Abstract
Previous results indicated that translation of four mitochondrion-encoded genes and one nucleus-encoded gene (COX4) is repressed in mutants (pgs1Δ) of Saccharomyces cerevisiae lacking phosphatidylglycerol and cardiolipin. COX4 translation was studied here using a mitochondrially targeted green fluorescence protein (mtGFP) fused to the COX4 promoter and its 5′ and 3′ untranslated regions (UTRs). Lack of mtGFP expression independent of carbon source and strain background was established to be at the translational level. The translational defect was not due to deficiency of mitochondrial respiratory function but was rather caused directly by the lack of phosphatidylglycerol and cardiolipin in mitochondrial membranes. Reintroduction of a functional PGS1 gene under control of the ADH1 promoter restored phosphatidylglycerol synthesis and expression of mtGFP. Deletion analysis of the 5′ UTRCOX4 revealed the presence of a 50-nucleotide fragment with two stem-loops as a cis-element inhibiting COX4 translation. Binding of a protein factor(s) specifically to this sequence was observed with cytoplasm from pgs1Δ but not PGS1 cells. Using HIS3 and lacZ as reporters, extragenic spontaneous recessive mutations that allowed expression of His3p and β-galactosidase were isolated, which appeared to be loss-of-function mutations, suggesting that the genes mutated may encode the trans factors that bind to the cis element in pgs1Δ cells.
Cardiolipin (CL) and its precursor phosphatidylglycerol (PG) are anionic phospholipids whose synthesis and distribution are limited primarily to the mitochondrial inner membrane of eukaryotic cells (25). As the major anionic phospholipid in the mitochondrial inner membrane, CL is specifically associated with a number of mitochondrial proteins, many of which also require CL for their optimal functions (4, 12, 16, 26, 32, 55). Together with PG, CL has been postulated or demonstrated to participate in many critical mitochondrial processes, such as maintaining membrane permeability and integrity (25, 33), solute transport (4, 31, 47), oxidative phosphorylation (35, 72), protein and phospholipid import (2, 13), and mitochondrion-mediated apoptosis (46). Due to its highly unsaturated fatty acyl chains, CL is considered an “antioxidant” and the primary lipid peroxidation target in mitochondria. Reduction in CL levels by oxidative damage or other means is correlated with negative aspects of several physiological states, such as Barth's syndrome (67), ischemia/reperfusion (41), and aging (50, 60).
Saccharomyces cerevisiae mutants disrupted in the PGS1 (encodes phosphatidylglycerophosphate synthase) (7) or CRD1 (encodes CL synthase) (8, 30, 64) gene have been created, and both manifest growth defects on nonfermentable carbon sources, indicating a central role for these lipids in oxidative phosphorylation. Yeast mutants (crd1Δ) deficient only in CL display a 7- to 10-fold increase in PG that may partially complement some of the CL functions, thereby masking the full effect of lack of these anionic phospholipids. Despite complementation by PG, CL has been shown to be essential under stress conditions (73) or for efficient coupling between complexes III and IV of the respiratory chain (52, 72). Yeast mutants (pgs1Δ) deficient in both PG and CL display more severe defects in mitochondrial function, such as slow growth on a fermentable carbon source and total inability to respire on nonfermentable carbon sources. One of the reasons for the lack of growth of pgs1Δ cells on nonfermentable carbon sources appears to be due to an initial reduction in the level of respiratory components (48) and eventual loss of mitochondrial DNA (mtDNA) (74).
In yeast, cytochrome c oxidase (complex IV) is a multisubunit complex consisting of 11 subunits (19). Subunits I, II, and III (Cox1p to Cox3p), comprising the catalytic core of the complex, are encoded by the mitochondrial genome, with the remainder, including subunit IV (Cox4p), encoded by the nuclear genome. In pgs1Δ cells, four of mitochondrion-encoded proteins (Cox1p to Cox3p and Cobp, a subunit of cytochrome bc1 (complex III)) and one nucleus-encoded protein (Cox4p) were reported missing (48). Although previous results were consistent with a block in translation of COX4 mRNA, rapid degradation of the unassembled components of complex IV (11) in the absence of the essential structural component, CL (57), was not completely excluded.
If lack of expression of nucleus-encoded Cox4p is due to a block in translation in pgs1Δ cells, how can the cytoplasmic translational machinery “sense” changes in mitochondrial phospholipid composition and repress translation of certain proteins that are targeted to mitochondria, where they are assembled into oxidative complexes together with the mitochondrion-encoded subunits? Cross talk between mitochondria and the nucleus is well documented (5, 15, 34, 51, 54). Mitochondrial stress such as mtDNA mutations and inhibition of respiration or mitochondrial gene expression can serve as signals transmitted to the nucleus to induce the expression of specific nuclear genes that in turn help mitochondria cope with stress (3, 14, 53). Most of this type of interorganelle signaling studied to date has involved regulation at the transcriptional level, but the mRNAs of some nucleus-encoded mitochondrial proteins are translated while bound to the mitochondrial membrane, possibly making their translation sensitive to mitochondrial membrane lipid composition. However, COX4 was not identified within the subset of nuclear genes which displayed a high value for mitochondrial localization of mRNA (MLR) in a genome-wide DNA microarray analysis (45), even though import of Cox4p into mitochondria is greatly enhanced cotranslationally compared to posttranslationally in a homologous yeast in vitro system (1, 17, 18). The MLR value for COX4 is 26.7 (on a scale of 100), similar to POR1 (encodes mitochondrial porin), with a MLR value of 19, suggesting that the majority of COX4 mRNA may be translated on free ribosomes in the cytoplasm.
In this study, we focused on the mechanism of repression of nucleus-encoded Cox4p expression in response to changes in mitochondrial anionic phospholipid content. Cox4p mRNA contains an intron in the 5′ untranslated region (UTR) (56) and encodes a cytoplasmically translated protein product with an amino-terminal mitochondrial targeting sequence. Our results establish suppression of Cox4p translational in response to lack of mitochondrial PG and CL (pgs1Δ cells) and support a novel cross talk mechanism, which regulates cytoplasmic translation of COX4 mRNA through binding of a nucleus-encoded trans-acting repressor protein(s) to a sequence in the 5′ UTR of the mature mRNA.
MATERIALS AND METHODS
Strains and growth conditions.
The S. cerevisiae strains used in this study are listed in Table 1. YPH499, YPH500, and DL1 are wild-type yeast strains with respect to the PGS1 locus. YZD1 and YCD4 are pgs1Δ strains lacking PG and CL. HMD22, DFS168, and DL1[rho−] are strains deficient in mitochondrial respiratory function. Yeast cells were grown at 30°C either in rich medium containing 1% yeast extract and 2% peptone (YP) or in complete synthetic medium (CSM) with selective amino acid dropouts (as indicated), both supplemented with either 2% dextrose (D), 2% sucrose (S), or 0.5% ethanol-3% glycerol (EG). Escherichia coli strain DH5α was grown in LB medium (1% tryptone, 0.5% yeast extract, 1% NaCl, pH 7.4) at 37°C with ampicillin (100 μg/ml) as a selection marker for plasmid preparation.
TABLE 1.
Yeast strains and plasmids
| Plasmid or strain | Characteristics or genotype | Source or reference |
|---|---|---|
| Strains | ||
| YPH499 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ63 ura3-52 | 61 |
| YPH500 | MATα ade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ63 ura3-52 | 61 |
| DL1 | MATahis3-11 15 leu2-3 12 ura3-251 328, 372 | 66 |
| YZD1 | MATapgs1::TRP1, derivative of YPH499 | 65 |
| YCD4 | MATapgs1::HIS3, derivative of DL1 | 7 |
| DL1[rho] | MATarho− derivative of DL1 | 59 |
| HMD22 | MATaura3-52 leu2-3,112 lys2 his3HindIII arg8::hisG cox2::ARG8m | 6 |
| DFS168 | MATaleu2-3,112 ura3Δ his4-519 arg8::URA3 cox3::ARG8m | 62 |
| YMS1 | MATa, lrg1 derivative of YZD1 | This work |
| YMS2 | MATa, lrg2 derivative of YZD1 | This work |
| Plasmids | ||
| pCG1 | PCOX4-mtGFP LEU2 CEN | This work |
| pCG2 | Derivative of pCG1, 342-bp intron deleted | This work |
| pCG3 | Derivative of pCG1, 102-bp 5′ UTR plus 342-bp intron deleted | This work |
| pCG4 | Derivative of pCG3, 31 bp deleted from the 3′ end of the 5′ UTR | This work |
| pCG5 | Derivative of pCG3, 56 bp deleted from the 3′ end of the 5′ UTR | This work |
| pCG6 | Derivative of pCG3, 81 bp deleted from the 3′ end of the 5′ UTR | This work |
| pCG7 | Derivative of pCG3, 28 bp deleted from the 5′ end of the 5′ UTR | This work |
| pCG8 | Derivative of pCG3, 53 bp deleted from the 3′ end of the 5′ UTR | This work |
| pCH1 | PCOX4-HIS3 LEU2 CEN | This work |
| pCL1 | PCOX4-lacZ URA3 2μm and ColE1 origin | 73 |
| pAHO1 | PADH1-HO URA3 2μm | This work |
| pAGS1 | PADH1-PGS1 URA3 2μm | This work |
Plasmids.
