Abstract
The C/EBPα transcription factor regulates growth and differentiation of several tissues during embryonic development. Several hypotheses as to how C/EBPα inhibits cellular growth in vivo have been derived, mainly from studies of tissue culture cells. In fetal liver it has been proposed that a short, centrally located, 15-amino-acid proline-histidine-rich region (PHR) of C/EBPα is responsible for the growth-inhibitory function of the protein through its ability to interact with CDK2 and CDK4, thereby inhibiting their activities. Homozygous CebpaΔPHR/ΔPHR (ΔPHR) mice, carrying a modified cebpa allele lacking amino acids 180 to 194, were born at the Mendelian ratio, reached adulthood, and displayed no apparent adverse phenotypes. When fetal livers from the ΔPHR mice were analyzed for their expression of cell cycle markers, bromodeoxyuridine incorporation, cyclin-dependent kinase 2 kinase activity, and global gene expression, we failed to detect any cell cycle or developmental differences between the ΔPHR mice and their control littermates. These in vivo data demonstrate that any C/EBPα-mediated growth repression via the PHR as well as the basic region is dispensable for proper embryonic development of, and cell cycle control in, the liver. Surprisingly, control experiments performed in C/EBPα null fetal livers yielded similar results.
C/EBPα is a key transcription factor mediating differentiation in several organ systems, including adipose, lung, and liver tissues, as well as within the hematopoietic system (4, 10, 38, 40). C/EBPα promotes differentiation by coordinating the two key events for terminal differentiation, i.e., the upregulation of lineage-specific gene products and exit from the cell cycle.
The capacity of C/EBPα to promote growth arrest has been extensively studied over the years, and several models as to how this is accomplished have been put forward (for a recent review see reference 11). These include C/EBPα-mediated (i) stabilization of p21 (32), (ii) recruitment of retinoblastoma (Rb) protein (33), (iii) repression of E2F activity (2, 10, 24, 28, 39), (iv) inhibition of cyclin-dependent kinase 2 (CDK2)/CDK4 activity (37), and (v) recruitment of SWI/SNF complexes (19). Whereas models i and ii appear to be difficult to reconcile with experiments performed in p21 (18) and Rb (8) null cell lines, respectively, it is more difficult to distinguish between models iii to v. The multiple models for C/EBPα-mediated growth inhibition likely reflect, in part, differences in testing systems and, at the organism level, that C/EBPα may promote growth arrest by various means in different cell types (Fig. 1A).
FIG. 1.
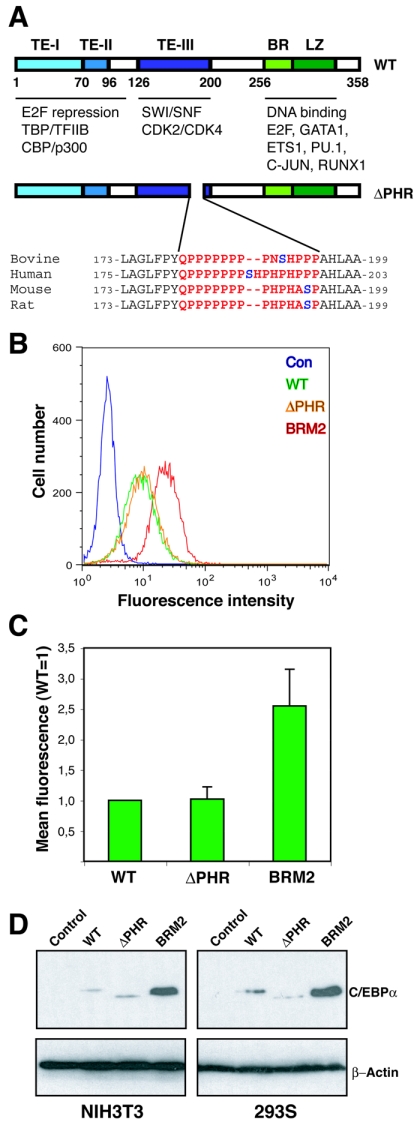
In vitro growth repression of continuously expressed C/EBPαΔPHR variant proteins. (A) Schematic representation of the C/EBPα protein showing the three transactivation elements (20), the DNA binding basic region (BR), and the leuzine zipper dimerization domain (LZ). Protein interaction partners are indicated below the protein. The ΔPHR variants (rat numbering) are also shown. The PHR sequences from four mammals are highlighted in red, and the putatively phosphorylated serine is shown in blue. (B) The eGFP fluorescence of NIH 3T3 cultures transduced with retroviruses derived from Phoenix-E cultures transfected with pBabePuro (Con), pBabePuro-C/EBPα-eGFP (WT), pBabePuro-C/EBPαΔPHR-eGFP (ΔPHR), or pBabePuro-C/EBPα-BRM2-eGFP (BRM2) was determined by flow cytometry after 14 days of culturing at subconfluency (approximately 15 cell divisions). (C) The mean eGFP fluorescence (autofluorescence subtracted from NIH 3T3 cells) was plotted from three independent experiments. Errors bars indicate standard deviations. (D) C/EBPα levels in NIH 3T3 (retrovirally transduced with pBabePuro derivatives) or HEK293S cultures (transfected with pcDNA3 derivatives) expressing C/EBPα (WT), C/EBPαΔPHR (ΔPHR), or C/EBPαBRM2 (BRM2). Controls (Con) indicate pBabePuro (NIH 3T3) or pcDNA3 (HEK293). Cells were kept subconfluent under selective conditions for 3 weeks, and lysates were prepared and subjected to Western blotting using a C/EBPα-specific antibody. A β-actin antibody was used to control for even loading. TE-I, TE-II, and TE-III, transactivation elements I, II, and III, respectively.
Different regions of C/EBPα have been defined as growth-inhibitory regions (Fig. 1A). According to the E2F repression model, C/EBPα recruits E2F through the non-DNA binding surface of its basic region and promotes repression via its N-terminal region by an unknown mechanism (10). In the CDK2/CDK4 inhibition model, C/EBPα interacts with and inhibits the activity of CDK2/CDK4 through a 15-amino-acid region located in the central part of the protein (37). Finally, according to the SWI/SNF recruitment model, C/EBPα interacts with SWI/SNF components through a centrally located 75-amino-acid region overlapping with the CDK2/CDK4 binding region (19, 22).
Mouse models have been extremely helpful in elucidating C/EBPα functions. C/EBPα null mice die within the first few hours after birth from glycogen deficiency and hyperammonemia and show several signs of hyperplasia in lung and liver tissues, strongly implying that C/EBPα is involved in growth regulation in these tissues during late embryonic development (4, 14, 38). Previously, we generated a knock-in mouse line expressing a C/EBPα allele (BRM2) deficient in E2F repression. Whereas the E2F repression-deficient mutants of C/EBPα had no impact on liver development and homeostasis (24), they led to severe hypotrophy of adipose tissues and to the almost complete absence of neutrophilic granulocytes in young mice. In older mice the hematopoietic phenotype progressed with incomplete penetrance to either a myeloproliferative or a myeloid leukemic phenotype, suggesting that the accumulation of secondary mutations led to the phenotypic progression (23, 24). These findings strongly imply that C/EBPα uses different mechanisms in different tissues to promote cell cycle exit.
In this context, the recent demonstration that C/EBPα is complexed with CDK2/CDK4 in liver extracts from young mice is particularly intriguing, as it suggests that CDK inhibition is the main growth-inhibitory activity mediated by C/EBPα in the developing liver (9).
