Abstract
After their release from host cells, most retroviral particles undergo a maturation process, which includes viral protein cleavage, core condensation, and increased stability of the viral RNA dimer. Inactivating the viral protease prevents protein cleavage; the resulting virions lack condensed cores and contain fragile RNA dimers. Therefore, protein cleavage is linked to virion morphological change and increased stability of the RNA dimer. However, it is unclear whether protein cleavage is sufficient for mediating virus RNA maturation. We have observed a novel phenotype in a murine leukemia virus capsid mutant, which has normal virion production, viral protein cleavage, and RNA packaging. However, this mutant also has immature virion morphology and contains a fragile RNA dimer, which is reminiscent of protease-deficient mutants. To our knowledge, this mutant provides the first evidence that Gag cleavage alone is not sufficient to promote RNA dimer maturation. To extend our study further, we examined a well-defined human immunodeficiency virus type 1 (HIV-1) Gag mutant that lacks a functional PTAP motif and produces immature virions without major defects in viral protein cleavage. We found that the viral RNA dimer in the PTAP mutant is more fragile and unstable compared with those from wild-type HIV-1. Based on the results of experiments using two different Gag mutants from two distinct retroviruses, we conclude that Gag cleavage is not sufficient for promoting RNA dimer maturation, and we propose that there is a link between the maturation of virion morphology and the viral RNA dimer.
During or soon after the assembly of most retroviral particles, viral protease (PR) cleaves the viral proteins. Gag polyproteins are the main structural proteins in the virion (55). In all retroviruses except spumaviruses, cleavage of Gag polyproteins generates mature proteins including the matrix (MA), capsid (CA), and nucleocapsid (NC) (28, 52, 61). Additionally, Gag polyproteins generate other cleavage products, which vary in number and size depending on the virus. For example, Gag cleavage also generates p12 in murine leukemia virus (MLV) and p2, p1, and p6 in human immunodeficiency virus type 1 (HIV-1) (28).
The newly generated virion undergoes a maturation process after PR-mediated Gag polyprotein cleavage. Morphologically, the virus particle changes from immature to mature (25, 54, 59); the immature virion has a translucent center, whereas the mature virion has a condensed core (35, 54, 62). During the maturation process, viral proteins undergo changes that are detectable by biochemical and structural analyses. For example, cross-linking experiments indicate that the cleaved CA alters points of protein-protein contact (12, 21); mutational and structural analyses define a major conformational change at the N terminus of the mature HIV-1 CA protein after Gag cleavage (57). It is thought that in the mature virion, MA proteins are associated with the lipid membrane, CA proteins define the outer shell of the core structure, and NC proteins are bound to the viral genomic RNA inside the core (15, 61). During virus maturation, properties of the viral RNA are also altered: in the immature virion, the two copies of RNA form a more fragile dimer that is less stable than that in the mature virion; during maturation, the dimeric RNA acquires a higher thermostability (16, 18, 50).
The virion maturation process can be blocked by abolishing the PR activity. PR-deficient (PR−) mutant virions have immature morphology and fragile RNA dimers. Many aspects of virion maturation are currently unknown; for example, it is unclear which factors mediate the maturation of the viral RNA dimer. It is possible that protein processing releases mature viral proteins to promote the stabilization of viral RNA dimers. With its proposed nucleic acid chaperone activity, NC protein is considered the leading candidate to play a role in RNA dimer stabilization (13, 41, 48). However, it is possible that factors other than protein processing are also required for virion RNA maturation.
In this report, we describe a mutation in MLV CA that caused the virion to present an immature morphology and the RNA dimer to have a lower thermostability despite the cleavage of Gag polyproteins. This observation suggests that viral protein processing alone is not sufficient to promote viral RNA maturation. To extend these results, we also examined genomic RNA dimers isolated from a well-characterized HIV-1 PTAP mutant that produces immature virions without major defects in Gag cleavage (9, 23). We found that RNA dimers from the PTAP-mutant-derived virions had a lower thermostability than that of wild-type virions.
MATERIALS AND METHODS
Plasmids and plasmid construction.
Standard cloning techniques were used for plasmid construction (44). The general structures of all plasmids constructed were characterized by restriction enzyme mapping, whereas all regions of the plasmids that have undergone PCR amplification were further characterized by DNA sequencing.
MLV gag-pol expression constructs were derived from pLGPS (31, 32), which contains a truncated 5′ long terminal repeat (LTR) and gag-pol, as well as deletions of most of the packaging signal, env, and a 3′ LTR. Plasmid pMLVCAΔ12 was constructed by PCR-mediated mutation: a DNA fragment containing the deletion was generated by overlapping PCR, digested with BsrGI and XhoI, and inserted into BsrGI-XhoI-digested pLGPS. The PR mutant pMLVPR− contains an inactivating mutation (D27A) in the PR active site, and was generated by inserting a ClaI-PflmI-digested PCR fragment containing the mutation into ClaI-PflmI digested pLGPS. Similarly, a mutated ClaI-PflmI-digested PCR fragment was cloned into pMLVCAΔ12 to generate the CA and PR double-mutant pMLVCAΔ12/PR−.
HIV-1 vectors were derived from pON-T0 (a kind gift from Olga Nikolaitchik), an NL4-3-based HIV-1 vector that contains the two LTRs, cis-acting elements essential for viral replication; expresses functional gag-pol, tat, and rev; and has disabling deletions in vif, vpu, vpr, and env. Additionally, pON-T0 contains a functional mouse thy-1.2 gene (thy), internal ribosomal entry site, and a mutated green fluorescent protein gene (gfp). The previously described HIV-1 vectors pNL4-3-PTAP−, pNL4-3-PR−, and pNL4-3- PTAP−/PR− (23) contained inactivating mutations in the PTAP motif (PTAP to LIRL) and/or PR active site (D25N). The SphI-SbfI fragments containing mutations in HIV-1 gag-pol were isolated and subcloned into SphI-SbfI-digested pON-T0 to generate plasmids pT0-PTAP−, pT0-PR−, and pT0-PTAP−/PR−.
Plasmid pSV-A-MLV-env expresses amphotropic MLV env (27). MLV vector pSR2-2GFP (6) contains all the cis-acting elements essential for viral replication and encodes the hygromycin phosphotransferase B gene (hygro) and gfp. Plasmid pCMVRΔ8.2 expresses all of the HIV-1-encoded proteins except Env and lacks many cis-acting elements, including part of the packaging signal (34). Plasmid pHCMV-G expresses the vesicular stomatitis virus G glycoprotein (VSV-G) (63).
