Abstract
Chinese hamster ovary (CHO) cells select specific replication origin sites within the dihydrofolate reductase (DHFR) locus at a discrete point during G1 phase, the origin decision point (ODP). Origin selection is sensitive to transcription but not protein synthesis inhibitors, implicating a pretranslational role for transcription in origin specification. We have constructed a DNA array covering 121 kb surrounding the DHFR locus, to comprehensively investigate replication initiation and transcription in this region. When nuclei isolated within the first 3 h of G1 phase were stimulated to initiate replication in Xenopus egg extracts, replication initiated without any detectable preference for specific sites. At the ODP, initiation became suppressed from within the Msh3, DHFR, and 2BE2121 transcription units. Active transcription was mostly confined to these transcription units, and inhibition of transcription by alpha-amanitin resulted in the initiation of replication within transcription units, indicating that transcription is necessary to limit initiation events to the intergenic region. However, the resumption of DHFR transcription after mitosis took place prior to the ODP and so is not on its own sufficient to suppress initiation of replication. Together, these results demonstrate a remarkable flexibility in sequence selection for initiating replication and implicate transcription as one important component of origin specification at the ODP.
In their replicon model (29), Jacob et al. proposed that replication initiation involves the recognition of a cis-acting DNA sequence or replicator by a trans-acting positive regulatory factor called the initiator. Since then, this model has been validated in bacteria and virus systems, but in eukaryotes a more complex model has been proposed to explain the diversity of replicator structures (22, 66). With the exception of budding yeast, a specific sequence element that defines the replication origin in eukaryotes has not been identified (2, 6, 7, 21, 23). Instead, it appears that a complex combination of primary DNA sequence and epigenetic factors dictates where replication initiates, and a unique combination of these factors may define the position of each initiation site (22). Although replication in most eukaryotic organisms does not initiate at random with respect to DNA sequence, the size and distribution of these sites are highly variable (for a review, see reference 23). In gene-dense regions, replication can initiate at very defined sites (1, 5), while in large intergenic regions multiple initiation sites are found throughout large (5- to 50-kb) initiation zones (11, 13, 28, 39, 45, 50).
A large (50-kb) region downstream of the dihydrofolate reductase (DHFR) gene in Chinese hamster ovary (CHO) cells is the most extensively studied initiation zone, yet the precise distribution of initiation sites within this region has still not been resolved. High-resolution mapping of the locations of small nascent DNA strands within a 12-kb region detected two very specific initiation sites (35), while two-dimensional gel electrophoresis (2D-gel) analysis of replication intermediates (13, 45) identified DNA structures representing replication bubbles within over 20 different fragments throughout the initiation zone. One possibility is that there are multiple discrete sites of initiation that would be revealed by a more comprehensive analysis of small nascent strands or by more recently developed methods of visualizing origins on DNA fibers (5, 50). Regardless of the precise pattern of initiation sites within the zone, the majority of initiation activity appears to be confined to this zone, as no comparably significant initiation has been detected elsewhere throughout approximately 270 kb. Since the initiation pattern at the DHFR locus is similar to several other large intergenic regions in metazoa (11, 20, 28, 39, 50), it is important to understand the mechanism that confines initiation sites to this region.
The act of transcription itself appears to play a role in the specification of origins at the CHO DHFR locus. Of the sites that have been interrogated at this locus, significant initiation activity has been confined to the intergenic region (although some very weak activity has been detected within a downstream, weakly transcribed gene) (12). Deletion of either the promoter (62) or the transcription terminator (31) of the DHFR gene results in a reduction in initiation activity throughout the locus. In the case of a promoter deletion, this is accompanied by the appearance of weak initiation activity within the body of the DHFR gene, resulting in a broadening of the initiation zone to include the silent transcription unit. In the case of the terminator deletion, transcription continues into the intergenic locus, and initiation throughout this extended transcription unit becomes suppressed (44). When integrated at ectopic sites, replication can initiate within either DHFR gene or nongene sequences, unless an active promoter is placed upstream (38). These observations provide strong genetic evidence for the suppression of initiation within active transcription units. Interestingly, almost all eukaryotic replication origins have been localized outside of active transcription units (22, 23, 43).
By incubating nuclei from CHO cells in Xenopus egg extracts, we have established a cell-free system that recapitulates the physiological initiation pattern of DNA replication at the CHO DHFR locus (24, 73). However, physiological origin specification requires nuclei prepared late in G1 phase. With early-G1-phase nuclei, replication appears to initiate at sites distributed throughout the entire locus (70). Hence, at a distinct point during G1 phase, CHO nuclei experience a transition (the origin decision point [ODP]) that selects which of many potential chromosomal sites will function as an origin of replication in the upcoming S phase. Intriguingly, several transcription inhibitors were shown to prevent the selection of origins at the ODP (33). This result is consistent with the hypothesis that initiation of replication is prevented from taking place within active transcription units, and it suggested that the ODP could represent the resumption of transcription within this region after global transcriptional shutdown during mitosis (15, 22, 23). However, the DHFR gene is transcribed from an E2F-regulated promoter that is presumed to be repressed until after the restriction point (9, 18, 49, 68), whereas the ODP takes place prior to the restriction point and is independent of mitogen stimulation (33, 71). Moreover, our previous results interrogated only 17 specific sites throughout the 120 kb surrounding the initiation zone. Hence, we did not have the resolution to conclude that initiation of replication was excluded from transcription units. Surprisingly, despite the amount of effort that has been invested in the CHO DHFR locus, very little of this region has previously been sequenced, and unique probes have only been available for a few scattered regions.
We have now sequenced 121 kb surrounding the DHFR initiation zone and have constructed DNA arrays containing either plasmid or PCR products that, after the removal of repetitive sequences, cover 78.9% of the sequence. We detected initiation activity throughout the intergenic region, with little or no activity within any of the three previously defined transcription units. When pre-ODP nuclei were incubated in Xenopus egg extracts, replication initiated without preference for any site, genic or intergenic; however, with post-ODP nuclei, initiation resembled the pattern seen in vivo. Nuclear run-on analysis with nuclei isolated at different times after mitosis demonstrated that transcription is confined to these previously described genes and that the same concentrations of alpha-amanitin that inhibited transcription of these genes also allowed initiation to take place within transcription units. Surprisingly, transcriptional activity was detected throughout G1 phase with little change at the ODP. These results demonstrate that replication can initiate at a remarkably flexible number of sites within mammalian chromatin, including within sites of ongoing transcription in pre-ODP nuclei. However, at the ODP, initiation of replication is suppressed within transcribed regions. Hence, transcription is necessary but not sufficient for this suppression.
MATERIALS AND METHODS
Sequencing of the DHFR locus.
An initial shotgun sequencing starting with cosmids H2, SC26, and II45 (40, 46) was contracted out to Genemed Synthesis, Inc. (South San Francisco, Calif.). Cosmids were partially digested with AluI and HaeIII and inserted into the EcoRV site of pBluescript SK(+). This provided an initial set of unaligned sequences from these cosmids. Since these cosmids were cloned from CHOC 400 genomic DNA (gDNA), they contained one of the amplicon junctions in this cell line, which was previously described in detail (51). To complete and align these cosmid sequences, we obtained end sequences from (i) subclones of shotgun plasmids that harbored inserts larger than 3 kb, and (ii) existing plasmid subclones whose positions but not sequences were known (earliest-labeled fragment hybridization [ELFH] probes) (37), as well as unpublished plasmids DG-1, DG-3, and DG-5. A published sequence (GenBank accession no. AF02817) from SC26 also aided sequence alignment. Sequences were assembled with software SeqMan (DNAStar). Next, gaps between these three cosmids were filled by sequencing plasmids pNeoS13, pneoS21, and pneoX9 (4, 24, 25) and pB6-1, pB6-7-1, pB13-6-1, and p13-7-1 (8). PCR products from CHOC 400 genomic DNA were cloned to generate pNOT11, pNOT21, pNOT31, pNOT32, pNOT33, and pNOT34, which were then sequenced to fill the remaining gaps. Details of the sequencing strategy will be made available upon request.
Array assembly. (i) Design of the PCR probes.
We essentially followed the approach of Rinn et al. (61). RepeatMasker (http://www.repeatmasker.org/) was used to identify repetitive sequences. A total of 78,810 bp of the 121,368-bp sequenced region were identified as nonrepetitive (i.e., 35% repetitive and 65% nonrepetitive). Initially, 90 PCR primer sets that would amplify fragments of the size of 300 to 700 bp were designed using MacVector (Accelrys, Inc.). The primers were selected such that their annealing temperatures were between 55°C and 65°C and they had an average G+C content of 45%. An additional 60 primers were designed manually, by visual inspection of sequences not chosen by MacVector. The sequences of these PCR primers sets will be provided on request.
(ii) PCRs.
PCRs were performed with 25 ng of CHOC 400 genomic DNA and 0.4 μM each forward and reverse primer. Control reactions were performed with forward or reverse primers alone during the first round of PCR, and only primer sets that were negative under these conditions were used. The reaction mixture contained 0.2 mM each deoxynucleoside triphosphate, 1.5 mM MgCl2, 1× buffer B (Fisher Scientific), and 1 U of Taq in a 25-μl reaction mixture. PCR product sizes were verified on a 2% agarose gel, and then products were gel purified in a 1.5% agarose gel with a Gel CleanUp kit (Eppendorf). These PCR products were then used as a template (5 ng) for a second round of PCR using the same primer sets to generate probes for the array. The second round of PCR amplification enabled the amplification of large quantities of PCR product free of contamination from genomic DNA and nonspecific amplification products. These PCR products were then diluted to 10 ng/μl in 10:0.1 TE (10 mM Tris-HCl [pH 8.0], 0.1 mM EDTA) and were printed onto Hybond-N+ membranes (Amersham).
