Abstract
Transforming growth factor β (TGF-β) induces apoptosis in a variety of cells. We have previously shown that TGF-β1 rapidly induces apoptosis in the FaO rat hepatoma cell line. We have now studied the effect of TGF-β1 on the expression of different members of the Bcl-2 family in these cells. We observed no detectable changes in the steady-state levels of Bcl-2, Bcl-XL, and Bax. However, TGF-β1 induced caspase-dependent cleavage of BAD at its N terminus to generate a 15-kDa truncated protein. Overexpression of the 15-kDa truncated BAD protein enhanced TGF-β1-induced apoptosis, whereas a mutant BAD resistant to caspase 3 cleavage blocked TGF-β1-induced apoptosis. Overexpression of Smad3 dramatically enhanced TGF-β1-induced cleavage of BAD and apoptosis, whereas antisense Smad3 blocked TGF-β1-induced apoptosis and BAD cleavage. These results suggest that TGF-β1 induces apoptosis through the cleavage of BAD in a Smad3-dependent mechanism.
Transforming growth factor β (TGF-β) family members regulate a broad spectrum of cellular processes, including cellular growth, differentiation, and apoptosis (37). The TGF-β family members signal through a heteromeric receptor complex consisting of both type I (TβRI) and type II (TβRII) serine/threonine kinase receptor subunits. Activated type I receptor phosphorylates the downstream signal transducers, the receptor-regulated Smad's (R-Smad's) (14, 15, 23, 31). This induces binding of the R-Smad to the common mediator, Smad4. Following binding to Smad4, the complex translocates into the nucleus to activate transcription of various target genes (32). TGF-β induces apoptosis in various cell types, including primary hepatocytes (2, 12, 16), hepatoma cell lines (10, 11, 29, 40), prostate epithelial cancer cells (24), B cells (1, 6), and hematopoietic cells (18). However, the biochemical mechanisms that are responsible for mediating this death process are still poorly understood. Recently, it has been shown that Daxx, a Fas-receptor-associated protein that mediates the activation of JNK and programmed cell death induced by Fas, physically interacts with TβRII and is involved in mediating TGF-β-induced apoptosis (35, 46). Another report has shown that ARTS, a protein product of the human septin H5/PNUTL2/CDCrel2b gene, acts to enhance cell death induced by TGF-β (27). ARTS localizes to mitochondria and undergoes a mitochondrion-to-nucleus translocation during TGF-β-induced apoptosis.
The Bcl-2 family proteins serve as critical regulators of pathways involved in apoptosis, acting to either inhibit or promote cell death (26, 33). Posttranscriptional modifications, together with the modulation of the levels of expression and subcellular localization of inhibitors (Bcl-2 and Bcl-XL) and promoters (BAX, BAD, BID, and BIK), determine how cells respond to apoptotic stimuli (22, 26, 33). In interleukin-3 (IL-3)-dependent lymphoid cells, BAD, a proapoptotic member of the Bcl-2 family of proteins is a key regulator of apoptosis (19). The function of BAD is regulated by reversible phosphorylation. Deprivation of survival factors induces BAD dephosphorylation by the specific serine/threonine phosphatase PP1α (38), resulting in dissociation of BAD from 14-3-3 proteins and translocation to the mitochondria, where it interacts with Bcl-XL and Bcl-2 and antagonizes their antiapoptotic functions. BAD is cleaved by a caspase(s) at its N terminus to generate a 15-kDa truncated protein after IL-3 deprivation-induced apoptosis in murine myeloid precursor 32Dcl3 cells (13). The 15-kDa truncated BAD is a more potent inducer of apoptosis than the wild-type (WT) protein, whereas a mutant BAD resistant to caspase 3 cleavage is a weaker apoptosis inducer (13).
Recently, we have reported that TGF-β1 rapidly induces apoptosis through the activation of Cdc2, Cdk2, and caspases in the rat hepatoma cell line FaO (5, 10, 11). Our present results show that BAD is cleaved 6 h after TGF-β1 treatment of FaO cells. Similar to previously described reports (10, 11), a caspase 3-resistant mutant BAD showed less proapoptotic activity than the WT protein, whereas overexpression of truncated BAD accelerates TGF-β1-dependent apoptosis. Furthermore, we find that overexpression of Smad3 enhances TGF-β1-dependent BAD cleavage and apoptosis and antisense Smad3 blocks its activity. These results show that TGF-β1 enhances cleavage of full-length BAD during TGF-β1-mediated apoptosis in a Smad3-dependent manner.
MATERIALS AND METHODS
Cell culture, transfection, and reporter assays.
FaO rat hepatoma cells were maintained at 37°C in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), penicillin, and streptomycin (100 μg/ml). The 293-derived PHOENIX E (kind gift of Lisa Choy, University of California) or GP-293 packaging cells were maintained in DMEM supplemented with 10% heat-inactivated FBS. FaO stable cells were transfected with 4× SBE-luc (47) in six-well plates using Lipofectin (Life Technology, Rockville, Md.) according to the manufacturer's instructions. After transfection, cells were treated with 5 ng of TGF-β1/ml for 24 h in media. All assays were performed in triplicate, and data shown are means (± standard error [SE]) of three independent transfections.
Plasmids.
The pLXSP WT BAD-hemagglutinin (HA), pLXSP DM56/61 BAD-HA, and pLXSP tBAD68 BAD-HA retroviral constructs were prepared as described previously (13). For antisense Smad3 construct, the complete coding sequence of Smad3 was obtained by reverse transcription-PCR from FaO rat hepatoma cells. Antisense Smad3 vector was generated by cloning reverse-oriented Smad3 cDNA into pDONA1, and endogenous Smad3 protein levels were analyzed by Western blotting.
Retrovirus infection and generation of stable cell lines.
To generate retroviruses, Phoenix E packaging cells were plated at 106 cells/60-mm-diameter tissue culture dish and transfected with the pLXSP retroviral vector (empty or containing HA-WT BAD, HA-DM56/61 BAD, or tBAD68 cDNA) by the calcium phosphate method. In the case of antisense rat Smad3 retroviral vector, GP-293 packaging cells were plated at 106 cells/100-mm-diameter tissue culture dish and cotransfected with the pDONA1 retroviral vector (empty or containing AS-Smad3) and pVSV-G encoding envelop protein. Immediately before transfection, cells were treated with 25 μM chloroquine to increase transfection efficiency. At 6 h posttransfection, the medium was replaced with fresh DMEM containing 10% FBS, and cells were grown for an additional 24 h. The conditioned medium containing recombinant retroviruses was collected and filtered through 0.45-μm-pore-size polysulfonic filters. Samples (1.5 ml each) of these supernatants were applied immediately to FaO cells, which had been plated 18 h before infection at a density of 105 cells/60-mm-diameter tissue culture dish. Polybrene (Sigma, St. Louis, Mo.) was added to a final concentration of 8 μg/ml, and the supernatants were incubated with the cells for 12 h. After infection, the cells were placed in fresh growth medium and cultured as usual. Selection with 2 μg of puromycin/ml or 600 μg of neomycin/ml was initiated 24 h after infection. After 15 days, individual clones picked from plates of the recombinant retrovirus-infected cells were transferred to 24 microtiter wells and expanded to generate cell clones stably expressing WT, mutant, truncated BAD, or antisense Smad3.