Cell culture, transfection, and infection.
D17 is a dog osteosarcoma cell line permissive to MLV infection (43), and 293T is a human embryonic kidney cell line (11, 38). As previously described, 293T/SR2 is a cell line that consists of a pool of SR2-2GFP-provius-containing 293T cells (7).
D17 and 293T cells were maintained at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 6% calf serum (D17) or 10% fetal calf serum (293T) and penicillin (50 U/ml) plus streptomycin (50 mg/ml). Hygromycin selection was performed at a final concentration of 120 μg/ml.
DNA transfection was performed using the calcium phosphate method (44). To generate MLV virions, 293T cells were cotransfected with MLV gag-pol expression constructs, pSR2-2GFP, and pSV-A-MLV-env at a 2:2:1 weight ratio. To generate HIV-1 virions, 293T cells were transfected with pON-T0 or its derivatives, pCMVRΔ8.2, and pHCMV-G at a 3:3:1 weight ratio. Supernatants were harvested from the transfected 293T cells 2 days after transfection, and cellular debris was removed by filtration through a 0.45-mm filter. MLV stocks were used to infect D17 cells, and viral titers were determined by the numbers of hygromycin-resistant D17 cell colonies. HIV-1 virions were used to infect 293T cells; 3 days after infection, cells were stained with allophycocyanin-conjugated α-Thy1 antibody (eBioscience) as previously described (42). Generally, more than 80% of the 293T cells were infected by HIV-1 vectors in all experiments. Virions were collected from these infected 293T cells and used for further analyses.
Western blotting analyses, RT assays, virion RNA analyses, and transmission electron microscopy (EM).
Virions derived from MLV and HIV-1 vectors were concentrated by centrifugation at 25,000 rpm for 90 min at 4°C using a Surespin 630 (Sorvall) rotor and a Discovery 100S Sorvall ultracentrifuge. Virion proteins were examined by Western blotting analyses as previously described (17). To detect MLV proteins, polyclonal rabbit anti-MLV-CA and anti-MLV-MA antibodies were used (kind gifts of the AIDS Vaccine Program, Science Applications International Corporation —Frederick). AIDS patient sera (obtained from the National Institutes of Health AIDS Reagent Program) were used to detect HIV proteins. Reverse transcriptase (RT) activity was measured by exogenous RT assays using standard procedures (6).
Viral RNA was extracted from cell-free virus particles as previously described (17). The amounts of SR2-2GFP vector RNA encapsidated by MLV proteins were determined by quantitative real-time RT PCR using primers and probe derived from hygro sequences and an ABI 7700 sequence detector (PerkinElmer) as previously described (7, 17). Plasmid pSR2-2GFP was used as a template for standard curves for SR2-2GFP RNA detection in the real-time RT PCR. RNA samples isolated from mock-transfected SR2/293T cells were used as negative controls.
To analyze the thermostability of the virion RNA dimer, RNAs were isolated from virus particles and analyzed by nondenaturing Northern analyses as previously described (18). Briefly, RNA samples were incubated at specified temperatures for 10 min, quenched on ice, and separated by electrophoresis using 1% agarose gels in TBE (89 mM Tris, [pH 8.3], 89 mM boric acid, 2.5 mM EDTA). These gels were then soaked in 6% formaldehyde for 30 min at 65°C and the RNAs were transferred onto nylon membranes. The RNAs were cross-linked to the wet membranes using a UV Stratalinker 1800 (Stratagene) for 2 min prior to hybridization. For MLV and HIV-1 Northern analyses, the membranes were probed with 32P-labeled hygro and gag riboprobes, respectively. A hygro-containing PCR DNA fragment was generated using plasmid pSR2-2GFP as template and primer sets T7SR2Hyg1655 (5′-TAATACGACTCACTAGTGCTAGAGTCGAC CTGCAGCCC-3′) and SR2Gyh2640 (5′-CGCTTCTGCGGGCGATTTG-3′). Similarly, an HIV-1 gag-containing PCR fragment was generated using plasmid pON-T0 and primer sets T7HIVGag790 (5′-TAATACGACTCACTAGTGATGGGTGCGAGA GCGTCGGTA-3′) and GagVIH2071 (5′-CAGTACAATCTTTCATTTGG-3′). The 986-bp hygro-containing PCR fragment or the 1280-bp gag-containing PCR fragment served as the template for T7 transcription, using the MAXIscript T7 kit (Ambion) in the presence of [α-32P]UTP (3,000 Ci/mmol) to generate the hygro or gag riboprobe.
EM analyses were performed as previously described (7). Transfected cells were harvested 48 h after transfection, washed once with phosphate-buffered saline, pelleted by low-speed centrifugation, and resuspended and fixed in 2% glutaraldehyde in cacodylate buffer (0.1 M, pH 7.4).
RESULTS
Analyses of the effects of a conserved 12-amino-acid deletion mutation on viral replication in gammaretroviruses.
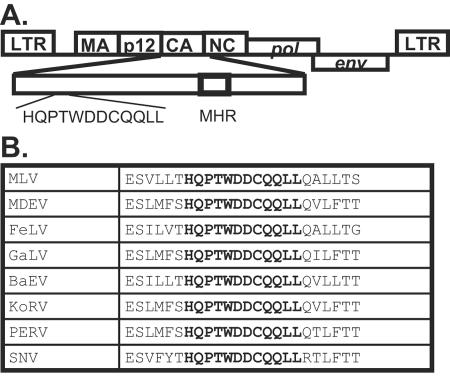
During amino acid sequence alignment, we observed a highly conserved motif (HQPTWDDCQQLL) near the N terminus of CA of many gammaretroviruses (Fig. 1). To determine the possible function of this motif, we modified the MLV gag-pol expression construct pLGPS (31, 32) to generate a deletion mutant, pMLVCAΔ12, that lacked these 12 amino acids (amino acids 263 to 274 in MLV Gag or amino acids 48 to 59 in MLV CA). To examine the phenotypes of this deletion mutant, we transfected pMLVCAΔ12 into 293T cells along with the MLV vector pSR2-2GFP (6) and the amphotropic MLV env-expressing construct pSV-A-MLV-env. Supernatants were harvested from the transfected cells; a portion was used for biochemical analyses and the other portion was used to infect D17 cells. Because the cotransfected MLV vector pSR2-2GFP encodes hygro, viral titers were determined by the numbers of hygromycin-resistant cell colonies as previously described (6).
