Metazoan organisms are subject to invasion by a wide range of microbial pathogens and have, as a result, evolved a range of defensive measures. While scientific attention historically has focused on adaptive immune responses, such as antibodies and cytotoxic T cells, it has of late become increasingly apparent that innate immunity also plays a key role in shielding animals from infection (62). Unlike adaptive immunity, innate immunity acts immediately and thus can prevent an infection from getting started. Innate immunity may be particularly effective at shielding animals from infection by opportunistic or zoonotic pathogens. However, because innate immune responses are largely invariant, microorganisms that are common pathogens of a given animal species will frequently have evolved mechanisms that confer resistance to relevant innate immune responses in that species. Indeed, comparative analyses of a host innate immune response and the microbial countermeasures to that response can provide key insights into the biological mechanisms underlying these antagonistic processes.
A wide range of innate immune responses have been identified; many of these rely on the recognition of a “pathogen-associated molecular pattern” (PAMP) that is characteristic of a given type of pathogen (62). In this paper, I will review our current understanding of one protein family, the apolipoprotein B mRNA editing enzyme catalytic polypeptide 3 (APOBEC3) proteins, which can confer innate immunity to a wide range of exogenous retroviruses. Moreover, individual APOBEC3 family members can also block the replication of hepatitis B virus, a distant relative of retroviruses, and inhibit the replication of retrotransposons, endogenous transposable elements related to retroviruses that can disrupt the integrity of host cell genomes.
APOBEC3 PROTEIN FAMILY
The genomes of humans and other primates encode at least five, and possibly up to seven, APOBEC3 proteins, all of which are encoded by a single gene cluster on chromosome 22 (20). This gene cluster appears to be unique to primates, as the genomes of several other mammalian species, such as mice, cats, and cows, encode only a single APOBEC3 protein (31, 36). The fact that the single APOBEC3 gene found in these other vertebrates is at the syntenic chromosomal location relative to the primate APOBEC3 gene cluster (66) indicates that the primate APOBEC3 gene family appeared relatively recently in vertebrate evolution due to gene duplication. This finding, together with the observation that the rate of nonsynonymous nucleotide substitutions has been significantly higher than the rate of synonymous substitutions during the evolution of several members of the APOBEC3 gene family, suggests that primate APOBEC3 genes have been under significant selective pressure (47a, 75). As discussed below, it seems possible that the evolutionary pressure for amplification and diversification of the primate APOBEC3 genes arose from the appearance of viral mechanisms that are able to neutralize the APOBEC3-mediated inhibition of retroviral replication.
The APOBEC3 protein family was so named because all APOBEC3 proteins display significant homology to APOBEC1 and to two other human gene products, APOBEC2 and activation-induced deaminase (AID) (20). APOBEC1, the founding member of this extended protein family, edits a single C residue to U on the mRNA encoding apolipoprotein B (ApoB), thereby introducing a premature stop codon and inducing production of a truncated form of ApoB that has a different biological function (66). While the role of APOBEC2 remains unclear, AID functions in activated B cells, where it randomly edits dC residues to dU in the immunoglobulin gene locus (5, 68). This causes immunoglobulin gene diversification, which is followed by selection for gene variants that express antibodies with particularly high affinity for their target antigen. Importantly, while APOBEC1 edits an mRNA molecule, AID edits the single-stranded DNA that arises during the melting of a DNA duplex during active transcription (68). Nevertheless, APOBEC1 and AID, as well as APOBEC2, share a conserved cytidine deaminase active site (CDA), with the consensus sequence His-X-Glu-X23-28-Pro-Cys-X2-4-Cys, which is also conserved in all members of the APOBEC3 protein family (20, 66). A single copy of this active site is found in the 199-amino acid (aa) human APOBEC3A (hA3A) and 190-aa APOBEC3C (hA3C) gene products, while two active sites are found in human APOBEC3B (hA3B, 382 aa), APOBEC3F (hA3F, 373 aa), and APOBEC3G (hA3G, 384 aa). Presumably, the APOBEC3 family members that contain two CDAs arose due to the tandem duplication of an ancestral gene that contained a single CDA. The other two APOBEC3 family members, APOBEC3D and APOBEC3E, do not appear to give rise to any protein product, although APOBEC3D does contain a predicted open reading frame (20). In general, the human APOBEC3 proteins show ∼50% sequence homology to each other at the amino acid level. Consistent with the presence of one or two consensus CDAs in each APOBEC3 family member, four of the related gene products, i.e., hA3B, hA3C, hA3F, and hA3G, have now been shown to function as cytidine deaminases (see below).
HUMAN IMMUNODEFICIENCY VIRUS AND THE APOBEC3 PROTEINS
The initial characterization and sequencing of the human APOBEC3 gene family led to the proposal that these genes encoded a novel set of orphan cytidine deaminases, based on their similarity to APOBEC1 and AID (20). However, the subsequent observation that APOBEC3 proteins can function as innate inhibitors of retroviral replication came from unrelated studies focused on the pathogenic lentivirus human immunodeficiency virus type 1 (HIV-1) and, more specifically, on the role and mechanism of action of the HIV-1 virion infectivity factor (Vif) gene product, an ∼192-aa cytoplasmic protein.
Mutants of HIV-1 lacking a functional vif gene fail to replicate in primary CD4+ T cells or macrophages and are also unable to replicate in a subset of CD4+ T-leukemia cell lines, such as CEM (16, 56). These cells types are referred to as “nonpermissive.” In contrast, a small number of “permissive” T-cell lines replicate Vif-deficient and wild-type HIV-1 with equal efficiency. One such permissive T-cell line, termed CEM-SS, is a subclone of the nonpermissive cell line CEM.
By fusing nonpermissive and permissive cells to form heterokaryons, it was demonstrated that the nonpermissive phenotype is dominant, thus suggesting that nonpermissive cells express an inhibitor that selectively blocks the replication of Vif-deficient HIV-1 (33, 55). Based on this insight, Sheehy et al. (53) used a cDNA subtraction strategy to identify mRNAs expressed in CEM that are absent in the closely related permissive CEM-SS subclone. One of these CEM-derived cDNAs, encoding hA3G, was found to be sufficient to confer the nonpermissive phenotype when expressed in CEM-SS cells. Moreover, Northern analysis demonstrated that hA3G mRNA was expressed at readily detectable levels in nonpermissive primary cells or cell lines but at low or undetectable levels in permissive cells.
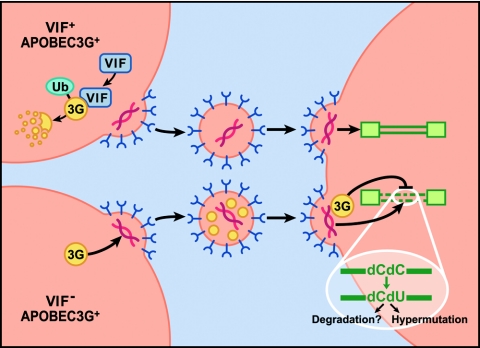
Previous work looking at the properties of Vif-deficient HIV-1 grown in nonpermissive cells had demonstrated that Vif-deficient and wild-type HIV-1 infected these cells with equal efficiency and gave rise to the same level of apparently normal progeny viral particles. However, the mutant virions were then unable to induce productive infections when added to either permissive or nonpermissive CD4+ cells (16, 56, 65). Similarly, artificial expression of hA3G in HIV-1 producer cells was found to render Vif-deficient progeny virions defective yet did not affect virus production (53). It is now known that hA3G is selectively incorporated into Vif-deficient HIV-1 virions during their morphogenesis in producer cells (Fig. 1). Upon infection of a subsequent susceptible cell, hA3G remains associated with the reverse transcription complex during virion uncoating and then edits dC residues to dU on the nascent proviral minus strand during the first step of reverse transcription (18, 34, 70, 74). Thus, hA3G is similar to AID in that it edits single-stranded DNA, not RNA. Editing by hA3G occurs in a graded frequency across the HIV-1 genome, with more extensive editing occurring in regions of the genome that are reverse transcribed first (70).
