Abstract
TGIF (TG-interacting factor) represses transforming growth factor β (TGF-β)-activated gene expression and can repress transcription via a specific retinoid response element. Mutations in human TGIF are associated with holoprosencephaly, a severe defect of craniofacial development with both genetic and environmental causes. Both TGF-β and retinoic acid signaling are implicated in craniofacial development. Here, we analyze the role of TGIF in regulating retinoid responsive gene expression. We demonstrate that TGIF interacts with the ligand binding domain of the RXRα retinoid receptor and represses transcription from retinoid response elements. TGIF recruits the general corepressor, CtBP, to RXRα, and this recruitment is required for full repression by TGIF. Interaction between TGIF and RXRα is reduced by the addition of retinoic acid, consistent with a role for TGIF as an RXRα transcriptional corepressor. We created a Tgif null mutation in mice and tested the sensitivity of mutant mice to increased levels of retinoic acid. Tgif mutant embryos are more sensitive to retinoic acid-induced teratogenesis, and retinoid target genes are expressed at a higher level in tissues from Tgif null mice. These results demonstrate an important role for TGIF as a transcriptional corepressor, which regulates developmental signaling by retinoic acid, and raises the possibility that TGIF may repress other RXR-dependent transcriptional responses.
TGIF (TG-interacting factor) is a member of the TALE (three-amino-acid loop extension) superfamily of homeodomain proteins (3, 4). In the TALE superfamily, the first two helices are separated by a loop, which is likely to affect interactions with other proteins but not alter DNA binding properties (4, 31, 34). Homeodomain proteins regulate numerous developmental processes and do so by activating or repressing gene expression through interactions either with DNA or with other DNA-bound proteins (8, 24, 25). Functional analysis has demonstrated that TGIF is a transcriptional repressor which interacts with general corepressor proteins, including CtBP, mSin3, and histone deacetylases (27, 40, 46, 48).
Retinoic acid binds to members of the nuclear receptor family of transcription factors: the retinoid X receptors (RXRs), which bind only 9-cis retinoic acid, and the retinoic acid receptors (RARs), which bind several forms of retinoic acid (1, 14, 19). Binding of retinoic acid to RAR or RXR induces a conformational change in the receptors which alters the balance between coactivator and corepressor interactions, allowing a switch from repression to activation of target gene expression (2, 5, 9). The receptors can both homo- and heterodimerize and then interact with bipartite DNA binding sites. The relative spacing of half sites within a retinoid response element can determine which RAR/RXR heterodimer or homodimer it binds (18, 35, 44). TGIF was isolated by its ability to bind adjacent to a retinoid response element (RXRE) from the rat cellular retinol binding protein II (CrbpII) gene (3). Binding of TGIF to this specific RXRE decreased activation of gene expression. However, most retinoic acid response elements do not have TGIF sites, and in vivo, retinoid responsive genes are not known to be regulated by TGIF.
Transforming growth factor β (TGF-β) binds to and activates a cell surface receptor complex, which phosphorylates and activates the intracellular mediators of TGF-β signaling, the Smad proteins. Receptor-activated Smads complex with Smad4, enter the nucleus and bind to DNA, either directly or via interactions with other factors (13, 21, 22, 51). Transcriptional activation by Smads relies on their interaction with general coactivators, such as p300/CBP (23). TGIF is a transcriptional corepressor for TGF-β-activated Smads, which competes with coactivators for Smad interaction, and recruits corepressors, resulting in active repression of TGF-β/Smad target genes (47).
The human TGIF gene was mapped to the minimal critical region at 18p11.3, which was deleted in a panel of holoprosencephaly (HPE) patients (11). HPE is a prevalent human genetic disease affecting craniofacial development, with an incidence of up to 1:250 conceptions but, due to a high level of intrauterine lethality, a much lower frequency at birth (12, 29). The primary defect in HPE is a failure of the ventral forebrain to divide, with concomitant defects in midline structures which can result in cyclopia and proboscosis (10, 37). Many genetic loci have been implicated in HPE, including those encoding the morphogen Sonic Hedgehog (Shh) and the transcription factors Zic2, Six3, and TGIF (29, 36). As with other HPE mutations, loss of TGIF is only partially penetrant with respect to the HPE phenotype, suggesting that the genetics of HPE are complex. HPE can also be caused by environmental factors, and only around half of HPE can be attributed to known genetic lesions (36, 45). In both mice and humans, prenatal exposure to retinoic acid has been shown to cause developmental defects, including HPE (17, 42). Thus, it may be the interplay of genetic and environmental factors that causes HPE.
Here we test whether TGIF is a general repressor of retinoic acid-dependent transcriptional activation. We demonstrate that TGIF represses transcription from multiple different retinoid response elements by interacting specifically with the RXRα retinoic acid receptor, and that it recruits the corepressor CtBP to RXRα. To test whether TGIF loss of function results in increased sensitivity to retinoic acid, we created Tgif null mice. We show that Tgif−/− mice are more sensitive to retinoic acid signals. Retinoic acid target genes show increased expression, and in utero exposure to exogenous retinoic acid caused an increased rate of developmental abnormalities in the mutant embryos. This work clearly demonstrates for the first time an in vivo role for TGIF in control of retinoid-regulated gene expression.
MATERIALS AND METHODS
Plasmids.
The DR1 and DR5 luciferase reporters were created by first ligating a double-stranded oligonucleotide containing a TATA element into pGL2-basic. Oligonucleotides containing either the DR1 element (GTACGCTGTCACAGGTCACAGGTCACAGGTCACAGGTCAC) or a DR5 response element (GATCAGGGTTCACCGAAAGTTCACTCGCT) were then inserted upstream of the TATA sequence (upper strands only are shown, the RAR/RXR half sites are underlined). All expression constructs were created in pCMV5 with either a Flag or T7 epitope tag and contained the entire coding sequences of TGIF, RARα or RXRα, PPARγ, or CtBP. Deletion mutants were made by PCR and were ligated into pCMV5 with Flag or T7 epitope tags. The ΔAF1 RXRβ and RXRγ constructs were created by PCR in T7-pCMV5. The estrogen response element (ERE) reporter was created in pGL3 (Promega) and contained two EREs. The TGIF small interfering RNA (siRNA) was created in pSUPER and contained the same target sequence as previously reported (39).
Luciferase assays.
HepG2 cells were maintained in Dulbecco's minimal essential medium with 10% fetal bovine serum and transfected using Exgen 500 (MBI Fermentas) according to the manufacturer's instructions. Cells were transfected with the appropriate luciferase reporter, phCMVRLuc, and the indicated expression constructs. After 48 h, firefly luciferase activity was assayed using a luciferase assay kit (Promega) and Renilla luciferase was assayed with 0.09 μM coelenterazine (Biosynth) using a Berthold LB953 luminometer. For experiments with the siRNA vector, luciferase activity was assayed 60 h after transfection. Retinoic acid (RA) (9-cis-RA [9C-RA] or all-trans-RA [AT-RA]; Sigma) was added for the final 24 h or as indicated.
Immunoprecipitation and Western blotting.
COS1 cells were maintained in Dulbecco's minimal essential medium with 10% bovine growth serum (HyClone) and were transfected using LipofectAmine (Invitrogen). At 36 h after transfection, cells were lysed by sonication in 75 mM NaCl, 50 mM HEPES, pH 7.8, 20% glycerol, 0.1% Tween 20, 0.5% NP-40 with protease and phosphatase inhibitors. Immunocomplexes were precipitated with Flag M2-agarose (Sigma). Following sodium dodecyl sulfate-polyacrylamide gel electrophoresis, proteins were electroblotted to Immobilon-P (Millipore) and incubated with antisera specific for Flag (Sigma) or T7 (Novagen). Proteins were visualized with horseradish peroxidase-conjugated goat anti-mouse or anti-rabbit immunoglobulin (Pierce) and enhanced chemiluminescence (Amersham Pharmacia Biotech). Endogenous protein complexes were precipitated using a mouse monoclonal RXRα-specific antibody (a kind gift of C. Rochette-Egly), and TGIF was detected using a rabbit polyclonal TGIF-specific antiserum (47).
