Abstract
The human cytomegalovirus (HCMV) US12 gene family is a group of predicted seven-transmembrane, G-protein-coupled receptor-related proteins, about which little is known. Specific rabbit polyclonal antibodies detected US17 and US18 beginning 54 and 36 h after infection, respectively, with expression of both proteins dependent on viral DNA synthesis. While US14 and US18 are expressed exclusively in the cytoplasm, we unexpectedly found abundant expression of US17 in both the cytoplasm and nucleoplasm. N- and C-terminally tagged versions of US17 were readily detected in the cytoplasm of transfected mammalian cells, but not in nuclei, suggesting that nuclear localization involves other viral proteins or an infection-triggered cellular process. There was no specific colocalization between US17 and other nuclear expressed HCMV-encoded proteins (IE-2, DNA polymerase processivity factor, and pp28/UL99). To determine whether the observed nuclear localization might be the product of a process by which a soluble C-terminal segment of the full-length protein is expressed, we constructed a recombinant virus that incorporates a synthetic epitope at its N terminus, which in conjunction with the antipeptide antibody that targets its predicted cytoplasmic C-terminal segment, enables simultaneous independent detection of both termini. In cells infected with the recombinant, the US17 N and C termini had limited colocalization, with the N-terminal segment not detected in nuclei, supporting the segmentation hypothesis. Consistent with this, a fragment with an apparent molecular size of 10 kDa was detected by immunoblotting. We have identified the first viral example of a seven-transmembrane protein that is either segmented or expressed in nuclei. Further study will be required to learn the mechanism by which this occurs and the function of the nuclear localizing segment. This likely represents yet another mechanism by which a virus has hijacked or modified cellular regulatory pathways for its benefit.
Human cytomegalovirus (HCMV) is a member of the betaherpesvirus subfamily. HCMV congenital infection is a leading cause of birth defects, and the virus is also a major cause of morbidity and mortality in immunocompromised individuals. As part of its biology, HCMV infects a wide variety of cell types and tissues (reviewed in reference 22). The virus initially enters via the epithelium of the upper alimentary, respiratory, or genitourinary tract to establish infection. Replication in cytotrophoblasts, the placental cells that form the barrier between maternal and fetal circulation systems, may be important in transmission of the virus to the fetus (24). Granulocyte-macrophage progenitor cells in bone marrow and peripheral blood monocytes are the chief reservoirs for latent HCMV infection (32). Differentiation of latently infected monocytes into macrophages leads to reactivation of lytic infection (31). This diversity of cellular tropisms and biological activity is the outcome of complex interplay between the virus and host.
The 236-kb HCMV genome is predicted to encode at least 165 protein-encoding genes (7, 23), plus recently discovered genes that encode microRNAs (25). Although analysis of individual gene function in HCMV is incomplete, at least 117 genes are dispensable for productive replication in fibroblasts (8, 39). A wealth of information makes it clear that a large proportion of HCMV genetic complexity is committed not to fundamental replicative processes but to defining its tissue tropism; optimizing growth in a wide diversity of cell types and cell states; regulating its dissemination from cell-to-cell, tissue-to-tissue, and person-to-person; modulation of innate and acquired immunity; and otherwise contributing to pathogenesis (6). Many of the genes whose functions remain elusive likely play roles in these processes.
As part of analyzing the DNA sequence of the HCMV unique short (US) region, Weston and Barrell (37) (and extended by Chee et al. [4]) made the important observation that HCMV encodes several independent families of related genes, each family having from 2 to as many as 12 members. Sequences within each gene family are generally highly divergent. This strategy of gene duplication and divergence has been used throughout the betaherpesvirus subfamily, but there is little evidence of it in the alpha- and gammaherpesviruses. We are interested in identifying the diversity of function that would have justified such an expenditure of evolutionary capital. One of these gene families, the US12 family, is a set of 10 contiguous tandemly arranged genes (US12 through US21) encoding distantly related proteins that are each predicted to have seven transmembrane segments (7TM proteins) and to be related to G-protein-coupled receptors (GPCR) (26). US12 family members have been identified only in the cytomegaloviruses of higher primates (HCMV, chimpanzee cytomegalovirus [CCMV], and rhesus cytomegalovirus [RhCMV]).
The US12 gene family has been not been studied in detail. HCMV US18, US19, and US20 are all expressed in infected cells, with the major US18 transcript being expressed as a late gene and the US20 transcript as an early gene (12). Deletion or insertional inactivation of individual US12 family member genes had little or no effect on viral replication in fibroblasts (the US13 deletion resulted in a 10−1 to 10−2 reduction in titer), but US16 and US19 deletions resulted in enhanced replication in human microvascular endothelial cells (8, 39). These results indicate that US12 family members may have complementary functions, that their effects may be subtle under many conditions, and that they may operate in a cell-specific manner. By using a phylogenetic analysis, we found that the US12 gene family forms a clade of 7TM proteins that is related to but distinct from the recognized GPCR families (M. Lesniewski and P. E. Pellett, unpublished data). These potential GPCRs are only distantly related to the other known and predicted GPCRs encoded by HCMV: UL33 and UL78 (homologs in all betaherpesviruses), US27 (CCMV and HCMV), and US28 (HCMV, CCMV, and RhCMV). Thus far, there have been no reports of US12 family proteins having GPCR-like signaling activity. No US12 family members were detected in HCMV virions or dense bodies in a recent comprehensive and sensitive analysis of HCMV virion composition (34).
We have begun studying the localization, expression patterns, and physiological effects of US12 family members to gain insight into their function. In the course of these studies, we made the observation that in addition to being present in the cytoplasm, US17 localizes to infected-cell nuclei. This unexpected localization of a protein that is predicted to be a 7TM protein and thus tightly associated with cellular membranes suggests the possible involvement of this protein in a form of signaling or other biological activity that differs from mechanisms commonly used by GPCR.
MATERIALS AND METHODS
Virus and cells.
MRC-5, HeLa, and HEK 293T cells were from the American Type Culture Collection (ATCC) (Manassas, VA). HCMV strain AD169 was obtained from ATCC. Viruses were propagated in primary human foreskin fibroblasts (passage 17 to 25) or low-passage human lung fibroblasts (HLF) (obtained from the Centers for Disease Control and Prevention). Virus was harvested when the cytopathic effect was >90%. Titers were determined in triplicate by plaque assay on MRC-5 cells. Cells were grown in Earle's modified Eagle's medium containing 10% fetal bovine serum (FBS) (HyClone, Logan, UT), 2 mM l-glutamine, 50 units/ml penicillin, and 50 μg/ml streptomycin. Unless noted otherwise, cells were infected in the same medium, except the FBS concentration was 5% and antibiotics were omitted.
Antibodies.
The following mouse monoclonal antibodies (MAb) against HCMV proteins were used: MAb810 against IE-2 (UL122; Chemicon, Inc.), MAb 13-131 against the UL44/ICP36/DNA polymerase processivity factor (p52) (Applied Biotechnologies, Columbia, MD), anti-V5 MAb (Invitrogen, Carlsbad, CA), anti-FLAG MAb (Stratagene, La Jolla, CA), and MAb 13-130 against pp28 (the tegument protein UL99) (Applied Biotechnologies). Secondary antibodies for immunofluorescence experiments included polyclonal goat anti-rabbit (Alexa Fluor 488 conjugated) and goat anti-mouse (Alexa Fluor 568 conjugated) (Molecular Probes, Eugene, OR).