In order to test the importance of C/EBPα-mediated repression of CDK2/CDK4 activity, we initially generated an internal deletion allele of C/EBPα in which the 15-amino-acid proline- and histidine-rich region (PHR) previously reported to be necessary for C/EBPα-mediated growth repression in tissue culture cells was deleted (37). Surprisingly, the ΔPHR mutant affected cellular growth in vitro in the same manner as wild-type C/EBPα in several assays. Since these contradictory findings may reflect differences in design between our experiments and the ones previously reported (37), we decided to generate a knock-in mouse line in which the wild-type C/EBPα allele was replaced with the PHR deletion variant. However, consistent with our in vitro data, mice homozygous for the ΔPHR allele did not display any overt phenotype that could be related to the role for C/EBPα as a growth repressor. Our findings demonstrate that C/EBPα-mediated repression of CDK2/CDK4 activity is dispensable for the embryonic development of the liver and raise doubts as to the essential role of C/EBPα as an inhibitor of growth during fetal liver development.
MATERIALS AND METHODS
Cloning.
All expression constructs were generated in the context of the rat C/EBPα cDNA. The ΔPHR variant was generated using the QuikChange system (Stratagene, La Jolla, Calif.) and the rat C/EBPα cloned in pcDNA3 as a BamHI/EcoRI fragment as template. For the retroviral transduction experiments, the C/EBPα-encoding sequences were moved as BamHI/EcoRI fragments into the pbabePuro vector (17). C-terminal tagging with enhanced green fluorescent protein (eGFP) was accomplished by fusing the C/EBPα-encoding sequence as a BamHI/XbaI fragment to a XbaI/EcoRI eGFP sequence in the pbabePuro context. Similarly, the C-terminal tagging of C/EBPα by endoplasmic reticulum (ER) was generated by fusing a BamHI/AscI fragment of C/EBPα to an AscI/EcoRI fragment of ER, again in the pbabePuro backbone.
Adipocyte differentiation.
NIH 3T3 cells and the ecotrophic Phoenix-E retroviral packaging cell line (a kind gift from G. Nolan, Stanford University, San Francisco, Calif.) were grown in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum and antibiotics. Retroviral stocks, obtained by transiently transfecting the Phoenix-E cells with the pbabePuro-C/EBPα derivatives followed by filtration of culture supernatants, were added to subconfluent (30%) NIH 3T3 cells in the presence of 5 μg/ml polybrene (Sigma). Following 24 h of infection, infected cells were grown in puromycin (1 μg/ml) for 14 days to allow differentiation to occur. The extent of differentiation was quantified either by staining with Oil Red O (27) or by Northern blotting by measuring the upregulation of the late adipocyte marker aP2 (24)
Transactivation.
HEK293S cells were cultured in DMEM supplemented with 10% fetal bovine serum and antibiotics. Cells were transiently transfected and assayed for reporter gene expression as previously described (26). pTK81-4xC/EBPwtLuc (containing a tetramer of the C/EBP binding site from the human granulocyte colony-stimulating factor receptor) was a kind gift from Daniel Tenen (29).
Growth repression assay.
In the experiments involving GTP-tagged versions of C/EBPα, retrovirally transduced NIH 3T3 cells were kept subconfluent in selective medium with regular trypsinization and replating of the cells. After approximately 2 weeks, cells were harvested and subjected to fluorescence-activated cell sorting (FACS) analysis.
Untagged C/EBPα was assayed either by retroviral transduction of NIH 3T3 cells followed by puromycin selection or by transfection of HEK293S cells with pcDNA3-C/EBPα constructs followed by G418 selection (active concentration, 320 μg/ml). Cells were kept at subconfluency as described above for 3 weeks and assayed by Western blotting analysis.
For the ER-tagged C/EBPα versions, C/EBPα-ER clones were generated by retroviral transduction of NIH 3T3 cells followed by puromycin selection. Individual clones were obtained by limited dilution followed by quantification of expression levels by Western blot analysis. In order to assay the effects of the C/EBPα-ER on the cell cycle profile, individual clones were induced with 1 μM 4-hydroxy tamoxifen for 3 days and pulsed with 100 μM bromodeoxyuridine (BrdU) (Sigma) for 45 min before harvesting with trypsin. Cells were fixed in 70% methanol, incubated on ice for 30 min, and washed once with phosphate-buffered saline (PBS). Following addition of 2 N HCl and 30 min of incubation at room temperature, the cells were washed three times with PBS plus 10% fetal calf serum (FCS). Twenty-five microliters of anti-BrdU fluorescein isothiocyanate (Becton Dickinson) was added and incubated 1 h at room temperature in the dark. After washing twice with PBS plus 5% FCS, cells were incubated for 30 min at 37°C in the dark with 200 μl propidium iodide buffer (10 mM Tris, pH 7.5; 5 mM MgCl2; 50 μg/ml propidium iodide [Sigma]; 0.1% NaN3; 10 μg/ml RNase A) and analyzed on a FACSCalibur (Becton Dickinson).
Generation of a ΔPHR knock-in mouse strain.
We have previously described the construction of targeting constructs for the generation of C/EBPα knock-in mouse lines with point mutations in the basic region of the protein (24). These constructs encode hybrids between mouse (5′ end) and rat (3′ end) with the fusion point located in the MluI site at nucleotide 573. The ΔPHR targeting construct was generated in a similar manner, except that the fusion point between the murine and rat sequences was moved to the FseI site at nucleotide 543 (the MluI site is located in the deleted part of C/EBPα). This does not change the protein sequence.
Embryonic day 14.1 (E14.1) embryonic stem (ES) cells were maintained in knockout DMEM containing 15% ES serum replacement (Invitrogen) and leukemia inhibitory factor. Electroporation, selection, and identification of correctly targeted clones were performed as previously described (24). Two clones were injected into C57BL/6 blastocysts, and the resulting chimeras were tested for germ line transmission. The ΔPHR knock-in strain was subsequently maintained on a mixed background (C57BL/6, 129/Ola). The generation of the WTki and BRM2 knock-in strains has been described previously (24). The C/EBPα knockout mice (originally constructed in the Darlington laboratory [38] and obtained from Dan Tenen, Harvard Institutes of Medicine) were kept on a mixed background. All mice strains were routinely genotyped by Southern blotting using EcoRI and a 1.7-kb HindIII/EcoRI probe. This procedure yielded the following bands: wild type (WT) (9.0 kb), BRM2/ΔPHR (3.0 kb), and null (1.7 kb).
Western blotting, immunoprecipitation, and antibodies.
Cultured cells were harvested by trypsinization, washed with PBS, and lysed with 1× sodium dodecyl sulfate (SDS) loading buffer. Nuclear proteins were extracted from fetal or adult liver as well as from tissue culture cells according to the protocol from Timchenko and coworkers (32). C/EBPα-FLAG and mutant versions were immunoprecipitated (Flag antibody, M2; Sigma) from nuclear extracts isolated from HEK293S cells cotransfected with pcDNA3-C/EBPα-FLAG (WT, ΔPHR, or BRM2) and pCMV-E2F1 (a kind gift from Kristian Helin) expression constructs.
Western blots were probed with the following antibodies: C/EBPα (14AA, Sc-61), cyclin A (c19, Sc-596), and CDK2 (M2, Sc-163), all from Santa Cruz Biotechnology, and proliferating cell nuclear antigen (PCNA) (Ab-1; Oncogene), α-tubulin (DM1A; Sigma), and β-actin (AC 15; Abcam).
Mouse phenotypic analysis. (i) Kinase assays.
Fetal liver was isolated from embryos at E16.5 or E18.5. Liver was minced in ECB buffer (50 mM Tris-HCl, pH 7.5, 120 mM NaCl, 0.5% NP-40, and 1 mM EDTA) using a pestle (Kontes, N.J.), and protein concentration was measured. Three-hundred nanograms of lysate was added to washed anti-CDK2-beads (M2, Sc-163) or anti-FLAG (M2) in immunoprecipitation buffer (50 mM HEPES, pH 7.5, 150 mM EDTA, 2.5 mM EGTA, 10% glycerol, and 0.1% Tween 20) and incubated for 4 h with rotation. Beads were pelleted by centrifugation and washed three times in immunoprecipitation buffer. After the last wash, the immunoprecipitates were resuspended in kinase assay buffer (40 mM HEPES, pH 7.5, 80 mM MgCl2, 4 mM MnCl2, 2 mM EGTA, 20 μM ATP, 0.08 μg/μl histone H1 [Roche], and 20 μCi [γ-32P]ATP) and incubated for 30 min at 30°C in the presence or absence of 25 μM roscovitine. Following addition of SDS loading buffer, the samples were loaded on a SDS-15% polyacrylamide gel electrophoresis gel. All buffers were supplemented with the following inhibitors: 10 mM β-glycerophosphate, 1 mM NaF, 1 mM dithiothreitol, 0.1 mM Na2VO4, 2.5 ng/ml leupeptin, 2.5 ng/ml aprotinin, and 50 μg/ml phenylmethylsulfonyl fluoride.