(iii) Removal of primer-dimers.
To remove primer-dimers that were formed in certain PCRs, PCR products were selectively precipitated from the reaction mixture by the addition of a 0.1 volume of 3 M sodium acetate (pH 5.2) and a 0.7 volume of isopropanol (P. Bertone, personal communication). Tubes were thoroughly mixed and centrifuged at 14,000 rpm at 4°C for 30 min. The pellet was washed twice with 70% ethanol, air dried, and resuspended in 10:0.1 TE.
(iv) Printing of arrays.
PCR products were diluted to 10 ng/μl in 10:0.1 TE in a total volume of 100 μl. A total of 20 μl of bromophenol blue in 10:0.1 TE (1 mg/ml) was added to these tubes, after which the DNA was denatured at 96°C for 10 min and quick chilled on ice. The DNA was spotted on Hybond N+ membranes using a DNA replicator (VP Scientific), per the manufacturer's instructions. Probes were cross-linked to the membranes with a UV cross-linker (Stratalinker; Stratagene), after which the arrays were stored in sealed plastic bags for further use. A total of 10 ng (each) of a segment of lambda DNA was spotted at various locations on the array as a negative hybridization control. In addition, 10 ng (each) of the 17 probes used for origin mapping in prior experiments was also included in each array (37); results with these probes were reproducibly consistent with our previously reported results.
(v) Identification of probes that contain repetitive sequences.
Although the PCR probes were carefully designed to avoid the repetitive sequences that were identified by RepeatMasker, the probes were further scanned empirically for repetitive sequences, as follows. Arrays were hybridized to labeled genomic DNA from CHOC 400 cells, and the intensities were represented as counts per minute per base pair. These intensities were normalized to the weakest hybridizing probe. Probes whose normalized intensities (in counts per minute per base pair) were 20 times greater than the intensities of the weakest hybridizing probe (61) were categorized as containing repetitive sequences and not used for further analysis.
(vi) Plasmid array.
A total of 100 ng of each plasmid obtained from shot gun sequencing was spotted on a Hybond N+ membrane as described above. Plasmids containing highly repetitive sequences were identified by being screened with labeled CHOC 400 genomic DNA, as described above.
ELFH assay.
CHOC 400 cells were cultured and synchronized in G0/G1 by isoleucine starvation (71), after which the cells were blocked at the G1/S boundary by the addition of complete medium containing 10-μg/ml aphidicolin or 400 μM mimosine. After 14 h, the cells were released to S phase by replacing the medium with complete warm growth medium for 5 min (aphidicolin) or 20 min (mimosine), and cells were collected by trypsinization. Nuclei were prepared by digitonin permeabilization (73). Early replication intermediates were labeled as previously described (24, 73), except that 5 million nuclei in 35 μl of replication cocktail were labeled at 34°C for 1.5 min. The reaction was stopped by the addition of 470 μl of lysis buffer (50 mM Tris [pH 7.8], 10 mM EDTA, 0.4 M NaCl, 0.6% sodium dodecyl sulfate [SDS]) containing 200-μg/ml proteinase K. DNA was isolated and sonicated as previously described (24, 73). Typically, 2 × 107 cells resulted in 30 million cpm of labeled DNA. Nascent DNA strands at replication forks in asynchronously growing cells were similarly labeled. Radiolabeled DNA was heat denatured and hybridized to four independently printed PCR arrays (and/or one to three plasmid arrays) in hybridization bottles as previously described (73). To normalize for any variation in probe intensity due to the size of the probes, hybridization intensity, or AT content, total genomic DNA was random labeled according to the manufacturer's protocol (Invitrogen) and similarly hybridized. Hybridization was allowed to proceed for 14 h, after which the membranes were washed twice with 2× SSC (1× SSC is 150 mM NaCl and 15 mM sodium citrate [pH 7.0]) and 0.2% SDS for 20 min each wash and with 0.2× SSC-0.2% SDS for 15 minutes each. The signal intensities were measured by exposure to Molecular Dynamics phosphorimager screens. The sizes of nascent strands were determined as previously described (72), except that 0.7% gels were run overnight at 60 V.
Analysis of DNA replication in Xenopus egg extracts.
Xenopus egg extracts were prepared and handled as previously described (73), except that 40 nM geminin was added to all reactions prior to the addition of nuclei to extract, to inhibit in vitro assembly of prereplication complexes (pre-RCs) (16, 52). Analysis of replication was essentially as previously described (14, 73). Briefly, cells were synchronized at various time points during G1 phase by mitotic selection and permeabilized with digitonin. For experiments that evaluated the positions of aphidicolin-arrested forks, nuclei were incubated in Xenopus egg extract at 21°C for 90 min at 10,000 nuclei per μl of extract in the presence of 100 μg of aphidicolin/ml. After incubation in Xenopus extract, reactions were stopped by the addition of ice-cold hypotonic buffer (20 mM HEPES [pH 7.5], 5 mM KCl, 1.5 mM MgCl2). Nuclei were pelleted at full speed for 16 s in a Sorvall MC-12V, resuspended in hypotonic buffer, and incubated on ice for 10 min to remove aphidicolin and nucleotide pools. Nuclei were pelleted and resuspended in a labeling mixture that contained 50 μCi of [32P]dATP, and the labeling reaction was performed as previously described for 10 min at 12°C (73). For experiments that labeled nascent DNA directly, nuclei were incubated in Xenopus egg extract supplemented with 2.0-mCi/ml [32P]dATP at 21°C for 40 min at 25,000 nuclei per μl of extract. Ice-cold hypotonic buffer was added to stop the reaction, and nuclei were pelleted and resuspended in lysis buffer with proteinase K as described above. DNA isolation and hybridization were as described above.
Nuclear run-on hybridizations.
We followed the protocol of Hirayoshi and Lis (26), with modifications described by Chasin and colleagues (65). Subconfluent dishes of cells were chilled on ice, and cells were washed twice with ice-cold 1× SSC. Cells were removed from the culture plate using a cell scraper in 10 ml of 1× SSC per dish and collected by centrifugation (500 × g; 5 min). Cell pellets were resuspended in 1 ml of NP-40 lysis buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.1 mM EDTA, 0.5% [vol/vol] NP-40) per 107cells and incubated for 5 min on ice. Nuclei were then washed twice with NP-40 lysis buffer, resuspended in 100 μl freezing buffer (50 mM Tris-HCl [pH 8.3], 40% [vol/vol] glycerol, 5 mM MgCl2, 0.1 mM EDTA) per 107 cells, snap frozen in liquid nitrogen, and stored at −80°C in 100-μl aliquots. To perform run-on reactions, aliquots of nuclei were thawed, mixed with 100 μl of 2× reaction buffer (10 mM Tris-HCl [pH 8.0]; 5 mM MgCl2; 300 mM KCl; 0.5 mM each ATP, CTP; and GTP; and 150 μCi [α-P32]UTP [800 Ci/mmol]), and incubated at 30°C for 15 min. A total of 60 U of RNase-free DNaseI (Promega) were added, and the reaction mixture was incubated for an additional 30 min at 37°C. An equal volume of stop buffer (20 mM Tris-HCl [pH 7.4], 2% SDS, 10 mM EDTA, 200-μg/ml proteinase K) was added to the reaction mixture and further incubated at 42°C for 30 min. RNA was then extracted twice with water-saturated phenol:chloroform:isoamyl alchohol (25:24:1). NH4 acetate was added to 2.5 M, and RNA was precipitated with 3.5 volumes of ethanol. RNA pellets were resuspended in 100-μl diethyl pyrocarbonate-treated water, reprecipitated in ethanol, and resuspended again in 100 μl diethyl pyrocarbonate-treated water. Ten units of DNaseI and 10 μl of 10× reaction buffer (400 mM Tris-HCl [pH 8.0], 100 mM MgSO4, 10 mM CaCl2; Promega) was added and incubated at 37°C for 30 min to completely remove all contaminating DNA. Reactions were extracted once with phenol to remove DNaseI, and DNA was precipitated and resuspended in 100 μl of water. DNA was denatured at 65°C for 5 min, added to hybridization buffer, and hybridized as described above.
RESULTS
Construction of a DHFR array.
