Abstract
macroH2A (mH2A) is an unusual histone variant consisting of a histone H2A-like domain fused to a large nonhistone region. In this work, we show that histone mH2A represses p300- and Gal4-VP16-dependent polymerase II transcription, and we have dissected the mechanism by which this repression is realized. The repressive effect of mH2A is observed at the level of initiation but not at elongation of transcription, and mH2A interferes with p300-dependent histone acetylation. The nonhistone region of mH2A is responsible for both the repression of initiation of transcription and the inhibition of histone acetylation. In addition, the presence of this domain of mH2A within the nucleosome is able to block nucleosome remodeling and sliding of the histone octamer to neighboring DNA segments by the remodelers SWI/SNF and ACF. These data unambiguously identify mH2A as a strong transcriptional repressor and show that the repressive effect of mH2A is realized on at least two different transcription activation chromatin-dependent pathways: histone acetylation and nucleosome remodeling.
DNA is organized into chromatin in the cell nucleus. Chromatin exhibits a repeating structure, and its basic unit, the nucleosome, is composed of an octamer of the four core histones (two each of H2A, H2B, H3, and H4), around which two superhelical turns of DNA are wrapped. The structure of the histone octamer (6) and the nucleosome (25) was solved by X-ray crystallography. In addition to the conventional core histones, the cells express a very small amount of their nonallelic isoforms, the so-called histone variants. The small amount of the histone variants present in the cell suggests that these proteins may play regulatory roles. Indeed, the incorporation of the histone variants into the histone octamer brings new structural properties to the nucleosome, which in turn might be essential for the regulation of several vital processes of the cell. For example, the histone variant H2A.Z is implicated in both gene activation (32) and gene silencing (15). Recently, a role of H2A.Z in chromosome segregation was also suggested (31). Another histone variant, H2AX, is essential for repair and the maintenance of genomic stability (7, 8). Incorporation of the histone variant H2ABbd into the histone octamer confers lower stability of the H2ABbd nucleosomes (16). Since the residues of conventional H2A, which are targets for posttranslational modifications, are mutated in H2ABbd, one could expect the function of this histone to be regulated in a distinct way (10, 5).
macroH2A (mH2A) is an unusual histone variant with a size approximately threefold the size of the conventional H2A (29). The N-terminal domain of mH2A (H2A-like), which shows a high degree of homology with the conventional H2A, is fused to a large nonhistone region (NHR) known as the macro domain (1, 24, 29). The immunofluorescence studies indicate that mH2A is preferentially located on the inactive X chromosome (9, 12, 13, 27). The mH2A nucleosomes exhibit structural alterations in the vicinity of the dyad axis, abrogating the binding of transcription factors to their recognition sequences when the sequences are inserted close to the dyad (4). In addition, the presence of mH2A interferes with SWI/SNF nucleosome remodeling and movement to neighboring DNA segments (4). All these data suggest that mH2A could be involved in transcriptional repression, but the mechanism by which mH2A operates is unknown. Indirect data indicated that the NHR of mH2A could be responsible for the repression of transcription (30). It was also recently suggested that macro domains could possess enzymatic activities [poly(ADP-ribose) formation] and could bind monomeric ADP-ribose and polymers of poly(ADP-ribose) (1, 20). Furthermore, it was recently demonstrated that the macro domain of macroH2A1.1 but not macroH2A1.2 was able to bind the SirT1 metabolite O-acetyl-ADP-ribose (23); however, the consequences of this property on the function of macroH2A and on chromatin structure are not known.
This work summarizes our studies on the effect of mH2A1.2 on transcription. We show that the presence of mH2A inhibits the Gal4-VP16- and p300-dependent histone acetylation and transcription from mH2A nucleosomal arrays. Importantly, this effect was determined only by the NHR of mH2A, since arrays reconstituted with conventional H2A fused to the mH2A NHR (H2A-NHR arrays), but not the H2A-like arrays, exhibited the same behavior. In addition, the chromatin remodeling machines SWI/SNF and ACF were unable to both remodel and mobilize nucleosomes reconstituted with the H2A-NHR fusion protein. These data suggest that the property of mH2A to affect transcriptional regulation resides mainly in its nonhistone region.
MATERIALS AND METHODS
Preparation of DNA probes and reconstitution of nucleosomes.
The 152-bp EcoRI-RsaI DNA fragments containing the Xenopus borealis 5S RNA gene were derived from plasmid pXP-10 (17) by PCR amplification. DNA was 3′ radiolabeled at the EcoRI side by [α-32P]ATP and Klenow enzyme. The 255-bp and 241-bp DNA probes, containing the strongly positioning sequence 601 (33) at the middle or at 8 bp from the 3′ end, respectively, were prepared by PCR amplification of plasmids pGEM3Z-601 and p199-1 (a kind gift from J. Widom and B. Bartholomew) using[γ-32P]ATP-labeled 5′ primer. The 154-bp fragment containing the five Gal4-VP16 binding sites was derived from plasmid pG5ML by PCR amplification using the following primers: 5′-CGA ATC TTT AAA CTC GAG TGC ATG CCT GCA and 5′-AAA GGG CCA AAT CGA TAG CGA GTA TAT ATA GGA CTG GGG ATC. All DNA probes were purified on 6% native polyacrylamide gel electrophoresis.
Nucleosome reconstitutions were performed by salt gradient dialysis as described previously (17). Briefly, ∼100 ng of radiolabeled DNA (5 × 105 cpm), was mixed with 2 μg of nonlabeled ∼180-bp average sequence chicken erythrocytes DNA in 100-μl volume, together with 0.8% (wt/wt) of preassembled histone octamers in high-salt buffer: 10 mM Tris (pH 7.4), 1 mM EDTA, 5 mM β-mercaptoethanol, and 2 M NaCl. Then, the solution was stepwise dialysed at 4°C for 2 h/step against decreasing concentrations (1.2 M, 1.0 M, 0.8 M, 0.6 M, and 0.1 M) of NaCl in the same buffer, followed by dialysis overnight against 10 mM Tris (pH 7.4), 0.25 mM EDTA, and 10 mM NaCl. Nucleosome formation was assessed by a 5% polyacrylamide electrophoretic mobility shift assay (EMSA) run in 0.3× Tris-borate-EDTA buffer.
