Abstract
The immunoglobulin kappa light chain (Igκ) locus is regulated in a lineage- and stage-specific manner during B-cell development. The highly restricted timing of V to J gene recombination at the pre-B-cell stage is under the control of two enhancers, the intronic enhancer (κEi) and the 3′ enhancer (κE3′), flanking the constant exon. E2A transcription factors have been indicated to be directly involved in the regulation of Igκ locus activation. In this study, we utilize E2A-deficient pre-B cells to directly investigate the mechanism of E2A-mediated Igκ activation. We demonstrate that Igκ germ line transcription is severely impaired and recombination is blocked in the absence of E2A. Reconstitution of E2A−/− pre-B cells with inducible human E2A (E47R) is sufficient to promote chromatin modification of Igκ and rescue Igκ germ line transcription and Jκ gene recombinase accessibility. Furthermore, we show that increased E2A recruitment to κEi and κE3′ correlates with activation of Igκ in pre-B cells and that recruitment of E2A to κE3′ is in part dependent on the transcription factor IRF-4. Inhibition of IRF-4 expression in pre-B cells leads to a significant reduction of Igκ germ line transcription and enhancer acetylation. In the absence of E2A, increased IRF-4 expression is not sufficient to promote Igκ enhancer chromatin modification or transcription, suggesting that the sequential involvement of IRF-4 and E2A is necessary for the activation of the Igκ locus. Finally, we provide genetic evidence in the mouse that E2A gene dosage can influence the development of pre-B cells during the phase of Igκ gene activation.
Each unique B lymphocyte antigen receptor gene is created by somatic recombination within the immunoglobulin (Ig) heavy and light chain loci. Heavy chain gene segment recombination precedes light chain gene segment recombination during B-cell development, ensuring that a single functional heavy chain is produced before light chain locus recombination begins. Light chain locus recombination is then activated by signaling through a successfully expressed pre-B-cell receptor. Mice and humans have two light chain genes, κ and λ, that can potentially undergo recombination to generate a functional light chain gene. However, nearly all mature B cells express only one functional κ or λ light chain expressed from a single allele. It has been demonstrated with mice that the κ genes are activated and undergo recombination before λ, which contributes significantly to the preferential usage of the κ light chain in mature B cells (95% of mature B cells in mouse are κ+) (1, 28). Igκ light chain locus activation is mediated by cis-acting regulatory elements which include two enhancers, the intronic enhancer (κEi) and the 3′ enhancer (κE3′), located in the J-C intron and 3′ of the Cκ exon, respectively. Deletion of either κEi or κE3′ negatively impacts the frequency of κ recombination, resulting in a reduction in the ratio of κ- versus λ-expressing B cells, while deletion of both enhancers blocks κ recombination altogether (13). Therefore, there is a degree of functional overlap between κEi and κE3′. In addition, each κ enhancer also mediates enhancer-specific, nonredundant effects which are required for normal κ recombination and expression (8, 13, 35). The activation of Igκ locus recombination in pre-B cells correlates with the expression of sterile germ line transcripts that originate from an initiation site immediately 5′ of the Jκ1 exon and from a second promoter approximately 3 kb 5′ of Jκ1 (31, 34). Deletion of the region 5′ of Jκ1 that contains the germ line transcript initiation sites severely impairs Igκ recombination in cis, indicating that germ line transcription may be involved in regulating recombinase accessibility of Jκ genes (4).
Kappa locus activation in pre-B cells is regulated by lineage-specific and ubiquitously expressed trans-acting factors. Molecular studies have implicated multiple transcription factors, such as E2A, NF-κB, Pax-5, Pu.1, SpiB, and IRF-4, in the regulation of Igκ locus activation (30). Strong evidence indicating that the ubiquitously expressed basic helix-loop-helix transcription factor E2A plays an important role in the regulation of Igκ has accumulated. Ectopic expression of E2A and the RAG recombinases in a nonlymphoid cell line is sufficient to induce a diverse repertoire of Igκ recombination (29). These data indicate that E2A expression is sufficient to promote chromatin modification and recombinase accessibility of the κ locus. E2A DNA binding sites (E-boxes) have been identified within both κEi and κE3′, and chromatin immunoprecipitation (ChIP) studies confirm that E2A directly associates with these two enhancers in pre-B cells (10, 21, 25). Moreover, mutation of just one of two critical E-boxes, E1 or E2, in κEi is sufficient to impair κ recombination on the targeted allele in pre-B cells, while mutation of both sites results in an even more severe block in κ recombination, thus directly demonstrating the functional importance of E2A in the regulation of Igκ (14). While these studies demonstrate the central importance of E2A in regulating Igκ transcription and recombination during B-cell development, it is not clear how E2A activity is regulated to ensure the proper timing of κ locus activation and recombination.
E2A expression is upregulated upon B-cell lineage commitment and is highly expressed in both pro-B and pre-B cells (36). Additionally, it has been shown that E-boxes in κEi and κE3′ are already occupied in pro-B cells (33). Thus, it is likely that additional regulators are required to modulate E2A activity at Igκ in order to promote the proper timing of Igκ gene recombination during B-cell development. Interferon regulatory factor (IRF)-4 and related gene family member IRF-8 are required for the pro-B-cell-to-pre-B-cell transition and Igκ locus activation (17). IRF-4, also known as Pip or NF-EM5, was originally characterized as a factor recruited by the transcription factor Pu.1 to a composite DNA binding site found in κE3′ and the Igλ enhancers Eλ2-4 and Eλ3-1 (6, 26). The ability of IRF-4 to bind to the composite site is dependent on Pu.1 and requires Pu.1 for transcriptional activity at κE3′ (6). However, recent studies show that Pu.1 and the related Ets family transcription factor SpiB are not required for the induction of Igκ germ line transcription, suggesting that IRF-4 may regulate Igκ through another mechanism in pre-B cells (32). It has been shown that IRF-4 can cooperatively interact with E2A at κE3′, resulting in enhanced E2A DNA binding and strong E2A/IRF-4 transcriptional synergy (22, 23). Additionally, IRF-4 expression increases as pro-B cells transition into pre-B cells in the bone marrow and in Ab-MuLV-transformed pre-B cells treated with the v-Abl inhibitor STI-571 (20). These studies indicate that the interaction of IRF-4 with E2A may play a central role in the regulation of the Igκ locus and suggest that IRF-4 may be a likely candidate regulator of E2A activity at κE3′.
We previously developed E2A-deficient pre-B-cell lines to study E2A-dependent gene regulation in B cells (9). In this study, we use these cell lines to directly investigate the causal links between E2A and Igκ activation and recombination. We show that E2A expression is required for Igκ germ line transcription and recombination and that retroviral expression of inducible human E2A cDNA in E2A−/− pre-B cells can promote histone acetylation at the Igκ enhancers and germ line promoters. Chromatin immunoprecipitation assays revealed that E2A is recruited to the Igκ enhancers in a manner that correlates with the induction of κ germ line transcription. We found that knockdown of IRF-4 transcript impaired E2A recruitment to κE3′ and severely impaired κ germ line transcription and enhancer histone acetylation. These data reveal that proper activation of the κ locus in pre-B cells is in part dependent on IRF-4-mediated recruitment of E2A to κE3′ and support the hypothesis that a synergistic interaction between these two factors plays an important role in regulating κE3′ function in B lymphocytes.
MATERIALS AND METHODS
Mice and pre-B-cell lines.