FIG. 1.
Schematic representation of the mechanisms of action of APOBEC3G and the HIV-1 Vif protein. See text for detailed discussion. Ub, ubiquitin; 3G, APOBEC3G.
Editing of the HIV-1 DNA minus strand by hA3G can be remarkably efficient, with up to 20% of all minus-strand dC residues being deaminated to dU. This extensive editing can have two possible effects on the HIV-1 provirus. One possibility is that reverse transcription proceeds normally except that the introduced dU residues template incorporation of A residues into the proviral plus strand, thus resulting in G-to-A hypermutation of the HIV-1 provirus (Fig. 1) (18, 34, 70, 74). While the resultant provirus can then be integrated normally into the target cell genome, this hypermutation renders the provirus defective. Alternatively, editing mediated by hA3G may destabilize HIV-1 reverse transcripts, leading to their degradation (Fig. 1) (56, 65). This phenomenon remains poorly understood, although it has been suggested that it might result from the action of cellular DNA repair enzymes, such as the uracil DNA glycosylase UNG (50).
The ability of hA3G to inhibit Vif-deficient HIV-1 replication prompted investigators to ask whether other members of the human APOBEC protein family could exhibit the same phenotype. In fact, none of the human cytidine deaminases outside the APOBEC3 subfamily, i.e., APOBEC1, APOBEC2, and AID, affect HIV-1 replication (3, 67, 76). Although hA3A was found to be inactive against HIV-1, hA3F proved to be almost as potent an inhibitor of Vif-deficient HIV-1 as hA3G (3, 27, 45, 67, 76). While hA3G and hA3F both reduce the infectivity of Vif-deficient HIV-1 virions 10- to >20-fold, hA3B was found to inhibit Vif-deficient HIV-1 infectivity 5- to 10-fold, while hA3C inhibited Vif-deficient HIV-1 only ∼2-fold (3, 11, 26, 45).
Analyses of HIV-1 proviruses produced from virions generated in the presence of the inhibitory APOBEC3 proteins revealed an interesting phenomenon. While proviruses generated from virions produced in the presence of hA3G were almost invariably edited at dC residues found in the sequence context CC* (where the asterisk indicates the edited dC residue), proviruses produced from virions generated in the presence of hA3F or hA3B were edited at dC residues in the sequence context TC* (3, 11, 18, 27, 67). Finally, hA3C induced editing at dC residues located 3′ to either dC or dT, although only inefficiently (26). Interestingly, in the case of the APOBEC3 proteins containing two CDAs, the carboxy-terminal CDA determines the sequence preference of the protein, i.e., a chimeric protein that contains the amino-terminal CDA from hA3G and the carboxy-terminal CDA from hA3F preferentially edits the sequence TC* (17). As discussed in more detail below, these sequence preferences have implications for the question of whether APOBEC3 proteins indeed edit HIV-1 in infected patients.
While the above studies were performed largely by using Vif-deficient HIV-1 mutants, research using wild-type HIV-1 showed that Vif expression made HIV-1 resistant to inhibition by not only hA3G but also hA3F and hA3C (3, 26, 67, 76). In contrast, wild-type HIV-1 was as sensitive as Vif-deficient HIV-1 to inhibition by hA3B (3, 11, 69). Analyses of the human tissue expression pattern of several APOBEC3 genes have revealed that hA3B mRNA is expressed at very low levels in human cells that are targets for HIV-1 infection in vivo (3, 11), such as peripheral blood mononuclear cells (but see reference 69 for a contrary position). In contrast, hA3G and hA3F mRNA was readily detectable in primary lymphoid cells and, indeed, in almost all human tissues examined (3, 27, 67). It appears possible that HIV-1 Vif has evolved to block the inhibitory activity of the two APOBEC3 proteins, i.e., hA3G and hA3F, that are expressed in the CD4+ cells that HIV-1 normally infects in vivo. In contrast, Vif has apparently not been selected for the ability to block the inhibitory action of hA3B, a human APOBEC3 protein that it would not normally encounter in vivo.
MECHANISM OF ACTION OF HIV-1 VIF
Comparative analysis of HIV-1 virions produced in hA3G-expressing cells revealed that hA3G is incorporated specifically into virions produced in the absence of Vif (Fig. 1). In contrast, hA3G virion incorporation is undetectable, or at least strongly inhibited, in the presence of Vif (8, 22, 37, 54, 59). The level of hA3G expressed in virion producer cells was also greatly reduced in the presence of Vif, thus suggesting that Vif compromises the stability of hA3G. Indeed, pulse-chase experiments revealed that the half-life of hA3G is greatly reduced in the presence of HIV-1 Vif. Moreover, this destabilization could be rescued by treatment of cells with proteasome inhibitors, thus implying that Vif was targeting hA3G for ubiquitination followed by proteasomal degradation (30, 37, 39, 54, 59). In fact, Vif was found to induce the polyubiquitination of hA3G when both proteins were coexpressed in the presence of a proteasome inhibitor (Fig. 1).
Subsequent analyses revealed that Vif interacts with hA3G in coexpressing cells and recruits hA3G to an E3 ubiquitin ligase complex consisting of the cellular proteins Cullin5, ElonginB, and ElonginC (71). The latter interaction is mediated by a highly conserved sequence motif in Vif, termed a suppressor of cytokine signaling (SOCS) box, which binds to ElonginC (38, 72). Mutation of the Vif SOCS box was found to block recruitment of the E3 ubiquitin ligase and to relieve the inhibition of hA3G function by HIV-1 Vif. The mechanism employed by Vif to destabilize hA3G is similar to the mechanisms of action of cellular proteins, termed “F-box” proteins, which act as a specific bridge between an E3 ubiquitin ligase complex and a protein targeted for ubiquitination and proteasomal degradation.
While HIV-1 Vif can also reduce the virion incorporation, and cellular expression level, of hA3F in coexpressing cells (29), Vif has no effect on hA3B incorporation into progeny virion particles or on hA3B stability (11). These disparate effects are explained by the observation that HIV-1 Vif interacts specifically with hA3F in coexpressing cells and promotes the degradation of hA3F via the ubiquitin-proteasomal pathway yet fails to detectably interact with hA3B (11, 29, 67).
OTHER EXOGENOUS VIRUSES AND THE APOBEC3 PROTEINS
HIV-1 entered the human population within the last century, so the ability of members of the human APOBEC3 protein family to inhibit the replication of HIV-1 is presumably not the driving force behind the evolution of these antiretroviral factors. In fact, humans are not naturally subject to infection by many retroviruses, as the only known exogenous human retroviruses are HIV-1 and HIV-2, which are zoonoses that crossed into the human population only recently, and human T-cell leukemia virus type I (HTLV-I) and HTLV-II, which are certainly human retroviruses but are also rare and geographically restricted. Of course, it could be argued that the relative paucity of exogenous retroviruses in the human population is a priori evidence that the APOBEC3 proteins are doing their job effectively. However, the fact that the human genome contains over 230,000 endogenous retroviruses (23) suggests that infection of our primate ancestors by exogenous retrovirus families that are now generally extinct could have been a source of selective pressure that favored the evolution and amplification of the primate APOBEC3 gene cluster.
These considerations raise two questions: first, whether human APOBEC3 proteins can inhibit the replication of nonhuman, potentially zoonotic retroviruses, and second, whether APOBEC3 proteins encoded by other species, in particular nonhuman primates and mice, can inhibit the replication of HIV-1 and/or of retroviruses that are commonly found in these nonhuman species.
Human immunodeficiency virus type 2.
