primer 38R-Xba (Table 1). UL38-45PCR F1 and UL38-63PCR F1 introduce a SalI site immediately upstream of amino acids 45 and 63 in the UL38 ORF. The PCR products were purified and cloned into pGEM-T Easy to generate pUL38-45PCR-T Easy and pUL38-63PCR-T Easy, respectively. An oligonucleotide, UL38NLS-45PCR F3, was designed that specifies the nuclear localization signal (NLS) of the simian virus 40 (SV40) large T antigen (sequence, PKKKRKV) (10) fused to the N terminus of the UL38 ORF truncated at amino acid 45 (Table 1). A PCR was performed using primers UL38NLS-45PCR F3 and 38R-Xba, with pUL38-45PCR-T Easy as the template. The PCR fragment was purified and cloned into pGEM-T Easy to generate pUL38-45NLS-T Easy. The UL38 insert was isolated by digestion with EcoRI and XbaI and ligated into EcoRI/XbaI-digested pFBpCI to generate pUL38-45NLSFBpCI. The SalI/XbaI fragment from pUL38-63PCR-T Easy was then purified and ligated into SalI/XbaI-digested pUL38-45NLSFBpCI to generate pUL38-63NLSFBpCI.
(iv) VP19C-green fluorescent protein (GFP) fusion.
DNA fragments containing increasing numbers of residues from the 5′ end of the VP19C ORF were produced by PCR using the forward primer UL38ATG and the reverse primers UL38AA46, UL38AA56, UL38AA66, UL38AA76, UL38AA83, and UL38AA118 (Table 1). The appropriate PCR products were purified and cloned into pGEM-T Easy. Individual colonies were grown and screened by digestion with EcoRI. The UL38 inserts were isolated by digestion with EcoRI and SalI and ligated into EcoRI/SalI-digested pEGFP-N1 (Clontech) to generate p46AA-UL38-GFP, p56AA-UL38-GFP, p66AA-UL38-GFP, p76AA-UL38-GFP, p83AA-UL38-GFP, and p118AA-UL38-GFP. To delete sequences from the N terminus, PCR was carried out with p66AA-UL38-GFP DNA as a template and using the forward primers UL38-24 NTERM and UL38-33 NTERM and the reverse primer GFP UPSTREAM. The resulting PCR products were cloned into pGEM-T Easy, and then the inserts were isolated by digestion with EcoRI and NotI and ligated into EcoRI/NotI-digested p66AA-UL38-GFP.
All plasmids were checked by diagnostic restriction enzyme digestion and DNA sequencing.
Cells.
BHK-21 clone 13 cells were grown in Glasgow minimal essential medium (MEM) supplemented with 10% tryptose phosphate broth and 10% newborn calf serum. UL38RSC cells (36) were grown in Dulbecco's MEM supplemented with 10% fetal calf serum and 500 μg/ml Geneticin G418 (Invitrogen) at every fourth pass. U2OS cells were grown in Dulbecco's MEM supplemented with 10% fetal calf serum. Sf21 (Spodoptera frugiperda) cells were cultured in TC100 medium supplemented with 5% fetal calf serum.
Viruses.
The HSV-1 UL38-minus mutant vΔ38YFP was propagated on UL38RSC cells (36).
Baculoviruses.
AcUL38 (expressing full-length VP19C), AcUL19 (VP5), and AcUL26.5 (preVP22a) have been described previously (23, 33). AcAB3.12 (expressing VP23, VP26, and the UL26 protease) was made by cloning the ORFs into the triple expression transfer plasmid pAcAB3 (BD Biosciences, Pharmingen) and recombining them into AcPAK6 (1). Recombinant baculoviruses expressing other VP19C proteins were produced using the BAC-TO-BAC Baculovirus Expression System (Invitrogen) following the manufacturer's protocol. Briefly, pUL38-45FB1, pUL38-63FB1, pUL38FBpCI, pUL38-45NLSFBpCI, and pUL38-63NLSFBpCI were transfected into competent DH10Bac Escherichia coli cells. White colonies were selected on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)-containing agar plates, and the recombinant bacmid DNAs were isolated and transfected into SF21 cells. The progeny baculoviruses were screened for the presence of inserts by PCR. Positive isolates were designated UL38-45FBac, UL38-63FBac, UL38FBacpCI, UL38-45NLSFBacpCI, and UL38-63NLSFBacpCI and were grown to high titer.
Plasmid transfection and baculovirus transduction.
BHK cells for immunofluorescence were grown on 13-mm glass coverslips and transfected with plasmid DNAs using Lipofectin PLUS reagent (Invitrogen) according to the manufacturer's instructions. After 3 h of incubation at 37°C, the transfection reagent was replaced with fresh culture medium and incubation was continued for a further 16 h before the cells were fixed for immunofluorescence.
BHK cells for virus rescue were grown directly in 24-well tissue culture dishes and transfected with plasmid DNA as described above. At 5 h posttransfection, the cells were infected with vΔ38YFP. After 1 h at 37°C, unabsorbed virus was inactivated by incubating the cells in Tris-glycine, pH 3, for 45 s (27). The culture medium was replaced, and the cells were incubated for a further 40 h at 37°C before being harvested into the medium and frozen at −70°C prior to titration on UL38RSC cells.
U2OS cells were overlaid with 50 PFU/cell (titrated on Sf21 cells) of the appropriate baculovirus and incubated at 20°C. After 1 h, the inoculum was removed and the cells were infected with 5 PFU/cell of vΔ38YFP. Incubation was continued at 37°C for 16 h, and the cells were harvested and processed for electron microscopy.
Immunofluorescence.
Transfected BHK cells were fixed for 1 h in methanol at −20°C, washed in phosphate-buffered saline (PBS), and incubated for 10 min in PBS containing 5% newborn calf serum and 0.05% Tween 20 (solution A). The coverslips were drained, inverted onto 20 μl of primary antibody, and incubated at room temperature for 45 min. After three washes in solution A, the coverslips were drained and inverted onto 20 μl of secondary antibody for a further 45 min. The coverslips were washed four times in solution A and twice in deionized water before being mounted in Mowiol 4-88 (Harco) plus 2.5% DABCO (Sigma) and examined on a Zeiss LSM 510 confocal microscope.
Western blot analysis.
Proteins separated by electrophoresis on 10% sodium dodecyl sulfate-polyacrylamide gels were transferred to Hybond ECL nitrocellulose membranes and analyzed using the enhanced-chemiluminescence method (Amersham). Antibody dilutions and membrane washes were carried out in PBS-T (PBS containing 0.05% [vol/vol] Tween 20 [Sigma]). Before addition of the first antibody, the membrane was blocked in PBS-T containing 5% milk powder. Protein A-peroxidase (Sigma) was diluted 1:1,000 in PBS-T.
Antibodies.