Mice carrying the null E2A allele E2Agal and κo-GFP mice have been described previously (15, 37). The E2A-deficient pre-B-cell lines were derived from Abelson murine leukemia virus (Ab-MuLV)-transformed bone marrow of a single E2AloxP/loxP mouse (24). Stable E2A−/− cell lines were established by transduction with the MSCV-puro (Stratagene) retrovirus expressing Cre recombinase as previously described (9). Ab-MuLV pre-B cells transduced with MSCV-puro containing antisense Cre served as the wild-type control for the E2A−/− cell lines. Clonal populations of E2A-deficient pre-B cells were derived from the Cre-transduced cells after puromycin selection. Human E47 estrogen receptor fusion protein reconstituted pre-B cells (E47R) were derived by transducing E2A−/− pre-B-cell lines with the MIGR1-E47R retrovirus (9). IRF-4 knockdown pre-B-cell lines were derived by introducing one of two independent mouse expression arrest short hairpin RNA interference (shRNAi) constructs (Open Biosystems) into a wild-type Abelson virus-transformed pre-B-cell line by retroviral transduction. IRF-4 mRNA knockdown was verified by reverse transcriptase PCR (RT-PCR) after puromycin selection. All cell lines were maintained at 37°C in RPMI 1640 media (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (HyClone), 100 U/ml penicillin (Gibco), 100 μg/ml streptomycin (Gibco), and 50 μM 2-mercaptoethanol (J. T. Baker).
STI-571 and tamoxifen preparation.
STI-571 (Gleevec; Novartis) was prepared as a 10-mM stock solution (1,000×) in water containing 10 mM HCl, sterile filtered, and stored in aliquots at −20°C until use. Tamoxifen (Sigma) was prepared as a 1-mM stock solution (1,000×) in cell-culture-grade dimethyl sulfoxide (DMSO) (Sigma) and stored at −20°C until use.
Gene expression analysis.
Total RNA was isolated from cells by use of TRIzol (Invitrogen) followed by isopropanol precipitation. Purified RNA was treated with RNase-free DNase I (Sigma), and random primed cDNA was made using Moloney murine leukemia virus reverse transcriptase (Invitrogen). Quantitative RT-PCR was performed with a Roche LightCycler and a FastStart DNA master SYBR green I kit (Roche). Semiquantitative RT-PCR was performed on serial dilutions of cDNA. Primers for RT-PCR are as follows: for E2A, E7 (CTAGCCCCTCAACGCCTGTG) and E15 (CGGTGCCAACAGCGTGGCT); for Igκ constant exon, AL2 (GATGTCTTGTGAGTGGCCCTC) and AL3 (CCAAAGACATCAATGTCAAGTGGAAG); for Igκ germ line transcript, IgK-GL 5′ (GAGGGGGTTAAGCTTTCGCCTACCCAC) and IgK-GL 3′ (CTGTATCTTTGCCTTGGAGAGTGCCAGAATCTGG); for IRF-4, IRF4#3 (GGCTTCACAATCTTCAAGGTGGAC) and IRF4#4 (CACACTTTCCTGTCGGGCTTAGAC); for SpiB, SpiB for (CTTCCAGTTCTCCTCCAAGCACAAG) and SpiB rev (TGGTAGGTGAGTTTGCGTTTGACC); for Spi1/PU.1, PU.1 for (CCTTATCAAACCTTGTCCCCAGCC) and PU.1 rev (ACCTCGCCTGTCTTGCCGTAGTTG); for RelA, relA for (GCAAGCCATTAGCCAGCGAATC) and relA rev (TTGAGAAAAGGAGCCTCGTGCC); for Pax-5, Pax-5a (CCGCCAAAGGATAGTGGAACTTG) and Pax-5b (CACAGTGTCATTGTCACAGACTCGC); for RAG1, YZ215 (TGCAGACATTCTAGCACTCTGG) and YZ216 (ACATCTGCCTTCACGTCGAT); for RAG2, YZ247 (CAGTGGGTCATAACATAGCC) and YZ248 (CTGGGTTCAGGGACATCTCC); and for EF1α, YZ95 (AGTTTGAGAAGGAGGCTGCT) and YZ96 (CAACAATCAGGACAGCACAGTC).
LM-PCR assay for RAG-mediated double-strand break.
Ligation-mediated PCR (LM-PCR) assays were conducted as previously described (20). Briefly, genomic DNA was prepared from cells and 1 μg of DNA was ligated overnight at 16°C with the BW linker. Signal ends were amplified by touchdown PCR using the linker-specific primer BW-1H and the Jκ-specific primer IgK-GL 5′. Touchdown PCR consisted of 30-s denaturation at 95°C; 30-s annealing at 68°C for 5 cycles, 65°C for 5 cycles, 62°C for 5 cycles, and 60°C for 20 cycles; and 45-s extension at 72°C. Specific PCR product was visualized by Southern blotting with the oligonucleotide probe Jk1-Jk2 (TGCTCTGTTCCTCTTCAGTG).
Mononucleosome chromatin immunoprecipitation assays.
Preparation of mononucleosomes and immunoprecipitations (IP) with antisera against acetyl-H3 and acetyl-H4 modified histones (Upstate Biotechnology) were performed as previously described (19). Briefly, nuclei were purified from Ab-MuLV-transformed pre-B cells and chromatin was partially digested with micrococcal nuclease (Worthington). Mononucleosomes were purified on a sucrose gradient, and immunoprecipitations were performed with antiserum against acetyl-H3 or acetyl-H4 modified histones. Normal rabbit serum was used as a negative IP control (Upstate Biotechnology). Quantitative PCR analysis of input and immunoprecipitated H3, H4, and control chromatin was performed using a Roche LightCycler and a FastStart DNA master SYBR green I kit (Roche). The primers used in this assay are as follows: for the intronic enhancer, IgKIE#2a (CTCTTGAAACTACTTTAGAGTCATTAAG) and IgKIE#2b (GAGTTCTTTACCAAGAAAAACAATAG); for the 3′ enhancer, IgK3E#3a (TGATCAAGAAGACCCTTTTGAGGAAC) and IgK3E#3b (GGTAGGGAGCAGGTGTATGAGGCTT); for the κo promoter, IgKGL#1a (CCACCTTCTTCCTCAATGATC) and IgKGL#1b (CTGACATATTGTGCTTGGTTG); for the Jκ1 promoter, IgKGLs#1a (GAGGCTGTCAGATTCCTTGCAG) and IgKGLs#1b (GGGCTGGAGAGGTGGCTCAG); for glucose-6-phosphate dehydrogenase (G6PD), G6PD#1 (GGGTCAGCTCAGTCAAAGCACA) and G6PD#2 (TAGTTGCCGCTGCCAAACAC); and for 3′ of the trypsinogen 4 gene (T4D), T4D#1 (GCCAGTACCTCTTAGACAA) and T4D#2 (GGGAGAGTAGAATTGTGTTA). All PCRs were performed in triplicate. The acetylation status of each site was determined by the IP bound signal/input signal. Each cell line was analyzed by at least two independent IP to confirm reproducibility. G6PD and T4D were used as positive and negative controls, respectively.
Immunoprecipitation of dual-affinity-tagged E2A.
Chromatin preparation and immunoprecipitation of dual-affinity-tagged E2A/DNA complexes were preformed as previously described (9). Enrichment of chromatin-immunoprecipitated DNA fragments was determined by quantitative PCR analysis using a Roche LightCycler and a FastStart DNA master SYBR green I kit (Roche), and the data are presented as ratio of immunoprecipitated DNA over input DNA. The PCR primers are as follows (Igκ intronic and 3′ enhancer primers are the same as those used for the mononucleosome ChIP assay): for the Ig heavy chain Eμ, Eu for (TCAGAACCAGAACACCTGCAGCA) and Eu rev (GGTGGGGCTGGACAGAGTGTTTC); for the mb-1 promoter, mb-1 for (CCACGCACTAGAGAGAGACTCAA) and mb-1 rev (CCGCCTCACTTCCTGTTCAGCCG); and for the CD19 promoter, CD19 for (CCTAATGCTATCCCCAGATGATA) and CD19 rev (TAAATATTTTTCAGATGAGTGGG).
Flow cytometry.
Bone marrow was prepared in fluorescence-activated cell sorter (FACS) buffer (1× phosphate-buffered saline, pH 7.4, plus 5% bovine calf serum) and stained on ice with CD43 phycoerythrin-, IgM biotin-, and B220 allophycocyanin-conjugated antibodies (BD Pharmingen), followed by streptavidin-Cy5-phycoerythrin staining (BD Pharmingen). Cells were run on a FACSCalibur and analyzed with CellQuest software (Beckman Coulter). Three hundred thousand events were collected for each sample, and dead cells were excluded by forward and side scatter gating.