Plasmid pCG1 was derived from plasmid pRS315 (61). A 1.2-kb DNA fragment including PCOX4 (promoter for COX4) and encoding the 5′ UTRCOX4 along with the first 25 codons of COX4 (encodes the mitochondrial targeting sequence) and a 500-bp DNA fragment including the downstream 3′ UTRCOX4 were isolated by PCR and fused to 5′ and 3′ of the open reading frame (ORF) of the green fluorescent protein (GFP) gene, respectively. The resulting plasmid, pCG1 (PCOX4-mtGFP), encodes a green fluorescent protein coded for import into the mitochondria (mtGFP). Plasmid pCG2 was constructed by deleting from plasmid pCG1 the DNA fragment corresponding to the 342-bp 5′ intron of the COX4 gene (56). Plasmids pCG3 to pCG8 are a series of deletions of pCG2 in the region encoding the 5′ UTRCOX4. Plasmid pCG3 contains a large deletion of the 5′ UTR (102-bp sequence upstream of the intron). Plasmids pCG4 to pCG8 were created by further deleting from plasmid pCG2 a 31-, 56-, or 81-bp fragment from the 3′ end or a 28- or 53-bp fragment from the 5′ end of the 109-bp sequence preceding the intron, respectively. An identical set of plasmids lacking the 3′ UTRCOX4 was also constructed. For the construction of plasmid pCH1 (PCOX4-HIS3 LEU2 ARS/CEN), the ORF of HIS3 was fused directly to the start codon of COX4 under control of PCOX4 and the 5′ UTRCOX4; these plasmids also lacked the 3′ UTRCOX4. Therefore, unlike plasmid pCG1, where the GFP is targeted to mitochondria by the classic Cox4p mitochondrial targeting sequence, the HIS3 gene product from plasmid pCH1 remains in the cytoplasm. Plasmid pCL1 (PCOX4-lacZ URA3 2μm) was created as reported before (73) and contains the same upstream regulatory information preceding lacZ as in plasmid pCH1. Plasmid pAHO1 (PADH1-HO URA3 2μm) is a derivative of pVT100u-mtGFP (68) and was used in mating type switching experiments. HO DNA was amplified using primers 5′-AAAAAGCTTATGCTTTCTGAAAACACGACTATTC-3′ and 5′-TTTTCTAGATTAGCAGATGCGCGCACCTGC-3′ from the plasmid pGAL1-HO (24), and the HindIII-NotI (sites introduced on primers) fragment was isolated and subcloned between the HindIII-NotI cleavage sites of pVT100u-mtGFP, replacing the segment encoding mtGFP. Plasmid pAGS1 (PADH1-PGS1 URA3 2μm) was also derived from pVT100u-mtGFP and was constructed directly through yeast homologous recombination (22). First, the PGS1 gene was amplified by PCR from strain DL1 genomic DNA by using sense primer 5′-ttttctgcacaatatttcaagctataccaagcatacaatcaactccaaGCTTAATAGCATACTCAGGATAACAT-3′ and antisense primer 5′-ggagacttgaccaaacctctggcgaagaagtccaaagctggatcctcTACAGGCGACATACTATGATAGAATA-3′ (PGS1 homology regions are in uppercase, and regions homologous to plasmid pVT100u-mtGFP are in lowercase). The amplified fragment was purified and transformed into strain DL1 together with HindIII- and XbaI-digested plasmid pVT100u-mtGFP by using the Alkali-Cation yeast kit (Q-Biogene). Transformants containing circularized recombinant plasmid were selected on CSMD-Ura plates, and plasmids were isolated from yeast directly using the E.Z.N.A yeast plasmid kit (Omega Bio-Tek, Inc.). DNA sequencing of the plasmids confirmed proper gene organization.
Isolation of mitochondria and preparation of mitoplasts.
Mitochondria were isolated from Zymolyase-lysed yeast cells by differential centrifugation as previously reported (20). Mitoplasts were prepared from isolated mitochondria by digitonin treatment followed by osmotic shock. In brief, mitochondria were resuspended at 5 mg/ml in SEM buffer (0.25 M sucrose, 0.5 M EDTA, 10 mM MOPS [morpholinepropanesulfonic acid], pH 7.2) and treated with digitonin at a ratio of digitonin to protein of 0.3 mg/mg. Mitochondria were then diluted 10-fold into hypotonic buffer (20 mM HEPES, pH 7.4, and 1 mM ATP) and incubated on ice for 30 min. Mitoplasts were recovered by centrifugation at 14,000 × g for 10 min at 4°C and resuspended in SEM buffer.
Western blotting analysis.
Forty micrograms of mitochondrial protein was subjected to sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis. Following electrophoresis, proteins were transferred to nitrocellulose sheets electrophoretically. Sheets were blocked in 5% bovine serum albumin overnight; probed with primary antibodies against GFP, porin (Molecular Probes), or Arg8p (kindly provided by Thomas Fox, Cornell University); and detected using horseradish peroxidase-coupled secondary antibodies. Nitrocellulose membranes were then visualized by SuperSignal West Pico chemiluminescence (Amersham Pharmacia Biotech) and quantified using a Bio-Rad Fluro-Max system.
Northern blotting analysis.
Total RNA was isolated from 10 ml of mid-log-phase yeast cells grown in YPD by using the RNeasy minikit (QIAGEN, Inc.). Fifteen micrograms of RNA was subjected to 1.2% agarose formaldehyde gel electrophoresis (59). RNA was transferred to positively charged nylon membranes (Roche Diagnostics) and UV cross-linked for one cycle. [32P]DNA probes were synthesized from PCR-amplified templates by using a random-primed DNA labeling kit (Roche Diagnostics). Primers used to generate template for probe synthesis were as follows: COX4 (sense, 5′-CTTTCACTACGTCAATCTAT-3′; antisense, 5′-GTGATGGTGGTCATCATTTG-3′), mtGFP (sense, 5′-AGTAAAGGAGAACTTTTCAC-3′; antisense, 5′-GTATAGTTCATCCATGCCA-3′), and TCM1 (sense, 5′-TCTCACAGAAAGTACGAAGC-3′; antisense, 5′-TTCAGAGATGGAACCACCGT-3′). Finally, hybridization was done using Rapid-Hyb (Amersham) according to the manufacturer's instructions.
RT-PCR and Q-RT-PCR analyses.
Isolated RNA was treated with DNase I to eliminate DNA contamination. For reverse transcriptase PCR (RT-PCR) analysis, a Superscript one-step RT-PCR kit (Invitrogen) was used with primers (sense, 5′-TACGAAGATTAGAATTTTTTTCA-3′; antisense, 5′-GGAACGGTACCCTC TTTAGC-3′) against COX4 mRNA. For real time quantitative RT-PCR (Q-RT-PCR), mtGFP and TCM1 transcripts were analyzed on the Smart Cycler (Cepheid) using a QuantiTect SYBR Green RT-PCR kit according to the manufacturer's instructions. One microgram of total RNA was used in Q-RT-PCR with primers for mtGFP (sense, 5′-GAACTTTTCACTGGAGTGGT-3′; antisense, 5′-CATGGAACAGGTAGCTTTCC-3′) and primers for TCM1 (sense, 5′-TCTCACAGAA AGTACGAAGC-3′; antisense, 5′-ACAATGGTGGTCATACCAGC-3′). PCR products were confirmed by melting curve analysis. Standard curves were generated from PCR amplification of template dilutions. Final data were normalized to those for TCM1 and presented as percentages of the wild-type levels.
RNA electrophoretic mobility shift assay.
Synthesis of the 32P-labeled 104-nucleotide (nt) RNA probe representing the mature 5′ UTRCOX4 was accomplished using the RiboScribe T7 RNA probe synthesis kit (Epicentre). The DNA template used for probe synthesis was amplified from plasmid pCG2 by PCR with primers 5′-cagcgtaatacgactcactatagggagaATACGAAGATTAGAATTTTTTTCATAC-3′ (sense; the T7 promoter is underlined) and 5′-ATTTCAAAATAATCTTATTTCCTGTT-3′ (antisense). A control RNA was also created with approximately the same length but missing 81 nt in the middle of the 5′ UTRCOX4. This RNA was transcribed from template amplified from plasmid pCG6 using the same sense primer as above and the antisense primer 5′-GATATCTAGAGCTACACAAAGTTCT-3′. Crude yeast extract was isolated from mid-log-phase wild-type and pgs1Δ cells grown in YPD by vortexing with glass beads in homogenization buffer (50 mM NaHPO4, pH 7.4, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 5% glycerol) and centrifuged twice at 1,500 × g for 5 min at 4°C. RNA binding assay was done by mixing 5 μg of total yeast extract and 0.5 μl of labeled RNA probe in 10 μl of binding buffer (10 mM HEPES, pH 7.6, 3 mM MgCl2, 100 mM KCl, 5 mM EDTA, 2 mM dithiothreitol, 5% glycerol, 0.5% NP-40, 1.5 μg/μl heparin, 0.2 μg/μl yeast tRNA) in the absence or presence of 20 μg of protease K and incubating at room temperature for 15 min. The reaction mixture was then loaded on a 5% polyacrylamide gel and run in TBE (45 mM Tris-borate and 1 mM EDTA) for 30 min at 4°C at 250 V. After electrophoresis, the gel was dried for 1 h at 80°C, exposed to a storage phosphorimaging screen, and analyzed using a Bio-Rad phosphorimager.
Isolation of spontaneous mutations.