FIG. 1.
Location and sequence of the 12-amino-acid motif. (A) Location and sequence of the motif in MLV (amino acid 48 to 59 of MLV CA). MHR, major homology region. (B) Comparison of sequences in the motif among several gammaretroviruses. MDEV, Mus dunni endogenous virus; FeLV, feline leukemia virus; GaLV, gibbon ape leukemia virus; BaEV, baboon endogenous virus; KoRV, Phascolarctos cinereus (koala) retrovirus; PERV, porcine endogenous virus; SNV, spleen necrosis virus. The 12-amino-acid motif is shown in bold letters.
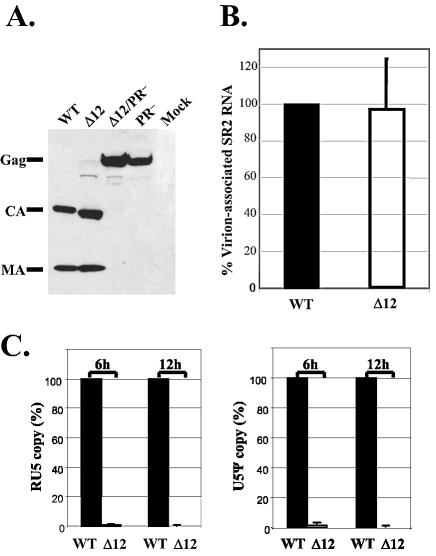
In four sets of independent experiments, transfection with pLGPS, which encodes the wild-type MLV gag-pol, generated viral titers between 5 × 104 and 8.5 × 104 CFU/ml. In contrast, transfection with pMLVCAΔ12 did not yield any detectable viral titers, indicating that deletion of the 12 amino acids severely affected the generation of infectious virions. Western analyses and RT assays were performed on the cell-free supernatants to examine the production of wild-type and mutant virions. A representative Western blotting analysis is shown in Fig. 2A. This and other western analyses revealed that pLGPS- and pMLVCAΔ12-transfected cells generated similar amounts of virions; furthermore, Gag polyproteins were cleaved in both types of virions. Results from RT assays indicated that within each experiment, pLGPS- and pMLVCAΔ12-derived supernatants generated similar levels of RT activity, which is in agreement with the western analyses (data not shown).
FIG. 2.
Analyses of the protein and RNA contents of cell-free virions and viral DNA generated by infection. (A) Western analysis of the cell-free virion proteins. Cell-free virions were harvested 2 days after transfection and examined by Western blotting using rabbit anti-MLV MA and CA antibodies. (B) Comparison of the amounts of MLV vector RNA packaged in cell-free virions. RNAs isolated from virions with similar levels of RT activity were used in the real-time RT PCR analysis; primers and probes in hygro were used to detect SR2-2GFP vector RNA. The amount of SR2-2GFP RNA packaged in pLGPS-derived virions was set to 100% in each experiment. Data from three independent experiments are shown as means ± standard errors. (C) Synthesis of vector DNA upon infection. Quantitative real-time PCR analysis of the early (RU5; left panel) and late (U5Ψ; right panel) viral DNA products generated by viral infection. Viral DNAs from D17 cells infected by viral stocks with similar levels of RT activity 6 h and 12 h postinfection were used in the real-time PCR analysis. RU5 and U5Ψ copy numbers synthesized by pLGPS-derived virions were set to 100% in each experiment. Data from five independent sets of experiments are shown as means ± standard deviations. WT, pLGPS; Δ12, pMLVCAΔ12; Δ12/PR−, pMLVCAΔ12/PR−; PR−, pMLVPR−; mock, mock transfection.
The ability of pLGPS- and pMLVCAΔ12-derived virions to package RNA derived from MLV vector pSR2-2GFP was analyzed by quantitative real-time RT PCR using primer and probe annealed to hygro as previously described (17). Data from three independent experiments are summarized in Fig. 2B; as indicated, these two types of virions packaged similar amounts of vector RNA.
Many CA mutants have defects in reverse transcription (1, 14, 22, 53). To explore whether reverse transcription was affected in the deletion mutant, we examined the reverse transcription products generated by infection of the pLGPS- and pMLVCAΔ12-derived virions by quantitative real-time PCR (17). Results from five sets of independent experiments indicated that pMLVCAΔ12-derived viruses have drastic defects in the generation of both early and late reverse transcription products (Fig. 2C and D).
Taken together, the 12-amino-acid deletion in MLV CA did not have an observable effect on virion assembly and release, Gag polyprotein cleavage, and the specific packaging of vector RNA; however, this deletion did cause a block(s) prior to or at the initiation of reverse transcription.
EM analyses of virion morphology.
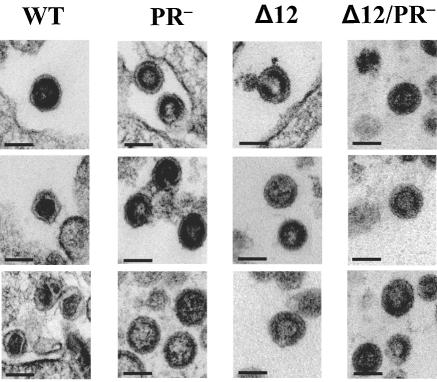
The cleaved CA proteins constitute the outer shell of the core of a mature virion; therefore, mutations in CA could cause aberrant morphology of the virion core. To further characterize the 12-amino-acid deletion, we examined the morphology of the virions produced by 293T cells transfected with pLGPS or pMLVCAΔ12; representative EM pictures are shown in Fig. 3. As expected, we found that most of the pLGPS-derived particles were mature virions (73%). However, most of the pMLVCAΔ12-derived particles were immature virions (83%) (Table 1), which were reminiscent of PR− virions. The observation that the CA mutant virions presented immature virion morphology led us to directly compare CA and PR mutant virions.
FIG. 3.
Electron micrographs of virions derived from MLV wild-type or mutant gag-pol expression constructs. Virions were derived from pLGPS (WT), pMLVPR− (PR−), pMLVCAΔ12 (Δ12), and pMLVCAΔ12/PR− (Δ12/PR−). Scale bars represent 100 nm (all panels).
