Abstract
We developed a mammalian plasmid replicon with a formerly uncharacterized origin of DNA synthesis, 8xRep*. 8xRep* functions efficiently to support once-per-cell-cycle synthesis of plasmid DNA which initiates within Rep*. By characterizing Rep*'s requirements for acting as an origin, we have uncovered several striking properties it shares with DS, the only other well-characterized, licensed, mammalian plasmid origin of DNA synthesis. Rep* contains a pair of previously unrecognized Epstein-Barr nuclear antigen 1 (EBNA1)-binding sites that are both necessary and sufficient in cis for its origin activity. These sites have an essential 21-bp center-to-center spacing, are bent by EBNA1, and recruit the origin recognition complex. The properties shared between DS and Rep* define cis and trans characteristics of a mammalian, extrachromosomal replicon. The role of EBNA1 likely reflects its evolution from cellular factors involved in the assembly of the initiation machinery.
The defining properties of origins of DNA synthesis in mammalian chromosomes are not known. Although some discrete sites have been mapped biochemically, genetic analyses of these sites have been limited and in some cases indicate that these sites may be unnecessary for the initiation of DNA synthesis within a zone encompassing them (4, 35, 45; for a review of discrete origins and initiation zones, see reference 13). Relocation of DNAs 1.2 to 5.8 kbp in length encompassing three mapped sites to other places in the genome has demonstrated that these ectopic origins still function (5, 41, 50). The origin recognition complex (ORC) binds at or near sites at which DNA synthesis initiates in Saccharomyces cerevisiae and Drosophila melanogaster and appears to perform similarly in mammals (1, 11, 25). However, this complex displays little or no sequence specificity when isolated from Drosophila or human cells (52, 60). It is not clear, therefore, the extent to which DNA sequence might contribute to defining an origin of DNA synthesis in mammalian chromosomes. One successful approach to understand mammalian biology has been to characterize viruses that replicate in mammalian cells and have evolved to coopt their molecular machinery. Studies of the human virus, Epstein-Barr virus (EBV), for example, have documented properties of one origin of DNA synthesis, oriP, that replicates once per cell cycle (2, 65), a hallmark of mammalian, licensed chromosomal synthesis.
oriP supports the initiation of DNA synthesis on the viral plasmid within a region mapped to include DS, a cis-acting sequence shown genetically to be required for DNA synthesis (22, 63). DS is composed of two pairs of binding sites for EBV's encoded Epstein-Barr nuclear antigen 1 (EBNA1) protein, the only viral trans-acting factor required for replication of DNAs containing oriP (42, 51, 66). One pair of these EBNA1-binding sites suffices to support DNA synthesis, albeit less efficiently than do both pairs (24, 36, 64). EBNA1 recruits the ORC and MCM complexes to DS in support of DNA synthesis (17, 20, 53, 54). EBNA1 binds the pairs of sites in DS with a specific, required spacing in order to support DNA synthesis and bends that DNA (8, 24). Additional cellular proteins, such as E2F1-4, Nbs1, and telomere-associated proteins, also bind near DS, but their contribution to the function of DS is unclear (19, 43).
Any general properties essential to DS as an origin of DNA synthesis have not been identified because no similar example has been available for comparison. We have developed 8xRep* as a second origin of DNA synthesis with which to compare oriP and have identified characteristics it shares with oriP. Rep* was initially identified as a DNA sequence in EBV's genome that could substitute functionally for DS albeit inefficiently and unstably (33). Measurements of ORC binding are consistent with Rep*'s contributing to origin function in the context of the viral genome (54). Just as multiples of elements within DS increase its efficiency, multiples of Rep* do, too (54). We have found that an octamer of Rep* supports DNA synthesis as efficiently as does DS, which has allowed a detailed characterization of Rep*. We found unexpectedly that Rep* contains a previously unrecognized pair of binding sites for EBNA1 and shares certain properties with DS that are likely critical for their shared support of DNA synthesis. Rep* is a bona fide, but inefficient, replicator. Its multimerization yields an efficient origin of replication and may serve well as a model for chromosomal zones of replication.
MATERIALS AND METHODS
Plasmids.
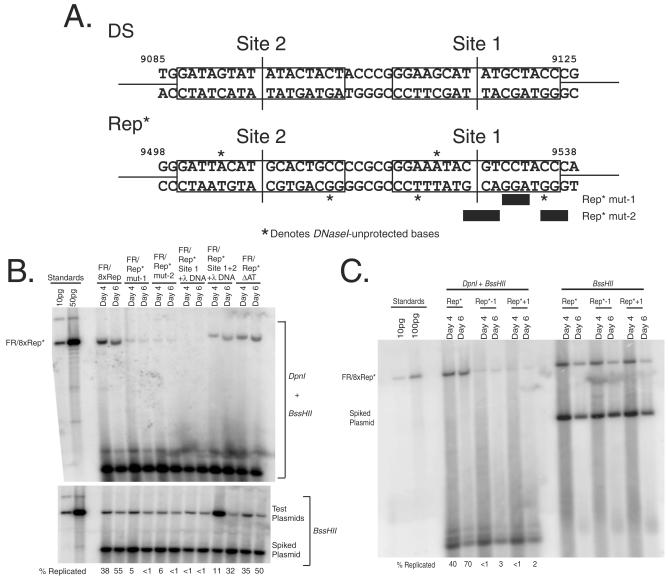
Plasmid derivatives of Rep* (see Fig. 5B) were obtained by the QuikChange method from Stratagene and then multimerized to eight copies. The lambda DNA fragment (present in FR-λ-Luc) does not support replication detectably (33). Plasmid “FR/Rep* mut-1” in Fig. 5B contains eight copies of Rep* with mutations from GTCCTACCCAGGA to GTttcACCCgaaA, plasmid “FR/Rep* mut-2” contains eight copies of Rep* with mutations from ACGTCCTACCCAG to AtacCCTAtttAG, plasmid “FR/Rep* Site 1+λ DNA” contains eight copies of the fusion of EBNA1-binding site 1 of Rep* (EBV nucleotides [nt] 9517 to 9541) with 290 bp of lambda DNA, plasmid “FR/Rep* Site 1+2+λ DNA” contains eight copies of the fusion of both EBNA1-binding sites of Rep* with 290 bp of lambda DNA, and plasmid “FR/Rep* ΔAT” contains eight copies of Rep* with a deletion of the AT element (EBV nt 9565 to 9573). The plasmids “Rep*+1” and “Rep*−1” in Fig. 5C contain FR and eight copies of Rep*-spacing mutants. The 1-bp addition (insertion of C between nt 9517 C and nt 9518 G, Fig. 6A) or the 1-bp deletion (the nt 9518 G is deleted; see Fig. 5A) of the spacing between EBNA1-binding sites was made by the QuikChange method from Stratagene. All other plasmids are described in the supplemental material.
FIG. 5.
A pair of properly spaced EBNA1-binding sites are necessary and sufficient for the origin function of Rep*. (A) Shown are the mapped EBNA1-binding sites in Rep* compared to a pair of EBNA1-binding sites of DS. The nucleotides within the binding sites that were unprotected by EBNA1 from DNase I cleavage were found only in Rep* but not in DS and are denoted by asterisks. Two sets of transversion mutations in site 1 of Rep* were made, and these bases changed are denoted by bars beneath the Rep* sequence. (B) Two EBNA1-binding sites of Rep* are both necessary and sufficient for its origin activity. BJAB/EBNA1 cells were transfected with the test plasmids, and 2 × 107 cells were harvested at 4 and 6 days posttransfection. The extracted plasmids were analyzed as described in Fig. 3. (C) The 21-bp spacing between EBNA1-binding sites is essential for its origin activity. Rep*−1 has nt 9518 deleted from between the two EBNA1-binding sites; Rep*+1 inserts a C between nt 9517 to 9518 between the two EBNA1-binding sites. The plasmids containing FR and eight copies of either Rep*−1 or Rep*+1 were tested as described in panel B.