Plasmid pCH1 was introduced into strain YZD1 by transformation, and transformants were selected on CSMD-Leu medium. Cells were then plated to CSMD-His containing 50 mM 3-aminotriazole (3AT) to eliminate weak growth due to leaky expression of HIS3. After 7 to 10 days, colonies that appeared were picked for subsequent analysis.
Mating switch and mating analysis.
To switch mating type, plasmid pAHO1 (URA3) was transformed into yeast cells (YPH499, YZD1, YMS1, and YMS2) containing plasmid pCH1 (LEU2). Transformants were selected on CSMD-Ura-Leu plates and grown in YPD medium for a few days to allow for expression of HO activity. Loss of plasmid pAHO1 from some of the cells was subsequently selected on CSMD-Leu plates containing 0.1% 5-fluoro-orotic acid. Colonies formed on these plates were then crossed with strain YZD1 containing plasmid pCL1 (URA3). To determine the complementation between strain YMS1 and YMS2, YMS1 derived from the mating switch experiment (bearing plasmids pCH1 and pAHO1) was screened for cells that had lost plasmid pCH1. The selected YMS1 cells bearing pAHO1 were then mated with YMS2 carrying plasmid pCH1. Diploid cells formed due to mating were selected on CSMD-Ura-Leu double-dropout plates.
Other methods.
Phosphatidylglycerophosphate synthase activity was determined at 30°C using isolated mitochondria or mitoplasts as previously described (7). Phospholipids were extracted from whole cells, and the extracted lipids were analyzed by thin-layer chromatography (48). β-Galactosidase activity was assayed in cell lysates as previously described (58), using o-nitrophenyl-β-galactoside as a substrate. Protein concentration was determined throughout using the Bradford method with bovine serum albumin as the standard.
RESULTS
Effect of carbon source and mitochondrial function on the expression of mtGFP.
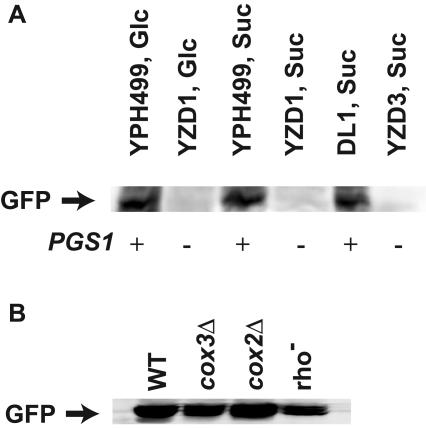
It was shown previously that Cox4p was barely detectable in a pgs1Δ background, and evidence was presented that the reduced COX4 expression occurred through inhibition at the level of translation, since message levels were near normal (48). Translational control is a mechanism widely used by cells to control their gene expression, and much of this regulation is conferred by the respective 5′ and/or 3′ UTRs of mRNAs expressed by the regulated genes (69, 70). In this study, we explored the possibility that cis-acting elements residing in the 5′ and/or 3′ UTRs of COX4 mRNA may be responsible for the translational control of COX4 expression. Cox4p is unstable when not assembled with mitochondrially encoded subunits of cytochrome c oxidase (40), which are also missing in pgs1Δ cells (48), and rapid degradation of Cox4p could occur either during or after synthesis. To circumvent this problem, mtGFP, a soluble monomeric foreign protein in yeast, was used as a reporter protein. mtGFP was placed under the control of PCOX4 and its 5′ and 3′ UTRs to test gene expression regulated by cis-acting elements in COX4 mRNA. Plasmid pCG1 expressing the above mtGFP construct was transformed into two different pgs1Δ strains (YZD1 and YCD4) and their respective PGS1 parental strains and grown in either YPS (to eliminate glucose repression of mitochondrial function) or YPD medium. Mitochondria were isolated, and mtGFP expressed was probed by Western blotting analysis. The results (Fig. 1A) showed that mtGFP was expressed well in PGS1 cells but was barely detectable in pgs1Δ cells, independent of the carbon source used and the cell background in which the pgs1Δ was generated; proper mitochondrial targeting of GFP was apparent in all cases by the presence of only the mature processed form. This result is consistent with the previously determined expression pattern of endogenous Cox4p in these cells (48). Furthermore, the lack of detection of mtGFP in a pgs1Δ background rules out increased instability of Cox4p in a pgs1Δ background as the basis for lack of detection and definitively demonstrates translational regulation.
FIG. 1.
mtGFP expression in response to different strain backgrounds and carbon sources. Mitochondria were isolated from mid-log-phase-grown cells and subjected to Western blotting analysis as described in Materials and Methods. mtGFP was detected by antibody against GFP. (A) Expression of mtGFP in YPH499 (wild type), YZD1 (pgs1Δ), DL1 (wild type), and YCD4 (pgs1Δ) grown in CSM with either glucose (Glc) or sucrose (Suc) as a carbon source. (B) Expression of mtGFP in cells with dysfunctional mitochondria, i.e., DFS168 (cox3Δ), HMD22 (cox2Δ), and DL1[rho−], grown in CSM with glucose as a carbon source. WT, wild type.
To determine if the lack of expression is a direct result of PG and CL deficiency or a secondary effect due to respiratory deficiency or loss of mtDNA that is caused by lack of these lipids, the expression of the fusion construct was compared in the three respiration-deficient strains DFS168 (cox3Δ), HMD22 (cox2Δ), and DL1[rho−] (mtDNA lesions introduced by growth on ethidium bromide) and in wild-type cells. Unlike pgs1Δ cells, all three respiration-deficient strains displayed significant mtGFP expression (Fig. 1B) although there was some reduction in the cox3Δ and rho− strains relative to the wild type. These results establish that lack of Cox4p is not due to respiratory deficiency.
Supplementation of PG and CL to mitochondria through a plasmid-borne copy of PGS1 restores mtGFP expression.
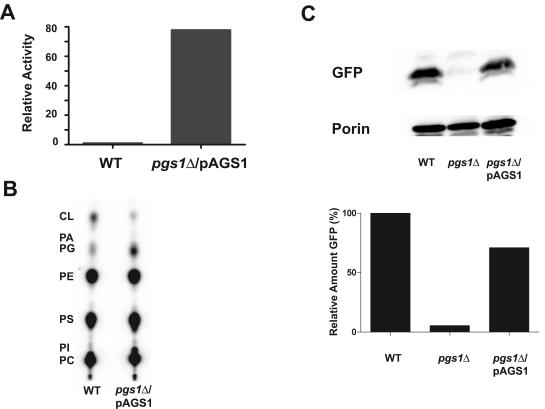
To further study dependence of mtGFP expression on anionic phospholipids, PG and CL were restored to mitochondria of pgs1Δ cells by supplying a PGS1 gene under control of the constitutive PADH1. Introduction of plasmid pAGS1 (PADH1-PGS1) resulted in a ∼60-fold increase in phosphatidylglycerophosphate synthase activity compared with that in wild-type cells (Fig. 2A) and restoration of PG content in pgs1Δ cells as revealed by total phospholipid analysis (Fig. 2B). Although the PG level was significantly elevated, the CL content, for some reason, was only slightly increased. In addition, pgs1Δ cells carrying this plasmid remained unable to grow on a nonfermentable carbon source (YPEG medium), indicating that the lack of PG and CL caused permanent damage to cells, which prevented the return of respiratory competence even after supplementation with PG. Consistent with this observation is the report that some yeast cells deficient in PG and CL lose mtDNA (74). We confirmed the lack of mitochondrially encoded mRNA transcripts by RT-PCR analysis of YZD1 (pgs1Δ) cells in contrast to wild-type cells, consistent with YZD1 cells lacking most if not all their mtDNA (data not shown). Another possible explanation for the observed phenotype is that Pgs1p supplied via this plasmid was not properly targeted to the mitochondrial inner membrane. Therefore, although the PG level was increased in whole cells, CL was not restored to wild-type levels because CL synthase is exclusively an enzyme of the mitochondrial inner membrane (8). This possibility was ruled out by preparing mitoplasts that are comprised mainly of mitochondrial inner membrane. Removal of the mitochondrial outer membrane did not decrease phosphatidylglycerophosphate synthase activity in the mitoplast fraction (data not shown), ruling out Pgs1p mislocalization. Despite the apparent lack of CL and mitochondrial respiratory function, the expression of mtGFP was restored to 70% of wild-type levels in pgs1Δ cells in which PG synthesis was restored (Fig. 2C); detection of porin, which is a nucleus-encoded mitochondrial outer membrane protein, was used as an internal control. The data further strongly support the conclusion that expression of the mtGFP reporter of COX4 gene expression is dependent on the presence of anionic phospholipids in the mitochondrial membrane and independent of mitochondrial respiratory function.
FIG. 2.
mtGFP expression is increased upon restoration of PG and CL through a plasmid-borne copy of PGS1. (A) Mitochondria were isolated from mid-log-phase cells grown in CSMD, and phosphatidylglycerophosphate synthase activity was assayed using 50 μg of mitochondrial protein as described in Materials and Methods. Enzyme activity in YZD1 (pgs1Δ) carrying the PGS1 gene under control of PADH1 (pAGS1) is expressed relative to activity in YPH500 (wild type [WT]) cells; YZD1 alone has no detectable activity. (B) Total yeast phospholipid (32P labeled) was extracted from YPH500 (WT) and YZD1 (pgs1Δ) carrying plasmid pAGS1 (PGS1). Phospholipid analysis was performed as described in Materials and Methods, relative to mobility of standards CL, PG, phosphatidic acid (PA), phosphatidylethanolamine (PE), phosphatidylserine (PS), phosphatidylinositol (PI), and phosphatidylcholine (PC). (C) Expression of mtGFP in mitochondria isolated from YPH500 (WT), YZD1 (pgs1Δ), and YZD1/pAGS1 as described for Fig. 1. The upper panel shows a Western blot probed with anti-GFP antibody and antiporin antibody, and the lower panel shows the quantification of the Western blot using the Bio-Rad Quantity One program. GFP was normalized to the porin control, with wild-type cell levels set to 100%.