Adenoviral infections.
Recombinant adenoviruses expressing Smad1, Smad2, Smad3, Smad4, Smad5, Smad7, or β-galactosidase were kindly provided by Kohei Miyazono (The Cancer Institute, Tokyo, Japan) and were used at a multiplicity of infection (MOI) of 200 with single viruses as described by Fujii et al. (20).
DNA fragmentation assay.
FaO cells were treated with lysis buffer (10 mM Tris-Cl, pH 7.4, 10 mM NaCl, 10 mM EDTA, 0.5% sodium dodecyl sulfate [SDS], and 0.1 mg of proteinase K/ml) and were incubated at 50°C for 2 h. The lysate was extracted successively with phenol, phenol-chloroform (1:1), and chloroform and was then precipitated with 2.5 volumes of ice-cold ethanol. The DNA was resuspended in Tris-EDTA buffer supplemented with 100 μg of RNase A/ml. DNA samples were electrophoretically separated on 2% agarose gel for 2 h at 50 V.
TUNEL assay.
FaO cells were plated at 5 × 104 cells/eight-well chamber slide (Nalge Nunc Int., Rochester, N.Y.) and incubate for 24 h prior to infection with recombinant adenoviruses. At 1 h postinfection, the medium was replaced with fresh DMEM containing 10% FBS and cells were grown for an additional 24 h. The cells were treated with TGF-β1 (5 ng/ml) for 6 h and fixed with 4% paraformaldehyde (pH 7.4) for 10 min. Apoptotic cells were assessed by measuring DNA fragmentation in a standard terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay according to the instructions with the kit (In situ Cell Death Detection Kit, POD; Roche Molecular Biochemicals, Indianapolis, Ind.).
Immunoblot analysis.
Whole-cell extracts were obtained in a 1% Triton X-100 lysis buffer (50 mM Tris-Cl, pH 8.0, 150 mM sodium chloride, 1 mM EDTA, 1 mM EGTA, 2.5 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 1 mM β-glycerophosphate, 1 μg of leupeptin/ml, and 1 mM phenylmethylsulfonyl fluoride). Western blotting was performed using anti-BAD (N-19; Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.), anti-Bcl-2 (N-19, Santa Cruz Biotechnology, Inc.), anti-Bcl-XS/L (S-18; Santa Cruz Biotechnology, Inc.), anti-Bax (P-19; Santa Cruz Biotechnology, Inc.), anti-Bid (M-20; Santa Cruz Biotechnology, Inc), anti-cytochrome c (7H8.2C12; PharMingen, San Diego, Calif.), anti-Smad3 (Zymed, San Francisco, Calif.), anti-FLAG(M2; Santa Cruz Biotechnology, Inc.), and anti-HA (Y-11; Santa Cruz Biotechnology) antibodies. Protein samples were heated at 95°C for 5 min and analyzed by SDS-16% polyacrylamide gel electrophoresis (PAGE).
Assessment of mitochondrial transmembrane potential.
Changes in mitochondria membrane potential were determined by staining cells with the fluorescence probe dihydrorhodamine 123 (Molecular Probes, Eugene, Oreg.). Cells were incubated in phosphate-buffered saline (PBS) containing 10 μM dihydrorhodamine 123 (Rh 123) for 30 min at 37°C in the dark and analyzed in a FACScalibur flow cytometry (Becton Dickinson, San Jose, Calif.). For control purposes, the protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP) (50 μM; Sigma) was used. The fluorescence was excited with an Argon laser (excitation wavelength, 488 nm) and analyzed in FL-1 (wavelength, 520 nm; photomultiplier tube [PMT] voltage, 437 V). At least 2 × 104 events were acquired in list mode and analyzed with CELLQuest software (Becton Dicknson).
Analysis of cytochrome c release.
For mitochondria cytochrome c release assay, FaO cells were scraped off in isotonic isolation buffer (10 mM HEPES, 1 mM EDTA, 250 mM sucrose, pH 7.6), collected by centrifugation at 2,500 × g for 5 min at 4°C, and resuspended in hypotonic isolaton buffer (10 mM HEPES, 1 mM EDTA, 50 mM sucrose, pH 7.6). Cells were disrupted by passing through a 27-gauge needle 5 to 10 times and checked for cracked cells by trypan blue staining. Hypertonic isolation buffer (10 mM HEPES, 1 mM EDTA, 450 mM sucrose, pH 7.6) was added to balance the buffer's tonicity. Samples were centrifuged at 1,000 × g (2,100 rpm) at 4°C for 10 min. Supernatants were recovered and centrifuged again at 100,000 × g. The mitochondrion pellet proteins were extracted in isotonic isolation buffer, and the supernatant contained the cytosolic protein extract. Protein concentration of lysates was determined using a protein assay kit (Bio-Rad, Hercules, Calif.) according to the manufacturer's instructions. After electrophoresis separation of 50 μg of protein per condition in SDS-16% polyacrylamide, gels were transferred by semidry transfer (Bio-Rad Labs, Richmond, Calif.) to nitrocellulose membranes. Immunoblots were blocked in TBS-T (10 mM Tris-HCl, 150 mM NaCl, pH 7.5, 0.05% Tween 20) containing 5% nonfat dried milk and incubated overnight with the primary antibody (monoclonal anti-cytochrome c diluted 1:1,000 in TBS-T-5% BSA). After washing, membranes were incubated with peroxide-conjugated anti-mouse immunoglobulin (1:3,000 in TBS-T-0.5% nonfat dried milk) for 1 h and the blot was developed with the ECL kit (Pierce Chemical Co., Rockford, Ill.).
RESULTS
Cleavage of proapoptotic BAD during TGF-β1-induced apoptosis in Fao hepatoma cells.
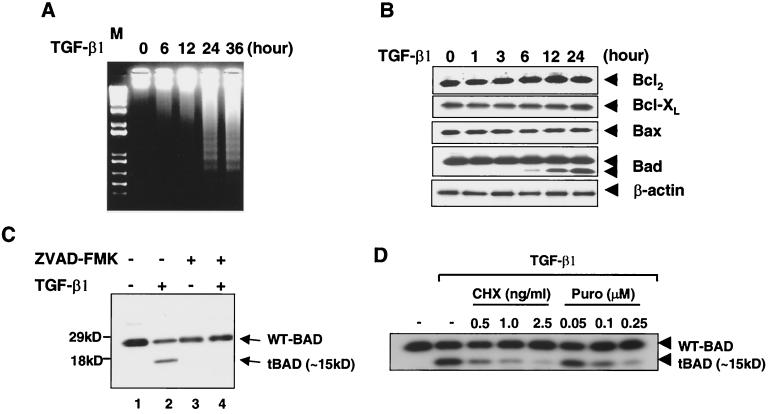
Treatment of FaO rat hepatoma cells with TGF-β1 induced a 180- to 200-bp internucleosomal DNA cleavage as early as 6 h after TGF-β1 treatment, with optimal laddering occurring by 24 h (Fig. 1A). To characterize the mechanism of TGF-β1-mediated apoptosis, the steady-state levels of several Bcl-2 family proteins were measured by Western blotting analysis. We detected no changes in expression of Bcl-2, Bcl-XL, or Bax over the TGF-β1 treatment time course and did not detect any cleaved products of these proteins (Fig. 1B). However, a ∼15-kDa cleavage product of BAD was increased over the TGF-β1 treatment time course (Fig. 1B). A longer exposure of the blot showed that truncated BAD could be detected as early as 6 h after TGF-β1 using an antibody against the C terminus of BAD (C-20; amino acids 185 to 204). A decrease in the full-length form of BAD protein level was also observed after TGF-β1 treatment in a short exposure of the blot of Fig. 1B (data not shown). In Fig. 1C, this decrease in the full-length form of BAD protein is clearly observed in a short exposure of the blot. An antibody specific for the N terminus of BAD (K-17) did not recognize the 15-kDa cleavage product (data not shown).
