Abstract
Gcn5 is a conserved histone acetyltransferase (HAT) found in a number of multisubunit complexes from Saccharomyces cerevisiae, mammals, and flies. We previously identified Drosophila melanogaster homologues of the yeast proteins Ada2, Ada3, Spt3, and Tra1 and showed that they associate with dGcn5 to form at least two distinct HAT complexes. There are two different Ada2 homologues in Drosophila named dAda2A and dAda2B. dAda2B functions within the Drosophila version of the SAGA complex (dSAGA). To gain insight into dAda2A function, we sought to identify novel components of the complex containing this protein, ATAC (Ada two A containing) complex. Affinity purification and mass spectrometry revealed that, in addition to dAda3 and dGcn5, host cell factor (dHCF) and a novel SANT domain protein, named Atac1 (ATAC component 1), copurify with this complex. Coimmunoprecipitation experiments confirmed that these proteins associate with dGcn5 and dAda2A, but not with dSAGA-specific components such as dAda2B and dSpt3. Biochemical fractionation revealed that ATAC has an apparent molecular mass of 700 kDa and contains dAda2A, dGcn5, dAda3, dHCF, and Atac1 as stable subunits. Thus, ATAC represents a novel histone acetyltransferase complex that is distinct from previously purified Gcn5/Pcaf-containing complexes from yeast and mammalian cells.
The association of eukaryotic DNA with histones to form nucleosomes provides opportunities to regulate DNA function. For example, the strong interactions between histones and DNA can prevent access to promoter elements by the transcription machinery (12, 25). Chromatin remodeling complexes help to overcome the obstacle of nucleosomes. These enzymes utilize the energy of ATP to slide nucleosomes and alter histone-DNA contacts, leaving promoter elements accessible to proteins involved in transcription regulation (10, 23). In addition, histones are exposed to a variety of posttranslational modifications, including phosphorylation, ubiquitylation, methylation, ADP-ribosylation, and acetylation. These covalent modifications can dramatically affect gene transcription (9, 11, 30).
The different posttranslational modifications of histones, as well as the protein complexes that catalyze these reactions, have been extensively studied (9). Many groups have investigated the effect of histone acetylation in transcription activation. Nucleosome arrays containing acetylated histones show reduced compaction and increased transcription in vitro (14). A strong connection between histone acetylation and transcription activation came with the discovery that the yeast transcriptional adaptor Gcn5 was highly homologous to a nuclear histone acetyltransferase (HAT) from Tetrahymena thermophila named p55 (5). Recombinant Gcn5 efficiently acetylates core histones. However, Gcn5 incorporation into multisubunit complexes potentiates its nucleosomal HAT activity and expands its lysine specificity (11). The conserved Gcn5 protein is the catalytic subunit of the yeast HAT complexes SAGA, SLIK/SALSA, ADA, and A2 (7). Numerous studies of the SAGA complex revealed that the presence of multiple subunits allows this complex to carry out its coactivator functions, which include nucleosome acetylation, recruitment by activators, and TATA-binding protein binding (4, 34). In addition to Saccharomyces cerevisiae, Gcn5-containing complexes have been purified from mammalian nuclear extracts. The mammalian STAGA, TFTC, and PCAF complexes show transcription coactivator functions and contain subunits which are homologous to the yeast Ada, Spt, and TAF proteins as well as the ATM-related protein Tra1 (7, 26, 28, 29, 42).
Gcn5 plays critical roles during development. Gcn5-null mice lack specific mesoderm-derived structures and die during embryogenesis. However, deletion of its homologue Pcaf results in viable animals, presumably due to compensation by Gcn5. In addition, animals carrying deletions in both genes die earlier than Gcn5-null animals (46, 47). Furthermore, mutations in Arabidopsis Gcn5 result in viable plants that show phenotypes in several developmental processes, such as cell elongation, leaf expansion, and flower development (40).
Flies contain a single homologue of Gcn5 (dGcn5), which is expressed throughout development (38). Previous studies identified two homologues of the Ada2 protein in Drosophila melanogaster, which is a Gcn5 partner in all known complexes from yeast to humans (18, 27). The two Ada2 proteins contain a ZZ domain, which is a putative zinc binding domain, a SANT domain, and three conserved regions called Ada boxes (27). We previously showed that one of the homologues, dAda2B, is present in a SAGA-like complex, which includes dGcn5, dAda3, dSpt3, and dTra1. On the other hand, dAda2A is only known to be associated with dGcn5 and dAda3. Both of these complexes show a preference for H3 and H4 acetylation (18). A recent report described a dAda2B mutant and demonstrated the importance of dSAGA during Drosophila development. The mutant animals showed reduced acetylation and died during early pupal stages (32). The potential importance of dAda2A complexes is unknown. However, the presence of two homologues of Ada2 has now also been reported in plants and mammals, suggesting two distinct conserved functions of these proteins (1, 40).
To investigate the functions of the dAdA2A protein, we sought to isolate protein complexes of which it is part and to identify other proteins associated with dAda2A. We describe here the identification of the 700-kDa ATAC (Ada two A containing) complex. We show that in addition to dAda2A, dAda3, and dGcn5, ATAC also contains host cell factor (dHCF) and the uncharacterized SANT-domain-containing protein cg9200, which we have named Atac1 (ATAC component 1).
MATERIALS AND METHODS
Plasmids.
The plasmids generated in this study are listed in Table 1. For expression in S2 cells, cDNAs were inserted into plasmids derived from pRmHa3 (6) or pMT (Invitrogen). pACXT-T7-dHCF-FLAG was a kind gift from Angus Wilson (24). For expression in Escherichia coli, cDNAs were inserted into pQE12 (QIAGEN). Primer sequences, restriction sites used, and maps of vectors are available upon request.
TABLE 1.
Plasmids generated in this study
| Construct name | Vector | Insert (accession no. or reference) |
|---|---|---|
| pRmHa3-NTAP | pRmHa3 | Sequence coding for N-terminal TAP tag (31) |
| pRmHa3-TAP-dGcn5 | pRmHa3-NTAP | cDNA for dGcn5 (18) |
| pRmHa3-CFL2 | pRmHa3 | Two FLAG tag sequences for C-terminal tagging |
| pRmHa3-CHA2FL2 | pRmHa3 | Two Hemagglutinin and two FLAG tag sequences for C-terminal tagging |
| pRmHa3-dAda3-FL2 | pRmHa3-CFL2 | cDNA for dAda3 (18) |
| pRmHa3-Atac1-HA2FL2 | pRmHa3-CHA2FL2 | cDNA for cg9200 (NM_135244) |
| pMT-CTAP | pMT | Sequence coding for C-terminal TAP tag (31) |
| pMT-dAda2A-TAP | pMT-CTAP | cDNA for dAda2A (18) |
| pQE12-Atac1 | pQE12 | cDNA for cg9200 (NM_135244) |
| pQE12-dHCF-C | pQE12 | cDNA coding for amino acids 1001 to 1260 of dHCF (24) |
Transfections and generation of stable lines.