RESULTS
E2A is required for immunoglobulin kappa light chain germ line transcription and recombination.
We have previously generated stable E2A−/− pre-B-cell lines from a mouse carrying a conditional E2A knockout allele (9). E2A−/− pre-B cells are phenotypically indistinguishable from wild-type control cell lines and express the B-lineage-specific transcription factors EBF and Pax-5 (9) as well as immunoglobulin heavy chain Cμ and the surrogate light chain genes VpreB and λ5 (data not shown). These observations demonstrate that the loss of E2A at the pre-B-cell stage does not alter B-cell lineage identity or abolish the expression of select pre-B-cell-specific genes. Ab-MuLV pre-B cells can be induced to differentiate and initiate immunoglobulin light chain recombination (20, 31). Therefore, we sought to directly investigate the role that E2A plays in the regulation of immunoglobulin light chain gene expression and recombination with our E2A−/− pre-B-cell lines.
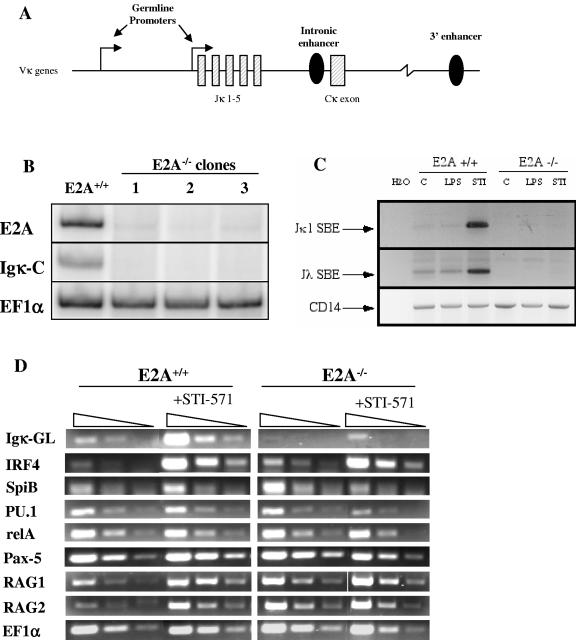
It has previously been shown that recombinase accessibility of the κ locus correlates with germ line transcription that originates from two promoters located 5′ of the Jκ1 exon (31) (Fig. 1A). We first assayed immunoglobulin kappa light chain gene transcription by RT-PCR in E2A−/− pre-B cells by using primers specific for the κ constant exon. We found that the loss of Igκ transcript correlated with the loss of E2A expression in three independently derived clonal E2A−/− pre-B-cell populations (Fig. 1B). Next, we asked if immunoglobulin light chain recombination was dependent on E2A. Previous studies have shown that Ab-MuLV pre-B cells can be induced to rearrange their immunoglobulin light chain genes by treatment with lipopolysaccharide (LPS) or the small molecule inhibitor of v-Abl, STI-571 (20). Wild-type and E2A−/− pre-B cells were treated with LPS or STI-571, and light chain recombination was measured by LM-PCR. The accumulation of Jκ1 and Jλ signal ends (Fig. 1C) and coding joints (data not shown) could be readily detected in E2A sufficient pre-B cells treated with STI-571 but not in E2A−/− pre-B cells. These results demonstrate that Ig light chain recombination is impaired in the absence of E2A.
FIG. 1.
Igκ germ line transcription and recombination is dependent on E2A. (A) Schematic of the 3′ region of the murine Igκ locus, highlighting the germ line transcript promoters, intronic enhancer, and 3′ enhancer. (B) E2A and Igκ expression in E2A-deficient Ab-MuLV pre-B cells. Primers for murine E47 (E2A) and Igκ constant exon (Igκ-C) were used to evaluate expression of these genes in E2A+/+ cells and three independent E2A−/− clones. EF1α serves as a positive loading control. (C) E2A-deficient Ab-MuLV pre-B cells fail to initiate light chain recombination. E2A+/+ or E2A−/− cells untreated (C) or treated with LPS or STI-571 were assayed for the production of RAG-mediated Jκ1 or Jλ signal break ends (SBE) by LM-PCR. CD14 gene amplification serves as a positive control for DNA loading and quality. (D) Expression of transcripts involved in Igκ gene regulation in E2A-deficient pre-B cells. E2A+/+ and E2A−/− cells were incubated for 5 h at 37°C with or without 10 μM STI-571, and the relative abundance of various transcripts was measured by semiquantitative RT-PCR. The expression levels of Igκ germ line transcript (Igκ-GL), RAG1, RAG2, and transcription factors known to regulate Igκ expression were examined. Each sample is presented as a series of threefold serial dilutions. EF1α was used to verify equivalent cDNA loading and quality between samples.
Multiple transcription factors have been directly implicated in the regulation of immunoglobulin light chain gene transcription and recombination. Altered expression of one or more of these factors might contribute to the dramatic loss of transcription and recombination observed with E2A−/− pre-B cells. RT-PCR was used to evaluate the expression levels of genes previously shown to be involved in Igκ regulation. Consistent with previous findings (20), STI-571-treated E2A+/+ pre-B cells showed accumulation of κ germ line transcript as well as IRF-4, SpiB, RAG1, and RAG2 transcripts (Fig. 1D). However, κ germ line transcript was undetectable in untreated E2A−/− pre-B cells and was weakly induced by STI-571 in the absence of E2A. We did not observe a reduction in IRF-4, Pax-5, RelA, RAG1, or RAG2 transcript levels between the STI-571-treated E2A+/+ and E2A−/− cells. Transcript levels of SpiB and PU.1 were each approximately two- to threefold lower in STI-571-treated E2A−/− pre-B than levels in wild-type cells. These results show that the loss of E2A perturbs κ germ line transcription but not the expression of genes known to be involved in immunoglobulin kappa transcription and recombination.
Human E47 rescues Igκ germ line transcription and recombination in E2A−/− pre-B cells.
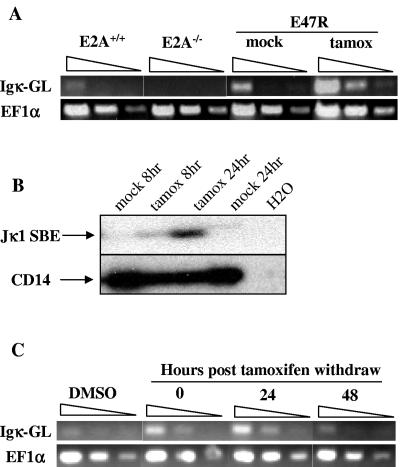
The correlation between E2A deficiency and defective Igκ germ line transcription suggests that E2A may play a direct role in regulating the activation state of Igκ genes. We further investigated the potential of E2A to activate Igκ by examining germ line transcription and recombination in E2A−/− pre-B cells reconstituted with tamoxifen-inducible human E47 cDNA (E47R). We have previously shown that in the absence of tamoxifen E47R-reconstituted pre-B cells show no detectable E2A DNA binding and that DNA binding can be restored by culturing the cells in the presence of tamoxifen (9). E47R-reconstituted pre-B cells were treated with tamoxifen, and Igκ germ line transcript levels were examined by RT-PCR. Tamoxifen induced κ germ line transcription in E47R pre-B cells but had no effect on κ transcript levels in wild-type or E2A−/− cells (Fig. 2A). Next, we examined Igκ recombinase accessibility in E47R cells by LM-PCR and found that tamoxifen treatment caused the accumulation of Jκ1 signal ends, while mock treatment of E47R cells did not (Fig. 2B). Since these data show that E47R is sufficient to activate Igκ germ line transcription and promote Jκ recombinase accessibility, we then asked if E2A is required to maintain Igκ gene activation in pre-B cells. E47R pre-B cells were treated with tamoxifen for 6 h, washed, and cultured in the absence of tamoxifen. Increased κ germ line transcription was observed upon removal of tamoxifen (t = 0) and was maintained for up to 24 h but by 48 h had declined to the level observed for mock-treated cells (Fig. 2C). These results demonstrate that E2A is necessary to activate and maintain Igκ germ line transcription and is involved in promoting recombinase accessibility of the Jκ genes in our pre-B-cell lines.