(ii) Partial hepatectomy.
Partial hepatectomy was performed on 8- to 12-week-old mice as described previously (16).
(iii) BrdU labeling.
CebpaΔPHR/+, CebpaWTki/+, CebpaBRM2/+, or Cebpa+/− heterozygotes were mated, and pregnant females were injected 16.5 or 18.5 days after mating with 100 μg BrdU/g of bodyweight. Fetal liver was harvested after 1.5 h, fixed overnight in 4% paraformaldehyde-PBS, paraffin embedded, and sectioned at 8 μm. BrdU incorporation was assayed using a monoclonal anti-BrdU (clone BU-33; Sigma) as the primary antibody and a fluorescein isothiocyanate-conjugated goat anti-rat immunoglobulin G (Jackson laboratories) as the secondary antibody.
(iv) Histology.
Tissues were dissected, fixed, paraffin embedded, sectioned, and subsequently stained with hematoxylin-eosin.
(v) DNA microarrays.
RNA isolated from E16.5 fetal liver of two wild-type and two CebpaΔPHR/ΔPHR embryos was labeled according to the one-cycle eukaryotic target labeling protocol from Affymetrix (Santa Clara, Calif.). The samples were hybridized to Affymetrix Mouse Genome 430 2.0 GeneChips, and data were analyzed by the perfect match-only model from dChip, version 1.3 (released 23 March 2005) (15). Detection calls were determined by using MAS5.0, and a gene must express 100% present calls within a replicate group to be considered expressed. Replicate groups were compared using the “compare samples” tool in dChip requiring a change of >2-fold in either direction and a P value of <0.05. Hierarchical clustering was done by dChip as previously described (3, 6).
(vi) Flow cytometry.
Bone marrow cells were obtained by flushing femurs with DMEM supplemented with 10% FCS and 20 mM K-HEPES (pH 7.5), whereas thymic and splenic cells were isolated by mechanical disruption of the tissue. Cells were stained as described previously (24) with the indicated combinations of conjugated antibodies (or relevant isotype controls) against cell surface markers and subsequently analyzed on a FACSCalibur.
(vii) Serial replating of bone marrow cells.
Bone marrow cells were harvested as described above, and 10.00 cells (per 35-mm dish, three dishes per mouse) were seeded in methylcellulose-based medium (M3434; Stem Cell Technologies) containing erythropoietin, interleukin-3, interleukin-6, and stem cell factor. After 7 days, the number of colonies was quantified, cells were harvested, and again 10,000 cells were replated in fresh M3434 and cultured for an additional week. This procedure was repeated for several weeks.
RESULTS
Deletion of the CDK2/CDK4 interaction domain of C/EBPα has no effect on growth inhibition in vitro.
Several regions of C/EBPα have been proposed to be involved in mediating growth repression via their interactions with various components of the cell cycle machinery (Fig. 1A). Recently, a small and relatively poorly conserved region of C/EBPα has been postulated to be essential for the growth-repressive potential of C/EBPα through its ability to interact with, and repress the activity of, CDK2/CDK4.
In the initial report by Wang and coworkers, the CDK2/CDK4 inhibitory domain of C/EBPα was pinpointed to a 15-amino-acid sequence (QPPPPPPPPHPHASP; 180 to 194 of the rat C/EBPα protein) containing S193 (37). This serine was later shown to be dephosphorylated in liver tumors as well as in hepatoma cell lines through a phosphatidylinositol 3 kinase/Akt-mediated dephosphorylation, and mutation of S193A disabled the growth-inhibitory effect of C/EBPα (35). Later it was demonstrated that introduction of the S193A mutant actually stimulated growth by a mechanism involving sequestering of inhibitory Rb (36).
To gain further insights into the role of the PHR of C/EBPα in growth control, we generated an allele of C/EBPα lacking this domain (Fig. 1A). Initially, we tested the growth-inhibitory potential of WT C/EBPα and C/EBPαΔPHR by tagging them with green fluorescent protein (GFP). We transduced NIH 3T3 cells with retroviruses encoding these alleles of C/EBPα and kept the resulting pool of transduced cells at subconfluency for approximately 15 generations. We have previously demonstrated that this approach allows for a semiquantification of the growth-inhibitory potential of a given C/EBPα allele, as its expression in the selected pool inversely correlates with its ability to inhibit growth (24). In contrast to what we have previously observed with the E2F repression-deficient BRM2 allele, we did not notice any differences between the WT and the ΔPHR alleles in this assay (Fig. 1B and C).
In order to exclude the possibility that the “tagging” per se of C/EBPα interfered with its ability to repress growth, we performed similar experiments (i.e., growth at subconfluency for >15 generations) with the untagged proteins and assayed their expression levels by Western blotting. Again, neither in NIH 3T3 cells nor in HEK293S cells did deletion of the CDK2/CDK4 interaction domain have any significant effect on the accumulation of C/EBPα, whereas the BRM2 mutation resulted in a marked increased in C/EBPα levels (Fig. 1D). These findings suggest that the PHR domain is dispensable for growth repression, at least in our systems.
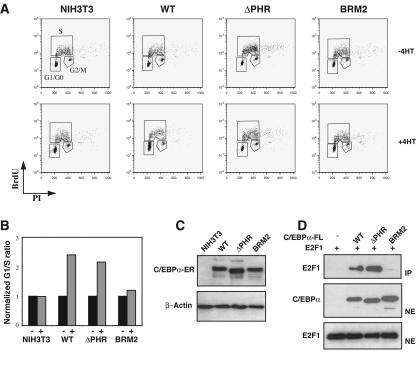
In an attempt to more specifically address the role of C/EBPα in growth repression, we generated NIH 3T3 clones expressing fusions of C/EBPα and the ligand-binding domain of the estrogen receptor (Fig. 2C). Addition of 4-hydroxytamoxifen to these cell lines induces the nuclear translocation of the fusion proteins. When NIH 3T3 cells expressing either wild-type C/EBPα or the ΔPHR variant as ER fusion proteins were induced with 4-hydroxytamoxifen, cells accumulated in GO/G1 (Fig. 2A). In contrast, cells expressing the E2F repression-deficient C/EBPα-BRM2-ER failed to accumulate in GO/G1 under similar conditions. Although some variation was observed between individual clones, analysis of several clones yielded similar G1/S ratios with no discernible differences between wild-type C/EBPα and the C/EBPαΔPHR protein (Fig. 2B). In summary, in none of our in vitro experiments did deletion of the CDK2/CDK4 interaction domain have any significant effect on C/EBPα-mediated growth repression in vitro.
FIG. 2.
In vitro growth repression of C/EBPα-ER fusions. (A) NIH 3T3 clones expressing C/EBPα (WT), C/EBPαΔPHR (ΔPHR), or C/EBPαBRM2 (BRM2). Nuclear localization of C/EBPα-ER or derivatives was accomplished by addition of 1 μM 4-hydroxy tamoxifen (4HT) for 72 h. Following BrdU labeling and propidium iodide staining (see Materials and Methods), cells were subjected to FACS analysis in order to quantify the distribution of cells in the different phases of the cell cycle. The experiments were performed three times. (B) A histogram plot showing the G1/S ratios derived from the experiments shown in panel A. (C) A Western blot of lysates derived from the cells shown in panel A probed with a C/EBPα-specific antibody. A β-actin antibody was used to control for even loading. (D) FLAG-tagged wild-type or mutant C/EBPα was immunoprecipitated from nuclear extracts from cells cotransfected with pcDNA3-C/EBPα-FLAG (WT, ΔPHR, or BRM2) and pCMV-E2F1. Western blot analysis shows E2F1 in the immunoprecipitate (IP) and E2F1 and C/EBPα in the nuclear extracts (NE).