S2 cells (37) were maintained at 25°C in Drosophila Schneider's medium, containing 10% fetal bovine serum and penicillin/streptomycin (Invitrogen). Cells were transfected with Effectene (QIAGEN), according to the manufacturer's protocol. Stable cell lines were generated by cotransfection with pCoHygro (Invitrogen). Selection was carried out for a month in medium containing 0.25 mg/ml hygromycin (Invitrogen).
Preparation of nuclear extracts.
Nuclear extracts were prepared as previously described (18). For affinity purifications, 8 liters of cells was grown to a density of 2 × 106 cells/ml and induced for 1 day with 0.5 mM CuSO4.
Generation of polyclonal antibodies.
Total RNA was purified from 12- to 18-h Oregon R embryos using TRIzol (Invitrogen). The cDNA for cg9200 was generated by reverse transcription-PCR, using total RNA from Oregon R embryos and the SuperScript first-strand synthesis system (Invitrogen), followed by PCR using Pfu Turbo polymerase (Stratagene). This cDNA was subsequently inserted into pQE12 (QIAGEN). A 3′ fragment of dHcf (corresponding to amino acids 1001 to 1260) was amplified using pACXT-T7-dHCF-FLAG as a template and inserted into pQE12. The C-terminally His-tagged recombinant proteins were expressed in E. coli and purified over Ni-nitrilotriacetic acid (NTA) agarose (QIAGEN) under denaturing conditions, as described by the manufacturer. Purified proteins were dialyzed twice against 20 mM HEPES, pH 7.4, 10% glycerol, 150 mM NaCl, and 1 mM phenylmethylsulfonyl fluoride (PMSF) and were used to immunize rats and rabbits. An amino-terminal dAda2B fragment corresponding to amino acids 1 to 241 (18) was used to immunize Guinea pigs. To generate antibodies against dAda2A, rats were immunized with the synthetic peptide EKTRDQNSSVPSATKDANRC, previously conjugated to keyhole limpet hemocyanin (Pocono Rabbit Farm and Laboratory, Inc.).
Antibodies against dGcn5, dAda3, dSpt3, dAda2B (rat), and dAda2A (rabbit) were previously described (18).
Coimmunoprecipitations, Western blots, and HAT assays.
One microgram of nuclear extract was incubated overnight with 2.5 μl of the corresponding rabbit antiserum (or preimmune bleed) at 4°C. To precipitate the immunocomplexes, 10 μl protein A-Sepharose (Amersham Biosciences) was added to the reaction mixture. After 2 h at 4°C, the beads were washed three times for 10 min in wash buffer (20 mM Tris-Cl, pH 8, 5 mM MgCl2, 10% glycerol, 300 mM NaCl, 0.1% Tween 20, 1 mM PMSF, 1 μg/ml pepstatin A, and 1 μg/ml leupeptin), and the antibody-protein complexes were eluted by heating for 5 min at 95°C in sodium dodecyl sulfate-containing loading buffer.
For anti-FLAG immunoprecipitations, cells were transfected and induced for 1 day with 0.5 mM CuSO4. The cells were subsequently washed in phosphate-buffered saline and lysed for 30 min at 4°C in 50 mM Tris-Cl, pH 8,150 mM NaCl, 1% NP-40, 1 mM PMSF, 1 μg/ml pepstatin A, and 1 μg/ml leupeptin. The NaCl concentration of the extract was subsequently adjusted to 300 mM. Two micrograms of whole-cell extract was incubated overnight with 10 μl M2-agarose beads (Sigma). The immunoprecipitates were washed three times for 10 min in wash buffer and heated for 5 min at 95°C in sodium dodecyl sulfate loading buffer to elute immunoprecipitated proteins.
The following antibodies were used for Western blots: peroxidase antiperoxidase (1:5,000; Sigma), dGcn5 (rabbit, 1:3,000), dAda3 (rabbit 1:5,000), dAda2A (rat, 1:1,000; rabbit, 1:2,000), dHCF (rabbit, 1:5,000; rat, 1:2,500), Atac1 (rabbit, 1:3,000; rat, 1:1,000), and dAda2B (rat, 1:1,000; guinea pig, 1:1,000).
HAT reactions were performed as described previously (8), except that the assays were performed in the presence of 10 μl protein A beads (after immunoprecipitations).
Affinity purifications and mass spectrometry.
Tandem affinity purification (TAP) was performed as previously described (33), starting with 200 mg of nuclear extracts from stable lines that express TAP-dGcn5, dAda2A-TAP, TAP alone, or wild-type cells.
For anti-FLAG purifications, approximately 200 mg of nuclear extract from cells expressing Atac1-HA2FL2 or wild-type S2 cells (negative control) were bound to 120 μl M2-agarose beads (Sigma) overnight at 4°C. The beads were washed three times for 10 min in S2 extraction buffer (20 mM HEPES, pH 7.4, 10% glycerol, 0.35 M NaCl, 1 mM MgCl2, 0.1% Triton X-100, 1 mM PMSF, 1 μg/ml pepstatin A, and 1 μg/ml leupeptin). The complexes were eluted off the beads by competing with 120 μl of 1 mg/ml of triple FLAG peptide (Sigma) in S2 extraction buffer for 30 min at room temperature. Affinity-purified complexes were resolved in denaturing polyacrylamide gels and visualized by silver staining.
MudPIT analysis of the affinity-purified complexes was carried out as previously described (19).
Fractionation of nuclear extracts.
Approximately 100 mg nuclear extract was incubated overnight with 5 ml of Ni-NTA-agarose beads (QIAGEN). The beads were washed with 25 ml S2 extraction buffer and 25 ml Mono Q buffer (20 mM Tris-Cl, pH 8, 5 mM MgCl2, 10% glycerol, 0.1% Tween 20, and 1 mM PMSF) containing 100 mM NaCl and 10 mM imidazole. Elution was carried out in Mono Q buffer containing 100 mM NaCl and 300 mM imidazole.
The imidazole eluate was applied to a Mono Q HR 5/5 column (Amersham Biosciences). Proteins were fractionated by a 100 to 500 mM NaCl gradient in Mono Q buffer. Five-hundred-microliter fractions were collected and analyzed by Western blotting. The fractions corresponding to the dAda2A complex (29 to 33) were pooled and concentrated, using an Amicon Ultra 4 (Millipore), to a final volume of 500 μl. This concentrated material was applied to a Superose 6 HR 10/30 column (Amersham Biosciences). Five-hundred-microliter fractions were collected and analyzed by Western blotting.
Indirect immunofluorescence.
Polytene chromosomes were dissected and stained as described previously (18). The following antibodies were used: dAda3 (rabbit, 1:100), dHCF (rabbit, 1:100; rat, 1:100), Atac1 (rabbit, 1:50). We used the following secondary antibodies: Alexa Fluor 594-conjugated goat anti-rabbit immunoglobulin G (1:500) and Alexa Fluor 488-conjugated goat anti-rat immunoglobulin G (1:500) (Molecular Probes). DNA was visualized with 4′-6′-diamidino-2-phenylindole dihydrochloride (DAPI).
RESULTS
dHCF and the uncharacterized protein cg9200 associate with TAP-dGcn5.