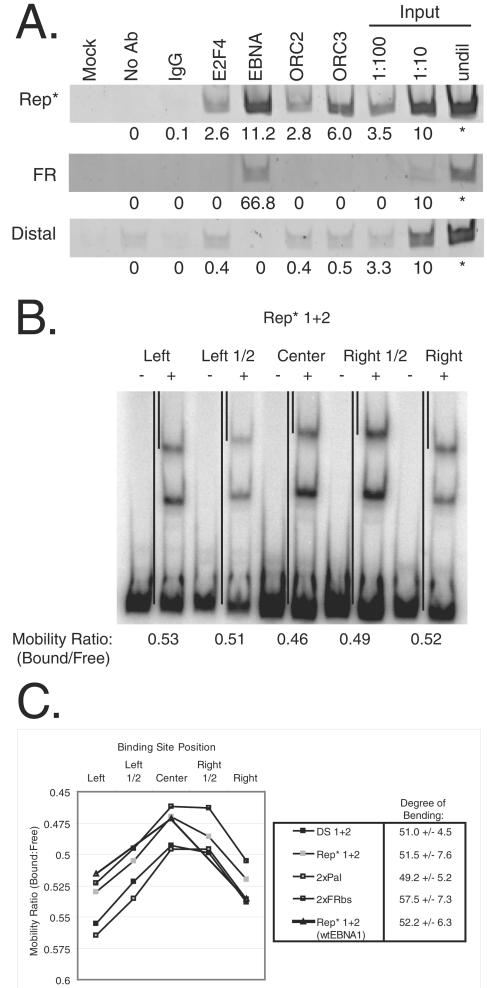
FIG. 6.
ORC is recruited to 8xRep* that is bound and bent by EBNA1. (A) ORC2 and -3 are recruited to Rep*. BJAB/EBNA1 cells stably harboring FR/8xRep* plasmids were cross-linked, and protein-DNA interactions were assessed by ChIP. Chromatin from 107 cell equivalents were precipitated with antibodies against rat immunoglobulin G (IgG), human E2F4, EBNA1, human ORC2, and human ORC3, as denoted above each lane. The mock sample contained neither chromatin nor antibodies, and the No Ab sample contained chromatin but was in the absence of antibodies. Immunoprecipitated chromatin or serial dilutions of input chromatin from 2 × 106 cells were analyzed by PCR with specific primers for FR, Rep*, a region distal to FR and Rep*, and DHFR (data not shown) and visualized by using ethidium bromide on a 4% polyacrylamide gel. Signal intensities for each immunoprecipitation were quantified and normalized by subtracting the background intensity observed from the No Ab sample. Values for these normalized signals compared to the input samples are listed below its respective band. (B and C) One pair of EBNA1-binding sites from either DS (“DS 1+2”), Rep* (“Rep* 1+2”), the EBNA1-DNA cocrystal (“2xPal”), or the most common EBNA1-binding site of FR (“2xFRbs”) were positioned at the left end, the center of the left half, the center, the center of the right half, or the right end of equal length probes. (B) A total of 10 fmol of each of the five probes was incubated with either 0 or 50 nM dnEBNA1/Softag1, electrophoresed on a 4% polyacrylamide gel, detected, and analyzed as described in Fig. 4A for EMSA. The gel image for the Rep* probes is shown and is representative of the other three probes tested. Black lines between the lanes of reactions with the binding sites positioned at either end and at the center denote the mobility of the bound and free probe. The mobility ratio (mobility of bound probe/mobility of free probe) of these reactions is given below the gel image. (C) The mobility ratios of all four probes are compared graphically for two dimers bound. These results are representative of three independent experiments. The mobility ratios of two dimers of wtEBNA1 bound to the Rep* probe are also compared. The average degree of bend is given to the right of each probe tested.
Cell lines and transfections.
Descriptions of 293 and BJAB cell lines stably expressing EBNA1 and cell transfection are given in the supplemental material and reference 44.
Long- and short-term replication assay and Southern blotting.
The colony formation assay to measure the plasmid replication efficiency in long term and the DpnI-based transient plasmid replication assay were performed as previously described with some modifications (32, 64). The details can be found in the supplemental material and reference 34.
TaqMan real-time PCR, CsCl density gradient analysis of cells labeled with BrdU.
Real-time PCR assay to measure the rate of loss of Rep* plasmids from established cells, cell labeling with bromodeoxyuridine (BrdU), DNA extraction, gradient DNA fractionation, and dot blotting are described in the supplemental material and reference 46.
Plasmid isolation and restriction enzyme digestions for the two-dimensional gel.
Enriched extrachromosomal DNAs were obtained from ∼3 × 108 exponentially growing BJAB cells that express EBNA1 and carry 20 to 30 copies of FR/8xRep* plasmids as detailed in the supplementary experimental procedures.
BND cellulose fractionation and two-dimensional gel electrophoresis.
These assays were performed as described previously (15, 27) with some modifications as outlined in the supplemental material.
Production of dnEBNA1/Softag1, electrophoretic mobility shift assay (EMSA), and DNase I footprinting.
These assays are described in detail in the supplemental material and references 3 and 59.
ChIP.
Formaldehyde cross-linked chromatin from BJAB/EBNA1 cells stably harboring ∼10 copies of a FR/8xRep* plasmid was analyzed for protein-DNA interactions by chromatin immunoprecipitation (ChIP) essentially as described previously (61). See the supplemental material for a description of alterations to the protocol.
Circular permutation assay.
One pair of EBNA1-binding sites from either DS (“DS 1+2”), Rep* (“Rep* 1+2”), the 18-bp palindromic binding site previously cocrystallized with a derivative of EBNA1 (GGTAGCATATGCTACC; “2xPal”), or the most common EBNA1-binding site of FR (GATAGCATATGCTACC; “2xFRbs”) were inserted in phase to one another into a unique XbaI site located between two head-to-tail copies of a 121-bp multiple cloning site in the pBend2 vector (30). Probes of identical size (176 bp) were prepared by digestion with one of five different enzymes (e.g., BamHI, KpnI, PvuII, SpeI, and MluI) to position the two EBNA1-binding sites at the left end, the center of the left half, the center, the center of the right half, or the right end of the probe with respect to the Watson strand. Probes were purified on a 4% polyacrylamide gel and were labeled, incubated with either 0 or 50 nM of a derivative of EBNA1, and analyzed as described for the EMSA.
RESULTS
8xRep* is an efficient origin of replication.