Generation of rabbit antipeptide polyclonal antibodies.
Peptides corresponding to epitopes predicted by computational analysis were synthesized in our institution's Biotechnology Core Facility, conjugated with keyhole limpet hemocyanin (KLH), and then sent to Rockland Immunochemicals (Gilbertsville, PA) for generation of antibodies. The amino acid sequences selected were from near the C termini of US14 (CA268RWDQMFSYLAKLG) and US17 (CE249DSLDKLI), and near the N terminus of US18 (CT4ASVSEHHESPTVTI). The N-terminal cysteine residues were added to allow conjugation to KLH (Imject Maleimide Activated mcKLH kit; Pierce Biotechnology, Rockford, IL). Total immunoglobulin G was purified from rabbit serum by using protein G Sepharose (fast-flow) affinity columns (Amersham Biosciences).
Cloning FLAG-, GFP-, and V5-tagged US14, US17, and US18 for mammalian cell expression.
Full-length coding sequences of US14, US17, and US18 open reading frames were amplified by PCR from HCMV(AD169) DNA using oligonucleotide primers and high-fidelity Taq DNA polymerase (Invitrogen, Carlsbad, CA). The 5′ primers were GTGAATTCATGGAGACAGTTTCCACGC (US14), GTGAATTCATGTCTCCGAACTCAGAGGCC (US17), and GTGAATTCATGGGCGACACCGCC (US18). The 3′ primers were CTGCCTCGAGGGCAGCCTTGCTCTG (US14), CTCTCGAGGGAGGCCGCGTTGGT (US17), and CTCTCGAGCAACAAGCTGAGGAGACTCA (US18). The 5′ primers included an EcoRI site upstream (underlined) of the ATG initiation codon (italic), while 3′ primers had an XhoI sequence (underlined) downstream from the C-terminal amino acid. Amplimers were cloned in frame as EcoRI/XhoI fragments into pCMVTag2B (N-terminal FLAG tag; Stratagene) and a derivative of pcDNA3.1 that enables construction of C-terminal green fluorescent protein (GFP) fusion proteins (provided courtesy of Achut Marlur). The clones were verified by DNA sequencing. US17 tagged with the V5 epitope at its N terminus was generated using the 5′ primer GGATCCATGGGTAAGCCTATCCCTAACCCTCTCC and 3′ primer CTCGAGTTACGCCATGGTTCGCGTGAGGTTTCTCTGTA from an HCMV(Towne) bacterial artificial chromosome (BAC) recombinant virus into which we had previously inserted the V5 epitope immediately downstream of the US17 initiation codon (described below). The resulting fragment was sequenced and then transferred to pcDNA3.1 at the BamHI and XhoI sites.
Transient expression.
MRC-5, HeLa and HEK 293T cells were plated at 3 × 105 cells per well in six-well plates the day before transfection (Lipofectamine 2000; Invitrogen) in Dulbecco's modified Eagle's medium containing 10% FBS, with no extra glutamine or antibiotics.
Generation of V5-tagged US17 recombinant virus.
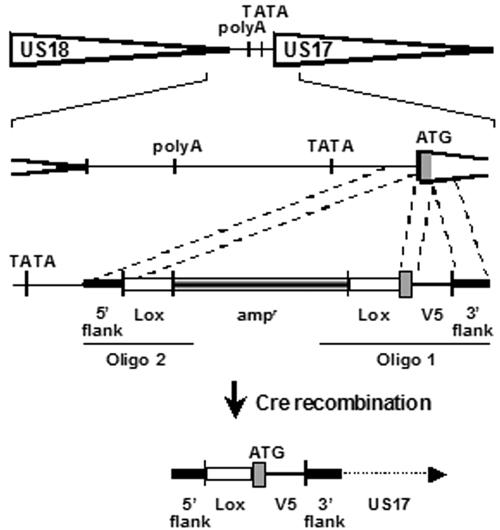
A BAC recombinant HCMV was constructed that incorporates the synthetic V5 epitope at the US17 N terminus and has the GFP gene that is present in the parental HCMV Towne BAC deleted (to reduce fluorescence channel bleed from the highly expressed GFP) (20) (the parental virus was kindly provided by Hua Zhu). The BAC was constructed in three main steps. First, the V5 epitope was added using the strategy illustrated in Fig. 8, in which the insertional sequence with an ampicillin resistance selection marker was generated by PCR using oligonucleotides that correspond to the regions shown in the figure. The bacterial recombination step was done in DY 380 cells, which enable efficient recombination from short homologous flanking sequences due to Red recombination through defective prophage (17). In the second step, LoxP/cre recombination was used to remove the ampicillin selection marker upstream of the US17 gene. In the third step, a similar strategy was used to remove the GFP gene from the BAC. The final construct was verified by whole-genome restriction enzyme analysis, DNA sequencing across the mutagenized locus, and protein expression.
FIG. 8.
Construction of HCMV BAC incorporating a synthetic epitope at the N terminus of US17. As described in Materials and Methods, a segment of DNA was obtained; this DNA segment contained an ampicillin resistance gene flanked by sequences harboring LoxP sites, plus a V5 synthetic epitope for insertion at the N terminus of US17, and flanking sequences to guide recombinational insertion into the parental HCMV BAC. The V5 epitope was inserted as a segment of 14 amino acids between the US17 initiation codon and the second US17 residue. The locations and contents of the synthetic oligonucleotides used to generate the insertion cassette (Oligo 1 and Oligo 2) are indicated in the diagram. The final construct harbors a 34-bp LoxP site just upstream of the US17 translation initiation codon and the V5 epitope in frame with US17, immediately downstream of the initiation codon.
Immunofluorescence microscopy.
HLF (2 × 104) were grown overnight in eight-well Labtek chamber slides (Nunc, Naperville, IL) and then infected at a low multiplicity of infection (MOI) (0.01). Infected cells were prepared for immunofluorescence by fixing them with 4% paraformaldehyde in phosphate-buffered saline (PBS), incubating in 50 mM ammonium chloride to quench autofluorescence, permeabilizing in PBS containing 0.2% Triton X-100 and normal goat serum for 15 min, and then incubating for 1 h in blocking buffer (10% normal goat serum and 5% glycine in PBS). Primary antibodies were diluted in blocking buffer prior to staining cells for 1 h at room temperature. After the cells were washed three times with PBS, they were allowed to react for 1 h with similarly prepared secondary antibody. After rinsing, cells were mounted using Vectashield with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA). Confocal images were captured with a Leica TCS-SP digital confocal microscope.
Immunoblots.