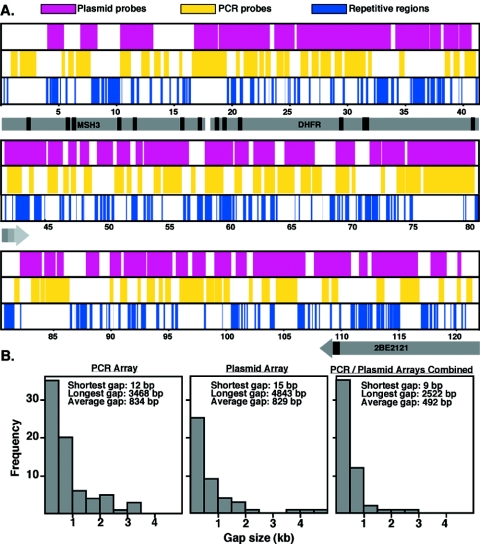
A DNA array was constructed containing all the nonrepetitive sequences across 121 kb surrounding the CHO DHFR gene and parts of the Msh3 and 2BE2121 genes (Fig. 1). Since sequence information was available only for the DHFR cDNA and a small segment of the intergenic region, the complete sequence was assembled by shotgun sequencing of several available cosmids and plasmids (DDBJ/EMBL/GenBank accession number BR000241). Repetitive sequences at this locus were identified using RepeatMasker, and PCR primers were designed to amplify 156 fragments of nonrepetitive DNA ranging in size from 300 to 700 bp. The PCR products were amplified from CHOC 400 genomic DNA and spotted on nylon membranes, along with positive (previously analyzed segments of DNA) (37) and negative (lambda DNA) control fragments. PCR probes containing repetitive DNA sequences not eliminated in silico were identified by hybridizing radiolabeled genomic DNA to the array. A clear demarcation was identified between the majority of probes whose ratio of counts per minute per base pair varied within 10 fold of the known single-copy probes and a few that hybridized >20 times more strongly. After elimination of these probes, 134 nonrepetitive probes (mean size = 440 kb) encompassed 75% of the unique sequences in the 121-kb locus (Fig. 1A). The majority of gaps in sequence were <1 kb in length (Fig. 1B); however, there were 19 gaps that were >1 kb and 3 gaps that were >3 kb. Some of the sequences within these gaps were interrogated using an array consisting of plasmid probes (Fig. 1B; discussed below). Between both arrays, 78.9% of the entire sequence was represented, with only five gaps larger than 1 kb and none larger than 3 kb.
FIG. 1.
Distribution of PCR and plasmid probes used to assemble arrays. (A) Map positions of the sequences determined in silico containing repeats using RepeatMasker and the plasmid and PCR probes remaining after eliminating probes containing repeats, as described in the text. Gray arrows represent the transcription units of Msh3, DHFR, and 2BE2121 genes. The arrowhead indicates the transcription termination site for the gene (the Msh3 termination site is not within the sequence); multiple termination sites for DHFR (63) are indicated by different shades of gray. Promoter elements for DHFR and Msh3 are located in a G+C-rich region near 18 kb. Positions of exons are indicated as black boxes. (B) Histograms illustrate the number and sizes of gaps with arrays constructed from PCR products and plasmids.
A broad replication initiation zone at the DHFR locus.
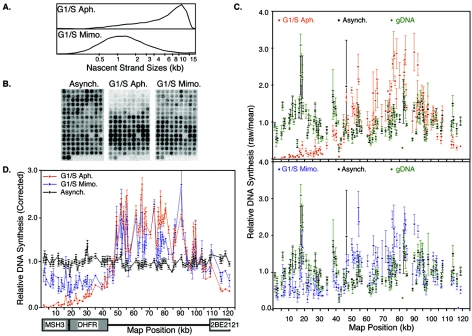
To evaluate the pattern of initiation at the DHFR intergenic region, we performed the ELFH assay. In this assay (24, 73), cells are first synchronized at the onset of S phase by culturing G1- or G0-synchronized cells into medium containing aphidicolin or mimosine. Aphidicolin allows replication to initiate and short nascent strands to accumulate near their sites of initiation but selectively inhibits the processive elongation of replication forks (24, 73). Mimosine is a Fe2+ and Zn2+ chelator whose mechanism of action remains to be elucidated but is an effective agent for synchronizing cells at or very near the initiation of replication (25, 36, 48, 53, 55). After removal of these inhibitors, cells were permeabilized with digitonin and briefly labeled in an in vitro replication cocktail containing high-specific-activity [32P]dATP to label the nascent strands emanating from the arrested replication forks. When this DNA was analyzed on alkaline agarose gels, labeled nascent DNA from aphidicolin-arrested cells revealed a broad distribution of sizes from 1 to 20 kb, while those from mimosine-arrested cells were primarily in the 500- to 2,500-bp range (Fig. 2A), consistent with previous results (13, 72). Labeled nascent strands isolated from these nuclei were sheared and hybridized simultaneously to four independently printed arrays (Fig. 2B). Nascent strands labeled in an identical manner but from asynchronously growing cells, which contain replication forks distributed randomly throughout the genome (24, 73), served as a control, and gDNA radioactively labeled by random prime labeling served as a hybridization reference, described below. The mean hybridization from four independently printed arrays controls for potential printing errors and constitutes one data set or one experiment.
FIG. 2.
Hybridization pattern of the earliest-labeled nascent strands to an array of PCR probes. (A) Nascent DNA strands in nuclei prepared from cells that were synchronized at the G1/S boundary with either aphidicolin (G1/S Aph.) or mimosine (G1/S Mimo.) were labeled in an in vitro replication cocktail and then separated by size on alkaline agarose gels. Gels were scanned with a phosphorimager, and each lane was traced with Image Quant software. Positions of molecular weights, determined from markers included in the same gel, are shown on the x axis. (B) Nascent strands prepared as in panel A, as well as those prepared similarly from asynchronously growing cells (Asynch.), were hybridized to PCR arrays. Shown are examples of array images after being scanned with a phosphorimager. (C) In each experiment, nascent strands were hybridized to four independently printed arrays to control for printing errors. Raw phosphorimager values for each probe were normalized to the mean of all probes on each array (raw/mean). Shown are the mean values from the four arrays hybridized in a single experiment. Error bars, representing the standard error of the mean, illustrate the reproducibility between independently printed batches of arrays. (Top) Comparison of aphidicolin-synchronized (red) to asynchronous (black) samples; (bottom) mimosine-synchronized (blue) compared to asynchronous (black) samples. The values for labeled gDNA (green), hybridized to the same set of arrays, provide a reference for variability between probes due to hybridization efficiencies. (D) Values for each probe hybridized with nascent strands were divided by the corresponding values with gDNA to correct for differences in probe hybridization efficiency. Shown are the means of two independent experiments synchronizing with mimosine (blue) and three independent experiments synchronizing with aphidicolin (red). Error bars indicate the standard deviation between experiments for each probe. A schematic diagram of the transcription units is provided below the x axis. A higher dynamic range of signals was consistently obtained with nascent strands from aphidicolin-arrested cells (red) compared to mimosine-arrested cells (blue). The reasons for this difference are not clear (similar total counts per minute were labeled) but could indicate that mimosine synchronizes cells at an earlier point in S phase, when fewer DHFR origins have fired relative to the population of earliest replicating origins, resulting in a lower signal-to-noise ratio. Consistent with this interpretation, the total counts per minute across all DHFR probes on arrays hybridized with mimosine-arrested forks were consistently two-thirds of those with aphidicolin-arrested forks, despite the similar total counts per minute labeled.
Figure 2C shows an example of the raw data from a single experiment. A high degree of reproducibility was observed between signals from independently printed arrays, and the dynamic range of signals was much greater with replication intermediates labeled in nuclei from either aphidicolin- or mimosine-synchronized cells, compared to asynchronous cells or gDNA. Since all sequences are represented at an equimolar ratio within gDNA, probe-to-probe variability obtained with this reference target resulted from differences in probe size, AT richness, hybridization, or labeling efficiency. To correct for these variables in the experimental samples, the values for each probe from hybridizations with replication intermediates were divided by the corresponding values obtained with gDNA. Figure 2D shows the average values from two independent experiments with mimosine-synchronized cells and three independent experiments with aphidicolin-synchronized cells. Results revealed that replication intermediates from asynchronously growing cells were distributed evenly across the entire locus, consistent with a random positioning of replication forks at the time of radiolabeling and the absence of significant replication fork pausing sites. In contrast, sites labeled in both aphidicolin- and mimosine-synchronized cells were distributed throughout the region between the transcription units and were clearly underrepresented within transcription units. However, there did appear to be slightly higher levels of initiation activity within the body of the 2BE2121 gene than within the Msh3 and DHFR genes, consistent with results from 2D-gel analysis that have detected weak initiation activity within the 2BE2121 gene (12).
Several sequence analyses of the 121-kb region were performed to determine if there were any unique features of the intergenic sequence that could distinguish it from the transcription units. Previous analysis of an initiation zone in Drosophila melanogaster (28) revealed peaks of AT richness coinciding with peaks of initiation activity. Sliding 200-bp windows in 10-bp steps were analyzed for AT content, revealing two sharp peaks of G+C-rich DNA, one in the bidirectional DHFR-Msh3 promoter region (17) and one at nucleotide positions 82.7 to 85.2 kb. Otherwise, uniform variation in AT content was observed throughout the remainder of the sequence. We also did not detect any matches to a previously reported consensus sequence for mammalian replication origins (57), although we detected occurrences of ≥16/20 matches throughout the entire 121-kb region. We also did not identify any DNA sequence characteristics that were unique to the intergenic region.
As shown in Fig. 1, the PCR-amplified probes represented in our array naturally contained gaps that were difficult to fill, due to the presence of repetitive sequences. However, one advantage of using CHOC 400 to analyze origin distribution is that the approximately 1,000 copies of the DHFR locus overshadowed the more moderately repetitive sequences, allowing us to interrogate a greater percentage of the DHFR locus. Hence, we constructed an array consisting of overlapping plasmid clones obtained as an outcome of shotgun sequencing. To eliminate probes that contained highly repetitive elements (>1,000 copies per cell), this panel was hybridized to labeled CHOC 400 genomic DNA, and probes that deviated >20 fold from the average counts per minute per base pair were removed from the array, leaving a panel of 122 overlapping plasmid probes with an average insert size of 970 bp, spanning 69.2% of the 121-kb sequenced region. When one considers the coverage of both the PCR and plasmid arrays, 78.9% (95,780 bp) of the total sequenced region (121,368 bp) was covered, leaving very few gaps (Fig. 1B).