While limited compared to the work performed with HIV-1, research using HIV-2 has shown that HIV-2 Vif, like HIV-1 Vif, can block the incorporation of hA3G and hA3F into HIV-1 virion particles and reduce the expression of both hA3G and hA3F in virion producer cells (67). Like HIV-1 Vif, HIV-2 Vif has no effect on the incorporation of hA3B into HIV-1 virion particles or on the intracellular level of hA3B expression (11). Therefore, it appears that HIV-2 Vif is functionally similar to HIV-1 Vif. Surprisingly, however, it has also been reported that an HIV-2 proviral clone lacking a functional vif gene can replicate effectively in a human T-cell line, termed H9, that expresses hA3G mRNA and that is nonpermissive for Vif-deficient HIV-1 (44). In contrast, primary CD4+ T-cells were found to be nonpermissive for both Vif-deficient proviral clones. These data suggest that HIV-2 may be somewhat resistant to inhibition by low levels of hA3G, even in the absence of a functional Vif protein.
Simian immunodeficiency viruses.
Nonhuman primates also encode a cluster of APOBEC3 genes, and research so far has focused on APOBEC3G variants derived from African green monkeys (agmA3G), rhesus macaques (macA3G), and chimpanzees (cpzA3G). The only simian immunodeficiency viruses (SIVs) that have been investigated so far are SIVs isolated from chimpanzees (SIVcpz) and African green monkeys (SIVagm) as well as an SIV recovered from a captive rhesus macaque (SIVmac) that actually represents a transmission in captivity of an SIV from a sooty mangabey (SIVsm). As HIV-2 is a zoonosis that arose from transmission of SIVsm to humans, one might predict that SIVmac would be similar in phenotype to HIV-2.
A summary of data generated using these reagents (4, 16a, 35, 36, 49) is shown in Table 1. Several important points emerge from these data. First, Vif-deficient HIV-1 is sensitive to all of the primate APOBEC3G proteins analyzed. Second, the HIV-1 and SIV Vif proteins all confer resistance to the APOBEC3G protein expressed by their cognate species. Third, wild-type HIV-1 is sensitive to both agmA3G and macA3G, thus implying that these primates should be nonpermissive for HIV-1. This is indeed observed, although whether APOBEC3 proteins play a role in this phenomenon remains to be established. Finally, it is interesting that the Vif protein from SIVmac is able to overcome all the APOBEC3 proteins analyzed, including hA3G, while SIVagm Vif acts only on the two monkey APOBEC3 proteins. Based on this finding, one would predict that human APOBEC3 proteins would prevent zoonotic infections by SIVagm but not by SIVmac or, more to the point, its close relative SIVsm. Indeed, while SIVagm has not crossed over into the human population, SIVsm has done so repeatedly to give rise to the various HIV-2 isolates. While this correlation is compelling, it should be noted that there are other determinants of lentiviral species tropism that may be as important, or more important, in determining which SIV variants have the potential to become zoonosis. Nevertheless, it seems likely that the inability of SIVagm Vif to neutralize the innate resistance to retroviral infection provided by hA3G would tend to make humans nonpermissive for SIVagm. Of note, a recent analysis of the biological activity of the vif gene products encoded by a wide range of SIVs (16a) showed that the Vif proteins encoded by SIVcpz and SIVsm are unique among SIV Vif proteins in being able to rescue the replication of a Vif-deficient HIV-1 virus in the nonpermissive human T-cell line H9. In contrast, Vif proteins derived from SIVs isolated from red-capped mangabeys, Sykes monkeys, mandrills, L'Hoest's monkeys, sun-tailed monkeys, and DeBrazza's monkeys shared the inability of SIVagm Vif to rescue the infectivity of Vif-deficient HIV-1 virions released by nonpermissive H9 cells (16a). Strikingly, all these SIVs also share with SIVagm the fact that they have never been detected in humans.
TABLE 1.
Species specificity of primate immunodeficiency virus Vif proteinsa
| Vif protein(s) | Result with:
|
|||
|---|---|---|---|---|
| Human APOBEC3G | Chimpanzee APOBEC3G | African green monkey APOBEC3G | Rhesus macaque APOBEC3G | |
| HIV-1 wild type | + | + | − | − |
| HIV-1ΔVif | − | − | − | − |
| HIV-1ΔVif + HIV-1 Vif | + | + | − | − |
| HIV-1ΔVif + SIVcpz Vif | + | + | − | − |
| HIV-1ΔVif + SIVagm Vif | − | − | + | + |
| HIV-1ΔVif + SIVmac Vif | + | + | + | + |
The ability of the indicated primate immunodeficiency virus Vif protein(s) to rescue the infectivity of a Vif-deficient HIV-1 proviral clone (HIV-1ΔVif) in trans, in the presence of the indicated primate APOBEC3G protein, is indicated. +, resistant; −, sensitive.
As hA3G and agmA3G are 77% identical at the amino acid level, it is of interest to ask why hA3G is nonresponsive to SIVagm Vif and, similarly, why agmA3G is nonresponsive to HIV-1 Vif. In fact, this lack of response is due to the fact that HIV-1 Vif is unable to bind to agmA3G in vivo (4). This inability to interact was in turn mapped to a single amino acid difference between hA3G and agmA3G, an aspartic acid at position 128 in hA3G that is a lysine in agmA3G (4, 35, 49). Switching these two charged residues reversed the observed specificity, i.e., agmA3G bearing an aspartic acid at residue 128 was fully responsive to HIV-1 Vif. Therefore, it appears that a single residue difference in hA3G versus agmA3G may play a role in blocking the zoonotic infection of humans by SIVagm.
One final difference between SIVmac and SIVagm on the one hand and HIV-1 on the other is that both SIVmac and SIVagm appear highly sensitive to inhibition by both hA3B and hA3C (69). In the case of hA3C, sensitivity was seen only with Vif-deficient variants of SIVmac and SIVagm. However, as in the case of HIV-1, both wild-type and Vif-deficient SIVmac and SIVagm were inhibited by hA3B, with the degree of inhibition being significantly more complete than the moderate, 5- to 10-fold inhibition seen with HIV-1 (3, 11).
Foamy viruses and other complex retroviruses.
Retroviruses can be divided broadly into complex and simple variants (10). Simple retroviruses encode the canonical Gag, Pol, and Env proteins and little or nothing else. Examples of simple retroviruses include murine leukemia virus (MLV), avian leukemia virus, and a range of other oncogenic animal retroviruses. Complex retroviruses encode not only Gag, Pol, and Env but also at least two other viral proteins, one of which is a transcriptional activator of the viral long terminal repeat (LTR) promoter. There are three distinct families of complex retroviruses, namely, the lentiviruses, the T-cell leukemia viruses, and the foamy viruses.
As discussed above, the lentiviruses HIV-1, HIV-2, and SIV encode a protein termed Vif that neutralizes the inhibitory activity of APOBEC3 proteins expressed in their normal target cells in their normal host species. Many nonprimate lentiviruses, such as feline immunodeficiency virus and visna-maedi virus, an ungulate lentivirus, also encode Vif proteins, but nothing is currently known about their function. One lentivirus, equine infectious anemia virus (EIAV), does not encode an obvious Vif homolog, and EIAV has been shown to be sensitive to inhibition by hA3G (34). How EIAV avoids inhibition by the APOBEC3 gene product(s) that are presumably expressed by equine cells remains unknown.
While HTLV-I can productively infect human lymphoid cells that express readily detectable levels of hA3G and hA3F, little is currently known about how HTLV-I avoids inhibition by these host proteins. However, it has been reported that HTLV-I is resistant to inhibition by levels of hA3G that block Vif-deficient HIV-1 infectivity (42). While no mechanism for this resistance has been proposed, HTLV-I does not encode an obvious Vif homolog and HTLV-I infection does not reduce hA3G protein expression.
The third complex retrovirus family consists of the foamy viruses. Foamy viruses infect almost all vertebrate species, including a wide range of nonhuman primates and apes, yet are only rarely detected in humans (28). The few instances where human foamy virus infection could be demonstrated involved either animal handlers or African bush meat hunters, and no further transmission to human contacts has been documented.