FIG. 1.
Expression of Bcl-2 family proteins during TGF-β1-induced apoptosis in FaO rat hepatoma cells. (A) Cellular DNA fragmentation after TGF-β1 treatment. Genomic DNAs were extracted from control and TGF-β1-treated cells at 6, 12, 24, and 36 h. Extracted DNAs were resolved on agarose gel. (B) The effect on Bcl2, Bcl-XL, Bax, and BAD protein levels in FaO cells was determined. Cells were incubated with 5 ng of TGF-β1/ml. Cell lysates were made, and equivalent amounts of cellular proteins were separated by SDS-15% PAGE, blotted, and then probed with the appropriate antibody. Blots were shown are typical of at least three individual experiments. (C) Effect of Z-VAD fmk (50 μM) on levels of endogenous full-length BAD and 15-kDa cleaved form of BAD in cells treated with 5 ng of TGF-β1/ml for 14 h. Western blot analyses were performed with an anti-BAD (C-20) antibody. Results are representative of three separate experiments. (D) Effect of cycloheximide and puromycin on levels of endogenous full-length BAD and the 15-kDa cleaved form of BAD in cells treated with 5 ng of TGF-β1/ml for 14 h. Western blot analyses were performed with an anti-BAD (C-20) antibody.
It was shown previously that treatment of 32Dc13 myeloid precursor cells with the broad-range caspase inhibitor Z-VAD fmk prevented the generation of the 15-kDa cleaved form of BAD (13). Therefore, we also assessed the possible involvement of a caspase(s) in the cleavage of BAD in FaO cells. Pretreatment of FaO cells with the Z-VAD fmk abrogated the generation of the 15-kDa cleaved form of BAD (Fig. 1C), suggesting that caspase(s) also play a role in BAD cleavage after TGF-β1 treatment of FaO cells. To test whether TGF-β-induced cleavage of BAD is dependent on new protein synthesis, FaO cells were pretreated with protein synthesis inhibitors (cycloheximide or puromycin) for 1 h and then treated with TGF-β1 for 24 h. Pretreatment with either cycloheximide or puromycin inhibited TGF-β-induced cleavage of BAD, indicating that de novo protein synthesis is required for the generation of the 15-kDa cleaved form of BAD by TGF-β1.
Differential effects of WT, DM56/61, and truncated BAD on TGF-β1-induced apoptosis in FaO cells.
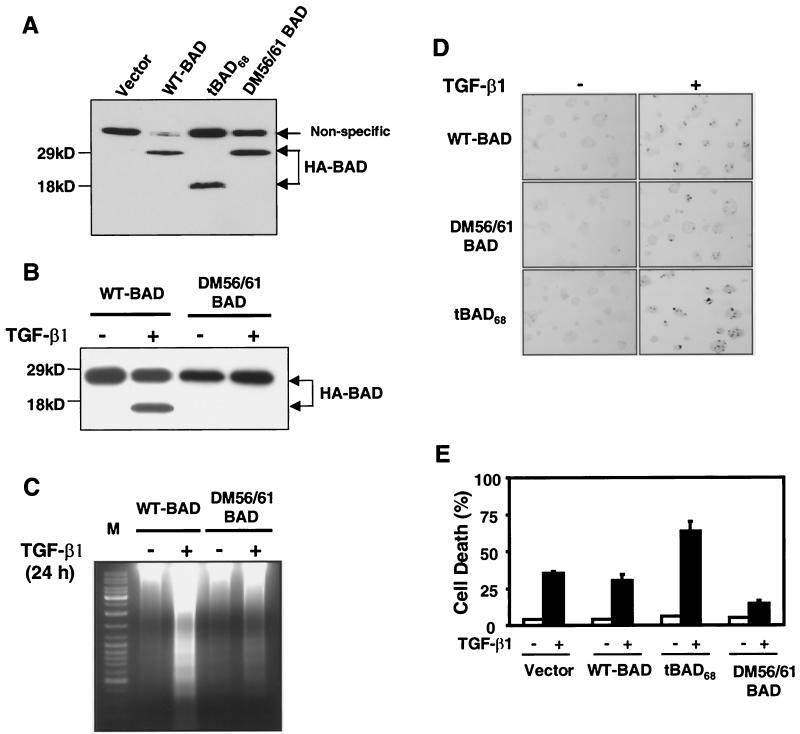
To assess the effect of double mutant DM56/61 (D56E and D61E) and truncated BAD on TGF-β1-induced apoptosis, stable cell lines expressing DM56/61 and truncated BAD were generated by infecting retroviruses encoding the HA-tagged WT BAD, DM56/61, or tBAD68. The puromycin-resistant colonies were pooled, and expression levels of HA-tagged WT BAD, DM56/61 BAD, or t-BAD68 were examined by Western blotting with an antibody raised against the HA epitope (Fig. 2A). These stable lines expressed similar levels of HA-tagged proteins. WT BAD cells and vector-infected cells showed similar levels of TGF-β1-induced apoptosis as estimated by the TUNEL assay. Overexpression of DM56/61 BAD blocked TGF-β1-induced apoptosis, generation of the 15-kDa cleaved form of BAD by TGF-β1, and TGF-β1-induced DNA fragmentation, whereas overexpression of tBAD68 led to an 80% increase in TGF-β1-induced apoptosis (Fig. 2B to E).
FIG. 2.
Differential effects of WT, DM56/61, and truncated BAD on FaO cell viability before and after TGF-β1 treatment. (A) Expression of WT BAD, DM56/61 BAD, and tBAD68 proteins in the FaO-infected cells was determined by a Western blot analysis using anti-HA antibody. (B) Apoptotic progression in FaO cells stably expressing WT BAD, DM56/61 BAD, and tBAD68 treated with TGF-β1. The cells were treated with TGF-β1 for 12 h. Cell lysates were made, and equivalent amounts of cellular proteins were separated by SDS-15% PAGE, blotted, and then probed with the anti-HA antibody. Levels of the C-terminal HA-tagged 15-kDa cleaved form of BAD in full-length BAD (WT-BAD) and DM56/61-BAD expressing cells were examined. Western blot analysis was performed with an anti-BAD (C-20) antibody. Results are representative of three separate experiments. (C) Cellular DNA fragmentation after TGF-β1 treatment. Genomic DNAs were extracted from the WT-BAD and DM56/61-BAD cells after TGF-β1 treatment at 24 h. Extracted DNAs were resolved on agarose gel. (D) TUNEL procedure was carried out and pictures were taken under a light microscope (magnification, ×200). (E) TUNEL-positive apoptotic cells at the respective incubation time were counted, and the percentage of apoptotic cells was graphed. Similar results were achieved in three separate experiments with comparable outcomes.