Protein expression and purification.
The recombinant Xenopus laevis full-length histone protein was produced and purified as previously described (26). For the production of the recombinant H2A-NHR protein, the coding sequences for the H2A protein (from M1 to P118) and for the NHR domain of human macroH2A1.2 (from R118 to N371) were individually amplified by PCR and fused in the pET30a vector to form the coding sequence of the chimera H2A-NHR protein.
After expression in Escherichia coli, the recombinant chimera NHR-H2A was purified to homogeneity as described for the conventional histones.
Nucleosome mobilization experiments.
Nucleosomes (final concentration, 30 to 50 nM) were mixed with SWI/SNF or ACF, as indicated, in buffer containing 10 mM Tris (pH 7.4), 1 mM dithiothreitol, 100 μg/ml bovine serum albumin, 5% glycerol, 0.02% (vol/vol) Nonidet P-40, 2.5 mM MgCl2, and 1 mM ATP. After incubation for 45 min, or the time indicated, the reaction was stopped by 0.05 units of apyrase, 10 mM EDTA (final concentration), and 1 μg of plasmid DNA. Nucleosome sliding was analyzed by a 5% polyacrylamide-bisacrylamide (29:1) EMSA. Nucleosome borders were mapped by limit digestion with exonuclease III (3 to 5 U/ml) for 15 min, and DNA fragments were analyzed by denaturing acrylamide-urea gel electrophoresis. The remodeling of the 152-bp 5S nucleosomal particles was assessed by DNase I footprinting. Briefly, 0.5 units of DNase I (Invitrogen) were added to the arrested reaction mixture for 2 min. DNA digestion was stopped by the addition of 20 mM EDTA (final concentration), 1 μg of proteinase K, and 0.1% sodium dodecyl sulfate (SDS) (final concentration). DNA partial digests were recovered by phenol extraction and ethanol precipitation and analyzed by 8% polyacrylamide-8 M urea sequencing gel electrophoresis in 1× Tris-borate-EDTA buffer at a constant power of 65 W.
Transcription experiments.
Chromatin arrays containing either conventional H2A or H2A-like, H2A-NHR histones were assembled using the pG5ML array template and recombinant Acf1, ISWI, and NAP-1 as previously described (2, 18). The p300 and Gal4-VP16 transcriptions and the histone acetyltransferase (HAT) assays were performed according to previously described protocols (2). The chromatin arrays reconstituted with the conventional H2A or the variant histones were soluble under the conditions used for transcription (data not shown). For the analysis of the transcription elongation through nucleosomes, polymerase II (Pol II) elongation complex assembly was ligated to either the DNA or the conventional or variant nucleosomes, and transcriptions were carried out as described previously (21).
RESULTS
mH2A represses Gal4-VP16- and p300-dependent histone acetylation and transcription.
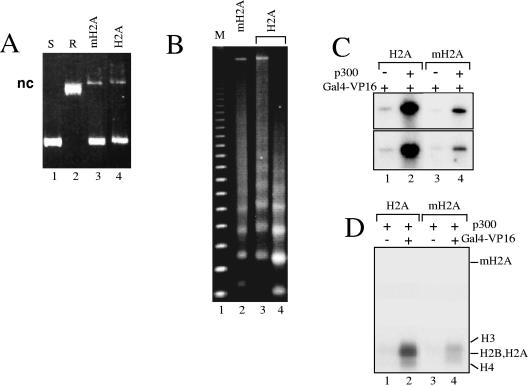
To analyze the effect of mH2A1.2 on transcription, we assembled mH2A nucleosomal arrays according to previously described protocols (18) using recombinant assembly factors and histone proteins (Fig. 1 and see Fig. 3). The assembly was carried out on a pG5ML array plasmid, which contains a promoter sequence flanked by five 208-bp repeats of the 5S RNA sea urchin gene. The reconstituted chromatins containing conventional H2A or histone variant mH2A were analyzed by both supercoiling assay (Fig. 1A) and digestion with micrococcal nuclease (MNase) (Fig. 1B). The supercoiling assay shows a very efficient assembly of the chromatin samples (Fig. 1A, lanes 3 and 4). The clear 200-bp repeats observed upon digestion of both samples with micrococcal nuclease indicate a proper structural organization of the nucleosomal arrays (Fig. 1B, lanes 2 to 4). Then, the conventional and mH2A nucleosomal arrays were used for transcription. The two templates, however, exhibited completely different behaviors in the transcription assay (Fig. 1C). In both cases, the presence of histones abolished the basal transcription (results not shown), and the addition of Gal4-VP16 resulted in a detectable transcription (Fig. 1C, lanes 1 and 3). The presence of p300 in the reaction mixture leads to a dramatic increase of the transcription from the conventional nucleosomal arrays but to a very small increase of the transcription from the mH2A arrays (Fig. 1C, compare lane 1 with lane 2 and lane 3 with lane 4). In the different experiments, the increase of the transcription from conventional arrays was found to be 10 to 12 times higher than that from mH2A arrays. These results clearly show that mH2A is a very efficient repressor of transcription. How does mH2A affect transcription? Since the efficiency of transcription from the chromatin templates was strongly dependent on the HAT p300 and thus on histone acetylation (Fig. 1) (2, 3), one obvious reason for the transcription repression could be the inability of p300 to acetylate histones within the mH2A nucleosomal arrays. This was tested by HAT assays, and we indeed found that the acetylation of histones by p300 was considerably reduced within the mH2A reconstituted chromatin (Fig. 1D, compare lane 2 with lane 4), thus confirming our hypothesis.