Cells were extracted at room temperature in a buffer consisting of 0.5% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 0.25% sodium deoxycholate, 10 mM Tris (pH 7.5), 5 mM MgCl2, 150 mM NaCl, and 5 mM dithiothreitol, plus a protease inhibitor cocktail (Roche Applied Science, Indianapolis, Ind.). The extract was mixed with sodium dodecyl sulfate (SDS) sample buffer (6×) (Tris-HCl [pH 6.8], 0.2% 2-mercaptoethanol, 30% glycerol, 10% SDS) at room temperature and, without boiling, was separated in a 15% SDS-polyacrylamide gel. After electrical transfer to a nitrocellulose membrane (0.2-μm pore size; Schleicher & Schuell, Keene, NH), the blot was preincubated with rocking for 1 h at room temperature in blocking buffer consisting of 5% fat-free milk powder in PBS containing 0.05% Tween 20 and reacted overnight with appropriately diluted rabbit antibody in a buffer consisting of 0.05% Tween 20 and 1% normal goat serum (Caltag, Burlingame, CA) in PBS. After the blot was rinsed three times at room temperature in PBS with 0.05% Tween 20, the reaction was detected using horseradish peroxidase-conjugated goat anti-rabbit antibody (Pierce Biotechnology) and chemiluminescence substrate (Pierce Biotechnology).
RESULTS
Generation and specificity of antibodies to US17 and US18.
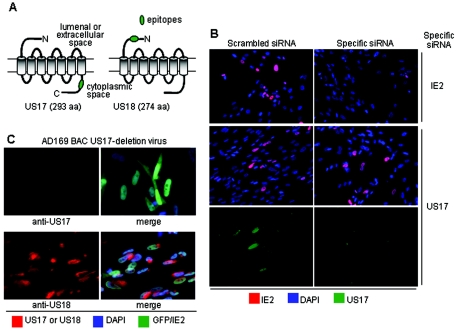
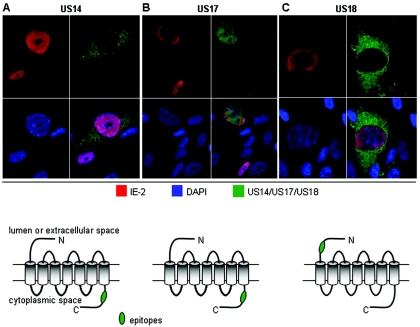
We generated antipeptide antibodies that target the N-terminal domain of US18 and the C-terminal domain of US17 (Fig. 1A). The peptides were chosen on the basis of their predicted antigenicity and solubility, lack of sequence similarity with other members of the US12 family, and having no nonspecific matches in translated versions in GenBank. Antibody specificity was assessed in a variety of ways, including lack of reactivity of preimmune serum with infected cells (data not shown), and use of low multiplicities of infection (no reactivity in nonplaque regions of infected-cell monolayers), RNA silencing, and deletion viruses. For the RNA silencing experiments (Fig. 1B), we transfected cells with cocktails of synthetic small interfering RNAs (siRNAs) against IE-2, US17, US18, and a scrambled-sequence negative control. This form of siRNA does not stimulate interferon production (15). As can be seen in the controls transfected with the scrambled sequence, siRNA transfection by itself had no discernible effect on the expression of any of the genes studied. siRNA against US17 led to substantial reduction of US17 expression, with no effect on IE-2 or US18 (not shown). An analogous result was obtained with an siRNA directed at US18 (data not shown). To extend the specificity analysis further, we tested the reactivities of antibodies with HCMV(AD169) recombinants containing individual deletions of US17 and US18 (kindly provided by Thomas Shenk) (39). As in the siRNA experiments, the antipeptide antibodies reacted with the appropriate specificity. Thus, for the US17 deletion virus, the US17 antibody did not react, while the US18 antibody did (Fig. 1C). The analogous result was obtained with the US18 deletion virus (data not shown).
FIG. 1.
Generation and specificity of antibodies to US17 and US18. (A) Antipeptide antibodies were generated that target the indicated N-terminal and C-terminal domains of US18 and US17, respectively. aa, amino acids. (B) Use of siRNA to assess antibody specificity. IE-2 and US17 expression were inhibited by treatment with specific siRNAs (as indicated to the right). Cells in the scrambled siRNA column were treated with a nonspecific siRNA, which had no effect on IE-2 or US17 expression. IE-2 was stained red, nuclei were stained blue with DAPI, and US17 was stained green with a rabbit polyclonal antibody directed against the C-terminal segment. Nonconfocal images were obtained at ×20 magnification. (C) Lack of reactivity of US17 antiserum with cells infected with HCMV deleted for US17. HLF infected with an HCMV BAC recombinant from which the US17 gene was deleted were reacted with a monoclonal antibody against the IE-2 protein and rabbit antibodies against either US17 (top pair of panels) or US18 (bottom pair of panels). The BAC recombinant expresses GFP, so infected cells are dually positive for GFP and green fluorescence from the IE-2 staining. The anti-rabbit immunoglobulin G secondary antibody provided the red fluorescence. The right panels show green and red fluorescence merged, and the left panels show red fluorescence alone. Nonconfocal images were obtained at ×20 magnification.
US17 and US18 expression kinetics.
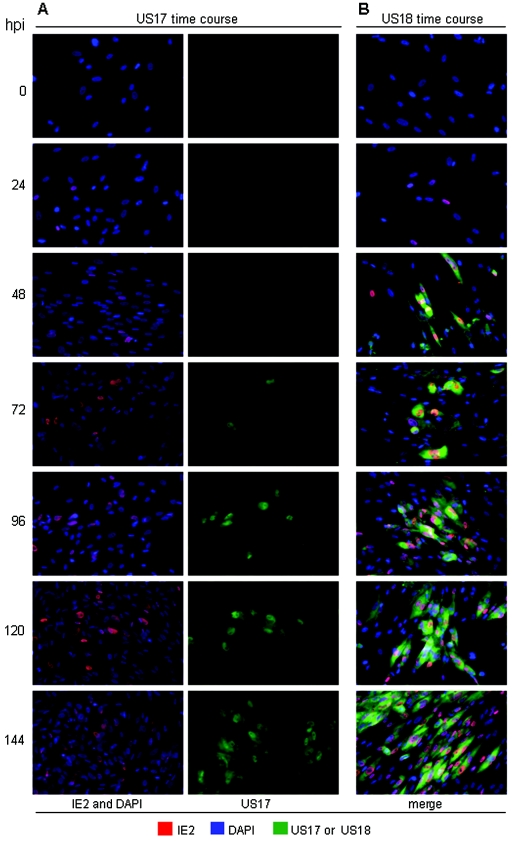
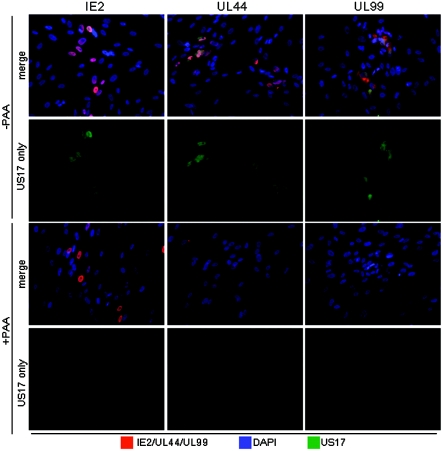
To learn the expression kinetics of these proteins, we studied their expression as a function of time after infection and in the presence of phosphonoacetic acid (PAA), an inhibitor of viral DNA synthesis. In these experiments, we simultaneously stained cells for the HCMV IE-2 protein, which is abundantly expressed in infected-cell nuclei by 24 h after infection, thus marking essentially all infected cells. US17 expression was reliably initially detected at 54 h after infection (data not shown, but the 72-h time point was positive in Fig. 2A), and US18 was detected at 36 h after infection (Fig. 2B). Both proteins continued to accumulate through the course of infection. In the presence of PAA, neither US17 nor US18 was expressed (Fig. 3). Under these conditions, IE-2 (an immediate-early or alpha gene product) and the DNA polymerase processivity factor (a beta gene encoded by UL44) were still expressed, but UL99 (a late gene) was not. The dependence of US17 and US18 expression on viral DNA synthesis indicates that they are late genes.