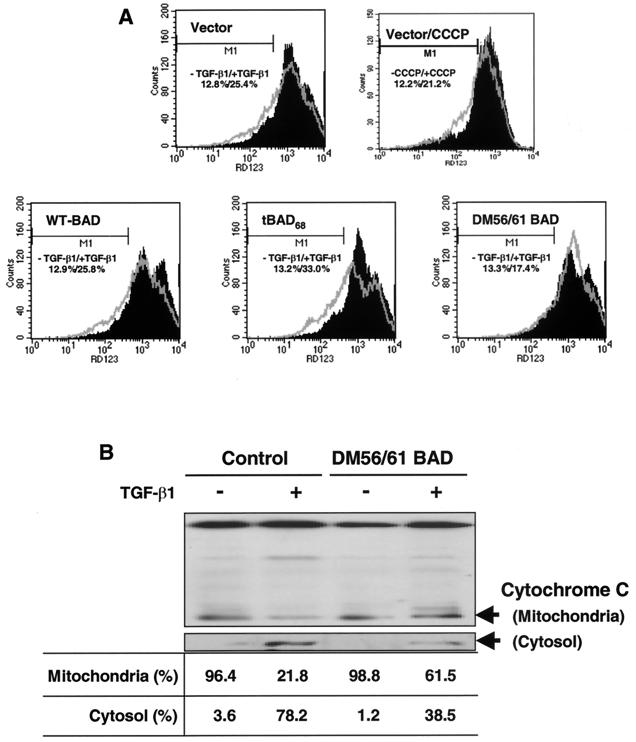
Truncated BAD acts to regulate mitochondrial membrane potential (Δψm) (26). We examined the changes in the mitochondrial membrane potential of the cells expressing WT BAD, DM56/61 BAD, or tBAD68. We used Rh123 as a fluorescent probe to detect changes in the mitochondrial membrane potential by flow cytometry. Increased Rh123 accumulation reflects an increase in mitochondrial membrane potential. Cells were incubated in the absence or presence of TGF-β1 (5 ng/ml). For 12 h after TGF-β1 treatment, Rh123 fluorescence was reduced in the control (vector) and WT BAD-expressing FaO cells (Fig. 3A). Rh123 fluorescence was further reduced in tBAD68-expressing FaO cells. However, these changes in mitochondrial membrane potential were not seen in cells overexpressing DM56/61 BAD.
FIG. 3.
Changes in mitochondrial transmembrane potential (Δψm) and the release of cytochrome c induced by TGF-β1 in cells expressing WT, DM56/61, and truncated BAD. (A) Cells incubated for 12 h in the absence (control) or presence of 5 ng of TGF-β1/ml were detached by trypsinization. After 30 min of incubation with Rh123 (5 μM), the intracellular fluorescence intensity was measured in a FACScan flow cytometer. Results representative of experiments run more than three times are shown. For a positive control, CCCP, an uncoupling agent, was incubated with cells for 10 min. (B) Release of cytochrome c from mitochondria to cytosol in the control and DM56/61 BAD cells. After incubation of cells for 12 h in the absence (−) or presence (+) of 5 ng of TGF-β1/ml, mitochondria were separated from the cytosol and cytochrome c content was analyzed by Western blotting as described in Materials and Methods. The ratio between mitochondria and cytosol is shown at the bottom.
In view of these results, we studied whether the loss in mitochondrial membrane potential (Δψm) could be coincident with the release of cytochrome c. After incubation of the cells for 24 h in the absence or presence of 5 ng of TGF-β1/ml, mitochondria were separated from cytosol and cytochrome c content was analyzed by Western blotting as described in Materials and Methods. TGF-β1-induced cytoplasmic accumulation of cytochrome c has been demonstrated in several systems (5, 9), but the involvement of BAD has not been examined. As shown in Fig. 3B, cytochrome c content decreased considerably in the mitochondria, corresponding to increased levels in the cytosol. However, cytochrome c release induced by TGF-β1 was markedly reduced in DM56/61 BAD-expressing FaO cells (Fig. 3B).
Smad3 enhances TGF-β1-mediated apoptosis and increases cleavage of BAD.
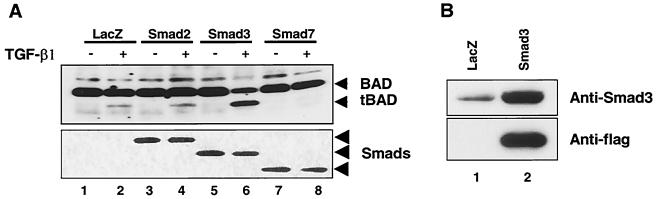
To investigate whether Smad proteins are directly involved in TGF-β1-mediated BAD cleavage, we infected FaO cells with adenoviruses carrying Smad cDNAs. Expression of FLAG-tagged Smads was confirmed by anti-FLAG immunoblotting (Fig. 4C). Without TGF-β1 treatment, apoptotic cell death was not observed in the cells expressing Smad proteins. Neither Smad1 nor Smad5, which are activated by bone morphogenetic protein (BMP), enhanced TGF-β1-induced apoptosis (Fig. 4A and B), although Smad proteins were expressed in the infected cells. Smad4, the common partner Smad in mammals, also did not enhance the level of apoptosis induced by TGF-β1. Overexpression of Smad2, which is activated by TGF-β1 and activin pathways, slightly enhanced TGF-β1-mediated apoptosis. However, Smad3, which is also activated by TGF-β1 and activin pathways, significantly enhanced apoptosis induced by TGF-β1 (Fig. 4A and B). Next, we examined cleavage of BAD in cells expressing Smad2, Smad3, or Smad7, which inhibits TGF-β1 signaling mediated by Smad2-Smad4 and Smad3-Smad4 complexes. Cleavage of Bad was not observed in cells expressing Smad2 and Smad3 without TGF-β1 treatment, whereas treatment with TGF-β1 led to the generation of the 15-kDa cleaved form of BAD in cells expressing β-galactosidase, Smad2, and Smad3 (Fig. 5A). When the FaO cells were infected with Smad3 adenovirus, the level of Smad3 protein was markedly enhanced compared to control cells, and generation of the 15-kDa BAD cleavage product was dramatically enhanced after treatment with 5 ng of TGF-β1/ml (Fig. 5A and B). However, Smad7 inhibited TGF-β1-induced cleavage of BAD. These results suggest that Smad3 plays a critical role in TGF-β1-dependent apoptosis and BAD cleavage during TGF-β1-induced apoptosis.
FIG. 4.