FIG. 1.
macroH2A interferes with both p300- and Gal4-VP16-dependent transcription and histone acetylation. Chromatin was assembled on the pG5ML array DNA by using recombinant Drosophila Acf1, ISWI, and the nucleosome assembly protein 1, an equimolar mixture of either conventional H2A or variant histone macroH2A, and the three remaining core histones H2B, H3, and H4. (A) DNA supercoiling assay for the assembly of chromatin. The DNA samples were run on 1% agarose gels and stained with ethidium bromide. Lane 1, supercoiled pG5ML DNA (S); lane 2, topoisomerase I-relaxed pG5ML DNA (R); lanes 3 and 4 show the plasmid DNA isolated from the chromatin samples assembled with macroH2A or conventional H2A, respectively. “nc” designates nick DNA. (B) Micrococcal nuclease digestion of the assembled macroH2A (lane 2) and conventional H2A (lanes 3 and 4) chromatin samples. The chromatin samples were digested with either 0.2 mU MNase (lanes 2 and 3) or 0.5 mU MNase (lane 4) for 10 min at 22°C, and the DNA was analyzed on 1.2% agarose gels. Lane 1, a 123-bp DNA ladder marker (M). (C) p300- and Gal4-VP16-dependent transcription of conventional H2A (lanes 1 and 2) and histone variant mH2A (lanes 3 and 4) nucleosomal arrays. The arrays were incubated with GAL4-VP16 alone or with both Gal4-VP16 and p300. The results from two independent experiments are shown. (D) HAT assays with nucleosomal arrays assembled with either conventional H2A (lanes 1 and 2) or macroH2A (lanes 3 and 4) nucleosomal arrays. All reaction mixtures contained p300 while Gal4-VP16 was present in mixtures for reactions 2 and 4 only. The positions of the histones are indicated.
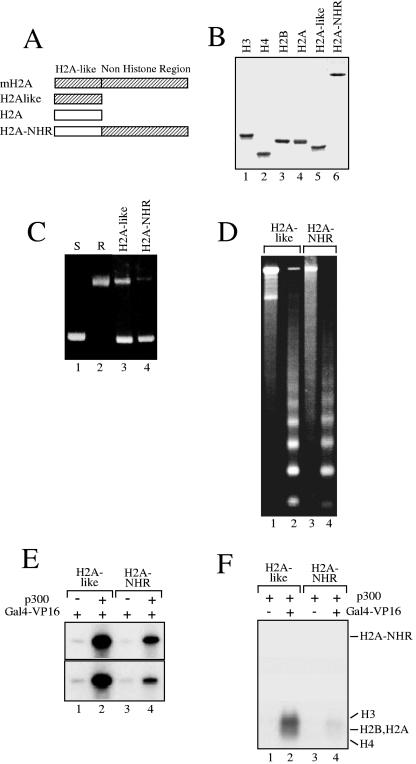
FIG. 3.
The NHR of the histone variant macroH2A is involved in both the repression of p300- and Gal4-VP16-dependent transcription and the inhibition of histone acetylation. Chromatin was assembled on the pG5ML array DNA as described in the legend for Fig. 1 but with the histone-like domain of mH2A (H2A-like) or with the fusion (H2A-NHR) of conventional H2A with the NHR of mH2A. (A) Schematic of the proteins used in the chromatin assembly experiments. (B) 18% SDS electrophoresis of the recombinant conventional core histones, the fusion H2A-NHR, and the H2A-like nucleosomal template. (C) Supercoiling assay for DNA isolated from chromatin assembled with the H2A-like (lane 3) or H2A-NHR (lane 4) nucleosomal template. (D) Micrococcal nuclease analysis of chromatin assembled with either the H2A-like (lanes 1 and 2) or H2A-NHR (lanes 3 and 4) nucleosomal template. The digestion was performed with 0.1 mU MNase (lanes 1 and 3) or with 0.5 mU MNase (lanes 2 and 4) for 10 min at 22°C. DNA was then extracted from the digested samples and analyzed on 1.2% agarose gels. (E) p300- and Gal4-VP16-dependent transcription of H2A-like (lanes 1 and 2) and H2A-NHR (lanes 3 and 4) nucleosomal templates. The results from two independent experiments are shown. (F) HAT assays of H2A-like (lanes 1 and 2) and H2A-NHR (lanes 3 and 4) nucleosomal arrays. Note the complete inhibition of histone acetylation in the H2A-NHR templates.
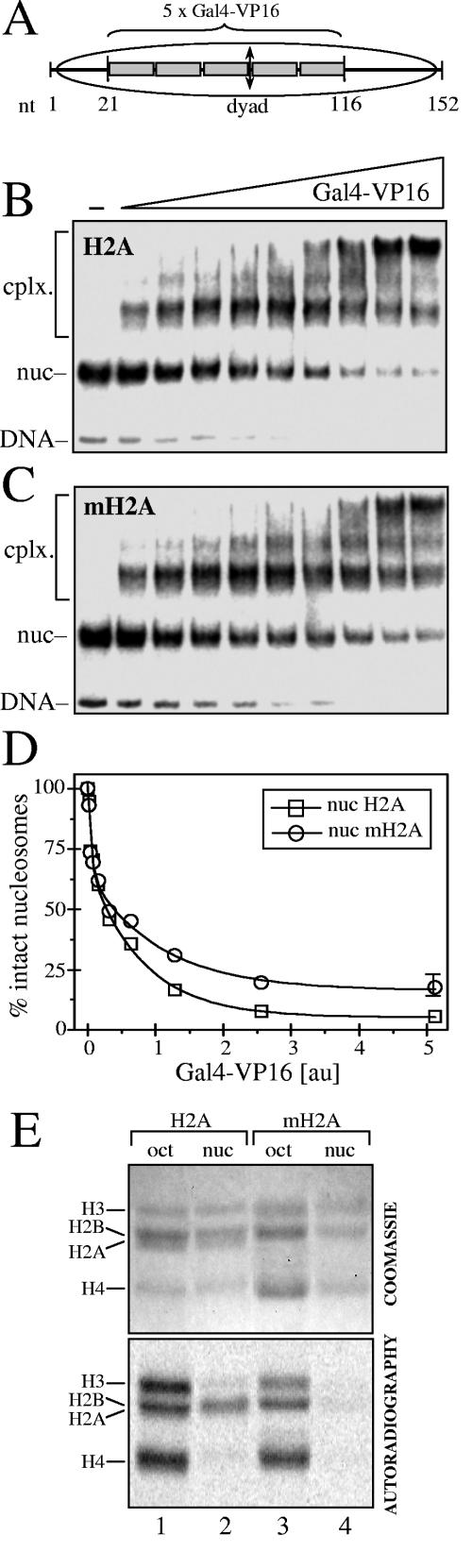
It is well documented that p300 is recruited to the promoter through Gal4-VP16 (2, 3, 22). In addition, the presence of mH2A interferes with the binding of the transcription factor NF-κB when its recognition sequence is inserted in the vicinity of the nucleosome dyad axis (4). This suggests that part of the inhibition of both transcription and histone acetylation could be associated with some possible impairment of Gal4-VP16 binding to its recognition sequence within the mH2A nucleosomes. To check this, we have reconstituted conventional and mH2A nucleosomes using a 154-bp DNA fragment derived from the pG5ML vector (used for reconstitution of the 5S nucleosome array transcription template) and containing the five Gal4-VP16 binding sites. Then, the binding of Gal4-VP16 to both nucleosomal templates and naked DNA was studied by EMSA (Fig. 2 and unpublished data). In agreement with the reports in the literature (14), we found that compared to naked DNA, a much larger amount of Gal4-VP16 was necessary for its binding to the nucleosomes (results not shown). The efficiencies of the binding of Gal4-VP16 to both conventional and mH2A nucleosomes were, however, not significantly different (Fig. 2B to D). Indeed, the quantification of the EMSA results demonstrated only a relatively small preference of Gal4-VP16 binding to conventional nucleosomes compared to mH2A nucleosomes (Fig. 2D). This could reflect the interference of the binding of Gal4-VP16 with two of the five binding sites. This suggests that the strong inhibition of both transcription and histone acetylation of mH2A nucleosomal arrays could not be explained by a lack of binding of Gal4-VP16 to the arrays. In agreement with this, we found that p300 was able to acetylate only very poorly the histones of the GAL4-VP16-bound mH2A nucleosomes (Fig. 2E).