FIG. 2.
Inducible human E47 rescues Igκ germ line transcription and recombination. (A) E47R is sufficient to activate Igκ germ line transcription. Total RNA was isolated from E2A+/+ or E2A−/− Ab-MuLV pre-B cells treated with 1 μM tamoxifen (tamox) and E47R-reconstituted Ab-MuLV B cells treated with 0.1% DMSO or 1 μM tamoxifen for 6 h. Semiquantitative RT-PCR was performed to determine the relative level of Igκ germ line transcription (Igκ-GL). EF1α serves as a positive loading control, and results are presented as threefold serial dilutions. (B) E47R promotes Igκ recombination. E47R-reconstituted pre-B cells were treated with 1 μM tamoxifen or 0.1% DMSO for 8 or 24 h. Genomic DNA was prepared from the cells, and the accumulation of Jκ1 signal break ends (SBE) was determined by LM-PCR. PCR amplification of the CD14 gene serves as a control for DNA loading and quality. (C) Igκ germ line transcription is dependent on E2A. E47R-reconstituted pre-B cells were treated with 1 μM tamoxifen for 6 h, washed, and cultured without tamoxifen. Total RNA was prepared at the indicated times after the removal of tamoxifen. Mock-treated cells were cultured in the presence of 0.1% DMSO for the entire duration of the experiment. Relative abundance of Igκ germ line transcript and EF1α transcript was determined by RT-PCR.
E2A deletion perturbs Igκ enhancer histone acetylation.
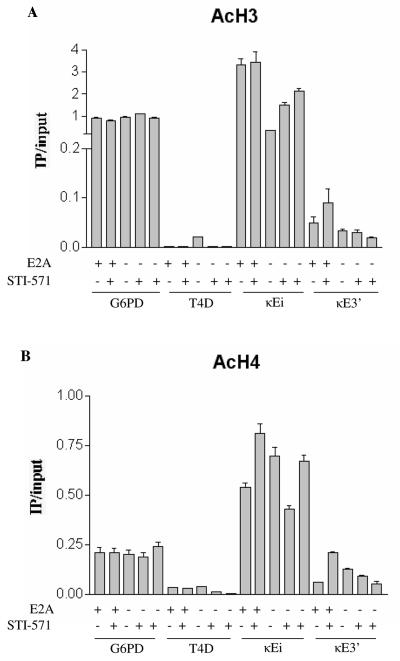
Since E2A has been implicated in mediating chromatin remodeling, we asked what effect E2A deficiency has on chromatin modification at the Igκ enhancers (5, 18). We purified mononucleosomes from E2A+/+ or E2A−/− pre-B cells and performed IPs to enrich for acetylated H3 or H4 histones. The enrichment of DNA sequences associated with acetylated H3 or H4 at κEi and κE3′ was determined by quantitative real-time PCR. We examined Igκ enhancer acetylation for three E2A−/− clones, one untreated and two treated with STI-571. We found that the H3 acetylation at κEi was decreased in E2A−/− pre-B cells (Fig. 3A). We also observed that in the presence of STI-571, H3 acetylation at κEi was marginally lower in two independent E2A−/− clones than in E2A+/+ cells. Reduced H3 acetylation was also observed to occur at κE3′ in untreated E2A−/− pre-B cells and in both E2A−/− clones treated with STI-571 (Fig. 3A). H4 acetylation was also reduced at κEi and κE3′ in STI-571-treated E2A−/− pre-B cells compared to the level in STI-571-treated E2A+/+ cells (Fig. 3B). These results indicate that E2A deficiency can perturb Igκ enhancer histone acetylation when the locus is activated but also show that the maintenance of H3 and H4 acetylation at κEi and κE3′ is not entirely dependent on E2A. The reduction in H3 and H4 acetylation we observed was specific to the Igκ enhancers, since enrichment of the housekeeping gene G6PD remained constant between all samples. We observed a consistent increase in H3 acetylation at κE3′ and H4 acetylation at κEi and κE3′ upon STI-571 treatment of E2A+/+ pre-B cells. However, we did not observe this increase with the two E2A−/− clones treated with STI-571, indicating that E2A is required to promote hyperacetylation of the Igκ enhancers after STI-571 treatment.
FIG. 3.
E2A gene status influences Igκ enhancer histone acetylation. (A) Chromatin modification of the Igκ enhancers in E2A−/− pre-B cells. Mononucleosomes were prepared from E2A+/+ pre-B-cell lines treated for 5 h with or without 10 μM STI-571, and acetylated H3 histones were immunoprecipitated. Relative enrichment of H3 acetylation sequences was determined by quantitative PCR and plotted as IP DNA over input DNA. Enrichment of sequences corresponding to the Igκ intronic (κEi) and 3′ (κE3′) core enhancers was determined. Enrichment of the housekeeping gene G6PD and the silent trypsinogen locus T4D served as positive and negative internal controls, respectively. Two independently derived E2A−/− clones treated with STI-571 were analyzed. (B) Mononucleosomes prepared from the pre-B cells described for panel A were immunoprecipitated and analyzed for H4 acetylated histones at various loci. Relative enrichment of acetylated H4 was plotted as IP/input. Acetyl-H4 enrichment at the G6PD and T4D loci and the Igκ enhancers (κEi and κE3′) was determined by quantitative PCR. Two independently derived E2A−/− clones treated with STI-571 were analyzed.
E47R can promote Igκ histone acetylation.
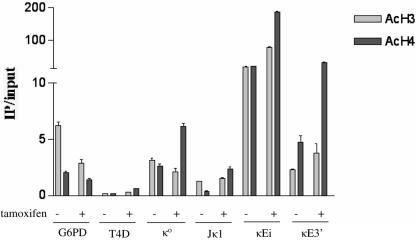
Since we observed that E47R promotes Igκ germ line transcription and recombinase accessibility, we asked if E47R could promote chromatin modification of Igκ. Tamoxifen-treated E47R cells show a moderate increase in histone H3 acetylation and a large increase in H4 acetylation at both κEi and κE3′ compared to levels for mock-treated E47R pre-B cells (Fig. 4). Therefore, E47R is capable of promoting Igκ enhancer histone acetylation in a manner that correlates with the induction of germ line transcription and recombinase accessibility. We also examined the two Igκ germ line promoters to determine if E47R activation could influence histone acetylation at these regulatory elements as well. The κo promoter, located approximately 3.5 kb upstream of Jκ1, showed a significant increase in H4 acetylation, while H3 acetylation decreased slightly in E47R cells treated with tamoxifen (Fig. 4). The Jκ1 promoter, located immediately upstream of the Jκ1 exon, also exhibited a significant increase in H4 acetylation and little change in H3 acetylation in response to tamoxifen treatment (Fig. 4). The increases in histone acetylation at the Igκ enhancers and promoters upon tamoxifen treatment was not due to a global increase in histone acetylation, since G6PD acetylation was slightly decreased in tamoxifen-treated E47R pre-B cells. These data demonstrate that increased H4 acetylation and to a lesser extent H3 acetylation may occur at the Igκ enhancers in a manner that is dependent on E47R activity.
FIG. 4.
E47R promotes histone acetylation of the Igκ enhancers and germ line promoters. Mononucleosomes were prepared from E47R-reconstituted pre-B cells treated with 0.1% DMSO or 1 μM tamoxifen for 24 h. Enrichment of acetylated histone H3 or acetylated histone H4 at the 5′ κo and Jκ1 proximal Igκ germ line promoters and the intronic (κEi) and 3′ (κE3′) enhancers was examined. Relative acetylation was determined by quantitative PCR and plotted as IP/input. The active G6PD and silent T4D loci serve as positive and negative controls, respectively.
Increased E2A recruitment to the κ enhancers upon STI-571 treatment.