The 5′ UTRCOX4 controls the translation of reporter genes.
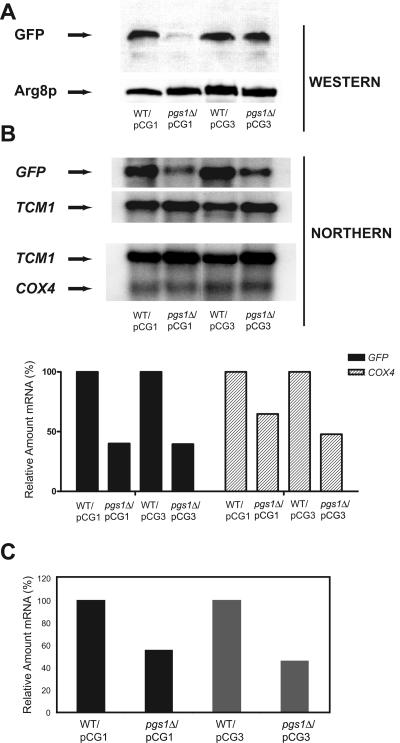
Next, we tested whether the 5′ UTR or the 3′ UTR controls reporter gene expression. For this purpose, a construct was made in which the 342-nt intron and the 102-nt fragment upstream of the intron were deleted from the 5′ UTRCOX4. Expression of this construct (pCG3) yields an mRNA with a very short 5′ UTR (∼20 nt). The level of mtGFP expressed from this construct in pgs1Δ cells was comparable to that in wild-type cells (Fig. 3A), while a construct with the full-length 5′ UTR (pCG1) barely expressed mtGFP in pgs1Δ cells; detection of Arg8p, which is a nucleus-encoded mitochondrial matrix protein (29), was used as a control. This indicates that the 5′ UTRCOX4 contains a negative cis-acting element that inhibits downstream gene expression in pgs1Δ cells. A parallel set of plasmid constructs lacking the 3′ UTRCOX4 gave the same expression pattern as observed in Fig. 3A, ruling out an involvement of the 3′ UTRCOX4 in translational regulation (data not shown).
FIG. 3.
Expression of mtGFP is controlled at the translational level by a cis element located in the 5′ UTRCOX4. Cells were grown to mid-log phase in CSMD-Leu medium. Mitochondria and RNA were isolated as described in Materials and Methods. (A) mtGFP present in mitochondria isolated from YPH500/pCG1 (wild type [WT]), YZD1/pCG1 (pgs1Δ), YPH500/pCG3 (WT), and YZD1/pCG3 (pgs1Δ) was detected by Western blotting analysis as described in Materials and Methods. Plasmid pCG1 contains the full-length 5′ UTRCOX4, and plasmid pCG3 lacks most of the 5′ UTRCOX4 upstream of mtGFP. Mitochondrial matrix protein Arg8p was used as a loading control. (B) Northern blotting analysis using RNAs isolated from YPH500/pCG1, YZD1/pCG1, YPH500/pCG3, and YZD1/pCG3. The Northern blotting analysis was carried out by hybridization with probes specific for mtGFP, COX4, and TCM1. TCM1 mRNA levels were used as a loading control. The upper panel shows the Northern blotting analysis of mRNAs transcribed from mtGFP, COX4, and TCM1 as indicated by an arrow. The lower panel is the quantification of the mtGFP (plasmid-encoded) and COX4 (genome-encoded) mRNAs normalized to TCM1 by using the Bio-Rad Quantity One program. The mRNA levels in WT cells were set to 100%. (C) Analysis of mtGFP mRNA levels in YPH500/pCG1, YZD1/pCG1, YPH500/pCG3, and YZD1/pCG3 by Q-RT-PCR as described in Materials and Methods. The results were normalized to TCM1 mRNA levels, and the mRNA levels in wild-type cells were set to 100%.
In order to confirm that the decrease in mtGFP expression was at the translational level, a Northern blotting analysis was performed on mRNA extracts from these strains with probes against the ORFs of both mtGFP (plasmid) and COX4 (chromosome) (Fig. 3B). The level of mtGFP mRNA in pgs1Δ cells was only ∼40% of that in the wild type (normalized to TCM1 message) no matter which construct (pCG1 [full length] or pCG3 [truncated]) was used. The chromosomally derived COX4 mRNA was also reduced in pgs1Δ cells (50 to 65% of wild-type level), consistent with the reported reduction in COX4 transcription by intergenomic signaling (9) in response to lack of mtDNA. This result is also in agreement with data obtained using Q-RT-PCR, which showed about a 50% reduction of mtGFP mRNA for both constructs (pCG1 and pCG3) in pgs1Δ cells (Fig. 3C). However, the reduction in mRNA levels cannot account for the difference between mtGFP expression from the full-length construct pCG1 and that from the truncated construct pCG3 in pgs1Δ cells. Given that there is no correlation between the level of mRNAs and their respective protein expression, the regulation of mtGFP expression occurs at the translational rather than at the transcriptional level. This result also agrees with the presence of significant levels of COX4 mRNA derived from chromosomal transcription (Fig. 3B) and the lack of Cox4p expression in pgs1Δ cells (48), thus fully supporting the conclusion that expression of COX4 under control of the native PCOX4 and 5′ UTRCOX4 is subject to translational regulation by its 5′ UTRCOX4 rather than transcriptional control.
The 5′ intron of COX4 is properly spliced.
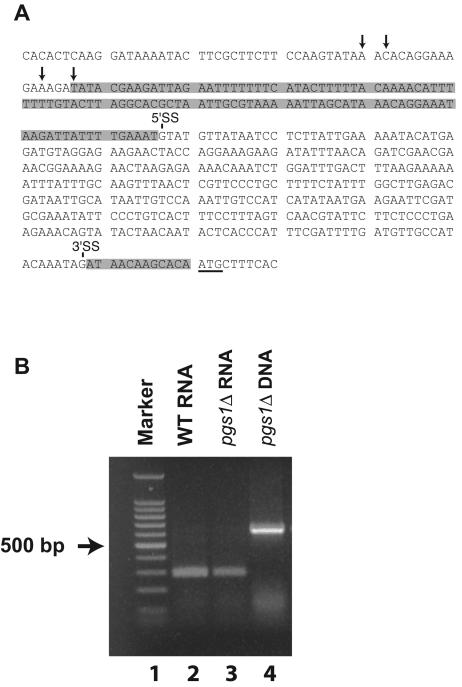
The next question addressed was what features in the 5′ UTRCOX4 confer the regulatable translational property of the COX4 gene. The 5′ UTRCOX4 contains a 342-nt intron (Fig. 4A) with several small untranslated ORFs (56). Upstream untranslated ORFs have been reported to function as cis elements that can inhibit translation of genes (39, 42). The existence of untranslated ORFs in the 5′ intron of COX4 mRNA could certainly repress translation from the primary ORF if the intron was not removed properly by splicing in a pgs1Δ background. To determine if proper slicing occurs, an RT-PCR analysis was performed on COX4 mRNA isolated from wild-type and pgs1Δ cells. With the primers designed, a spliced mRNA should give a band at approximately 300 bp, while the unspliced mRNA should give a band which is 342 bp larger (same as that of the amplified genomic DNA). The result in Fig. 4B shows that bands of the same size (∼300 bp) were amplified from both wild-type and pgs1Δ RNAs, indicating that the intron was correctly spliced in the pgs1Δ background.
FIG. 4.
The 5′ intron of COX4 mRNA is properly spliced in pgs1Δ cells. (A) 5′ UTR of COX4 gene, with arrows indicating the possible transcription start sites. The DNA segment encoding the 342-nt intron is marked with 5′ and 3′ splicing sites (SS). The segment encoding the 5′ UTR of mature COX4 mRNA is shaded. The ATG start codon is underlined. (B) RT-PCR analysis of the 5′ UTR of COX4 mRNA. Cells were grown to mid-log phase in YPD medium. RNA was isolated from YPH500 (wild-type [WT]) cells, and RNA and DNA were isolated from YZD1 (pgs1Δ) cells. RT-PCR was performed as described in Materials and Methods. Lanes: 1, Promega 100-bp step ladder; 2, RNA from YPH500; 3, RNA from YZD1; 4, DNA from YZD1.
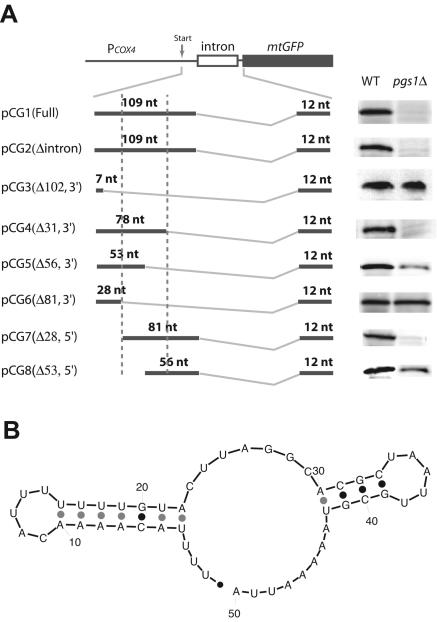
Deletion analysis of 5′ UTRCOX4.