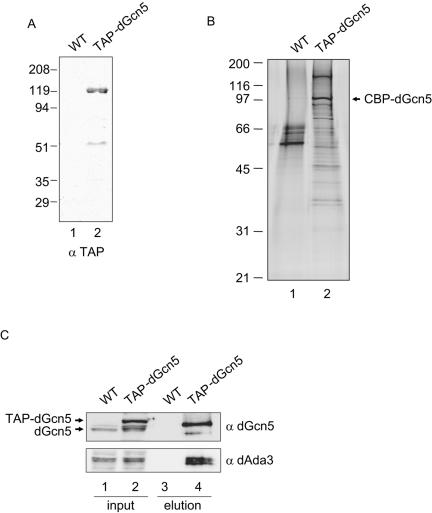
To identify new components of dGcn5-containing complexes, we took an affinity purification approach. To this end, we generated an S2 cell line that expresses TAP-dGcn5 under control of the inducible metallothionein promoter (6). We tested nuclear extracts from these cells (TAP-dGcn5) and from wild-type (WT) cells by Western blotting, using an antibody that recognizes the protein A moiety of the TAP tag, and confirmed that the full-length tagged protein was expressed (Fig. 1A, lane 2). We also observed a faster-migrating polypeptide of ∼51 kDa, corresponding to a degradation product of TAP-dGcn5. We used nuclear extracts from that stable cell line to purify dGcn5-containing complexes, following the TAP procedure (31, 33). As a negative control, we subjected nuclear extracts from wild-type S2 cells to the same procedure. After elution from the calmodulin beads, the purified fractions were resolved in a denaturing gel and silver stained (Fig. 1B). From our previous work, we knew that dGcn5 is a component of at least two distinct HAT complexes (18). Therefore, we expected to see multiple bands on the gel. We observed a number of proteins that bound specifically to the anti-FLAG resin (Fig. 1B, compare lanes 1 and 2). Since dGcn5 was overexpressed, we anticipated seeing a strong band corresponding to the calmodulin binding peptide fused to dGcn5 (CBP-dGcn5). We further confirmed the presence of dGcn5 and dAda3 in the purified material by Western blotting, using antibodies that recognize these proteins (Fig. 1C). Both dGcn5 and dAda3 are only present in that TAP-dGcn5 purified fraction but not in the material purified from wild-type cells (lanes 3 and 4). Moreover, CBP-dGcn5 (lane 4, dGcn5 blot) has an electrophoretic mobility intermediate between TAP-dGcn5 and untagged dGcn5 (lane 2, dGcn5 blot).
FIG. 1.
Affinity purification of dGcn5-containing complexes. (A) Thirty micrograms of nuclear extracts from WT cells (lane 1) or cells that stably express TAP-tagged dGcn5 (TAP-dGcn5, lane 2) were analyzed by Western blotting using an antibody that recognizes the protein A moiety of the TAP tag (α TAP). The migration of the molecular weight standards is shown to the left. (B) Silver stain gel showing the purified material from WT (lane 1) and TAP-dGcn5 extracts (lane 2). The migration of the tagged protein is shown with an arrow (CBP-dGcn5). (C) The affinity-purified material from the TAP-dGcn5 cells (lane 4) and the mock purification (WT, lane 3) were analyzed by Western blotting using antibodies against dGcn5 (α dGcn5) and dAda3 (α dAda3). The inputs correspond to 30 μg of nuclear extract from wild type (lane 1) or tagged cells (lane 2). The migration of TAP-dGcn5 and endogenous dGcn5 are indicated with arrows.
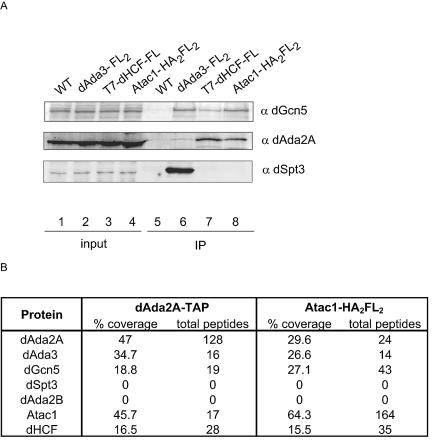
We subjected this purified TAP-dGcn5 sample to MudPIT analysis. MudPIT is an unbiased and sensitive mass spectrometry procedure that allows the identification of proteins in a complex mixture without prior electrophoretic separation (41). This technique has been successfully exploited for the identification of subunits in transcription regulatory complexes from different organisms, such as mammalian Mediator (36), yeast SWI/SNF, SAGA (20), TFIID (35), and Drosophila Tip60 (17). MudPIT analysis identified 337 peptide sequences for dGcn5, 53 for dAda3, 33 for dAda2B, and 17 for dAda2A (Fig. 2A). This result confirmed that the affinity purification was successful and indicated that different types of dGcn5-containing complexes were present in the purified material. In addition to known proteins that interact with dGcn5, we obtained peptides for two proteins which had not been previously found to be associated with dGcn5 (Fig. 2A>).
FIG. 2.
MudPIT analysis identifies peptides for dHCF and the uncharacterized protein cg9200 (Atac1). (A) Results from MudPIT, showing the number of nonredundant spectra for each protein (total peptides) and the amino acid sequence coverage (% coverage). (B) Primary structure of Atac1 gene product. The peptides identified for this protein are shown in boldface type. The sequence underlined indicates the SANT domain.
One of the novel dGcn5-associated proteins was dHCF. dHCF is a 1,500-amino-acid protein with a predicted molecular mass of 160 kDa (24). This protein is conserved among higher eukaryotes, and its human homologue HCF-1 has been studied in detail. This abundant protein was originally identified as a component of the VP16-induced complex, formed upon infection of human cells by the herpes simplex virus (44). The N-terminal region of this protein contains a Kelch domain, while the C terminus harbors two fibronectin III repeats. Another interesting feature of this 2,035-amino-acid protein is that it gets processed after translation to generate two polypeptides that remain noncovalently bound (43). Although dHCF lacks the conserved repeats required for the processing of mammalian HCF-1, it is still cleaved to generate two fragments of approximately 120 and 72 kDa that remain associated (24).
We also obtained three peptides corresponding to the uncharacterized protein cg9200 (NP_609088.1). When we examined the primary structure of this 356-amino-acid protein by means of the Protein Families (Pfam) database, we recognized a SANT domain located in the center of its amino acid sequence (Fig. 2B). Members of the SANT family include Swi3, which is a component of the yeast chromatin remodeling complex SWI/SNF, and Ada2, which is a subunit of the yeast HATs SAGA, SLIK, ADA, and A2 (7). The SANT domain is required for the function of SWI/SNF in vivo and for stimulation of the HAT activity of SAGA (2, 39). We have named this protein Atac1 (ATAC component 1).
dHCF and Atac1 are components of the ATAC complex.