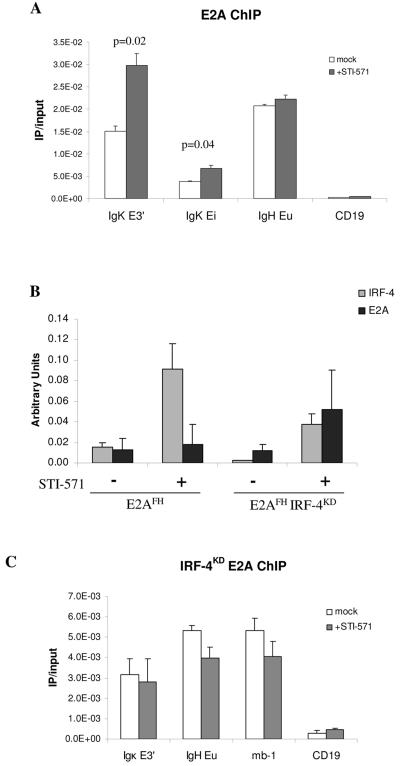
We directly examined E2A association with the Igκ enhancers by E2A ChIP with an Ab-MuLV-transformed pre-B-cell line from a mouse carrying dual-affinity-tagged E2A (E2AFH) (10). E2A ChIP was performed on chromatin from E2AFH pre-B cells treated with or without STI-571 for 6 h, and the relative enrichment of sequences corresponding to κEi and κE3′ was determined by quantitative PCR. We observed significant enrichment of E2A at the Igκ enhancers in untreated cells and found that upon STI-571 treatment E2A association doubled at the 3′ enhancer and increased approximately 70% at the intronic enhancer (Fig. 5A). This increase in E2A association was specific to the Igκ enhancers, since we observed no significant change in the enrichment of sequence corresponding to the Ig heavy chain intronic enhancer between STI-571-treated and untreated cells. We also consistently found that E2A association with κE3′ was significantly greater than with κEi.
FIG. 5.
IRF-4 expression promotes E2A association with κE3′ in pre-B cells. (A) Increased E2A association with the Igκ enhancers upon locus activation. A pre-B-cell line carrying a dual-affinity-tagged E2A (E2AFH) was treated with or without 10 μM STI-571 for 6 h. Relative E2A association with the Igκ intronic (κEi) and 3′ (κE3′) enhancers was determined by quantitative PCR. E2A enrichment at the Ig heavy chain (IgH) intronic enhancer (Eμ) and CD19 promoter serve as positive and negative controls for E2A DNA binding, respectively. Results are presented as ratio of IP to input DNA and are the averages of two independent IP ± standard deviations. (B) IRF-4 and E2A gene expression in E2AFH and E2AFH IRF-4KD pre-B cells with or without STI treatment. Total RNA was prepared from a portion of the cells used as described for panel A, and the relative expression levels of IRF-4 and E2A mRNA were determined by quantitative RT-PCR. IRF-4 and E2A transcript levels were normalized to the housekeeping gene EF1α. (C) IRF-4 expression influences E2A association with κE3′ and Ig heavy chain Eμ. Dual-affinity-tagged E2A was immunoprecipitated from E2AFH/FH IRF-4KD pre-B-cell lines treated for 6 h with or without 10 μM STI-571. Relative E2A associations with κE3′, Ig heavy chain Eμ, and the mb-1 promoter were determined by quantitative PCR and are presented as ratio of IP to input DNA. E2A enrichment at the CD19 promoter is a negative control.
IRF-4 promotes E2A DNA binding to κE3′ in vivo and regulates Igκ locus activation.
Since IRF-4 has been shown to promote E2A DNA binding at κE3′ in vitro (22) and IRF-4 expression is induced upon STI-571 treatment (Fig. 1D), we speculated that the increased E2A association with κE3′ observed after STI-571 treatment may be dependent on IRF-4. To test this hypothesis, we inhibited IRF-4 expression by stably transducing the E2AFH cell line with a nshRNAi IRF-4 knockdown construct (IRF-4KD). Quantitative RT-PCR revealed that the IRF-4 mRNA level was reduced approximately sixfold in the untreated E2AHF cell line (Fig. 5B). We observed that STI-571 treatment induced IRF-4 mRNA levels in both the parental and knockdown cell lines but that the absolute level of IRF-4 mRNA was approximately 60% lower in the knockdown cell line than in the parental cell line (Fig. 5B). Next, we asked if the STI-571-dependent increase in E2A association with κE3′ could be inhibited in the E2AFH IRF-4KD cells. Since previously published works suggest that IRF-4 may influence E2A DNA binding at Ig heavy chain Eμ, we also analyzed the mb-1 promoter to provide an additional independent locus that is known to associate with E2A (10, 23). Quantitative PCR analysis of E2A ChIP DNA fragments from E2AFH IRF-4KD cells showed that E2A association with κE3′ failed to increase after STI-571 treatment (Fig. 5C). E2A association with Ig heavy chain Eμ or the mb-1 promoter in the presence of STI-571 was only slightly decreased compared to results with untreated cells. Quantitative RT-PCR analysis of the cell lines verified that the failure to observe increased E2A association with κE3′ in the IRF-4KD line was not due to a reduction in E2A gene expression (Fig. 5B). These data provide evidence indicating that IRF-4 can promote E2A association with κE3′ in the context of the endogenous locus.
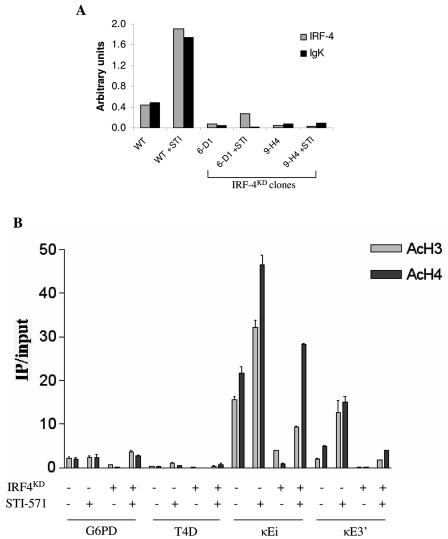
Next, we examined if IRF-4 knockdown had an effect on Igκ germ line transcription and chromatin modification. Two independent shRNAi IRF-4 knockdown constructs were stably expressed in an Ab-MuLV-transformed pre-B-cell line, and IRF-4 and Igκ transcript levels were determined by quantitative RT-PCR. The shRNAi constructs 6-D1 and 9-H4 suppressed IRF-4 mRNA levels 6-fold and 10-fold, respectively, in untreated cells (Fig. 6A). STI-571 treatment increased IRF-4 mRNA levels in the parental cell line and in clone 6-D1, but the absolute IRF-4 mRNA level in clone 6-D1 was still sevenfold lower than that of the parental cell line. We observed that Igκ transcript levels were also significantly reduced for both untreated and STI-571-treated IRF-4 knockdown cells (Fig. 6A). These data show that Igκ germ line transcription is dependent on the expression of IRF-4.
FIG. 6.
IRF-4 is required for Igκ germ line transcription and promotes enhancer histone acetylation. (A) IRF-4 RNAi perturbs Igκ transcription. Pre-B-cell lines were stably transduced with one of two IRF-4 shRNAi constructs (6-D1 or 9-H4). Wild-type (WT) or IRF-4 knockdown cell lines were treated with or without 10 μM STI-571 for 6 h. The relative expression levels of IRF-4 and Igκ transcripts were determined by quantitative RT-PCR. IRF-4 and Igκ transcript levels were normalized to EF1α. (B) Effect of IRF-4 knockdown on Igκ enhancer chromatin modification. Mononucleosomes were prepared from chromatin of wild-type or IRF-4KD pre-B-cell lines treated with or without STI-571 for 6 h. Enrichment of sequences associated with acetyl-H3 or acetyl-H4 histones at κEi or κE3′ was determined by ChIP. Relative acetylation was plotted as IP/input. The G6PD and T4D loci serve as internal controls for an active and a silent locus, respectively.