FIG. 2.
The presence of mH2A does not affect the binding of Gal4-VP16 to the nucleosome. (A) Schematic of the nucleosomes used in Gal4-VP16 binding studies. A 152-bp DNA fragment, derived from the pG5ML vector and containing the five Gal4-VP16 sites inserted in the E4 promoter, was PCR amplified and used to reconstitute both conventional and mH2A nucleosomes. The positions of the five Gal4- VP16 binding sites and the nucleosome dyad are designated. nt, nucleotide. (B) Binding of Gal4-VP16 to conventional H2A nucleosomes. Increasing amounts of Gal4-VP16 were added to the solution containing conventional nucleosomes, and Gal4-VP16 binding was assessed by EMSA. The positions of free DNA, nucleosomes (nuc), and Gal4-VP16 nucleosome complexes (cplx.) are designated on the left part of the figures. (C) Data are presented as described for panel B but for macroH2A nucleosomes. (D) Quantification of the data presented in panels B and C. (E) HAT assays with either conventional H2A (lane 2) or macroH2A (lane 4) mononucleosomes. Acetylation of the histone mixtures consisting of conventional histones or containing mH2A is shown in lanes 1 and 3, respectively. All reaction mixtures contained p300 and Gal4-VP16. The positions of the histones are indicated.
The NHR of mH2A is responsible for the repression of transcription and the inhibition of histone acetylation.
A major feature of mH2A is the presence of a long C-terminal extremity (NHR domain) fused to the histone-like domain (H2A-like) which is highly homologous to H2A (Fig. 3A). To determine whether the inhibition of transcription and of p300-dependent histone acetylation observed with mH2A chromatin templates could be attributed to one specific domain of mH2A, recombinant histone proteins corresponding to H2A-like, H2A-NHR (fusion of the NHR domain of mH2A to the conventional H2A) proteins were purified (Fig. 3B) and used for the reconstitution of chromatin on the pG5ML plasmid. Supercoiling (Fig. 3C) and micrococcal nuclease (Fig. 3D) assays showed an efficient reconstitution and proper structural organization of the reconstituted H2A-like and H2A-NHR chromatin templates. These chromatin templates were then used in a transcription assay (Fig. 3E). p300-mediated and Gal4-VP16-dependent transcription from the H2A-like chromatin templates (Fig. 3E, lanes 1 to 2) showed a transcription level similar to the transcription level from conventional nucleosomal arrays. In contrast, the transcription level from the H2A-NHR chromatin templates was repressed about 10-fold (Fig. 3E, lanes 3 to 4) compared to the transcription level from wild-type or H2A-like chromatin templates. This level of repression was similar to what was obtained with the mH2A chromatin template (Fig. 1C), suggesting that the repression of the p300-mediated and Gal4-VP16-dependent transcription observed in the presence of mH2A could be attributed to the NHR domain of mH2A. Since the repression of transcription in the presence of mH2A is associated with a reduced level of p300-dependent acetylation of histones of the chromatin templates, we next tested whether this effect is associated with the presence of NHR (Fig. 3F). Indeed, the level of p300-mediated histone acetylation within the H2A-like chromatin template was very similar to the level of acetylation of conventional nucleosomes (Fig. 3F, lanes 1 to 2), whereas the acetylation level of histones within the H2A-NHR chromatin template (Fig. 3F, lanes 3 to 4) was very low, suggesting that the repression of p300 HAT activity within the mH2A chromatin template is the consequence of the presence of the NHR domain of mH2A.
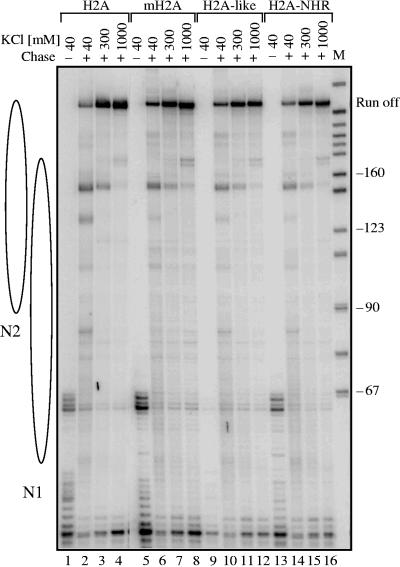
The repression of transcription observed in the presence of mH2A or of the fusion H2A-NHR could be the consequence of a lower initiation level or because of an inhibition of the elongation of Pol II transcription through the nucleosome. To differentiate between these two possibilities, we carried out transcription elongation experiments by using conventional H2A-like, H2A-NHR, and mH2A nucleosomes (Fig. 4). Briefly, the four types of nucleosomes were reconstituted and ligated to Pol II elongation complexes immobilized on beads as described previously (21). The transcription elongation reaction was carried out in the presence of 40 mM, 300 mM, or 1 M KCl, and the nascent RNA was pulse-labeled (21). At 40 mM KCl, the nucleosomal templates efficiently blocked the elongation reaction (Fig. 4, lanes 2, 6, 10, and 14). The 10 to 25% of transcripts observed at this KCl concentration roughly reflect the presence of free DNA in the different template solutions (results not shown) (21). The nucleosome-specific pausing patterns were similar for the four different templates (Fig. 4, lanes 2, 6, 10, and 14). An increase of the ionic strength to 300 mM KCl destabilizes the nucleosomes, and a further increase of the KCl concentration to 1 M results also in a partial removal of H2A-H2B and mH2A-H2B dimers. This, in turn, results in much more efficient transcript elongation on the nucleosomal templates (Fig. 4, lanes 3, 4, 7, 8, 11, 12, 15, and 16). The elongation efficiencies of polymerase II were, however, very similar for the four different types of chromatin templates (Fig. 4, lanes 3, 4, 7, 8, 11, 12, 15, 16). This demonstrates that the observed inhibition of transcription from mH2A and H2A-NHR templates is associated with the initiation but not with the elongation of transcription. Therefore, the NHR domain of mH2A is responsible for the inhibition of the initiation of the Pol II transcription reaction.