Mouse monoclonal antibody mAb02040, raised against purified VP19C, was diluted 1:1,000 in solution A (immunofluorescence) or PBS-T (Western blotting). Rabbit antisera rAb184 and rAb186, against purified VP5 and VP23, respectively, were diluted 1:1,000 in solution A. Secondary antibodies, goat anti-mouse (GAM) fluorescein isothiocyanate (FITC) conjugate and goat anti-rabbit (GAR) tetramethyl rhodamine isothiocyanate (TRITC) conjugate (Sigma), were diluted 1:100 in solution A.
Electron microscopy.
Cells were harvested and pelleted in embedding capsules (TAAB). The cell pellets were fixed with 2.5% glutaraldehyde and 1% osmium tetroxide, dehydrated through a graded alcohol series, and embedded in Epon 812 resin (TAAB). Thin sections were cut using a LEICA ULTRACUT E, stained with uranyl acetate and lead citrate, and examined on a JEOL 100S electron microscope.
RESULTS
Generation and analysis of insertional mutants in VP19C.
Fifty clonally distinct mutants containing 5-amino-acid insertions in the UL38 ORF were generated by transposon-based insertional mutagenesis into pUL38FbpCI, which expresses VP19C from the HCMV IE promoter. The insertions were distributed throughout the ORF with an average spacing of ∼9 amino acids and a maximum separation of 38. Two pairs of insertions (in242A-in242B and in358A-in358B) were at different sites in the same codon. In these cases, the mutant proteins differed only in the identities of the inserted amino acids (Table 2). One insertion (inSTOP) disrupted the TGA stop codon, resulting in the addition of 5 amino acids at the end of the protein.
TABLE 2.
Locations and sequences of five amino acid insertions into VP19C
| AA no.a | AA sequenceb |
|---|---|
| 16 | ggCGRSGst |
| 23 | ppMRPHPtt |
| 24 | ptAAAPTtr |
| 26 | trCGRTRdt |
| 37 | vlLRPHLrp |
| 51 | lpSAAAPrg |
| 80 | tpCGRTPnd |
| 84 | dtDAAATeq |
| 86 | eqVRPQQal |
| 102 | alMRPHLig |
| 106 | spAAASPrh |
| 107 | prAAAPRhh |
| 113 | rqCGRSQvi |
| 143 | hlVRPHLah |
| 182 | raVRPHAgl |
| 183 | agAAAAGlv |
| 193 | aaALVAAca |
| 200 | daLRPHArd |
| 207 | avAAAAVra |
| 217 | rgCGRSGtr |
| 234 | amCGRTMvh |
| 235 | mvAAAMVht |
| 242A | phAAAPHev |
| 242B | phCGRTHev |
| 243 | heCGRNEvm |
| 248 | fgCGRIGgl |
| 286 | avAAAAVil |
| 289 | lpVRPHPac |
| 310 | flLRPHLyl |
| 327 | cvCGRSVyv |
| 328 | vyAAAVYvl |
| 329 | yvMRPHVik |
| 330 | viNAAAIks |
| 338 | rgCGRSGle |
| 343 | alVRPHLer |
| 350 | lrCGRIRit |
| 358A | hgCGRNGte |
| 358B | hgAAAHGte |
| 365 | ppVRPHPap |
| 368 | pnCGRTNrn |
| 390 | agCGRTGqv |
| 396 | pqCGRTQfa |
| 426 | lgAAALGmm |
| 436 | clLRPHLhr |
| 438 | hrMRPHRte |
| 439 | rtDAAATer |
| 449 | pvAAAPVvi |
| 459 | rpGAAAPge |
| 463 | wrVRPQRac |
| STOP | caCAAAA |
AA, amino acid. The number denotes the last unchanged amino acid before the insertion.
The five inserted amino acids are shown in uppercase, and the unchanged flanking residues are in lowercase.
To analyze the effects of the mutations on the properties of VP19C, their abilities to complement growth of the HSV-1 UL38 deletion mutant vΔ38YFP (36) were examined. DNA of each insertional mutant was transfected into BHK cells, which were incubated at 37°C for 5 h before being infected with 2 PFU/cell of vΔ38YFP. After 1 h, the unabsorbed input virus was neutralized by an acid wash. Incubation was continued at 37°C for 40 h, at which time the progeny virus was harvested and titrated on UL38RSC rabbit skin cells expressing VP19C. Figure 1 shows the relative yield obtained for each of the insertional mutants compared to that obtained with the wild-type control.
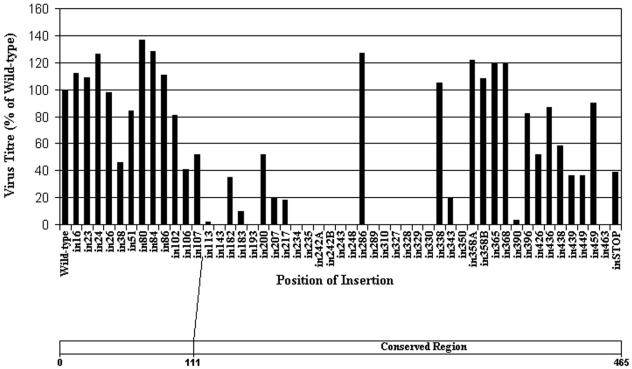
FIG. 1.
Functional analysis of VP19C insertional mutants. (Top) Complementation of growth of the VP19C-minus mutant vΔ38YFP by transfected plasmids expressing the VP19C insertional mutants was carried out in BHK cells. The progeny virus was titrated on UL38RSC cells. The results are shown as percentages of the titer obtained with the wild-type VP19C control plasmid. (Bottom) Scale drawing of the UL38 ORF showing the extent of the poorly conserved N-terminal sequences (Fig. 7a).
Eighteen of the 50 insertional mutants had little or no effect on the ability of VP19C to support virus growth, producing yields of over 80% of the wild-type VP19C control level. The remaining 32 showed lower levels of complementation, with two producing yields of <10% and 16 being completely negative for virus growth. The results shown in Fig. 1 suggest that VP19C can be divided into three broadly defined regions, with the majority of the severely disabling insertions (<10% of wild-type yield) occurring in the central region of the protein, between in113 and in350. The yields obtained with all 12 of the insertions into the first 107 amino acids were >40% of the wild-type level, with only in38 in the middle of this region and in106 and in107 at its C terminus giving <80%. Similarly, all but 2 (in390 and in463) of the 14 insertions into the C-terminal 107 amino acids gave complementation levels of >30%. In contrast, 16 of the 18 insertions that gave complementation values of <10% mapped in the central region of the protein between amino acids 113 and 350, and of the 24 insertions in this region, only 4 gave values of >30%.
Interaction of capsid proteins.