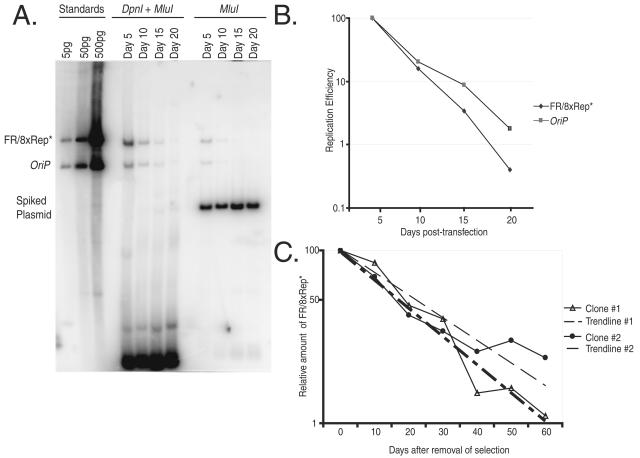
We constructed a plasmid containing FR and eight copies of Rep* and compared its ability to replicate stably under selection for 3 to 4 weeks in cells expressing EBNA1 with that of DS in the presence of FR. We find that 8xRep* replicates extrachromosomally as efficiently as DS (see Table S1 and Fig. S1 in the supplemental material). Newly introduced oriP plasmids are lost precipitously from cells before they are established by an epigenetic event, and this establishment takes 2 to 3 weeks after transfection (39). Once established, they are lost at rates of 2 to 4% per generation (32). Because measurements of these rates reflect the efficiencies of both DNA synthesis and segregation and provide a sensitive assay to detect subtle differences in the replication activity, both of these rates for FR/8xRep* were determined. The rate of establishment of FR/8xRep* plasmids was measured in BJAB/EBNA1 cells. An equal number of molecules of FR/8xRep* and oriP plasmids were introduced into the cells separately in the absence of selection, and the numbers of newly introduced, synthesized FR/8xRep* DNAs and oriP DNAs were determined by Southern blotting every 5 days. The rates of loss of these DNAs were similar and their levels at day 20 were each ca. 1% of those at day 5, indicating that FR/8xRep* is established as efficiently as is oriP (Fig. 1A and B). Two established 293/EBNA1 clones were used to assay the rate of loss of 8xRep* DNA. These cells were grown for 60 days after the removal of selection, and the copy numbers of FR/8xRep* were measured every 10 days by real-time PCR (Fig. 1C). After 2 months of growth in the culture without selection, 5 to 10% of the plasmid DNAs still remained. FR/8xRep* plasmids were thus lost at 2.6 and 3.8% per cell division, a finding similar to that of oriP plasmids (32, 57, 66). 8xRep* supports replication of plasmids carrying FR in cis and with EBNA1 in trans as efficiently as does DS as measured by their establishment, extrachromosomal maintenance, and rate of loss in the absence of selection in human cells.
FIG. 1.
FR/8xRep* is established and replicates with similar efficiencies as oriP in transfected cells and established clones. (A) Equal amounts of oriP and FR/8xRep* plasmids were transfected into BJAB/EBNA1 cells. At the indicated time points, the plasmids were extracted from 2 × 107 cells. To minimize the variations of the recovery of oriP and FR/8xRep* plasmids, the cells transfected with either oriP or FR/8xRep* were combined and processed in the same tube. To test the completeness of DpnI digestion, 10 ng of pPur plasmid was added as “spiked DNA” during extraction. For the Southern analysis, 40% of the samples was digested with both MluI and DpnI, and 10% was digested with MluI. As standards, 5, 50, and 500 pg of linearized oriP and FR/8xRep* plasmids were loaded on the left of the blot. (B) Graphic representation of two independent time course experiments described in panel A. The replication efficiency was plotted versus the days after transfection. For each independent experiment, the level of replicated FR/8xRep* plasmid or oriP plasmid at the first time point was set to 100%, and the replication efficiency of this plasmid at later time points was set relative to this initial time point. The numbers are the average of two independent experiments. (C) The rate of loss of FR/8xRep* plasmids from established clones was 2.6 to 3.8% per cell generation. Two independent clones of 293/EBNA1 cells stably harboring FR/8xRep* plasmids were grown in the absence of selection for a period of 60 days. At the indicated times, the total cellular DNA was extracted from each clone and the number of the FR/8xRep* plasmids per cell was determined by real-time PCR. The number of FR/8xRep* plasmids at the first time point was set to 100, and the number of DNA molecules per cell at later time points was set relative to this initial time point. The rate of loss can be calculated by the equation N/N0 = e(−λ*t), where N0 and N are the relative amounts of plasmid DNA at a initial and later time points, t is the time in days, and λ is the rate of loss.
8xRep* is a licensed mammalian replicator.
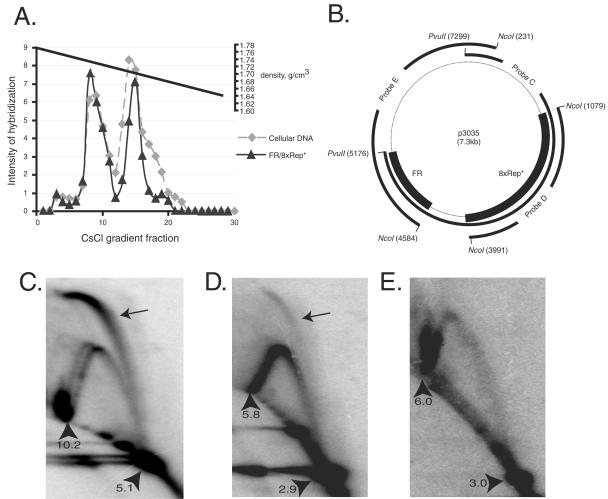
OriP DNA is licensed as evidenced by its replicating once per cell cycle (2, 65). We performed a density labeling experiment with a clone of BJAB/EBNA1 cells that carries about 10 copies of FR/8xRep* DNA to determine whether it is licensed as well. We monitored the synthesis of FR/8xRep* and its host cell DNA by measuring the incorporation of BrdU and separated the differently substituted DNAs on a CsCl density gradient. The amount of FR/8xRep* DNA or cellular DNA in each fraction was determined by nucleic acid hybridization “dot blots,” which indicated that each DNA incorporated BrdU with identical kinetics, demonstrating that the replication of FR/8xRep* occurs synchronously and to the same extent as does cellular DNA and is therefore licensed (Fig. 2A).
FIG. 2.
8xRep* is a licensed replicator and DNA synthesis initiates within it. (A) FR/8xRep* plasmids replicate once per cell cycle in established clones. A BJAB/EBNA1 cell clone carrying FR/8xRep* plasmids was labeled with BrdU for 8 h. Total cellular DNA was isolated from 2 × 107 cells and separated on a CsCl density gradient. One-third of each fraction was dotted onto a nylon filter in a volume of 30 μl and hybridized to the pPur or the EBNA1 gene (which is integrated and serves as a measure of chromosomal DNA), and the dot blot was exposed to a phosphorimager screen. The average intensity of hybridization of the last six points was set as a background, and the relative intensity of each point was plotted versus the density gradient fraction number. The densities of each fraction, as determined by the refractive index, were plotted in the upper part of the graph. Note that the densities of LL and HL DNA are 1.70 and 1.74 g/cm3, respectively. No DNA can be detected in the fractions with densities near 1.76 g/cm3, at which HH DNA would migrate. (B to E) Neutral/neutral two-dimensional analysis of replication intermediates of FR/8xRep* plasmids stably maintained in BJAB/EBNA1 cells. The low-molecular-weight DNA extracted by the method of Hirt from 2 × 108 cells was digested with a restriction enzyme, passed through a BND-cellulose column to enrich for replicative intermediates, and then subjected to two-dimensional gel electrophoresis. (B) The plasmid map of FR/8xRep* (plasmid number p3035). FR and 8xRep* are indicated by a bold line. Probes C, D, and E are used in panels C, D, and E, respectively, and they are indicated by bold lines. (C) The 5.1-kb PvuII fragment of the FR/8xRep* plasmid analyzed by two-dimensional gel as indicated in Fig. 2B contains FR at one end and 8xRep* in the middle (probe C). The blot was probed with this PvuII fragment. (D) The 2.9-kb NcoI fragment of the FR/8xRep* plasmid as indicated in Fig. 3B contains 8xRep* but not FR, and the blot was probed with 8xRep* fragment (probe D). (E) The 3.0-kb NcoI fragment of the FR/8xRep* plasmid contains FR at one end but not 8xRep*, and the blot was probed with this 3.0-kb NcoI fragment (probe E). The arrowheads in panels C to E indicate the positions of 1x and 2x spots measured in Kbp; the 2x spots in panels C and D contain the pronounced termination signals due to the inclusion of FR sequences; the arrows in panels C and D indicate the presence of replication bubbles.
DNA synthesis initiates within Rep* as determined by two-dimensional gel analysis.