FIG. 4.
The NHR of mH2A does not affect polymerase II elongation through nucleosomal templates. A 245-bp DNA fragment was used to reconstitute nucleosomes with either conventional H2A or the fusion H2A-NHR. The templates, a mixed population of two positioned nucleosomes, N1 and N2, were then ligated to the elongation Pol II complex immobilized on beads as described previously (21). The Pol II elongation complex was allowed to transcribe the nucleosomal DNA, and the nascent RNA was pulse labeled. The transcription was performed in the presence of either 40 mM, 300 mM, or 1 M KCl, and the labeled RNA was extracted and analyzed. The RNA isolated from the transcription reactions of conventional H2A (lanes 1 to 4), mH2A (lanes 5 to 8), H2A-like (lanes 9 to 12), and H2A-NHR (lanes 13 to 16) nucleosomal templates was analyzed on an 8% denaturing polyacrylamide gel. The transcriptions from preformed stalled elongation complexes are also shown (lanes 1, 5, 9, and 13). M, a marker for the molecular mass of the transcripts.
The NHR of mH2A induces structural alterations within the nucleosome.
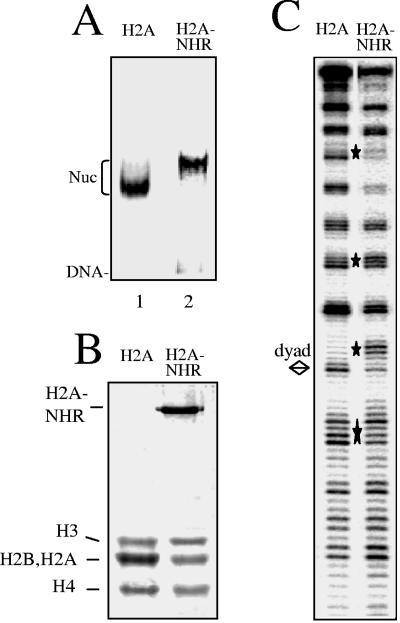
To further examine how the NHR of mH2A could affect transcription initiation, we first determined the consequences of the presence of the NHR on the structure of the nucleosomes. Conventional H2A and H2A-NHR fusion proteins were produced, purified, and used for the reconstitution of nucleosomes on a radioactively end-labeled 152-bp 5S DNA gene (Fig. 5A and B). In order to study the structural consequences of the incorporation of the NHR into the reconstituted particles, we performed DNase I footprinting (Fig. 5C). The DNase I cleavage patterns of the control and H2A-NHR nucleosomes show the 10-bp repeat characteristic of the nucleosome particle. Some pronounced alterations were, however, detected in the DNase I cleavage of the H2A-NHR essentially around the dyad axis. The DNase I cleavage pattern of particles reconstituted with the H2A-like domain of mH2A was essentially the same as that for conventional H2A (for detail, see Fig. 7 of reference 4). We attribute the observed alterations in the DNase I cleavage pattern of the H2A-NHR particle to the presence of the NHR domain. These alterations would reflect some changes in the DNA structure in proximity to the nucleosome dyad and/or some inaccessibility of the enzyme to the nucleosomal DNA. Similar perturbations in the DNase I footprinting were reported for mH2A (4) and H2ABbd (5) nucleosomes, and these perturbations were associated with the inability of the remodeling factors to remodel these variant particles, suggesting that the presence of H2A-NHR within the nucleosome could affect its remodeling.
FIG. 5.
The presence of NHR of mH2A results in alterations in the structure of the H2A-NHR nucleosomes. (A) EMSA of the reconstituted H2A and H2A-NHR nucleosomes (Nuc). (B) 18% SDS-polyacrylamide gel electrophoresis of the histones isolated from H2A (lane 1) and H2A-NHR (lane 2) nucleosomes. The positions of the histones are shown on the left part of the figure. Note that the H2A and H2B Xenopus laevis histones comigrate under the electrophoresis conditions. (C) DNase I footprinting of conventional H2A and H2A-NHR nucleosomes reconstituted on a 152-bp fragment comprising the 5S DNA Xenopus borealis gene. The digestion products were analyzed on an 8% denaturing polyacrylamide gel. The bottom strand of the nucleosomal DNA was P32 labeled. The diamond designates the dyad axis of the nucleosome. Stars indicate the alterations of the H2A-NHR nucleosome DNase I digestion pattern.
FIG. 7.