Electrophoretic mobility shift assays.
For electrophoretic mobility shift assays, 0.75 ng of radiolabeled double-stranded oligonucleotide probe containing a perfect TGIF binding site (ACTCTGCCTGTCAAGCGAGG) was incubated with 1.5 μl of mini nuclear extract from COS1 cells transfected with either a control vector or a TGIF expression plasmid, together with 1 μg poly(dI-dC) · poly(dI-dC) and 500 ng single-stranded DNA with or without specific competitor oligonucleotides. Reaction mixtures were incubated on ice in binding buffer (20 mM HEPES, pH 7.9, 50 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 20% glycerol, 1 mM dithiothreitol) for 30 min and separated on a 4% polyacrylamide gel in 0.5× Tris-borate-EDTA. Gels were dried and visualized on a PhosphorImager.
Tgif gene disruption.
A lambda phage 129/SvJ genomic library was screened for mouse Tgif, and positive clones were analyzed by restriction enzyme digestion and sequencing. We removed most of the second and third exons of the Tgif gene and fused the coding sequence for the green fluorescent protein (GFP) in frame with the ninth codon of the Tgif gene. Correct integration of this targeting vector removes all Tgif coding sequence after the ninth codon and results in expression from the Tgif promoter of a fusion of the first 9 amino acids of Tgif with GFP. The Neo gene is expressed from a separate promoter, downstream of the GFP 3′ untranslated region. The targeting construct had a 900-bp short arm upstream of the second coding exon and a 9-kb EcoRI genomic fragment as the long arm 3′ of the final exon. 129/Sv embryonic stem cells were electroporated, and clones were selected with G418. Positive clones were identified by PCR and confirmed by Southern blotting. Mouse embryo fibroblasts (MEFs) were isolated from embryonic day 13.5 (E13.5) embryos and were passaged in culture three times prior to use for chromatin immunoprecipitation analysis. Procedures were approved by the Animal Care and Use Committee of the University of Virginia.
DNA and RNA analyses.
DNA was isolated and purified from tail biopsy or yolk sac using the Promega Wizard purification kit or by HotShot (43). DNA for Southern blotting was purified from an adult liver by standard proteinase K digestion and phenol-chloroform extraction methods. RNA was isolated from E13.5 embryos using Trizol reagent. Northern and Southern blotting were carried out by standard techniques. Array analysis was carried out using GE Pathway Finder Arrays (SuperArray Bioscience), with 32P labeled probes, according to the manufacturer's instructions.
Real-time PCR.
Total RNA was extracted from testes from wild-type or mutant animals (Trizol extraction followed by Rneasy mini kit purification). RNA was used for cDNA synthesis. The cDNA was used as template in a quantitative real-time PCR (SYBR green; SuperArray Bioscience) using a Bio-Rad MyiQ thermocycler (annealing temperature, 56°C). All of the real-time values were averaged and compared using the cycle threshold (CT) method, where the amount of target RNA (2−−ΔΔCT) was normalized to the endogenous beta-actin (forward primer, 5′-CACACCCGCCACCAGTTCG-3′; reverse primer, 5′-GACCCATTCCCACCATCACACC-3′) reference (ΔCT) and related to the amount of targets in wild-type cells (ΔΔCT), which was set as the calibrator at 1.0. Commercially available gene-specific primers validated for real-time PCR experiments have been used (RT2 gene expression assays; SuperArray Bioscience). The values represent the means ± standard errors of the means of the results from triplicate experiments.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation was performed essentially as described previously (15, 32). Briefly, wild-type or mutant MEFs at passage three were treated with 9C-RA for 16 h or left untreated and were cross-linked with 1% formaldehyde for 10 min at 37°C. Following chromatin isolation, DNA was sheared by sonication to between 200 and 1,000 bp in length. Immunoprecipitations were carried out using 2 μl of a polyclonal TGIF antiserum (47) or of preimmune serum. The oligonucleotides used for PCR are described in reference 32, and PCR was carried out for 31 cycles (RARβ) and 35 cycles (GAPDH) with an annealing temperature of 62°C. For input controls, 10% of the input chromatin was purified without immunoprecipitation, and a portion of this was subjected to PCR under the same conditions.
Retinoic acid treatment.
Tgif heterozygotes were intercrossed, and pregnant females were treated by gavage at E7.5 with 7.5 mg of all-trans-retinoic acid (Sigma)/kg of body weight in 200 μl of corn oil, a dose known to cause HPE in mice (42). Embryos were then excised at E10.5 and genotyped by PCR from yolk sac DNA.
RESULTS
TGIF represses RXR-mediated transcription.
To analyze the effect of TGIF on retinoid-regulated gene expression, we tested the effects of TGIF on retinoic acid-responsive luciferase reporters in HepG2 cells. We first used a reporter containing a DR1 retinoid response element. The DR1 RXRE has five consensus RAR/RXR half sites, each separated by 1 bp, and can be bound by both RAR/RXR heterodimers (which repress this element) and RXR homodimers (which activate it). HepG2 cells were transfected with the DR1-TATA-luc reporter and an RXRα expression construct, with or without a TGIF expression plasmid, and cells were either treated with increasing amounts of 9C-RA or left untreated. The DR1 reporter was activated in the presence of transfected RXRα (Fig. 1A). Coexpression of TGIF clearly repressed RXR-mediated activation of the DR1 luciferase reporter, and this repression was partially overcome by the addition of 9C-RA. We observed fourfold repression of the DR1 reporter by TGIF in the absence of added ligand, and this dropped to just over twofold repression at the maximum dose of 9C-RA (Fig. 1A). Next we tested the effect of expressing increasing amounts of TGIF on the activity of the DR1 reporter in the absence or presence of two concentrations of 9C-RA. As shown in Fig. 1B, we observed a dose-dependent increase in repression of this element by TGIF in the presence of retinoic acid, whereas even the lowest amount of TGIF repressed reporter activity in the absence of added retinoic acid.
FIG. 1.
Regulation of RXR-dependent transcription by TGIF. HepG2 cells were transfected with a DR1-TATA-luc luciferase reporter, together with expression vectors encoding RXRα and TGIF, as indicated (−, not used). 9C-RA was added 24 h prior to analysis, as indicated. In panel A, + indicates 10−7 M 9C-RA, and the titration of increasing 9C-RA was 10−9 M, 10−8 M, and 10−7 M. In panel B, increasing amounts of TGIF were cotransfected as indicated (in ng per duplicate). Luciferase activity was assayed 40 h after transfection and is presented, in arbitrary units, as the mean ± standard deviation of results from duplicate transfections.
TGIF represses via a DR5 RARE.
Most known retinoid-responsive elements consist of a canonical DR5 element, in which two direct repeats of the retinoic acid receptor binding site are separated by 5 base pairs. Since relatively few DR1 response elements have been identified, we were interested to know whether TGIF could also repress via a DR5 RARE. Additionally, the DR1 element has an adjacent partial match to a TGIF binding site, and it has been suggested that TGIF represses by binding directly to this DNA sequence (3). However, most retinoic acid response elements, including the DR5 reporters used here, do not have TGIF binding sites. We created luciferase reporters containing either 1, 2, or 4 copies of the RARE from the RARβ gene. When we used a reporter with four copies of the DR5 element, together with high levels of either 9C-RA or AT-RA, coexpression of TGIF was unable to repress activity. However, activation of the four-copy DR5 reporter by RXRα in the absence of added RA was repressed fourfold by coexpression of TGIF (Fig. 2A). To test whether TGIF was able to better repress when fewer copies of the RARE were present, we analyzed reporters with one or two copies of the RARE. The single RARE reporter was activated by RA in the presence of RXRα, and TGIF repressed its activity to close to the basal level (Fig. 2B). Similar results were obtained with a reporter containing two copies of the RARβ DR5 element, although this reporter was much better activated by RXRα and 9C-RA (Fig. 2B). Thus, TGIF is able to inhibit activation of a DR5 retinoid response element in the presence of RXRα and limiting retinoic acid, but once the level of RA is increased, this repression can be overcome. These results suggest that TGIF limits the effectiveness of signaling by low levels of retinoic acid.