In order to ensure the formation of functional capsids and hence infectious virions, VP19C must interact correctly with the other triplex protein, VP23, and with the major capsid protein, VP5. Since these interactions are essential, they can be assumed to have formed correctly in those VP19C mutants that complemented vΔ38YFP. Therefore, only the 18 mutants that gave less than 10% of the yield of wild-type VP19C and were considered to be severely disabled were examined. Previous studies have shown that VP19C is a nuclear protein. VP23 and VP5, when expressed in the absence of other virus proteins, are uniformly distributed throughout the cell but become predominantly nuclear in the presence of VP19C (25). Furthermore, immunofluorescence analysis of vΔ38YFP-infected BHK cells revealed that VP23 remained distributed throughout the cell (data not shown), demonstrating that its nuclear localization was entirely dependent on the presence of VP19C.
To examine these interactions, BHK cells were cotransfected with plasmids expressing the VP19C mutants and with pE18 (expressing VP23) (20) or with pE19 (VP5) (25), and the distributions of the proteins were determined by immunofluorescence. One of the mutant VP19Cs (in330) was no longer able to locate to the nucleus, making it impossible to determine its effect on the distribution of the other proteins (Fig. 2 and 3).
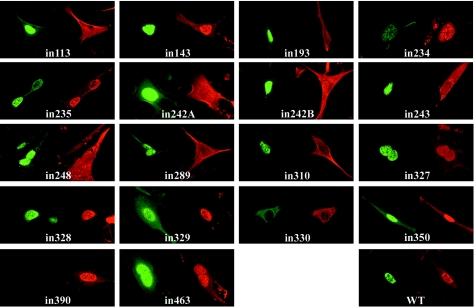
FIG. 2.
Influence of VP19C insertional mutants on the distribution of VP23. BHK cells were cotransfected with pE18 (expressing VP23) and either pUL38FBpCI (wild-type [WT] VP19C) or one of the VP19C insertional mutants as indicated. VP19C was detected with the monoclonal antibody mAb02040 and visualized using FITC GAM (green). VP23 was detected with the antiserum rAb186 and visualized using TRITC-conjugated GAR (red).
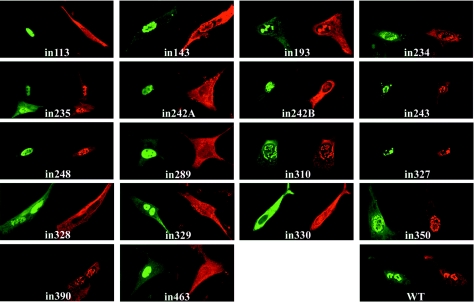
FIG. 3.
Influence of VP19C insertional mutants on the distribution of VP5. BHK cells were cotransfected with pE19 (expressing VP5) and either pUL38FBpCI (wild-type [WT] VP19C) or one of the VP19C insertional mutants as indicated. VP19C was detected with the monoclonal antibody mAb02040 and visualized using FITC GAM (green). VP5 was detected with the antiserum rAb184 and visualized using TRITC-conjugated GAR (red).
When the interaction between VP23 and the remaining 17 severely disabled VP19C mutants was examined in this assay, two distinct patterns were seen (Fig. 2). Nine mutants (in143, in234, in235, in327, in328, in329, in350, in390, and in463) exhibited a wild-type pattern, with VP19C and VP23 colocalizing to the nucleus. In the case of in390, the VP19C antibody was unable to bind to the mutant protein. However, VP23 was clearly transported to the nucleus, implying that the insertion had disrupted the antibody-binding site but had not interfered with the ability of VP19C to interact with VP23 or to localize to the nucleus. In the remaining eight mutants (in113, in193, in242A, in242B, in243, in248, in289, and in310), VP19C localized to the nucleus as expected but VP23 remained in the cytoplasm, implying that the inserted sequences had disrupted the interaction between VP19C and VP23.
Seven severely disabled VP19C mutants (in113, in242A, in242B, in289, in328, in329, and in463) were unable to transport VP5 to the nucleus (Fig. 3). Of these, four (in113, in242A, in242B, and in289) also failed to transport VP23 to the nucleus, while the other three (in328, in329, and in463) interacted successfully with VP23. The results of the VP23 and VP5 analyses are summarized in Table 3.
TABLE 3.
Intracellular distribution of severely disabled VP19C insertional mutants
| VP19C mutant | Localizes to nucleusa | Takes VP23 to nucleusa | Takes VP5 to nucleusa |
|---|---|---|---|
| in113 | + | − | − |
| in143 | + | + | + |
| in193 | + | − | + |
| in234 | + | + | + |
| in235 | + | + | + |
| in242A | + | − | − |
| in242B | + | − | − |
| in243 | + | − | + |
| in248 | + | − | + |
| in289 | + | − | − |
| in310 | + | − | + |
| in327 | + | + | + |
| in328 | + | + | − |
| in329 | + | + | − |
| in330 | − | − | − |
| in350 | + | + | + |
| in390 | + | + | + |
| in463 | + | + | − |
+, yes; −, no.
Role of the VP19C N terminus.
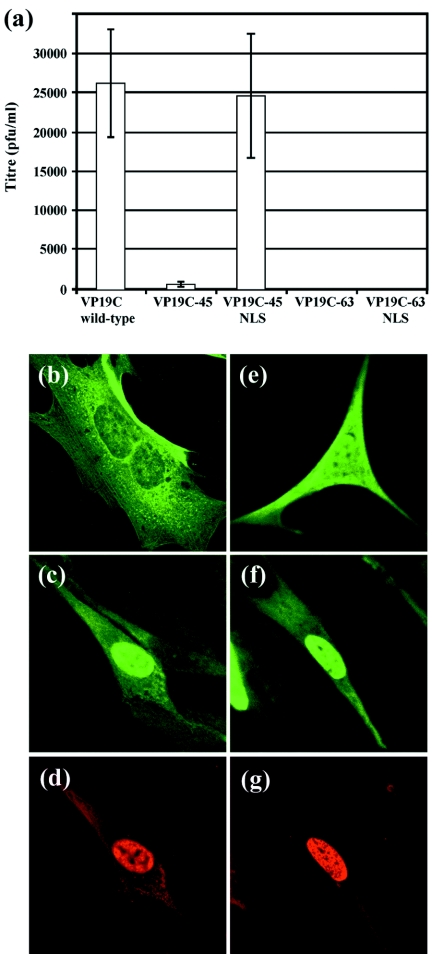
Studies examining HSV-1 capsid formation using a recombinant-baculoviruses-based model system, had shown that the N-terminal 90 amino acids of VP19C were not absolutely required for capsid assembly (30). The results of the complementation analysis (Fig. 1) are consistent with this conclusion, as all of the insertions into the N-terminal 107 codons of the UL38 ORF were able to support virus growth. To investigate the roles of these N-terminal sequences further, plasmids pUL38-45FBpCI and pUL38-63FBpCI, expressing VP19C proteins lacking the N-terminal 45 (VP19C-45) and 63 (VP19C-63) amino acids, respectively, were constructed and tested for the ability to complement growth of vΔ38YFP. As shown in Fig. 4a, the yield obtained with VP19C-45 was approximately 50-fold lower than for the VP19C-WT control, while removal of the first 63 amino acids completely abolished complementation, thereby demonstrating that the N-terminal sequences of VP19C are important for production of wild-type levels of infectious virus.