We next tested the ability of different C/EBPα variants to interact with CDK2 and E2F1 using coimmunoprecipitation analysis. In order to maximize the chances of detecting these interactions, we coexpressed FLAG-tagged C/EBPα with either E2F1 or CDK2. Despite numerous attempts, we failed to detect any CDK2 in the precipitates when nuclear extracts from cells transfected with FLAG-tagged wild-type C/EBPα or the ΔPHR and BRM2 variants were immunoprecipitated with an antibody directed against the FLAG epitope (data not shown). One potential reason could be the relatively harsh conditions needed for extracting C/EBPα proteins from the nucleus (0.42 M NaCl), which in our hands could lead to destruction of C/EBPα-CDK2 complexes. Thus, we have been able to verify neither the existence of a C/EBPα-CDK2 complex nor its destruction by deleting the PHR domain. In contrast, when the ability of FLAG-tagged wild-type C/EBPα to interact with E2F1 was tested in a similar setting, both wild-type C/EBPα and the ΔPHR variant, but not the BRM2 variant, readily coimmunoprecipitated E2F1 (Fig. 2D).
Deletion of the CDK2/CDK4 interaction domain reduces in vitro adipocyte differentiation.
We and others have previously demonstrated that transduction of NIH 3T3 cells with retroviruses encoding C/EBPα is sufficient to initiate and sustain adipocyte differentiation (5, 22, 24). Moreover, as this system fails to induce the endogenous C/EBPα gene, it is ideally suited for analyzing the importance of various functional regions of C/EBPα in in vitro adipogenesis (5, 22, 24).
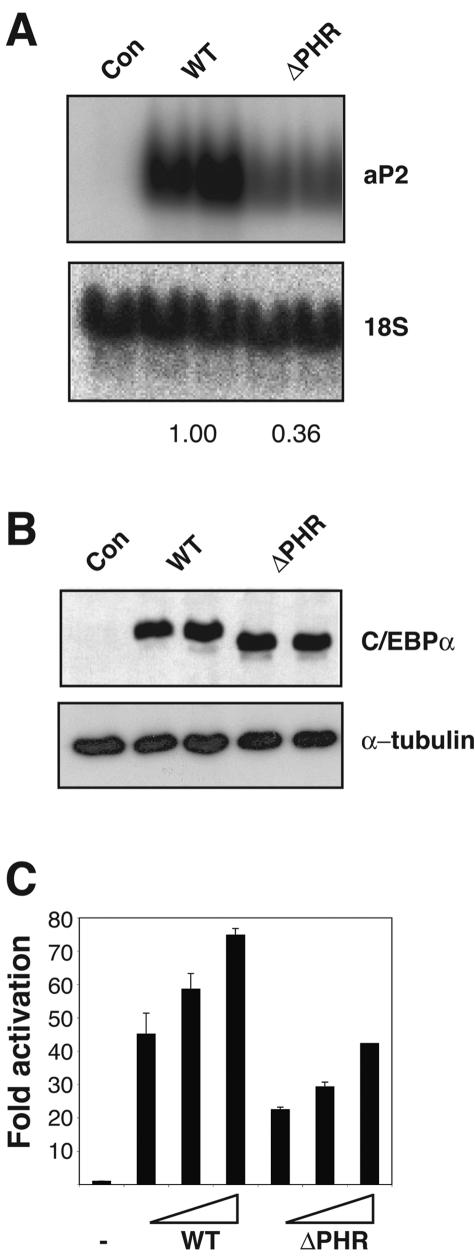
Transduction of NIH 3T3 cells with retroviruses encoding either wild-type C/EBPα or C/EBPαΔPHR leads to adipocyte differentiation as assayed by the late adipogenic marker aP2. Although both proteins sustained adipogenesis, the ΔPHR variant was markedly impaired, as illustrated by the almost threefold lower upregulation of the adipocyte marker transcript (Fig. 3A). The finding that wild-type C/EBPα and the C/EBPαΔPHR proteins accumulated to similar extents in the transduced cells suggests that the reduced adipogenic potential conferred by the ΔPHR variant was not due to differences in protein stability and/or expression levels (Fig. 3B). As the PHR is located within the transactivation element III (spanning amino acids 126 to 200 of the protein) of C/EBPα, we next compared the transactivation potential of the wild-type C/EBPα to that of the ΔPHR variant. We found a 2.5-fold reduction when assayed on a minimal promoter construct containing four consecutive C/EBP sites in transient transfection experiments in HEK293S cells (Fig. 3C). This observation suggests that the reduced adipogenic potential of the ΔPHR mutant in vitro is due to a reduction in target gene activation.
FIG. 3.
In vitro adipogenesis. (A) NIH 3T3 cells, transduced with retroviruses from Phoenix-E cultures transfected with either pBabePuro (Con) expressing C/EBPα (WT) or C/EBPαΔPHR (ΔPHR), were allowed to differentiate into adipocytes. Northern blot analysis used probes against the late adipogenic marker aP2 and 18S rRNA (for normalization). The numbers below the blots indicate the extent of differentiation relative to the wild type (derived from two independent sets of experiments). (B) Western blot analysis of the cultures from panel A probed with a C/EBPα-specific antibody. An α-tubulin antibody was used to control for even loading. (C) HEK293S cells were transiently transfected with the 1-μg reporter (pTK81-4xC/EBPwtLuc) and internal control (pcH110) and increasing amounts (10, 20, and 30 ng) of either pcDNA3-C/EBPα (WT) or pcDNA3-C/EBPαΔPHR (ΔPHR). After approximately 48 h, luciferase activities were measured and normalized to the internal β-galactosidase control. Each value represents the average of two independent experiments. Error bars indicate standard deviations.
Generation and analysis of knock-in mice expressing C/EBPαΔPHR.
We were puzzled by the apparent contradictions between our in vitro results and those published by Wang and coworkers (35-37). Although it is likely that these differences to a large extent may be explained by differences in experimental design, they certainly underline the importance of developing an in vivo system for testing the importance of the PHR domain. Thus, we generated C/EBPαΔPHR/+ knock-in mice in which the Cebpa allele was replaced with the ΔPHR variant using our previously described targeting strategy (24).
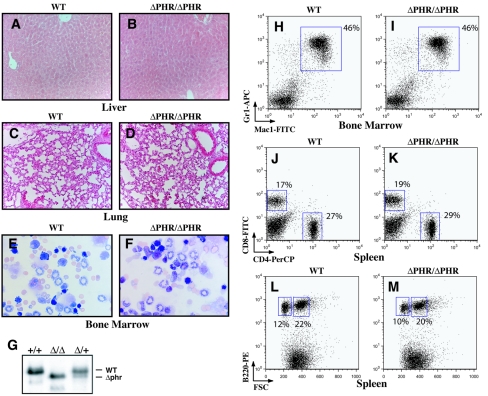
Homozygous ΔPHR knock-in mice were born and weaned at the Mendelian ratio, demonstrating that the CDK2/CDK4 interaction domain is dispensable for embryonic survival (data not shown). Expression of the ΔPHR variant of C/EBPα in the liver was comparable to its wild-type counterpart, demonstrating the feasibility of the knock-in approach (Fig. 4G). Genetic manipulation of the cebpa locus has previously been shown to affect the development of several organs and cell types (4, 24, 38). The lack of phenotypic consequences in liver (Fig. 4A and B), lung (Fig. 4C and D), and fat (data not shown and T. Å. Pedersen, O. Bereshchenko, P. Kirstetter, E. Kurz, B. T. Porse, and C. Nerlov, unpublished data) tissues in adult (8 to 12 weeks old) homozygous ΔPHR knock-in mice demonstrate that the potential CDK2/CDK4 interaction is dispensable for C/EBPα-mediated development of these organs.