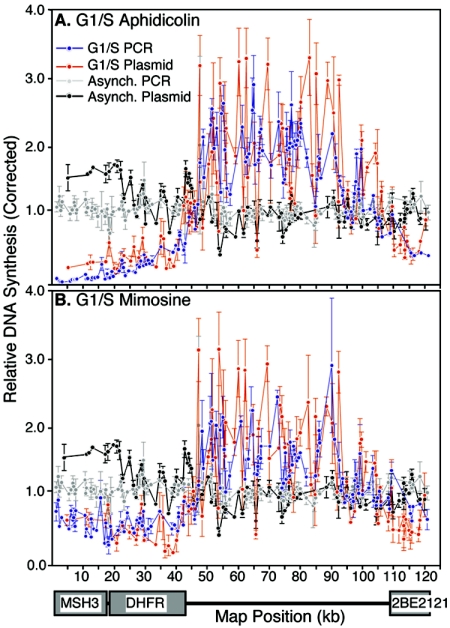
The experiments shown in Fig. 2 were repeated, hybridizing labeled nascent DNA from aphidicolin- and mimosine-arrested cells to this panel of plasmid probes (Fig. 3). These results also revealed a nearly uniform distribution of earliest-labeled nascent strands throughout the intergenic region and excluded from the known transcription units. Overlaying the plasmid and PCR arrays revealed a similar pattern of initiation from both arrays, supporting the conclusion that initiation is largely, if not exclusively, confined to the intergenic region.
FIG. 3.
Hybridization pattern of the earliest labeled nascent strands to an array of plasmid probes. Nascent strands from cells synchronized at the G1/S border with aphidicolin (A) or mimosine (B) were labeled as in Fig. 2 and hybridized to a plasmid array (red). Values for each probe were corrected to the corresponding values from arrays hybridized with randomly labeled genomic DNA, as in Fig. 2. Shown are the means from two independent experiments and the standard deviation. To illustrate the coverage obtained by using both PCR and plasmid arrays, the results of the PCR array from Fig. 2C (blue) were plotted alongside those from the plasmid arrays (red).
Potential initiation sites are ubiquitous.
Using an array of 17 site-specific probes (5 clustered near ori-β, 4 at other sites within the intergenic region, 2 in the 2BE2121 gene, and 6 in the DHFR gene), we previously showed that initiation at the DHFR locus becomes confined to 9 intergenic probes at the ODP. This was demonstrated by introducing nuclei isolated from CHOC 400 cells at different times during G1 phase into Xenopus egg extracts that had been rendered deficient in their ability to build pre-RCs, due to the addition of a nondegradable form of geminin (16, 52). Since replication can only initiate at functionally assembled pre-RCs, these extracts provide a convenient readout for the chromosomal positions of hamster pre-RCs that are available for initiation by the extract at each G1-phase time period. Replication intermediates were distributed equally near each of the 17 probes with nuclei isolated prior to the ODP. However, the use of so few probes precluded us from concluding that they were distributed evenly throughout the entire locus in pre-ODP nuclei. One or a few initiation sites within and outside of the DHFR and 2BE2121 genes could also result in an even distribution of replication forks throughout these probes. The availability of a high-resolution DNA array allowed us to address this question more comprehensively.
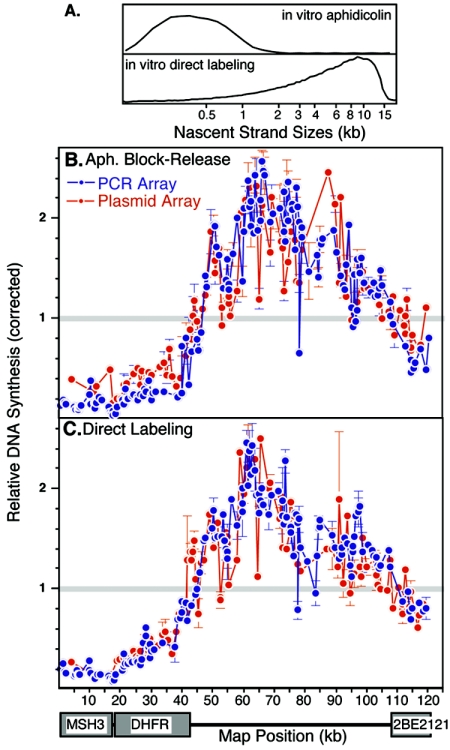
Two different labeling schemes were used to map the sites of initiation of replication in Xenopus egg extracts. In the first, nuclei isolated at various times after mitosis were introduced into geminin-supplemented extracts that also contained aphidicolin to arrest in vitro-generated replication forks near their sites of initiation. After we allowed sufficient time to initiate replication in all nuclei, nuclei were then washed free of aphidicolin-supplemented extract and replication forks were extended in a replication cocktail containing [32P]dATP. Under these conditions, labeled nascent strands were <1 kb (Fig. 4A) (14, 32), suggesting that forks are much more efficiently arrested in the cell-free system than they are in vivo. In the second method, we labeled replication intermediates directly in geminin-supplemented extracts that contained [32P]dATP but no aphidicolin, which labels nascent DNA starting from the first nucleotides to be synthesized but captures a population of nascent strands ranging in size from 1 to 20 kb (Fig. 4A). In both cases, labeled nascent DNA was hybridized to both PCR and plasmid arrays as in Fig. 2 and 3. An example of results using each method with nuclei isolated 5 h after mitosis is shown in Fig. 4. With both methods, the initiation pattern closely resembles that seen when replication initiates in vivo (Fig. 2 and 3), except that slightly more initiation activity was detected within the 2BE2121 gene and there appeared to be a more even distribution of initiation sites within the intergenic region in vitro than in vivo. Both PCR and plasmid arrays gave similar results, consistent with a broad distribution of initiation sites throughout the intergenic region.
FIG. 4.
Sites of initiation of replication in Xenopus egg extracts. Nuclei from cells synchronized at 5 h after mitosis were introduced into a Xenopus egg extract, and nascent strands were either arrested with aphidicolin for 90 min and then labeled after release (in vitro aphidicolin) or were labeled directly in extract for 40 min (i.e., a 20-min lag period plus 20 min of label incorporation). (A) The sizes of nascent strands were determined as in Fig. 2. (B) Nascent strands were hybridized to both PCR (blue) and plasmid (red) arrays and analyzed as in the results shown in Fig. 3. Shown are the means of three independent experiments in which nascent strands were hybridized to four independently printed arrays. A gray line is drawn at 1.0, to indicate the mean of all probes in each array.
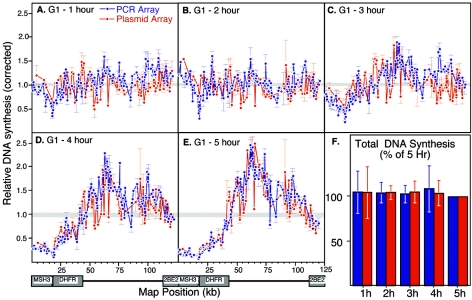
To examine the dynamics of potential origin sites over the course of G1 phase, we performed similar analyses after in vitro initiation within nuclei isolated at hourly intervals after mitosis. As shown in Fig. 5, initiation sites were distributed throughout the entire 121-kb region with nuclei isolated 1 or 2 h after mitosis (Fig. 5A and B), whereas initiation became focused to the intergenic region with nuclei that were isolated at later times (Fig. 5C through E). Within each experiment, the same amount of total initiation activity was detected across the panel of probes on the array (Fig. 5F), indicating that this focusing involved the redistribution of a similar total number of initiation sites with each preparation of nuclei. Total chromosomal DNA synthesis, as measured by total radiolabeling incorporated during the labeling time or during the course of incubation of nuclei in egg extracts, was also the same with each preparation of nuclei (70, 73; data not shown). We conclude that there is remarkable flexibility in the number of potential sites that can function as replication origins both within and outside of transcription units. Since no new pre-RC assembly can occur in these extracts, the simplest interpretation is that functional pre-RCs are assembled at sites distributed throughout the entire locus and that some event occurring at the ODP suppresses the use of pre-RCs within transcription units. This suppression is clearly stronger within the Msh3 and DHFR transcription units than within the 2BE2121 gene.
FIG. 5.
Initiation is redistributed to the intergenic region at the ODP. (A to E) Nuclei from cells synchronized at hourly intervals after mitosis were introduced into Xenopus egg extracts, and the sites of initiation were mapped by the direct labeling method shown in Fig. 4. As in Fig. 4, results with both PCR (blue) and plasmid (red) arrays are coplotted, and a gray line indicates the mean of all probes in each experiment. Shown are mean values from two independent experiments and the standard deviation. (F) The total counts per minute across all DHFR probes for each time point were normalized to the same total for nuclei at 5 h after mitosis. Shown are the mean values from two independent experiments.
Transcription precedes origin specification during G1 phase.