With the intention of confirming interactions between dHCF, Atac1, and dGcn5, we transfected S2 cells with the plas- mids pACXT-T7-dHCF-FLAG and pRmHa3-Atac1-HA2FL2, which code for C-terminally FLAG-tagged dHCF and Atac1, respectively, and prepared whole-cell extracts from these cells. Coimmunoprecipitation experiments using antibodies directed to the FLAG epitope confirmed the association of these two proteins with dGcn5 (Fig. 3A, top panel, lanes 7 and 8). As a positive control for the immunoprecipitation reactions, we used extracts from cells expressing C-terminally FLAG-tagged dAda3 (Fig. 3A, top panel, lane 6). To determine if dHCF and Atac1 were components of the ATAC complex and not dSAGA, we repeated the anti-FLAG immunoprecipitations and probed the Western blots with antibodies against dAda2A (Fig. 3A, middle panel). We observed that dHCF and Atac1 coimmunoprecipitated dAda2A (lanes 7 and 8). Neither protein coimmunoprecipitated the dSAGA component dSpt3 (lanes 7 and 8). However, dSpt3 could associate with dAda3 (lane 6). This suggests that dHCF and Atac1 are components of the ATAC complex and not dSAGA.
FIG. 3.
Tagging of Atac1, dHCF, and dAda2A confirms their association. (A) S2 cells were transfected with pRmHa3-dAda3-FL2, pRmHa3-Atac1-HA2FL2, and pACXT-T7-dHCF-FLAG plasmids. After a 1-day induction, whole-cell extracts were prepared and incubated with M2-agarose beads. Untransfected S2 cells (WT) were used as a negative control. The immunoprecipitated material was analyzed by Western blotting using antibodies against dGcn5 (α dGcn5, rabbit), dAda2A (α dAda2A, rat), and dSpt3 (α dSpt3, rabbit). Lanes 1 to 4 correspond to 40 μg of whole-cell extract (2% input). Lanes 5 to 8 correspond to the immunoprecipitated material (IP). (B) Protein complexes from a stable line expressing Atac1-HA2FL2 were affinity purified using anti-FLAG-agarose beads. dAda2A-containing complexes were purified from dAda2A-TAP-expressing cells, according to the TAP protocol. These complexes were analyzed by MudPIT. The number of nonredundant spectra for each protein (total peptides) and the amino acid sequence coverage (% coverage) are shown.
To further confirm the interactions of dHCF and Atac1 with dAda2A, we established cell lines that stably express dAda2A-TAP and Atac1-HA2FL2. We prepared nuclear extracts from these cells, and the epitope-tagged complexes were purified according to the TAP procedure (for dAda2A-TAP) or using anti-FLAG-agarose beads (for Atac1-HA2FL2). We also subjected negative control extracts to identical purification procedures (see Materials and Methods). The complexes were analyzed by mass spectrometry (Fig. 3B). MudPIT analysis of the TAP-purified dAda2A material identified 128 peptides for dAda2A, 19 for dGcn5, and 16 for dAda3. In addition to those three known subunits of the ATAC complex, we obtained 17 peptides for Atac1 and 28 for dHCF. Affinity purification of Atac1-containing complexes followed by MudPIT studies exhibited similar results. Besides recovering 164 peptides for the tagged protein, we identified 24 peptides for dAda2A, 14 for dAda3, 43 for dGcn5, and 35 for dHCF. Moreover, neither of the two affinity purifications revealed peptides for components of dSAGA, such as dSpt3 and dAda2B. From these results, we conclude that Atac1 and dHCF are novel subunits of the ATAC complex.
Polyclonal antibodies against dHCF and Atac1 coimmunoprecipitate HAT activity.
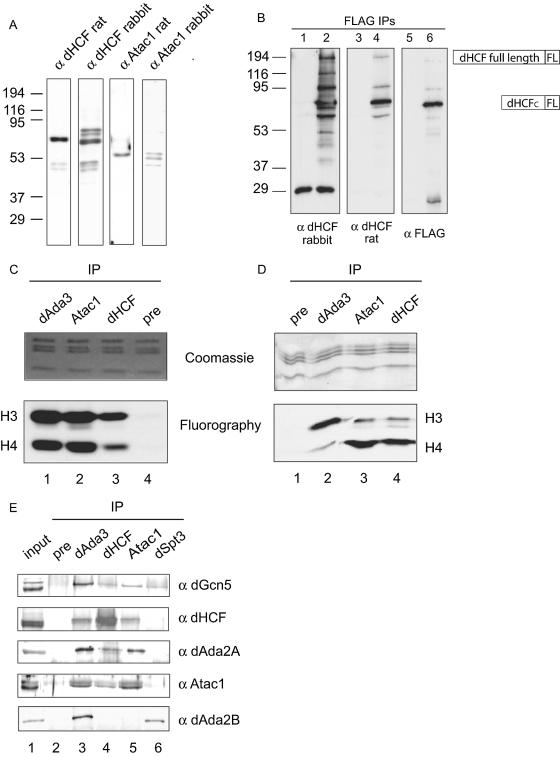
To further characterize the association of Atac1 and dHCF with the dAda2A complex, we raised polyclonal antibodies against recombinant full-length Atac1 and a C-terminal fragment of dHCF in rats and rabbits, and Western blots of nuclear extracts indicated that these antibodies were specific for the antigens they were raised against (Fig. 4A). Similar Western blots performed with preimmune bleeds showed no signal (data not shown). Both rat and rabbit antibodies against dHCF detected a strong band of approximately 70 kDa, corresponding to the C-terminal dHCF fragment. This is consistent with published results, which showed that full-length dHCF gets proteolytically cleaved to yield two polypeptides of approximately 120 and 70 kDa (24). However, we did not see a band of 160 kDa corresponding to the full-length protein, which suggests that the majority of dHCF gets processed after translation. We also observed other minor bands in the dHCF blots. These protein species may result from proteolytic cleavage of the full-length protein at other sites within the central region of the polypeptide. Antibodies directed to Atac1 detect a specific band of approximately 55 kDa, which occasionally appears as a doublet. This suggests that this protein may undergo posttranslational modification that results in two species with different electrophoretic mobilities.
FIG. 4.
Polyclonal antibodies against Atac1 and dHCF coimmunoprecipitate HAT activity and components of the ATAC complex. (A) Thirty micrograms of nuclear extracts from S2 cells were used in Western blots to determine the specificity of the antibodies against Atac1 (α Atac1) and dHCF (α dHCF). The primary antibody used is shown at the top of each blot. (B) One milligram of whole-cell extract from wild-type cells or cells expressing T7-dHCF-FLAG was immunoprecipitated with M2-agarose beads. The immunoprecipitated (IP) material was analyzed by Western blotting using antibodies against dHCF (α dHCF, rabbit and dHCF rat) and FLAG (α FLAG). Lanes 1, 3, and 5 correspond to 40, 40, and 20% FLAG beads from wild-type cells, respectively. Lanes 2, 4, and 6 correspond to 40, 40, and 20% FLAG beads from T7-dHCF-FLAG cells, respectively. (C and D) One milligram of nuclear extract was immunoprecipitated with rabbit antibodies against dAda3, Atac1, dHCF, or a preimmune bleed (pre). Immunoprecipitation reactions were subjected to HAT assays using radiolabeled acetyl coenzyme A and core histones (C) or nucleosomes (D) as substrates. The top panels show the protein gels stained with Coomassie blue to show total histone substrate. The bottom panels show fluorographies of the same gels to show the acetylated histones. The migrations of H3 and H4 are indicated. (E) Antibodies against dAda3, dHCF, Atac1, dSpt3, and a rabbit preimmune bleed (pre) were incubated with 1 mg of nuclear extract from S2 cells for immunoprecipitation reactions. The immunoprecipitated material was analyzed by Western blotting using antibodies against dGcn5 (α dGcn5, rabbit), dAda2A (α dAda2A, rat), dHCF (α dHCF, rat), Atac1 (α Atac1, rat), and dAda2B (α dAda2B, guinea pig). Lane 1 corresponds to 2% of the input.