TABLE 1.
Electron microscopy analyses of virions derived from wild-type and mutant gag-pol expression constructs
| Construct | No. of virions
|
||
|---|---|---|---|
| Mature | Immature | Total | |
| pLGPS | 73 | 27 | 100 |
| pMLVCAΔ12 | 17 | 83 | 100 |
| pMLVPR− | 0 | 100 | 100 |
| pMLVCAΔ12/PR− | 0 | 100 | 100 |
We generated two PR mutants, each containing an inactivating mutation (D27A) in the active site of PR; pMLVPR− and pMLVCAΔ12/PR− were derived from pLGPS and pMLVCAΔ12, respectively. The phenotypes of both PR mutants were examined; as expected, the mutants did not generate detectable viral titers (data not shown), Gag polyproteins were not processed (Fig. 2), and EM analysis demonstrated that both mutants generated only immature particles (Fig. 3, Table 1).
Analyses of the thermostability of virion RNA dimers.
Earlier studies indicated that immature virus particles isolated from PR-deficient mutants contain unstable viral genomic RNA dimers (16, 18), which dissociate into monomers at a lower temperature compared with the RNA dimers from wild-type viruses. However, it was not clear whether protein processing per se is sufficient for promoting RNA dimer maturation. Analyses of the pMLVCAΔ12-derived virions revealed that, although their Gag polyproteins were processed, these particles were largely immature in morphology. Therefore, this mutant provides an excellent tool to examine factors required for viral RNA dimer maturation. If Gag processing and the release of the mature NC protein are sufficient for RNA dimer maturation, then pMLVCAΔ12-derived virions should contain mature dimers. In contrast, if other factors are needed for dimer maturation, then it is possible that pMLVCAΔ12-derived virions contain immature dimers.
We compared the thermostability of the viral RNA dimers isolated from virions derived from wild-type and mutant gag-pol expression constructs. To avoid complications from the contamination of the transfected vector DNA in these experiments, we used 293T/SR2 cells, which are 293T cells that express proviruses derived from vector pSR2-2GFP. 293T/SR2 cells were transfected with pLGPS, pMLVCAΔ12, pMLVPR−, or pMLVCAΔ12/PR−, supernatants were collected, and cell-free virion RNAs were isolated and analyzed by nondenaturing Northern blotting as previously described (18).
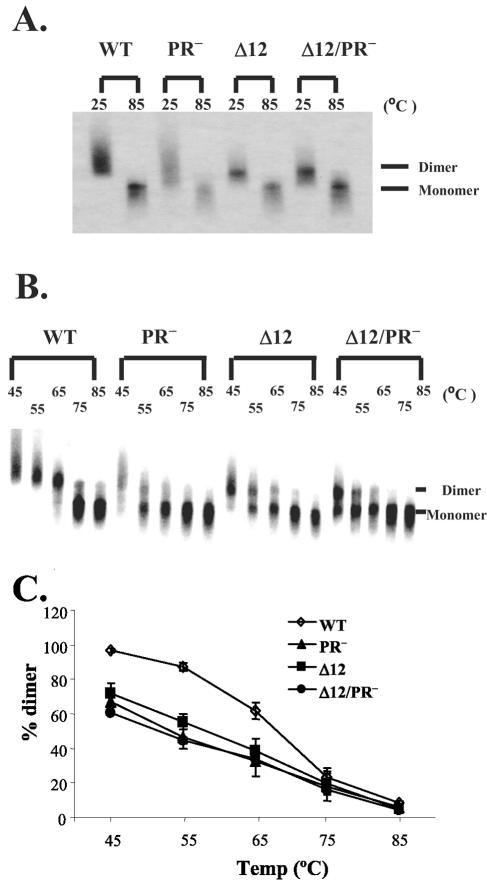
As shown in a representative Northern analysis in Fig. 4A, cell-free virion RNAs derived from cells transfected with wild-type or mutant gag-pol expression constructs were in dimer form at 25°C; these dimers dissociated into monomers after the RNA samples were heated to 85°C. The thermostability of these dimeric RNA samples was analyzed by heating the RNAs to different temperatures, performing nondenaturing Northern analyses, and quantifying the RNA by Phosphorimaging analysis. A representative Northern analysis is shown in Fig. 4B, and data summarized from three independent experiments are shown in Fig. 4C. RNA isolated from pLGPS-derived virions remained mostly as dimers after heating to 65°C, but dissociated to monomers after heating to 75°C. In contrast, cell-free virion RNA isolated from pMLVPR−-derived virions had a lower thermostability; nearly half of the RNA became monomers at 55°C, and most of the RNA was in monomer form after the RNA samples were heated to 65°C. RNA isolated from virions derived from pMLVCAΔ12 or pMLVCAΔ12/PR− had a thermostability indistinguishable from that of RNA isolated from pMLVPR−-derived virions and was different from that of RNA isolated from pLGPS-derived virions. The melting temperature, in which half of the viral RNA remained dimeric, was higher than 65°C for pLGPS-derived virion RNA, and was close to 55°C for the pMLVCAΔ12-, pMLVPR−-, or pMLVCAΔ12/PR−-derived virion RNA (Fig. 4C).
FIG. 4.
Nondenaturing Northern analyses of cell-free virion RNAs from MLV. (A and B) Representative nondenaturing Northern blots of cell-free virion RNAs. RNAs were either not heated (25°C) or heated for 10 min at the indicated temperatures prior to electrophoresis; riboprobes containing hygro sequences were used in these analyses. (C) Thermostability of the virion RNA dimer from wild type or mutant MLV particles. The proportions of dimeric and monomeric RNAs in the Northern analyses were quantified by a PhosphoImager. Data from three independent experiments are shown as means ± standard deviations.
Taken together, our data demonstrate that virions generated by pMLVCAΔ12 had cleaved Gag proteins but displayed immature virion morphology and immature RNA dimers. These observations suggest that Gag cleavage alone is not sufficient to promote RNA maturation. We speculate that the maturation of the virion morphology is linked to the increased stability of the virion RNA dimer.
Examining the thermostability of RNA dimers from an HIV-1 gag mutant.