The fact that the ODP represents an elimination of potential sites of initiation selectively within the transcription units raises the question of where and when DHFR transcription resumes after mitosis. It is known that transcription is largely silent in mammalian cells during mitosis and resumes at some point in early G1 phase (10, 56). Although there is a lag between nuclear membrane formation after mitosis and the entry of RNA polymerase II (Pol II) into the nucleus (H. Kimura, personal communication), we found that green fluorescent protein-tagged RNA polymerase II reentered the nucleus very early in G1 phase in CHO cells (C. Kumagai, unpublished data), consistent with recently published results (10, 56). We have also found that total transcription, as measured by the incorporation of bromouridine or by total incorporation of radiolabeled uridine in nuclear run-on assays, resumed as early as 1 h after mitosis (not shown). However, it is generally presumed that DHFR transcription begins at the G1/S boundary after the phosphorylation of retinoblastoma protein relieves repression of E2F-regulated genes (9, 18, 49, 68). In contrast, the ODP is upstream and independent of restriction point control (33, 71). Most studies of DHFR gene induction, however, have been performed with cells released from quiescence, and at least one study has suggested that DHFR is not cell cycle regulated in proliferating cells (19). To determine when DHFR transcription resumes relative to the ODP, we performed nuclear run-on experiments with nuclei isolated before and after the ODP. Since isolated nuclei incubated in a transcription cocktail can continue using template-engaged RNA polymerase but cannot initiate de novo transcription, labeled RNA reflects the positions of active RNA polymerase complexes.
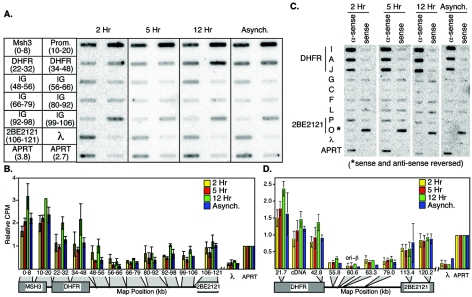
Because CHO DHFR is a weakly transcribed gene (65), it was very difficult to detect nuclear run-on products using the small probes present on our PCR array, even though the DHFR gene in CHOC 400 cells is present at 1,000 copies, and even after growing CHOC 400 cells for several weeks in 400-μg/ml methotrexate, the drug used to select for a high DHFR copy number. To overcome this obstacle, we pooled groups of PCR-generated nonrepetitive probes representing ∼10-kb regions across the sequenced region and immobilized these pooled probes on a slot blot (Fig. 6A). Radiolabeled nuclear run-on products were prepared using nuclei isolated from cells synchronized at 2 and 5 h after mitosis (pre- and post-ODP), 12 h after mitosis (S phase), and from asynchronous cells. To compare results from different run-on reactions, equal numbers of total counts per minute from run-on products were used; a cDNA probe from the hamster adenine phosphoribosyltransferase (APRT) gene, which is robustly expressed throughout the cell cycle (47), was included on the slot blot. After quantitative analysis of the hybridized filter, the relative counts per minute for each pool of PCR products were normalized to the corresponding value for the APRT gene (Fig. 5B). When Fig. 6B is compared to Fig. 2 to 5, it is clear that there is an inverse relationship between transcription and origin activity near the 3′ end of the DHFR gene, with the probes in the region from nucleotide position 56 to 79 kb being the most active initiation sites and the most weakly transcribed. Weak transcription activity above the background detected with the negative control lambda DNA probe was detected from 80 to 106 kb. Consistently, this region also showed lower activity for replication initiation (Fig. 2 to 5). However, although transcription within the 2BE2121 gene was also weak, suppression of replication initiation activity was considerably more pronounced in this region (Fig. 2 to 5). Moreover, transcription was clearly detected prior to the ODP.
FIG. 6.
Nuclear run-on analysis of transcription during G1 phase. (A) Labeled transcription products from nuclear run-on reactions with nuclei synchronized at 2, 5, or 12 h, as well as from asynchronous (Asynch.) cells, were hybridized to pools of PCR probes covering approximately 10 kb per pool. The arrangement of these probes is schematically illustrated on the left. APRT is transcribed throughout interphase (47) and serves as an internal control for each hybridization (separate 3.8- and 2.7-kb APRT probes). (B) Quantification of the data in panel A, with the relative counts per minute for the larger APRT probe adjusted to 1.0 for each hybridization. The x axis includes a schematic diagram of the probe positions indicated in panel A, the map position boundaries of the probes pooled together in each data point, and gray boxes to indicate the positions of the pooled probe boundaries on the schematic. Shown are the means of three independent experiments and the standard deviation. (C) To verify the polarity of transcripts synthesized in nuclear run-on reactions, labeled products were also hybridized to previously described (24) single-stranded (M13 phage) probes from specific locations along the length of the DHFR locus. For genes, antisense refers to the DNA strand predicted to be transcribed, whereas for intergenic probes, antisense refers to the orientation of the predicted transcribed strand for the DHFR gene. Note that the sense and antisense strands of probe O were inadvertently spotted in the reverse order, as indicated. (D) Quantification of the data in panel C, with the relative counts per minute for the double-stranded APRT probe adjusted to 1.0 as in panel B. Here, gray triangles point to the indicated map position of the single probe on the schematic. Probe C at 60.6 kb is at the position of ori-β as it has been most precisely defined (35). Shown are the means of three independent experiments and the standard error of the mean. The x axis indicates the map positions of probes, and the gray triangles indicate these positions on the schematic map.
To verify that the RNA synthesized in nuclear run-on reactions was truly representative of the sense strand of transcription units, we performed parallel hybridizations to a panel of nine pairs of single-stranded probes from across the DHFR locus. Within the DHFR and 2BE2121 transcription units, significant signal above the negative-control probe (a single-stranded segment of lambda phage DNA) was detected only with the probes representing the antisense strand, while probes within the intergenic region detected very little transcription from either strand. 2BE2121 again showed weaker activity than DHFR or Msh3 (note that probes from the weakly transcribed region downstream of 2BE2121 were not included in this analysis). Most importantly, in both sets of hybridizations, the amount of transcription from these genes was almost identical between nuclei isolated at 2 or 5 h after mitosis and from asynchronous cells, and there was a small but consistent increase in transcription of the DHFR and Msh3 genes during S phase. We conclude that the basal level of transcription associated with actively proliferating cells resumes shortly after mitosis prior to origin specification and does not substantially increase at the ODP. Moreover, whether nuclei were isolated in the pre-ODP or post-ODP stages of G1 phase, transcription continued and even appeared to be more robust after introduction into Xenopus egg extracts (not shown). Since no origin specification was detected within nuclei at 2 h postmitosis, these results strongly suggest that the resumption of transcription is not sufficient to restrict initiation to the intergenic region.
Origin specification requires transcription.
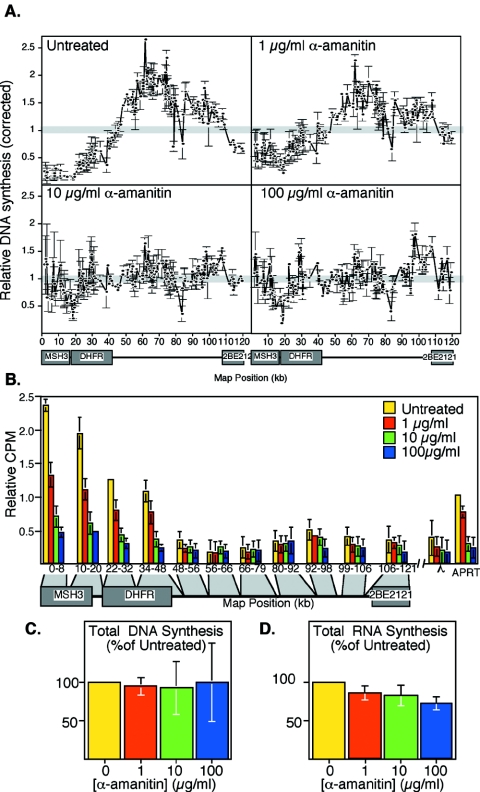
The genomic regions within which initiation of replication becomes excluded coincide with the known transcription units, with the greatest amount of suppression within the more highly expressed DHFR and Msh3 genes. These data, together with data summarized in the introduction, strongly suggest that although transcription is not sufficient to suppress initiation of replication, it is likely to play some role. To more directly address whether transcription is necessary for focusing initiation to the intergenic region, we examined the effect of inhibiting RNA Pol II transcription on the specification of origin sites at the ODP. Alpha-amanitin binds to the large subunit of Pol II and inhibits transcription by Pol II considerably more effectively than rRNA polymerases I and III (69). CHOC 400 cells were treated with 0-, 1-, 10-, or 100-μg/ml alpha-amanitin at 2 h after mitosis and then incubated an additional 3 h. Nuclei were then isolated, and aliquots of those nuclei were subjected to nuclear run-on assays to measure the amount of DHFR transcription, while the remainder of the nuclei were introduced into Xenopus egg extract to examine the sites of initiation of DNA replication. As illustrated in Fig. 7, the focusing of initiation sites to within the intergenic region at the ODP was strongly prevented between 1- and 10-μg/ml alpha-amanitin (Fig. 7A), and DHFR and Msh3 gene transcription was strongly inhibited within the same range of alpha-amanitin concentrations. Since we have previously shown that origin selection does not require protein synthesis (33), the requirement for transcription is not due to an indirect requirement for the expression of a gene product during this time. These results implicate a pretranslational role for transcription in the suppression of initiation of DNA replication that occurs at the ODP.
FIG. 7.