The structures of replication intermediates of FR/8xRep* DNA maintained extrachromosomally in BJAB/EBNA1 cells were analyzed by neutral/neutral two-dimensional gel electrophoresis. Replication bubbles resulting from DNA initiating within the middle third of a DNA fragment containing 8xRep* were clearly detected in fragments of two separate digests of the replicative intermediates (Fig. 2C and D). In our analyses, the slower-migrating portions of the bubble signals in both digests were robust and sharp, whereas the lower portions were either weak or undetectable. As a control, the blot for the NcoI fragment was probed with the 3.0-kb plasmid DNA that is opposite to the Rep*-containing NcoI fragment, and no replication bubble structure was detected (Fig. 2E). It has been shown that replication intermediates containing initiation events at different locations on different molecules comigrate to the greatest extent toward the 2x spot (55). We thus interpret our two-dimensional gels to mean that DNA replication initiates within any one of multiple copies of Rep* on different molecules during each cell cycle. The strong signals arising from the 2x spots in Fig. 2C and E are caused by the replication-stalling effects at FR (21).
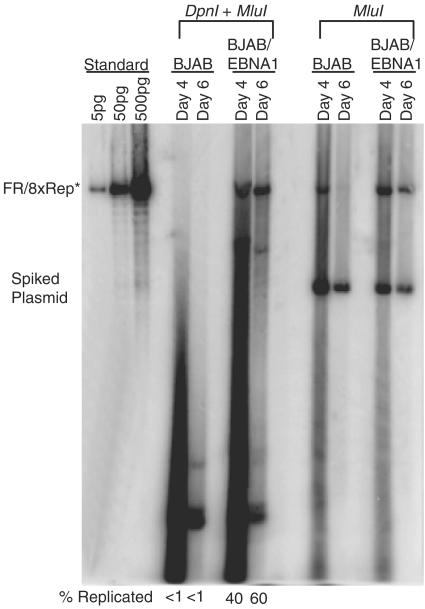
FR/8xRep* plasmids replicate only in cells expressing EBNA1.
The origin activity of DS is dependent on its binding to EBNA1. Sequence analysis did not identify any EBNA1-binding sites within Rep* (33). We therefore tested whether Rep* could replicate in the presence or absence of EBNA1 in two cell recipients. We introduced FR/8xRep* plasmids into BJAB and 293 cells expressing or not expressing EBNA1. Unexpectedly, FR/8xRep* only replicated in cells expressing EBNA1 (Fig. 3; see Fig. S2 in the supplemental material; data not shown). In the case of BJAB cells, 40 and 60% of the total FR/8xRep* DNA at days 4 and 6 was resistant to digestion with DpnI in cells expressing EBNA1, whereas the level of the DpnI-resistant DNA was too low to be detectable in the absence of EBNA1 (Fig. 3); in the case of 293 cells, 10 and 50% of the total FR/8xRep* DNA was resistant to digestion with DpnI at 2 and 4 days after transfection in cells expressing EBNA1 (see Fig. S2 in the supplemental material). EBNA1 might be required for the detection of replicated FR/8xRep* plasmids because it is needed to maintain all extrachromosomal replicons derived from EBV by binding to FR. These results therefore did not prove a role for EBNA1 in the synthesis of FR/8xRep*, even though the short duration of these experiments makes it likely that EBNA1 contributes to their synthesis, as well as to their maintenance. This latter likelihood was strengthened by findings in 293 cells in which a vector containing only 8xRep* was shown to replicate at least 3- to 10-fold above background at 2 and 4 days posttransfection only in the presence of EBNA1 (Fig. S2 in the supplemental material).
FIG. 3.
Replication of FR/8xRep* plasmids are dependent on EBNA1. BJAB and BJAB/EBNA1 cells were transfected with FR/8xRep* plasmids as indicated. At 4 and 6 days posttransfection, plasmids were isolated from 1.5 × 107 cells. For Southern blotting, 90% of each sample was digested with DpnI and MluI, and 10% was digested with MluI only as indicated at the top. To monitor the recovery of plasmids and the completeness of DpnI digestion, the pPUR backbone plasmid was used as the “spiked plasmid” DNA. The percentages of replicated DNA refer to the ratio of the level of DpnI-resistant plasmid over the level of MluI-linearized plasmid. The numbers were normalized against the level of the spiked DNA in each sample and are indicated below the blot.
Rep* contains two EBNA1-binding sites.
The apparent dependence of FR/8xRep* and 8xRep* on EBNA1 for their synthesis led us to determine whether EBNA1 could bind Rep*. An EMSA demonstrated that two EBNA1-binding sites are present within Rep* (Fig. 4A). The specificity of EBNA1 binding was ensured by addition of excess poly(dI-dC) DNA to each reaction. In addition, a 14-kbp EcoRI-XbaI restriction fragment near OriLytR that is frequently used to initiate DNA synthesis in the Raji strain of EBV was screened by EMSA for site-specific EBNA1-binding (40). Using Rep* as a positive control and the same derivative of EBNA1 as was used to identify the EBNA1-binding sites of Rep*, 30 overlapping 600-bp fragments from this 14-kbp region were screened, and no EBNA1-binding sites were found (Chen-Yu Wang, unpublished data). The concentration of the derivative of EBNA1 required to bind 50% of the DS and Rep* probes with two dimers of protein was determined and used to calculate the relative affinity of EBNA1 for DS 1+2 and Rep* 1+2. The apparent affinity of EBNA1 for DS 1+2 was ∼1.6-fold higher than for Rep* 1+2.
FIG. 4.
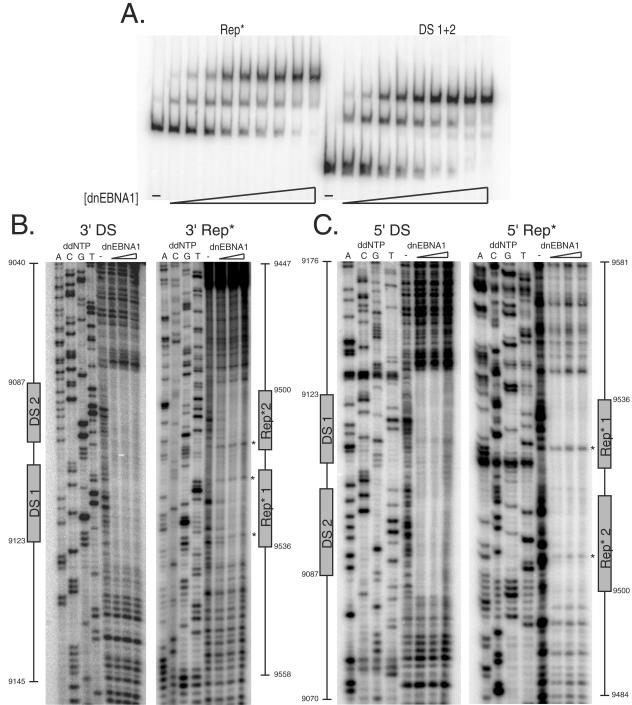
Rep* contains two EBNA1-binding sites with 21-bp center-to-center spacing. (A) A total of 10 fmol of end-labeled probe was incubated either in the absence or presence of increasing concentrations of dnEBNA1/Softag1 in a total volume of 20 μl of 1× EMSA buffer for 10 min at room temperature. Samples were electrophoresed through a 4% polyacrylamide gel at 300 V at 4°C for 1.5 to 2 h. (B and C) Two 156-bp double-stranded DNA probes were amplified by PCR to position one pair of EBNA1-binding sites from DS or Rep* in the center of the probe. Probes were end labeled and purified, and one end was cleaved to retain an end label on one strand. Probes were then purified and resuspended in distilled H2O. A total of 50 fmol of probe was incubated at room temperature for 10 min in 50 μl of 1× EMSA buffer with either 0, 80, 160, or 320 nM dnEBNA1/Softag1 prior to a 60-s digestion with DNase I. Dideoxynucleotide (ddNTP) sequencing reactions were run for each strand footprinted using the same primers as used for PCR amplification of the probes. Nucleotide and EBNA1-binding site positions are indicated graphically adjacent to the gel images.