Work characterizing the foamy virus replication cycle has generally focused on a proviral clone that is originally of chimpanzee origin, termed primate foamy virus (PFV) (28). PFV encodes Gag, Pol, and Env proteins as well as Tas, a transcriptional transactivator of the PFV LTR promoter, and a fifth protein termed Bet. Bet is an ∼53-kDa cytoplasmic protein that shows no evident sequence homology to HIV-1 Vif and does not contain an obvious SOCS box, the motif used by Vif to recruit an E3 ubiquitin ligase to hA3G and hA3F. Despite this lack of sequence similarity, Bet significantly enhances the infectivity of PFV or HIV-1 virions produced in the presence, but not the absence, of either hA3G or agmA3G (47). This increased infectivity correlated with a marked reduction in the incorporation of either hA3G or agmA3G into progeny virion particles. While PFV Bet, like HIV-1 Vif, was able to interact specifically with hA3G and hA3F in coexpressing cells, PFV Bet differed from HIV-1 Vif in that it did not destabilize either protein.
Similar studies performed using a distantly related feline foamy virus (FFV) showed that inactivation of the FFV Bet gene resulted in both G-to-A hypermutation of FFV passed in primary cat cells and a dramatic reduction in FFV virion infectivity (31). Moreover, FFV Bet bound the feline APOBEC3 protein in coexpressing cells and reduced the packaging of APOBEC3 into FFV virions. However, FFV Bet again had little or no effect on cellular APOBEC3 protein expression. Together, these data demonstrate that foamy virus Bet proteins bind APOBEC3 proteins expressed in their normal target cells and reduce their incorporation into progeny viral particles. This prevents hypermutation of PFV and FFV proviruses during reverse transcription and enhances virion infectivity. However, Bet differs from Vif in that it does not induce the degradation of target APOBEC3 proteins but, rather, simply sequesters them away from progeny virions. One can therefore view the Bet proteins as a primitive ortholog of HIV-1 Vif. Presumably due to the fact that stoichiometric levels of Bet would be required to fully prevent inhibition of virion infectivity by host APOBEC proteins, Bet is expressed at extremely high levels in foamy virus infected cells (28).
An interesting aspect of the life cycle of foamy viruses that distinguishes them from all other retroviruses is that foamy virus reverse transcription occurs in the producer cells so that mature foamy virus particles, unlike all other retroviral particles, actually contain a DNA genome (28). This implies that editing of foamy virus reverse transcripts by APOBEC3 proteins must also occur in the virus producer cells rather than early after infection of new target cells, as seen with HIV-1.
Murine leukemia virus.
Although the mouse genome contains only one APOBEC3 gene, this gene is widely expressed in mouse tissues (12, 41) and it is therefore of interest to ask how MLV avoids inhibition by this antiretroviral defense factor. Importantly, murine APOBEC3 (mA3) is a potent inhibitor of both wild-type and Vif-deficient HIV-1 when expressed in human cells (3, 67). While mA3 fails to interact with HIV-1 Vif, thus explaining its lack of response to Vif, it is efficiently packaged into HIV-1 virions and induces hypermutation of HIV-1 proviruses during reverse transcription (3, 12, 36, 67).
Analyses of the effects of the various human APOBEC3 proteins and mA3 on the infectivity of MLV showed that MLV was essentially unaffected by levels of mA3 that almost entirely blocked HIV-1 infectivity (3, 12, 25). In contrast, MLV was as sensitive as Vif-deficient HIV-1 to hA3G (3, 12, 18, 25, 34). MLV was also moderately inhibited by hA3B, although it proved to be resistant to hA3F, hA3C, and hA3A (3, 12). An analysis of MLV virions produced in the presence of these APOBEC3 proteins showed that MLV packaged both hA3G and hA3B but did not incorporate either hA3F or, more importantly, the cognate mA3 protein which, it will be recalled, is effectively incorporated into HIV-1 virions (12, 25, 36, 67). As discussed in more detail below, this packaging correlated with the ability of these APOBEC3 proteins to bind specifically the MLV Gag protein in vitro. Thus, hA3G and hA3B both bind MLV Gag and are incorporated into MLV virions, while mA3 and hA3F fail to bind to MLV Gag and are not packaged into progeny virion particles (12). It therefore appears that MLV is able to replicate normally in cells expressing mA3 because MLV Gag has been evolutionarily selected to be resistant to mA3 binding. In contrast, MLV Gag has not evolved the ability to avoid binding by hA3G, and MLV remains fully sensitive to this heterologous resistance factor. It remains to be established whether the many other simple retroviruses that infect mammals also use this mechanism to avoid inhibition by their normal host's APOBEC3 protein(s).
Hepatitis B virus.
Although hepatitis B virus (HBV) is not a retrovirus, it does share with retroviruses several key steps in its replication cycle. Most importantly, HBV virions initially package an RNA that is then reverse transcribed, in the virion producer cells, to generate a largely double-stranded DNA genome. In the latter regard, HBV is therefore similar to foamy viruses but differs from all other retroviruses (28). HBV does differ from all retroviruses in that this viral DNA is not integrated into the genome of target cells but, rather, persists as a circular episome.
Evidence that hA3G can inhibit HBV replication was first presented by Turelli et al. (63), who showed potent inhibition of HBV replication in hA3G-expressing cells and also demonstrated a specific interaction between the HBV core antigen and hA3G. However, these workers saw little evidence of HBV DNA editing and found that mutations of the hA3G CDAs that blocked the ability of hA3G to inhibit HIV-1 infectivity had no effect on inhibition of HBV infectivity. Similarly, Rosler et al. (46) showed that both hA3G and hA3F could interfere with the production of replication-competent HBV nucleocapsids, apparently by interfering with some step in HBV virion morphogenesis, but did not induce editing of HBV DNA. Contrary data have been reported by Suspène et al. (60), who reported that hA3G, hA3F, hA3B, and hA3C were all selectively incorporated into HBV capsids, where they edited HBV DNA. However, because the latter report (60) used a novel PCR technique designed to selectively amplify DNA containing G-to-A mutations, it is not clear how frequently hypermutation of HBV reverse transcripts actually occurred. While this controversy remains to be resolved, it is clear that HBV variants that are G-to-A hypermutated can be recovered from HBV-infected patients, albeit at low frequency (60). As hA3G and hA3F are expressed in the human liver (27, 67), it remains unclear at present how HBV is able to replicate effectively in this tissue.
APOBEC3 PROTEINS, ENDOGENOUS RETROVIRUSES, AND RETROTRANSPOSONS
All vertebrate genomes contain large numbers of endogenous retroviruses and retrotransposons (23). Endogenous retroviruses derive from exogenous retroviruses that infected germ cells over millions of years of evolution. The human genome contains ∼230,000 endogenous retroviruses, and the large majority of these appear to be defective. Nevertheless, many endogenous retroviruses are actively transcribed, particularly in dividing cells (52), and several endogenous retroviruses retain open reading frames that can give rise to proteins and even to viral particles.
Retrotransposons can be divided into two families, depending on whether they contain an LTR (23). LTR retrotransposons are closely related to retroviruses and undergo a similar life cycle. However, LTR retrotransposons encode only Gag and Pol, and the virus-like particles (VLPs) produced by LTR retrotransposons are assembled in the cell cytoplasm and undergo reverse transcription in the same cell before being transported to the nucleus for “proviral” integration (23). While the human genome contains only a limited number of LTR retrotransposons, several families exist in mice, including the intracisternal A particle (IAP) and MusD families, which are found at ∼1,000 and ∼100 copies per murine genome, respectively. Interestingly, while MusD lacks any evidence of an env gene, the IAP family retains a highly defective env sequence (40), thus suggesting that IAPs, despite being able to function as LTR retrotransposons, actually originated as endogenous retroviruses. LTR retrotransposons are also found in many other vertebrate and nonvertebrate species, including yeast, which encodes several different LTR retrotransposons, including the Ty1 family (23).