Effect of overexpression of Smad's on TGF-β1-dependent apoptosis. FaO cells were infected with adenoviruses carrying Smad1, Smad2, Smad3, Smad4, and Smad5 at an MOI of 300 and treated with 5 ng of TGF-β1/ml. Adenovirus carrying β-galatosidase (MOI of 800) was used as a control. (A) A TUNEL procedure was carried out and pictures were taken under a light microscope. Magnification, ×200. (B) TUNEL-positive apoptotic cells at the respective incubation time were counted, and the percentage of apoptotic cells was graphed. (C) Expression of Smad's was confirmed by anti-FLAG immunoblotting. Similar results were achieved in three separate experiments with comparable outcomes.
FIG. 5.
Effect of Smad's on generation of ∼15-kDa cleaved form of BAD induced by TGF-β1. (A) FaO cells were infected with adenoviruses carrying Smad2, Smad3, and Smad7 at an MOI of 300 and treated with 5 ng of TGF-β1/ml for 24 h. Adenovirus carrying β-galatosidase (MOI of 800) was used as a control. Cell lysates were made and equivalent amounts of cellular proteins were separated by SDS-15% PAGE, blotted, and then probed with an anti-BAD (C-20) antibody. Expression of Smad's was confirmed by anti-FLAG immunoblotting (bottom). Results are representative of three separate experiments. Expression of Smad's was confirmed by anti-FLAG immunoblotting (bottom). Results are representative of three separate experiments. (B) Level of Smad3 in FaO cells infected with adenoviruses carrying β-galactosidase and Smad3. Cell lysates were made 24 h after adenovirus infection, and equivalent amounts of cellular proteins were separated by SDS-15% PAGE, blotted, and then probed with either anti-Smd3 antibody or anti-flag antibody.
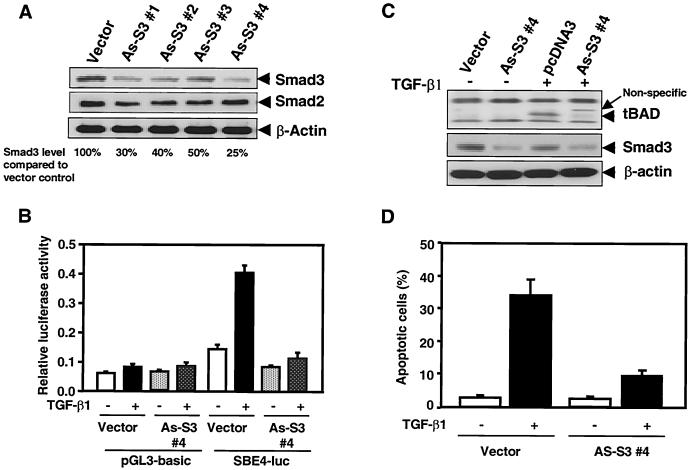
To confirm the critical role of Smad3 in the generation of 15-kDa cleaved form of BAD by TGF-β1, we generated the FaO cell lines stably expressing antisense Smad3. As shown in Fig. 6A, each stable transfectant expressed a ∼50 to 75% decreased level of endogenous Smad3, whereas antisense Smad3 did not effect expression of endogenous Smad2 protein. To test whether this decrease of Smad3 affects TGF-β1 signaling, we transfected SBE4-luc, which contains four SBE (Smad binding element) sites in tandem, into control cells and antisense Smad3-expressing cells (clone 4) (47). TGF-β1 treatment induced the SBE4-luc reporter activity 2.5-fold in control cells. However, TGF-β1-induced transactivation was markedly decreased in antisense Smad3-expressing cells (Fig. 6B). Expression of antisense Smad3 significantly inhibited the apoptosis induced by TGF-β1 (Fig. 6D) and markedly decreased the generation of 15-kDa cleaved form of BAD by TGF-β1 (Fig. 6C).
FIG. 6.
Effect of antisense-Smad3 on SBE4-luc reporter activity, apoptosis, and BAD cleavage induced by TGF-β1. (A) Expression of endogenous Smad3 in control and antisense-Smad3 transfectant clones. Total lysates were analyzed by SDS-PAGE. Western blotting was performed using anti-Smad3 or anti-Smad2 antibodies. Anti-β-actin was used as a control. (B) Either pGL3-basic (control) or SBE4-luc (47) was transfected into either control cells (vector) or antisense Smad3 cells (As-S3#4). Luciferase activity was measured 24 h after TGF-β1 stimulation. Data shown are means of triplicate measurements from one representative transfection. (C) Generation of 15-kDa cleaved form of BAD induced by TGF-β1 was examined in control and antisense-Smad3-expressing cells. Cell lysates were made, and equivalent amounts of cellular proteins were separated by SDS-15% PAGE, blotted, and then probed with an anti-BAD (C-20) antibody. Results are representative of three separate experiments. (D) The TUNEL assay was performed with the cells treated with 5 ng of TGF-β1/ml for 24 h and TUNEL-positive apoptotic cells at the respective incubation time were counted, and the percentage of apoptotic cells was graphed. Similar results were achieved in three separate experiments with comparable outcomes.
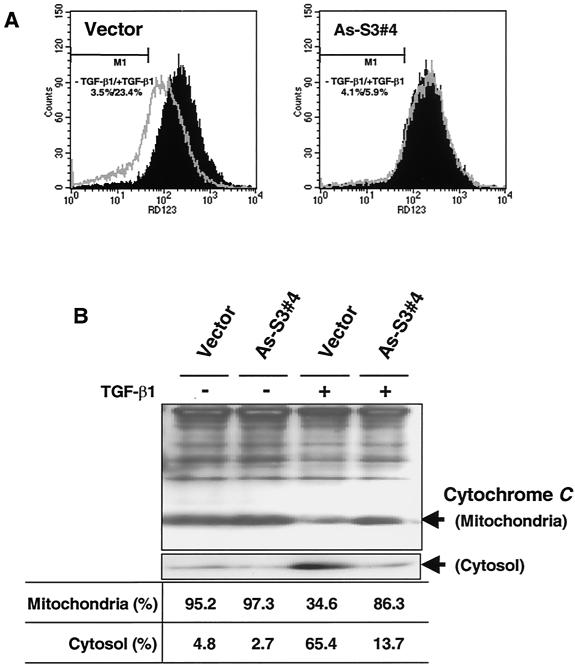
We also examined the changes in the mitochondrial membrane potential (Δψm) of the cells expressing antisense Smad3. As shown in Fig. 7A, after TGF-β1 treatment, Rh123 fluorescence decreased in control cells (pDON1) but not in antisense Smad3 cells. This suggests that TGF-β1 decreases mitochondrial membrane potential through a Smad3-dependent pathway.
FIG. 7.
Changes in mitochondrial transmembrane potential (Δψm) and release of cytochrome c induced by TGF-β1 in cells expressing antisense-Smad3. (A) The cells incubated for 12 h in the absence (control) or presence of 5 ng of TGF-β1/ml were detached by trypsinization. After 30 min of incubation with Rh123 (5 μM), the intracellular fluorescence intensity was measured in a FACScan flow cytometer. Results representative of experiments run more than three times are shown. (B) Release of cytochrome c from mitochondria to cytosol in the control and antisense-Smad3 cells. After incubating cells for 12 h in the absence (−) or presence (+) of 5 ng of TGF-β1/ml, mitochondria were separated from cytosol and cytochrome c content was analyzed by Western blotting as described in Materials and Methods. The ratio between mitochondria and cytosol is shown at the bottom.