To extend our observations and test our hypothesis that virion and RNA dimer maturation are linked, we examined the RNA dimer stability of a well-characterized HIV-1 gag mutant in which the PTAP motif in p6 was changed to LIRL (23). This mutant can generate virions, albeit at a lower level than wild type in some cells, and can cleave the Gag polyproteins but does not form a condensed core (9, 23).
We modified HIV-1 vector pON-T0 to contain the PTAP-to-LIRL mutation; the resulting vector is termed pT0-PTAP−. The NL4-3-based pON-T0 contains all of the cis-acting elements essential for virus replication and encodes gag-pol, tat, rev, and a functional thy marker gene. We also generated two additional pON-T0-derived vectors containing a mutation in PR (pT0-PR−) or dual mutations in PTAP and PR (pT0-PTAP−/PR−).
To avoid possible complications from contaminated transfected DNA, we generated the virus using a two-step method. We first transfected into 293T cells the vector plasmids along with two helper plasmids, pCMVRΔ8.2 and pHCMV-G, which express HIV-1 proteins and VSV G protein, respectively. We harvested the resulting viruses and infected fresh 293T cells to generate infected cell pools. Using this approach, we generated multiple 293T cell pools, each having more than 80% of the cells infected with a particular T0 virus as determined by flow cytometry analysis of Thy expression. Cell-free supernatants were harvested from these infected producer cell pools; virion morphology was analyzed by EM, viral proteins were examined by Western blotting, and virion RNAs were isolated and analyzed by Northern analyses.
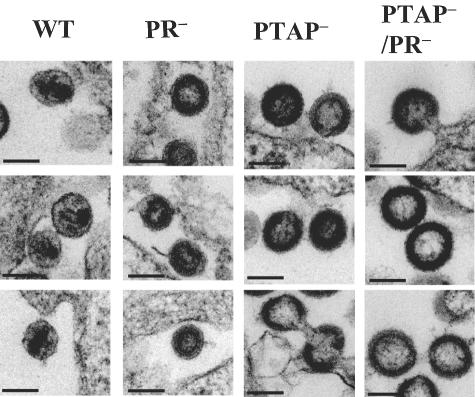
The morphology of the various mutant virions was confirmed by EM. Representative electron micrographs are shown in Fig. 5. We observed both mature and immature virions in particles generated by ON-T0, which contained wild-type Gag/Gag-Pol. Consistent with the literature, we observed immature virions in particles generated by T0-PTAP−, T0-PR− or T0-PTAP−/PR−, and virions tethered to one another or to the cell from T0-PTAP− and T0-PTAP−/PR samples.
FIG. 5.
Electron micrographs of virions derived from HIV-1 wild-type or mutant vectors. Virions were derived from pON-T0 (WT), pT0PR− (PR−), pT0PTAP− (PTAP−), or pT0PTAP− (PTAP−/PR−). Scale bars represent 100 nm (all panels).
We also examined the Gag polyprotein processing by Western analyses; a representative analysis is shown in Fig. 6. This and other western analyses demonstrated that virions from ON-T0 or T0-PTAP− contained mostly processed Gag products. This is generally consistent with previous studies although we did not observe the slight accumulation of p41 and p25 Gag processing intermediates in these mutant virions (9, 23). In contrast, virions from T0-PR− or pT0-PTAP−/PR− had unprocessed Gag.
FIG. 6.
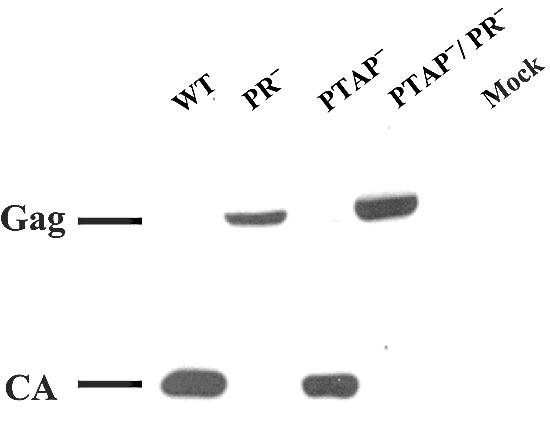
Western analysis of the cell-free virion proteins from wild-type or mutant HIV-1 vectors. Abbreviations are the same as described in the legend to Fig. 5. Mock, mock-transfected samples.
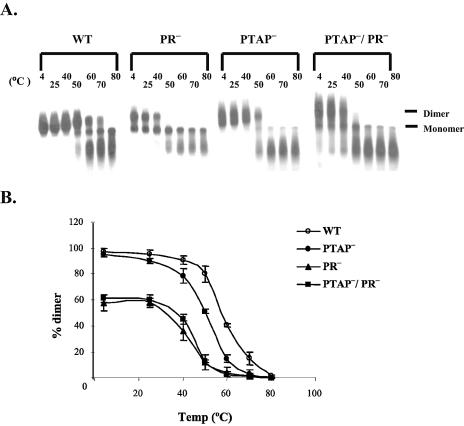
We then examined the thermostability of the dimeric virion RNA by nondenaturing Northern blotting using a probe generated from a DNA fragment containing gag sequences (18). A representative Northern blot is shown in Fig. 7A; results from three independent sets of experiments were quantified by phosphorimaging analyses and averaged (Fig. 7B). The virion RNA from T0 (wild type) had the following thermostability profile: most RNAs were dimeric after 50°C incubation, approximately half remained dimeric after 60°C, and most RNAs became monomeric after 70°C and completely dissociated into monomers after 80°C incubation. For PTAP−-derived virion RNA, approximately half remained dimeric after 50°C incubation, and most became monomeric after 60°C and completely dissociated into monomers after 70°C incubation. RNAs isolated from PR−- and PTAP−/PR−-derived virions had a similar thermostability profile: RNAs were partially monomeric even when the samples were kept at 4°C at all times, most of the remaining dimers dissociated at 50°C, and very few dimers were detected after 60°C incubation. Therefore, the thermostability of the ON-PTAP−-derived RNA dimer had an intermediate phenotype: it was more stable than those from PR− viruses and yet distinctly less heat stable than that of the wild-type virus.
FIG. 7.
Nondenaturing Northern analyses of cell-free virion RNAs from HIV-1. (A) Representative nondenaturing Northern blots of cell-free virion RNAs. RNAs were incubated at the indicated temperatures for 10 min prior to electrophoresis, and 32P-labeled gag riboprobes were used in these analyses. (B) Thermostability of the virion RNA dimers from wild-type or mutant HIV-1 particles. Data from three independent experiments are shown as means ± standard deviations. Abbreviations are the same as described in the legend to Fig. 5.