The two EBNA1 binding sites of Rep* are spaced 21 bp center to center.
In order to identify the sequences bound by EBNA1 within Rep* and determine their relative positioning, we used DNase I footprinting to reveal which phosphodiester bonds were protected by EBNA1 from cleavage. The derivative of EBNA1 protected the DNA of DS containing the known EBNA1-binding sites and several upstream and downstream bases flanking them (Fig. 4B and C). Similarly, the derivative protected two sites within Rep* that strikingly were positioned 21 bp apart center to center just as in DS (Fig. 4B and C). Five bases were found to be unprotected from DNase I cleavage within the region protected by EBNA1 on Rep*, whereas no similar sites were detected in DS (denoted by asterisks in Fig. 4B and C and 5A). The identity of both sites was also confirmed by competitive EMSA, with the apparent affinity of the EBNA1-binding sites being as follows: DS 1 > Rep* 1 > DS 2 ≈ Rep* 2 (data not shown). These two EBNA1-binding sites of Rep* were found to have some sequence similarity to other natural EBNA1-binding sites (compared in Fig. 5A) but were sufficiently different to preclude their identification in silico.
Both EBNA1-binding sites within Rep* with 21-bp center-to-center spacing are necessary and sufficient for its origin activity.
The binding sites for EBNA1 within Rep* map to its center. Examination of the sequence surrounding those sites revealed an AT-rich element which might be important along with the EBNA1-binding sites for Rep*'s function. Mutations in this region as depicted in Fig. 5A were constructed to test if one or both EBNA-1 binding sites were required, if the AT-rich element was required, and if the DNA surrounding these sites was required for Rep*'s function. These five mutations were multimerized to eight copies, cloned into a vector containing FR, and tested in BJAB/EBNA1 cells for their support of replication (Fig. 5B). When the EBNA1-binding site 1 is inactivated by 6-nt transversions, eight copies of site 2 failed to support plasmid replication; similarly, eight copies of the fusion of only site 1 with lambda DNA failed to support plasmid replication. These results indicated that both copies of EBNA1-binding sites within Rep* are necessary to support EBNA1-mediated replication. Furthermore, the AT-rich element localized 24 bp downstream of site 1 could be deleted without affecting the replication activity of 8xRep*. Lastly, the sequences in Rep* other than the EBNA1-binding sites 1+2 could be replaced with lambda DNA. This last result indicates that the paired EBNA1-binding sites are the only cis-acting elements in Rep* necessary for its replication activity.
Because the two EBNA1-binding sites in Rep* are spaced by 21 bp just as in DS, we determined whether this spacing was also critical for its origin activity as it is for DS (8). The spacing between the two EBNA1-binding sites in Rep* was changed to either 20 or 22 bp. Plasmids containing FR and eight copies of each spacing mutant of Rep* failed to support replication in a short-term replication assay (Fig. 5C).
The human ORC is recruited to 8xRep*.
DNA synthesis in eukaryotes relies on ORC as a key regulator for once-per-cell-cycle origin usage. ORC is recruited to the lamin B2 chromosomal origin (50) and to the DS extrachromosomal origin as demonstrated by ChIP (17, 53, 54). We used ChIP to determine whether ORC is recruited to 8xRep* in BJAB/EBNA1 cells. Chromatin immunoprecipitated by antibodies to E2F4, EBNA1, ORC2, or ORC3 was amplified by PCR with primer sets specific for the dihydrofolate reductase (DHFR) promoter, a region adjacent to FR, a region partially encompassing Rep*, and a region distal to FR and Rep*. PCR products from each primer set were normalized to a serial dilution of input DNA in order to compare signals across primer sets. E2F4 is known to bind the DHFR promoter (61), and the selection of the DHFR promoter region by the E2F4 antibody served as a positive control (data not shown). DNA containing either FR or Rep* was enriched by precipitating EBNA1 as expected due to the presence of EBNA1-binding sites. Rep*, but not FR nor the region distal to FR and Rep*, was enriched by precipitating E2F4, ORC2, and ORC3 (Fig. 6A).
EBNA1 bends Rep* 1+2, the crystallized 16-bp palindromic binding site, and the most common EBNA1-binding site of FR.
EBNA1 can bend DS (8). We used a circular permutation assay (62) to determine whether EBNA1 also bends Rep*. Oligonucleotides containing a pair of EBNA1-binding sites from DS (DS 1+2), Rep* (Rep* 1+2), the 16-bp palindromic binding site previously cocrystallized with a derivative of EBNA1 (2xPal) (12), or the most common EBNA1-binding site of FR (2xFRbs) were inserted between two head-to-tail copies of a multiple cloning site in the pBend2 vector (30). Digestion of these constructs with different restriction enzymes produced probes for EMSA of equal length where the binding sites were positioned at different locations in the probe. The degree of bending can be determined by comparing the ratio of mobility of the shifted probe to the free probe when the binding sites are in the center of the probe (uM) to when they are positioned at either end (uE) using the following equation: uM/uE = cos(a/2), where a is the degree of bend produced (58). Figure 6B shows a gel image using probes from Rep* that is representative of the other probes tested. The degree of bending for both DS 1+2 and Rep* 1+2 was approximately 50o when placed in the context of pBend2. EBNA1 bent Rep* 1+2 in the context of its flanking viral DNA approximately 75° as determined by three independent experiments (data not shown), as was previously found for DS (8). The degree of bending for both 2xPal and 2xFRbs when the two binding sites were in phase was similar to that observed for DS 1+2 and Rep* 1+2 (compared in Fig. 6C). We have also tested a full-length derivative of EBNA1 with a shortened Gly-Ala domain (“wtEBNA1”) expressed from plasmid p1553 described previously (47) for its ability to bend Rep* 1+2 by the same assay. We observed that this full-length derivative of EBNA1 bent DNA to the same extent, as did the DNA-binding and dimerization domain, indicating that the N-terminal half of EBNA1 does not contribute significantly to DNA bending (compared in Fig. 6C).
DISCUSSION
8xRep* is an efficient, licensed origin of extrachromosomal replication.
Rep* was identified serendipitously in an extensive genetic screen of sequences flanking DS for cis elements that mediate EBNA1-dependent transcription (33). FR/Rep* supported plasmid synthesis ∼10% as well as FR/DS (oriP) plasmids at 96 h after transfection. From this measurement, it was estimated that Rep* supports extrachromosomal replication ∼45% as efficiently as DS per cell division (33). Multimerization of Rep* eight times coupled with FR in cis and EBNA1 in trans generated a plasmid that is synthesized, maintained, and established as efficiently as is an oriP plasmid (see Table S1 and Fig. S1 in the supplemental material; see Fig. 1). In addition, a density-labeling experiment demonstrated that a FR/8xRep* plasmid replicates once per cell cycle, indicating that the origin function of Rep* is licensed (Fig. 2A). The neutral/neutral two-dimensional gel analyses of Rep* indicate that Rep* functions as a replicator and acts physically as an origin of bidirectional replication (OBR) (Fig. 2C to E). The increase of replication activity afforded by the additional copies of Rep* per replicon indicates that each one functions as an independent replication unit. Our previous (33) and current measurements for the replication efficiencies of Rep* multimers compared to DS are: 1xRep*, 45%; 3xRep%, 83%; and 8xRep*, 100%. Thus, the addition of each copy of Rep* provides a multiplicative increase in the efficiency of replication [for example, 3xRep*, 83% ≈ 1 − (1 to 45%)3]. A single copy of Rep* is an inefficient replicator; its multimerization yields an efficient origin. We propose that 8xRep* represents a tractable model for mammalian zones of initiation (35).