FIG. 4.
Analysis of a knock-in mouse line expressing the C/EBPαΔPHR variant. Representative hematoxylin-eosin-stained sections of liver (A and B) (original magnification, 20×) and lung (C and D) (original magnification, 20×) tissues from 8- to 12-week-old ΔPHR/ΔPHR (B and D) and +/+ littermate controls (A and C). Representative cytospins of bone marrow cells (E and F) (original magnification, 40×). (G) Western blot showing the expression of C/EBPα in wild-type (+/+), ΔPHR/+ (Δ/+), and ΔPHR/ΔPHR (Δ/Δ) mice in adult liver. Representative FACS analyses of WT and ΔPHR homozygotes showing the distributions of Mac1+, Gr1+ granulocytes in the bone marrow (H and I), splenic CD4− CD8+ and/or CD4+ CD8− T cells (J and K), and splenic B220+ B cells (L and M). The sizes of the various populations are indicated next to the gates. FITC, fluorescein isothiocyanate; APC, allophycocyanin; PerCP, peridinin chlorophyll protein; PE, phycoerythrin.
C/EBPα null mice completely lack neutrophilic granulocytes, whereas mice homozygous for the E2F-interaction-deficient BRM2 allele initially suffer from neutropenia that later progresses to either a myeloproliferative or a myeloid leukemic phenotype (23, 24, 40). These findings prompted us to analyze the hematopoietic development in the ΔPHR knock-in mice. No gross morphological differences were observed when bone marrow cytospins derived from ΔPHR homozygotes were compared to their wild-type littermates (Fig. 4E and F). In addition, flow cytometric analysis (FACS) failed to pick up any differences in the cellular distributions of granulocytes (Mac1+, Gr1+) in either bone marrow (Fig. 4H and I) or spleen (data not shown), T cells (CD4− CD8+ or CD4+ CD8−) in spleen (Fig. 4J and K), or B cells (B220+) in spleen (Fig. 4L and M). These findings demonstrate that the CDK2/CDK4 interaction domain of C/EBPα is dispensable for differentiation along these hematopoietic lineages.
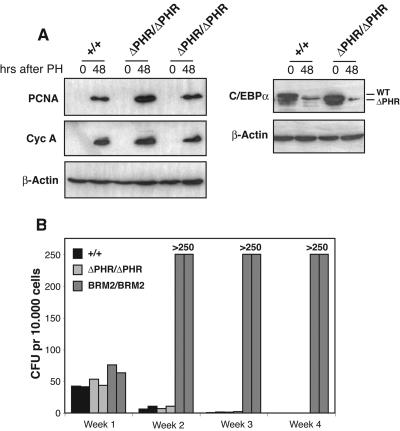
To gain further insights into the putative functional roles of the PHR in vivo, we next conducted two types of experiments. First, since modulation of C/EBPα activity in the liver has been shown to be instrumental in order for postmitotic cells to reenter the cell cycle, we tested whether livers from adult (8 to 12 weeks old) wild-type and ΔPHR/ΔPHR mice responded similarly to partial hepatechtomy. As shown in Fig. 5A, we observed a robust upregulation of two cell cycle-related proteins, proliferating cell nuclear antigen (PCNA) and cyclin A, 48 h after partial hepatechtomy in mice of both genotypes, accompanied by a comparable downregulation of C/EBPα levels. These findings suggest that deletion of the PHR neither promoted nor interfered with the ability of postmitotic liver cells to reenter the cell cycle. Secondly, as we have previously shown that abrogation of the ability of C/EBPα to inhibit E2F activity results in an increase in the proliferative capacity of myeloid progenitors (23), we next analyzed the functional consequences of deleting the PHR in a serial replating experiment (Fig. 5B). In this assay, bone marrow cells were seeded in semisolid medium, allowing outgrowth of all myeloid cell types, and replated on a weekly basis. Bone marrow cells from ΔPHR/ΔPHR mice displayed the same limited proliferative potential as their wild-type counterparts and ceased to form colonies at the third replating. In contrast, bone marrow cells derived from BRM2/BRM2 mice (carrying the E2F repression deficient version of C/EBPα) had strongly increased replating potential and continued to proliferate well beyond the fourth replating (Fig. 5B) (23 and data not shown). These findings are inconsistent with the PHR having any functional role in regulating either growth or differentiation in the myeloid compartment.
FIG. 5.
Analysis of the proliferative capacity of ΔPHR homozygous mice. (A) Western blots of nuclear extracts derived from +/+, ΔPHR/+, and ΔPHR/ΔPHR mice before and 48 h after partial hepatechtomy (PH). The blots were probed with antibodies against PCNA, cyclin A, C/EBPα, and β-actin. (B) Serial replating of bone marrow cells seeded in semisolid M3434 medium as described in Materials and Methods. Each bar represents the average of an experiment performed in triplicate.
No evidence for the involvement of the CDK2/CDK4 interaction domain in C/EBPα-mediated growth repression in the liver during embryonic development.
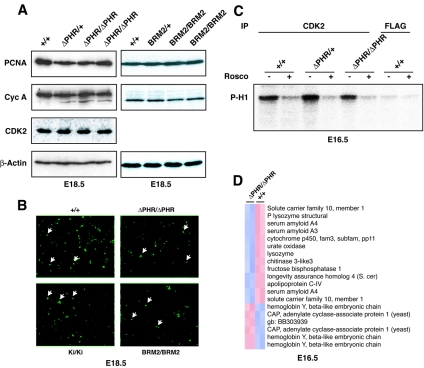
Given the suggested importance of the CDK2/CDK4 interaction domain for C/EBPα-mediated growth repression during development of the fetal liver, we were surprised to find that the ΔPHR homozygotes were born at the Mendelian ratio. It has been reported that C/EBPα null embryos had significantly increased expression levels of several cell cycle-related proteins, such as PCNA and cyclin A, in the developing liver correlating with higher numbers of cycling cells (4, 32, 33). To test whether deletion of the CDK2/CDK4 interaction domain resulted in a similar phenotype, nuclear extracts were isolated from wild-type, ΔPHR/+, and ΔPHR/ΔPHR livers harvested at E18.5, and the levels of relevant proteins were assayed by Western blotting. In contrast to the previously reported findings in C/EBPα null embryos, we observed no upregulation of either PCNA, cyclin A, or CDK2 (Fig. 6A). In order to more directly assay the number of cells in the S phase, we performed an in vivo BrdU incorporation assay. Consistent with the protein expression data, comparable numbers of BrdU-positive cells were observed in the livers derived from embryos of the various genotypes (Fig. 6B).
FIG. 6.
In vivo growth regulation during liver development in the ΔPHR homozygous mice. (A) Western blots of nuclear extracts isolated from fetal livers of +/+, ΔPHR/+, ΔPHR/ΔPHR, BRM2/+, and BRM2/BRM2 embryos were probed with antibodies raised against PCNA, cyclin A, CDK2, and β-actin. (B) BrdU incorporation in fetal liver (original magnification, 20×). A few labeled nuclei are indicated by arrows. The percentage of BrdU-labeled nuclei in the wild-type (+/+) mice, the ΔPHR homozygotes (ΔPHR/ΔPHR), the Wtki homozygotes (Ki/Ki), and the BRM2 homozygotes (BRM2/BRM2) ranged between 5 and 7% (approximately 600 nuclei were counted). (C) Phosphorylation of histone H1 (P-H1) was assayed using anti-CDK2- and anti-FLAG-precipitated liver lysates isolated from +/+, ΔPHR/+, or ΔPHR/ΔPHR embryos. The CDK2 inhibitor roscovitine (Rosco) was added in the lanes marked with a plus sign. (D) Transcriptional gene profiling of total fetal liver RNA derived from two ΔPHR/ΔPHR mice and two +/+ littermate controls. Hierarchical clustering of the 14 genes displays altered gene expression. Pearson correlation coefficients were >0.988 for the individual pairwise comparisons. S. cer, Saccharomyces cerevisiae; gb, GenBank.