FIG. 2.
Repression of DR5-mediated transcription by TGIF. HepG2 cells were transfected with DR5-TATA-luc luciferase reporters, together with expression vectors encoding RXRα and TGIF, as indicated. +, present; −, absent. The DR5 reporters contained either 1, 2, or 4 tandem copies of the DR5 element as indicated. (A) AT-RA or 9C-RA was added 24 h prior to analysis, as indicated, to a final concentration of 10−7 M. The boxed inset in panel A shows the DR5-TATA-luc activity in the absence of added retinoic acid. (B) Cells were transfected with a control reporter lacking DR5 elements (TATA-luc) or with reporters containing 1 or 2 DR5 elements, together with RXRα and TGIF expression constructs, as indicated. 9C-RA was added as indicated 24 h prior to lysis. (C) Nuclear extracts from COS1 cells transfected with a TGIF or control vector were incubated with a radiolabeled probe containing a single consensus TGIF site (CTGTCAA). Unlabeled competitor oligonucleotides, either the consensus TGIF site or the DR5 RARE, were added as indicated. Triangles represent a titrations of a 5-, 20-, 80-, 320-, and 530-fold excesses of unlabeled competitor. The TGIF shifted band and free probe are indicated. (D) Cells were transfected with the DR1 or DR5 (two copy) reporters together with a control vector (pSUPER) or one expressing a TGIF-specific hairpin RNA (siTGIF) and treated with 9C-RA, as indicated, 24 h prior to analysis. Three days after transfection, luciferase activity was assayed. (E) Cells were transfected with a control vector, a TGIF expression plasmid, or a TGIF siRNA vector, together with a luciferase reporter in which transcription is activated by an estrogen response element (ERE). Estrogen (E2) was added, as indicated, 24 h prior to analysis.
Purified recombinant TGIF has been shown to bind to the CRBPII RXRE, which contains a close match to the TGIF consensus binding site (3). However, the DR5 element does not contain a TGIF site, suggesting that binding of TGIF to DNA is unlikely to account for the observed repression. To test whether TGIF binds directly to the DR5 RARE, we performed electrophoretic mobility shift assays using nuclear extracts from transfected COS1 cells. We incubated extracts from COS1 cells transfected with TGIF with a radiolabeled probe containing a consensus TGIF binding site (CTGTCAA) (3) and included increasing amounts of an unlabeled double-stranded oligonucleotide with either a TGIF site or the DR5 element. As shown in Fig. 2C, TGIF bound to the probe containing its own site, and this binding was efficiently competed by the unlabeled TGIF competitor but not by the DR5 element, even with the maximum amount of competitor. Together, these data demonstrate that TGIF does not bind to DR5 retinoic acid response elements but is able to repress transcription via these elements.
To test whether endogenously expressed TGIF represses retinoid signaling, we used a previously published siRNA vector directed against TGIF (39). As shown in Fig. 2D, cotransfection of the TGIF siRNA vector resulted in about a twofold increase in the activity of both the DR1 and DR5 luciferase reporters, suggesting that endogenous TGIF contributes to repression of a retinoid response. In contrast, we did not observe repression of an estrogen-responsive luciferase reporter by overexpressed TGIF, nor did the TGIF siRNA vector increase the activity of this reporter, suggesting that TGIF does not repress all nuclear receptor-mediated transcription (Fig. 2E).
TGIF interacts with RXRα.
Since TGIF was able to repress both the DR1 and DR5 elements in an RXRα-dependent manner but did not bind to a DR5 element, we considered the possibility that the ability of TGIF to repress was due to an interaction with RXRα. To test whether TGIF interacts with retinoic acid receptors, we transfected COS1 cells with expression vectors encoding T7 epitope-tagged RARα or RXRα with and without a Flag-TGIF expression construct. Protein complexes were collected on anti-Flag agarose and analyzed by Western blotting for the presence of coprecipitating retinoic acid receptors. As shown in Fig. 3A, RXRα clearly coprecipitated with TGIF, and this interaction was reduced by overnight treatment of the cells with 9C-RA prior to lysis. We observed only a weak interaction of TGIF with RARα, which was also abolished in cells treated with 9C-RA for 16 h (Fig. 3A). To further examine the retinoid regulation of the interaction of TGIF with RXRα, we coexpressed both proteins in COS1 cells and either left them untreated or treated them with 9C-RA for up to 6 h. As shown in Fig. 3B, we observed little effect of 9C-RA treatment at 1 or 2 h, but the interaction of TGIF with RXRα was clearly diminished at 4 and 6 h.
FIG. 3.
TGIF interacts with RXRα. (A) COS1 cells were transfected with T7 epitope-tagged RARα or RXRα expression constructs, together with a Flag-tagged TGIF or control vector, and cells were treated with 9C-RA for 1 or 16 h (10−7 M) prior to lysis as indicated. Protein complexes were isolated on anti-Flag agarose and analyzed by Western blotting (WB) for the presence of coprecipitating RARα or RXRα. A portion of the lysates was subjected to analysis by direct Western blotting to monitor protein expression (below). +, present. (B) Cells were transfected as described for panel A, and 9C-RA was added for 1, 2, 4, or 6 h as indicated. Protein complexes were analyzed as described for panel A. (C) COS1 cells either treated for 16 h with 9C-RA (+) or left untreated (−) were lysed by sonication. Protein complexes were precipitated with a RXRα-specific monoclonal antibody and Western blotted with a TGIF-specific antiserum (above). The expression of TGIF and RXRα was monitored in the lysates by direct Western blotting (below). Arrowheads in panel C indicate specific bands; the bar indicates the heavy chain. The positions of molecular mass markers (in kDa) are shown to the left of each panel. IP, immunoprecipitation.
To test whether TGIF and RXRα interact, when expressed at endogenous levels, we immunoprecipitated RXRα from COS1 cells that had been treated with 9C-RA or left untreated. When immunoprecipitates were probed with a TGIF-specific antiserum, TGIF was readily detectable in RXRα precipitates from untreated cells (Fig. 3C). Importantly, as with the overexpressed proteins, TGIF interaction was clearly reduced with RA treatment, suggesting that the interaction is specific. This demonstrates a retinoid-regulated interaction of endogenous TGIF and RXRα. Taken together, these results suggest that TGIF can repress transcription from multiple retinoic acid response elements, including the common DR5 element, and that this repression is mediated by the retinoid regulatable interaction of TGIF with RXRα.
Identification of the interaction domains in TGIF and RXRα.
To identify which domains of TGIF and RXRα mediate the interaction, we first created a series of T7-tagged RXRα expression constructs (Fig. 4C) and tested interaction with TGIF in COS1 cells. Each of the RXRα expression constructs was coexpressed with Flag-TGIF or a control vector, and complexes were isolated on anti-Flag agarose. As shown in Fig. 4A, all constructs containing the ligand binding domain (LBD) of RXRα interacted with TGIF, whereas deletion of this region of the protein completely abolished interaction. The activation function 2 core, which is encompassed by the LBD, was also dispensable for interaction. Importantly, we detected a clear interaction with the RXRα LBD construct, which lacks the DNA binding domain, suggesting that the observed interaction is independent of DNA binding. We next tested interaction of a series of Flag-tagged TGIF expression constructs (Fig. 4D) with RXRα. For this analysis, we used the RXRα expression construct lacking the AF1 domain (ΔAF1), since this showed a clear interaction and was well separated from the immunoglobulin heavy chain. As shown in Fig. 4B, the RXRα ΔAF1 efficiently coprecipitated with all constructs containing the TGIF homeodomain but failed to interact with a construct expressing amino acids 108 to 272, which lacks the homeodomain. Together, these data demonstrate that the TGIF homeodomain interacts with the LBD of RXRα.