We wanted to further confirm if the strong band and the extra bands seen in the dHCF blots consisted of different dHCF species. To this end, we took advantage of cells expressing C-terminally FLAG-tagged dHCF and performed anti-FLAG immunoprecipitations with extracts from these tagged cells and wild-type cells (as a negative control). The immunoprecipitated material was analyzed by Western blotting, using antibodies against dHCF (rat) and dHCF (rabbit), and compared to the pattern of bands obtained using the anti-FLAG antibodies (Fig. 4B). dHCF sera obtained from rats and rabbits, as well as FLAG antibodies, recognized a strong band with a migration of ∼70 kDa in the immunoprecipitated material from tagged cells (lanes 2, 4, and 6) but not in the controls (lanes 1, 3, and 5). These antibodies also recognized several other protein species in the immunoprecipitates, including full-length dHCF (migration close to the 194-kDa marker, detected in lanes 2, 4, and 6). It is clear from these results that the dHCF antibody generated in rabbits is more sensitive than the one raised in rats, since additional bands can be detected in the blot probed with the rabbit antibody (lane 2), which correspond to different forms of dHCF. Since tagged dHCF is not expressed at its endogenous levels, we cannot conclude that these different forms we observe in the immunoprecipitations (lanes 2, 4, and 6), including the full-length protein, are present under normal growth conditions or arise as a consequence of expressing dHCF under control of the actin promoter. Our results indicate that dHCF gets rapidly processed in the cell and one of the main stable products of this cleavage is a 70-kDa C-terminal polypeptide. Our experiments suggest that there may be other forms of dHCF arising from cleavage at different sites in the full-length protein, which could be explained by the fact that dHCF, unlike its human counterpart HCF-1, does not possess the conserved processing repeats (24).
We set out to test whether the new antibodies were capable of pulling down HAT activity from nuclear extracts. We carried out immunoprecipitations with antibodies against dAda3, Atac1, dHCF, or a preimmune bleed (negative control) and subjected the immunoprecipitated material to HAT reactions, using core histones or nucleosomes as substrates (Fig. 4C and D). We found that Atac1 and dHCF were present in HAT complexes that acetylate H3 and H4 (Fig. 4C, lower panel, lanes 2 and 3). This pattern, identical to that of the dAda3 immunoprecipitate (lane 1), is a trait of Gcn5-containing complexes when assayed on free histones. When we assayed the HAT activity of the immunoprecipitates on nucleosomes, we observed that Atac1 and dHCF associated with H3 and H4 HAT activity (Fig. 4D, lower panel, lanes 3 and 4). However, the substrate preference was reversed, displaying an H4 signal higher than H3. The nucleosomal HAT activity of dAda3 immunoprecipitates targeted mostly H3 (lane 2), suggesting that, although dAda3 is part of ATAC, the majority of Ada3 in the cell is associated with a complex that preferentially acetylates nucleosomal H3. We anticipate that this dAda3-containing complex is dSAGA. The finding that ATAC displays a strong nucleosomal H4 activity was unexpected, since the yeast Gcn5-containing complexes SAGA, SLIK, and ADA preferentially acetylate nucleosomal H3 in vitro. However, experiments done with yeast gcn5Δ cells indicate that Gcn5 is responsible for global acetylation levels of both H3 and H4. Gcn5 mutants display reduced levels of histone H4 acetylation on lysines 5, 8, 12, and 16 (50). An alternative explanation for the unexpected nucleosomal HAT activity of ATAC is the presence of an additional catalytic polypeptide in the complex. MudPIT analysis of purified ATAC revealed peptides for a protein with a putative HAT domain (data not shown), which could be responsible for the H4 activity.
Antibodies directed to dHCF and Atac1 coimmunoprecipitate the ATAC complex.
Using dHCF and Atac1 antibodies, we further investigated interactions of dHCF and Atac1 with the ATAC complex. We incubated nuclear extracts with antibodies against dAda3, dHCF, Atac1, or a preimmune serum. The immunoprecipitated material was recovered on protein A-agarose beads and analyzed by Western blotting, using dGcn5, dAda2A, dHCF, Atac1, and dAda2B antisera (Fig. 4E). We observed that dSpt3 serum could only immunoprecipitate dGcn5 and dAda2B but not any of the other proteins tested (lane 6). This is consistent with dSpt3 and dAda2B being subunits of dSAGA but not ATAC. dHCF and Atac1 associated with each other (dHCF blot, lane 5; Atac1 blot, lane 4) and with dAda2A (lanes 4 and 5) but not with dAda2B (lanes 4 and 5). Although dHCF immunoprecipitation was very efficient (dHCF blot, lane 4), it did not pull down comparable amounts of dAda2A or dGcn5 (dAda2A and dGcn5 blots, lanes 4). This likely reflects the fact that the abundant dHCF protein can be found in other protein complexes besides ATAC (see below).
dHCF, Atac1, and dAda2A cofractionate as a 700-kDa multisubunit complex.
Since our data indicate that dHCF and Atac1 are subunits of ATAC, we tested whether these two new components cofractionate after a series of chromatographic steps. We first obtained a HAT-enriched fraction by incubating a nuclear extract with Ni-NTA agarose beads, followed by imidazole elution. We observed that many Drosophila HAT complexes bound to Ni-agarose in the same way yeast HAT complexes do (data not shown). We next applied the imidazole eluate to a Mono Q anion-exchange column and eluted the bound proteins with a linear gradient of NaCl. We analyzed the column fractions by Western blotting (Fig. 5A). We observed a sharp peak corresponding to the ATAC complex (dAda2A blot, fractions 29 to 33). These fractions also showed signals for Atac1 and dHCF (Fig. 5A, bottom blots). Note the paucity of dAda2B in these fractions, which eluted from the column earlier. In addition to eluting with the ATAC complex, dHCF elutes in two other peaks at fractions 11 and 19, which indicates that dHCF is found in additional complexes. This is consistent with the finding that human HCF-1 associates with a Sin3 histone deacetylase complex and a Set1/Ash2 histone methyltransferase complex (45, 48). We assume that one or more of these alternative HCF complexes also exists in flies. Although dHCF and dAda2B partially overlap (fraction 19), MudPIT analysis of affinity-purified dSAGA complex confirmed that dHCF is not a stable component of this complex (data not shown). These findings demonstrated that dGcn5, dAda2A, dHCF, and Atac1 stably associated and coeluted during anion-exchange fractionation.
FIG. 5.
dHCF, Atac1, and dAda2A are components of a 700-kDa complex. (A) Nuclear extract was incubated with Ni-NTA-agarose, eluted with imidazole, and applied to a Mono Q anion-exchange column. The bound proteins were resolved with a linear gradient of 100 to 500 mM NaCl, and odd fractions were analyzed by Western blotting, using antibodies against dAda2B (α dAda2B, rat), dGcn5 (α dGcn5, rabbit), dAda2A (α dAda2A, rabbit), dHCF (α dHCF, rabbit), and Atac1 (α Atac1, rabbit). I, 10 μl of imidazole eluate (input); FT, 10 μl of Mono Q flowthrough. (B) Fractions 29 to 33 were pooled, concentrated, and loaded onto a Superose 6 gel filtration column. Molecular mass standards were run under the same conditions, and their migration is indicated with arrows. I, 10 μl of pooled MonoQ fractions (input).