Almost all eukaryotic genomes also contain non-LTR retrotransposons. The most prevalent non-LTR retrotransposon in humans is the long interspersed element 1 (LINE-1) family, which comprises nearly 17% of the human genome (23). While almost all LINE-1 retrotransposons are defective, up to 100 or so remain retrotransposition competent and can therefore cause genetic lesions. Of note, LINE-1 nucleoprotein particles differ from both retroviral virions and LTR retrotransposon VLPs in that reverse transcription occurs in the nucleus.
The close similarity of exogenous retroviruses and endogenous LTR retrotransposons suggested that APOBEC3 proteins might also be able to block these retrotransposition events. In fact, both hA3G and, to a lesser extent, mA3 have been shown to inhibit retrotransposition of the IAP and MusD family of retrotransposons (14). The observed inhibition was ∼5-fold in the case of IAP and up to 20-fold in the case of MusD. Moreover, this inhibition was associated with G-to-A hypermutation of LTR-retrotransposon DNA. An analysis of endogenous LTR-retrotransposon sequences in mice suggested that hypermutation had also occurred in vivo and likely contributed to making the majority of IAP and MusD retrotransposons defective (14).
While APOBEC3 proteins are found only in mammalian species, it is of interest to ask whether they could also function in a nonmammalian context, not least because a positive result would imply that APOBEC3 protein function either did not require a specific cofactor or that this cofactor was highly conserved. To this end, two groups asked whether human APOBEC3 proteins can inhibit retrotransposition by Ty1 retrotransposons in yeast (13, 51). In fact, both hA3G and hA3F can effectively block Ty1 retrotransposition in yeast cells, and this inhibition again correlated with G-to-A hypermutation of Ty1 DNA. Moreover, the Ty1 Gag protein was found to interact specifically with hA3G, and hA3G was selectively packaged into Ty1 virus-like particles (13). Interestingly, this inhibition was not rescued by expression of HIV-1 Vif, which implies that Vif, unlike hA3G, does require a host cell cofactor for its function. Most probably, this Vif cofactor is a component of the E3 ubiquitin ligase that is recruited by Vif in human cells to induce hA3G degradation.
So far, it has not been possible to address the question of whether APOBEC3 proteins have any effect on human endogenous retroviruses, as these are generally defective. While hA3G has been shown to have no effect on LINE-1 retrotransposition (64), it remains to be determined whether LINE-1 is also unaffected by other human APOBEC3 proteins.
SPECIFIC PACKAGING OF APOBEC3 PROTEINS INTO RETROVIRAL PARTICLES
A key aspect of the mechanism of action of hA3G and the other APOBEC3 proteins is their specific incorporation into retroviral virions, as well as HBV virions and LTR-retrotransposon VLPs. Only after incorporation can the APOBEC3 proteins interfere with virion or VLP function, by inducing deamination of dC residues present on reverse transcripts and/or by interfering with aspects of virion or VLP morphogenesis. The APOBEC3 proteins, and hA3G in particular, inhibit a wide range of retroviruses and LTR retrotransposons, so it is important to understand how the APOBEC3 proteins are able to target all of these disparate viruses or genomic parasites, i.e., to understand what defines the PAMP shared by these viruses and virus-like elements that is recognized by the APOBEC3 protein family.
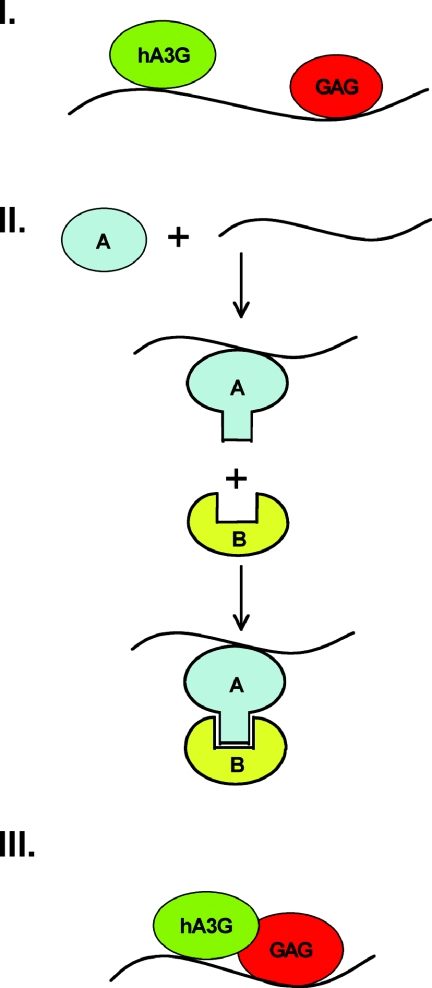
This question has been partly addressed by work analyzing the packaging of hA3G into HIV-1 virions. This research demonstrated that hA3G binds to the nucleocapsid (NC) domain of the Gag polyprotein during virion assembly (1, 6, 24, 32, 48, 73). As a result, HIV-1 Gag derivatives lacking NC, which can still assemble into VLPs, fail to package hA3G (48, 73). Interestingly, the Gag-hA3G interaction is dependent on the presence of RNA (24, 48, 61, 73). While viral RNA may be ideal, nonspecific cellular RNA seems almost as effective at mediating the Gag-hA3G interaction. Of note, a key role of the NC domain of Gag is to recruit the viral RNA genome into virion particles, and NC displays both specific and nonspecific RNA binding properties (2). Similarly, hA3G has also been shown to bind RNA nonspecifically (70). Based on these considerations, one can propose three models to explain the role of RNA in mediating the Gag-hA3G interaction (Fig. 2). In model I, both proteins bind to a single RNA molecule either specifically or nonspecifically and do not, in fact, interact with each other specifically. In model II, one of the proteins binds RNA, resulting in a conformational shift that triggers a specific Gag-hA3G interaction. In model III, Gag, hA3G and RNA form a ternary complex in which all three components bind to each other (Fig. 2).
FIG. 2.
Potential models to explain the critical role of RNA in mediating the formation of Gag-hA3G complexes. In model II, which suggests that RNA binds to either Gag or hA3G and induces a conformational shift that permits complex formation, the identity of the protein undergoing the conformational shift is left open, i.e., A could be either hA3G or Gag. RNA is shown as a wavy line for simplicity, and RNA binding could be either specific or nonspecific. See text for detailed discussion.
Analysis of hA3G packaging into MLV virion particles allows model I to be dismissed. Specifically, as noted above, both hA3G and mA3 are packaged into HIV-1 virions, while only hA3G is packaged into MLV virions (12). An analysis of Gag binding shows that HIV-1 Gag binds both hA3G and mA3, while MLV Gag binds only to hA3G in vitro, again in an RNA-dependent manner. This result explains the virus tropism of mA3 but, more importantly in this context, also demonstrates that the Gag-APOBEC3 interaction is not simply bridged by RNA, as proposed in model I, but must instead involve both protein-RNA interactions and a specific protein-protein interaction, as proposed in models II and III. If model I were indeed correct, then all Gag proteins would be able to bind all APOBEC3 proteins equally well, which is not observed.