Alpha-amanitin inhibition of transcription and origin selection during G1 phase. (A) CHOC 400 cells were treated with the indicated concentrations of alpha-amanitin at 2 h after mitosis, nuclei were collected at 5 h, and the sites of initiation of replication in Xenopus egg extracts were mapped as in Fig. 5. (B) Aliquots of the same populations of cells shown in panel A were analyzed for transcription by nuclear run-on assays as in Fig. 6. (C) The total counts per minute hybridized to all DHFR probes for each time point in panel A are expressed as a percentage of that for nuclei isolated from untreated cells, as in Fig. 5F. (D) The total counts per minute incorporated into high-molecular-weight RNA in each of the experiments shown in panel B are expressed as a percentage of that for untreated cells. Since the vast majority of RNA synthesis in nuclear run-on reactions is ribosomal and mediated by RNA polymerases I and III, which are poorly inhibited by alpha-amanitin, the total counts per minute hybridized in the experiments shown in panel B were very similar. Shown are the mean values from two independent experiments and the standard deviation.
DISCUSSION
There is mounting evidence that the initiation of replication is not determined exclusively by DNA sequences but by a combination of factors that work both upstream and downstream of pre-RC formation and may be unique to each origin (22). We have used a comprehensive DNA array approach that reveals a remarkable flexibility of initiation sites throughout a 121-kb region surrounding the CHO DHFR gene. Prior to the ODP, potential sites of initiation were distributed throughout the entire locus. At the ODP, sites with the potential to initiate DNA replication that were within transcribed regions were suppressed. We and others have previously demonstrated that the selection of origin sites at the DHFR locus is profoundly affected by transcriptional activity (31, 33, 62). Here, we demonstrate that the inhibition of transcription prevents any detectable origin selection at the ODP. However, transcription itself was not sufficient to prevent initiation of DNA replication; pre-ODP nuclei actively transcribed the DHFR gene, yet replication initiated within the body of the transcription unit until cells passed through the ODP. Together with our previous results (33) and recent genetic analyses (62), we conclude that transcription is necessary but not sufficient to prevent initiation of replication within transcription units and that transcription is one component of origin selection at the ODP.
The contribution of DNA sequence in the context of an initiation zone.
Our previous results demonstrated that potential sites for the initiation of replication become more focused to the DHFR intergenic region at the ODP. However, the resolution of these prior studies, which interrogated 17 independent sites throughout a 110-kb region, could not distinguish whether initiation within pre-ODP nuclei was taking place at many or only a few additional sites. For example, we have demonstrated that a very specific site within the DHFR gene is highly preferred as an initiation site when topoisomerase II-condensed metaphase chromosomes are introduced into Xenopus egg extracts (37). Activation of just this one additional site could substantially skew the relative distribution of replication forks among these 17 probes. By sequencing the entire DHFR locus, we have provided a novel tool with which to probe initiation in this region, allowing us to demonstrate that there is little or no site preference for initiation throughout the entire 121 kb when replication initiates within pre-ODP nuclei. Thus, under these conditions, it appears that virtually any sequence can function as a site for the initiation of replication. Indeed, it is possible that initiation within the DHFR locus in pre-ODP nuclei is as flexible as initiation in early Xenopus embryos, which initiate replication at random with respect to DNA sequence (27, 42). Although we cannot yet rule out the possibility that many specific sites are spaced at frequent intervals (every few kilobases), DNA sequence specificity is all but ruled out.
In post-ODP nuclei and in vivo, initiation is clearly suppressed within the transcription units, but our results indicate that initiation can take place at a large number of closely spaced sites within the intergenic region. At first glance, this may seem to contradict the precise identification of ori-β and ori-β′ initiation sites (35). However, these results can be reconciled by considering the resolution of the different studies. These specific sites were identified by interrogating the presence of a few specific sequences in preparations of 0.8- to 1.5-kb nascent DNA strands. Our analysis is more comprehensive but is a lower-resolution study that might not distinguish differences in initiation activity within a few kilobases, due to the sizes of the probes (300 to 1,500 bp) and nascent strands (10 to 20 kb for aphidicolin and 500 to 2,500 bp for mimosine). Moreover, Fig. 2 and 3 suggest some discontinuity in the frequency of site usage within the intergenic region. In fact, when we have hybridized smaller (0.8- to 1.5-kb) nascent strands to our arrays, more-prominent peaks could be observed, including those at ori-β and ori-β′, but no sites within the intergenic region were completely silent (under these conditions, many peaks were represented by single probes, rendering these data too preliminary to report). Hence, our results are more consistent with 2D-gel analyses that detect some initiation activity in regions that appear silent by the directed PCR method (13). At present, then, the only remaining discrepancy in the initiation pattern at this locus is whether the sites surrounding these peaks are truly silent, as the directed PCR studies suggest, or whether they are used at a lower frequency. Higher-resolution arrays should resolve this long-standing controversy.
Regardless, the large number of sites that can function as replication origins raises the question as to the role of DNA sequences within the context of such a relaxed initiation zone. In fact, deletion of ori-β or even larger segments of the DHFR intergenic region appears to stimulate the frequency of usage of other sequences in the intergenic region (45), suggesting that redundancy is part of a mechanism to ensure that initiation takes place somewhere within this region. How is it, then, that specific sequences within a small DHFR ori-β fragment are required for initiation activity when this fragment is moved to ectopic sites (3)? Indeed, this seems almost impossible to reconcile with the results of 2D-gel studies that detect replication bubbles at many sites within the same specific fragment (30). At present, the most parsimonious explanation is that, even within the context of relaxed replicator selection in a large initiation zone, there are localized effects of specific sequences that can influence the frequency with which particular sites are utilized. In this view, transcription contributes to the global exclusion of initiation from transcription units, while other features of chromatin, DNA methylation, topology, and DNA sequence composition contribute to the relative frequency with which replication will initiate at particular sites (22). A challenge for the future is to determine whether the locations of these sites serve some function or whether they are simply the most permissive sites for initiation in that region. Since it is now clear that duplicating genomes once per cell cycle does not require site-specific initiation (23), any putative function for specific origin sites must transcend the basic need to duplicate the genome.
Replication and transcription.
One obvious conclusion of our analysis is that replication initiation is largely suppressed within the transcription units of the DHFR locus. This is consistent with a recent genetic study showing that deletion of the DHFR promoter resulted in the initiation of replication at many sites within the gene (62). We have previously suggested a model in which pre-RCs in the path of a moving RNA polymerase are destabilized upon collision with the transcriptional machinery (15, 22, 23). This model accounted for all prior observations, including the occasional replication origin found within transcription units (34, 64), provided that transcription does not occur prior to the initiation of replication at any given origin. In fact, a large inhibitor study revealed that all but one inhibitor of general transcription also inhibited the ODP (33). The one exception was a protein kinase A inhibitor, which likely inhibits transcription through the CREB pathway that does not regulate DHFR transcription (75). However, our results reported here indicate that this model is an oversimplification. First, transcription of the genes within the DHFR locus is active prior to the exclusion of transcription units as sites of initiation at the ODP. No significant increase in the number of RNA polymerase molecules loaded onto DHFR transcription units was detected at the ODP. Hence, the resumption of transcription during G1 phase is not sufficient to inactivate pre-RCs. Furthermore, the DHFR gene is not transcribed very efficiently; the approximately 1,000 copies were transcribed at a frequency similar to that of the single-copy APRT gene (Fig. 6). The 2BE2121 gene was transcribed even less efficiently. Hence, it is possible that only a minority of the gene copies are active in any given cell.
Nonetheless, both the inhibition of transcription (Fig. 7) and deletion of the DHFR promoter (62) have profound affects on origin specification. Inhibitor studies show that the effect does not require translation of RNA (33), and genetic studies suggest that the mechanism linking transcription and origin specification acts in cis at the DHFR locus rather than through a trans-acting, noncoding RNA. Since a sufficient fraction of the total amplified DHFR promoters are occupied with transcription factors to generate an in vivo footprint (54, 67), it is possible that some aspect of the transcriptionally active state, rather than the act of transcription itself, renders the entire transcription unit inaccessible to replication initiation factors. Alternatively, it is possible that only those gene copies that are actively engaged in transcription are competent to initiate replication and that those are also the copies for which transcription has suppressed intragenic initiation. This would imply that transcription plays both positive and negative roles in origin selection. Whatever the nature of this component, the transcriptionally active state on its own is clearly not sufficient to alter the usage of replication origin sites in pre-ODP nuclei. Some additional event, possibly cooperating with the transcriptionally active state, restricts initiation to the intergenic region.
The use of arrays to probe initiation.
The remarkable flexibility in the DNA sequences that can function as replication origins in mammalian cells underscores the need for origin mapping approaches that are not biased toward the evaluation of any particular sites predicted to be origin or nonorigin regions. Presently, three such methods have been described. One method is to map the sites of initiation along the length of stretched DNA fibers (5, 50). This method has the distinct advantage of detecting the frequency of multiple initiation events on the same DNA fiber, but it is currently a very demanding and time-consuming method. Another method is to evaluate the presence of many individual sequences positioned at 1-kb intervals in preparations of small nascent DNA strands (58). Carrying out this method one primer set at a time is also extremely laborious. In principle, hybridization of the entire population of small nascent strands to a complete-coverage DNA array should provide a much more convenient and rapid means of evaluating origin usage. Unfortunately, this method has not succeeded in any eukaryotic organism. In fact, the genome-wide identification of origins using DNA arrays has only been indirectly determined in budding yeast by identifying sites of origin replication complex binding (60) or earliest-replicating DNA (59, 74) and in Drosophila by hybridizing earliest-labeled DNA to a high-resolution array of a chromosome arm (43). Here, we present the first similar approach in mammalian cells, hybridizing earliest-labeled G1/S DNA to an array covering a defined 121-kb region. As discussed above, our attempts to hybridize purified small nascent strands to this array have not yet provided conclusive results, a problem that we hope to solve with higher-resolution arrays. However, starting with the amplified DHFR locus in CHOC 400 cells provides the sensitivity afforded by simpler model organisms, while at the same time allowing us to work through the methodology in mammalian cells. By taking advantage of cell lines with fewer and fewer DHFR amplicons (41), we hope to systematically work our way toward the goal of rapidly mapping the distribution of origins at single-copy loci in mammalian cells.