FIG. 2.
Time course of US17 (A) and US18 (B) expression during infection. HLF were infected at a MOI of 0.01 with HCMV(AD169) and then fixed and stained at the indicated times after infection (hours after infection [hpi]). Nuclei were stained with DAPI (blue), HCMV IE-2 protein was detected with a mouse monoclonal antibody (red), and US17 and US18 were detected with rabbit antibodies (green). Nonconfocal images were obtained at ×20 magnification.
FIG. 3.
Dependence of US17 expression on viral DNA synthesis. Cells were stained for the presence of US17 and US18 (green), and representative viral immediate-early (IE-2), early (UL44), and late (UL99) gene products (red) 96 h after infection with HCMV(AD169) at an MOI of 0.01 in the absence (−) and presence (+) of PAA (100 μg/ml). Nonconfocal images were obtained at ×20 magnification.
Nuclear localization of US17 but not of US14 or US18.
To better understand the potential roles of US17 and US18, we wanted to learn where they are expressed in infected cells. To do this, we examined infected cells by confocal microscopy, simultaneously staining for IE-2 (to mark infected cells red), a nuclear stain (DAPI [blue]), and an antibody against either US17 or US18 (green). The micrographs were collected as 1-μm sections and are displayed here as single unstacked sections. US18 was absent from the nucleus and was consistently detected throughout the cytoplasm of infected cells, with a somewhat higher concentration in the vicinity of the nucleus (Fig. 4). Unexpectedly, in addition to the expected cytoplasmic fluorescence, we found strong US17 reactivity in infected-cell nuclei (Fig. 4). This was unexpected, because all US12 family members are predicted to have multiple membrane-spanning segments that would normally preclude their entry into the nucleoplasm. As made clear in Z-series confocal images, the US17 staining is not simply staining the nuclear envelope but is staining a substantial volume of the nucleoplasm (data not shown).
FIG. 4.
Intracellular distribution of US14, US17, and US18. Rabbit polyclonal antibodies against the indicated N- and C-terminal peptide sequences were used to examine HLF 72 h after infection of HLF at an MOI of 0.01 with HCMV(AD169). Nuclei were stained with DAPI (blue), HCMV IE-2 protein was detected with a mouse monoclonal antibody (red), and US14, US17, and US18 were detected with rabbit antibodies (green). Confocal images were obtained at a magnification of ×63.
The US17 antibody is directed against the C-terminal domain of the protein, and the US18 antibody is directed against the N-terminal domain (Fig. 1A). Because it is possible that the US17 antibody is reacting with a C-terminal fragment of the protein that is able to translocate to infected-cell nuclei, we examined the reactivity of another US12 family member (US14) for which we had generated an antipeptide antibody against its C-terminal domain; this antibody reacted only in the cytoplasm of infected cells (Fig. 4). Thus, while the US17 nuclear localization may not be unique to this protein, it is not a shared property across the entire US12 family. We note that in the earlier work by Guo and Huang (12), a polyclonal antibody against full-length bacterially expressed US18 gave a staining pattern that may have included nuclear staining, but this cannot be said with certainty because confocal microscopy was not used, the image is at low magnification, and no nuclear counterstain was used.
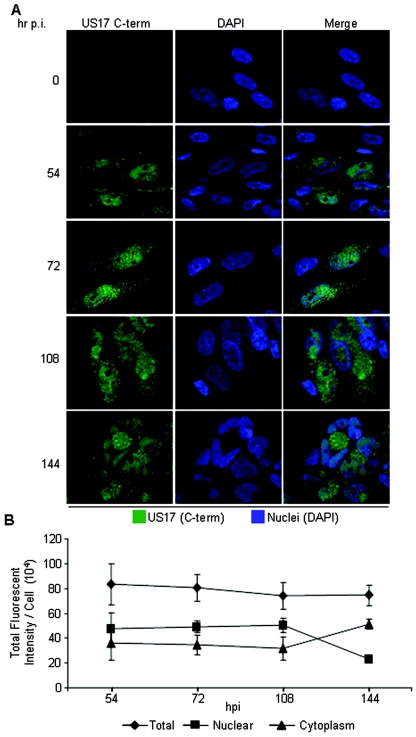
US17 nuclear localization is a dynamic process, with nuclear localization occurring shortly after the initial cytoplasmic appearance, followed by US17 appearing to be more abundant in nuclei at intermediate stages of infection and then redistributing to more abundant cytoplasmic distribution at late times (Fig. 5A). To quantitate this, we measured the fluorescence intensity of US17 in nuclei and cytoplasm at various time points following infection. On a per cell basis, the overall abundance of US17 did not change over the time span studied, but the ratio of nuclear to cytoplasmic protein inverted from predominantly nuclear to greater cytoplasmic abundance by 144 h after infection (Fig. 5B). In some cells (more so at earlier times), over 80% of the total US17 fluorescence is nuclear. We quantified total fluorescence intensity in the nuclear and cytoplasmic compartments. As can be readily seen in the photographs, in the fibroblasts used here, nuclei generally occupied less than half of the cellular cross section. Thus, even though the overall levels of US17 in nuclei are only modestly greater than in the cytoplasm (Fig. 5B), in most cells the nuclear concentration of US17 is markedly higher than in the cytoplasm.
FIG. 5.
Time course of nuclear versus cytoplasmic distribution of US17. (A) HLF infected with HCMV(AD169) were monitored for US17 expression using the rabbit antipeptide antibody. hr p.i., hours postinfection; C-term, C-terminal segment. (B) Total fluorescence intensities of cytoplasmic and nuclear US17 were measured in 5 to 13 infected cells at each time point using ImagePro 5 software (Media Cybernetics, Inc., Silver Spring, MD), and graphed as the means ± standard deviations (error bars). Confocal images of 1.0-μm thickness (the best plane) were obtained at a magnification of ×63 and were captured using the same photomultiplier tube voltage for each color throughout the time course. hpi, hours postinfection.
Dependence of US17 nuclear localization on infection.