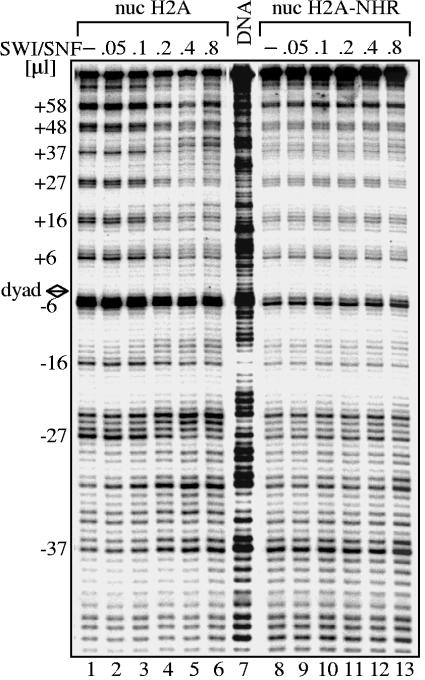
The NHR of mH2A interferes with SWI/SNF nucleosome remodeling. Conventional H2A and H2A-NHR were reconstituted on a radioactively end-labeled 152-bp DNA fragment containing the Xenopus borealis 5S RNA gene. Increasing amounts of SWI/SNF were added to the nucleosome (nuc) solutions, and the remodeling reaction was carried out for 40 min at 30°C. After digestion with DNase I, DNA was extracted and subjected to an 8% sequencing gel. The position of the DNase I cleavage repeat is indicated on the left part of the figure. The DNase I digestion pattern of free DNA is shown in lane 7. The diamond designates the dyad axis of the nucleosome.
The NHR domain of mH2A inhibits the remodeling of nucleosomes by SWI/SNF and ACF.
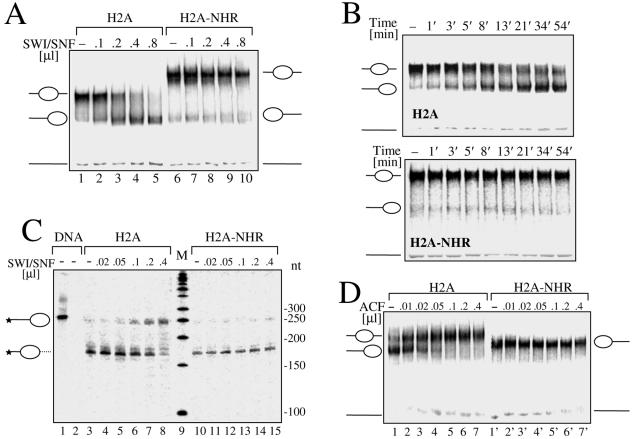
In a recent study, we have demonstrated that the H2A-like domain was able to interfere with SWI/SNF-induced nucleosome mobilization (4). To test whether the NHR domain of mH2A is also able to affect nucleosome remodeling, nucleosomes containing the conventional H2A or the H2A-NHR histone were reconstituted on a 255-bp 5′-end-labeled 601 sequence, which, according to the reported data, should give rise mostly to centrally positioned nucleosomes if conventional histones are used for reconstitution (19). Since both particles show different migration properties, it was important to map precisely the position of the variant H2A-NHR nucleosomal particle on this DNA fragment. Exonuclease III digestion of conventional (Fig. 6C, lane 3) and H2A-NHR (Fig. 6C, lane 10) nucleosomes clearly indicates that both nucleosomal particles are on the same starting central position on the 601 DNA fragment. The incubation of these centrally positioned nucleosomes with increasing amounts of SWI/SNF in the presence of ATP results in a very clear SWI/SNF-dependent sliding of conventional nucleosomes (Fig. 6A, lanes 2 to 5), whereas no mobilization of the H2A-NHR nucleosomes could be observed (Fig. 6, lanes 7 to 10). The kinetics of SWI/SNF-induced sliding of conventional H2A and H2A-NHR nucleosomes (Fig. 6B) confirm this result. No mobilization of the H2A-NHR nucleosomes could be observed after a 54-min incubation period in the presence of 0.5 μl of SWI/SNF (Fig. 6B; H2A-NHR), whereas most of the conventional H2A nucleosomes moved to the DNA ends (Fig. 6B; H2A). Exonuclease III digestion of nucleosomal templates after incubation with different amounts of SWI/SNF (Fig. 6C) confirms that the H2A-NHR nucleosomes are not mobilized by SWI/SNF (lanes 11 to 15), whereas the conventional H2A nucleosomes move to the DNA ends as expected (lanes 4 to 8). To further characterize the mobility properties of H2A-NHR nucleosomes, we used the ACF remodeling factor, which promotes histone octamer sliding from the end to the center of DNA. End-positioned conventional H2A and H2A-NHR nucleosomes reconstituted on the end-positioned sequence 601 241-bp DNA fragment (19) were incubated with increasing amounts of ACF (Fig. 6D). Conventional nucleosomes were efficiently mobilized (Fig. 6C, lanes 1 to 7), whereas no sliding of the H2A-NHR nucleosomes could be detected (lanes 8 to 14).
FIG. 6.
The NHR of mH2A interferes with SWI/SNF and ACF nucleosome mobilization. Conventional H2A and NHR-H2A nucleosomes were reconstituted by using a 255-bp fragment containing the centrally positioned sequence 601 (19). (A) SWI/SNF mobilization of H2A and NHR-H2A nucleosomes. Both types of nucleosomes were incubated for 45 min at 30°C in the presence of increasing amounts of SWI/SNF and ATP. Mobilization of the histone octamer was revealed by EMSA on 5.5% native polyacrylamide gels. The center- and end-positioned nucleosomes and free DNA are indicated on the left part of the figure. (B) Time course of the SWI/SNF-induced mobilization of conventional H2A and H2A-NHR nucleosomes. The nucleosome solutions were supplemented with ATP and 0.5 μl of SWI/SNF and incubated for the indicated time. The nucleosome mobilization was arrested by apyrase treatment, and the reaction mixtures were stored on ice until they were loaded on the gel. The central- and end-positioned nucleosomes and free DNA are indicated. (C) Mapping of H2A and H2A-NHR nucleosome positions after treatment with SWI/SNF. Both types of particles were incubated for 45 min at 30°C in the presence of increasing amounts of SWI/SNF as indicated. Then, the mobilization reaction was arrested by apyrase treatment, the samples were digested with exonuclease III, and the digestion products were run on an 8% denaturating gel. Stars indicate the radioactively labeled end of the DNA used for the reconstitution. (D) ACF is unable to mobilize H2A-NHR nucleosomes. Conventional H2A and H2A-NHR nucleosomes were reconstituted by using a 241-bp fragment containing the end-positioned sequence 601. Reconstituted conventional H2A and H2A-NHR nucleosomes were incubated for 45 min at 30°C with increasing amounts of ACF in the presence of ATP and run on a 5.5% native acrylamide gel. The center- and end-positioned nucleosomes and free DNA are indicated. nt, nucleotide; M, a marker for the molecular mass of the transcripts.