To further confirm cofractionation of ATAC components and to determine the apparent molecular weight of the ATAC complex, we applied Mono Q fractions 29 to 33 to Superose 6 gel filtration chromatography. We analyzed the column fractions by Western blotting, using antibodies against dAda2A, dHCF, and Atac1 (Fig. 5B). We observed that these three proteins are components of a complex with an apparent molecular mass of approximately 700 kDa (fractions 12 and 13). These fractions containing the ATAC complex also displayed HAT activity (data not shown)
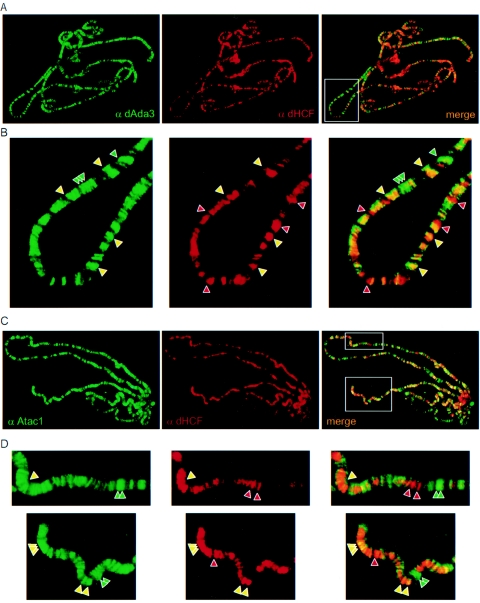
dHCF colocalizes with dAda3 and Atac1 on polytene chromosomes.
Previous experiments from our lab showed that the dAda2A complex and dSAGA have different patterns of chromatin binding (18). To further explore the interactions among the newly identified ATAC subunits and to support our biochemical evidence, we analyzed the localization of Atac1, dHCF, and dAda3 on polytene chromosomes. For this purpose, we dissected salivary glands from third-stage larvae and stained the polytene chromosomes with antibodies against dAda3 (rabbit) and dHCF (rat). We observed high concentrations of dHCF in many chromosomal regions. This is consistent with our biochemical data, which reflect that dHCF is a component of multiple complexes. When we compared the distribution of dAda3 and dHCF proteins on the chromosomes (Fig. 6A), we observed that many loci containing dAda3 also showed signal for dHCF. These bands corresponded to the ATAC complex recruited to multiple chromatin regions. Conversely, some loci stained exclusively for dAda3 or dHCF. These bands correspond to either dSAGA, which lacks dHCF, or dGcn5-independent dHCF-containing complexes.
FIG. 6.
dHCF colocalizes with Atac1 and dAda3 on polytene chromosomes. (A) Polytene chromosomes were stained with antibodies against dAda3 (α dAda3, rabbit, green) and dHCF (α dHCF, rat, red). The green- and red-stained images were overlapped to generate the merge panel (yellow). (B) Magnification of the boxed region from panel A. Green and red arrowheads indicate chromosome sites that are bound exclusively by dAda3 or dHCF, respectively. Yellow arrowheads point to chromosomal loci occupied by dAda3 and dHCF (C) Polytene chromosomes were stained with antibodies against Atac1 (α Atac1, rabbit, green) and dHCF (α dHCF, rat, red). (D) Magnification of the boxed regions from panel C. The arrowheads point to chromosome sites that are bound by Atac1 (green), dHCF (red), or both (yellow).
We also stained chromosomes with antibodies against Atac1 (rabbit) and dHCF (rat) (Fig. 6B). Atac1 and dHCF colocalized in many chromosomal regions, supporting the fact that these two proteins function together in the multisubunit ATAC complex. In addition, we found areas which only stained for dHCF, reinforcing our idea that dHCF functions in additional complexes, independently of dGcn5 and dAda2A. In some cases, we observed Atac1 in regions that lacked dHCF. This may suggest that Atac1 may also be contained in unidentified additional complexes independent of the ATAC complex. In fact, on the anion-exchange column, Atac1 began to elute a couple fractions before dAda2A which, in principle, could represent another complex (Fig. 5A). A more likely possibility is that additional Atac1-containing complexes were not present in the Ni-agarose fraction applied to the anion-exchange column.
The polytene stains confirmed the association of dAda3 with dHCF and the association of dHCF with Atac1. They further illustrated that dHCF can be bound to chromatin independent of dAda3 and Atac1, supporting the observation that dHCF is in other complexes in addition to ATAC. And finally, the polytene stains provided the insight that some Atac1 is chromatin bound independent of dHCF, suggesting the possibility of additional Atac1-containing complexes in addition to ATAC.
DISCUSSION
This work describes the affinity purification of Drosophila ATAC (Ada two A containing) HAT complex for the first time. We purified dGcn5-containing complexes and subjected them to MudPIT analysis. We discovered two new proteins that copurified with TAP-dGcn5: Atac1 (cg9200) and dHCF. We generated antibodies against these two proteins and utilized them in a number of biochemical experiments that allowed us to demonstrate that Atac1 and dHCF are stable components of the ATAC complex but not of dSAGA. To gain insight into the function of Atac1, we looked for homologues in other species. We found that Atac1 shares 35% identity with a human protein named zzz3 (NP_056349, 903 amino acids), which contains both a SANT and ZZ domain. However, no function has been assigned to this protein yet.
Many chromatin remodelers and modifiers harbor a SANT domain in one of their subunits. Some examples include the yeast complexes SAGA, ADA, NuA4, RPD3, SWI/SNF, RSC, and ISW1/2 (3). This 50-amino-acid motif was shown to be critical for acetylation by SAGA and remodeling by SWI/SNF. For example, SAGA complex purified from strains carrying a deletion in the SANT domain of Ada2 is intact but shows a drastic reduction in nucleosomal acetylation (2, 39). There are a few cases where two SANT domains are present in the same complex. The nuclear receptor corepressor SMRT (silencing mediator of retinoid and thyroid receptors), which contains a pair of SANT domains, exists in a HDAC3-containing repressor complex. While the N-terminal SANT1 binds and activates HDAC3, its adjacent motif SANT2 is required for histone interaction. These two motifs are not functionally interchangeable (49). We are intrigued by the fact that the ATAC HAT complex also contains two SANT domains: one in dAda2A and the other one in Atac1. We consider the possibility that the former, like its yeast homologue, is involved in enhancing dGcn5 catalytic activity. Atac1, on the other hand, could utilize its SANT domain for binding to nucleosomes or for other protein-protein interactions.