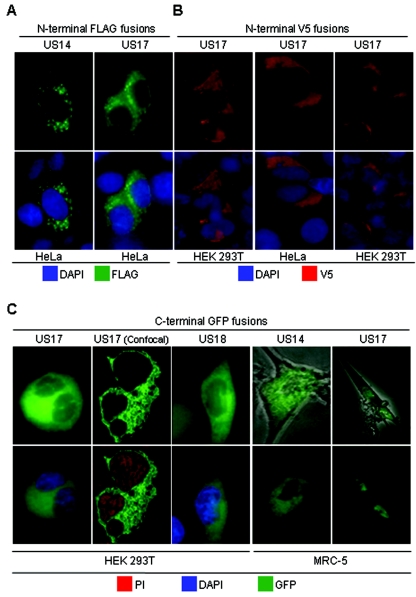
To test the dependence of US17 nuclear localization on infection, we transfected mammalian cells with constructs in which the protein is fused at its N terminus to either the FLAG or V5 epitope or at its C terminus to GFP. Using an anti-FLAG MAb, US17 was detected in the cytoplasm but not the nuclei of transiently transfected HeLa cells from 24 to 72 h after transfection (a 48-h time point is shown in Fig. 6A); a similar result was obtained for an analogous US14 construct. Likewise, only cytoplasmic localization was detected for the N-terminal V5 fusion in HeLa and HEK 293T cells (Fig. 6B). The C-terminal GFP fusion was likewise detected only cytoplasmically in HEK 293T cells, as were analogous US14 and US18 constructs (Fig. 6C) (US14 data not shown). Native GFP expressed from a similar vector was diffusely distributed throughout cells (data not shown). Because neither HeLa nor HEK 293T cells are highly permissive for HCMV replication, we did a similar experiment using MRC-5 cells, which allow efficient HCMV replication. Similar to the result in the other cell types, C-terminally GFP-tagged US14 and US17 were not expressed in MRC-5 cell nuclei. These results indicate that in the absence of infection, US17 does not translocate to nuclei. We were unable to detect US17 expression in transfected cells using the same antipeptide antibody used in our other experiments, although it did react with extracts of transfected cells in immunoblots (not shown), suggesting that the epitope of the antipeptide antibody is inaccessible or in a nonreactive conformation in the absence of infection.
FIG. 6.
Dependence of US17 nuclear localization on infection. The indicated mammalian cells were transfected with constructs in which US14, US17, and US18 are fused at their N terminus to the FLAG (A) or V5 (B) epitope or at their C terminus to GFP (C). Labeling was with the indicated antibodies and stains. Confocal images were obtained 24 h after transfection at a magnification of ×40 at 2.0× zoom, and the nonconfocal images were obtained at ×20 magnification. PI, propidium iodide.
These experiments involved the use of constructs in which additional amino acids were incorporated into the amino acid sequences of the studied proteins, either as a few additional residues in the form of N-terminal synthetic epitopes or as C-terminal GFP fusions. While such constructs have proven their value in many experiments, it remains a possibility that these sequence alterations could lead to protein misfolding, instability, or improper routing through the synthesis and processing machinery, as has sometimes been observed (10, 18). Our results indicate that the incorporated sequences did not lead to gross instability. At this level of analysis, the intracellular distributions of US14 and US18 are similar to their patterns in infected cells.
Intranuclear localization of US17 relative to other viral proteins that transit the nucleus.
We studied the intranuclear localization of US17 relative to three virally encoded proteins: IE-2, UL44, and UL99.
As previously shown by others, IE-2 is expressed almost exclusively in nuclei, beginning as early as 2 h after infection (1, 30, 38). As shown in Fig. 4, by 72 h, the protein is diffusely distributed throughout the nucleus, except for nucleoli, and is more abundant at the nuclear periphery than at the center. Both IE-2 and US17 are expressed throughout much of the nucleus (except for nucleoli), but IE-2 concentrations are highest toward the nuclear periphery (Fig. 4). In contrast, US17 distribution is often more uniform. The lack of coincidental concentration maxima suggests that these proteins may not have specific strong interactions.
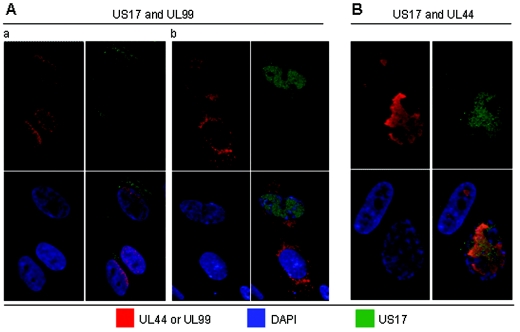
UL99 is a tegument protein (pp28) that previously had been detected in the nucleus only if its membrane-anchoring myristoylation sequence is mutagenized (14, 28, 29). The latter result indicates that the protein has intrinsic ability to translocate specifically to the nucleus. We repeatedly detected UL99 in the nuclei of cells that are in the early stages of infection, with a subsequent shift to an almost exclusively cytoplasmic localization. This suggests that HCMV manipulates the myristoylation process, leading to altered intracellular distribution of UL99 during the course of infection. US17 nuclear localization has an apparently reciprocal relationship with UL99 (Fig. 7A). As mentioned above, in the early stages of infection, UL99 is expressed transiently in the nucleus, after which it apparently translocates completely to the cytoplasm; at 72 h after infection, both localizations are present. In such cells, we did not observe nuclear US17 expression during the times UL99 was present in the nucleus (Fig. 7A, compare panels a and b). Because of the apparent inverse association between US17 and UL99 nuclear localization, we examined the expression pattern of UL99 cells infected with a virus deleted for US17. The time course of UL99 expression and nuclear transit in these cells was the same as in cells infected with virus expressing US17 (data not shown), indicating that the nuclear localization and egress of the two proteins are not directly linked.
FIG. 7.
Localization of US17 relative to other viral proteins that localize to infected-cell nuclei. (A) US17 and UL99. Panels a and b show examples of UL99 that are either predominantly nuclear or cytoplasmically located; the images shown are from the same slide. (B) US17 and UL44. Confocal images were obtained at a magnification of ×63, 72 h after infection of HLF at an MOI of 0.01 with HCMV(AD169).
UL44 encodes the DNA polymerase processivity factor and is a component of viral DNA replication complexes. It is detected initially as discrete dots that appear to coalesce and then ultimately fill much of the nuclear interior, with little staining at the nuclear margins (Fig. 7B). The most extensive overlapping signal is between US17 and UL44 (Fig. 7B), but as for IE-2, there was little evidence for coincidental concentration maxima. It thus appears that US17 localizes to the viral replication compartment in the nucleoplasm but without specific association with the proteins analyzed here.
Segmented expression of US17.