FIG. 4.
TGIF interacts with the RXRα ligand binding domain. (A) COS1 cells were transfected with expression constructs encoding T7 epitope-tagged RXRα deletion constructs, together with a Flag-tagged TGIF or control vector. Protein complexes were isolated on anti-Flag agarose and analyzed by Western blotting (WB) for the presence of coprecipitating T7-tagged RXRα constructs. (B) COS1 cells were transfected with T7 epitope-tagged ΔAF1 RXRα, together with a series of Flag-tagged TGIF expression constructs. Protein complexes were isolated on anti-Flag agarose and analyzed by Western blotting for the presence of coprecipitating RXRα. A portion of the lysates was subjected to analysis by direct Western blotting to monitor protein expression (below). (C) The RXRα deletion constructs used in panel A are shown schematically, together with the amino acids present in each construct. TGIF interaction is indicated to the right (+, interaction; −, none). AF1, activation function 1; DBD, DNA binding domain; LBD, ligand binding domain; AF2, activation function 2. (D) The TGIF deletion constructs are shown schematically, together with the amino acids in each and their interaction with RXRα. The two major conserved domains of vertebrate TGIFs are indicated. HD + 20, homeodomain plus 20 amino acid extension; RD2, repression domain 2; IP, immunoprecipitation.
Interaction of TGIF with RXR containing heterodimers.
To further examine the specificity of the interaction of TGIF with other nuclear receptors, we generated T7-tagged versions of RXRβ and RXRγ, both lacking the AF1 domain. COS1 cells were transfected with ΔAF1 RXRα, -β, or -γ expression vectors together with Flag-TGIF or a control vector, and proteins were collected on Flag-agarose. As shown in Fig. 5A, all three RXR isoforms interacted with TGIF, consistent with the high degree of homology between their ligand binding domains. Next, we further tested interaction with RARα and with a more distantly related RXR partner, PPARγ. COS1 cells were transfected with T7-RARα and Flag-TGIF with or without a T7-RXRα ΔAF1 expression plasmid. As shown in Fig. 5B, little interaction of RARα with TGIF was observed in the absence of coexpressed RXRα, whereas the interaction was clearly detectable in its presence. We next performed similar experiments using T7-tagged PPARγ, although here we used a TGIF expression plasmid encoding only the amino-terminal 116 amino acids, since expression of the transfected PPARγ was relatively low and this TGIF construct is also expressed at a lower level. As with RARα, only a low level of interaction of PPARγ with TGIF was detected, but this was clearly increased in the presence of coexpressed RXRα (Fig. 5C). Together, these data suggest that TGIF interacts preferentially with retinoid X receptors, and this can result in the incorporation of TGIF into heterodimeric RXR-containing nuclear receptor complexes.
FIG. 5.
TGIF and RXR cocomplexes. (A) COS1 cells were transfected with T7-tagged ΔAF1 RXRα, β, and γ constructs with or without Flag-TGIF. Complexes were isolated on Flag agarose and analyzed for coprecipitating T7-tagged RXRs. (B and C) COS1 cells were transfected with expression plasmids encoding T7-tagged RARα (B) or PPARγ (C), together with T7-RXRα ΔAF1 and Flag-TGIF, as indicated. Complexes were collected on Flag agarose and analyzed for the presence of coprecipitating T7-tagged proteins. A portion of the lysates was analyzed by direct Western blotting (below). IP, immunoprecipitation; WB, Western blotting; +, present.
Recruitment of corepressors to RXRα.
TGIF interacts with transcriptional corepressors, including CtBP (27). To test whether TGIF recruits CtBP to RXRα, COS1 cells were transfected with expression plasmids encoding T7 tagged RXRα ΔAF1, TGIF (1 to 116), and a Flag-CtBP expression construct. As shown in Fig. 6A, RXRα ΔAF1 was clearly detectable in Flag immunoprecipitates when the TGIF construct was also present, whereas little or no RXRα ΔAF1 coprecipitated with Flag-CtBP in its absence. We next performed luciferase assays using the DR1 reporter together with either a control plasmid or one expressing wild-type or mutant TGIF. The mutant TGIF plasmid encodes a protein with a single serine-to-cysteine mutation (S28C) in the CtBP binding site, which dramatically reduces interaction of CtBP with TGIF (27). Following transfection, cells were treated with a fixed concentration of RA over a 24-h time course. As shown in Fig. 6B, coexpression of TGIF repressed DR1 activity at all time points, whereas the S28C mutant form of TGIF was clearly compromised in its ability to repress transcription from this reporter. The S28C mutation in TGIF did not affect interaction of TGIF with RXRα (Fig. 6C), suggesting that the reduced repression is not due to altered interaction with RXRα. Taken together, these data suggest that TGIF recruits CtBP to RXRα to mediate repression of RXR-dependent transcriptional responses.
FIG. 6.
TGIF recruits CtBP to RXRα. (A) COS1 cells were transfected with expression plasmids encoding T7-tagged RXRα ΔAF1 and TGIF (amino acids 1 to 116) and Flag-tagged CtBP, as indicated. Proteins were precipitated on Flag agarose and analyzed for the presence of coprecipitating T7-RXRα ΔAF1. A portion of the lysates was analyzed by direct Western blotting (below). (B) HepG2 cells were transfected with a DR1-TATA-luc luciferase reporter, and expression vectors encoding RXRα and TGIF or a mutant form of TGIF (S28C), as indicated. 9C-RA was added for the indicated times prior to analysis. Luciferase activity is presented, in arbitrary units, as the mean ± standard deviation of the results from duplicate transfections. (C) COS1 cells were transfected with T7-RXRα ΔAF1 and a control vector or one encoding Flag-TGIF or the S28C mutant form of TGIF. Flag immunoprecipitates were analyzed for the presence of T7-RXRα ΔAF1, and expression of transfected proteins in the lysates is shown below. IP, immunoprecipitation; WB, Western blotting; +, present.
Disruption of the Tgif gene.
Mutations in human TGIF are associated with HPE (11), and in mice, prenatal exposure to retinoids causes anterior and neural tube defects as well as HPE (17, 42). If TGIF represses retinoic acid-regulated gene expression, its loss would be expected to mimic increased retinoic acid signaling. It is therefore possible that TGIF mutations contribute to HPE by deregulating retinoic acid-responsive gene expression. To test whether loss of TGIF function results in increased sensitivity to retinoic acid, we created a Tgif null mutation in mice. The mouse Tgif gene contains three coding exons with all but the first five codons present in the final two exons, which are separated by an intron of less than 1 kb (Fig. 7A). We therefore decided to replace most of coding exon 2 and all of exon 3 with sequences for the green fluorescent protein (GFP) and a neomycin resistance marker (Fig. 7A). GFP was fused in frame with the ninth codon of the Tgif gene, such that GFP is expressed from the native Tgif promoter. Targeted 129/SvJ embryonic stem cells were used to create Tgif mutant mice, and germ line transmission of the targeted allele was verified by breeding to wild-type C57BL/6J mice. All three expected genotypes were observed in the offspring obtained by intercrossing two Tgif heterozygotes as revealed by PCR assay of tail biopsy DNA. Analysis of the wild-type and mutant Tgif loci by restriction enzyme digestion and Southern blotting demonstrated that the targeting vector had integrated correctly, removing coding exons 2 and 3 (Fig. 7A and B). To confirm that we had created a null allele, total RNA was isolated from E13.5 embryos from intercrosses of heterozygous Tgif mutants. Northern analysis revealed that Tgif mRNA was absent from homozygous mutants and was present at about half the level in the heterozygotes as in the wild types (Fig. 7C). Additionally, GFP was observed in a reciprocal expression pattern. We also tested expression of the related Tgif2 and observed no significant change in expression level in the mutants at this stage of development. These results demonstrate that we have created a true null allele of the murine Tgif gene.