To narrow down the element that regulates COX4 expression at the translational level, a series of deletions of the 109-nt fragment within the mature 5′ UTRCOX4 were created. As can be seen from Fig. 5A, a 31-nt deletion into this sequence from its 3′ end (pCG4) had no effect on the expression of mtGFP, while an additional 25-nt deletion (total of 56 nt) into the region (pCG5) gave partial expression in pgs1Δ cells. Another 25-nt deletion (total of 81 nt) further into this region (pCG6) gave full expression. Similarly, a 28-nt deletion from the 5′ end (pCG7) had no effect, while another 25-nt deletion (total of 53 nt) deeper into the sequence (pCG8) gave partial expression. Taken together, these results indicate that the 50-nt region in the middle of the 109-nt fragment functions as a minimum sequence that inhibits translation of COX4 mRNA in a pgs1Δ background. A parallel set of plasmids lacking the 3′ UTRCOX4 gave the same results (data not shown). Interestingly, the RNA secondary structure of the 50-nt sequence predicted by MFOLD at 30°C shows an RNA structure with two stem-loops with a total folding free energy (ΔG°30) of −12.0 kcal/mol (Fig. 5B). The first stem-loop is located in the first half of the 50-nt sequence, while the other stem-loop is situated in the second half of this 50-nt region. When each stem-loop was deleted individually, a partial expression of downstream mtGFP was observed, and when both were deleted together, full expression was achieved, consistent with a role for both stem-loops in controlling downstream expression in pgs1Δ cells.
FIG. 5.
Deletion analysis defines a 50-nt sequence with two stem-loops that function as a minimum cis-acting element. (A) Western blotting analysis of mtGFP expressed under control of different lengths of the 5′ UTRCOX4 in YPH500 (wild-type) and YZD1 (pgs1Δ) cells. Mitochondria were isolated and subjected to Western blotting analysis for mtGFP as described in Materials and Methods. The 342-bp intron is shown as an empty box, and mtGFP is indicated as a solid box. The arrow indicates the transcriptional start site. The mature mRNAs are shown by two boldface lines connected by a jagged line indicating splicing. The length of the 5′ UTR of the resulting RNAs is labeled in the graph. The sequence between the two vertical dashed lines indicates the 50-nt RNA fragment that is responsible for inhibition of mtGFP translation. (B) The 50-nt RNA fold as predicted by MFOLD 2.3 from the M. Zuker group (http://www.bioinfo.rpi.edu/applications/mfold/old/rna/form1-2.3.cgi) at 30°C with default parameters. The first stem-loop is located between nt 4 and 22 and the second stem-loop is located between nt 30 and 42 of the sequence.
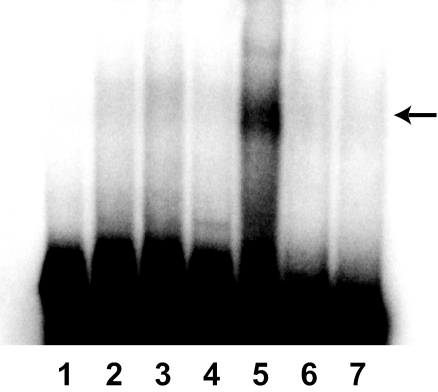
RNA gel mobility shift assay.
The role of the 5′ UTRCOX4 in COX4 translation was further supported by results obtained from an RNA electrophoresis mobility shift assay using an in vitro T7 promoter-synthesized 104-nt 5′ UTRCOX4 RNA and cell lysates isolated from both wild-type and pgs1Δ cells. Lysate from pgs1Δ cells caused retardation of the 5′ UTRCOX4 RNA fragment, forming a slowly migrating band that was absent when a lysate isolated from wild-type cells was used (Fig. 6). Addition of protease K to the shift assay totally abolished the shifted band, supporting participation by protein factors in this process. A similar length of RNA without the stem-loop structures was also used in a control experiment in which no shift was detected no matter which cell lysate was used, indicating the importance of the stem-loops in binding of protein factors. This result demonstrated that binding of a trans factor(s) to the 5′ UTRCOX4 occurs only in pgs1Δ cells and suggests that the formation of a complex between the stem-loop cis element and protein trans factors specifically in a pgs1Δ background results in translational arrest of COX4 mRNA in these cells.
FIG. 6.
A factor(s) in cell lysate from pgs1Δ cells binds to the 5′ UTR of COX4 mRNA. Cell extracts were prepared from mid-log-phase YPH500 (wild-type) and YZD1 (pgs1Δ) cells grown in YPD. A 104-nt RNA probe representing the 5′ UTRCOX4 and a control RNA without the predicted stem-loops (but of the same length) were transcribed from the T7 promoter and labeled with [α-32P]UTP, and an electrophoretic mobility shift assay was done as described in Materials and Methods. The arrow indicates the shift that occurs only in pgs1Δ cells. Lane 1, free probe. Lane 2, control RNA plus wild-type lysate. Lane 3, control RNA plus pgs1Δ lysate. Lane 4, COX4 RNA plus wild-type lysate. Lane 5, COX4 RNA plus pgs1Δ lysate. Lane 6, COX4 RNA plus wild-type lysate plus protease K. Lane 7, COX4 RNA plus pgs1Δ lysate plus protease K.
Isolation and analysis of bypass mutations.
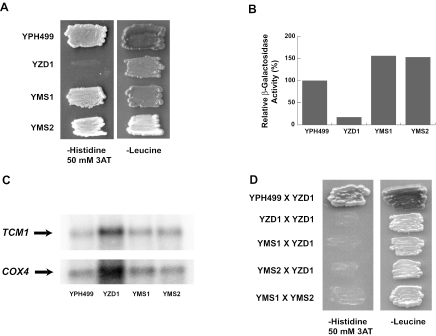
To better understand the mechanism underlying the lack of translation of COX4, yeast genetics were employed to isolate mutants in which translational repression is bypassed. Such mutants would result from mutation either directly in the translational control machinery or in proteins related to the regulatory machinery. Factors, once identified, could shed light on the possible mechanism of regulation. To identify such factors, a selectable system was generated. Given the fact that the 5′ UTRCOX4 controls its downstream gene expression, a HIS3 selectable marker was placed under control of PCOX4 and 5′ UTRCOX4 (plasmid pCH1). The effectiveness of this system was demonstrated by lack of growth on histidine dropout plates of pgs1Δ (pgs1Δ his3) cells carrying this plasmid (data not shown), indicating lack of HIS3 expression under PCOX4 and 5′ UTRCOX4 control in pgs1Δ cells. In comparison, wild-type cells (PGS1 his3) harboring this fusion construct grew well on these plates.
The derepression of HIS3 expression from plasmid pCH1 was then selected for by growth of pgs1Δ his3 cells plated on histidine dropout plates. To eliminate possible weak growth resulting from leaky expression of HIS3, 50 mM 3AT was added to the plates. 3AT is a specific competitive inhibitor of imidazoleglycerol-phosphate dehydratase encoded by the HIS3 gene (44). To overcome the inhibitory effect of this concentration of 3AT in the plate, cells have to express a significant amount HIS3 gene product under control of PCOX4 and UTRCOX4 in order to survive. After incubation at 30°C for 7 to 10 days, a few colonies appeared (the mutation frequency was approximately 10−7). The growth of the colonies was then confirmed to be due to spontaneous mutations rather than physical adaptation. For this purpose, two colonies (named YMS1 and YMS2) were picked and tested in two different ways. First they were grown in a nonselective (for his3) CSMD-Leu (plasmid marker was LEU2) medium for ∼100 generations, and then cell growth was reconfirmed on CSMD-His plates with 50 mM 3AT (Fig. 7A). Second, a multicopy plasmid (pCL1) that has lacZ under control of PCOX4 and 5′ UTRCOX4 was transformed into the cells, and β-galactosidase activity was measured. As indicated in Fig. 7B, wild-type cells had a high level of expression of β-galactosidase, while β-galactosidase expression in pgs1Δ cells was very low even though pCL1 is a high-copy-number plasmid; the two mutants had an expression level of β-galactosidase even higher than that of wild-type cells. Since the pCL1 plasmid was newly introduced into the cells, we can safely rule out the possibility that mutations within plasmid pCH1 allowed for HIS3 expression and thereafter cell growth. Because pgs1Δ cells have lost the ability to express mtDNA-encoded proteins, the derepression of the translational inhibition resulted from chromosomal mutations. We also tested by Northern blotting analysis if the chromosomal mutations promote COX4 expression by increasing transcription from the COX4 gene (Fig. 7C). The data obtained indicated that the level of COX4 mRNA transcripts was not significantly affected by these mutations. Therefore, both mutations appeared to affect COX4 expression again at the translational level.
FIG. 7.