Since inhibiting IRF-4 expression dramatically impaired Igκ germ line transcription, we hypothesized that IRF-4 may play a role in regulating Igκ chromatin modification. IRF-4 knockdown clone 6D-1 was further analyzed to determine levels of H3 and H4 histone acetylation at κEi and κE3′. Mononucleosomes were prepared from parental cells and IRF-4KD cells treated with or without STI-571, and acetylated H3 or acetylated H4 histones were immunoprecipitated. We found that knockdown of IRF-4 significantly reduced both H3 and H4 acetylation at both κ enhancers (Fig. 6B). In addition, STI-571-mediated H3 and H4 hyperacetylation of κEi and κE3′ was significantly reduced in the IRF-4KD cell line. These results suggest that IRF-4 facilitates Igκ germ line transcription through a mechanism that involves promoting Igκ enhancer histone acetylation.
Regulation of the Igκ locus by E2A during normal pre-B-cell development.
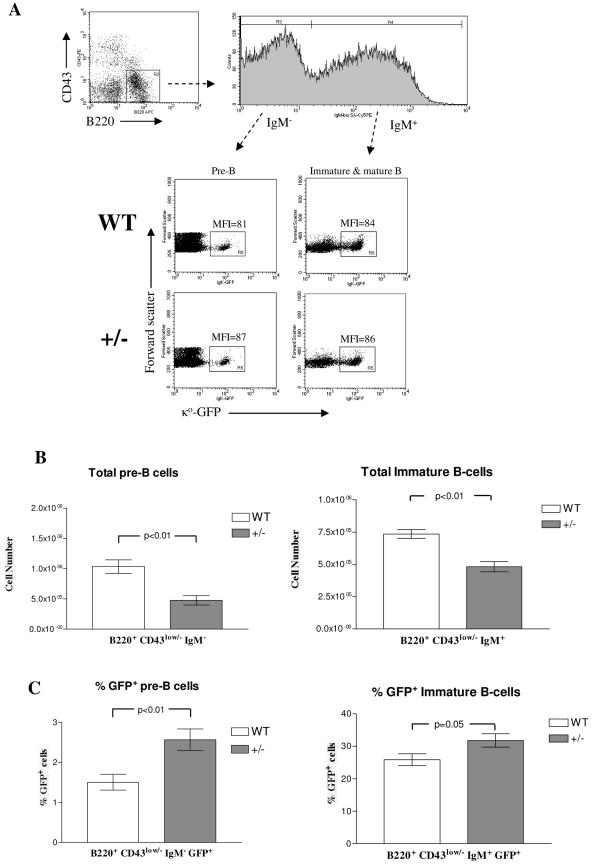
Finally, we sought to understand how E2A regulates Igκ locus activation and germ line transcription during normal B-cell development. E2A heterozygous mice were crossed with mice carrying a κo-GFP knockin allele, in which κo germ line transcription is conveniently revealed by a green fluorescent protein (GFP) marker inserted into the Jκ1 exon (15). We first asked if a reduction in E2A gene dose has an effect on the rate of monoallelic Igκ germ line transcription in pre-B cells. Bone marrow from 2- to 3-month-old E2A+/+ Igκ+/G or E2A+/− Igκ+/G mice was isolated and examined for GFP expression in B220+ CD43low/− IgM− small pre-B cells. We observed no significant difference in GFP mean fluorescence intensity with pre-B cells from E2A+/+ mice and pre-B cells from E2A+/− mice, suggesting that E2A expression level does not influence the rate of germ line transcription from an active κo-GFP allele (Fig. 7A).
FIG. 7.
E2A+/− Igκ+/G mice have a greater proportion of GFP+ pre-B cells than E2A+/+ Igκ+/G mice. (A) Expression level of Igκ germ line transcript is not influenced by E2A gene status. Bone marrow was collected from E2A+/+ (WT) or E2A+/− (+/−) mice carrying one κo-GFP allele and analyzed by FACS. The expression levels of Igκ germ line transcript in E2A WT and heterozygous mice were determined by calculating GFP mean fluorescence intensity (MFI) among GFP+ (R5) pre-B cells (B220+ CD43low/− IgM−) and immature/mature bone marrow B cells (B220+ CD43low/− IgM+). FACS plots are representative of 12 WT and 9 heterozygous mice analyzed. APC, allophycocyanin; bio, biotin; SA, streptavidin; PE, phycoerythrin. (B) Total numbers of pre-B and immature bone marrow B cells were calculated from 12 E2A+/+ Igκ+/G and 9 E2A+/− Igκ+/G mice and plotted. (C) Percentages of GFP+ pre-B cells and immature bone marrow B cells from each of these mice were also calculated and plotted.
Next, we asked if E2A expression could influence the frequency of pre-B cells expressing the κo-GFP allele. We found that the total number of pre-B cells and immature B cells was reduced in E2A+/− mice (Fig. 7B). This result is consistent with previous studies showing that E2A heterozygous mice have a partial block in B-cell development and reduced numbers of pro-B cells and pre-B cells (2, 11, 27, 37). A closer examination of the pre-B-cell populations in E2A+/+ and E2A+/− mice revealed that there was a significantly greater proportion of GFP+ pre-B cells in E2A heterozygous mice (Fig. 7C). The percentage of GFP+ pre-B cells was 1.5% ± 0.2% (n = 12) in E2A+/+ mice and 2.6% ± 0.3% (n = 9) in E2A+/− mice. Interestingly, the total numbers of GFP+ pre-B cells per tibia in E2A+/+ (1.4 × 104 ± 0.17 × 104) mice and E2A+/− (1.4 × 104 ± 0.15 × 104) mice were equivalent. When we examined the immature-B-cell population of the bone marrow, we found that there was only a slight increase in the percentage of GFP+ cells in the E2A heterozygous mice. These results suggest that E2A gene dosage can influence the frequency of pre-B cells expressing κ germ line transcripts during B-cell development in vivo.
DISCUSSION
Igκ germ line transcription and recombination are regulated by multiple regulatory elements which include the intronic enhancer, 3′ enhancer, and germ line promoters (4, 13, 35). E2A directly interacts with both of the kappa enhancers, but the importance of these interactions and the mechanism by which E2A regulates Igκ is only beginning to be rigorously explored (10). Our data demonstrate that E2A is absolutely required for facilitation of Igκ locus activation and recombination. E2A−/− pre-B cells retain B-cell lineage identity and express the B-cell-specific transcription factors Pax-5 and EBF (9). Among the transcripts we examined, only PU.1 and SpiB expression levels were decreased in E2A−/− cells treated with STI-571. However, it is unlikely that reduced expression of PU.1 and SpiB can account for the severe decrease in Igκ germ line transcription, since it has been demonstrated that PU.1−/−/SpiB−/− pro-B cells are capable of expressing Igκ germ line transcript (32). These results strongly indicate that the defect in Igκ transcription is due to the absence of E2A interactions with cis regulatory elements on the κ locus rather than a defect in the expression of a secondary gene.
Interestingly, RAG expression was not perturbed in our E2A-deficient cell lines. Previous studies have shown that the RAG locus contains an enhancer, Erag, which is regulated in part by E2A and is required for optimal RAG expression in early B-cell progenitors (12). E2A binds to Erag and can activate transcription from an Erag-containing reporter construct. Although E2A regulates the RAG locus, similar levels of RAG transcript were observed with STI-571-treated E2A+/+ and E2A−/− pre-B-cell lines. These results suggest that E2A may play a role in regulating Erag at an earlier stage of B-cell development or that E2A is not absolutely essential for the maintenance of RAG transcription once the locus is active. These data also show that the block in Igκ and Igλ recombination in our E2A−/− pre-B-cell lines is not due to a deficiency in RAG expression.
It is known that E2A binds directly to the Igκ intronic enhancer (10, 21). The E-boxes, E1 and E2, located in the Igκ intronic enhancer have been shown to be essential for efficient κ recombination in vivo (14). Evidence suggests that E2A may play a role in regulating Igκ chromatin structure, since E2A has been shown to interact directly with histone acetyl transferases and the SAGA chromatin remodeling complex (3, 18). Upon analysis of the intronic enhancer, we were surprised to find that histone acetylation was not dramatically altered by E2A gene status in untreated cells. Interestingly, we found that upon STI-571 treatment histone hyperacetylation of the Igκ 3′ enhancer was perturbed in the absence of E2A. This suggests that E2A may play a role in regulating κ enhancer chromatin modification upon activation of the locus. While we show that maintenance of intronic enhancer acetylation is not entirely dependent on E2A, these results do not rule out the possibility that E2A is required to initiate chromatin modification of the Igκ enhancers at an earlier point in B-cell development. These results also raise the possibility that E2A regulates intronic enhancer function through a mechanism that may involve but is not entirely dependent on histone acetylation. The induction of κ germ line transcription by STI-571 also correlates with a significant increase in histone acetylation at the 3′ enhancer in E2A+/+ pre-B cells. E2A−/− pre-B cells treated with STI-571 express little κ germ line transcript and show no increase in 3′ enhancer acetylation (Fig. 1D and 3A and B). These data suggest that E2A activity may be developmentally regulated at the 3′ enhancer and that E2A plays an important role in regulating the activity of this enhancer.