FIG. 7.
A Tgif null mutation in mice. (A) The targeting vector, Tgif locus, and targeted allele are shown, with restriction enzyme sites and predicted sizes of restriction fragments. Coding exons are shown in black, and noncoding exons are shown in gray. The arrows indicate the positions of PCR primers used for genotype analyses. The hatched gray bar indicates the position of the probe used for Southern blotting. (B) Genomic DNA from mice of the indicated genotypes was subjected to restriction enzyme digestion and Southern blotting. (C) RNA from mice of the indicated genotypes was subjected to Northern analysis with probes for Tgif, GFP, Tgif2, and Gapdh. (D) Heterozygous Tgif mutants were intercrossed, and the genotypes of the offspring at 21 days after birth were determined by PCR from genomic DNA. The positions of the PCR primers are indicated by arrows in panel A. The numbers of mice of each genotype and the percentage of the total are shown, together with the number of litters and average litter size.
We intercrossed heterozygous Tgif mutants and analyzed the frequencies of each genotype in the offspring from these crosses at postnatal day 21. Although there was a slight decrease in the number of heterozygous and homozygous mutants relative to the wild-type littermates, this deviation from the expected genotype ratios was not statistically significant (Fig. 7D), and we did not observe any defects in Tgif mutant mice. Both heterozygous and homozygous Tgif mutants appeared normal and were fertile, suggesting that in this mixed strain background, even homozygous loss of Tgif does not significantly affect viability.
Increased expression of retinoid target genes in Tgif null mice.
To test whether there was any change in gene expression in Tgif null mice compared to wild types, we first used small-scale nitrocellulose-based DNA arrays, containing around 100 genes (GE Arrays). Since there was no apparent phenotype in the null mice, we tested embryos as well as several different tissues from adult mice, including the testis (where TGIF expression is relatively high) and liver. Of the tissues tested, we observed the greatest changes in gene expression in the testis, so we used this tissue for further analysis. The gene which showed the greatest relative change, and was clearly expressed in both the wild type and mutant, was cathepsin D, which was increased 3.1-fold in the mutant compared to the wild type. However, the arrays used were designed to test a few genes involved in each of several signaling pathways and did not include well-characterized RA target genes. We therefore decided to test the expression of several known RA target genes by real-time reverse transcription-PCR (RT-PCR) (Fig. 8A) as well as to confirm the change in cathepsin D expression observed with the arrays. Using real-time RT-PCR, we observed about a threefold increase in cathepsin D expression in the mutant, confirming the array data. In addition, expression of RARβ, Hoxb1, and two Stra (stimulated by RA) (38) genes was increased in the mutant by about twofold (Fig. 8A). A third Stra gene tested (Stra13) did not change significantly in the mutant compared to the wild type. These data suggest that although there is no obvious phenotype in the Tgif null mice, there is derepression of RA target genes, at least in some tissues.
FIG. 8.
Analysis of endogenous gene expression in Tgif null cells. (A) RNA was isolated from wild-type or mutant testes and subjected to real time RT-PCR analysis. Expression of the indicated genes was analyzed in triplicate together with actin, and results are presented as expression relative to actin for the wild type and mutant, with expression of each gene arbitrarily set equal to 1 for the wild-type samples. (B) Wild-type or Tgif null mouse embryo fibroblasts were treated with (+) or without (−) RA for 16 h, as indicated. Chromatin was cross-linked with formaldehyde and subjected to immunoprecipitation with a TGIF-specific antiserum (α-TGIF) or the preimmune serum (Pre-imm). Precipitates were analyzed by PCR for the presence of a region of the RARβ promoter which spans the RARE or part of the GAPDH promoter. In addition, the input chromatin samples were analyzed with the same oligonucleotide pairs.
To test whether TGIF is recruited to the promoter of an RA target gene, we chose the RARβ gene, since this has a well-characterized DR5 element (the one used in our DR5 luciferase reporters), and its expression was increased in the mutant testis. Wild-type and mutant MEFs were isolated from E13.5 embryos and passaged three times in culture, and RA was added for the final 16 h to one-half of each primary MEF culture. Chromatin was cross-linked with formaldehyde and subjected to immunoprecipitation with a TGIF-specific antiserum (47) or preimmune serum. Following reversal of the cross-links, DNA was analyzed by PCR with oligonucleotides designed to detect the RARβ promoter (spanning the DR5 RARE) or a region of the GAPDH promoter. As shown in Fig. 8B, TGIF was present at the RARβ promoter in wild-type MEFs cultured without added RA, but this association was clearly decreased by the addition of RA. Importantly, no signal was detected in Tgif null MEFs or in any of the precipitates using the preimmune serum. Additionally, the GAPDH signal was not detected to a significant level in any of the precipitates, further confirming the specificity of the association of TGIF with the RARβ promoter. Together these data suggest that TGIF is recruited to the RARβ promoter and that loss of TGIF results in increased expression of RA target genes, including RARβ.
Increased sensitivity to retinoic acid.
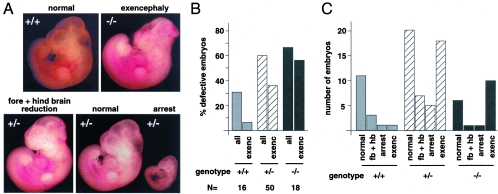
Retinoic acid is known to play an important role in anterior structure patterning and neural tube development during embryonic development, and in utero treatment with retinoic acid results in defects in anterior structure formation in the embryo (42). Based on the observed repression of retinoic acid-regulated reporters in cell culture and the increased expression of RA target genes in tissues from Tgif null mice, we reasoned that the Tgif mutant embryos may be more sensitive to increased levels of retinoic acid. To test this possibility, we intercrossed Tgif heterozygous mutants and treated the pregnant females with retinoic acid at 7.5 days of gestation. Embryos were isolated 3 days later and analyzed for both genotype and gross morphological defects. We observed defects which fell into three groups: failure of neural tube closure, developmental arrest, and a reduction of the fore- and hindbrain. Examples of these defects are shown in Fig. 9A. We observed relatively few severe defects in wild-type embryos, and only 31% of all wild-type embryos had any defect (Fig. 9B). In most cases, these were a developmental delay or brain reduction. Both the Tgif heterozygous and homozygous mutants exhibited increased rates of retinoid-induced teratogenesis, rising to 60% and 67% of heterozygotes and homozygotes, respectively (Fig. 9B). The rate of exencephaly was greater than 36% and 56% for these genotypes, whereas only one wild-type embryo (6%) had exencephaly (Fig. 9B and C). Taken together, these data demonstrate that the Tgif null embryos are more sensitive to increased levels of retinoic acid, suggesting that, during embryonic development, TGIF plays an important role in regulating the magnitude of the response to retinoic acid.
FIG. 9.
Retinoic acid teratogenesis in Tgif mutant embryos. Pregnant females, from heterozygous intercrosses, were treated with retinoic acid at gestational day 7.5. Embryos were examined for developmental defects at E10.5. (A) Embryos representative of each defect are shown. A normal wild type embryo and a homozygous mutant with exencephaly (severe open neural tube) are shown above. Below are three heterozygous mutants, one with a reduced fore and hind brain (left) and normal and arrested littermates (right). (B) The percentage of defective embryos (all defects) of each Tgif genotype is shown, together with the percentage with exencephaly. The total number of embryos of each genotype analyzed is shown below. (C) The numbers of embryos with each defect and the numbers of normal embryos are shown for each Tgif genotype. Defects: fb + hb, reduced fore and hind brain; arrest, severe developmental arrest or delay; exenc, exencephaly.