The OBR of ARS1 in yeast has been mapped to within 4 or 5 bp away from the ORC binding site by using a replication initiation point assay (11). However, studies of the amplified replicons in metazoans indicate that OBRs in higher eukaryotes can be separate from the binding sites of initiators. For example, in the chorion gene locus of Drosophila, replication bubbles have been detected by two-dimensional gel electrophoresis ∼10 kbp downstream of the replicator, the ACE3 element (25). We interpret our two-dimensional gel analyses to indicate that replication initiates within 150 bp of the EBNA1-binding site, given that 8xRep* is composed of eight independent replication units, each 300 bp in length. This finding means that the OBR and the replicator can be coincident for some origins in metazoans, such as the EBNA1-dependent DS or Rep*, and, perhaps, the chromosomal lamin B2 origin (1).
An essential cis requirement of EBNA1-dependent replicators is a pair of properly spaced EBNA1-binding sites.
Twenty-six EBNA1-binding sites had previously been identified in the EBV genome: 20 in FR, 4 in DS, and 2 in the Q locus (51). Rep* contains a pair of formerly unrecognized binding sites, which differ from an established consensus sequence at six or seven positions in the 16-bp sequence (6). Based upon the EBNA1/DNA cocrystal structure, the critical nucleotides for base-specific hydrogen-bonding at positions ± 7 and 8 are present in the Rep* EBNA1-binding sites (12). An additional hydrogen bond between EBNA1 and the DNA thought to be important occurs between Gly 463 and a guanine at position ± 4. However, several EBNA1-binding sites, including Rep* 1 and 2, have an adenine present at this position in one of their half-sites. Additional hydrogen bonding requirements between EBNA1 and DNA as assessed by the cocrystal structure can be fulfilled with any nucleotide present. However, many of the bases not contacted directly are conserved between binding sites, and may be critical to yield an appropriate overall conformation. Among these now 28 sites, only DS and Rep* support replication. DS contains three telomeric protein-binding sites (19). In addition, two-thirds of the sequence in DS is a palindrome. The Rep* sequence has neither of these features. It is striking that when its two EBNA1-binding sites are embedded in lambda DNA and multimerized eight times, they support replication, as well as do 8xRep* and oriP (Fig. 5B). This observation indicates that neither the telomeric protein-binding sites nor the palindromic sequence in DS is necessary in general for extrachromosomal origins of DNA synthesis. The telomeric protein binding sites do enhance the replication efficiency of a pair of EBNA1-binding sites from DS (36, 64), presumably by the cooperative binding of TRF2 and EBNA1 (19). Therefore, the need to polymerize Rep* to equalize its replication efficiency with DS may reflect the need to increase the apparent affinity of EBNA1 for a replicon by the addition of more of its binding sites.
DNase I footprinting, competitive EMSA, and sequence analysis of Rep* have shown that its two EBNA1-binding sites are spaced by 21 bp center to center, just as are the pairs of sites in DS (Fig. 4B and C and data not shown). It has been demonstrated that deletion or insertion of only 1 bp between the paired EBNA1-binding sites abolishes the replication activity of DS (8). Our findings that EBNA1-binding sites in Rep* are also spaced by two full turns of B-form DNA and that this 21-bp spacing is essential for its origin activity (Fig. 5C) demonstrate the generality of an EBNA1-dependent replicator requiring a precise arrangement of the EBNA1-recognition elements. Rep* is not identical to DS, however. Rep* has five bases within its binding sites that are unprotected from DNase I cleavage which are not found in DS 1+2 (Fig. 4B and C, denoted in Fig. 5A).
A single-molecule analysis of the entire EBV genome observed that initiation of DNA synthesis can occur at many locations throughout the genome in addition to the canonical DS origin (49). Our identification of a previously unrecognized pair of EBNA1-binding sites that serves as an origin of DNA synthesis raises the question of whether additional pairs of binding sites could be scattered throughout the EBV genome. A 14-kb region that is frequently used to initiate DNA synthesis in the Raji strain of EBV, termed Raji ori, was screened by EMSA for site-specific EBNA1-binding with 30 overlapping 600-bp fragments, and no such sites were found (Chen-Yu Wang, unpublished data). Therefore, it is likely that, in addition to EBNA1-dependent origins such as DS and Rep*, EBNA1-independent origins exist within the EBV genome.
ORC recruitment and EBNA1-induced DNA bending are features shared in trans by EBNA1-dependent replicators and are likely essential.
ORC is an integral participant in all known licensed DNA replication. In lower eukaryotes, ORC appears to bind DNA directly (10, 18, 38). In higher eukaryotes, how ORC associates with chromosomal origins of DNA synthesis is not known. However, ORC has also been shown to be recruited to the viral plasmid origin DS and, moreover, that the dynamics of ORC localization at DS throughout the cell cycle matches that of the cell (53). In fact, one pair of EBNA1-binding sites from DS has been shown to be sufficient to recruit ORC (29). We have also shown that members of ORC associate with 8xRep* by ChIP in BJAB cells expressing EBNA1 (Fig. 6A). Antibodies to ORC2 and ORC3 selectively immunoprecipitated DNA fragments containing Rep*, but not those containing a region distal to Rep* and FR. Licensed DNA synthesis from 8xRep*, like that of DS and chromosomal origins, relies on the recruitment of ORC.
Rep* also has an E2F-binding site that partially overlaps Rep* site 1 and the bases internal to Rep* 1+2, whereas oriP is flanked by two E2F-binding sites (43). Antibodies to E2F4 did selectively immunoprecipitate chromatin containing Rep*, indicating that this E2F site is occupied by E2F4 despite an apparent steric hindrance by DNA-bound EBNA1 (Fig. 6A). This finding probably means that not all of the pairs of EBNA1-binding sites of 8xRep* are occupied by EBNA1 and that multimerization of Rep* may allow greater overall EBNA1 occupancy and therefore origin usage. However, neither E2F site is required for the function of oriP (63), while any specific role for the E2F site in Rep* is unknown.
A second feature that is shared between Rep* and DS in trans is their being bent by EBNA1. Bashaw and Yates have demonstrated by using a gel shift assay that one dimer of EBNA1 can bend its recognition site by approximately 45° (8). We have confirmed this finding and have shown that EBNA1 bends all binding sites tested, including the binding sites of EBNA1-dependent origins (Rep* and DS) and the plasmid maintenance element (FR) (Fig. 6B and C). Moreover, wild-type, intact EBNA1 bends DNA similarly to the DNA-binding and dimerization domain alone, indicating that the N-terminal portion of the protein does not contribute significantly to DNA bending (Fig. 6C). We propose that the structural distortion of the DNA molecules bound by EBNA1 is created by a simple bend at both binding sites which are additive. This proposal is probable because the DNA molecules assayed were linear and relatively short (176 bp) and would not form supercoiled structures. Moreover, the EBNA1/DNA cocrystal structure demonstrated a slight bend of the DNA toward the protein (12). Bending of DNA at origins of DNA synthesis by initiator proteins is common in many viral models, including lambda phage (67), adenovirus (48), simian virus 40 (14), and papillomavirus (23, 26). The binding of S. cerevisiae ORC to the ARS1 origin has also been shown to induce a highly bent structure via biochemical and biophysical methods (16, 37), indicating that the bending of origins of DNA synthesis is also common to lower and higher eukaryotes.