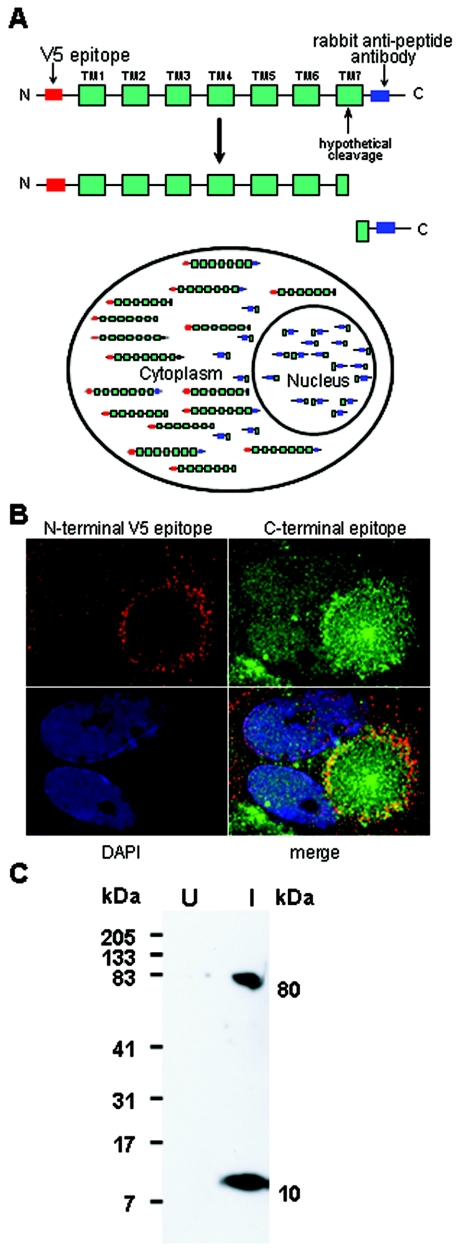
A possible explanation for the US17 nuclear localization is that the protein is expressed in a segmented manner, by any of several candidate transcriptional, translational, or posttranslational mechanisms, with a soluble C-terminal segment being able to migrate to the nucleus. To test this hypothesis, we constructed an HCMV BAC that incorporates a V5 synthetic epitope at the US17 N terminus (Fig. 8). As shown in the diagram (Fig. 9A), in combination with the rabbit antipeptide antibody against the C-terminal segment of the protein, this enables simultaneous independent detection of both ends of the protein. As shown in the micrographs, the US17 N and C termini have markedly different intracellular localizations. The N-terminal segment localizes predominantly to the periphery of structures that correspond to the cytoplasmic virion maturation complexes described by Sanchez et al. (27), in obvious contrast to the localization of the C-terminal segment in nuclei and throughout the maturation complex. The only significant colocalization is at the periphery of the maturation complex; in this region, the patterns are more of tight adjacency, rather than precise coincidence (Fig. 9B). These results support the segmentation hypothesis. While it is possible that the V5 epitope may have altered the localization of the N-terminal segment relative to the native sequence, we note that reactivity of the rabbit antibody against the C-terminal segment of US17 was similar to the pattern seen with the native sequence.
FIG.9.
Differential localization of US17 N and C termini in infected cells. (A) Schematic representation of a model in which the N and C termini of US17 are expressed as part of the same primary translation product and subsequently processed to N- and C-terminal fragments, of which only the C-terminal fragment is able to migrate to the nucleus. The N- and C-terminal fragments, seven transmembrane segments (green boxes), V5 epitope (red boxes), and rabbit antipeptide antibody (blue boxes) are shown. (B) HLF infected with a recombinant HCMV that expresses the V5 epitope at its N terminus. Ninety-six hours after infection, cells were stained as indicated in Materials and Methods. The field shown represents a fairly common pattern; the red ring from the V5 epitope staining outlines a cytoplasmic virus assembly compartment. (C) Immunoblot of HCMV-infected HLF probed with the rabbit antipeptide antibody against the US17 C-terminal segment. Immunoblots of uninfected (U) and infected (I) (multiplicity of infection of 0.5 for 96 h) HLF were prepared as described in Materials and Methods. The best extractions were obtained when all processing of infected cells was done at room temperature, and specimens were not boiled prior to electrophoresis. The sizes of the protein standards and the calculated sizes of the US17 segments are shown at the sides of the blot.
Consistent with this result, in an immunoblot of an infected-cell extract (Fig. 9C), we detected two species reactive with the US17 antibody, one with an apparent molecular size of 80 kDa that likely represents full-length US17 (its apparent size is larger than the 31.9 kDa calculated from the amino acid sequence, possibly because of its extremely hydrophobic nature or because it forms multimers that were not dissociated during the room temperature extraction), and another with an apparent molecular size of 10 kDa. The latter species has a size consistent with it being the product of proteolytic cleavage in the C-terminal domain of the protein.
DISCUSSION
While there has been much progress toward identification of HCMV gene function, there is still much to learn. Of the 165 genes recently enumerated by Dolan and colleagues in the low-passage isolate Merlin (7), functions have been ascribed to only about half. For 16 of 57 genes that are either essential for growth or whose deletion leads to a severe growth defect in the types of cells examined, no additional functional information is available (8, 39). The US12 family constitutes approximately 5% of the HCMV genetic content, yet little is known of the function of US12 family members other than each having seven predicted transmembrane domains and sequence features that suggest that they are related to GPCRs, their being nonessential for virus growth in cultured cells, and deletion of some US12 family members having modest effects on virus replication. The extensive expenditure of evolutionary capital on the US12 family suggests important biological roles. To begin to better understand their function, our initial objective was to identify the intracellular localization and expression pattern of three US12 family proteins. In the course of this work, we made the unexpected and novel observation that US17 localizes to infected-cell nuclei.
Generation of polyclonal antibodies against US12 family members.
It is obviously important to have specific and sensitive means to identify the localization, expression, processing, and transport patterns of proteins. Although it is possible to use various molecular tags (e.g., GFP) to study the synthesis and processing of such proteins, as noted above, such tags can lead to alterations in the patterns of expression and intracellular transport and may affect ligand binding or signaling. In addition, as we observed here, processing and transport can differ between viral proteins expressed in the absence of infection and in the context of infection. This leads to the additional complexity of needing to generate recombinant viruses expressing the tagged molecules (and revertants). The obvious alternative is to generate specific antibodies. Raising useful antibodies against multiple hydrophobic proteins, such as GPCRs, is difficult. Challenges include identification of sufficiently antigenic epitopes for stimulating antibody generation, the necessity that the target epitope is accessible in cells, and the requirement that these epitopes not be highly hydrophobic because of practicalities of peptide synthesis, purification, and solubilization. Even with these possibilities in mind, of 10 peptides we chose for raising antibodies to various segments of these proteins, 3 antibodies resulted that are specific and experimentally useful.
Kinetics of US17 and US18 expression.
In time course experiments, US17 was detected beginning as early as 54 h after infection and US18 was detected at 36 h after infection, and both were abundant to at least 144 h after infection. Expression of both proteins was dependent on viral DNA synthesis; thus, they are expressed as late proteins. For US18, these results are consistent with the transcript time course and kinetic class observed in Northern blotting experiments by Guo and Huang (12) (US17 transcription has not been studied in detail) but differ from a gene array result in which early transcripts were identified across US17 and US18 (3). Together, these results indicate that expression of US12 family members is regulated in a complex manner that may be unique for each gene.
US17 is expressed in infected-cell nuclei.
To a first approximation, US14, US17, and US18 have cytoplasmic distributions that are similar to those of other membrane proteins (11, 33). In a detailed analysis of their intracellular distribution, we have found an association of US17 and US18 predominantly with trans-Golgi and early endosome markers (S. Das and P. E. Pellett, unpublished data). The key observation reported here is that, as revealed by confocal microscopy, US17 additionally localizes to the interior of infected-cell nuclei, with a distribution distinct from those of other viral proteins that are present in or transit the nucleus, while US14 and US18 are present only in the cytoplasm. Thus, nuclear localization is not a shared property of all US12 family members; this is a strong indication that US12 family members are likely to embody a spectrum of functional diversity.