DISCUSSION
We set out to determine whether TGIF regulates normal development by modulating the response to retinoic acid signaling. We show that TGIF interacts with RXRα, recruits the general corepressor, CtBP, and inhibits retinoid responsive transcription, without the need for a TGIF binding site on DNA. In mice, the combination of a Tgif null mutation and in utero exposure to retinoic acid results in an increased frequency of developmental defects, and RA target genes are upregulated in the Tgif null animal, even in the absence of added RA.
Tgif as an RXR-specific corepressor.
TGIF was originally identified in a screen for proteins which could bind a specific DR1 RXRE from the rat CrbpII gene (3). Overexpressed TGIF was shown to reduce activation of reporter gene expression in the presence of RA and transfected RXR. The rat and mouse CrbpII RXREs have a single 6/7 match to the consensus TGIF binding site, together with four retinoid receptor binding sites. However, this TGIF site is not conserved in the human CRBPII gene, and relatively few retinoid response elements have adjacent TGIF binding sites, suggesting that this is unlikely to be a general regulatory mechanism. Importantly, TGIF sites are not found associated with canonical DR5 retinoic acid response elements. It has since been shown that TGIF is a transcriptional repressor, which can recruit multiple general corepressors, such that its recruitment to a DNA element would actively repress transcription (27, 40, 48). Here we show that TGIF is unable to bind directly to the canonical DR5 retinoic acid response element but that increased TGIF expression represses transcription from both DR1 and DR5 elements. In addition, we show by chromatin immunoprecipitation that endogenous TGIF can associate with a region of the RARβ promoter which contains the canonical DR5 element used in the transcriptional reporters. This suggests that TGIF is a general regulator of RA-responsive gene expression.
We show that TGIF interacts with RXRα, and with RXRβ and γ, suggesting that TGIF is an RXR transcriptional corepressor. Importantly, the interaction of TGIF with RXRα was readily detectable with endogenously expressed proteins. The interaction with TGIF is mediated by the RXRα LBD, which in RARα is relatively divergent at the primary amino acid level (RXRα and RARα share 31% identity over a 187-amino-acid region of their LBDs). We did see some interaction with transfected RARα, but this was clearly less than that with RXRα, and we cannot rule out that this is bridged by endogenous RXRα. Thus, TGIF interacts at least preferentially with RXR. In support of the idea that RXR bridges or enhances interaction of TGIF with other nuclear receptors, we show that coexpression of RXRα increases interaction of TGIF with both RARα and PPARγ. Thus, TGIF may target multiple RXR-containing nuclear receptor complexes via its interaction with RXR. In our reporter assays, we see specific repression of both DR1 and DR5 retinoid reporters. The DR1 reporter is more sensitive to repression by TGIF, consistent with the fact that this is activated by RXR homodimers and not RAR/RXR heterodimers. In contrast, the DR5 element, which can only be activated by RAR/RXR heterodimers, is less sensitive to repression by TGIF. TGIF is able to effectively repress the RXR-dependent activation of DR1 and DR5 elements, and it appears that this repression can be at least partially overcome by increased retinoic acid levels, consistent with a retinoid-induced switch from corepressor to coactivator binding.
The interaction of TGIF with RXRα is sensitive to the addition of retinoic acid. Within 4 h of the addition of high levels of retinoic acid, we observed a clear decrease in the interaction, and overnight exposure reduced interaction even more dramatically. Similarly, we observed an RA-sensitive association of TGIF with the RARβ promoter in mouse embryo fibroblasts. Again, this fits with a switch in RXR from corepressor to coactivator binding in the presence of ligand. Consistent with this, the interaction with RXRα is mediated by the LBD, which undergoes a significant structural alteration in the presence of ligand (7). It will be of interest to determine where within the LBD the TGIF interaction occurs and how this fits with structural changes induced by ligand binding. Taken together, our data suggest that TGIF represses RXR-dependent transcription and that this repression can be alleviated by retinoic acid, consistent with a role for TGIF as a RXR-specific corepressor. In this context, we demonstrate that TGIF can recruit CtBP to RXRα and that a single point mutation in TGIF, which abolishes interaction with CtBP (27), clearly decreased repression of a retinoid-responsive reporter by TGIF. RXR is a heterodimeric partner for many nuclear receptors, in addition to the RARs (16). Since TGIF was recruited to an RXRα-PPARγ complex by coexpression of RXRα, it is possible that TGIF plays a role in regulating RXR-dependent transcriptional responses other than those activated by retinoids. It will now be of interest to determine the extent of such regulation by TGIF.
Tgif and embryonic development.
We created a Tgif null mouse to determine the in vivo effects of TGIF loss of function. So far, we have not observed any obvious phenotypic abnormalities, in agreement with the detailed examination of another line of Tgif null mice which was recently reported (41). It is possible that in combination with a second mutation, or when the mutation is present on a different genetic background, a phenotype will be apparent. However, it should be noted that a second TGIF (TGIF2) is present, which may partially compensate for loss of TGIF (26). Here we show that, although our Tgif null mice do not have any gross abnormalities, RA target gene expression is increased relative to the wild type, at least in some tissues. Changes in gene expression observed in whole embryos were relatively small, consistent with the lack of a developmental phenotype, but by analyzing several tissues from the adult mice, we were able to demonstrate significant changes in RA target gene expression in the testis, where TGIF expression is high, suggesting that in vivo, TGIF can regulate RA-responsive gene expression. To examine the potential importance of the regulation of retinoid signaling by TGIF in embryogenesis, we treated embryos with teratogenic doses of RA. Tgif null embryos are more sensitive to treatment in utero with retinoic acid, further suggesting that, in vivo, TGIF is an important regulator of retinoic acid signaling. A similar analysis of RA teratogenesis in Tgif mutant embryos (41) did not reveal any difference in defects between heterozygous and homozygous Tgif mutant embryos. Our data are not in significant conflict with this, as we did not observe a dramatic difference between the rate of defects in heterozygous and homozygous mutants. However, the frequency of defects and, particularly, of exencephaly was clearly greater in both heterozygotes and homozygotes than in the wild-type embryos, suggesting that loss of even one copy of TGIF can sensitize an embryo to excess RA. Since in the absence of added RA, we do not see a significant level of defects in the Tgif null mice in this strain background, we suggest that loss of TGIF results in a small but significant increase in retinoic acid signaling in the embryo. In the presence of a second mutation, or environmental insult, this may result in developmental defects, either by further increasing the response to RA or by affecting a second pathway which regulates similar developmental processes. However, since loss of TGIF appears to sensitize mice to RA signaling, it is possible that, in combination with mutations in other pathways, this could contribute to developmental defects, including HPE.
HPE is a prevalent human disorder affecting craniofacial development, in which the primary defect is a failure of the ventral forebrain to divide correctly. The causes of HPE are complex and can be both genetic and environmental (36, 45). From the diversity of mutations associated with HPE, it appears that several developmental pathways are important for this disease. Mutations which cause HPE show incomplete penetrance, suggesting that a single genetic lesion is not enough to cause the disease. HPE may be caused by a combination of two or more mutations or possibly the interplay of a predisposing mutation with environmental factors. Interestingly, there is some evidence for the polygenic nature of HPE in humans. Two HPE patients with SHH mutations were also shown to have mutations of TGIF. Both had at least one phenotypically normal parent who had only a SHH mutation but was wild type for TGIF, suggesting that it is the combination of SHH and TGIF mutations that causes HPE (28). The Tgif mutants described here, or by Shen and Walsh (41), do not exhibit HPE, even when the mutation is in the homozygous state. It is noteworthy that despite the association of heterozygous SHH mutations with HPE in humans, heterozygous Shh mutations do not cause HPE in mice (6). Thus, for both TGIF and SHH, the genetic lesion which causes the human disease does not do so in mice. However, unlike Tgif, homozygous Shh null mutations cause HPE in mice. Interestingly, even the combination of a Tgif mutation and a heterozygous mutation in Shh did not appear to dramatically affect viability (41), suggesting that, at least in mice, these two mutations do not synergize.