We then tested whether H2A-NHR nucleosomes could be remodeled by SWI/SNF. Conventional H2A and H2A-NHR nucleosomes were formed using a radioactively end-labeled 152-bp DNA fragment containing the Xenopus borealis 5S RNA gene. The samples were incubated with increasing amounts of SWI/SNF and digested with DNase I (Fig. 7). The perturbation of the 10-bp cleavage pattern of the conventional H2A nucleosomes in the presence of SWI/SNF shows that these nucleosomes are efficiently remodeled (Fig. 7, lanes 1 to 6). In contrast, no perturbation of the cleavage pattern could be detected in the presence of SWI/SNF for the H2A-NHR nucleosome (Fig. 7, lanes 1 to 7), demonstrating that this particle cannot be remodeled by this complex. Therefore, the NHR domain of macroH2A interferes with nucleosome remodeling by SWI/SNF.
DISCUSSION
The data reported in this work demonstrate that in vitro mH2A is an efficient repressor of p300- and Gal4-VP16-dependent Pol II-activated transcription. We found that this property of mH2A resides mainly in its NHR domain. Indeed, our experiments show that mH2A and the fusion H2A-NHR were able to impede both Gal4-VP16-dependent Pol II-activated transcription and histone acetylation as well as nucleosome remodeling by SWI/SNF and ACF. Since the presence of mH2A was found to affect weakly the efficiency of Gal4-VP16 binding to the mH2A nucleosomes, the contribution of this effect to the repression of transcription is expected to be small. Bearing in mind that Gal4-VP16 is responsible for the recruitment of p300 to the promoter (2, 3, 22) and that Gal4-VP16 is able to invade mH2A nucleosomes, one could hypothesize that the NHR domain of mH2A is involved in the impediment of histone tail acetylation by p300. The mechanism of this impediment is presently unknown. NHR does not exhibit histone deacetylase activity (1), suggesting that the involvement of NHR in the impediment of histone acetylation is rather steric.
Interestingly, the H2A-like domain of mH2A does not affect p300- and Gal4-VP16-dependent Pol II-activated transcription (this work) but interferes with SWI/SNF nucleosome mobilization (4). Thus, mH2A exhibits some redundancy in function with respect to nucleosome remodeling since each individual domain of mH2A (either H2A-like or NHR, when fused to H2A) was able to impair nucleosome remodeling.
We speculate that in vivo mH2A could contribute to the repression of transcription by affecting at least two different pathways: histone acetylation and chromatin remodeling. Since these two events, i.e., histone acetylation and nucleosome remodeling, are essential for the activation of transcription, it appears that mH2A could be viewed as a major stopper of transcriptional activation. Interestingly, the efficiencies of Pol II passage through conventional H2A, mH2A, and fusion H2A-NHR nucleosomes were essentially the same for the three types of particles. This suggests that the presence of a positioned single mH2A nucleosome on the promoter of specific genes could be sufficient to impede transcription activation by repressing the initiation of transcription.
Our data suggest that the interference of the NHR domain with histone acetylation through steric hindrance would be one of the reasons for this repression. In addition, as shown in a recent report, NHR specifically interacts with HDAC1,2 (11). Consequently, the NHR domain could interfere with the ability of HAT to acetylate the histones of the promoter associated with the macroH2A nucleosome, and in addition, it could recruit histone deacetylase, which further abrogates the possibility of histone acetylation.
One cannot exclude, however, that the presence of several mH2A nucleosomes, some of which reside on the gene coding region, would affect transcription more efficiently. Indeed, the structure of chromatin domains which contain mH2A could be distinct from the 30-nm fiber canonical structure, which in turn might be more refractive to transcription.
Our finding that mH2A behaves as a major stopper of Pol II activation of transcription in vitro raises several questions, since to fulfill such function in vivo, mH2A should be localized specifically on the promoter of transcriptionally inactive genes. The presence of mH2A on such genes would repress transcription. For the transcriptional activation of these genes, the repressive function of mH2A should be eliminated. This could be achieved by the specific removal of mH2A from the promoter and its replacement by conventional H2A by an mH2A-specific histone chaperone as recently described for the histone variant H2A.Z (28). The identification of genes for which expression is controlled by mH2A as well as the understanding of the mechanism of specific deposition and removal of mH2A from these genes remains a challenge for future studies.
Acknowledgments
This work was supported by CNRS, INSERM, Région Rhône-Alpes, and grants from the Ministère de la Recherche (ACI Biologie cellulaire Moléculaire et Structurale, BCM0070, and ACI Interface Physique-Chimie-Biologie: Dynamique et réactivité des Assemblages Biologiques [DRAB], 2004, no. 04 2 136, ANR Projet no. NT05-1_41978).
D.A. is on leave from the Institute of Solid State Physics, BAS, Sofia, Bulgaria.