Isolation and analysis of spontaneous mutations that confer high expression of a reporter gene placed after the 5′ UTRCOX4. (A) Spontaneous mutants (YMS1 and YMS2) of YZD1 (pgs1Δ)/pCH1 can grow on CSMD-His plates plus 50 mM 3AT. The pgs1Δ cells carrying plasmid pCH1 were plated onto CSMD-His plates with 50 mM 3AT. Cells were incubated at 30°C for 7 to 10 days before the colonies appeared. Two colonies (named YMS1 and YMS2) were picked and patched along with YPH499/pCH1 (wild type) and YZD1/pCH1 (pgs1Δ) as controls onto CSMD-His plates with 50 mM 3AT to test for His3p expression and on CSMC-Leu plates to verify the presence of plasmid pCH1. Cells were allowed to grow at 30°C for 3 days at the time the photograph was taken. (B) High-level expression of β-galactosidase activity under control of the 5′ UTRCOX4 in the mutants. A multicopy plasmid, pCL1, was introduced into the indicated strains. β-Galactosidase activity was assayed in extracts from these cells as described in Materials and Methods. (C) Levels of COX4 transcripts from chromosomes in YMS1 and YMS2. mRNAs were isolated from YPH499 (wild type), YZD1 (pgs1Δ), YMS1 (pgs1Δ), and YMS2 (pgs1Δ). Levels of COX4 and TCM1 mRNAs were determined by Northern blotting analysis as described in Materials and Methods. The amounts of COX4 mRNA normalized to TCM1 mRNA were as follows: YPH499, 100%; YZD1, 58%; YMS1, 82%; and YMS2, 69%. (D) YMS1 and YMS2 carry recessive mutations that belong to different complementation groups. YMS1, YMS2, YPH499, and YZD1 (pgs1Δ) were switched to the opposite mating type and mated with the original YZD1 (pgs1Δ) cells (see Materials and Methods). YMS1 was also mated to YMS2 as described in Materials and Methods to determine complementation. Diploid cells carrying plasmids pAHO1 and pCH1 were selected on CSMD-His-Leu double-dropout plates. Growth of diploid cells derived from mating were patched to CSMD-His plates with 50 mM 3AT to test for complementation and to CSMC-Leu plates to verify the presence of plasmid pCH1.
To test whether these suppressor mutations were dominant or recessive, strains YMS1 and YMS2 were backcrossed with the parental pgs1Δ strain. For this purpose, the wild-type cells, the pgs1Δ cells, and the two suppressor mutant cells (MATa ura3 leu2), all carrying plasmid pCH1 (LEU2), were first switched to the opposite mating type and then mated with an original pgs1Δ strain (MATa ura3 leu2) carrying plasmid pCL1 (URA3) (see Materials and Methods). Figure 7D shows the growth properties on a CSMD-His plate containing 50 mM 3AT of the diploid cells resulting from these matings. As can been seen, YMS1 and YMS2 mutants lost their ability to grow in the absence of histidine once mated with the pgs1Δ parental cells. Because pgs1Δ cells or cells derived from pgs1Δ appear not to contain mtDNA (74), this result indicated that the mutants contain genomic recessive mutations that are most likely due to loss of function. In order to determine if the recessive mutations belong to the same complementation group, strains YMS1 and YMS2 were mated to each other (see Materials and Methods), and plasmid pCH1 expression was examined in the diploid cells. Figure 7D shows that the resulting haploid cells cannot grow on histidine dropout 50 mM 3AT plates, indicating that the mutated genes (tentatively named lipid responsive gene [LRG] 1 and 2) in the two recessive mutants belong to different complementation groups.
DISCUSSION
This study was initiated when pgs1Δ cells were found to apparently exhibit regulation of COX4 expression primarily at the translational level (48). First, translational regulation was definitively established by the presence of COX4 and reporter gene (mtGFP) mRNA and the lack of normally stable reporter protein. Evidence that the 5′ UTR contains a cis-acting element responsible for repression of COX4 mRNA translation in a pgs1Δ background came from 5′ UTR deletion and reporter expression studies. When most of the 5′ UTR was deleted from COX4 mRNA, reporter gene expression, as indicated by mtGFP expression detected by Western blotting or His3p expression shown by cell growth on histidine dropout plates (data not shown), returned to wild-type cell levels. These results demonstrate that the 5′ UTR functions as a negative regulator of translation. Further deletion analysis confined the 5′ cis element to a 50-nt segment. Secondary structure prediction of the 50-nt sequence presents an RNA structure containing two stem-loops, with each comprising half of the 50-nt sequence. The additive effect of these two halves of the 50-nt segment, taken together with deletion analysis, is consistent with both stem-loops being important for the efficient repression of COX4 gene expression at the translational level in a pgs1Δ background.
There are several underlying mechanisms by which a stem-loop structure can affect translation of various genes. Usually, translation initiation is down-regulated with a stable 5′ stem-loop structure (ΔG° more negative than −50 kcal/mol) which stalls the migration of 40S ribosomal subunit (37, 38). However, less stable secondary structures (ΔG° lower than −30 kcal/mol) alone cannot confer the ability to stall the 40S ribosomal subunit (36). The predicted stem-loop structure in COX4 mRNA has a ΔG° of −12.0 kcal/mol, which is insufficient to make stable hairpins. In wild-type cells, these hairpins may be readily unwound by helicase eIF4A in the cap-binding complex and therefore would have no effect on translation. However, in pgs1Δ cells possible binding of cytoplasmic (trans-acting) factors induced by lack of PG and CL may stabilize the hairpins, as has been observed for binding of iron regulatory proteins to iron-responsive elements in the coordinately regulated cycle for iron metabolism (21, 49, 63). The complex formed may represent a large energy obstacle for the ribosomal subunit to overcome, thereby repressing translation. Accordingly, elimination of complex formation by either deletion of the cis-acting element (5′ UTRCOX4) or mutation of the trans-acting factors (two recessive suppressor mutants) might suppress the repressive effect and result in the observed reporter gene expression.
This hypothesis was further supported by results obtained from an RNA electrophoretic mobility shift assay using an in vitro-synthesized 5′ UTRCOX4 RNA and cell lysates isolated from both wild-type and pgs1Δ cells. A lysate from pgs1Δ cells caused retardation of the 5′ UTRCOX4 RNA fragment, forming a slowly migrating band that was not observed with lysates from wild-type cells. Protease sensitivity of the observed gel shift and the lack of a gel shift of an RNA element lacking the above-described 50-nt segment strongly support the binding of a protein trans factor(s) present only in pgs1Δ cells to a specific sequence in the 5′ UTRCOX4. Mutations were isolated that in a pgs1Δ background suppressed the repression of COX4 translation. These were shown to be genomic and not in the reporter plasmid used to isolate the mutations. Subsequent mating of two independently isolated mutants (YMS1 and YMS2) with the parental pgs1Δ strain and with each other demonstrated that the mutations were recessive and in two different gene loci, suggesting loss-of-function mutations consistent with a mutation in trans elements functional only in a pgs1Δ background.
One question naturally arises as to how the trans factors are activated in response to a defect affecting mitochondrial lipid composition. As phospholipids in the mitochondrial inner and possibly outer membranes (10, 27), PG and/or CL is important not only in mitochondrial respiratory function but to all proteins that interact with mitochondrial membranes and rely on anionic phospholipids for their function. It is highly possible that some of the proteins in or on the mitochondrial membranes transmit a signal, triggered by lack of PG and CL, back to the nucleus for cross talk regulation of COX4 gene expression. Such a signal could induce the expression of nuclear proteins in the classic cross talk pathways (54) and possibly, in this case, the putative trans factors that bind to the 5′ cis element, resulting in translational inhibition.
Interestingly, the proposed mechanism by which the COX4 translation is inhibited is reminiscent of the unfolded protein response mediated by Ire1p in the endoplasmic reticulum to inhibit unfolded protein translation or induce nuclear genes encoding endoplasmic reticulum folding catalysts and chaperones for remedying protein misfolding under conditions of endoplasmic reticulum stress (23, 28, 43, 71). Ire1p is a type I transmembrane serine/threonine protein kinase with endoribonuclease activities that is activated by unfolded proteins in the endoplasmic reticulum. It has been implicated as the membrane receptor that transmits the stress signal from the endoplasmic reticulum to the nucleus. Activated Ire1p functions as a site-specific RNase that splices HAC1 mRNA that encodes Hac1p, a basic leucine zipper transcription factor that binds to the unfolded protein response element in the nucleus and results in the induction of target nuclear genes to correct protein-folding defects. A similar receptor may exist in mitochondrial membranes and may be activated by PG and CL deficiency to transmit a signal to the nucleus.
In summary, our results are consistent with the existence of two stem-loops as a cis-acting element in the 5′ UTRCOX4 and a novel cross talk pathway between mitochondria and the nucleus that is responsive to PG and CL deficiency in S. cerevisiae. It remains a question whether this pathway works for other genes in yeast or exists in other organisms. Unfortunately, we have not been able to pinpoint any RNA sequence that is similar to the 5′ UTRCOX4 in yeast or in other organisms. In yeast, activation of this pathway in pgs1Δ cells results in binding of trans factors to the cis element and repression of mRNA translation. Disruption of this protein-RNA complex formation eliminated its inhibitory effect and thereby restored ability of cells to express downstream genes. An exhaustive analysis of the isolated mutants and further identification of the trans factors are required for understanding the details of the novel cross talk pathway and the regulatory cycle. Experiments to identify the genes whose apparent loss of function restores translation of COX4 mRNA in pgs1Δ cells are under way.
Acknowledgments
This work was supported Public Health Service grant GM56389 (to W.D.) from the National Institute of General Medical Sciences.
We thank Thomas Fox and Kelvin Morano for providing strains, plasmids, and antibody used in this work.