The best characterized role of TGIF is as a transcriptional corepressor of TGF-β-activated, Smad-dependent gene expression (47). In response to TGF-β, an activated Smad complex can interact with transcriptional corepressors, such as TGIF, c-Ski or SnoN, which displace coactivators and limit the extent of transcriptional activation (49). There is evidence for a role for TGF-β signaling pathways in HPE. For example, mice with heterozygous deletions of both Smad2 and Nodal or of Smad2 and Smad3, which result in decreased TGF-β responses, have craniofacial defects similar to HPE (20, 30). Since TGIF is a Smad corepressor, any model in which loss of Tgif causes HPE by affecting TGF-β signaling is to some degree contradictory to the finding that mutations which decrease TGF-β signaling also cause HPE. Our results demonstrate that TGIF represses retinoid signaling in vivo and that loss of TGIF sensitizes embryos to neural tube defects. We suggest, therefore, that human TGIF mutations contribute to HPE by altering retinoid-regulated transcription. Loss of a repressor of retinoid signaling and exposure to excess retinoic acid would be expected to have similar outcomes. In contrast, it is decreased activation of the TGF-β pathway, not increased activation, as would be expected with loss of TGIF, that contributes to HPE. An interaction of RARγ with Smad3 has been reported, and RAR overexpression was shown to increase the transcriptional activity of Smad complexes, suggesting a Smad coactivator function for RARs (33). Since TGIF plays roles in both TGF-β and retinoid signaling, it is possible that its role is to regulate cross talk between these pathways. We have, however, so far been unable to demonstrate a specific effect of TGIF on the interaction of these transcriptional regulatory pathways. It is also possible that TGIF might affect other points of cross talk between the Smad proteins and RXR-containing nuclear receptor complexes, such as the cooperative action of RXR-VDR heterodimers and the Smads (50). However, we have been unable to demonstrate the formation of a Smad-TGIF-RXR cocomplex. We therefore suggest that TGIF is a transcriptional corepressor for both TGF-β-activated Smads and RXR-containing nuclear receptor complexes but that it plays predominantly independent roles in each of these pathways.
In summary, we have demonstrated that a Tgif mutation, in combination with prenatal exposure to retinoic acid, causes defects in neural tube development. Our results suggest a role for TGIF as a general inhibitor of retinoic acid-regulated gene expression during embryogenesis, and we show that TGIF can specifically repress RXR-dependent transcriptional activation via retinoid response elements. These data suggest that mutations in TGIF, which cause HPE in humans, may do so by disrupting RA signaling.
Acknowledgments
We thank J. Massagué, A. E. Sutherland, F. Rastinejad, and L. F. Pemberton for critical reading of the manuscript. We thank R. M. Evans and A. Yen for providing RAR and RXR cDNAs, D. Mangelsdorf for the RXRβ and γ cDNAs, and C. Rochette-Egly for the RXRα-specific antibody. We thank M. W. Mayo for advice on chromatin immunoprecipitation analysis.
This work was supported in part by research grant no. 1-FY01-243 from the March of Dimes (to D.W.) and in part by a grant from the NIH, HD39926 (to D.W.).
REFERENCES
- 1.Allenby, G., R. Janocha, S. Kazmer, J. Speck, J. F. Grippo, and A. A. Levin. 1994. Binding of 9-cis-retinoic acid and all-trans-retinoic acid to retinoic acid receptors alpha, beta, and gamma. Retinoic acid receptor gamma binds all-trans-retinoic acid preferentially over 9-cis-retinoic acid. J. Biol. Chem. 269:16689-16695. [PubMed] [Google Scholar]
- 2.Bastien, J., and C. Rochette-Egly. 2004. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene 328:1-16. [DOI] [PubMed] [Google Scholar]
- 3.Bertolino, E., B. Reimund, D. Wildt-Perinic, and R. Clerc. 1995. A novel homeobox protein which recognizes a TGT core and functionally interferes with a retinoid-responsive motif. J. Biol. Chem. 270:31178-31188. [DOI] [PubMed] [Google Scholar]
- 4.Burglin, T. R. 1997. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 25:4173-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambon, P. 1996. A decade of molecular biology of retinoic acid receptors. FASEB J. 10:940-954. [PubMed] [Google Scholar]
- 6.Chiang, C., Y. Litingtung, E. Lee, K. E. Young, J. L. Corden, H. Westphal, and P. A. Beachy. 1996. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 383:407-413. [DOI] [PubMed] [Google Scholar]
- 7.Egea, P. F., A. Mitschler, N. Rochel, M. Ruff, P. Chambon, and D. Moras. 2000. Crystal structure of the human RXRalpha ligand-binding domain bound to its natural ligand: 9-cis retinoic acid. EMBO J. 19:2592-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gehring, W. J., M. Affolter, and T. Burglin. 1994. Homeodomain proteins. Annu. Rev. Biochem. 63:487-526. [DOI] [PubMed] [Google Scholar]
- 9.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 10.Golden, J. A. 1998. Holoprosencephaly: a defect in brain patterning. J. Neuropathol. Exp. Neurol. 57:991-999. [DOI] [PubMed] [Google Scholar]
- 11.Gripp, K. W., D. Wotton, M. C. Edwards, E. Roessler, L. Ades, P. Meinecke, A. Richieri-Costa, E. H. Zackai, J. Massague, M. Muenke, and S. J. Elledge. 2000. Mutations in TGIF cause holoprosencephaly and link NODAL signalling to human neural axis determination. Nat. Genet. 25:205-208. [DOI] [PubMed] [Google Scholar]
- 12.Hayhurst, M., and S. K. McConnell. 2003. Mouse models of holoprosencephaly. Curr. Opin. Neurol. 16:135-141. [DOI] [PubMed] [Google Scholar]
- 13.Heldin, C.-H., K. Miyazono, and P. ten Dijke. 1997. TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature 390:465-471. [DOI] [PubMed] [Google Scholar]
- 14.Heyman, R. A., D. J. Mangelsdorf, J. A. Dyck, R. B. Stein, G. Eichele, R. M. Evans, and C. Thaller. 1992. 9-cis retinoic acid is a high affinity ligand for the retinoid X receptor. Cell 68:397-406. [DOI] [PubMed] [Google Scholar]
- 15.Hoberg, J. E., F. Yeung, and M. W. Mayo. 2004. SMRT derepression by the IkappaB kinase alpha: a prerequisite to NF-kappaB transcription and survival. Mol. Cell 16:245-255. [DOI] [PubMed] [Google Scholar]
- 16.Khorasanizadeh, S., and F. Rastinejad. 2001. Nuclear-receptor interactions on DNA-response elements. Trends Biochem. Sci. 26:384-390. [DOI] [PubMed] [Google Scholar]
- 17.Lammer, E. J., D. T. Chen, R. M. Hoar, N. D. Agnish, P. J. Benke, J. T. Braun, C. J. Curry, P. M. Fernhoff, A. W. Grix, Jr., I. T. Lott, et al. 1985. Retinoic acid embryopathy. N. Engl. J. Med. 313:837-841. [DOI] [PubMed] [Google Scholar]
- 18.Leid, M., P. Kastner, R. Lyons, H. Nakshatri, M. Saunders, T. Zacharewski, J. Y. Chen, A. Staub, J. M. Garnier, S. Mader, et al. 1992. Purification, cloning, and RXR identity of the HeLa cell factor with which RAR or TR heterodimerizes to bind target sequences efficiently. Cell 68:377-395. [DOI] [PubMed] [Google Scholar]