FIG. 4.
Roles of the VP19C N-terminal sequences. (a) Complementation of growth of the VP19C-minus mutant vΔ38YFP by transfected plasmid pUL38-45FBpCI (expressing VP19C-45), pUL38-63FBpCI (VP19C-63), pUL38-45NLSFBpCI (VP19C-45NLS), or pUL38-63NLSFBpCI (VP19C-63NLS) was carried out in BHK cells. The progeny virus was titrated on UL38RSC cells. The error bars indicate the standard errors of the means. (b to g) Intracellular localization of N-terminal mutants of VP19C. BHK cells were transfected singly with plasmids (b) pUL38-45FBpCI (expressing VP19C-45) and (e) pUL38-63FBpCI (VP19C-63) or were cotransfected with (c and d) pUL38-45NLSFBpCI (VP19C-45NLS) and pE18 (VP23) or with (f and g) pUL38-63NLSFBpCI (VP19C-63NLS) and pE18. VP19C (b, c, e, and f) was detected with the monoclonal antibody mAb02040 and visualized using FITC-conjugated GAM (green). VP23 (d and g) was detected with the antiserum rAb186 and visualized using TRITC-conjugated GAR (red).
To determine the effects of the N-terminal truncations on the behavior of VP19C, the distributions of VP19C-45 and VP19C-63 in transfected cells were examined. In marked contrast to the pattern seen with VP19C-WT (Fig. 2 and 3), neither VP19C-45 (Fig. 4b) nor VP19C-63 (Fig. 4e) was specifically nuclear. These results suggested that these sequences contain an NLS. Since the nucleus is the site of capsid assembly, the smaller amounts of the mutant VP19C entering this compartment could account for the reduction in virus yield seen in the complementation assay. To determine whether this was the case, the SV40 large T antigen NLS (10) was fused to the N-termini of VP19C-45 and VP19C-63. The resulting proteins, VP19C-45NLS (Fig. 4c) and VP19C-63NLS (Fig. 4f), both localized efficiently to the nucleus, and in addition, both transported VP23 (Fig. 4d and g) and VP5 (not shown) to the nucleus, confirming that the exogenous NLS was functioning as expected. When the abilities of these constructs to support virus growth were tested, VP19C-45NLS was found to complement vΔ38YFP as effectively as VP19C-WT (Fig. 4a), thereby demonstrating that the reduction in growth caused by the removal of the N-terminal 45 amino acids was primarily a result of the defect in nuclear localization. In contrast, no complementation occurred with VP19C-63NLS (Fig. 4a), suggesting that amino acids 45 to 63 have an additional role in the formation of infectious virus.
Mapping the VP19C NLS.
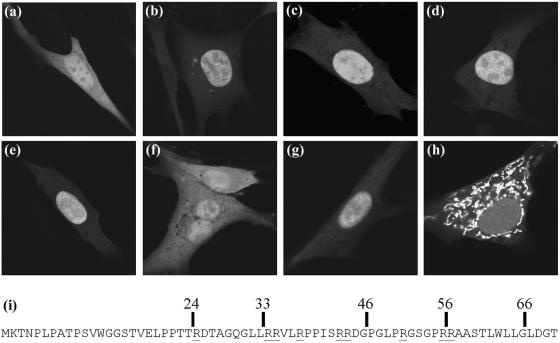
Although VP19C is transported to the nucleus in the absence of other viral proteins, no NLS was identified when the complete amino acid sequence was submitted to the NLS prediction program (http://cubic.bioc.columbia.edu/predictNLS/ and http://psort.nibb.ac.jp/form2.html). To confirm that the N-terminal region of VP19C did contain an NLS, increasing amounts of sequence from the N terminus were fused to the GFP, and the intracellular distributions of the chimeric proteins were determined. In cells expressing GFP alone, the fluorescence was uniformly distributed between the cytoplasm and the nucleus (Fig. 5a). However, fusing GFP to the N-terminal 118 (not shown), 83, 76, 66, or 56 amino acids of VP19C converted it to a nuclear protein (Fig. 5b to e). In contrast, the distribution seen with the N-terminal 46 amino acids resembled the GFP control (Fig. 5f). Thus, it is clear that the NLS is located within the first 56 residues of VP19C. Examination of the amino acid sequence in this region revealed a high concentration of arginine residues extending from amino acids 25 to 57 (Fig. 5i). Since stretches of basic amino acids are characteristic of many NLSs, the entire arginine-rich sequence from amino acids 24 to 66 was fused to GFP. This also resulted in nuclear localization of GFP (Fig. 5g), confirming that the NLS mapped in this region. In an attempt to define the NLS more precisely, amino acids 33 to 66 were also fused to GFP. However, the resulting chimera showed an unexpected and distinctive cytoplasmic pattern that resembled the distribution expected for mitochondria (Fig. 5h). Submission of the VP19C sequence to the subcellular localization site prediction program iPSORT (http://biocaml.org/ipsort/iPSORT/#predict) identified amino acids 32 to 40 (LLRRVLRPP) as a potential mitochondrial localization signal. Given what is known about the properties and functions of VP19C, it seems unlikely that this mitochondrial localization signal is functional in the context of the intact protein. Presumably, the NLS, which occupies the same region, is dominant and the mitochondrial signal becomes apparent only after the increasingly large deletions have rendered the NLS inactive. Although further analysis is required to determine the precise composition of the VP19C NLS, the results presented here map it unambiguously to an arginine-rich sequence between residues 24 and 56.
FIG. 5.
Mapping the VP19C NLS. BHK cells were transfected with plasmids expressing the GFP protein fused in frame to the N-terminal 83 (b), 76 (c), 66 (d), 56 (e), or 46 (f) amino acids of VP19C. (g and h) GFP is fused to amino acids 24 to 66 and 33 to 66 of VP19C. (a) GFP control. (i) N-terminal 70 amino acids of VP19C with the positions of various truncations indicated. Arginine residues are underlined.
Nature of the growth defect associated with VP19C-63NLS.