Analysis of E47R-reconstituted pre-B cells revealed that E2A activity can promote histone acetylation of the Igκ enhancers. In addition, we also observed that H4 acetylation of the Igκ germ line promoters was increased by E47R. Sequence analysis of the κo and Jκ1 germ line promoters revealed that both contain potential E2A DNA binding sites, but we did not detect significant E2A association with either promoter by E2A ChIP (data not shown). Therefore, it is possible that the increase in H4 acetylation observed at the germ line promoters is an indirect consequence of E47R activity. We also noticed that the overall level of Igκ enhancer acetylation was significantly greater in E47R-reconstituted pre-B cells than in the parental E2A−/− pre-B-cell line or E2A sufficient cells. In addition, uninduced E47R-reconstituted cells also exhibit a higher basal level of Igκ germ line transcript than E2A+/+ cells. Increased Igκ gene activity in E47R pre-B cells may be a result of quantitative differences in the level of retrovirally expressed E47R versus endogenous E2A expression. Furthermore, it is possible that E47R, which was created from the human E2A isoform E2-5 cDNA, may function at Igκ in a manner that is qualitatively different from the endogenous mouse E2A protein, thereby resulting in increased Igκ germ line transcription and enhancer acetylation. Nevertheless, E47R can complement E2A deficiency at Igκ and we observe that enhancer hyperacetylation correlates with increased κ germ line transcription in E47R cells just as it does in E2A+/+ pre-B cells treated with STI-571.
Chromatin immunoprecipitation of E2A revealed that there is a significant increase in E2A association with the Igκ enhancers that correlates with Igκ locus activation. These data suggest that there may be a threshold of E2A association with Igκ that must be achieved in order to fully activate the locus. Such a mechanism may explain why tamoxifen activation of retrovirally expressed E47R or ectopic E2A expression in nonlymphoid cells alone is sufficient to activate Igκ (7, 29). Interestingly, we also found that there was a significantly stronger association of E2A with the 3′ enhancer than with the intronic enhancer in our pre-B-cell lines. This suggests that E2A association with κE3′ may be particularly important for κ locus activation in pre-B cells.
It has been shown that IRF-4 directly binds to κE3′ and is required for Igκ gene activation and recombination in pre-B cells (17). E2A recruitment to κE3′ in pre-B cells correlates with the STI-571-induced increase in IRF-4 expression. Conversely, suppression of IRF-4 transcript by RNAi prevented increased E2A association with κE3′. These observations provide evidence that supports previous in vitro findings showing that IRF-4 promotes E2A DNA binding at κE3′ (22). Our results also suggest that IRF-4 works in parallel with E2A at Igκ in pre-B cells, since the induction of IRF-4 transcription by STI-571 is not dependent on E2A and is not sufficient to rescue Igκ germ line transcription in E2A−/− pre-B cells (Fig. 1D). Based on our findings, we propose that IRF-4 promotes the recruitment of E2A to κE3′ in pre-B cells and that E2A/IRF-4 synergy at κE3′ plays a critical role in driving κ locus activation.
IRF-4 deficiency also impaired Igκ germ line transcription and enhancer histone acetylation. We found that IRF-4 deficiency has a greater impact on Igκ enhancer histone acetylation than E2A deficiency, suggesting that IRF-4 plays a broader role in regulating Igκ chromatin modification, possibly through its interaction with Pu.1. We also found that histone hyperacetylation of the intronic enhancer was decreased in IRF-4KD cells. Since IRF-4 does not directly bind to this enhancer, it is likely that more-distal IRF-4 DNA binding sites must influence the intronic enhancer. It has recently been shown that the κ intronic and 3′ enhancers can directly interact through DNA looping (16). It is possible that the disruption of IRF-4 associations at the 3′ enhancer disrupts looping of the enhancers, thereby perturbing intronic enhancer histone acetylation and activation of the Igκ locus. Alternately, it is possible that IRF-4 is required for the expression of a factor that is required for κEi histone acetylation. Further studies should help to clarify the mechanism by which IRF-4 influences κEi chromatin modification.
Our analysis of κo-GFP mice revealed that the percentage of GFP+ pre-B cells in the bone marrow of E2A+/− Igκ+/G mice was greater than that for E2A wild-type mice. Previous studies have shown that the GFP+ pre-B cells are actively engaged in V-J recombination and are the precursors of immature B cells (15). Thus, the accumulation of GFP+ pre-B cells in E2A+/− mice may indicate that Igκ recombination is partially blocked or inefficient. An E2A-dependent reduction in Igκ recombination efficiency could result from reduced recombinase accessibility of the κ locus or reduced RAG expression. BCR transgenic E2A heterozygous mice have been shown to have significantly lower RAG expression than their E2A wild-type counterparts (27). Impaired RAG expression in our E2A heterozygous mice could result in reduced light chain recombination efficiency. In addition, insufficient E2A expression may negatively impact kappa locus recombinase accessibility by perturbing chromatin modification of the locus. These two mechanisms are not mutually exclusive and may both contribute to the pre-B phenotype we observed with the E2A+/− Igκ+/G mice.
In light of our data on the interaction of E2A and IRF-4, it would be of great interest to further test genetic interaction between these two molecules. Such studies will provide valuable insight into the underlying mechanisms of Igκ regulation and shed light on how a broadly expressed transcription factor such as E2A can be specifically directed to regulate the activity of a lineage-restricted locus such as Igκ.
Acknowledgments
We thank Mike Krangel and Annette Jackson for providing helpful discussions and technical support for the histone acetylation studies and Mary Elizabeth Jones and Jason Wojciechowski for critical reading of the manuscript.
This work was supported by National Institutes of Health grants CA072433 to Y. Zhuang and HL48702 and AI40227 to M. Schlissel.
We have no conflicting financial interests.