- 19.Levin, A. A., L. J. Sturzenbecker, S. Kazmer, T. Bosakowski, C. Huselton, G. Allenby, J. Speck, C. Kratzeisen, M. Rosenberger, A. Lovey, et al. 1992. 9-cis retinoic acid stereoisomer binds and activates the nuclear receptor RXR alpha. Nature 355:359-361. [DOI] [PubMed] [Google Scholar]
- 20.Liu, Y., M. Festing, J. C. Thompson, M. Hester, S. Rankin, H. M. El-Hodiri, A. M. Zorn, and M. Weinstein. 2004. Smad2 and Smad3 coordinately regulate craniofacial and endodermal development. Dev. Biol. 270:411-426. [DOI] [PubMed] [Google Scholar]
- 21.Massagué, J. 1998. TGFβ signal transduction. Annu. Rev. Biochem. 67:753-791. [DOI] [PubMed] [Google Scholar]
- 22.Massagué, J., A. Hata, and F. Liu. 1997. TGFβ signaling through the Smad pathway. Trends Cell Biol. 7:187-192. [DOI] [PubMed] [Google Scholar]
- 23.Massagué, J., and D. Wotton. 2000. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J. 19:1745-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGinnis, W., R. L. Garber, J. Wirz, A. Kuroiwa, and W. J. Gehring. 1984. A homologous protein-coding sequence in Drosophila homeotic genes and its conservation in other metazoans. Cell 37:403-408. [DOI] [PubMed] [Google Scholar]
- 25.McGinnis, W., M. S. Levine, E. Hafen, A. Kuroiwa, and W. J. Gehring. 1984. A conserved DNA sequence in homoeotic genes of the Drosophila Antennapedia and bithorax complexes. Nature 308:428-433. [DOI] [PubMed] [Google Scholar]
- 26.Melhuish, T. A., C. M. Gallo, and D. Wotton. 2001. TGIF2 interacts with histone deacetylase 1 and represses transcription. J. Biol. Chem. 276:32109-32114. [DOI] [PubMed] [Google Scholar]
- 27.Melhuish, T. A., and D. Wotton. 2000. The interaction of C-terminal binding protein with the Smad corepressor TG-interacting factor is disrupted by a holoprosencephaly mutation in TGIF. J. Biol. Chem. 275:39762-39766. [DOI] [PubMed] [Google Scholar]
- 28.Ming, J. E., and M. Muenke. 2002. Multiple hits during early embryonic development: digenic diseases and holoprosencephaly. Am. J. Hum. Genet. 71:1017-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muenke, M., and P. A. Beachy. 2000. Genetics of ventral forebrain development and holoprosencephaly. Curr. Opin. Genet. Dev. 10:262-269. [DOI] [PubMed] [Google Scholar]
- 30.Nomura, M., and E. Li. 1998. Smad2 role in mesoderm formation, left-right patterning and craniofacial development. Nature 393:786-790. [DOI] [PubMed] [Google Scholar]
- 31.Passner, J. M., H. D. Ryoo, L. Shen, R. S. Mann, and A. K. Aggarwal. 1999. Structure of a DNA-bound Ultrabithorax-Extradenticle homeodomain complex. Nature 397:714-719. [DOI] [PubMed] [Google Scholar]
- 32.Pavri, R., B. Lewis, T. K. Kim, F. J. Dilworth, H. Erdjument-Bromage, P. Tempst, G. de Murcia, R. Evans, P. Chambon, and D. Reinberg. 2005. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol. Cell 18:83-96. [DOI] [PubMed] [Google Scholar]
- 33.Pendaries, V., F. Verrecchia, S. Michel, and A. Mauviel. 2003. Retinoic acid receptors interfere with the TGF-beta/Smad signaling pathway in a ligand-specific manner. Oncogene 22:8212-8220. [DOI] [PubMed] [Google Scholar]
- 34.Piper, D. E., A. H. Batchelor, C.-P. Chang, M. L. Cleary, and C. Wolberger. 1999. Structure of a HoxB1-Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell 96:587-597. [DOI] [PubMed] [Google Scholar]
- 35.Rastinejad, F. 2001. Retinoid X receptor and its partners in the nuclear receptor family. Curr. Opin. Struct. Biol. 11:33-38. [DOI] [PubMed] [Google Scholar]
- 36.Roessler, E., and M. Muenke. 1998. Holoprosencephaly: a paradigm for the complex genetics of brain development. J. Inherit. Metab. Dis. 21:481-497. [DOI] [PubMed] [Google Scholar]
- 37.Rubenstein, J. L., and P. A. Beachy. 1998. Patterning of the embryonic forebrain. Curr. Opin. Neurobiol. 8:18-26. [DOI] [PubMed] [Google Scholar]
- 38.Sapin, V., P. Bouillet, M. Oulad-Abdelghani, B. Dastugue, P. Chambon, and P. Dolle. 2000. Differential expression of retinoic acid-inducible (Stra) genes during mouse placentation. Mech. Dev. 92:295-299. [DOI] [PubMed] [Google Scholar]
- 39.Seo, S. R., F. Lallemand, N. Ferrand, M. Pessah, S. L'Hoste, J. Camonis, and A. Atfi. 2004. The novel E3 ubiquitin ligase Tiul1 associates with TGIF to target Smad2 for degradation. EMBO J. 23:3780-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma, M., and Z. Sun. 2001. 5′TG3′ interacting factor interacts with Sin3A and represses AR-mediated transcription. Mol. Endocrinol. 15:1918-1928. [DOI] [PubMed] [Google Scholar]
- 41.Shen, J., and C. A. Walsh. 2005. Targeted disruption of Tgif, the mouse ortholog of a human holoprosencephaly gene, does not result in holoprosencephaly in mice. Mol. Cell. Biol. 25:3639-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sulik, K. K., D. B. Dehart, J. M. Rogers, and N. Chernoff. 1995. Teratogenicity of low doses of all-trans retinoic acid in presomite mouse embryos. Teratology 51:398-403. [DOI] [PubMed] [Google Scholar]
- 43.Truett, G. E., P. Heeger, R. L. Mynatt, A. A. Truett, J. A. Walker, and M. L. Warman. 2000. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). BioTechniques 29:52, 54. [DOI] [PubMed] [Google Scholar]
- 44.Umesono, K., K. K. Murakami, C. C. Thompson, and R. M. Evans. 1991. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell 65:1255-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallis, D. E., and M. Muenke. 1999. Molecular mechanisms of holoprosencephaly. Mol. Genet. Metab. 68:126-138. [DOI] [PubMed] [Google Scholar]
- 46.Wotton, D., P. S. Knoepfler, C. D. Laherty, R. N. Eisenman, and J. Massague. 2001. The Smad transcriptional corepressor TGIF recruits mSin3. Cell Growth Differ. 12:457-463. [PubMed] [Google Scholar]
- 47.Wotton, D., R. S. Lo, S. Lee, and J. Massague. 1999. A Smad transcriptional corepressor. Cell 97:29-39. [DOI] [PubMed] [Google Scholar]
- 48.Wotton, D., R. S. Lo, L. A. Swaby, and J. Massague. 1999. Multiple modes of repression by the smad transcriptional corepressor TGIF. J. Biol. Chem. 274:37105-37110. [DOI] [PubMed] [Google Scholar]
- 49.Wotton, D., and J. Massague. 2001. Smad transcriptional corepressors in TGF beta family signaling. Curr. Top. Microbiol. Immunol. 254:145-164. [PubMed] [Google Scholar]
- 50.Yanagisawa, J., Y. Yanagi, Y. Masuhiro, M. Suzawa, M. Watanabe, K. Kashiwagi, T. Toriyabe, M. Kawabata, K. Miyazono, and S. Kato. 1999. Convergence of transforming growth factor-beta and vitamin D signaling pathways on SMAD transcriptional coactivators. Science 283:1317-1321. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, Y., and R. Derynck. 1999. Regulation of Smad signaling by protein associations and signaling crosstalk. Trends Cell Biol. 9:274-279. [DOI] [PubMed] [Google Scholar]