Taken together, the results from the MLV capsid mutant and the HIV-1 PTAP− mutant indicated that the processing of viral Gag polyproteins is not sufficient to convert the immature viral RNA dimer into mature RNA dimer with higher stability. Rather, other factors are also required for the maturation of the viral RNA dimer.
DISCUSSION
Proteolytic cleavage of the viral Gag proteins is one of the requirements for the maturation of virion morphology and the increased stability of the RNA dimer. In this report, we examined the properties of two gag mutants, and found that although Gag polyproteins in these mutant virions were mostly processed, their particles had immature virion morphology and the thermostability of the RNA dimer was lower than that of wild type viruses. These results indicated that proteolytic cleavage of Gag is not sufficient for mediating the maturation of viral RNA dimer. We propose that the maturation of the virion morphology and RNA dimer are linked.
In the two mutants we examined, proteolytic cleavage affected the stability of the RNA dimers differently. Consistent with the literature, we observed that viral RNAs were almost entirely dimeric in the MLV PR− mutant (18) but present in both monomeric and dimeric forms in HIV-1 PR− mutant (16). The RNA dimer stability of MLV PR− mutant and MLV CA mutant were indistinguishable, indicating that proteolytic cleavage does not significantly promote RNA dimer stability in the MLV CA mutant. In contrast, the virion RNA property of HIV-1 PR− mutant and HIV-1 PTAP− mutant were quite different; most of the RNAs in PTAP− mutant were dimeric whereas the RNAs in PR− mutants were monmeric and dimeric. The monomeric RNA observed in the HIV-1 PR− deficient mutant could be monomer or dissociated dimer. Therefore, proteolytic processing of the HIV-1 PTAP− mutant could either promote RNA dimerization, or stabilize fragile dimers. It is possible that the different effects of proteolytic cleavage on RNA properties are specific to the mutants used. However, it is also possible that the different effects of Gag processing on virion RNA reflected distinct stages in MLV and HIV-1 RNA maturation. We speculate that MLV virion RNAs are immature in the PR− mutant; after proteolytic cleavage and other processes involved in virion maturation, these RNAs become mature dimers. HIV-1 virion RNAs are immature prior to proteolytic cleavage, become an intermediate form after proteolytic processing, and fully mature after the occurrence of other essential processes for virion maturation. Currently, we cannot distinguish whether the different effects of proteolytic cleavage are mutant-specific or virus-specific; analyses of additional Gag mutants are needed to distinguish these two possibilities.
The effect of CA mutation on retroviral replication.
Studies of several retroviruses indicate that the CA domains of Gag polyproteins play an important role in virus assembly (2, 3, 5, 10, 14, 24, 26, 29, 40, 46, 49, 56, 64), and the mature CA proteins are important in virion core formation and postentry events of viral replication (1, 10, 14, 20, 22, 39, 46, 53, 57, 58). Therefore, mutations in CA can generate defects in virus assembly and/or postentry events (8, 30, 51, 60, 61). The CA mutant we used in this report has a distinct phenotype: although virion production, protein processing, and RNA packaging are comparable to wild type, the CA mutant displays immature virion morphology and RNA dimer stability. To our knowledge, this is the first example that mutation in CA can cause a defect in viral RNA dimer maturation. It is most likely that other CA mutants also have RNA maturation defects, but such defects were not described previously because most of the analyses for CA mutant do not include RNA stability studies.
While the experiments described in this study were performed, the crystal structure of the hexameric amino-terminal domain of MLV CA was solved (33). The crystal structure revealed that part of the conserved 12 amino acids deleted in pMLVCAΔ12 was in a conserved α helical structure (α3), which was proposed to be part of the contact between different CA molecules during core formation. Additionally, this deletion included the Asp 54, which formed a salt bridge with Pro 1, and was critical for β1-β2 N-terminal hairpin loop formation (33). The deletion of these 12 amino acids most likely affected the CA-CA interactions, thus causing a defect in virion maturation.
In this report, we used two Gag mutants to demonstrate that although proteolytic cleavage occurred, viral RNAs were not promoted to the same thermostability as the wild type viruses. Although we did not observe major Gag cleavage defects in the two mutants examined, we could not rule out the possibility that these mutants have subtle defects in Gag cleavage, which ultimately affected RNA maturation. Nevertheless, our results demonstrated that factors other than proteolytic cleavage of most of the Gag polyproteins are required to fully convert the virion RNA to the mature form. Several other factors have been documented to affect the properties of virion RNA. It was shown that virion RNA is monomeric in a Rous sarcoma virus (RSV) MA mutant (37); experimental evidence indicated that Gag targeting (45), rather than other properties of Gag or viral RNA (19), resulted in this phenotype. The incorporation and correct processing of pol gene products are important for HIV-1 dimer maturation (4, 47, 48).
The effects of blocking individual cleavage sites of HIV-1 and MLV Gag via mutating the cleavage site had also been examined (36, 48). In HIV-1, blocking the p2-NC junction prevented the formation of the mature RNA dimer; because this mutant had an immature morphology, it was proposed that virion core formation is important for RNA dimer maturation (48). In MLV, blocking the processing of the p12-CA junction eliminates the virus infectivity but does not affect the RNA dimer stability. Interestingly, virions with p12-CA blocks have varied morphology: some are immature and others have an aberrant morphology with dark interior similar to mature virion but without the clear border surrounding the dark interior (36). Both of the mutants we studied in this report had an immature morphology and lacked a condensed core. We speculate that structural rearrangement of virion components is necessary for RNA maturation; this rearrangement often results in observable morphological differences, such as the formation of the condensed core or a condensed region (e.g., the MLV p12-CA processing mutant). Mutations in Gag can cause defects in this rearrangement and prevent the maturation of RNA dimers.
Acknowledgments
We thank Alan Rein and Rebecca Craven for discussions and insightful suggestions on experiments, Anne Arthur for expert editorial help, Alan Rein for critical reading of the manuscript, and Sook-Kyung Lee for generating 293T/SR2 cells.