In Fig. 3, we have shown that after TGF-β1 treatment the loss in mitochondrial membrane potential (Δψm) is coincident with the release of cytochrome c. To study whether cytochrome c release into the cytosol induced by TGF-β1 also depends on Smad3, we examined cytochrome c release in cells expressing antisense Smad3. TGF-β1 treatment activated the release of cytochrome c in control cells, whereas expression of antisense Smad3 markedly reduced the cytochrome c release (Fig. 7B).
DISCUSSION
BAD is a Bcl-2 family member belonging to a divergent subset that shares substantial sequence homology only within the BH3 amphipathic α-helical domain (45, 49). The activity of BAD is regulated by changes in phosphorylation and subcellular localization. Upon an increase in calcium influx or growth factor deprivation, phosphorylated BAD is rapidly dephosphorylated by the specific serine-phosphatase calcineurin (42) or PP1α (3, 38) and translocates to the mitochondrial outer membrane where, through its BH3 domain, it interacts with antiapoptotic Bcl-2 and Bcl-XL (25, 34, 45, 49).
Following deprivation of 32Dc13 myeloid precursor cells of IL-3, BAD is cleaved in the N terminus to generate two smaller products: one very similar in size to the full-length protein (26 kDa) and the second with a molecular mass of ∼15 kDa. In FaO cells, TGF-β1 treatment generates only the ∼15-kDa cleaved form of BAD (Fig. 1 and 2). The nature of the high-molecular-mass form of cleaved BAD is unclear. A previous study demonstrated that generation of the ∼15-kDa truncated form of BAD is blocked by the caspase inhibitor, Z-VAD fmk, suggesting that it is the product of caspase activity (13). In vitro studies have shown that BAD is cleaved by capases 2, 3, 7, 8, and 10 (13). Since TGF-β1 is also known to activate caspases 2, 3, 7, and 8 (4, 5, 7, 10), it is likely that caspases activated by TGF-β1 are responsible for the cleavage of BAD in FaO cells. TNF-α, TRAIL, or anti-CD95 antibody treatment also induced cleavage of endogenous BAD in human Jurkat T cells. Clearly cleavage of BAD enhances sensitivity to apoptosis since TGF-β1 induced cell death more rapidly and more potently in cells expressing truncated BAD than in cells overexpressing WT BAD, whereas cell death induced by TGF-β1 is reduced in cells expressing caspase-resistant mutant BAD.
Proapoptotic Bcl-2 family proteins, including Bax, BAD, and Bak among others, induce changes in mitochondrial membrane permeability, often accompanied by cytochrome c release in the cytosol with activation of downstream caspases (30, 41). In contrast, Bcl-XL, an antiapoptotic member of the Bcl-2 family, prevents cytochrome c release (17, 41). It has been demonstrated that Bcl-XL inhibits cytochrome c release through physical interaction with components of the voltage-dependent anion channel via a mechanism inhibited by several proapoptotic Bcl-2 proteins (17, 41). In IL-3-dependent lymphoid cells, IL-3 deprivation results in the dephosphorylation of BAD, facilitating its association with Bcl-2 and Bcl-XL. Phosphorylated BAD does not bind mitochondrial Bcl-2 or Bcl-XL and is sequestered in the cytosol by 14-3-3 (48), suggesting that phosphorylation regulates the exposure of the BH3 domain of BAD. It has been suggested that truncated BAD may be poorly phosphorylated at serine 155 since phosphorylation of serine 136 seems to be required for serine 155 phosphorylation. Truncated BAD has a similar or even higher affinity for Bcl-XL than WT BAD, and it is a more potent inducer of cytochrome c release than full-length WT BAD (13). Therefore, increased generation of truncated BAD induced by TGF-β1 may result in enhanced cytochrome c release by antagonizing antiapoptotic Bcl-XL. A recent study has shown that protein phosphatase 2A (PP2A) or a PP2A-like phosphatase catalyzes BAD dephosphorylation and regulates its proapoptotic activity in IL-3-dependent lymphoid cells by a mechanism requiring dissociation from 14-3-3 (8). The Bα regulatory subunit of PP2A interacts with the cytoplasmic domain of the type I TGF-β receptor and is a direct target for their kinase activity (21, 36). Therefore, induction of apoptosis induced by TGF-β1 in FaO cells may, in part, be mediated through dephosphorylation of BAD by PP2A activated by TGF-β1 as well as by increased cleavage of BAD. The regulation of BAD by PP2A inhibitors during TGF-β1-induced apoptosis is presently under investigation.
It is well known that Smad proteins are key components in TGF-β1 signaling for growth inhibition and apoptosis (28, 43). Although various observations indicate that Smad proteins are involved in TGF-β1 signaling for apoptosis, their role is still obscure and further studies are required to identify the specific molecular mechanisms involved. Smad3 overexpression induced apoptotic cell death in the presence of TGF-β1 in immortalized human normal lung epithelial cell lines (44), and overexpression of the dominant-negative form of Smad3 inhibits TGF-β1-dependent apoptosis in M1 and Hep3B cells (43). Constitutive expression of Smad2 also induced apoptotic cell death in the presence of TGF-β1 but less efficiently than by that of Smad3. Analogous to these results, we have now shown that that TGF-β1-dependent apoptosis of rat FaO cells is potently enhanced by expression of Smad3 and less efficiently by Smad2 and that this apoptosis can be blocked by antisense Smad3. Together, these results and our unpublished results in human hepatoma cells suggest that Smad3 may be an important mediator of TGF-β1-induced apoptosis in hepatoma cells in general.
In conclusion, we have demonstrated that TGF-β1 induces apoptosis through the cleavage of BAD in a Smad3-dependent mechanism in FaO cells. A recent report has implicated mitogen-activated protein (MAP) kinases in TGF-β1-induced apoptosis (39), and our unpublished results also suggest that inhibition of p38 MAP kinase activity by a specific inhibitor can block TGF-β1-induced apoptosis in FaO cells. These results suggest that TGF-β1-induced apoptosis may involve complex cross talk and cooperativity between the Smad signaling pathway and other signaling pathways, including the MAP kinase pathways. We are presently investigating whether inhibitors of MAP kinases inhibit cleavage of BAD induced by TGF-β1 and whether TGF-β1-induced BAD cleavage is specific for FaO cells or if this is a general mechanism underlying many cell systems where TGF-β1 promotes apoptosis.
Acknowledgments
We thank K. Miyazono and S. Kern for adenoviruses expressing Smads and SBE4-luc, respectively. We thank Anita Roberts for the critical reading of the manuscript.
This work was in part supported by a grant of the Korea Health 21 R & D Project, Ministry of Health & Welfare, Republic of Korea (HMP-00-B-20800-0035) (to K.S.C.) and by NIH grant PO1 CA78890 to B.C.