Additional work focusing on APOBEC3 protein packaging into retroviral virions or VLPs has shown that hA3B also interacts with the NC domain of HIV-1 and that this interaction is RNA dependent (11). Moreover, hA3G has been shown to also interact specifically with the Gag proteins expressed by PFV and, in an RNA-dependent fashion, by the Ty1 retrotransposon (13, 47). This result is striking, as PFV and Ty1 differ from HIV-1 and MLV in terms of the structure of the Gag protein motif used for genomic RNA binding. In both HIV-1 and MLV, the NC domain of Gag contains “CCHC” zinc finger motifs that mediate genomic RNA recognition and packaging (2). In contrast, both PFV and Ty1 Gag contain carboxy-terminal basic domains, rich in arginine and glycine residues, that mediate genomic RNA binding and incorporation (9, 58). While the NC domains of HIV-1 and MLV Gag are also quite basic, this result nevertheless implies that the mechanisms used by these different retroelements to package their RNA genome are likely to be at least partly distinct. Presumably, there are nevertheless aspects of the process that are conserved and that somehow form the PAMP recognized by hA3G and other APOBEC3 proteins. That this PAMP is not totally invariant is shown by the fact that mA3 is no longer able to bind MLV Gag and package into MLV virion particles (12). On the other hand, the fact that HIV-1 and PFV have dealt with the APOBEC3-mediated inhibition of viral infectivity by evolving the viral Vif and Bet proteins, respectively (8, 22, 31, 37, 47, 54, 59), rather than by modifying the process of virion assembly and RNA genome incorporation, suggests that this virion assembly associated retroviral PAMP is difficult to change without interfering with this key step in the viral life cycle.
Every retroviral Gag protein examined so far, as well as the distantly related Gag-like proteins encoded by Ty1 and the HBV core antigen, has been found to interact specifically with hA3G in vitro and/or by coimmunoprecipitation. It therefore seems likely that a wide range of other Gag proteins, including the frequently defective Gag proteins encoded by endogenous retroviruses, will be able to interact with hA3G. In fact, an analysis of hA3G expressed in dividing cells shows that it forms large (>700-kDa) protein complexes that fall apart when treated with RNase, a characteristic shared with Gag-hA3G complexes (7, 32). Consistent with the idea that these represent endogenous Gag-hA3G complexes, hA3G migrates at its predicted ∼40-kDa molecular mass on sucrose gradients when expressed in yeast cells that do not express Ty1 Gag but moves into a large protein complex when Ty1 Gag is coexpressed (13).
While the above hypothesis would appear to provide a reasonable explanation for the fact that hA3G forms part of a large, ribonucleoprotein complex in dividing cells, it has also been proposed that this complex might contain nonviral host proteins that sequester hA3G in an enzymatically inactive form that perhaps serves to prevent hA3G from introducing random mutations into the cellular genome (7). Of interest is the fact that, in resting CD4+ T cells, the majority of hA3G is not in the high-molecular-mass form, and it has been suggested that it is the low-molecular-mass form of hA3G that renders resting T-cells refractory to infection by HIV-1 (7). On the other hand, it is likely that resting T-cells, which are largely transcriptionally quiescent, do not express the levels of endogenous retrovirus Gag that are found in dividing cells (52). In this context, it is interesting that AID, another human cytidine deaminase, is found in a ribonucleoprotein complex that renders it enzymatically inactive and shows in vitro enzymatic activity only after RNase treatment (5). The issue of whether hA3G is also incorporated into specific ribonucleoprotein complexes that serve to sequester hA3G enzymatic activity or into Gag-containing complexes that result from endogenous retrovirus expression remains to be clarified, although these two proposals are not, in fact, mutually exclusive. It is important to remember that the hA3G expressed in dividing cells is effectively incorporated in newly formed retroviral particles, so either the hA3G protein is released from the large ribonucleoprotein complexes present in these cells or only newly synthesized hA3G is involved.
IMPLICATIONS FOR HIV-1 PATHOGENESIS AND TREATMENT
Wild-type HIV-1 is clearly resistant to inhibition by all human APOBEC3 proteins except hA3B, which does not appear to be expressed in the human tissues normally infected by HIV-1. It is, however, possible that the inhibition of hA3G and/or hA3F function by HIV-1 Vif may not always be complete, especially in cells that express unusually high levels of these proteins. Moreover, HIV-1 undergoes rapid sequence evolution in infected patients, presumably due largely to the error-prone viral reverse transcriptase but also potentially due to editing by hA3G and/or hA3F. If the vif gene were to be fully or partially inactivated due to mutation, then one might predict that hypermutated HIV-1 proviruses would arise.
Evidence in favor of both of these hypotheses exists. Modest overexpression of hA3G in cultured cells can inhibit the replication of wild-type HIV-1, and this correlates with the appearance of a low level of G-to-A changes in the resultant HIV-1 proviruses (36). Moreover, recent evidence suggests that individuals who express high levels of hA3G mRNA tend to have low levels of HIV-1 viremia after infection, and vice versa (21). If correct, these data would imply that even a modest inhibition of Vif function, either by interfering with binding to hA3G and/or hA3F or by blocking recruitment of the Cullin5-ElonginB-ElonginC E3 ubiquitin ligase, might significantly reduce HIV-1 virus loads in vivo. Moreover, if Vif were to be even slightly inhibited, one would predict that the rapidly replicating HIV-1 would begin to accumulate mutations that could eventually lead to a viral error catastrophe. On the other hand, a very low level of editing by hA3G or hA3F could also facilitate HIV-1 replication in vivo by promoting the appearance of mutants that are resistant to antiretroviral drugs.
The second hypothesis, that G-to-A hypermutated HIV-1 proviruses would be likely to appear over time in infected patients, has clearly been confirmed by several investigators (15, 19). Moreover, the G-to-A mutations that occur are almost all in the sequence contexts G*A and G*G (the complement of TC* and CC*), which is to say in the sequence contexts favored by hA3F and hA3G, respectively, for dC-to-dU editing (3, 11, 18, 27, 67). This result strongly suggests that hA3G and hA3F can indeed edit HIV-1 proviruses in vivo (27, 57).
A recent analysis of vif alleles present in HIV-1 infected patients, several of whom were long-term nonprogressors, suggests that partially or fully defective vif genes may be surprisingly prevalent, with 15 out of the 79 vif genes recovered being at least partially defective (57). Of interest, some vif alleles were able to block hA3G but not hA3F function, and vice versa. Consistent with the observed prevalence of attenuated viral vif genes, these authors recovered a number of hypermutated HIV-1 proviral sequences from their patient populations (57). Most strikingly, hypermutated proviruses recovered from patients infected with HIV-1 variants whose vif gene products were able to block specifically only hA3G or only hA3F function showed the hypermutation pattern predicted if only the nontargeted APOBEC3 protein remained active, i.e., patients who were infected by viruses able to block hA3G but not hA3F function showed selective hypermutation at G*A but not at G*G.
A FEW OPEN QUESTIONS
While there has been remarkable progress in understanding the mechanism of action of the APOBEC3 proteins on the one hand and of viral countermeasures, such as HIV-1 Vif, on the other, there are still a number of key issues that remain unresolved. A partial listing of these open questions follows.
(i) Why does hA3G incorporation into virions result in destabilization of reverse transcripts in subsequently infected cells (56, 65)? Is this indeed a result of the action of cellular DNA repair enzymes that seek to remove dU residues from the proviral DNA prior to formation of the second DNA strand (50)? If so, which cellular enzymes are involved?
(ii) Work analyzing the effect of hA3G on HBV replication has suggested that hA3G can inhibit HBV without inducing editing (46, 63). Moreover, one report has claimed that hA3G derivatives that lack a functional CDA can still inhibit HIV-1 infectivity (43). Finally, it has also been suggested that hA3G expressed in resting T cells blocks the infectivity of incoming HIV-1 virions without inducing editing (7). Can hA3G, and the other APOBEC3 proteins, indeed inhibit retroviruses independently of their editing function and, if so, by what mechanism?
(iii) What are the precise features of virion morphogenesis and RNA genome incorporation that cause the specific incorporation of hA3G and other APOBEC3 proteins into progeny retroviral and HBV virion particles and retrotransposon VLPs?
(iv) What molecular contacts mediate formation of the hA3G-Vif E3 ubiquitin ligase complex? Can this structure be resolved using X-ray crystallography and, if so, does this structure suggest potential small molecule inhibitors of complex formation?