REFERENCES
- 1.Arakawa, H., T. Shimizu, and S. Takeda. 1996. Re-evaluation of the probabilities for productive arrangements on the kappa and lambda loci. Int. Immunol. 8:91-99. [DOI] [PubMed] [Google Scholar]
- 2.Bain, G., E. C. Maandag, D. J. Izon, D. Amsen, A. M. Kruisbeek, B. C. Weintraub, I. Krop, M. S. Schlissel, A. J. Feeney, M. van Roon, et al. 1994. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell 79:885-892. [DOI] [PubMed] [Google Scholar]
- 3.Bradney, C., M. Hjelmeland, Y. Komatsu, M. Yoshida, T. P. Yao, and Y. Zhuang. 2003. Regulation of E2A activities by histone acetyltransferases in B lymphocyte development. J. Biol. Chem. 278:2370-2376. [DOI] [PubMed] [Google Scholar]
- 4.Cocea, L., A. De Smet, M. Saghatchian, S. Fillatreau, L. Ferradini, S. Schurmans, J.-C. Weill, and C.-A. Reynaud. 1999. A targeted deletion of a region upstream from the Jκ cluster impairs κ chain rearrangement in cis in mice and in the 103/bcl2 cell line. J. Exp. Med. 189:1443-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckner, R., T. P. Yao, E. Oldread, and D. M. Livingston. 1996. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 10:2478-2490. [DOI] [PubMed] [Google Scholar]
- 6.Eisenbeis, C. F., H. Singh, and U. Storb. 1995. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 9:1377-1387. [DOI] [PubMed] [Google Scholar]
- 7.Goebel, P., N. Janney, J. R. Valenzuela, W. J. Romanow, C. Murre, and A. J. Feeney. 2001. Localized gene-specific induction of accessibility to V(D)J recombination induced by E2A and early B cell factor in nonlymphoid cells. J. Exp. Med. 194:645-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorman, J. R., N. van der Stoep, R. Monroe, M. Cogne, L. Davidson, and F. W. Alt. 1996. The Ig(kappa) enhancer influences the ratio of Ig(kappa) versus Ig(lambda) B lymphocytes. Immunity 5:241-252. [DOI] [PubMed] [Google Scholar]
- 9.Greenbaum, S., A. S. Lazorchak, and Y. Zhuang. 2004. Differential functions for the transcription factor E2A in positive and negative gene regulation in pre-B lymphocytes. J. Biol. Chem. 279:45028-45035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenbaum, S., and Y. Zhuang. 2002. Identification of E2A target genes in B lymphocyte development by using a gene tagging-based chromatin immunoprecipitation system. Proc. Natl. Acad. Sci. USA 99:15030-15035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herblot, S., P. D. Aplan, and T. Hoang. 2002. Gradient of E2A activity in B-cell development. Mol. Cell. Biol. 22:886-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu, L. Y., J. Lauring, H. E. Liang, S. Greenbaum, D. Cado, Y. Zhuang, and M. S. Schlissel. 2003. A conserved transcriptional enhancer regulates RAG gene expression in developing B cells. Immunity 19:105-117. [DOI] [PubMed] [Google Scholar]
- 13.Inlay, M., F. W. Alt, D. Baltimore, and Y. Xu. 2002. Essential roles of the kappa light chain intronic enhancer and 3′ enhancer in kappa rearrangement and demethylation. Nat. Immunol. 3:463-468. [DOI] [PubMed] [Google Scholar]
- 14.Inlay, M. A., H. Tian, T. Lin, and Y. Xu. 2004. Important roles for E protein binding sites within the immunoglobulin kappa chain intronic enhancer in activating Vkappa Jkappa rearrangement. J. Exp. Med. 200:1205-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang, H. E., L. Y. Hsu, D. Cado, and M. S. Schlissel. 2004. Variegated transcriptional activation of the immunoglobulin kappa locus in pre-b cells contributes to the allelic exclusion of light-chain expression. Cell 118:19-29. [DOI] [PubMed] [Google Scholar]
- 16.Liu, Z., and W. T. Garrard. 2005. Long-range interactions between three transcriptional enhancers, active Vκ gene promoters, and a 3′ boundary sequence spanning 46 kilobases. Mol. Cell. Biol. 25:3220-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu, R., K. L. Medina, D. W. Lancki, and H. Singh. 2003. IRF-4,8 orchestrate the pre-B-to-B transition in lymphocyte development. Genes Dev. 17:1703-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Massari, M. E., P. A. Grant, M. G. Pray-Grant, S. L. Berger, J. L. Workman, and C. Murre. 1999. A conserved motif present in a class of helix-loop-helix proteins activates transcription by direct recruitment of the SAGA complex. Mol. Cell 4:63-73. [DOI] [PubMed] [Google Scholar]
- 19.McMurry, M. T., and M. S. Krangel. 2000. A role for histone acetylation in the developmental regulation of VDJ recombination. Science 287:495-498. [DOI] [PubMed] [Google Scholar]
- 20.Muljo, S. A., and M. S. Schlissel. 2003. A small molecule Abl kinase inhibitor induces differentiation of Abelson virus-transformed pre-B cell lines. Nat. Immunol. 4:31-37. [DOI] [PubMed] [Google Scholar]
- 21.Murre, C., P. S. McCaw, and D. Baltimore. 1989. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56:777-783. [DOI] [PubMed] [Google Scholar]
- 22.Nagulapalli, S., and M. L. Atchison. 1998. Transcription factor Pip can enhance DNA binding by E47, leading to transcriptional synergy involving multiple protein domains. Mol. Cell. Biol. 18:4639-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagulapalli, S., A. Goheer, L. Pitt, L. P. McIntosh, and M. L. Atchison. 2002. Mechanism of e47-Pip interaction on DNA resulting in transcriptional synergy and activation of immunoglobulin germ line sterile transcripts. Mol. Cell. Biol. 22:7337-7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan, L., J. Hanrahan, J. Li, L. P. Hale, and Y. Zhuang. 2002. An analysis of T cell intrinsic roles of E2A by conditional gene disruption in the thymus. J. Immunol. 168:3923-3932. [DOI] [PubMed] [Google Scholar]
- 25.Pongubala, J. M. R., and M. L. Atchison. 1991. Functional characterization of the developmentally controlled immunoglobulin kappa 3′ enhancer: regulation by Id, a repressor of helix-loop-helix transcription factors. Mol. Cell. Biol. 11:1040-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pongubala, J. M. R., S. Nagulapalli, M. J. Klemsz, S. R. McKercher, R. A. Maki, and M. L. Atchison. 1992. PU.1 recruits a second nuclear factor to a site important for immunoglobulin κ 3′ enhancer activity. Mol. Cell. Biol. 12:368-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quong, M. W., A. Martensson, A. W. Langerak, R. R. Rivera, D. Nemazee, and C. Murre. 2004. Receptor editing and marginal zone B cell development are regulated by the helix-loop-helix protein, E2A. J. Exp. Med. 199:1101-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rolink, A., M. Streb, and F. Melchers. 1991. The kappa/lambda ratio in surface immunoglobulin molecules on B lymphocytes differentiating from DHJH-rearranged murine pre-B cell clones in vitro. Eur. J. Immunol. 21:2895-2898. [DOI] [PubMed] [Google Scholar]
- 29.Romanow, W. J., A. W. Langerak, P. Goebel, I. L. Wolvers-Tettero, J. J. van Dongen, A. J. Feeney, and C. Murre. 2000. E2A and EBF act in synergy with the V(D)J recombinase to generate a diverse immunoglobulin repertoire in nonlymphoid cells. Mol. Cell 5:343-353. [DOI] [PubMed] [Google Scholar]
- 30.Schlissel, M. S. 2004. Regulation of activation and recombination of the murine Igkappa locus. Immunol. Rev. 200:215-223. [DOI] [PubMed] [Google Scholar]
- 31.Schlissel, M. S., and D. Baltimore. 1989. Activation of immunoglobulin kappa gene rearrangement correlates with induction of germline kappa gene transcription. Cell 58:1001-1007. [DOI] [PubMed] [Google Scholar]
- 32.Schweitzer, B. L., and R. P. DeKoter. 2004. Analysis of gene expression and Ig transcription in PU.1/Spi-B-deficient progenitor B cell lines. J. Immunol. 172:144-154. [DOI] [PubMed] [Google Scholar]
- 33.Shaffer, A. L., A. Peng, and M. S. Schlissel. 1997. In vivo occupancy of the kappa light chain enhancers in primary pro- and pre-B cells: a model for kappa locus activation. Immunity 6:131-143. [DOI] [PubMed] [Google Scholar]
- 34.Van Ness, B. G., M. Weigert, C. Coleclough, E. L. Mather, D. E. Kelley, and R. P. Perry. 1981. Transcription of the unrearranged mouse C kappa locus: sequence of the initiation region and comparison of activity with a rearranged V kappa-C kappa gene. Cell 27:593-602. [DOI] [PubMed] [Google Scholar]
- 35.Xu, Y., L. Davidson, F. W. Alt, and D. Baltimore. 1996. Deletion of the Ig kappa light chain intronic enhancer/matrix attachment region impairs but does not abolish V kappa J kappa rearrangement. Immunity 4:377-385. [DOI] [PubMed] [Google Scholar]
- 36.Zhuang, Y., A. Jackson, L. Pan, K. Shen, and M. Dai. 2004. Regulation of E2A gene expression in B-lymphocyte development. Mol. Immunol. 40:1165-1177. [DOI] [PubMed] [Google Scholar]
- 37.Zhuang, Y., P. Soriano, and H. Weintraub. 1994. The helix-loop-helix gene E2A is required for B cell formation. Cell 79:875-884. [DOI] [PubMed] [Google Scholar]