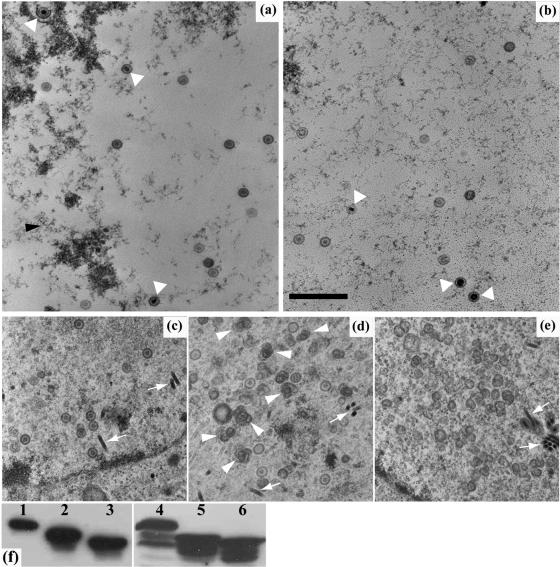
To determine at what stage during infection the block on replication due to deletion of the N-terminal 63 amino acids was occurring, duplicate 35-mm plates of U2OS cells (one containing a glass coverslip) were incubated with 50 PFU/cell of baculoviruses, UL38FBacpCI (expressing VP19C), UL38-45NLSFBacpCI (VP19C-45NLS), and UL38-63NLSFBacpCI (VP19C-63NLS), for 1 h at 37°C. The baculovirus inoculum was removed, the cells were infected with 5 PFU/cell of vΔ38YFP, and incubation was continued at 37°C. After 16 h, the coverslip from one of the duplicate plates was removed and processed for immunofluorescence microscopy, while the remaining cells were used for Western blotting. The second plate was harvested and processed for electron microscopy. Immunofluorescence showed similar numbers of cells expressing the three forms of VP19C (data not shown), and Western blotting (Fig. 6f) confirmed that all three proteins were expressed at similar levels. Examination of thin sections revealed the presence of capsids in the nuclei of 48% of the UL38FBacpCI-transduced and 57% of the UL38-45NLSFBacpCI-transduced samples. As expected for VP19C proteins that support productive infection, both DNA-containing and empty capsids were seen in cells expressing wild-type VP19C (Fig. 6a) and VP19C-45NLS (Fig. 6b). In contrast, no capsid-like particles of any type were seen in over 100 nuclei of the UL38-63NLSFBacpCI-transduced samples. Published studies had reported that capsid assembly in a baculovirus model was not prevented by the removal of up to 90 amino acids from the N terminus of VP19C, although it was less efficient than with full-length VP19C (30). Therefore, the failure to detect capsids in the UL38-63NLSFBacpCI-transduced U2OS cells was unexpected. To investigate this further, capsid assembly was analyzed using the much more efficient baculovirus model system. Duplicate plates of Sf21 cells (one containing a coverslip) were infected separately with 5 PFU/cell of baculovirus AcUL38 (expressing full-length VP19C), UL38-45FBac (VP19C-45), or UL38-63FBac (VP19C-63). All samples were also infected with 5 PFU/cell each of baculoviruses AcUL19 (VP5), AcUL26.5 (preVP22a), and AcAB3.12 (VP23, VP26, and the UL26 protease) to supply the remaining capsid proteins. As expected, characteristic herpesvirus B capsids were readily observed in cells expressing wild-type VP19C (Fig. 6c) and in smaller amounts in cells expressing VP19C-45 (Fig. 6d). The reduced number of intact capsids seen with VP19C-45 is probably a result of inefficient nuclear localization due to the absence of the NLS. However, in cells expressing VP19C-63, only incomplete capsid shells were seen (Fig. 6e). Once again, immunofluorescence (data not shown) and Western blotting (Fig. 6f) confirmed that all three proteins were being expressed at similar levels, thereby demonstrating that the block on particle formation was a direct result of the VP19C-63 deletion.
FIG. 6.
Effects of VP19C N-terminal mutations on capsid assembly. (a and b) Thirty-five-millimeter plates of U2OS cells were transduced with 50 PFU/cell of baculovirus UL38FBacpCI (a) or UL38-45NLSFBacpCI (b) and incubated at 20°C for 1 h before being infected with 5 PFU/cell of vΔ38YFP. Infection was continued at 37°C for 16 h. (c to e) Thirty-five-millimeter plates of Sf21 cells were infected with 5 PFU/cell each of baculoviruses AcUL19, AcUL26.5, and AcAB3.12 and with 5 PFU/cell of AcUL38 (c), UL38-45FBac (d), or UL38-63FBac (e) and incubated at 28°C for 48 h. At the appropriate times after infection, the cells were harvested and prepared for electron microscopy. The majority of capsids in panels a to d are B capsids and are unlabeled. In panels a and b, the C (DNA-containing) capsids are indicated by triangles. In panel d, concentrations of incomplete capsids are indicated with arrowheads. Only incomplete capsids are present in panel e, and they are not labeled. Baculovirus capsids (c to e) are indicated by arrows. Scale bar = 500 nm. (f) Expression of VP19C in the infected cells was confirmed by Western blotting. Protein samples were separated on a 10% sodium dodecyl sulfate-polyacrylamide gel and detected by enhanced chemiluminescence using mAb02040 and protein A-peroxidase. Lanes 1 and 2 show U2OS cell samples corresponding to panels a and b. Lane 3 shows a sample from U2OS cells transduced with UL38-63NLSFBacpCI and infected with vΔ38YFP. Lanes 4 to 6 show Sf21 cell samples corresponding to panels c to e.
DISCUSSION
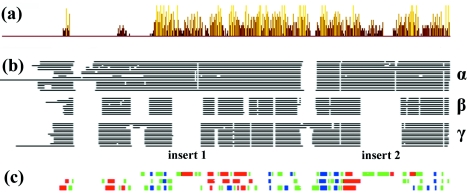
Members of the three subfamilies of mammalian and avian herpesviruses (Alpha-, Beta-, and Gammaherpesviridae) share a common capsid architecture, and in all examples analyzed to date, the triplex is composed of one copy of a VP19C homologue (the α-subunit) and two copies of a VP23 homologue (the β-subunit) (3, 6, 40). In HSV-1 and most other alphaherpesviruses, the α-subunit is considerably larger than the β-subunit. However, in beta- and gammaherpesviruses, the α-subunit is significantly smaller than VP19C (6, 13, 21) and is of a size similar to that of the β-subunit. Therefore, it seems likely that the additional sequences in the alphaherpesvirus VP19C homologues represent relatively recent evolutionary developments. Comparison of α-subunit sequences reveals that the N-terminal 111 amino acids of VP19C are not conserved in the other alphaherpesviruses (Fig. 7a). However, differences in the N-terminal region are not entirely responsible for the size variation among the different herpesvirus subfamilies. An alignment of sequences from alpha-, beta-, and gammaherpesviruses is in broad agreement with earlier findings (38) and indicates that the greater size of the alphaherpesvirus proteins is largely accounted for by two internal insertions, each of ∼40 to 50 residues (insert 1 and insert 2), as well as by extra sequences at the N terminus (Fig. 7b).