C/EBPα-mediated repression of E2F activity is another candidate pathway by which C/EBPα may regulate growth in the developing liver. To test this possibility, we performed identical experiments using the previously described BRM2 knock-in line that express an E2F repression-deficient C/EBPα allele (24). Similar to what was found for the homozygous ΔPHR embryos, homozygous BRM2 embryonic livers also expressed comparable levels of cell cycle-related genes as the wild-type and BRM2/+ controls (Fig. 6A). Moreover, when incorporation of BrdU was assayed in vivo, we failed to observe any increase in the number of cycling cells in the BRM2/BRM2 homozygotes as well as in the previously described WTki/WTki control strain (Fig. 6B) (24).
As deletion of a region similar to PHR was previously shown to disrupt binding to CDK2/CDK4 in vitro, we wished to address whether CDK2 activity was altered in embryonic livers of the homozygous ΔPHR mice (37). Liver extracts from E16.5 embryos were prepared, and the ability of the immunoprecipitated CDK2 to phosphorylate histone H1 was assayed in the presence or absence of the CDK inhibitor roscovitine (which inhibits CDKs 1, 2, 5, and 7 [1]). High levels of kinase activity, which could be inhibited by roscovitine, were observed in the CDK2 precipitates, whereas only very low background activity was detectable using a control antibody (Fig. 6C). However, again no differences were detected when fetal liver extracts from wild-type, ΔPHR/+, and ΔPHR/ΔPHR mice were compared.
In a final attempt to detect any differences between the wild-type and the homozygous ΔPHR embryos in terms of the development of the embryonic liver, we performed a DNA microarray experiment on RNA isolated from E16.5 fetal livers. A total of 14 genes (out of the 39.000 transcripts present on the Affymetrix 430 2.0 chip) displayed altered expression levels (Fig. 5D; >2-fold; P < 0.05). The deregulated genes mainly encode proteins involved in metabolic processes, and although this may reflect a function for the CDK2/CDK4-interacting PHR in the developing liver, neither the transcriptional profiling nor the remaining in vivo data presented above support any function for this part of C/EBPα in growth regulation during embryonic development of the liver.
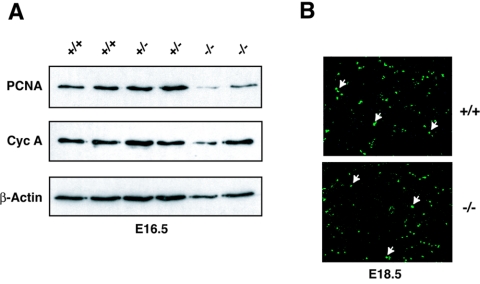
We were surprised by the findings that abrogation of two of the suggested growth-inhibitory functions of C/EBPα, i.e., E2F repression and CDK2 inhibition, failed to confer any aberrant cell cycle-related phenotypes in the developing liver. We therefore went back and reevaluated the initial observations by Timchenko and colleagues (32, 33), i.e., the finding that C/EBPα null embryos have increased levels of proliferation in the developing liver. Western blot analysis of nuclear extracts derived from E16.5 (Fig. 7A) and E18.5 (data not shown) fetal livers from wild-type, Cebpα+/−, and Cebpα−/− embryos revealed similar levels of both PCNA and cyclin A. Similarly, there was no general increase in BrdU incorporation in the developing liver of C/EBPα knockout embryos at either E16.5 (data not shown) or at E18.5 (Fig. 7B). These findings are inconsistent with C/EBPα playing a general role in regulating proliferation in the developing liver.
FIG. 7.
In vivo growth regulation during liver development in C/EBPα knockout mice. (A) Western blots of nuclear extracts isolated form fetal liver of +/+, +/−, and −/− embryos were probed with antibodies against PCNA, cyclin A (Cyc A), and β-actin. (B) BrdU incorporation in fetal liver (original magnification, 20×). A few labeled nuclei are indicated by arrows.
DISCUSSION
In the present work we have tested the role of the centrally located 15-amino-acid proline-histidine-rich region of C/EBPα with respect to C/EBPα-mediated growth regulation. This part of C/EBPα has attracted considerable attention, as its deletion was demonstrated to ablate the growth-inhibitory potential of C/EBPα in tissue culture experiments and that it did so through its inability to bind to and repress the activity of CDK2 and CDK4 (37). Subsequently, it was shown that in mouse-rat serine 193 (S193, suggested to correspond to S190 in humans) was critical for this activity, and although it is situated in a relatively loosely conserved region of C/EBPα, mutation of S193 has similar consequences in the context of the mouse and human C/EBPα sequences (35). Moreover, it was shown that the growth-repressive potential of C/EBPα could be released by phosphatidylinositol 3 kinase/Akt-mediated dephosphorylation (or mutation) of S193 (35).
Given the importance of C/EBPα in the development and maintenance of several organ systems and as a newly discovered tumor suppressor (7, 12, 21, 25, 30, 31), it is crucial to unravel the mechanism by which C/EBPα mediates growth repression and differentiation. In order to address the role of the PHR of C/EBPα with respect to this, we initially performed a series of in vitro growth repression experiments using both untagged, GFP-tagged, and ER-tagged versions of C/EBPα but failed to provide any evidence to support the role of this putatively critical region in mediating growth repression. However, in a C/EBPα-driven adipose differentiation system we did find that the ΔPHR mutant was significantly impaired in the induction of adipocyte differentiation correlating with a reduction in the transactivation potential of the protein. The functional implications of deleting the PHR has also been addressed in two myeloid differentiation systems; however, in neither case did deletion of this domain have any effect on differentiation (13, 39). Moreover, in the myeloid system it was also found that whereas the E2F repression-deficient BRM2 version of C/EBPα failed to inhibit G1/S phase transition, the ΔPHR variant did so to an extent similar to that of wild-type C/EBPα (39).
Given the inconsistencies between the data obtained by various groups regarding the function of the PHR of C/EBPα in vitro (which at least in part can be explained by differences in experimental setup, expression levels, etc.), we wished to move our analysis to a more robust system, i.e., the mouse. We have previously generated mice in which the wild-type allele of C/EBPα was replaced with point mutated versions (24), and we applied a similar strategy to introduce the ΔPHR variant into the wild-type cebpa locus. Mice homozygous for the ΔPHR allele were born at the Mendelian ratio and grew to adulthood with no apparent overt phenotype. In addition, no morphological changes or changes in cellular fate could be detected in tissues known to be affected in the C/EBPα null animals, such as liver, lung, and fat and within the hematopoietic system. To further address any functional role of the ΔPHR, we next performed an in vitro serial replating experiment aimed at detecting alterations in the proliferative capacity of myeloid progenitor cells. Here, ΔPHR/ΔPHR bone marrow cells displayed the same limited proliferative potential as their wild-type counterparts, whereas BRM2/BRM2 cells strongly enhanced proliferation of myeloid progenitors, supporting our previous conclusions that E2F repression is the main antimitotic effect of C/EBPα in the myeloid system (23, 24). In a final attempt to detect any phenotypic consequences related to cell cycle control in the adult ΔPHR/ΔPHR mice, we performed partial hepatechtomy. These experiments demonstrated a comparable robust upregulation of markers for cycling cells accompanied by downregulation of C/EBPα in both ΔPHR/ΔPHR and control mice.
To date, the only phenotypic alteration we have been able to detect in the ΔPHR homozygotes is the deregulated expression of a subset of SREBP-1-regulated lipogenic enzymes which, in turn, lead to increased triglyceride storage in the liver (T. Å. Pedersen, O. Bereshchenko, P. Kirstetter, E. Kurz, B. T. Porse, and C. Nerlov, unpublished data). These findings suggest a very specific function of the PHR of C/EBPα in adult liver homeostasis that is dissociated from any effect that this deletion may have on cell cycle control.