This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
REFERENCES
- 1.Alin, K., and S. P. Goff. 1996. Amino acid substitutions in the CA protein of Moloney murine leukemia virus that block early events in infection. Virology 222:339-351. [DOI] [PubMed] [Google Scholar]
- 2.Alin, K., and S. P. Goff. 1996. Mutational analysis of interactions between the Gag precursor proteins of murine leukemia viruses. Virology 216:418-424. [DOI] [PubMed] [Google Scholar]
- 3.Borsetti, A., A. Ohagen, and H. G. Gottlinger. 1998. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J. Virol. 72:9313-9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buxton, P., G. Tachedjian, and J. Mak. 2005. Analysis of the contribution of reverse transcriptase and integrase proteins to retroviral RNA dimer conformation. J. Virol. 79:6338-6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chazal, N., C. Carriere, B. Gay, and P. Boulanger. 1994. Phenotypic characterization of insertion mutants of the human immunodeficiency virus type 1 Gag precursor expressed in recombinant baculovirus-infected cells. J. Virol. 68:111-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheslock, S. R., J. A. Anderson, C. K. Hwang, V. K. Pathak, and W. S. Hu. 2000. Utilization of nonviral sequences for minus-strand DNA transfer and gene reconstitution during retroviral replication. J. Virol. 74:9571-9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheslock, S. R., D. T. Poon, W. Fu, T. D. Rhodes, L. E. Henderson, K. Nagashima, C. F. McGrath, and W. S. Hu. 2003. Charged assembly helix motif in murine leukemia virus capsid: an important region for virus assembly and particle size determination. J. Virol. 77:7058-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craven, R. C., A. E. Leure-duPree, R. A. Weldon, Jr., and J. W. Wills. 1995. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J. Virol. 69:4213-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demirov, D. G., J. M. Orenstein, and E. O. Freed. 2002. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J. Virol. 76:105-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorfman, T., A. Bukovsky, A. Ohagen, S. Hoglund, and H. G. Gottlinger. 1994. Functional domains of the capsid protein of human immunodeficiency virus type 1. J. Virol. 68:8180-8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DuBridge, R. B., P. Tang, H. C. Hsia, P. M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehrlich, L. S., B. E. Agresta, and C. A. Carter. 1992. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J. Virol. 66:4874-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng, Y. X., T. D. Copeland, L. E. Henderson, R. J. Gorelick, W. J. Bosche, J. G. Levin, and A. Rein. 1996. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc. Natl. Acad. Sci. USA 93:7577-7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitzon, T., B. Leschonsky, K. Bieler, C. Paulus, J. Schroder, H. Wolf, and R. Wagner. 2000. Proline residues in the HIV-1 NH2-terminal capsid domain: structure determinants for proper core assembly and subsequent steps of early replication. Virology 268:294-307. [DOI] [PubMed] [Google Scholar]
- 15.Freed, E. O. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 16.Fu, W., R. J. Gorelick, and A. Rein. 1994. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J. Virol. 68:5013-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu, W., and W. S. Hu. 2003. Functional replacement of nucleocapsid flanking regions by heterologous counterparts with divergent primary sequences: effects of chimeric nucleocapsid on the retroviral replication cycle. J. Virol. 77:754-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu, W., and A. Rein. 1993. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 67:5443-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garbitt, R. A., J. A. Albert, M. D. Kessler, and L. J. Parent. 2001. trans-Acting inhibition of genomic RNA dimerization by Rous sarcoma virus matrix mutants. J. Virol. 75:260-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gitti, R. K., B. M. Lee, J. Walker, M. F. Summers, S. Yoo, and W. I. Sundquist. 1996. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science 273:231-235. [DOI] [PubMed] [Google Scholar]
- 21.Hansen, M. S., and E. Barklis. 1995. Structural interactions between retroviral Gag proteins examined by cysteine cross-linking. J. Virol. 69:1150-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu, H. W., P. Schwartzberg, and S. P. Goff. 1985. Point mutations in the P30 domain of the gag gene of Moloney murine leukemia virus. Virology 142:211-214. [DOI] [PubMed] [Google Scholar]
- 23.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jowett, J. B., D. J. Hockley, M. V. Nermut, and I. M. Jones. 1992. Distinct signals in human immunodeficiency virus type 1 Pr55 necessary for RNA binding and particle formation. J. Gen. Virol. 73:3079-3086. [DOI] [PubMed] [Google Scholar]
- 25.Katoh, I., Y. Yoshinaka, A. Rein, M. Shibuya, T. Odaka, and S. Oroszlan. 1985. Murine leukemia virus maturation: protease region required for conversion from “immature” to “mature” core form and for virus infectivity. Virology 145:280-292. [DOI] [PubMed] [Google Scholar]
- 26.Kattenbeck, B., A. von Poblotzki, A. Rohrhofer, H. Wolf, and S. Modrow. 1997. Inhibition of human immunodeficiency virus type 1 particle formation by alterations of defined amino acids within the C terminus of the capsid protein. J. Gen. Virol. 78:2489-2496. [DOI] [PubMed] [Google Scholar]
- 27.Landau, N. R., K. A. Page, and D. R. Littman. 1991. Pseudotyping with human T-cell leukemia virus type I broadens the human immunodeficiency virus host range. J. Virol. 65:162-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leis, J., D. Baltimore, J. M. Bishop, J. Coffin, E. Fleissner, S. P. Goff, S. Oroszlan, H. Robinson, A. M. Skalka, H. M. Temin, et al. 1988. Standardized and simplified nomenclature for proteins common to all retroviruses. J. Virol. 62:1808-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang, C., J. Hu, J. B. Whitney, L. Kleiman, and M. A. Wainberg. 2003. A structurally disordered region at the C terminus of capsid plays essential roles in multimerization and membrane binding of the Gag protein of human immunodeficiency virus type 1. J. Virol. 77:1772-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mammano, F., A. Ohagen, S. Hoglund, and H. G. Gottlinger. 1994. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J. Virol. 68:4927-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, A. D., and C. Buttimore. 1986. Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Mol. Cell. Biol. 6:2895-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, A. D., J. V. Garcia, N. von Suhr, C. M. Lynch, C. Wilson, and M. V. Eiden. 1991. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J. Virol. 65:2220-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mortuza, G. B., L. F. Haire, A. Stevens, S. J. Smerdon, J. P. Stoye, and I. A. Taylor. 2004. High-resolution structure of a retroviral capsid hexameric amino-terminal domain. Nature 431:481-485. [DOI] [PubMed] [Google Scholar]