The nuclear localization was unexpected, given the highly hydrophobic nature of the protein and the absence of extensive membrane structures in the nucleoplasm. We also found that the nuclear localization of US17 is dependent on infection, inasmuch as the protein is detected only in the cytoplasm of transfected cells. This result was obtained both by antibody detection of transfected US17 sequences harboring N-terminal epitope tags (FLAG and V5 epitopes) and with a US17 sequence tagged at the C terminus with GFP, thus reducing the possibility that the failure to detect nuclear US17 was due to the epitope being inaccessible as expressed in uninfected cells. Interestingly, the rabbit antibody used throughout this paper did not react with US17 as expressed in transfected cells. In the absence of segmentation, the epitope recognized by the rabbit antibody may be masked, perhaps due to its proximity to the membrane.
US17 nuclear localization: possible mechanisms and functions.
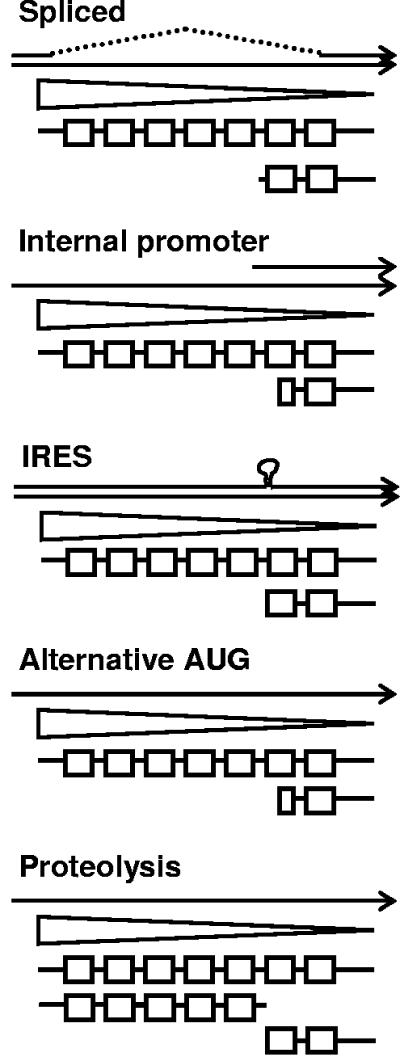
The two main mechanistic possibilities for nuclear translocation of US17 we considered were as follows. (i) The membrane-associated full-length molecule is expressed in or processed to a soluble form that may consist of a subsegment of the full-length protein. (ii) The intact protein is transported to the nucleoplasm. There are only a few examples of apparently intact 7TM or other membrane-associated proteins being detected in the nucleoplasm (as opposed to nuclear membranes), and the mechanism by which this occurs is unknown (5, 16, 19, 21). We considered the segmentation possibility to be the most likely and did an experiment employing a recombinant virus that expresses a version of US17 for which we can independently and simultaneously detect the N- and C-terminal regions. Although we could detect both ends of the molecule, we found very little colocalization of the two termini, indicating that the protein is indeed expressed in a segmented manner. In addition, a small fragment consistent with this hypothesis was detected by immunoblotting. Segmented expression of US17 might occur by any of several transcriptional (splicing or internal promoter), translational (alternative initiation codon or internal ribosomal entry sequence), or proteolytic means (Fig. 10). Additional study will be required to learn the mechanism by which this occurs and the segment boundaries. We have not identified conventional nuclear localization signal motifs in the US17 amino acid sequence, so its nuclear transport must be based on a noncanonical sequence or interaction with another protein that accesses the nuclear transport machinery.
FIG. 10.
Major hypotheses for US17 segmentation. mRNA species are shown as arrows, the US17 open reading frame is shown as a triangle, with the protein beneath (the boxes are the transmembrane segments). The various possibilities are not mutually exclusive. IRES, internal ribosome entry site.
The presence in nuclei of components of signaling cascades indicates that such mechanisms are in general use, and roles for such nucleus-localizing signaling molecules can be envisaged (reviewed in references 13 and 36). For example, nuclear localization of the receptors for type 1 parathyroid hormone and parathyroid hormone-related peptide, as well as the angiotensin II receptor, are linked to ligand-induced regulation of their target genes (9, 19, 35).
Perhaps pertinent, some membrane receptors are proteolytically cleaved in response to specific stimuli and then transported to the nucleus where they act as transcription factors. The process is known as regulated intramembrane proteolysis, is conserved across entities from bacteria to humans, and includes well-characterized examples that include the sterol response binding protein, Notch signaling, and processing of the amyloid precursor protein (reviewed in reference 2). We note the presence of candidate cleavage sites for the regulated intramembrane proteolysis-associated proteases S1P and S2P at the junction between the fifth and sixth transmembrane segments of US17 and its CCMV and RhCMV homologs. Cleavage here would generate a fragment of the size we have detected. We are exploring this and related possibilities.
In sum, we have made the novel observation of a virally encoded 7TM protein with a segmentally expressed nuclear localizing domain. Regardless of the mechanism by which it occurs, it is possible that the nuclear translocated segment of US17 plays a novel regulatory role during HCMV infection. This likely represents yet another mechanism by which a virus has hijacked or modified cellular regulatory pathways for its benefit.
Acknowledgments
We thank Amit Vasanji and Judy Drazba from the Lerner Research Institute Imaging Facility for assistance with confocal imaging, Achut Marlur for pcDNA3.1 plasmid, Pamela Petruna for cell and virus culture, Hua Zhu and Thomas Shenk for generously providing HCMV BACs, and Neal Copeland for γY380 cells.