Drosophila has only one orthologue that belongs to the HCF family of proteins, present in higher eukaryotes. This protein shares a high degree of conservation with human HCF-1. Analysis of the primary structure of dHCF does not reveal the presence of HCFpro repeats found in HCF-1. However, experiments done with S2 cells that express dHCF tagged at both ends demonstrated that this protein is cleaved after translation to generate two polypeptides that remain physically associated (24). By generating antibodies directed to the C terminus of dHCF, we were able to confirm those results with the endogenous protein. We could not detect full-length protein by Western blot analysis. Therefore, we suggest that most of the dHCF precursor gets processed. Additionally, we detect more than one form of dHCF using the antiserum that recognizes the C terminus. We believe that this reflects multiple processing sites in dHCF. This observation agrees with experiments done with HCF-1, which show that antibodies that recognize the N- or C-terminal subunits display several bands on Western blots (15).
When we looked at the amino acid sequence of the peptide hits from the MudPIT analysis of affinity-purified ATAC complex in more detail, we identified peptides corresponding to the N- and C-terminal subunits of dHCF. This finding indicates that both subunits are components of the ATAC HAT complex. Furthermore, we showed that antibodies against the dHCF C terminus coimmunoprecipitated the ATAC complex. Unfortunately, we were unable to obtain antibodies against the N-terminal fragment to perform similar experiments.
Our anion-exchange fractionation revealed that dHCF is present in at least two other complexes that elute at different salt concentrations. We ruled out the possibility of dHCF being present in other dGcn5-containing complexes for two reasons. First, immunoprecipitations using dSpt3 antibodies do not pull down dHCF. Second, we established S2 cell lines expressing tagged dSAGA-specific subunits, affinity purified complexes from these cells, and subjected them to MudPIT. We could not identify peptides for dHCF in those complexes (data not shown), which led us to conclude that dHCF is uniquely associated with the ATAC HAT complex. Since dHCF is an abundant protein, we were not surprised to find it in column fractions where the ATAC complex was not present. It is likely that dHCF is a component of other chromatin-modifying complexes. In fact, the N-terminal subunit of human HCF-1 forms a supercomplex that contains subunits of the Sin3 histone deacetylase complex and the Set1/Ash2 histone methyltransferase (45). In addition, immunopurifications with antibodies directed to the proto-oncoprotein MLL reveal associations of HCF-1, HCF-2, menin, and components of the Set1/Ash2 complex (48).
What could be the biological relevance of dHCF in a HAT complex? There is a strong link between HCF-1 and transcription. Experiments done with HCF-1 indicate that its N- and C-terminal subunits interact with cellular transcription activators (21, 22). This could be a mechanism by which the ATAC complex is recruited to specific promoters. In addition, we cannot rule out the possibility of the ATAC complex having a role during the cell cycle, since we know that mammalian HCF-1N and HCF-1C are crucial for G1 phase progression and proper cytokinesis (15). Furthermore, yeast cells carrying deletions in Gcn5 and Sas3 genes arrest at G2/M, indicating that H3 HATs are essential for cell cycle progression (13). Mammalian cells lacking Gcn5 exhibit a delay in the progression from G1 to S phase and present abnormal levels of expression of cell cycle-related genes (16).
We anticipate that there are additional unidentified subunits in the ATAC complex. First, the estimated molecular mass of the complex is 700 kDa. The sum of the molecular masses of the known subunits (dGcn5, dAda3, dAda2A, Atac1, and dHCF) is approximate 450 kDa, which suggests there could be few more subunits in this complex. Secondly, our MudPIT analyses from TAP-dGcn5, dAda2A-TAP, and Atac1-FLAG purifications have revealed unconfirmed candidate proteins, one of which contains a putative HCF binding motif, identified in transcription factors that interact with the β-propeller region of HCF-1 (21). We are currently tagging these proteins and raising antibodies against them to confirm interactions with dAda2A, dGcn5, dAda3, dHCF, and Atac1. The identification of all the components of this novel HAT complex should provide additional evidence to help uncover its function in cell growth and Drosophila development.
Acknowledgments
We thank the members of the Workman lab for helpful discussions. We thank Kenneth Lee for critical comments on the manuscript. We thank Angus Wilson for providing dHCF-expression plasmid and Winship Herr for suggestions on making dHCF antibodies.
REFERENCES
- 1.Barlev, N. A., A. V. Emelyanov, P. Castagnino, P. Zegerman, A. J. Bannister, M. A. Sepulveda, F. Robert, L. Tora, T. Kouzarides, B. K. Birshtein, and S. L. Berger. 2003. A novel human Ada2 homologue functions with Gcn5 or Brg1 to coactivate transcription. Mol. Cell. Biol. 23:6944-6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyer, L. A., M. R. Langer, K. A. Crowley, S. Tan, J. M. Denu, and C. L. Peterson. 2002. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol. Cell 10:935-942. [DOI] [PubMed] [Google Scholar]
- 3.Boyer, L. A., R. R. Latek, and C. L. Peterson. 2004. The SANT domain: a unique histone-tail-binding module? Nat. Rev. Mol. Cell Biol. 5:158-163. [DOI] [PubMed] [Google Scholar]
- 4.Brown, C. E., L. Howe, K. Sousa, S. C. Alley, M. J. Carrozza, S. Tan, and J. L. Workman. 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292:2333-2337. [DOI] [PubMed] [Google Scholar]
- 5.Brownell, J. E., J. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84:843-851. [DOI] [PubMed] [Google Scholar]
- 6.Bunch, T. A., Y. Grinblat, and L. S. Goldstein. 1988. Characterization and use of the Drosophila metallothionein promoter in cultured Drosophila melanogaster cells. Nucleic Acids Res. 16:1043-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrozza, M. J., R. T. Utley, J. L. Workman, and J. Cote. 2003. The diverse functions of histone acetyltransferase complexes. Trends Genet. 19:321-329. [DOI] [PubMed] [Google Scholar]
- 8.Eberharter, A., S. John, P. A. Grant, R. T. Utley, and J. L. Workman. 1998. Identification and analysis of yeast nucleosomal histone acetyltransferase complexes. Methods 15:315-321. [DOI] [PubMed] [Google Scholar]
- 9.Fischle, W., Y. Wang, and C. D. Allis. 2003. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15:172-183. [DOI] [PubMed] [Google Scholar]
- 10.Flaus, A., and T. Owen-Hughes. 2004. Mechanisms for ATP-dependent chromatin remodelling: farewell to the tuna-can octamer? Curr. Opin. Genet. Dev. 14:165-173. [DOI] [PubMed] [Google Scholar]
- 11.Grant, P. A., and S. L. Berger. 1999. Histone acetyltransferase complexes. Semin. Cell Dev. Biol. 10:169-177. [DOI] [PubMed] [Google Scholar]
- 12.Grant, P. A., D. E. Sterner, L. J. Duggan, J. L. Workman, and S. L. Berger. 1998. The SAGA unfolds: convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol. 8:193-197. [DOI] [PubMed] [Google Scholar]
- 13.Howe, L., D. Auston, P. Grant, S. John, R. G. Cook, J. L. Workman, and L. Pillus. 2001. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev. 15:3144-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howe, L., C. E. Brown, T. Lechner, and J. L. Workman. 1999. Histone acetyltransferase complexes and their link to transcription. Crit. Rev. Eukaryot. Gene Expr. 9:231-243. [DOI] [PubMed] [Google Scholar]