Acknowledgments
We thank J. Hamlin and L. Chasin for providing cosmids and plasmids with which to complete our sequencing analysis; M. Hawkins, J. Merrit, and A. Leskovar for technical assistance; Paul Bertone, L. Chasin, and J. Lis for helpful discussions; and J. Huberman for critical review of the manuscript.
This work was supported by NIH grant GM-57233 to D.M.G. T.S. was supported by a fellowship from the New York State DOH Breast Cancer Initiative.
REFERENCES
- 1.Abdurashidova, G., M. Deganuto, R. Klima, S. Riva, G. Biamonti, M. Giacca, and A. Falaschi. 2000. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science 287:2023-2026. [DOI] [PubMed] [Google Scholar]
- 2.Aladjem, M. I., and E. Fanning. 2004. The replicon revisited: an old model learns new tricks in metazoan chromosomes. EMBO Rep. 5:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altman, A. L., and E. Fanning. 2004. Defined sequence modules and an architectural element cooperate to promote initiation at an ectopic mammalian chromosomal replication origin. Mol. Cell. Biol. 24:4138-4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anachkova, B., and J. L. Hamlin. 1989. Replication in the amplified dihydrofolate reductase domain in CHO cells may initiate at two distinct sites, one of which is a repetitive sequence element. Mol. Cell. Biol. 9:532-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anglana, M., F. Apiou, A. Bensimon, and M. Debatisse. 2003. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell 114:385-394. [DOI] [PubMed] [Google Scholar]
- 6.Antequera, F. 2004. Genomic specification and epigenetic regulation of eukaryotic DNA replication origins. EMBO J. 23:4365-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biamonti, G., S. Paixao, A. Montecucco, F. A. Peverali, S. Riva, and A. Falaschi. 2003. Is DNA sequence sufficient to specify DNA replication origins in metazoan cells? Chromosome Res. 11:403-412. [DOI] [PubMed] [Google Scholar]
- 8.Carothers, A. M., G. Urlaub, N. Ellis, and L. A. Chasin. 1983. Structure of the dihydrofolate reductase gene in Chinese hamster ovary cells. Nucleic Acids Res. 11:1997-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, Y. C., S. Illenye, and N. H. Heintz. 2001. Cooperation of E2F-p130 and Sp1-pRb complexes in repression of the Chinese hamster dhfr gene. Mol. Cell. Biol. 21:1121-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, D., M. Dundr, C. Wang, A. Leung, A. Lamond, T. Misteli, and S. Huang. 2005. Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J. Cell Biol. 168:41-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dijkwel, P. A., L. D. Mesner, V. V. Levenson, J. d'Anna, and J. L. Hamlin. 2000. Dispersive initiation of replication in the Chinese hamster rhodopsin locus. Exp. Cell Res. 256:150-157. [DOI] [PubMed] [Google Scholar]
- 12.Dijkwel, P. A., J. P. Vaughn, and J. L. Hamlin. 1994. Replication initiation sites are distributed widely in the amplified CHO dihydrofolate reductase domain. Nucleic Acids Res. 22:4989-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dijkwel, P. A., S. Wang, and J. L. Hamlin. 2002. Initiation sites are distributed at frequent intervals in the Chinese hamster dihydrofolate reductase origin of replication but are used with very different efficiencies. Mol. Cell. Biol. 22:3053-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dimitrova, D., and D. Gilbert. 1998. Regulation of mammalian replication origin usage in Xenopus egg extracts. J. Cell Sci. 111:2989-2998. [DOI] [PubMed] [Google Scholar]
- 15.Dimitrova, D. S., and D. M. Gilbert. 1999. DNA replication and nuclear organization: prospects for a soluble in vitro system. Crit. Rev. Eukaryot. Gene Expr. 9:353-361. [DOI] [PubMed] [Google Scholar]
- 16.Dimitrova, D. S., T. A. Prokhorova, J. J. Blow, I. T. Todorov, and D. M. Gilbert. 2002. Mammalian nuclei become licensed for DNA replication during late telophase. J. Cell Sci. 115:51-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farnham, P. J., and R. T. Schimke. 1986. Murine dihydrofolate reductase transcripts through the cell cycle. Mol. Cell. Biol. 6:365-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farnham, P. J., and R. T. Schimke. 1985. Transcriptional regulation of mouse dihydrofolate reductase in the cell cycle. J. Biol. Chem. 260:7675-7680. [PubMed] [Google Scholar]
- 19.Feder, J. N., Y. G. Assaraf, L. C. Seamer, and R. T. Schimke. 1989. The pattern of dihydrofolate reductase expression through the cell cycle in rodent and human cultured cells. J. Biol. Chem. 264:20583-20590. [PubMed] [Google Scholar]
- 20.Gale, J. M., R. A. Tobey, and J. A. D'Anna. 1992. Localization and DNA sequence of a replication origin in the rhodopsin gene locus of Chinese hamster cells. J. Mol. Biol. 224:343-358. [DOI] [PubMed] [Google Scholar]
- 21.Gerbi, S. A., Z. Strezoska, and J. M. Waggener. 2002. Initiation of DNA replication in multicellular eukaryotes. J. Struct. Biol 140:17-30. [DOI] [PubMed] [Google Scholar]
- 22.Gilbert, D. M. 2004. In search of the holy replicator. Nat. Rev. Mol. Cell Biol. 5:848-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert, D. M. 2001. Making sense of eukaryotic DNA replication origins. Science 294:96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbert, D. M., H. Miyazawa, and M. L. DePamphilis. 1995. Site-specific initiation of DNA replication in Xenopus egg extract requires nuclear structure. Mol. Cell. Biol. 15:2942-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilbert, D. M., A. Neilson, H. Miyazawa, M. L. DePamphilis, and W. C. Burhans. 1995. Mimosine arrests DNA synthesis at replication forks by inhibiting deoxyribonucleotide matabolism. J. Biol. Chem. 270:9597-9606. [DOI] [PubMed] [Google Scholar]
- 26.Hirayoshi, K., and J. T. Lis. 1999. Nuclear run-on assays: assessing transcription by measuring density of engaged RNA polymerases. Methods Enzymol. 304:351-362. [DOI] [PubMed] [Google Scholar]
- 27.Hyrien, O., K. Marheineke, and A. Goldar. 2003. Paradoxes of eukaryotic DNA replication: MCM proteins and the random completion problem. Bioessays 25:116-125. [DOI] [PubMed] [Google Scholar]
- 28.Ina, S., T. Sasaki, Y. Yokota, and T. Shinomiya. 2001. A broad replication origin of Drosophila melanogaster, oriDα, consists of AT-rich multiple discrete initiation sites. Chromosoma 109:551-564. [DOI] [PubMed] [Google Scholar]
- 29.Jacob, F., S. Brenner, and F. Cuzin. 1964. On the regulation of DNA replication in bacteria. Cold Spring Harbor Symp. Quant. Biol. 28:329-348. [Google Scholar]
- 30.Kalejta, R., and J. Hamlin. 1996. Composite patterns in neutral/neutral two-dimensional gels demonstrate inefficient replication origin usage. Mol. Cell. Biol. 16:4915-4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalejta, R. F., X. Li, L. D. Mesner, P. A. Dijkwel, H. B. Lin, and J. L. Hamlin. 1998. Distal sequences, but not ori-beta/OBR-1, are essential for initiation of DNA replication in the Chinese hamster DHFR origin. Mol. Cell 2:797-806. [DOI] [PubMed] [Google Scholar]
- 32.Keezer, S. M. 2002. Ph.D. thesis. State University of New York Upstate Medical University, Syracuse.