REFERENCES
- 1.Allen, M. D., A. M. Buckle, S. C. Cordell, J. Lowe, and M. Bycroft. 2003. The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non-histone domain of macroH2A. J. Mol. Biol. 330:503-511. [DOI] [PubMed] [Google Scholar]
- 2.An, W., V. B. Palhan, M. A. Karymov, S. H. Leuba, and R. G. Roeder. 2002. Selective requirements for histone H3 and H4 N termini in p300-dependent transcriptional activation from chromatin. Mol. Cell 9:811-821. [DOI] [PubMed] [Google Scholar]
- 3.An, W., and R. G. Roeder. 2003. Direct association of p300 with unmodified H3 and H4 N termini modulates p300-dependent acetylation and transcription of nucleosomal templates. J. Biol. Chem. 278:1504-1510. [DOI] [PubMed] [Google Scholar]
- 4.Angelov, D., A. Molla, P. Y. Perche, F. Hans, J. Cote, S. Khochbin, P. Bouvet, and S. Dimitrov. 2003. The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol. Cell 11:1033-1041. [DOI] [PubMed] [Google Scholar]
- 5.Angelov, D., A. Verdel, W. An, V. Bondarenko, F. Hans, C. M. Doyen, V. M. Studitsky, A. Hamiche, R. G. Roeder, P. Bouvet, and S. Dimitrov. 2004. SWI/SNF remodeling and p300-dependent transcription of histone variant H2ABbd nucleosomal arrays. EMBO J. 23:3815-3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arents, G., R. W. Burlingame, B. C. Wang, W. E. Love, and E. N. Moudrianakis. 1991. The nucleosomal core histone octamer at 3.1 Å resolution: a tripartite protein assembly and a left-handed superhelix. Proc. Natl. Acad. Sci. USA 88:10148-10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bassing, C. H., H. Suh, D. O. Ferguson, K. F. Chua, J. Manis, M. Eckersdorff, M. Gleason, R. Bronson, C. Lee, and F. W. Alt. 2003. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell 114:359-370. [DOI] [PubMed] [Google Scholar]
- 8.Celeste, A., S. Difilippantonio, M. J. Difilippantonio, O. Fernandez-Capetillo, D. R. Pilch, O. A. Sedelnikova, M. Eckhaus, T. Ried, W. M. Bonner, and A. Nussenzweig. 2003. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell 114:371-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chadwick, B. P., C. M. Valley, and H. F. Willard. 2001. Histone variant macroH2A contains two distinct macrochromatin domains capable of directing macroH2A to the inactive X chromosome. Nucleic Acids Res. 29:2699-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chadwick, B. P., and H. F. Willard. 2001. A novel chromatin protein, distantly related to histone H2A, is largely excluded from the inactive X chromosome. J. Cell Biol. 152:375-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakravarthy, S., S. K. Gundimella, C. Caron, P. Y. Perche, J. R. Pehrson, S. Khochbin, and K. Luger. 2005. Structural characterization of the histone variant macroH2A. Mol. Cell. Biol. 25:7616-7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costanzi, C., and J. R. Pehrson. 1998. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 393:599-601. [DOI] [PubMed] [Google Scholar]
- 13.Costanzi, C., and J. R. Pehrson. 2001. MACROH2A2, a new member of the MACROH2A core histone family. J. Biol. Chem. 276:21776-21784. [DOI] [PubMed] [Google Scholar]
- 14.Cote, J., J. Quinn, J. L. Workman, and C. L. Peterson. 1994. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 265:53-60. [DOI] [PubMed] [Google Scholar]
- 15.Dhillon, N., and R. T. Kamakaka. 2000. A histone variant, Htz1p, and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol. Cell 6:769-780. [DOI] [PubMed] [Google Scholar]
- 16.Gautier, T., D. W. Abbott, A. Molla, A. Verdel, J. Ausio, and S. Dimitrov. 2004. Histone variant H2ABbd confers lower stability to the nucleosome. EMBO Rep. 5:715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayes, J. J., and K. M. Lee. 1997. In vitro reconstitution and analysis of mononucleosomes containing defined DNAs and proteins. Methods 12:2-9. [DOI] [PubMed] [Google Scholar]
- 18.Ito, T., M. E. Levenstein, D. V. Fyodorov, A. K. Kutach, R. Kobayashi, and J. T. Kadonaga. 1999. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 13:1529-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kagalwala, M. N., B. J. Glaus, W. Dang, M. Zofall, and B. Bartholomew. 2004. Topography of the ISW2-nucleosome complex: insights into nucleosome spacing and chromatin remodeling. EMBO J. 23:2092-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karras, G. I., G. Kustatscher, H. R. Buhecha, M. D. Allen, C. Pugieux, F. Sait, M. Bycroft, and A. G. Ladurner. 2005. The macro domain is an ADP-ribose binding module. EMBO J. 24:1911-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kireeva, M. L., W. Walter, V. Tchernajenko, V. Bondarenko, M. Kashlev, and V. M. Studitsky. 2002. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol. Cell 9:541-552. [DOI] [PubMed] [Google Scholar]
- 22.Kundu, T. K., V. B. Palhan, Z. Wang, W. An, P. A. Cole, and R. G. Roeder. 2000. Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol. Cell 6:551-561. [DOI] [PubMed] [Google Scholar]
- 23.Kustatscher, G., M. Hothorn, C. Pugieux, K. Scheffzek, and A. G. Ladurner. 2005. Splicing regulates NAD metabolite binding to histone macroH2A. Nat. Struct. Mol. Biol. 12:624-625. [DOI] [PubMed] [Google Scholar]
- 24.Ladurner, A. G. 2003. Inactivating chromosomes: a macro domain that minimizes transcription. Mol. Cell 12:1-3. [DOI] [PubMed] [Google Scholar]
- 25.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 26.Luger, K., T. J. Rechsteiner, and T. J. Richmond. 1999. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 119:1-16. [DOI] [PubMed] [Google Scholar]
- 27.Mermoud, J. E., C. Costanzi, J. R. Pehrson, and N. Brockdorff. 1999. Histone macroH2A1.2 relocates to the inactive X chromosome after initiation and propagation of X-inactivation. J. Cell Biol. 147:1399-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizuguchi, G., X. Shen, J. Landry, W. H. Wu, S. Sen, and C. Wu. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303:343-348. [DOI] [PubMed] [Google Scholar]
- 29.Pehrson, J. R., and V. A. Fried. 1992. macroH2A, a core histone containing a large nonhistone region. Science 257:1398-1400. [DOI] [PubMed] [Google Scholar]
- 30.Perche, P. Y., C. Vourc'h, L. Konecny, C. Souchier, M. Robert-Nicoud, S. Dimitrov, and S. Khochbin. 2000. Higher concentrations of histone macroH2A in the Barr body are correlated with higher nucleosome density. Curr. Biol. 10:1531-1534. [DOI] [PubMed] [Google Scholar]
- 31.Rangasamy, D., I. Greaves, and D. J. Tremethick. 2004. RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nat. Struct. Mol. Biol. 11:650-655. [DOI] [PubMed] [Google Scholar]
- 32.Santisteban, M. S., T. Kalashnikova, and M. M. Smith. 2000. Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell 103:411-422. [DOI] [PubMed] [Google Scholar]
- 33.Thastrom, A., P. T. Lowary, H. R. Widlund, H. Cao, M. Kubista, and J. Widom. 1999. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J. Mol. Biol. 288:213-229. [DOI] [PubMed] [Google Scholar]