REFERENCES
- 1.Ahn, J. H., and G. S. Hayward. 1997. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J. Virol. 71:4599-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, M. S., J. Ye, R. B. Rawson, and J. L. Goldstein. 2000. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100:391-398. [DOI] [PubMed] [Google Scholar]
- 3.Chambers, J., A. Angulo, D. Amaratunga, H. Guo, Y. Jiang, J. S. Wan, A. Bittner, K. Frueh, M. R. Jackson, P. A. Peterson, M. G. Erlander, and P. Ghazal. 1999. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J. Virol. 73:5757-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 5.Chen, R., Y. V. Mukhin, M. N. Garnovskaya, T. E. Thielen, Y. Iijima, C. Huang, J. R. Raymond, M. E. Ullian, and R. V. Paul. 2000. A functional angiotensin II receptor-GFP fusion protein: evidence for agonist-dependent nuclear translocation. Am. J. Physiol. Renal Physiol. 279:F440-F448. [DOI] [PubMed] [Google Scholar]
- 6.Davison, A. J., D. J. Dargan, and N. D. Stow. 2002. Fundamental and accessory systems in herpesviruses. Antivir. Res. 56:1-11. [DOI] [PubMed] [Google Scholar]
- 7.Dolan, A., C. Cunningham, R. D. Hector, A. F. Hassan-Walker, L. Lee, C. Addison, D. J. Dargan, D. J. McGeoch, D. Gatherer, V. C. Emery, P. D. Griffiths, C. Sinzger, B. P. McSharry, G. W. Wilkinson, and A. J. Davison. 2004. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 85:1301-1312. [DOI] [PubMed] [Google Scholar]
- 8.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, and F. Liu. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 100:14223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eggena, P., J. H. Zhu, K. Clegg, and J. D. Barrett. 1993. Nuclear angiotensin receptors induce transcription of renin and angiotensinogen mRNA. Hypertension 22:496-501. [DOI] [PubMed] [Google Scholar]
- 10.Ferguson, P. L., and W. F. Flintoff. 1999. Topological and functional analysis of the human reduced folate carrier by hemagglutinin epitope insertion. J. Biol. Chem. 274:16269-16278. [DOI] [PubMed] [Google Scholar]
- 11.Fraile-Ramos, A., A. Pelchen-Matthews, T. N. Kledal, H. Browne, T. W. Schwartz, and M. Marsh. 2002. Localization of HCMV UL33 and US27 in endocytic compartments and viral membranes. Traffic 3:218-232. [DOI] [PubMed] [Google Scholar]
- 12.Guo, Y. W., and E. S. Huang. 1993. Characterization of a structurally tricistronic gene of human cytomegalovirus composed of US18, US19, and US20. J. Virol. 67:2043-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jans, D. A., and G. Hassan. 1998. Nuclear targeting by growth factors, cytokines, and their receptors: a role in signaling? Bioessays 20:400-411. [DOI] [PubMed] [Google Scholar]
- 14.Jones, T. R., and S. W. Lee. 2004. An acidic cluster of human cytomegalovirus UL99 tegument protein is required for trafficking and function. J. Virol. 78:1488-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, D. H., M. Longo, Y. Han, P. Lundberg, E. Cantin, and J. J. Rossi. 2004. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat. Biotechnol. 22:321-325. [DOI] [PubMed] [Google Scholar]
- 16.Lee, D. K., A. J. Lanca, R. Cheng, T. Nguyen, X. D. Ji, F. Gobeil, Jr., S. Chemtob, S. R. George, and B. F. O'Dowd. 2004. Agonist-independent nuclear localization of the apelin, angiotensin AT1, and bradykinin B2 receptors. J. Biol. Chem. 279:7901-7908. [DOI] [PubMed] [Google Scholar]
- 17.Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56-65. [DOI] [PubMed] [Google Scholar]
- 18.Liu, X. Y., and L. H. Matherly. 2002. Analysis of membrane topology of the human reduced folate carrier protein by hemagglutinin epitope insertion and scanning glycosylation insertion mutagenesis. Biochim. Biophys. Acta 1564:333-342. [DOI] [PubMed] [Google Scholar]
- 19.Lu, D., H. Yang, G. Shaw, and M. K. Raizada. 1998. Angiotensin II-induced nuclear targeting of the angiotensin type 1 (AT1) receptor in brain neurons. Endocrinology 139:365-375. [DOI] [PubMed] [Google Scholar]
- 20.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marrache, A. M., F. Gobeil, T. Zhu, and S. Chemtob. 2005. Intracellular signaling of lipid mediators via cognate nuclear G protein-coupled receptors. Endothelium 12:63-72. [DOI] [PubMed] [Google Scholar]
- 22.Mocarski, E. S., Jr., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe, P. M. Howley, B. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 23.Murphy, E., D. Yu, J. Grimwood, J. Schmutz, M. Dickson, M. A. Jarvis, G. Hahn, J. A. Nelson, R. M. Myers, and T. E. Shenk. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. USA 100:14976-14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pereira, L., E. Maidji, S. McDonagh, and T. Tabata. 2005. Insights into viral transmission at the uterine-placental interface. Trends Microbiol. 13:164-174. [DOI] [PubMed] [Google Scholar]
- 25.Pfeffer, S., A. Sewer, M. Lagos-Quintana, R. Sheridan, C. Sander, F. A. Grasser, L. F. van Dyk, C. K. Ho, S. Shuman, M. Chien, J. J. Russo, J. Ju, G. Randall, B. D. Lindenbach, C. M. Rice, V. Simon, D. D. Ho, M. Zavolan, and T. Tuschl. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269-276. [DOI] [PubMed] [Google Scholar]
- 26.Rigoutsos, I., J. Novotny, T. Huynh, S. T. Chin-Bow, L. Parida, D. Platt, D. Coleman, and T. Shenk. 2003. In silico pattern-based analysis of the human cytomegalovirus genome. J. Virol. 77:4326-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez, V., E. Sztul, and W. J. Britt. 2000. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-Golgi-intermediate compartment. J. Virol. 74:3842-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silva, M. C., Q. C. Yu, L. Enquist, and T. Shenk. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 77:10594-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sinzger, C., M. Kahl, K. Laib, K. Klingel, P. Rieger, B. Plachter, and G. Jahn. 2000. Tropism of human cytomegalovirus for endothelial cells is determined by a post-entry step dependent on efficient translocation to the nucleus. J. Gen. Virol. 81:3021-3035. [DOI] [PubMed] [Google Scholar]
- 31.Smith, M. S., G. L. Bentz, P. M. Smith, E. R. Bivins, and A. D. Yurochko. 2004. HCMV activates PI(3)K in monocytes and promotes monocyte motility and transendothelial migration in a PI(3)K-dependent manner. J. Leukoc. Biol. 76:65-76. [DOI] [PubMed] [Google Scholar]
- 32.Soderberg-Naucler, C., K. Fish, and J. A. Nelson. 1999. Cytomegalovirus, p. 209-242. In R. Ahmed and I. S. Y. Chen (ed.), Persistent viral infections. John Wiley & Sons, New York, N.Y.
- 33.Somia, N. V., M. J. Schmitt, D. E. Vetter, D. Van Antwerp, S. F. Heinemann, and I. M. Verma. 1999. LFG: an anti-apoptotic gene that provides protection from Fas-mediated cell death. Proc. Natl. Acad. Sci. USA 96:12667-12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varnum, S. M., D. N. Streblow, M. E. Monroe, P. Smith, K. J. Auberry, L. Pasa-Tolic, D. Wang, D. G. Camp, K. Rodland, S. Wiley, W. Britt, T. Shenk, R. D. Smith, and J. A. Nelson. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watson, P. H., L. J. Fraher, B. V. Natale, M. Kisiel, G. N. Hendy, and A. B. Hodsman. 2000. Nuclear localization of the type 1 parathyroid hormone/parathyroid hormone-related peptide receptor in MC3T3-E1 cells: association with serum-induced cell proliferation. Bone 26:221-225. [DOI] [PubMed] [Google Scholar]
- 36.Wells, A., and U. Marti. 2002. Signalling shortcuts: cell-surface receptors in the nucleus? Nat. Rev. Mol. Cell Biol. 3:697-702. [DOI] [PubMed] [Google Scholar]
- 37.Weston, K., and B. G. Barrell. 1986. Sequence of the short unique region, short repeats, and part of the long repeats of human cytomegalovirus. J. Mol. Biol. 192:177-208. [DOI] [PubMed] [Google Scholar]
- 38.Xu, Y., K. S. Colletti, and G. S. Pari. 2002. Human cytomegalovirus UL84 localizes to the cell nucleus via a nuclear localization signal and is a component of viral replication compartments. J. Virol. 76:8931-8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu, D., M. C. Silva, and T. Shenk. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. USA 100:12396-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]