REFERENCES
- 1.Arsura, M., M. Wu, and G. E. Sonenshein. 1996. TGF β1 inhibits NF-B/Rel activity inducing apoptosis of B cells: transcriptional activation of IB. Immunity 5:31-40. [DOI] [PubMed] [Google Scholar]
- 2.Arsura, M., M. J. Fitzgerald, N. Fausto, G. E. Sonenshein. 1997. Nuclear factor-B/Rel blocks transforming growth factor β1-induced apoptosis of murine hepatocyte cell lines. Cell Growth Differ. 8:1049-1059. [PubMed] [Google Scholar]
- 3.Ayllon, V., C. Martinez-A, A. Garcia, X. Cayla, and A. Rebollo. 2000. Protein phosphatase 1 α is a Ras-activated BAD phosphatase that regulates interleukin-2 deprivation-induced apoptosis. EMBO J. 19:2237-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, T. L., S. Patil, R. K. Basnett, and P. H. Howe. 1998. Caspase inhibitor BD-fmk distinguishes transforming growth factor β-induced apoptosis from growth inhibition. Cell Growth Differ. 9:869-875. [PubMed] [Google Scholar]
- 5.Cain, K., and C. Freathy. 2001. Liver toxicity and apoptosis: role of TGF-β1, cytochrome c and the apoptosome. Toxicol. Letters 120:307-315. [DOI] [PubMed] [Google Scholar]
- 6.Chaouchi, N., L. Arvanitakis, M. T. Auffredou, D. A. Blanchard, A. Vazquez, and S. Sharma. 1995. Characterization of transforming growth factor-β1 induced apoptosis in normal human B cells and lymphoma B cell lines. Oncogene 11:1615-1622. [PubMed] [Google Scholar]
- 7.Chen, R. H., and T. Y. Chang. 1997. Involvement of caspase family proteases in transforming growth factor-β-induced apoptosis. Cell Growth Differ. 8:821-827. [PubMed] [Google Scholar]
- 8.Chiang, C.-W., G. Harris, C. Ellig, S. C. Masters, R. Subramanian, S. Shenolokar, B. E. Wadzinski, and E. Yang. 2001. Protein phosphatase 2A activates the proapoptotic function of BAD in interleukin-3-dependent lymphoid cells by a mechanism requiring 14-3-3 dissociation. Blood 97:1289-1297. [DOI] [PubMed] [Google Scholar]
- 9.Chipuk, J. E., B. Manjunatha, A. Y. Hsing, J. Ma, and D. Danielpour. 2001. Bcl-XL blocks transforming growth factor-β1-induced apoptosis by inhibiting cytochrome c release and not by directly antagonizing apaf-1dependent caspase activation in prostate epithelial cells. J. Biol. Chem. 276:26614-26621. [DOI] [PubMed] [Google Scholar]
- 10.Choi, K. S., I. K. Lim, J. N. Brady, and S.-J. Kim. 1998. ICE-like protease is involved in transforming growth factor-β-mediated apoptosis. Hepatology 27:415-421. [DOI] [PubMed] [Google Scholar]
- 11.Choi, K. S., Y. W. Eom, Y. Kang, M. J. Ha, H. Rhee, J. W. Yoon, and S.-J. Kim. 1999. Cdc2 and Cdk2 kinase activated by transforming growth factor-β1 trigger apoptosis through the phosphorylation of retinoblastoma protein in FaO hepatoma cells. J. Biol. Chem. 274:31775-31783. [DOI] [PubMed] [Google Scholar]
- 12.Christensen, J. G., A. J. Gonzales, R. C. Cattley, and T. L. Goldworthy. 1998. Regulation of apoptotis in mouse hepatocytes and alteration of apoptosis by nongenotoxic carcinogens. Cell Growth Differ. 9:815-825. [PubMed] [Google Scholar]
- 13.Condorelli, F., P. Salomoni, S. Cotteret, V. Cesi, S. M. Srivasula, E. S. Alnemri, and B. Calabretta. 2001. Caspase cleavage enhances the apoptotis-inducing effects of BAD. Mol. Cell. Biol. 21:3025-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Caestecker, M. P., E. Piek, and A. B. Roberts. 2000. Role of transforming growth factor-β signaling. J. Natl. Cancer Inst. 92:1388-1402. [DOI] [PubMed] [Google Scholar]
- 15.Derynck, R., Y. Zhang, and X. H. Feng. 1998. Smads: transcriptional activators of TGF-β responses. Cell 95:737-740. [DOI] [PubMed] [Google Scholar]
- 16.Fan, G., X. Ma, B. T. Kren, and C. J. Steer. 1996. The retinoblastoma gene product inhibits TGF-β1 induced apoptosis in primary rat hepatocytes and human HuH-7 hepatoma cells. Oncogene 12:1909-1919. [PubMed] [Google Scholar]
- 17.Finucane, D. M., E. Bossy-Wetzel, N. J. Waterhouse, T. G. Cotter, and D. R. Green. 1999. Bax-induced caspase activation and apoptosis via cytochrome c release from mitochondria is inhibitable by Bcl-XL. J. Biol. Chem. 274:2225-2233. [DOI] [PubMed] [Google Scholar]
- 18.Francis, J. M., C. M. Heyworth, E. Spooncer, A. Pierce, T. M. Dexter, and A. D. Whetton. 2000. Transforming growth factor-β1 induces apoptosis independently of p53 and selectively reduces expression of Bcl-2 in multipotent hematopoietic cells. J. Biol. Chem. 275:39137-39145. [DOI] [PubMed] [Google Scholar]
- 19.Franke, T. F., and L. C. Cantley. 1997. Apoptosis: a bad kinase makes good. Nature 390:116-117. [DOI] [PubMed] [Google Scholar]
- 20.Fujii, M., K. Takeda, T. Imamura, H. Aoki, T. K. Sampath, S. Enomoto, M. Kawabata, M. Kato, H. Ichijo, and K. Miyazono. 1999. Roles of bone morphogenetic protein type I receptors and Smad proteins in osteoblast and chondroblast differentiation. Mol. Biol. Cell. 10:3801-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griswold-Prenner, I., C. Kamibayashi, E. M. Maruoka, M. C. Mumby, R. Derynck. 1998. Physical and functional interactions between type I transforming growth factor β receptors and Bα, a WD-40 repeat subunit of phosphatase 2A. Mol. Cell. Biol. 18:6595-6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harada, H., B. Becknell, M. Wilm, M. Mann, L. J. Huang, S. S. Taylor, J. D. Scott, and S. J. Korsmeyer. 1999. Phosphorylation and inactivation of BAD by mitochondria-anchored protein kinase A. Mol. Cell 3:413-422. [DOI] [PubMed] [Google Scholar]
- 23.Heldin, C.-H., K. Miyazono, and P. ten Dijke. 1997. TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature 390:465-471. [DOI] [PubMed] [Google Scholar]
- 24.Hsing, A. Y., K. Kadomatsu, M. J. Bonham, and D. Danielpour. 1996. Regulation of apoptosis induced by transforming growth factor-β1 in nontumorigenic and tumorigenic rat prostatic epithelial cell lines. Cancer Res. 56:5146-5149. [PubMed] [Google Scholar]