(v) What process has driven the rapid evolution of the primate APOBEC3 protein family (20, 47a, 75)? Although APOBEC3 proteins are invariably ineffective against retroviruses that normally infect a given species, they appear to be effective against most heterologous retroviruses. Is this sufficient to select for APOBEC3 function, or are the APOBEC3 proteins important primarily due to their ability to inhibit retrotransposon function and hence prevent the gradual accumulation of genetic mutations (47a)? While the recent report of a knock-out mouse strain lacking the single mA3 gene (41) may permit these issues to be addressed, preliminary observation suggests that these mA3-deficient mice are phenotypically normal.
The final and most important question is whether the basic research described in this review will facilitate the identification of drugs that can selectively inhibit HIV-1 Vif function and thereby lead to a clear improvement in the prognosis of HIV-1-infected individuals. Only time will tell if this goal can be achieved.
Acknowledgments
The research from my laboratory mentioned in this review was supported by grants AI057099 and AI065301 from the National Institutes of Health.
REFERENCES
- 1.Alce, T. M., and W. Popik. 2004. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 279:34083-34086. [DOI] [PubMed] [Google Scholar]
- 2.Berkowitz, R. D., A. Ohagen, S. Hoglund, and S. P. Goff. 1995. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J. Virol. 69:6445-6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop, K. N., R. K. Holmes, A. M. Sheehy, N. O. Davidson, S.-J. Cho, and M. H. Malim. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392-1396. [DOI] [PubMed] [Google Scholar]
- 4.Bogerd, H. P., B. P. Doehle, H. L. Wiegand, and B. R. Cullen. 2004. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV-1 Vif. Proc. Natl. Acad. Sci. USA 101:3770-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bransteitter, R., P. Pham, M. D. Scharff, and M. F. Goodman. 2003. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl. Acad. Sci. USA 100:4102-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cen, S., F. Guo, M. Niu, J. Saadatmand, J. Deflassieux, and L. Kleiman. 2004. The interaction between HIV-1 Gag and APOBEC3G. J. Biol. Chem. 279:33177-33184. [DOI] [PubMed] [Google Scholar]
- 7.Chiu, Y.-L., V. B. Soros, J. F. Kreisberg, K. Stopak, W. Yonemoto, and W. C. Greene. 2005. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 435:108-114. [DOI] [PubMed] [Google Scholar]
- 8.Conticello, S. G., R. S. Harris, and M. S. Neuberger. 2003. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13:2009-2013. [DOI] [PubMed] [Google Scholar]
- 9.Cristofari, G., D. Ficheux, and J.-L. Darlix. 2000. The Gag-like protein of the yeast Ty1 retrotransposon contains a nucleic acid chaperone domain analogous to retroviral nucleocapsid proteins. J. Biol. Chem. 275:19210-19217. [DOI] [PubMed] [Google Scholar]
- 10.Cullen, B. R. 1991. Human immunodeficiency virus as a prototypic complex retrovirus. J. Virol. 65:1053-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doehle, B. P., A. Schäfer, and B. R. Cullen. 2005. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology 339:281-288. [DOI] [PubMed] [Google Scholar]
- 12.Doehle, B. P., A. Schäfer, H. L. Wiegand, H. P. Bogerd, and B. R. Cullen. 2005. Differential sensitivity of murine leukemia virus to APOBEC3-mediated inhibition is governed by virion exclusion. J. Virol. 79:8201-8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dutko, J. A., A. Schäfer, A. E. Kenny, B. R. Cullen, and M. J. Curcio. 2005. Inhibition of a yeast LTR retrotransposon by human APOBEC3 cytidine deaminases. Curr. Biol. 15:661-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esnault, C., O. Heidmann, F. Delebecque, M. Dewannieux, D. Ribet, A. J. Hance, T. Heidmann, and O. Schwartz. 2005. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433:430-433. [DOI] [PubMed] [Google Scholar]
- 15.Fitzgibbon, J. E., S. Mazar, and D. T. Dubin. 1993. A new type of G→A hypermutation affecting human immunodeficiency virus. AIDS Res. Hum. Retrovir. 9:833-838. [DOI] [PubMed] [Google Scholar]
- 16.Gabuzda, D. H., K. Lawrence, E. Langhoff, E. Terwilliger, T. Dorfman, W. A. Haseltine, and J. Sodroski. 1992. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J. Virol. 66:6489-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Gaddis, N. C., A. M. Sheehy, K. M. Ahmad, C. M. Swanson, K. N. Bishop, B. E. Beer, P. A. Marx, F. Gao, F. Bibollet-Ruche, B. H. Hahn, and M. H. Malim. 2004. Further investigation of simian immunodeficiency virus Vif function in human cells. J. Virol. 78:12041-12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haché, G., M. T. Liddament, and R. S. Harris. 2005. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J. Biol. Chem. 280:10920-10924. [DOI] [PubMed] [Google Scholar]
- 18.Harris, R. S., K. N. Bishop, A. M., Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 19.Janini, M., M. Rogers, D. R. Birx, and F. E. McCutchan. 2001. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4+ T cells. J. Virol. 75:7973-7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, and N. Navaratnam. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79:285-296. [DOI] [PubMed] [Google Scholar]
- 21.Jin, X., A. Brooks, H. Chen, R. Bennett, R. Reichman, and H. Smith. 2005. APOBEC3G/CEM15 (hA3G) mRNA levels associate inversely with human immunodeficiency virus viremia. J. Virol. 79:11513-11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kao, S., M. A. Khan, E. Miyagi, R. Plishka, A. Buckler-White, and K. Strebel. 2003. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 77:11398-11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kazazian, H. H., Jr. 2004. Mobile elements: drivers of genome evolution. Science 303:1626-1632. [DOI] [PubMed] [Google Scholar]
- 24.Khan, M. A., S. Kao, E. Miyagi, H. Takeuchi, R. Goila-Gaur, S. Opi, C. L. Gipson, T. G. Parslow, H. Ly, and K. Strebel. 2005. Viral RNA is required for the association of APOBEC3G with human immunodeficiency virus type 1 nucleoprotein complexes. J. Virol. 79:5870-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi, M., A. Takaori-Kondo, K. Shindo, A. Abudu, K. Fukunaga, and T. Uchiyama. 2004. APOBEC3G targets specific virus species. J. Virol. 78:8238-8244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langlois, M.-A., R. C. L. Beale, S. G. Conticello, and M. S. Neuberger. 2005. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 33:1913-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liddament, M. T., W. L. Brown, A. J. Schumacher, and R. S. Harris. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 14:1385-1391. [DOI] [PubMed] [Google Scholar]
- 28.Linial, M. L. 1999. Foamy viruses are unconventional retroviruses. J. Virol. 73:1747-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, B., P. T. N. Sarkis, K. Luo, Y. Yu, and X.-F. Yu. 2005. Regulation of APOBEC3F and human immunodeficiency virus type 1 Vif by Vif-Cul5-ElonB/C E3 ubiquitin ligase. J. Virol. 79:9579-9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, B., X. Yu, K. Luo, Y. Yu, and X.-F. Yu. 2004. Influence of primate lentiviral Vif and proteasome inhibitors on human immunodeficiency virus type 1 virion packaging of APOBEC3G. J. Virol. 78:2072-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Löchelt, M., F. Romen, P. Bastone, H. Muckenfuss, N. Kirchner, Y.-B. Kim, U. Truyen, U. Rösler, M. Battenberg, A. Saib, E. Flory, K. Cichutek, and C. Münk. 2005. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc. Natl. Acad. Sci. USA 102:7982-7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo, K., B. Liu, Z. Xiao, Y. Yu, X. Yu, R. Gorelick, and X.-F. Yu. 2004. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J. Virol. 78:11841-11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madani, N., and D. Kabat. 1998. An endogenous inhibitor of human immunodeficiency virus in human lymphocytes is overcome by the viral Vif protein. J. Virol. 72:10251-10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 35.Mangeat, B., P. Turelli, S. Liao, and D. Trono. 2004. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 279:14481-14483. [DOI] [PubMed] [Google Scholar]