REFERENCES
- 1.Ades, I. Z., and R. A. Butow. 1980. The products of mitochondria-bound cytoplasmic polysomes in yeast. J. Biol. Chem. 255:9918-9924. [PubMed] [Google Scholar]
- 2.Ardail, D., F. Lerme, and P. Louisot. 1992. Phospholipid import into mitochondria: possible regulation mediated through lipid polymorphism. Biochem. Biophys. Res. Commun. 186:1384-1390. [DOI] [PubMed] [Google Scholar]
- 3.Barath, Z., and H. Kuntzel. 1972. Cooperation of mitochondrial and nuclear genes specifying the mitochondrial genetic apparatus in Neurospora crassa. Proc. Natl. Acad. Sci. USA 69:1371-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battelli, D., M. Bellei, E. Arrigoni-Martelli, U. Muscatello, and V. Bobyleva. 1992. Interaction of carnitine with mitochondrial cardiolipin. Biochim. Biophys. Acta 1117:33-36. [DOI] [PubMed] [Google Scholar]
- 5.Biswas, G., O. A. Adebanjo, B. D. Freedman, H. K. Anandatheerthavarada, C. Vijayasarathy, M. Zaidi, M. Kotlikoff, and N. G. Avadhani. 1999. Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. EMBO J. 18:522-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonnefoy, N., N. Bsat, and T. D. Fox. 2001. Mitochondrial translation of Saccharomyces cerevisiae COX2 mRNA is controlled by the nucleotide sequence specifying the pre-Cox2p leader peptide. Mol. Cell. Biol. 21:2359-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, S. C., P. N. Heacock, C. J. Clancey, and W. Dowhan. 1998. The PEL1 gene (renamed PGS1) encodes the phosphatidylglycero-phosphate synthase of Saccharomyces cerevisiae. J. Biol. Chem. 273:9829-9836. [DOI] [PubMed] [Google Scholar]
- 8.Chang, S. C., P. N. Heacock, E. Mileykovskaya, D. R. Voelker, and W. Dowhan. 1998. Isolation and characterization of the gene (CLS1) encoding cardiolipin synthase in Saccharomyces cerevisiae. J. Biol. Chem. 273:14933-14941. [DOI] [PubMed] [Google Scholar]
- 9.Dagsgaard, C., L. E. Taylor, K. M. O'Brien, and R. O. Poyton. 2001. Effects of anoxia and the mitochondrion on expression of aerobic nuclear COX genes in yeast. Evidence for a signaling pathway from the mitochondrial genome to the nucleus. J. Biol. Chem. 276:7593-7601. [DOI] [PubMed] [Google Scholar]
- 10.de Kroon, A. I., D. Dolis, A. Mayer, R. Lill, and B. de Kruijff. 1997. Phospholipid composition of highly purified mitochondrial outer membranes of rat liver and Neurospora crassa. Is cardiolipin present in the mitochondrial outer membrane? Biochim. Biophys. Acta 1325:108-116. [DOI] [PubMed] [Google Scholar]
- 11.Dowhan, W., C. R. Bibus, and G. Schatz. 1985. The cytoplasmically-made subunit IV is necessary for assembly of cytochrome c oxidase in yeast. EMBO J. 4:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eble, K. S., W. B. Coleman, R. R. Hantgan, and C. C. Cunningham. 1990. Tightly associated cardiolipin in the bovine heart mitochondrial ATP synthase as analyzed by 31P nuclear magnetic resonance spectroscopy. J. Biol. Chem. 265:19434-19440. [PubMed] [Google Scholar]
- 13.Eilers, M., T. Endo, and G. Schatz. 1989. Adriamycin, a drug interacting with acidic phospholipids, blocks import of precursor proteins by isolated yeast mitochondria. J. Biol. Chem. 264:2945-2950. [PubMed] [Google Scholar]
- 14.Epstein, C. B., J. A. Waddle, W. ten Hale, V. Dave, J. Thornton, T. L. Macatee, H. R. Garner, and R. A. Butow. 2001. Genome-wide responses to mitochondrial dysfunction. Mol. Biol. Cell 12:297-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forsburg, S. L., and L. Guarente. 1989. Communication between mitochondria and the nucleus in regulation of cytochrome genes in the yeast Saccharomyces cerevisiae. Annu. Rev. Cell Biol. 5:153-180. [DOI] [PubMed] [Google Scholar]
- 16.Fry, M., and D. E. Green. 1981. Cardiolipin requirement for electron transfer in complex I and III of the mitochondrial respiratory chain. J. Biol. Chem. 256:1874-1880. [PubMed] [Google Scholar]
- 17.Fujiki, M., and K. Verner. 1993. Coupling of cytosolic protein synthesis and mitochondrial protein import in yeast. Evidence for cotranslational import in vivo. J. Biol. Chem. 268:1914-1920. [PubMed] [Google Scholar]
- 18.Fujiki, M., and K. Verner. 1991. Coupling of protein synthesis and mitochondrial import in a homologous yeast in vitro system. J. Biol. Chem. 266:6841-6847. [PubMed] [Google Scholar]
- 19.Geier, B. M., H. Schagger, C. Ortwein, T. A. Link, W. R. Hagen, U. Brandt, and G. Von Jagow. 1995. Kinetic properties and ligand binding of the eleven-subunit cytochrome-c oxidase from Saccharomyces cerevisiae isolated with a novel large-scale purification method. Eur. J. Biochem. 227:296-302. [DOI] [PubMed] [Google Scholar]
- 20.Glick, B. S., and L. A. Pon. 1995. Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 260:213-223. [DOI] [PubMed] [Google Scholar]
- 21.Gray, N. K., and M. W. Hentze. 1994. Regulation of protein synthesis by mRNA structure. Mol. Biol. Rep. 19:195-200. [DOI] [PubMed] [Google Scholar]
- 22.Gunyuzlu, P. L., G. F. Hollis, and J. H. Toyn. 2001. Plasmid construction by linker-assisted homologous recombination in yeast. BioTechniques 31:1246, 1248, 1250. [DOI] [PubMed] [Google Scholar]
- 23.Harding, H. P., Y. Zhang, A. Bertolotti, H. Zeng, and D. Ron. 2000. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell 5:897-904. [DOI] [PubMed] [Google Scholar]
- 24.Herskowitz, I., and R. E. Jensen. 1991. Putting the HO gene to work: practical uses for mating-type switching. Methods Enzymol. 194:132-146. [DOI] [PubMed] [Google Scholar]
- 25.Hoch, F. L. 1992. Cardiolipin and biomembrane function. Biochim. Biophys. Acta 1113:71-133. [DOI] [PubMed] [Google Scholar]
- 26.Hoffmann, B., A. Stockl, M. Schlame, K. Beyer, and M. Klingenberg. 1994. The reconstituted ADP/ATP carrier activity has an absolute requirement for cardiolipin as shown in cysteine mutants. J. Biol. Chem. 269:1940-1944. [PubMed] [Google Scholar]
- 27.Hovius, R., H. Lambrechts, K. Nicolay, and B. de Kruijff. 1990. Improved methods to isolate and subfractionate rat liver mitochondria. Lipid composition of the inner and outer membrane. Biochim. Biophys. Acta 1021:217-226. [DOI] [PubMed] [Google Scholar]
- 28.Iwawaki, T., A. Hosoda, T. Okuda, Y. Kamigori, C. Nomura-Furuwatari, Y. Kimata, A. Tsuru, and K. Kohno. 2001. Translational control by the ER transmembrane kinase/ribonuclease IRE1 under ER stress. Nat. Cell Biol. 3:158-164. [DOI] [PubMed] [Google Scholar]
- 29.Jauniaux, J. C., L. A. Urrestarazu, and J. M. Wiame. 1978. Arginine metabolism in Saccharomyces cerevisiae: subcellular localization of the enzymes. J. Bacteriol. 133:1096-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang, F., H. S. Rizavi, and M. L. Greenberg. 1997. Cardiolipin is not essential for the growth of Saccharomyces cerevisiae on fermentable or non-fermentable carbon sources. Mol. Microbiol. 26:481-491. [DOI] [PubMed] [Google Scholar]
- 31.Jiang, F., M. T. Ryan, M. Schlame, M. Zhao, Z. Gu, M. Klingenberg, N. Pfanner, and M. L. Greenberg. 2000. Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J. Biol. Chem. 275:22387-22394. [DOI] [PubMed] [Google Scholar]
- 32.Kadenbach, B., P. Mende, H. V. Kolbe, I. Stipani, and F. Palmieri. 1982. The mitochondrial phosphate carrier has an essential requirement for cardiolipin. FEBS Lett. 139:109-112. [DOI] [PubMed] [Google Scholar]
- 33.Kawasaki, K., O. Kuge, S. C. Chang, P. N. Heacock, M. Rho, K. Suzuki, M. Nishijima, and W. Dowhan. 1999. Isolation of a Chinese hamster ovary (CHO) cDNA encoding phosphatidylglycerophosphate (PGP) synthase, expression of which corrects the mitochondrial abnormalities of a PGP synthase-defective mutant of CHO-K1 cells. J. Biol. Chem. 274:1828-1834. [DOI] [PubMed] [Google Scholar]
- 34.Kirchman, P. A., S. Kim, C. Y. Lai, and S. M. Jazwinski. 1999. Interorganelle signaling is a determinant of longevity in Saccharomyces cerevisiae. Genetics 152:179-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koshkin, V., and M. L. Greenberg. 2000. Oxidative phosphorylation in cardiolipin-lacking yeast mitochondria. Biochem. J. 347:687-691. [PMC free article] [PubMed] [Google Scholar]