During embryonic development, C/EBPα has been shown to be a key regulator of cell cycle exit in the developing liver, and, conversely, liver regeneration is associated with downregulation of C/EBPα (4, 32). Livers isolated from C/EBPα null embryos or newborns have been reported to express increased levels of cell cycle-associated proteins such as cyclin A and PCNA accompanied by a significant increase in the numbers of cells incorporating BrdU (4, 32). These findings demonstrate that deletion of C/EBPα leads to an increase of cycling cells in the developing liver. Moreover, as it has been demonstrated that in embryonic and young livers C/EBPα binds to and represses the activity of CDK2, we expected a growth-related phenotype in the homozygous ΔPHR knock-in mice lacking the CDK2/CDK4-interacting PHR (9). Much to our surprise, PCNA and cyclin A levels were similar in ΔPHR homozygotes and their wild-type littermates in both E16.5 (data not shown) and E18.5 livers, and we observed no differences in the incorporation of BrdU. These findings were paralleled by the unaltered levels of CDK2 activity in the embryonic livers of wild-type, ΔPHR/+, and ΔPHR/ΔPHR mice and by the absence of any deregulation of genes associated with cell cycle progression. Thus, in summary, the phenotypic behavior of our ΔPHR mice lacking the putative CDK2/CDK4 interaction domain leads us to conclude that C/EBPα-mediated downregulation of CDK2/CDK4 activity is dispensable during embryonic development of the liver.
Our ΔPHR knock-in line was constructed before the demonstration that S193 was the key amino acid in the PHR, and an argument could be made as to whether deletion of the PHR mimics the phosphorylation-disabling S193A mutation. We believe it does so, as both the ΔPHR and the S193A alleles have comparable effects on the two key readouts of the in vitro systems, i.e., growth inhibition and CDK2/CDK4 interaction (35, 37).
We have also tested our previously described BRM2 knock-in line for any defect in C/EBPα-mediated growth inhibition during embryonic development of the liver, and similar to what we found for the ΔPHR homozygotes, we were unable to demonstrate any phenotype related to cell cycle control either in the developing or the adult liver (24). These findings provide clear genetic evidence that neither C/EBPα-mediated repression of CDK2/CDK4 nor C/EBPα-mediated repression of E2F activity is the mechanism by which C/EBPα regulates growth in the developing liver. Although these observations could suggest that other regions of C/EBPα may be responsible for C/EBPα-mediated growth repression in the developing liver, we were concerned about the original observation made in the C/EBPα knockout mouse (32, 33). Surprisingly, in our hands C/EBPα knockout fetal livers did not express increased levels of either PCNA or of cyclin A, and we did not observe any overall increase of BrdU incorporation. Given the fact that we used the same C/EBPα knockout strain as Timchenko and coworkers, the most likely explanation for these diverging results is differences in the genetic background on which the C/EBPα knockout allele was maintained.
One potential caveat when interpreting the antimitotic role of C/EBPα is the intertwined impact on growth regulation and differentiation that ablation of C/EBPα has in the developing liver. In particular, observations from Tomizawa and coworkers demonstrating the appearance of pseudoglandular structures accompanied by changes in cellular fates in C/EBPα null embryonic livers suggest that deletion of C/EBPα has a strong impact on liver cell differentiation (34). Thus, although our studies strongly question a general function of C/EBPα in hepatocyte growth arrest, the changes in cell fate observed upon ablation of C/EBPα preclude us from reaching a definitive conclusion as to the role of C/EBPα in growth regulation in the developing liver. In particular, despite the absence of any general upregulation of cell cycle markers in C/EBPα null livers, we cannot exclude the possibility that C/EBPα may have an antimitotic role in a small subset of cells in the developing liver.
Acknowledgments
This work was supported by the Danish Medical Research Council, The Danish Cancer Society, and the Association for International Cancer Research.
We thank Daniel Tenen for providing us with the C/EBPα knockout line.
REFERENCES
- 1.Bain, J., H. McLauchlan, M. Elliott, and P. Cohen. 2003. The specificities of protein kinase inhibitors: an update. Biochem. J. 371:199-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D'Alo, F., L. M. Johansen, E. A. Nelson, H. S. Radomska, E. K. Evans, P. Zhang, C. Nerlov, and D. G. Tenen. 2003. The amino terminal and E2F interaction domains are critical for C/EBP alpha-mediated induction of granulopoietic development of hematopoietic cells. Blood 102:3163-3171. [DOI] [PubMed] [Google Scholar]
- 3.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flodby, P., C. Barlow, H. Kylefjord, L. Ahrlund-Richter, and K. G. Xanthopoulos. 1996. Increased hepatic cell proliferation and lung abnormalities in mice deficient in CCAAT/enhancer binding protein alpha. J. Biol. Chem. 271:24753-24760. [DOI] [PubMed] [Google Scholar]
- 5.Freytag, S. O., D. L. Paielli, and J. D. Gilbert. 1994. Ectopic expression of the CCAAT/enhancer-binding protein alpha promotes the adipogenic program in a variety of mouse fibroblastic cells. Genes Dev. 8:1654-1663. [DOI] [PubMed] [Google Scholar]
- 6.Golub, T. R., D. K. Slonim, P. Tamayo, C. Huard, M. Gaasenbeek, J. P. Mesirov, H. Coller, M. L. Loh, J. R. Downing, M. A. Caligiuri, C. D. Bloomfield, and E. S. Lander. 1999. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286:531-537. [DOI] [PubMed] [Google Scholar]
- 7.Gombart, A. F., W. K. Hofmann, S. Kawano, S. Takeuchi, U. Krug, S. H. Kwok, R. J. Larsen, H. Asou, C. W. Miller, D. Hoelzer, and H. P. Koeffler. 2002. Mutations in the gene encoding the transcription factor CCAAT/enhancer binding protein alpha in myelodysplastic syndromes and acute myeloid leukemias. Blood 99:1332-1340. [DOI] [PubMed] [Google Scholar]
- 8.Hendricks-Taylor, L. R., and G. J. Darlington. 1995. Inhibition of cell proliferation by C/EBP alpha occurs in many cell types, does not require the presence of p53 or Rb, and is not affected by large T-antigen. Nucleic Acids Res. 23:4726-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iakova, P., S. S. Awad, and N. A. Timchenko. 2003. Aging reduces proliferative capacities of liver by switching pathways of C/EBPalpha growth arrest. Cell 113:495-506. [DOI] [PubMed] [Google Scholar]
- 10.Johansen, L. M., A. Iwama, T. A. Lodie, K. Sasaki, D. W. Felsher, T. R. Golub, and D. G. Tenen. 2001. c-Myc is a critical target for c/EBPα in granulopoiesis. Mol. Cell. Biol. 21:3789-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson, P. F. 2005. Molecular stop signs: regulation of cell-cycle arrest by C/EBP transcription factors. J. Cell Sci. 118:2545-2555. [DOI] [PubMed] [Google Scholar]
- 12.Kaeferstein, A., U. Krug, J. Tiesmeier, M. Aivado, M. Faulhaber, M. Stadler, J. Krauter, U. Germing, W. K. Hofmann, H. P. Koeffler, A. Ganser, and W. Verbeek. 2003. The emergence of a C/EBPα mutation in the clonal evolution of MDS towards secondary AML. Leukemia 17:343-349. [DOI] [PubMed] [Google Scholar]