FIG. 7.
Sequence comparison of VP19C homologues. We used the CLUSTAL W (http://www.ebi.ac.uk/clustalw/) multiple sequence alignment program (34) to align the triplex α-subunit sequences of 15 alphaherpesviruses, 9 betaherpesviruses, and 12 gammaherpesviruses identified by a BLAST search from the VIDA database (http://www.biochem.ucl.ac.uk/bsm/virus_database/VIDA.html) using the HSV-1, HCMV, and Kaposi’s sarcoma-associated herpesvirus triplex α-subunit sequences as input. The CLUSTAL W output was used as the basis for subsequent analyses. (a) Levels of sequence conservation among the triplex α-subunits in alphaherpesviruses were analyzed and plotted using the JalView (http://www.jalview.org/) multiple sequence editor program (4). (b) Schematic representation of the consensus alignment between alpha-, beta- and gammaherpesvirus triplex α-subunit sequences, showing the locations of additional sequences (insert 1 and insert 2 comprising residues 140 to 176 and 357 to 405, respectively, in HSV-1) in the alphaherpesvirus proteins. The consensus sequence was calculated using the program Consensus (http://www.bork.embl-heidelberg.de/Alignment/consensus.html). (c) Secondary-structure predictions for triplex α-subunits of alpha- (top), beta- (middle), and gammaherpesviruses (bottom). Secondary-structure predictions were carried out on each individual sequence using PredictProtein (28) (http://cubic.bioc.columbia.edu/predictprotein/). Structural elements present in >50% of examples were plotted relative to the alignments shown in Fig. 7b for each subfamily. The positions of α-helices are shown in red, β-sheet in blue, and coil in green.
Transposon-based insertional mutagenesis is a rapid and efficient method of screening a protein sequence for functional domains. However, the results of mutational analysis must be interpreted with caution, particularly when considering the behavior of individual mutants. In some cases, pairs of mutants with contrasting properties may be very close together. Thus, the plasmids containing insertions in286 and in289 differ markedly in their abilities to complement growth of vΔ38YFP. In addition, not all functions are equally sensitive to insertion. For example, although in38 maps within the sequences identified as containing a nuclear localization signal, the ability of this mutant to enter the nucleus was not affected (data not shown). Therefore, without detailed information on the structure of the target protein or specific assays to measure particular properties, it is usually not possible to make other than a general interpretation of results. Three-dimensional structural analysis of the capsid has shown that the component proteins interact in different ways at distinct locations in the capsid (42). Furthermore, the reconfiguration that occurs during capsid maturation involves domain movements leading to changes in the patterns of interaction (8, 37). This type of behavior means that the importance of particular regions of a protein may vary with maturation status and location. Nevertheless, the distribution of severely disabling mutations across VP19C was nonuniform, with 16 in the central region of VP19C but none in the N-terminal 107 residues and only 2 in the C-terminal 107 residues. The most N-terminal severely disabled mutant, in113, occurs almost immediately after the first evidence of conservation between the sequences of the alphaherpesvirus α-subunits (Fig. 7a). There is no equivalent distinction marking the boundary of the C-terminal 107 residues, although interestingly, its start at in358 coincides with the beginning of the second large insert unique to alphaherpesviruses (insert 2) (Fig. 7b). However, unlike the poorly conserved N-terminal region, insert 2 contains a severely disabling mutation, in390. Only one insertion (in143) mapped within the insert 1 locus, and it was also severely disabling, indicating that this inserted sequence is functionally important. The distribution of severely disabled mutants suggests that VP19C can be divided into three regions based on the ability to tolerated insertions and may suggest that the three regions have differing roles. However, this interpretation must be treated with caution, as the distribution of insertions is not uniform and clustering of mutations with similar phenotypes, such as in327 to -330, tends to distort the pattern. Statistical analysis by chi-square analysis indicates that, although the N-terminal region is more tolerant of insertions, the perceived difference between the central and C-terminal regions is marginal.
The sensitivity to insertion of the central region suggests that it may be important for folding of VP19C in a manner similar to that suggested for the central region (VP5ud) of the major capsid protein, VP5, which has been proposed to act as a folding nucleus for the more flexible N- and C-terminal regions (2). The large number of tolerated insertions in the C-terminal region is rather surprising, as deletional analysis had shown that removal of as few as 15 amino acids from the C terminus destroyed the ability of VP19C to support capsid assembly (30). When considered alongside the behavior of in463, which completely fails to support virus growth, this seems to indicate that sequences very near the C terminus of VP19C play essential roles in capsid assembly. However, the viability of most of the mutants in the C-terminal region suggests that it has considerable structural flexibility despite the secondary-structure predictions, which appear to show that conservation of secondary structure increases toward the C terminus (Fig. 7c).
It is difficult to discern any pattern in the interaction properties of the severely disabled insertional mutants. The 18 severely disabled mutants can be divided into four categories based on the ability to interact with VP5 and VP23 (Table 3). Five (including in242A and in242B, which are in the same codon) failed to transport either VP23 or VP5 to the nucleus, suggesting that these insertions may have altered the overall folding of the protein. These insertions (residues 113, 242, 289, and 330) are evenly spread across the central region and do not appear to demarcate any obvious domain, although they may lie close together in the folded protein. Four of the mutants interact with VP5 but not with VP23, while in three, this pattern is reversed. The specificity of these mutants suggests that the insertions do not cause global misfolding of the protein but have a more localized effect. The mutants that failed to take VP23 to the nucleus were located N terminal (between residues 193 and 310) to those that failed to bind VP5 (between 328 and 463), suggesting that the regions involved are particularly important for binding of VP23 and VP5, respectively. Since VP23 and VP5 are both essential for capsid assembly (5, 33, 35), it is not surprising that the mutants which fail to interact with one or both of them do not support virus growth. The nature of the defect in the remaining severely disabled mutants is unclear. Further analysis is required to determine whether they support the assembly of structurally normal capsids. However, examination of mutants in143, in234, in235, in327, in350, and in390 in a plasmid-based DNA-packaging assay (9) showed that they were unable to support DNA packaging in vΔ38YFP-infected cells (data not shown). This suggests either that assembly of the capsid shell was disrupted, presumably due to some subtle alteration in the interactions with VP23 or VP5, or that an interaction involving one of the other virion components had been affected. VP19C has been reported to interact with the UL25 protein (22). The tegument protein present at the vertices in mature virions is in contact with the peripentonal triplexes and may also interact with VP19C (41). Failure to bind tegument would be lethal but would not be expected to prevent DNA packaging, However, UL25 mutants show aberrant DNA packaging (12, 32), and disruption of this interaction might result in the type of phenotype seen here.