- 34.Naldini, L., U. Blomer, F. H. Gage, D. Trono, and I. M. Verma. 1996. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc. Natl. Acad. Sci. USA 93:11382-11388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohagen, A., R. B. Luftig, A. S. Reicin, L. Yin, K. Ikuta, T. Kimura, S. P. Goff, and S. Hoglund. 1997. The morphology of the immature HIV-1 virion. Virology 228:112-114. [DOI] [PubMed] [Google Scholar]
- 36.Oshima, M., D. Muriaux, J. Mirro, K. Nagashima, K. Dryden, M. Yeager, and A. Rein. 2004. Effects of blocking individual maturation cleavages in murine leukemia virus Gag. J. Virol. 78:1411-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parent, L. J., T. M. Cairns, J. A. Albert, C. B. Wilson, J. W. Wills, and R. C. Craven. 2000. RNA dimerization defect in a Rous sarcoma virus matrix mutant. J. Virol. 74:164-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reicin, A. S., A. Ohagen, L. Yin, S. Hoglund, and S. P. Goff. 1996. The role of Gag in human immunodeficiency virus type 1 virion morphogenesis and early steps of the viral life cycle. J. Virol. 70:8645-8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reicin, A. S., S. Paik, R. D. Berkowitz, J. Luban, I. Lowy, and S. P. Goff. 1995. Linker insertion mutations in the human immunodeficiency virus type 1 gag gene: effects on virion particle assembly, release, and infectivity. J. Virol. 69:642-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rein, A., L. E. Henderson, and J. G. Levin. 1998. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 23:297-301. [DOI] [PubMed] [Google Scholar]
- 42.Rhodes, T. D., O. Nikolaitchik, J. Chen, D. Powell, and W. S. Hu. 2005. Genetic recombination of human immunodeficiency virus type 1 in one round of viral replication: effects of genetic distance, target cells, accessory genes, and lack of high negative interference in crossover events. J. Virol. 79:1666-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riggs, J. L., R. M. McAllister, and E. H. Lennette. 1974. Immunofluorescent studies of RD-114 virus replication in cell culture. J. Gen. Virol. 25:21-29. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Scheifele, L. Z., R. A. Garbitt, J. D. Rhoads, and L. J. Parent. 2002. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc. Natl. Acad. Sci. USA 99:3944-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwartzberg, P., J. Colicelli, M. L. Gordon, and S. P. Goff. 1984. Mutations in the gag gene of Moloney murine leukemia virus: effects on production of virions and reverse transcriptase. J. Virol. 49:918-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shehu-Xhilaga, M., S. M. Crowe, and J. Mak. 2001. Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J. Virol. 75:1834-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shehu-Xhilaga, M., H. G. Kraeusslich, S. Pettit, R. Swanstrom, J. Y. Lee, J. A. Marshall, S. M. Crowe, and J. Mak. 2001. Proteolytic processing of the p2/nucleocapsid cleavage site is critical for human immunodeficiency virus type 1 RNA dimer maturation. J. Virol. 75:9156-9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Srinivasakumar, N., M. L. Hammarskjold, and D. Rekosh. 1995. Characterization of deletion mutations in the capsid region of human immunodeficiency virus type 1 that affect particle formation and Gag-Pol precursor incorporation. J. Virol. 69:6106-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stoltzfus, C. M., and P. N. Snyder. 1975. Structure of B77 sarcoma virus RNA: stabilization of RNA after packaging. J. Virol. 16:1161-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strambio-de-Castillia, C., and E. Hunter. 1992. Mutational analysis of the major homology region of Mason-Pfizer monkey virus by use of saturation mutagenesis. J. Virol. 66:7021-7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 53.Tang, S., T. Murakami, B. E. Agresta, S. Campbell, E. O. Freed, and J. G. Levin. 2001. Human immunodeficiency virus type 1 N-terminal capsid mutants that exhibit aberrant core morphology and are blocked in initiation of reverse transcription in infected cells. J. Virol. 75:9357-9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vogt, V. M. 1996. Proteolytic processing and particle maturation. Curr. Top. Microbiol. Immunol. 214:95-131. [DOI] [PubMed] [Google Scholar]
- 55.Vogt, V. M. 1997. Retroviral virions and genomes. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 56.von Poblotzki, A., R. Wagner, M. Niedrig, G. Wanner, H. Wolf, and S. Modrow. 1993. Identification of a region in the Pr55gag-polyprotein essential for HIV-1 particle formation. Virology 193:981-985. [DOI] [PubMed] [Google Scholar]
- 57.von Schwedler, U. K., T. L. Stemmler, V. Y. Klishko, S. Li, K. H. Albertine, D. R. Davis, and W. I. Sundquist. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 17:1555-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, C. T., and E. Barklis. 1993. Assembly, processing, and infectivity of human immunodeficiency virus type 1 gag mutants. J. Virol. 67:4264-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wiegers, K., G. Rutter, H. Kottler, U. Tessmer, H. Hohenberg, and H. G. Krausslich. 1998. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J. Virol. 72:2846-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Willems, L., P. Kerkhofs, L. Attenelle, A. Burny, D. Portetelle, and R. Kettmann. 1997. The major homology region of bovine leukaemia virus p24gag is required for virus infectivity in vivo. J. Gen. Virol. 78:637-640. [DOI] [PubMed] [Google Scholar]
- 61.Wills, J. W., and R. C. Craven. 1991. Form, function, and use of retroviral gag proteins. AIDS 5:639-654. [DOI] [PubMed] [Google Scholar]
- 62.Yeager, M., E. M. Wilson-Kubalek, S. G. Weiner, P. O. Brown, and A. Rein. 1998. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc. Natl. Acad. Sci. USA 95:7299-7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yee, J. K., A. Miyanohara, P. LaPorte, K. Bouic, J. C. Burns, and T. Friedmann. 1994. A general method for the generation of high-titer, pantropic retroviral vectors: highly efficient infection of primary hepatocytes. Proc. Natl. Acad. Sci. USA 91:9564-9568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, W. H., D. J. Hockley, M. V. Nermut, Y. Morikawa, and I. M. Jones. 1996. Gag-Gag interactions in the C-terminal domain of human immunodeficiency virus type 1 p24 capsid antigen are essential for Gag particle assembly. J Gen. Virol. 77:743-751. [DOI] [PubMed] [Google Scholar]