We conclude that the defining characteristics of an EBNA1-dependent replicator are the association of ORC with two EBNA1-binding sites with a bent, 21-bp center-to-center spacing. It is likely that DNA sequences are also involved in the recruitment of ORC to DS or Rep*. Because the C-terminal DNA-binding domain of EBNA1 inhibits the replication function of wild-type EBNA1 (31), the amino terminus likely contributes to the DNA synthesis of DS and Rep*. Intriguingly, it has been shown that the N terminus of EBNA1 can be functionally substituted with some common chromatin constituents, such as HMG-I(Y) and histone H1 (28). HMG-I(Y) and the amino terminus of EBNA1 each have AT-hook activities (7, 56). These activities bind AT-rich DNA and do not provide sequence-specific binding but could contribute to a structure recognized by ORC.
We propose that EBNA1's bending of its properly spaced pairs of sites yields a protein/DNA structure in the context of chromatin that can be recognized by ORC. In Drosophila, the Myb complex directly affects origin usage by site-specific, DNA binding within the ACE3 chorion loci (9). One extension of this proposal is that EBNA1 has evolved from one or more cellular ancestors that, like the Myb complex, bind and structure chromatin to define the origins of DNA synthesis.
Supplementary Material
Acknowledgments
We thank C. Fox and P. Lambert for help with two-dimensional gel electrophoresis, as well as S. Harkins-Perry and S. Bartley for extensive technical assistance on the ChIP. We thank P. Hua for the initial construction of 8xRep*, S. Adhya for the pBend2 vector, J. Hearing for pHEBO1.1 dpm 3+4 vector, S. Duellman for wtEBNA1, A. Schepers for antibodies to ORC2/3 and EBNA1, and R. R. Burgess and N. Thompson for antibodies to the Softag1 epitope. We thank Chen-Yu Wang for providing her findings on surveying Raji ori for EBNA1-binding sites by EMSA. Finally, we thank J. Yates and members of the Sugden laboratory for their critical reviews of the manuscript.
This study was supported by Public Health Service grants CA-22443, P30-CA-014520, and T32-CA-009135. B.S. is an American Cancer Society Research Professor.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abdurashidova, G., M. Deganuto, R. Klima, S. Riva, G. Biamonti, M. Giacca, and A. Falaschi. 2000. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science 287:2023-2026. [DOI] [PubMed] [Google Scholar]
- 2.Adams, A. 1987. Replication of latent Epstein-Barr virus genomes in Raji cells. J. Virol. 61:1743-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aiyar, A., and B. Sugden. 1998. Fusions between Epstein-Barr viral nuclear antigen-1 of Epstein-Barr virus and the large T-antigen of simian virus 40 replicate their cognate origins. J. Biol. Chem. 273:33073-33081. [DOI] [PubMed] [Google Scholar]
- 4.Altman, A. L., and E. Fanning. 2001. The Chinese hamster dihydrofolate reductase replication origin beta is active at multiple ectopic chromosomal locations and requires specific DNA sequence elements for activity. Mol. Cell. Biol. 21:1098-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altman, A. L., and E. Fanning. 2004. Defined sequence modules and an architectural element cooperate to promote initiation at an ectopic mammalian chromosomal replication origin. Mol. Cell. Biol. 24:4138-4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ambinder, R. F., W. A. Shah, D. R. Rawlins, G. S. Hayward, and S. D. Hayward. 1990. Definition of the sequence requirements for binding of the EBNA-1 protein to its palindromic target sites in Epstein-Barr virus DNA. J. Virol. 64:2369-2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aravind, L., and D. Landsman. 1998. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 26:4413-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bashaw, J. M., and J. L. Yates. 2001. Replication from oriP of Epstein-Barr virus requires exact spacing of two bound dimers of EBNA1 which bend DNA. J. Virol. 75:10603-10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beall, E. L., J. R. Manak, S. Zhou, M. Bell, J. S. Lipsick, and M. R. Botchan. 2002. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature 420:833-837. [DOI] [PubMed] [Google Scholar]
- 10.Bell, S. P., and B. Stillman. 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357:128-134. [DOI] [PubMed] [Google Scholar]
- 11.Bielinsky, A. K., and S. A. Gerbi. 1999. Chromosomal ARS1 has a single leading strand start site. Mol. Cell 3:477-486. [DOI] [PubMed] [Google Scholar]
- 12.Bochkarev, A., J. A. Barwell, R. A. Pfuetzner, E. Bochkareva, L. Frappier, and A. M. Edwards. 1996. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein, EBNA1, bound to DNA. Cell 84:791-800. [DOI] [PubMed] [Google Scholar]
- 13.Bogan, J. A., D. A. Natale, and M. L. Depamphilis. 2000. Initiation of eukaryotic DNA replication: conservative or liberal? J. Cell Physiol. 184:139-150. [DOI] [PubMed] [Google Scholar]
- 14.Borowiec, J. A., F. B. Dean, and J. Hurwitz. 1991. Differential induction of structural changes in the simian virus 40 origin of replication by T antigen. J. Virol. 65:1228-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brewer, B. J., and W. L. Fangman. 1987. The localization of replication origins on ARS plasmids in Saccharomyces cerevisiae. Cell 51:463-471. [DOI] [PubMed] [Google Scholar]
- 16.Chastain, P. D., 2nd, J. L. Bowers, D. G. Lee, S. P. Bell, and J. D. Griffith. 2004. Mapping subunit location on the Saccharomyces cerevisiae origin recognition complex free and bound to DNA using a novel nanoscale biopointer. J. Biol. Chem. 279:36354-36362. [DOI] [PubMed] [Google Scholar]
- 17.Chaudhuri, B., H. Xu, I. Todorov, A. Dutta, and J. L. Yates. 2001. Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 98:10085-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chuang, R. Y., L. Chretien, J. Dai, and T. J. Kelly. 2002. Purification and characterization of the Schizosaccharomyces pombe origin recognition complex: interaction with origin DNA and Cdc18 protein. J. Biol. Chem. 277:16920-16927. [DOI] [PubMed] [Google Scholar]
- 19.Deng, Z., L. Lezina, C. J. Chen, S. Shtivelband, W. So, and P. M. Lieberman. 2002. Telomeric proteins regulate episomal maintenance of Epstein-Barr virus origin of plasmid replication. Mol. Cell 9:493-503. [DOI] [PubMed] [Google Scholar]
- 20.Dhar, S. K., K. Yoshida, Y. Machida, P. Khaira, B. Chaudhuri, J. A. Wohlschlegel, M. Leffak, J. Yates, and A. Dutta. 2001. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell 106:287-296. [DOI] [PubMed] [Google Scholar]
- 21.Dhar, V., and C. L. Schildkraut. 1991. Role of EBNA-1 in arresting replication forks at the Epstein-Barr virus oriP family of tandem repeats. Mol. Cell. Biol. 11:6268-6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gahn, T. A., and C. L. Schildkraut. 1989. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell 58:527-535. [DOI] [PubMed] [Google Scholar]
- 23.Gillitzer, E., G. Chen, and A. Stenlund. 2000. Separate domains in E1 and E2 proteins serve architectural and productive roles for cooperative DNA binding. EMBO J. 19:3069-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrison, S., K. Fisenne, and J. Hearing. 1994. Sequence requirements of the Epstein-Barr virus latent origin of DNA replication. J. Virol. 68:1913-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heck, M. M., and A. C. Spradling. 1990. Multiple replication origins are used during Drosophila chorion gene amplification. J. Cell Biol. 110:903-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hegde, R. S., S. R. Grossman, L. A. Laimins, and P. B. Sigler. 1992. Crystal structure at 1.7 Å of the bovine papillomavirus-1 E2 DNA-binding domain bound to its DNA target. Nature 359:505-512. [DOI] [PubMed] [Google Scholar]
- 27.Huberman, J. A. 1997. Mapping replication origins, pause sites, and termini by neutral/alkaline two-dimensional gel electrophoresis. Methods 13:247-257. [DOI] [PubMed] [Google Scholar]
- 28.Hung, S. C., M. S. Kang, and E. Kieff. 2001. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc. Natl. Acad. Sci. USA 98:1865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Julien, M. D., Z. Polonskaya, and J. Hearing. 2004. Protein and sequence requirements for the recruitment of the human origin recognition complex to the latent cycle origin of DNA replication of Epstein-Barr virus oriP. Virology 326:317-328. [DOI] [PubMed] [Google Scholar]