The N-terminal regions of all three herpesvirus subfamilies are particularly poorly conserved in both length and sequence, with effectively no homology evident until after amino acid 111 of VP19C (Fig. 7a). This raises questions regarding the function of this highly variable region. Part of that function was revealed by the deletion of 45 residues from the N terminus of VP19C, which removed a nuclear localization signal. That the failure of this deleted protein to concentrate in the nucleus was entirely responsible for the 50-fold reduction in its ability to complement growth of a VP19C-minus mutant was demonstrated by the recovery in virus growth when an exogenous (SV40) nuclear localization signal was present. From this observation, it is evident that correct transport of the component proteins to the site of capsid assembly is an important function of VP19C. Although the precise sequence of the VP19C NLS has not been identified, it has been mapped to a region containing 33 amino acids, and database searching has revealed that it does not belong to any of the known classes of NLS. Interestingly, the pattern of arginine residues seen in the VP19C NLS is not present in the N-terminal sequences of any other alphaherpesvirus VP19C homologue, apart from that of the very closely related HSV-2. Furthermore, examination of triplex α-subunit sequences from all three herpesvirus subfamilies using the predictNLS program failed to identify any recognizable NLSs. Therefore, it is unclear whether a role in nuclear localization represents the usual function of the poorly conserved alphaherpesvirus N termini or where NLSs, if any are present, might be located in the α-subunits from beta- and gammaherpesviruses.
Since the functional NLS is contained within the first 56 amino acids of VP19C, the function of the rest of the poorly conserved N-terminal region remains unclear. Electron microscopic analysis failed to identify capsid formation by the VP19C-63 truncation, either in a baculovirus-based capsid assembly model or by complementation of vΔ38YFP. The incomplete capsid shells seen in baculovirus-infected cells confirm the fluorescence results showing that the VP19C-63 protein is able to interact with VP5 and VP23 and indicate that the defect in assembly is not a result of a general disruption of the protein's structure. The results presented here contrast with those of Spencer et al. (30), who showed that a mutant of VP19C (nd90) lacking the N-terminal 90 amino acids could support capsid assembly in a baculovirus model system. The reason for this difference is unclear. Although apparently intact capsids were formed with nd90, their assembly was inefficient, and the majority of structures seen resembled the incomplete particles shown in Fig. 5e. It is possible that slight differences in the baculovirus model systems used in the two studies account for the contrasting results. Alternatively, when exposed at the N terminus, amino acids 63 to 90 may have a disruptive effect on protein folding or interaction that is negated when they are absent. Whatever the reason, it is clear that sequences between residues 45 and 63 are necessary for efficient capsid formation, and further analysis will be needed to determine precisely what roles they play.
Acknowledgments
We thank D. McGeoch for critical reading of the manuscript and J. Mitchell for excellent technical assistance. pFBpCI was provided by R. Everett.
This work was supported by the United Kingdom Medical Research Council. W. E. Adamson was funded by an MRC Research Training studentship.
REFERENCES
- 1.Bishop, D. H. L. 1992. Baculovirus expression vectors. Semin. Virol. 3:253-264. [Google Scholar]
- 2.Bowman, B. R., M. L. Baker, F. J. Rixon, W. Chiu, and F. A. Quiocho. 2003. Structure of the herpesvirus major capsid protein. EMBO J. 22:757-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, D. H., H. Jiang, M. Lee, F. Y. Liu, and Z. H. Zhou. 1999. Three-dimensional visualization of tegument/capsid interactions in the intact human cytomegalovirus. Virology 260:10-16. [DOI] [PubMed] [Google Scholar]
- 4.Clamp, M., J. Cuff, S. M. Searle, and G. J. Barton. 2004. The Jalview Java alignment editor. Bioinformatics 20:426-427. [DOI] [PubMed] [Google Scholar]
- 5.Desai, P., N. A. Deluca, J. C. Glorioso, and S. Person. 1993. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. J. Virol. 67:1357-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibson, W., M. K. Baxter, and K. S. Clopper. 1996. Cytomegalovirus “missing” capsid protein identified as heat-aggregable product of human cytomegalovirus UL46. J. Virol. 70:7454-7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grunewald, K., P. Desai, D. C. Winkler, J. B. Heymann, D. M. Belnap, W. Baumeister, and A. C. Steven. 2003. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302:1396-1398. [DOI] [PubMed] [Google Scholar]
- 8.Heymann, J. B., N. Q. Cheng, W. W. Newcomb, B. L. Trus, J. C. Brown, and A. C. Steven. 2003. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat. Struct. Biol. 10:334-341. [DOI] [PubMed] [Google Scholar]
- 9.Hodge, P. D., and N. D. Stow. 2001. Effects of mutations within the herpes simplex virus type 1 DNA encapsidation signal on packaging efficiency. J. Virol. 75:8977-8986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalderon, D., W. D. Richardson, A. F. Markham, and A. E. Smith. 1984. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311:33-38. [DOI] [PubMed] [Google Scholar]
- 11.Lu, G. Y., Z. H. Zhou, M. L. Baker, J. Jakana, D. Y. Cai, X. C. Wei, S. X. Chen, X. C. Gu, and W. Chiu. 1998. Structure of double-shelled rice dwarf virus. J. Virol. 72:8541-8549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McNab, A. R., P. Desai, S. Person, L. L. Roof, D. R. Thomsen, W. W. Newcomb, J. C. Brown, and F. L. Homa. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 72:1060-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nealon, K., W. W. Newcomb, T. R. Pray, C. S. Craik, J. C. Brown, and D. H. Kedes. 2001. Lytic replication of Kaposi's sarcoma-associated herpesvirus results in the formation of multiple capsid species: isolation and molecular characterization of A, B, and C capsids from a gammaherpesvirus. J. Virol. 75:2866-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newcomb, W. W., F. L. Homa, D. R. Thomsen, F. P. Booy, B. L. Trus, A. C. Steven, J. V. Spencer, and J. C. Brown. 1996. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J. Mol. Biol. 263:432-446. [DOI] [PubMed] [Google Scholar]
- 15.Newcomb, W. W., F. L. Homa, D. R. Thomsen, Z. Ye, and J. C. Brown. 1994. Cell-free assembly of the herpes simplex virus capsid. J. Virol. 68:6059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newcomb, W. W., D. R. Thomsen, F. L. Homa, and J. C. Brown. 2003. Assembly of the herpes simplex virus capsid: identification of soluble scaffold-portal complexes and their role in formation of portal-containing capsids. J. Virol. 77:9862-9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newcomb, W. W., B. L. Trus, F. P. Booy, A. C. Steven, J. S. Wall, and J. C. Brown. 1993. Structure of the herpes simplex virus capsid: molecular composition of the pentons and the triplexes. J. Mol. Biol. 232:499-511. [DOI] [PubMed] [Google Scholar]