- 33.Keezer, S. M., and D. M. Gilbert. 2002. Sensitivity of the origin decision point to specific inhibitors of cellular signaling and metabolism. Exp. Cell Res. 273:54-64. [DOI] [PubMed] [Google Scholar]
- 34.Kelly, R. E., M. L. DeRose, B. W. Draper, and G. M. Wahl. 1995. Identification of an origin of bidirectional DNA replication in the ubiquitously expressed mammalian CAD gene. Mol. Cell. Biol. 15:4136-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobayashi, T., T. Rein, and M. DePamphilis. 1998. Identification of primary initiation sites for DNA replication in the hamster DHFR gene initiation zone. Mol. Cell. Biol. 18:3266-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krude, T. 1999. Mimosine arrests proliferating human cells before onset of DNA replication in a dose-dependent manner. Exp. Cell Res. 247:148-159. [DOI] [PubMed] [Google Scholar]
- 37.Lawlis, S. J., S. M. Keezer, J.-R. Wu, and D. M. Gilbert. 1996. Chromosome architecture can dictate site-specific initiation of DNA replication in Xenopus egg extracts. J. Cell Biol. 135:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin, H. B., P. A. Dijkwel, and J. L. Hamlin. 2005. Promiscuous initiation on mammalian chromosomal DNA templates and its possible suppression by transcription. Exp. Cell Res. 308:53-64. [DOI] [PubMed] [Google Scholar]
- 39.Little, R. D., T. H. Platt, and C. L. Schildkraut. 1993. Initiation and termination of DNA replication in human rRNA genes. Mol. Cell. Biol. 13:6600-6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Looney, J. E., and J. L. Hamlin. 1987. Isolation of the amplified dihydrofolate reductase domain from methotrexate-resistant Chinese hamster ovary cells. Mol. Cell. Biol. 7:569-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Looney, J. E., C. Ma, T. H. Leu, W. F. Flintoff, W. B. Troutman, and J. L. Hamlin. 1988. The dihydrofolate reductase amplicons in different methotrexate-resistant Chinese hamster cell lines share at least a 273-kilobase core sequence, but the amplicons in some cell lines are much larger and are remarkably uniform in structure. Mol. Cell. Biol. 8:5268-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lucas, I., M. Chevrier-Miller, J. M. Sogo, and O. Hyrien. 2000. Mechanisms ensuring rapid and complete DNA replication despite random initiation in Xenopus early embryos. J. Mol. Biol. 296:769-786. [DOI] [PubMed] [Google Scholar]
- 43.MacAlpine, D. M., H. K. Rodriguez, and S. P. Bell. 2004. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev. 18:3094-3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mesner, L. D., and J. L. Hamlin. 2005. Specific signals at the 3′ end of the DHFR gene define one boundary of the downstream origin of replication. Genes Dev. 19:1053-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mesner, L. D., X. Li, P. A. Dijkwel, and J. L. Hamlin. 2003. The dihydrofolate reductase origin of replication does not contain any nonredundant genetic elements required for origin activity. Mol. Cell. Biol. 23:804-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Milbrandt, J. D., J. C. Azizkhan, K. S. Greisen, and J. L. Hamlin. 1983. Organization of a Chinese hamster ovary dihydrofolate reductase gene identified by phenotypic rescue. Mol. Cell. Biol. 3:1266-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitchell, P. J., A. M. Carothers, J. H. Han, J. D. Harding, E. Kas, L. Venolia, and L. A. Chasin. 1986. Multiple transcription start sites, DNase I-hypersensitive sites, and an opposite-strand exon in the 5′ region of the CHO dhfr gene. Mol. Cell. Biol. 6:425-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mosca, P. J., H. B. Lin, and J. L. Hamlin. 1995. Mimosine, a novel inhibitor of DNA replication, binds to a 50 kDa protein in Chinese hamster cells. Nucleic Acids Res. 23:261-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noe, V., C. Alemany, L. A. Chasin, and C. J. Ciudad. 1998. Retinoblastoma protein associates with SP1 and activates the hamster dihydrofolate reductase promoter. Oncogene 16:1931-1938. [DOI] [PubMed] [Google Scholar]
- 50.Norio, P., and C. L. Schildkraut. 2004. Plasticity of DNA replication initiation in Epstein-Barr virus episomes. PLoS Biol. 2:E152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okuno, Y., P. J. Hahn, and D. M. Gilbert. 2004. Structure of a palindromic amplicon junction implicates microhomology-mediated end joining as a mechanism of sister chromatid fusion during gene amplification. Nucleic Acids Res. 32:749-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okuno, Y., A. J. McNairn, N. den Elzen, J. Pines, and D. M. Gilbert. 2001. Stability, chromatin association and functional activity of mammalian pre-replication complex proteins during the cell cycle. EMBO J. 20:4263-4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oppenheim, E. W., I. M. Nasrallah, M. G. Mastri, and P. J. Stover. 2000. Mimosine is a cell-specific antagonist of folate metabolism. J. Biol. Chem. 275:19268-19274. [DOI] [PubMed] [Google Scholar]
- 54.Pemov, A., S. Bavykin, and J. L. Hamlin. 1995. Proximal and long-range alterations in chromatin structure surrounding the Chinese hamster dihydrofolate reductase promoter. Biochemistry 34:2381-2392. [DOI] [PubMed] [Google Scholar]
- 55.Perry, C., R. Sastry, I. M. Nasrallah, and P. J. Stover. 2005. Mimosine attenuates serine hydroxymethyltransferase transcription by chelating zinc. Implications for inhibition of DNA replication. J. Biol. Chem. 280:396-400. [DOI] [PubMed] [Google Scholar]
- 56.Prasanth, K. V., P. A. Sacco-Bubulya, S. G. Prasanth, and D. L. Spector. 2003. Sequential entry of components of the gene expression machinery into daughter nuclei. Mol. Biol. Cell 14:1043-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Price, G. B., M. Allarakhia, N. Cossons, T. Nielsen, M. Diaz-Perez, P. Friedlander, L. Tao, and M. Zannis-Hadjopoulos. 2003. Identification of a cis-element that determines autonomous DNA replication in eukaryotic cells. J. Biol. Chem. 278:19649-19659. [DOI] [PubMed] [Google Scholar]
- 58.Prioleau, M. N., M. C. Gendron, and O. Hyrien. 2003. Replication of the chicken β-globin locus: early-firing origins at the 5′ HS4 insulator and the ρ- and βA-globin genes show opposite epigenetic modifications. Mol. Cell. Biol. 23:3536-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raghuraman, M. K., E. A. Winzeler, D. Collingwood, S. Hunt, L. Wodicka, A. Conway, D. J. Lockhart, R. W. Davis, B. J. Brewer, and W. L. Fangman. 2001. Replication dynamics of the yeast genome. Science 294:115-121. [DOI] [PubMed] [Google Scholar]
- 60.Ren, B., F. Robert, J. J. Wyrick, O. Aparicio, E. G. Jennings, I. Simon, J. Zeitlinger, J. Schreiber, N. Hannett, E. Kanin, T. L. Volkert, C. J. Wilson, S. P. Bell, and R. A. Young. 2000. Genome-wide location and function of DNA binding proteins. Science 290:2306-2309. [DOI] [PubMed] [Google Scholar]
- 61.Rinn, J. L., G. Euskirchen, P. Bertone, R. Martone, N. M. Luscombe, S. Hartman, P. M. Harrison, F. K. Nelson, P. Miller, M. Gerstein, S. Weissman, and M. Snyder. 2003. The transcriptional activity of human chromosome 22. Genes Dev. 17:529-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saha, S., Y. Shan, L. D. Mesner, and J. L. Hamlin. 2004. The promoter of the Chinese hamster ovary dihydrofolate reductase gene regulates the activity of the local origin and helps define its boundaries. Genes Dev. 18:397-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scotto, K. W., H. Yang, J. P. Davide, and P. W. Melera. 1992. Differential utilization of poly (A) signals between DHFR alleles in CHL cells. Nucleic Acids Res. 20:6597-6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tasheva, E. S., and D. J. Roufa. 1994. A mammalian origin of bidirectional DNA replication within the Chinese hamster RPS14 locus. Mol. Cell. Biol. 14:5628-5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Urlaub, G., P. J. Mitchell, C. J. Ciudad, and L. A. Chasin. 1989. Nonsense mutations in the dihydrofolate reductase gene affect RNA processing. Mol. Cell. Biol. 9:2868-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weinreich, M., M. A. Palacios DeBeer, and C. A. Fox. 2004. The activities of eukaryotic replication origins in chromatin. Biochim. Biophys. Acta 1677:142-157. [DOI] [PubMed] [Google Scholar]
- 67.Wells, J., P. Held, S. Illenye, and N. H. Heintz. 1996. Protein-DNA interactions at the major and minor promoters of the divergently transcribed dhfr and rep3 genes during the Chinese hamster ovary cell cycle. Mol. Cell. Biol. 16:634-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wells, J. M., S. Illenye, J. Magae, C.-L. Wu, and N. H. Heintz. 1997. Accumulation of E2F-4 · DP-1 DNA binding complexes correlates with induction of dhfr gene expression during the G1 to S phase transition. J. Biol. Chem. 272:4483-4492. [DOI] [PubMed] [Google Scholar]
- 69.Wieland, T., and H. Faulstich. 1991. Fifty years of amanitin. Experientia 47:1186-1193. [DOI] [PubMed] [Google Scholar]
- 70.Wu, J.-R., and D. M. Gilbert. 1996. A distinct G1 step required to specify the Chinese hamster DHFR replication origin. Science 271:1270-1272. [DOI] [PubMed] [Google Scholar]
- 71.Wu, J.-R., and D. M. Gilbert. 1997. The replication origin decision point is a mitogen-independent, 2-aminopurine-sensitive, G1-phase event that precedes restriction point control. Mol. Cell. Biol. 17:4312-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu, J.-R., S. Keezer, and D. Gilbert. 1998. Transformation abrogates an early G1-phase arrest point required for specification of the Chinese hamster DHFR replication origin. EMBO J. 17:1810-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu, J.-W., G. Yu, and D. M. Gilbert. 1997. Origin-specific initiation of mammalian nuclear DNA replication in a Xenopus cell-free system. Methods 13:313-324. [DOI] [PubMed] [Google Scholar]
- 74.Yabuki, N., H. Terashima, and K. Kitada. 2002. Mapping of early firing origins on a replication profile of budding yeast. Genes Cells 7:781-789. [DOI] [PubMed] [Google Scholar]
- 75.Zhang, X., D. T. Odom, S. H. Koo, M. D. Conkright, G. Canettieri, J. Best, H. Chen, R. Jenner, E. Herbolsheimer, E. Jacobsen, S. Kadam, J. R. Ecker, B. Emerson, J. B. Hogenesch, T. Unterman, R. A. Young, and M. Montminy. 2005. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. USA 102:4459-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]