- 13.Keeshan, K., G. Santilli, F. Corradini, D. Perrotti, and B. Calabretta. 2003. Transcription activation function of C/EBPα is required for induction of granulocytic differentiation. Blood 102:1267-1275. [DOI] [PubMed] [Google Scholar]
- 14.Kimura, T., V. M. Christoffels, S. Chowdhury, K. Iwase, H. Matsuzaki, M. Mori, W. H. Lamers, G. J. Darlington, and M. Takiguchi. 1998. Hypoglycemia-associated hyperammonemia caused by impaired expression of ornithine cycle enzyme genes in C/EBPα knockout mice. J. Biol. Chem. 273:27505-27510. [DOI] [PubMed] [Google Scholar]
- 15.Li, C., and W. H. Wong. 2001. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98:31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ludde, T., S. Kubicka, J. Plumpe, C. Liedtke, M. P. Manns, and C. Trautwein. 2001. Ras adenoviruses modulate cyclin E protein expression and DNA synthesis after partial hepatectomy. Oncogene 20:5264-5278. [DOI] [PubMed] [Google Scholar]
- 17.Morgenstern, J. P., and H. Land. 1990. A series of mammalian expression vectors and characterisation of their expression of a reporter gene in stably and transiently transfected cells. Nucleic Acids Res. 18:1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muller, C., M. Alunni-Fabbroni, E. Kowenz-Leutz, X. Mo, M. Tommasino, and A. Leutz. 1999. Separation of C/EBPα-mediated proliferation arrest and differentiation pathways. Proc. Natl. Acad. Sci. USA 96:7276-7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller, C., C. F. Calkhoven, X. Sha, and A. Leutz. 2004. The CCAAT enhancer-binding protein alpha (C/EBPα) requires a SWI/SNF complex for proliferation arrest. J. Biol. Chem. 279:7353-7358. [DOI] [PubMed] [Google Scholar]
- 20.Nerlov, C., and E. B. Ziff. 1994. Three levels of functional interaction determine the activity of CCAAT/enhancer binding protein-alpha on the serum albumin promoter. Genes Dev. 8:350-362. [DOI] [PubMed] [Google Scholar]
- 21.Pabst, T., B. U. Mueller, P. Zhang, H. S. Radomska, S. Narravula, S. Schnittger, G. Behre, W. Hiddemann, and D. G. Tenen. 2001. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPα), in acute myeloid leukemia. Nat. Genet. 27:263-270. [DOI] [PubMed] [Google Scholar]
- 22.Pedersen, T. A., E. Kowenz-Leutz, A. Leutz, and C. Nerlov. 2001. Cooperation between C/EBPα TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes Dev. 15:3208-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porse, B. T., D. Bryder, K. Theilgaard-Monch, M. S. Hasemann, K. Anderson, I. Damgaard, S. E. Jacobsen, and C. Nerlov. 2005. Loss of C/EBP alpha cell cycle control increases myeloid progenitor proliferation and transforms the neutrophil granulocyte lineage. J. Exp. Med. 202:85-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porse, B. T., T. A. Pedersen, X. Xu, B. Lindberg, U. M. Wewer, L. Friis-Hansen, and C. Nerlov. 2001. E2F repression by C/EBPα is required for adipogenesis and granulopoiesis in vivo. Cell 107:247-258. [DOI] [PubMed] [Google Scholar]
- 25.Preudhomme, C., C. Sagot, N. Boissel, J. M. Cayuela, I. Tigaud, S. de Botton, X. Thomas, E. Raffoux, C. Lamandin, S. Castaigne, P. Fenaux, and H. Dombret. 2002. Favorable prognostic significance of CEBPA mutations in patients with de novo acute myeloid leukemia: a study from the Acute Leukemia French Association (ALFA). Blood 100:2717-2723. [DOI] [PubMed] [Google Scholar]
- 26.Querfurth, E., M. Schuster, H. Kulessa, J. D. Crispino, G. Doderlein, S. H. Orkin, T. Graf, and C. Nerlov. 2000. Antagonism between C/EBPβ and FOG in eosinophil lineage commitment of multipotent hematopoietic progenitors. Genes Dev. 14:2515-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramirez-Zacarias, J. L., F. Castro-Munozledo, and W. Kuri-Harcuch. 1992. Quantitation of adipose conversion and triglycerides by staining intracytoplasmic lipids with Oil Red O. Histochemistry 97:493-497. [DOI] [PubMed] [Google Scholar]
- 28.Slomiany, B. A., K. L. D'Arigo, M. M. Kelly, and D. T. Kurtz. 2000. C/EBPα inhibits cell growth via direct repression of E2F-DP-mediated transcription. Mol. Cell. Biol. 20:5986-5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith, L. T., S. Hohaus, D. A. Gonzalez, S. E. Dziennis, and D. G. Tenen. 1996. PU. 1 (Spi-1) and C/EBP alpha regulate the granulocyte colony-stimulating factor receptor promoter in myeloid cells. Blood 88:1234-1247. [PubMed] [Google Scholar]
- 30.Smith, M. L., J. D. Cavenagh, T. A. Lister, and J. Fitzgibbon. 2004. Mutation of CEBPA in familial acute myeloid leukemia. N. Engl. J. Med. 351:2403-2407. [DOI] [PubMed] [Google Scholar]
- 31.Snaddon, J., M. L. Smith, M. Neat, M. Cambal-Parrales, A. Dixon-McIver, R. Arch, J. A. Amess, A. Z. Rohatiner, T. A. Lister, and J. Fitzgibbon. 2003. Mutations of CEBPA in acute myeloid leukemia FAB types M1 and M2. Genes Chromosomes Cancer 37:72-78. [DOI] [PubMed] [Google Scholar]
- 32.Timchenko, N. A., T. E. Harris, M. Wilde, T. A. Bilyeu, B. L. Burgess-Beusse, M. J. Finegold, and G. J. Darlington. 1997. CCAAT/enhancer binding protein alpha regulates p21 protein and hepatocyte proliferation in newborn mice. Mol. Cell. Biol. 17:7353-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Timchenko, N. A., M. Wilde, and G. J. Darlington. 1999. C/EBPα regulates formation of S-phase-specific E2F-p107 complexes in livers of newborn mice. Mol. Cell. Biol. 19:2936-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomizawa, M., S. Garfield, V. Factor, and K. G. Xanthopoulos. 1998. Hepatocytes deficient in CCAAT/enhancer binding protein alpha (C/EBP alpha) exhibit both hepatocyte and biliary epithelial cell character. Biochem. Biophys. Res. Commun. 249:1-5. [DOI] [PubMed] [Google Scholar]
- 35.Wang, G. L., P. Iakova, M. Wilde, S. Awad, and N. A. Timchenko. 2004. Liver tumors escape negative control of proliferation via PI3K/Akt-mediated block of C/EBP alpha growth inhibitory activity. Genes Dev. 18:912-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, G. L., and N. A. Timchenko. 2005. Dephosphorylated C/EBPα accelerates cell proliferation through sequestering retinoblastoma protein. Mol. Cell. Biol. 25:1325-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, H., P. Iakova, M. Wilde, A. Welm, T. Goode, W. J. Roesler, and N. A. Timchenko. 2001. C/EBPα arrests cell proliferation through direct inhibition of Cdk2 and Cdk4. Mol. Cell 8:817-828. [DOI] [PubMed] [Google Scholar]
- 38.Wang, N. D., M. J. Finegold, A. Bradley, C. N. Ou, S. V. Abdelsayed, M. D. Wilde, L. R. Taylor, D. R. Wilson, and G. J. Darlington. 1995. Impaired energy homeostasis in C/EBP alpha knockout mice. Science 269:1108-1112. [DOI] [PubMed] [Google Scholar]
- 39.Wang, Q. F., R. Cleaves, T. Kummalue, C. Nerlov, and A. D. Friedman. 2003. Cell cycle inhibition mediated by the outer surface of the C/EBPα basic region is required but not sufficient for granulopoiesis. Oncogene 22:2548-2557. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, D. E., P. Zhang, N. D. Wang, C. J. Hetherington, G. J. Darlington, and D. G. Tenen. 1997. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc. Natl. Acad. Sci. USA 94:569-574. [DOI] [PMC free article] [PubMed] [Google Scholar]