- 19.Newcomb, W. W., B. L. Trus, N. Q. Cheng, A. C. Steven, A. K. Sheaffer, D. J. Tenney, S. K. Weller, and J. C. Brown. 2000. Isolation of herpes simplex virus procapsids from cells infected with a protease-deficient mutant virus. J. Virol. 74:1663-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicholson, P., C. Addison, A. M. Cross, J. Kennard, V. G. Preston, and F. J. Rixon. 1994. Localization of the herpes simplex virus type 1 major capsid protein VP5 to the cell nucleus requires the abundant scaffolding protein VP22a. J. Gen. Virol. 75:1091-1099. [DOI] [PubMed] [Google Scholar]
- 21.O'Connor, C. M., B. Damania, and D. H. Kedes. 2003. De novo infection with rhesus monkey rhadinovirus leads to the accumulation of multiple intranuclear capsid species during lytic replication but favors the release of genome-containing virions. J. Virol. 77:13439-13447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogasawara, M., T. Suzutani, I. Yoshida, and M. Azuma. 2001. Role of the UL25 gene product in packaging DNA into the herpes simplex virus capsid: location of UL25 product in the capsid and demonstration that it binds DNA. J. Virol. 75:1427-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Preston, V. G., M. F. Al-Kobaisi, I. M. McDougall, and F. J. Rixon. 1994. The herpes simplex virus gene UL26 proteinase in the presence of the UL26.5 gene product promotes the formation of scaffold-like structures. J. Gen. Virol. 75:2355-2366. [DOI] [PubMed] [Google Scholar]
- 24.Rixon, F. J. 1993. Structure and assembly of herpesviruses. Semin. Virol. 4:135-144. [Google Scholar]
- 25.Rixon, F. J., C. Addison, A. McGregor, S. J. Macnab, P. Nicholson, V. G. Preston, and J. D. Tatman. 1996. Multiple interactions control the intracellular localization of the herpes simplex virus type 1 capsid proteins. J. Gen. Virol. 77:2251-2260. [DOI] [PubMed] [Google Scholar]
- 26.Rixon, F. J., and D. McNab. 1999. Packaging-competent capsids of a herpes simplex virus temperature-sensitive mutant have properties similar to those of in vitro-assembled procapsids. J. Virol. 73:5714-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenthal, K. S., M. D. Leuther, and B. G. Barisas. 1984. Herpes simplex virus binding and entry modulate cell surface protein mobility. J. Virol. 49:980-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rost, B., G. Yachdav, and J. Liu. 2004. The PredictProtein server. Nucleic Acids Res. 32:W321-W326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saad, A., Z. H. Zhou, J. Jakana, W. Chiu, and F. J. Rixon. 1999. Roles of triplex and scaffolding proteins in herpes simplex virus type 1 capsid formation suggested by structures of recombinant particles. J. Virol. 73:6821-6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spencer, J. V., W. W. Newcomb, D. R. Thomsen, F. L. Homa, and J. C. Brown. 1998. Assembly of the herpes simplex virus capsid: preformed triplexes bind to the nascent capsid. J. Virol. 72:3944-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steven, A. C., and P. G. Spear. 1997. Herpesvirus capsid assembly and envelopment, p. 312-351. In W. Chiu, R. M. Burnett, and R. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, N.Y.
- 32.Stow, N. D. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J. Virol. 75:10755-10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tatman, J. D., V. G. Preston, P. Nicholson, R. M. Elliott, and F. J. Rixon. 1994. Assembly of herpes simplex virus type 1 capsids using a panel of recombinant baculoviruses. J. Gen. Virol. 75:1101-1113. [DOI] [PubMed] [Google Scholar]
- 34.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomsen, D. R., L. L. Roof, and F. L. Homa. 1994. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J. Virol. 68:2442-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thurlow, J. K., F. J. Rixon, M. Murphy, P. Targett-Adams, M. Hughes, and V. G. Preston. 2005. The herpes simplex virus type 1 DNA packaging protein UL17 is a virion protein that is present in both the capsid and the tegument compartments. J. Virol. 79:150-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trus, B. L., F. P. Booy, W. W. Newcomb, J. C. Brown, F. L. Homa, D. R. Thomsen, and A. C. Steven. 1996. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J. Mol. Biol. 263:447-462. [DOI] [PubMed] [Google Scholar]
- 38.Trus, B. L., J. B. Heymann, K. Nealon, N. Q. Cheng, W. W. Newcomb, J. C. Brown, D. H. Kedes, and A. C. Steven. 2001. Capsid structure of Kaposi's sarcoma-associated herpesvirus, a gammaherpesvirus, compared to those of an alphaherpesvirus, herpes simplex virus type 1, and a betaherpesvirus, cytomegalovirus. J. Virol. 75:2879-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trus, B. L., F. L. Homa, F. P. Booy, W. W. Newcomb, D. R. Thomsen, N. Q. Cheng, J. C. Brown, and A. C. Steven. 1995. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J. Virol. 69:7362-7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu, X. K., C. M. O'Connor, V. Atanasov, B. Damania, D. H. Kedes, and Z. H. Zhou. 2003. Three-dimensional structures of the A, B, and C capsids of rhesus monkey rhadinovirus: insights into gammaherpesvirus capsid assembly, maturation, and DNA packaging. J. Virol. 77:13182-13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou, Z. H., D. H. Chen, J. Jakana, F. J. Rixon, and W. Chiu. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 73:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou, Z. H., W. Chiu, K. Haskell, H. Spears, J. Jakana, F. J. Rixon, and L. R. Scott. 1998. Refinement of herpesvirus B-capsid structure on parallel supercomputers. Biophys. J. 74:576-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou, Z. H., M. Dougherty, J. Jakana, J. He, F. J. Rixon, and W. Chiu. 2000. Seeing the herpesvirus capsid at 8.5 Å. Science 288:877-880. [DOI] [PubMed] [Google Scholar]
- 44.Zhou, Z. H., J. He, J. Jakana, J. D. Tatman, F. J. Rixon, and W. Chiu. 1995. Assembly of VP26 in herpes simplex virus 1 inferred from structures of wild-type and recombinant capsids. Nat. Struct. Biol. 2:1026-1030. [DOI] [PubMed] [Google Scholar]
- 45.Zhou, Z. H., S. J. Macnab, J. Jakana, L. R. Scott, W. Chiu, and F. J. Rixon. 1998. Identification of the sites of interaction between the scaffold and outer shell in herpes simplex virus-1 capsids by difference electron imaging. Proc. Natl. Acad. Sci. USA 95:2778-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou, Z. H., B. V. V. Prasad, J. Jakana, F. J. Rixon, and W. Chiu. 1994. Protein subunit structures in the herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J. Mol. Biol. 242:456-469. [DOI] [PubMed] [Google Scholar]