- 30.Kim, J., C. Zwieb, C. Wu, and S. Adhya. 1989. Bending of DNA by gene-regulatory proteins: construction and use of a DNA bending vector. Gene 85:15-23. [DOI] [PubMed] [Google Scholar]
- 31.Kirchmaier, A. L., and B. Sugden. 1997. Dominant-negative inhibitors of EBNA-1 of Epstein-Barr virus. J. Virol. 71:1766-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirchmaier, A. L., and B. Sugden. 1995. Plasmid maintenance of derivatives of oriP of Epstein-Barr virus. J. Virol. 69:1280-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirchmaier, A. L., and B. Sugden. 1998. Rep*: a viral element that can partially replace the origin of plasmid DNA synthesis of Epstein-Barr virus. J. Virol. 72:4657-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knutson, J. C., and D. Yee. 1987. Electroporation: parameters affecting transfer of DNA into mammalian cells. Anal. Biochem. 164:44-52. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi, T., T. Rein, and M. L. DePamphilis. 1998. Identification of primary initiation sites for DNA replication in the hamster dihydrofolate reductase gene initiation zone. Mol. Cell. Biol. 18:3266-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koons, M. D., S. Van Scoy, and J. Hearing. 2001. The replicator of the Epstein-Barr virus latent cycle origin of DNA replication, oriP, is composed of multiple functional elements. J. Virol. 75:10582-10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, D. G., and S. P. Bell. 1997. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol. Cell. Biol. 17:7159-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, J. K., K. Y. Moon, Y. Jiang, and J. Hurwitz. 2001. The Schizosaccharomyces pombe origin recognition complex interacts with multiple AT-rich regions of the replication origin DNA by means of the AT-hook domains of the spOrc4 protein. Proc. Natl. Acad. Sci. USA 98:13589-13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leight, E. R., and B. Sugden. 2001. Establishment of an oriP replicon is dependent upon an infrequent, epigenetic event. Mol. Cell. Biol. 21:4149-4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Little, R. D., and C. L. Schildkraut. 1995. Initiation of latent DNA replication in the Epstein-Barr virus genome can occur at sites other than the genetically defined origin. Mol. Cell. Biol. 15:2893-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, G., M. Malott, and M. Leffak. 2003. Multiple functional elements comprise a Mammalian chromosomal replicator. Mol. Cell. Biol. 23:1832-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lupton, S., and A. J. Levine. 1985. Mapping genetic elements of Epstein-Barr virus that facilitate extrachromosomal persistence of Epstein-Barr virus-derived plasmids in human cells. Mol. Cell. Biol. 5:2533-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maser, R. S., O. K. Mirzoeva, J. Wells, H. Olivares, B. R. Williams, R. A. Zinkel, P. J. Farnham, and J. H. Petrini. 2001. Mre11 complex and DNA replication: linkage to E2F and sites of DNA synthesis. Mol. Cell. Biol. 21:6006-6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Menezes, J., W. Leibold, G. Klein, and G. Clements. 1975. Establishment and characterization of an Epstein-Barr virus (EBC)-negative lymphoblastoid B-cell line (BJA-B) from an exceptional, EBV-genome-negative African Burkitt's lymphoma. Biomedicine 22:276-284. [PubMed] [Google Scholar]
- 45.Mesner, L. D., X. Li, P. A. Dijkwel, and J. L. Hamlin. 2003. The dihydrofolate reductase origin of replication does not contain any nonredundant genetic elements required for origin activity. Mol. Cell. Biol. 23:804-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meuth, M., and H. Green. 1974. Induction of a deoxycytidineless state in cultured mammalian cells by bromodeoxyuridine. Cell 2:109-112. [DOI] [PubMed] [Google Scholar]
- 47.Middleton, T., and B. Sugden. 1994. Retention of plasmid DNA in mammalian cells is enhanced by binding of the Epstein-Barr virus replication protein EBNA1. J. Virol. 68:4067-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mysiak, M. E., C. Wyman, P. E. Holthuizen, and P. C. van der Vliet. 2004. NFI and Oct-1 bend the Ad5 origin in the same direction leading to optimal DNA replication. Nucleic Acids Res. 32:6218-6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Norio, P., and C. L. Schildkraut. 2004. Plasticity of DNA replication initiation in Epstein-Barr virus episomes. PLoS Biol. 2:e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paixao, S., I. N. Colaluca, M. Cubells, F. A. Peverali, A. Destro, S. Giadrossi, M. Giacca, A. Falaschi, S. Riva, and G. Biamonti. 2004. Modular structure of the human lamin B2 replicator. Mol. Cell. Biol. 24:2958-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rawlins, D. R., G. Milman, S. D. Hayward, and G. S. Hayward. 1985. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell 42:859-868. [DOI] [PubMed] [Google Scholar]
- 52.Remus, D., E. L. Beall, and M. R. Botchan. 2004. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J. 23:897-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ritzi, M., K. Tillack, J. Gerhardt, E. Ott, S. Humme, E. Kremmer, W. Hammerschmidt, and A. Schepers. 2003. Complex protein-DNA dynamics at the latent origin of DNA replication of Epstein-Barr virus. J. Cell Sci. 116:3971-3984. [DOI] [PubMed] [Google Scholar]
- 54.Schepers, A., M. Ritzi, K. Bousset, E. Kremmer, J. L. Yates, J. Harwood, J. F. Diffley, and W. Hammerschmidt. 2001. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 20:4588-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schvartzman, J. B., M. L. Martinez-Robles, and P. Hernandez. 1993. The migration behavior of DNA replicative intermediates containing an internal bubble analyzed by two-dimensional agarose gel electrophoresis. Nucleic Acids Res. 21:5474-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sears, J., M. Ujihara, S. Wong, C. Ott, J. Middeldorp, and A. Aiyar. 2004. The amino terminus of Epstein-Barr virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J. Virol. 78:11487-11505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sugden, B., and N. Warren. 1988. Plasmid origin of replication of Epstein-Barr virus, oriP, does not limit replication in cis. Mol. Biol. Med. 5:85-94. [PubMed] [Google Scholar]
- 58.Thompson, J. F., and A. Landy. 1988. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 16:9687-9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thompson, N. E., T. M. Arthur, and R. R. Burgess. 2003. Development of an epitope tag for the gentle purification of proteins by immunoaffinity chromatography: application to epitope-tagged green fluorescent protein. Anal. Biochem. 323:171-179. [DOI] [PubMed] [Google Scholar]
- 60.Vashee, S., C. Cvetic, W. Lu, P. Simancek, T. J. Kelly, and J. C. Walter. 2003. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 17:1894-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weinmann, A. S., S. M. Bartley, T. Zhang, M. Q. Zhang, and P. J. Farnham. 2001. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol. 21:6820-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu, H. M., and D. M. Crothers. 1984. The locus of sequence-directed and protein-induced DNA bending. Nature 308:509-513. [DOI] [PubMed] [Google Scholar]
- 63.Yates, J., N. Warren, D. Reisman, and B. Sugden. 1984. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc. Natl. Acad. Sci. USA 81:3806-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yates, J. L., S. M. Camiolo, and J. M. Bashaw. 2000. The minimal replicator of Epstein-Barr virus oriP. J. Virol. 74:4512-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yates, J. L., and N. Guan. 1991. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J. Virol. 65:483-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yates, J. L., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313:812-815. [DOI] [PubMed] [Google Scholar]
- 67.Zahn, K., and F. R. Blattner. 1987. Direct evidence for DNA bending at the lambda replication origin. Science 236:416-422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.