- 36.Mariani, R., D. Chen, B. Schröfelbauer, F. Navarro, R. König, B. Bollman, C. Münk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 37.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nature Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 38.Mehle, A., J. Goncalves, M. Santa-Marta, M. McPike, and D. Gabuzda. 2004. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 18:2861-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mehle, A., B. Strack, P. Ancuta, C. Zhang, M. N. McPike, and D. Gabuzda. 2004. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 279:7792-7798. [DOI] [PubMed] [Google Scholar]
- 40.Mietz, J. A., Z. Grossman, K. K. Lueders, and E. L. Kuff. 1987. Nucleotide sequence of a complete mouse intracisternal A-particle genome: relationship to known aspects of particle assembly and function. J. Virol. 61:3020-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mikl, M. C., I. N. Watt, M. Lu, W. Reik, S. L. Davies, M. S. Neuberger, and C. Rada. 2005. Mice deficient in APOBEC2 and APOBEC3. Mol. Cell. Biol. 25:7270-7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Navarro, F., B. Bollman, H. Chen, R. König, Q. Yu, K. Chiles, and N. R. Landau. 2005. Complementary function of the two catalytic domains of APOBEC3G. Virology 333:374-386. [DOI] [PubMed] [Google Scholar]
- 43.Newman, E. N. C., R. K. Holmes, H. M. Craig, K. C. Klein, J. R. Lingappa, M. H. Malim, and A. M. Sheehy. 2005. Anti-viral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15:166-170. [DOI] [PubMed] [Google Scholar]
- 44.Ribeiro, A. C., A. Maia e Silva, M. Santa-Marta, A. Pombo, J. Moniz-Pereira, J. Goncalves, and I. Barahona. 2005. Functional analysis of Vif protein shows less restriction of human immunodeficiency virus type 2 by APOBEC3G. J. Virol. 79:823-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rose, K. M., M. Marin, S. L. Kozak, and D. Kabat. 2005. Regulated production and anti-HIV type 1 activities of cytidine deaminases APOBEC3B, 3F, and 3G. AIDS Res. Hum. Retrovir. 21:611-619. [DOI] [PubMed] [Google Scholar]
- 46.Rosler, C., J. Kock, M. Kann, M. H. Malim, H. E. Blum, T. F. Baumert, F. Von Weizsacker. 2005. APOBEC-mediated interference with hepadnavirus production. Hepatology 42:301-309. [DOI] [PubMed] [Google Scholar]
- 47.Russell, R. A., H. L. Wiegand, M. D. Moore, A. Schäfer, M. O. McClure, and B. R. Cullen. 2005. Foamy virus Bet proteins function as novel inhibitors of the APOBEC3 family of innate antiretroviral defense factors. J. Virol. 79:8724-8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47a.Sawyer, S. L., M. Emerman, and H. S. Malik. 2004. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2:1278-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schäfer, A., H. P. Bogerd, and B. R. Cullen. 2004. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology 328:163-168. [DOI] [PubMed] [Google Scholar]
- 49.Schröfelbauer, B., D. Chen, and N. R. Landau. 2004. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Natl. Acad. Sci. USA 101:3927-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schröfelbauer, B., Q. Yu, S. G. Zeitlin, and N. R. Landau. 2005. Human immunodeficiency virus type 1 Vpr induces the degradation of the UNG and SMUG uracil-DNA glycosylases. J. Virol. 79:10978-10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schumacher, A. J., D. V. Nissley, and R. S. Harris. 2005. APOBEC3G hypermutates genomic DNA and inhibits Ty1 retrotransposition in yeast. Proc. Natl. Acad. Sci. USA 102:9854-9859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seifarth, W., O. Frank, U. Zeilfelder, B. Spiess, A. D. Greenwood, R. Hehlmann, and C. Leib-Mösch. 2005. Comprehensive analysis of human endogenous retrovirus transcriptional activity in human tissues with a retrovirus-specific microarray. J. Virol. 79:341-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 54.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 55.Simon, J. H. M., N. C. Gaddis, R. A. M. Fouchier, and M. H. Malim. 1998. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat. Med. 4:1397-1400. [DOI] [PubMed] [Google Scholar]
- 56.Simon, J. H., and M. H. Malim. 1996. The human immunodeficiency virus type 1 Vif protein modulates the postpenetration stability of viral nucleoprotein complexes. J. Virol. 70:5297-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simon, V., V. Zennou, D. Murray, Y. Huang, D. D. Ho, and P. D. Bieniasz. 2005. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 1:e6. [DOI] [PMC free article] [PubMed]
- 58.Stenbak, C. R., and M. L. Linial. 2004. Role of the C terminus of foamy virus Gag in RNA packaging and Pol expression. J. Virol. 78:9423-9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12:591-601. [DOI] [PubMed] [Google Scholar]
- 60.Suspène, R., D. Guétard, M. Henry, P. Sommer, S. Wain-Hobson, and J.-P. Vartanian. 2005. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc. Natl. Acad. Sci. USA 102:8321-8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Svarovskaia, E. S., H. Xu, J. L. Mbisa, R. Barr, R. J. Gorelick, A. Ono, E. O. Freed, W.-S. Hu, and V. K. Pathak. 2004. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J. Biol. Chem. 279:35822-35828. [DOI] [PubMed] [Google Scholar]
- 62.Tosi, M. F. 2005. Innate immune responses to infection. J. Allergy Clin. Immunol. 116:241-249. [DOI] [PubMed] [Google Scholar]
- 63.Turelli, P., B. Mangeat, S. Jost, S. Vianin, and D. Trono. 2004. Inhibition of hepatitis B virus replication by APOBEC3G. Science 303:1829. [DOI] [PubMed] [Google Scholar]
- 64.Turelli, P., S. Vianin, and D. Trono. 2004. The innate antiretroviral factor APOBEC3G does not affect human LINE-1 retrotransposition in a cell culture assay. J. Biol. Chem. 279:43371-43373. [DOI] [PubMed] [Google Scholar]
- 65.von Schwedler, U., J. Song, C. Aiken, and D. Trono. 1993. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J. Virol. 67:4945-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wedekind, J. E., G. S. C. Dance, M. P. Sowden, and H. C. Smith. 2003. Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends Genet. 19:207-216. [DOI] [PubMed] [Google Scholar]
- 67.Wiegand, H. L., B. P. Doehle, H. P. Bogerd, and B. R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23:2451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoshikawa, K., I. M. Okazaki, T. Eto, K. Kinoshita, M. Muramatsu, H. Nagaoka, and T. Honjo. 2002. AID enzyme-induced hypermutation in an actively transcribed gene in fibroblasts. Science 296:2033-2036. [DOI] [PubMed] [Google Scholar]
- 69.Yu, Q., D. Chen, D., R. König, R. Mariani, D. Unutmaz, and N. R. Landau. 2004. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 279:53379-53386. [DOI] [PubMed] [Google Scholar]
- 70.Yu, Q., R. König, S. Pillai, K. Chiles, M. Kearney, S. Palmer, D. Richman, J. M. Coffin, and N. R. Landau. 2004. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 11:435-442. [DOI] [PubMed] [Google Scholar]
- 71.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X.-F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 72.Yu, Y., Z. Xiao, E. S. Ehrlich, X. Yu, and X.-F. Yu. 2004. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 18:2867-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zennou, V., D. Perez-Caballero, H. Göttlinger, and P. D. Bieniasz. 2004. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J. Virol. 78:12058-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang, J., and D. M. Webb. 2004. Rapid evolution of primate antiviral enzyme APOBEC3G. Hum. Mol. Genet. 13:1785-1791. [DOI] [PubMed] [Google Scholar]
- 76.Zheng, Y.-H., D. Irwin, T. Kurosu, K. Tokunaga, T. Sata, and B. M. Peterlin. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 78:6073-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]