- 36.Kozak, M. 1989. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol. Cell. Biol. 9:5134-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozak, M. 1990. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc. Natl. Acad. Sci. USA 87:8301-8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozak, M. 1986. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl. Acad. Sci. USA 83:2850-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Law, G. L., A. Raney, C. Heusner, and D. R. Morris. 2001. Polyamine regulation of ribosome pausing at the upstream open reading frame of S-adenosylmethionine decarboxylase. J. Biol. Chem. 276:38036-38043. [DOI] [PubMed] [Google Scholar]
- 40.Lemaire, C., S. Robineau, and P. Netter. 1998. Molecular and biochemical analysis of Saccharomyces cerevisiae cox1 mutants. Curr. Genet. 34:138-145. [DOI] [PubMed] [Google Scholar]
- 41.Lesnefsky, E. J., S. Moghaddas, B. Tandler, J. Kerner, and C. L. Hoppel. 2001. Mitochondrial dysfunction in cardiac disease: ischemia—reperfusion, aging, and heart failure. J. Mol. Cell Cardiol. 33:1065-1089. [DOI] [PubMed] [Google Scholar]
- 42.Lincoln, A. J., Y. Monczak, S. C. Williams, and P. F. Johnson. 1998. Inhibition of CCAAT/enhancer-binding protein alpha and beta translation by upstream open reading frames. J. Biol. Chem. 273:9552-9560. [DOI] [PubMed] [Google Scholar]
- 43.Liu, C. Y., M. Schroder, and R. J. Kaufman. 2000. Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J. Biol. Chem. 275:24881-24885. [DOI] [PubMed] [Google Scholar]
- 44.Mangus, D. A., N. Amrani, and A. Jacobson. 1998. Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol. Cell Biol. 18:7383-7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marc, P., A. Margeot, F. Devaux, C. Blugeon, M. Corral-Debrinski, and C. Jacq. 2002. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep. 3:159-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McMillin, J. B., and W. Dowhan. 2002. Cardiolipin and apoptosis. Biochim. Biophys. Acta 1585:97-107. [DOI] [PubMed] [Google Scholar]
- 47.Mende, P., F. J. Huther, and B. Kadenbach. 1983. Specific and reversible activation and inactivation of the mitochondrial phosphate carrier by cardiolipin and nonionic detergents, respectively. FEBS Lett. 158:331-334. [DOI] [PubMed] [Google Scholar]
- 48.Ostrander, D. B., M. Zhang, E. Mileykovskaya, M. Rho, and W. Dowhan. 2001. Lack of mitochondrial anionic phospholipids causes an inhibition of translation of protein components of the electron transport chain. A yeast genetic model system for the study of anionic phospholipid function in mitochondria. J. Biol. Chem. 276:25262-25272. [DOI] [PubMed] [Google Scholar]
- 49.Pantopoulos, K. 2004. Iron metabolism and the IRE/IRP regulatory system: an update. Ann. N. Y. Acad. Sci. 1012:1-13. [DOI] [PubMed] [Google Scholar]
- 50.Paradies, G., F. M. Ruggiero, G. Petrosillo, and E. Quagliariello. 1998. Peroxidative damage to cardiac mitochondria: cytochrome oxidase and cardiolipin alterations. FEBS Lett. 424:155-158. [DOI] [PubMed] [Google Scholar]
- 51.Parikh, V. S., M. M. Morgan, R. Scott, L. S. Clements, and R. A. Butow. 1987. The mitochondrial genotype can influence nuclear gene expression in yeast. Science 235:576-580. [DOI] [PubMed] [Google Scholar]
- 52.Pfeiffer, K., V. Gohil, R. A. Stuart, C. Hunte, U. Brandt, M. L. Greenberg, and H. Schagger. 2003. Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 278:52873-52880. [DOI] [PubMed] [Google Scholar]
- 53.Poyton, R. O. 1980. Cooperative interaction between mitochondrial and nuclear genomes: cytochrome c oxidase assembly as a model. Curr. Top. Cell Regul. 17:231-295. [DOI] [PubMed] [Google Scholar]
- 54.Poyton, R. O., and C. J. Dagsgaard. 2000. Mitochondrial-nuclear crosstalk is involved in oxygen-regulated gene expression in yeast. Adv. Exp. Med. Biol. 475:177-184. [DOI] [PubMed] [Google Scholar]
- 55.Robinson, N. C. 1993. Functional binding of cardiolipin to cytochrome c oxidase. J. Bioenerg. Biomembr. 25:153-163. [DOI] [PubMed] [Google Scholar]
- 56.Schneider, J. C., and L. Guarente. 1987. The untranslated leader of nuclear COX4 gene of Saccharomyces cerevisiae contains an intron. Nucleic Acids Res. 15:3515-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sedlak, E., and N. C. Robinson. 1999. Phospholipase A2 digestion of cardiolipin bound to bovine cytochrome c oxidase alters both activity and quaternary structure. Biochemistry 38:14966-14972. [DOI] [PubMed] [Google Scholar]
- 58.Shen, H., and W. Dowhan. 1998. Regulation of phosphatidylglycerophosphate synthase levels in Saccharomyces cerevisiae. J. Biol. Chem. 273:11638-11642. [DOI] [PubMed] [Google Scholar]
- 59.Shen, H., and W. Dowhan. 1997. Regulation of phospholipid biosynthetic enzymes by the level of CDP-diacylglycerol synthase activity. J. Biol. Chem. 272:11215-11220. [DOI] [PubMed] [Google Scholar]
- 60.Shigenaga, M. K., T. M. Hagen, and B. N. Ames. 1994. Oxidative damage and mitochondrial decay in aging. Proc. Natl. Acad. Sci. USA 91:10771-10778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steele, D. F., C. A. Butler, and T. D. Fox. 1996. Expression of a recoded nuclear gene inserted into yeast mitochondrial DNA is limited by mRNA-specific translational activation. Proc. Natl. Acad. Sci. USA 93:5253-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thomson, A. M., J. T. Rogers, and P. J. Leedman. 1999. Iron-regulatory proteins, iron-responsive elements and ferritin mRNA translation. Int. J. Biochem. Cell Biol. 31:1139-1152. [DOI] [PubMed] [Google Scholar]
- 64.Tuller, G., C. Hrastnik, G. Achleitner, U. Schiefthaler, F. Klein, and G. Daum. 1998. YDL142c encodes cardiolipin synthase (Cls1p) and is non-essential for aerobic growth of Saccharomyces cerevisiae. FEBS Lett. 421:15-18. [DOI] [PubMed] [Google Scholar]
- 65.Vaena de Avalos, S., Y. Okamoto, and Y. A. Hannun. 2004. Activation and localization of inositol phosphosphingolipid phospholipase C, Isc1p, to the mitochondria during growth of Saccharomyces cerevisiae. J. Biol. Chem. 279:11537-11545. [DOI] [PubMed] [Google Scholar]
- 66.Van Loon, A. P., E. Van Eijk, and L. A. Grivell. 1983. Biosynthesis of the ubiquinol-cytochrome c reductase complex in yeast. Discoordinate synthesis of the 11-kd subunit in response to increased gene copy number. EMBO J. 2:1765-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vreken, P., F. Valianpour, L. G. Nijtmans, L. A. Grivell, B. Plecko, R. J. Wanders, and P. G. Barth. 2000. Defective remodeling of cardiolipin and phosphatidylglycerol in Barth syndrome. Biochem. Biophys. Res. Commun. 279:378-382. [DOI] [PubMed] [Google Scholar]
- 68.Westermann, B., and W. Neupert. 2000. Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast 16:1421-1427. [DOI] [PubMed] [Google Scholar]
- 69.Wilkie, G. S., K. S. Dickson, and N. K. Gray. 2003. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci. 28:182-188. [DOI] [PubMed] [Google Scholar]
- 70.Yoon, H., S. P. Miller, E. K. Pabich, and T. F. Donahue. 1992. SSL1, a suppressor of a HIS4 5′-UTR stem-loop mutation, is essential for translation initiation and affects UV resistance in yeast. Genes Dev. 6:2463-2477. [DOI] [PubMed] [Google Scholar]
- 71.Zhang, K., and R. J. Kaufman. 2004. Signaling the unfolded protein response from the endoplasmic reticulum. J. Biol. Chem. 279:25935-25938. [DOI] [PubMed] [Google Scholar]
- 72.Zhang, M., E. Mileykovskaya, and W. Dowhan. 2002. Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J. Biol. Chem. 277:43553-43556. [DOI] [PubMed] [Google Scholar]
- 73.Zhang, M., X. Su, E. Mileykovskaya, A. A. Amoscato, and W. Dowhan. 2003. Cardiolipin is not required to maintain mitochondrial DNA stability or cell viability for Saccharomyces cerevisiae grown at elevated temperatures. J. Biol. Chem. 278:35204-35210. [DOI] [PubMed] [Google Scholar]
- 74.Zhong, Q., J. Gvozdenovic-Jeremic, P. Webster, J. Zhou, and M. L. Greenberg. 2005. Loss of function of KRE5 suppresses temperature sensitivity of mutants lacking mitochondrial anionic lipids. Mol. Biol. Cell 16:665-675. [DOI] [PMC free article] [PubMed] [Google Scholar]