- 15.Julien, E., and W. Herr. 2003. Proteolytic processing is necessary to separate and ensure proper cell growth and cytokinesis functions of HCF-1. EMBO J. 22:2360-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kikuchi, H., Y. Takami, and T. Nakayama. 2005. GCN5: a supervisor in all-inclusive control of vertebrate cell cycle progression through transcription regulation of various cell cycle-related genes. Gene 347:83-97. [DOI] [PubMed] [Google Scholar]
- 17.Kusch, T., L. Florens, W. H. Macdonald, S. K. Swanson, R. L. Glaser, J. R. Yates III, S. M. Abmayr, M. P. Washburn, and J. L. Workman. 2004. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306:2084-2087. [DOI] [PubMed] [Google Scholar]
- 18.Kusch, T., S. Guelman, S. M. Abmayr, and J. L. Workman. 2003. Two Drosophila Ada2 homologues function in different multiprotein complexes. Mol. Cell. Biol. 23:3305-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, K. K., L. Florens, S. K. Swanson, M. P. Washburn, and J. L. Workman. 2005. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol. Cell. Biol. 25:1173-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, K. K., P. Prochasson, L. Florens, S. K. Swanson, M. P. Washburn, and J. L. Workman. 2004. Proteomic analysis of chromatin-modifying complexes in Saccharomyces cerevisiae identifies novel subunits. Biochem. Soc. Trans. 32:899-903. [DOI] [PubMed] [Google Scholar]
- 21.Luciano, R. L., and A. C. Wilson. 2003. HCF-1 functions as a coactivator for the zinc finger protein Krox20. J. Biol. Chem. 278:51116-51124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luciano, R. L., and A. C. Wilson. 2000. N-terminal transcriptional activation domain of LZIP comprises two LxxLL motifs and the host cell factor-1 binding motif. Proc. Natl. Acad. Sci. USA 97:10757-10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lusser, A., and J. T. Kadonaga. 2003. Chromatin remodeling by ATP-dependent molecular machines. Bioessays 25:1192-1200. [DOI] [PubMed] [Google Scholar]
- 24.Mahajan, S. S., K. M. Johnson, and A. C. Wilson. 2003. Molecular cloning of Drosophila HCF reveals proteolytic processing and self-association of the encoded protein. J. Cell. Physiol. 194:117-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marmorstein, R. 2001. Structure and function of histone acetyltransferases. Cell. Mol. Life Sci. 58:693-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez, E., V. B. Palhan, A. Tjernberg, E. S. Lymar, A. M. Gamper, T. K. Kundu, B. T. Chait, and R. G. Roeder. 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 21:6782-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muratoglu, S., S. Georgieva, G. Papai, E. Scheer, I. Enunlu, O. Komonyi, I. Cserpan, L. Lebedeva, E. Nabirochkina, A. Udvardy, L. Tora, and I. Boros. 2003. Two different Drosophila ADA2 homologues are present in distinct GCN5 histone acetyltransferase-containing complexes. Mol. Cell. Biol. 23:306-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogryzko, V. V. 2001. Mammalian histone acetyltransferases and their complexes. Cell. Mol. Life Sci. 58:683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogryzko, V. V., T. Kotani, X. Zhang, R. L. Schlitz, T. Howard, X.-J. Yang, B. H. Howard, J. Qin, and Y. Nakatani. 1998. Histone-like TAFs within the PCAF histone acetylase complex. Cell 94:35-44. [DOI] [PubMed] [Google Scholar]
- 30.Peterson, C. L., and M. A. Laniel. 2004. Histones and histone modifications. Curr. Biol. 14:R546-R551. [DOI] [PubMed] [Google Scholar]
- 31.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 32.Qi, D., J. Larsson, and M. Mannervik. 2004. Drosophila Ada2b is required for viability and normal histone H3 acetylation. Mol. Cell. Biol. 24:8080-8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 34.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 35.Sanders, S. L., J. Jennings, A. Canutescu, A. J. Link, and P. A. Weil. 2002. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol. 22:4723-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sato, S., C. Tomomori-Sato, T. J. Parmely, L. Florens, B. Zybailov, S. K. Swanson, C. A. Banks, J. Jin, Y. Cai, M. P. Washburn, J. W. Conaway, and R. C. Conaway. 2004. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol. Cell 14:685-691. [DOI] [PubMed] [Google Scholar]
- 37.Schneider, I. 1972. Cell lines derived from late embryonic stages of Drosophila melanogaster. J. Embryol. Exp. Morphol. 27:353-365. [PubMed] [Google Scholar]
- 38.Smith, E. R., J. M. Belote, R. L. Schiltz, X. J. Yang, P. A. Moore, S. L. Berger, Y. Nakatani, and C. D. Allis. 1998. Cloning of Drosophila GCN5: conserved features among metazoan GCN5 family members. Nucleic Acids Res. 26:2948-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sterner, D. E., X. Wang, M. H. Bloom, G. M. Simon, and S. L. Berger. 2002. The SANT domain of Ada2 is required for normal acetylation of histones by the yeast SAGA complex. J. Biol. Chem. 277:8178-8186. [DOI] [PubMed] [Google Scholar]
- 40.Vlachonasios, K. E., M. F. Thomashow, and S. J. Triezenberg. 2003. Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell 15:626-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Washburn, M. P., D. Wolters, and J. R. Yates III. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19:242-247. [DOI] [PubMed] [Google Scholar]
- 42.Wieczorek, E., M. Brand, X. Jacq, and L. Tora. 1998. Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature 393:187-191. [DOI] [PubMed] [Google Scholar]
- 43.Wilson, A. C., M. Boutros, K. M. Johnson, and W. Herr. 2000. HCF-1 amino- and carboxy-terminal subunit association through two separate sets of interaction modules: involvement of fibronectin type 3 repeats. Mol. Cell. Biol. 20:6721-6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wysocka, J., and W. Herr. 2003. The herpes simplex virus VP16-induced complex: the makings of a regulatory switch. Trends Biochem. Sci. 28:294-304. [DOI] [PubMed] [Google Scholar]
- 45.Wysocka, J., M. P. Myers, C. D. Laherty, R. N. Eisenman, and W. Herr. 2003. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 17:896-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu, W., D. G. Edmondson, Y. A. Evrard, M. Wakamiya, R. R. Behringer, and S. Y. Roth. 2000. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat. Genet. 26:229-232. [DOI] [PubMed] [Google Scholar]
- 47.Yamauchi, T., J. Yamauchi, T. Kuwata, T. Tamura, T. Yamashita, N. Bae, H. Westphal, K. Ozato, and Y. Nakatani. 2000. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc. Natl. Acad. Sci. USA 97:11303-11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yokoyama, A., Z. Wang, J. Wysocka, M. Sanyal, D. J. Aufiero, I. Kitabayashi, W. Herr, and M. L. Cleary. 2004. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol. Cell. Biol. 24:5639-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu, J., Y. Li, T. Ishizuka, M. G. Guenther, and M. A. Lazar. 2003. A SANT motif in the SMRT corepressor interprets the histone code and promotes histone deacetylation. EMBO J. 22:3403-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, W., J. R. Bone, D. G. Edmondson, B. M. Turner, and S. Y. Roth. 1998. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 17:3155-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]