- 25.Kelekar, A., B. S. Chang, J. E. Harlan, S. W. Fesik, and C. B. Thompson. 1997. Bad is a BH3 domain-containing protein that forms an inactive dimmer with Bcl-XL. Mol. Cell. Biol. 17:7040-7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korsmeyer, S. J., A. Gross, H. Harada, J. Zha, K. Wang, X.-M. Yin, M. Wei, and S. Zinkel. 1999. Death and survival signals determine active/inactive conformations of pro-apoptotic BAX, BAD, and BID molecules. Cold Spring Harbor Symp. Quant. Biol. 64:343-350. [DOI] [PubMed] [Google Scholar]
- 27.Larisch-Bloch, S., Y. Yi, R. Lotan, H. Kerner, S. Eimerl, W. T. Parks, M. P. de Caestecker, D. Danielpour, N. Book-Melamed, R. Timberg, R. J. Lechleider, J. Orly, S.-J. Kim, and A. B. Roberts. 2000. A novel mitochondrial septin, ARTS, mediates TGF-β-induced apoptosis via its P-loop motif. Nat. Cell Biol. 2:915-921. [DOI] [PubMed] [Google Scholar]
- 28.Le Dai, J., R. K. Bansal, and S. E. Kern. 1999. G1 cell cycle arrest and apoptosis induction by nuclear Smad4/Dpc4: phenotypes reversed by a tumorigenic mutation. Proc. Natl Acad. Sci. USA 96:1427-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, J. K., and C. K. Chou. 1992. In vitro apoptosis in the human hepatoma cell line induced by transforming growth factor-β1. Cancer Res. 52:385-388. [PubMed] [Google Scholar]
- 30.Marzo, I., C. Brenner, N. Zamzami, J. M. Jürgensmeier, S. A. Susin, H. L. A. Vieira, M.-C. Prévost, Z. Xie, S. Matsuyama, J. C. Reed, and G. Kroemer. 1998. Bax and adenine nucleotide translocator cooperate in the mitochondrial control of apoptosis. Science 281:2027-2031. [DOI] [PubMed] [Google Scholar]
- 31.Massagué, J., and Y. G. Chen. 2000. Controlling TGF-β signaling. Genes Dev. 14:627-644. [PubMed] [Google Scholar]
- 32.Massagué, J., and D. Wotton. 2000. Transcriptional control by the TGF-β/Smad signaling system. EMBO J. 19:1745-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oltvai, Z. N., and S. Korsmeyer. 1994. Checkpoints of dueling dimers foil death wishes. Cell 79:189-192. [DOI] [PubMed] [Google Scholar]
- 34.Ottilie, S. L., L. Diaz, W. Horne, J. Chang, Y. Wang, G. Wilson, S. Chang, L. C. Fritz, and T. Oltersdorf. 1997. Dimerization properties of human BAD. Identification of a B domain and analysis of its binding to mutant BCL-2 and BCL proteins. J. Biol. Chem. 272:30866-30872. [DOI] [PubMed] [Google Scholar]
- 35.Perlman, R., W. P. Schiemann, M. W. Brooks, H. F. Lodish, and R. A. Weinberg. 2001. TGF-β-induced apoptosis is mediated by the adaptor protein Daxx that facilitates JNK activation. Nat. Cell Biol. 3:708-714. [DOI] [PubMed] [Google Scholar]
- 36.Petritsch, C., H. Beug, A. Balmain, and M. Oft. 2000. TGF-β inhibits p70 S6 kinase via protein phosphatase 2A to induce G(1) arrest. Genes Dev. 15:3093-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts, A. B., and M. B. Sporn. 1990. The transforming growth factor-βs, p. 419-472. In M. B. Sporn and A. B. Roberts (ed.), Peptide growth factors and their receptors, part I. Springer-Verlag, Heidelberg, Germany.
- 38.Salomoni, P., F. Condorelli, S. M. Sweeney, and B. Calabretta. 2000. Versatility of BCR/ABL-expressing leukemic cells in circumventing pro-apoptotic BAD effects. Blood 96:676-684. [PubMed] [Google Scholar]
- 39.Schiffer, M., M. Bitzer, I. S. D. Roberts, J. B. Kopp, P. ten Dijke, P. Mundel, and E. P. Böttinger. 2001. Apoptosis in podocytes induced by TGF-β and Smad7. J. Clin. Investig. 108:807-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shima, Y., K. Nakao, T. Nakashima, A. Kawakami, K. Nakata, K. Hamasaki, Y. Kato, K. Eguchi, and N. Ishii. 1999. Activation of caspase-8 in transforming growth factor-β-induced apoptosis of human hepatoma cells. Hepatology 30:1215-1222. [DOI] [PubMed] [Google Scholar]
- 41.Shimizu, S., M. Narita, and Y. Tsujimoto. 1999. Bcl-2 family proteins regulate the release of apoptogenic cytochrome c by the mitochondrial channel VDAC. Nature 399:483-487. [DOI] [PubMed] [Google Scholar]
- 42.Wang, H.-G., N. Pathan, I. M. Ethell, S. Krajewski, Y. Yamaguchi, F. Shibasaki, F. Mckeon, T. Bobo, T. F. Franke, and J. C. Reed. 1999. Ca2+-induced apoptosis through calicineurin dephosphorylation of BAD. Science 284:339-343. [DOI] [PubMed] [Google Scholar]
- 43.Yamamura, Y., X. Hua, S. Bergelson, and H. F. Lodish. 2000. Critical role of Smads and AP-1 complex in transforming growth factor-β-dependent apoptosis. J. Biol. Chem. 275:36295-36302. [DOI] [PubMed] [Google Scholar]
- 44.Yanagisawa, K., H. Osada, A. Masuda, M. Kondo, T. Saito, Y. Yatabe, K. Takagi, T. Takahashi, and T. Takahashi. 1998. Induction of apoptosis by Smad3 and down-regulation of Smad3 expression in response to TGF-β in human normal lung epithelial cells Oncogene 17:1743-1747. [DOI] [PubMed] [Google Scholar]
- 45.Yang, E., J. Zha, J. Jockel, L. H. Boise, C. B. Thompson, and S. J. Korsmeyer. 1995. Bad: a heterodimeric partner for Bcl-XL ans Bcl-2 displaces bax and promotes cell death. Cell 80:285-291. [DOI] [PubMed] [Google Scholar]
- 46.Yang, X., R. Khosravi-Far, H. Y. Chang, and D. Baltimore. 1997. Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell 89:1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zawel, L., J. L Dai, P. Buckhaults. S. Zhou, K. W. Kinzler, B. Vogelstein, and S. E. Kern. 1998. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol. Cell 1:611-617. [DOI] [PubMed] [Google Scholar]
- 48.Zha, J., H. Harada, K. E. Yang, J. Jockel, and S. J. Korsmeyer. 1996. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not Bcl-XL. Cell 87:619-628. [DOI] [PubMed] [Google Scholar]
- 49.Zha, J., H. Harada, K. Osipov, J. Jockel, G. Waksman, and S. J. Korsmeyer. 1997. BH3 domain of BAD is required for heteromerization with BCL-XL and pro-apoptotic activity. J. Biol. Chem. 272:24101-24104. [DOI] [PubMed] [Google Scholar]