Abstract
Chloroplast-encoded genes, like nucleus-encoded genes, exhibit circadian expression. How the circadian clock exerts its control over chloroplast gene expression, however, is poorly understood. To facilitate the study of chloroplast circadian gene expression, we developed a codon-optimized firefly luciferase gene for the chloroplast of Chlamydomonas reinhardtii as a real-time bioluminescence reporter and introduced it into the chloroplast genome. The bioluminescence of the reporter strain correlated well with the circadian expression pattern of the introduced gene and satisfied all three criteria for circadian rhythms. Moreover, the period of the rhythm was lengthened in per mutants, which are phototactic rhythm mutants carrying a long-period gene in their nuclear genome. These results demonstrate that chloroplast gene expression rhythm is a bona fide circadian rhythm and that the nucleus-encoded circadian oscillator determines the period length of the chloroplast rhythm. Our reporter strains can serve as a powerful tool not only for analysis of the circadian regulation mechanisms of chloroplast gene expression but also for a genetic approach to the molecular oscillator of the algal circadian clock.
Circadian rhythms, which are endogenous ∼24-h activity cycles seen in most likely all organisms from cyanobacteria to humans, are characterized by three salient criteria: (i) they persist under constant conditions; (ii) the phases are reset by external stimuli, especially day/night light cycles; and (iii) they show temperature compensation (i.e., the period length is relatively constant at different ambient temperatures) (8). The rhythms are generated by a circadian clock that consists of molecular machinery residing in individual cells (11, 17). DNA microarray analyses reveal that the circadian clock regulates the timing of gene expression in diverse cellular processes (16, 37). Thus, the circadian clock seems to be a fundamental cellular component that coordinates the temporal program of cellular events by regulating gene expression.
The chloroplast, which evolved from an endosymbiotic cyanobacterium (3), has its own genetic system that is similar to that of prokaryotes (1). Circadian regulation of gene expression is observed not only in the nuclear genome but also in the chloroplast genome. It was first described for the unicellular green alga Chlamydomonas reinhardtii. Endogenous fluctuations in chloroplast tufA, atpA, and atpB mRNA levels were observed on the first day under constant conditions (24, 39). Hwang and coworkers carried out a conclusive demonstration that tufA mRNA level oscillated robustly for 2 to 3 days under constant conditions (18). Thereafter, circadian regulation of the chloroplast psbD light-responsive promoter was found in a higher plant (31).
Several studies suggest the regulation mechanisms for chloroplast gene expression rhythm by factors encoded in the nuclear genome. The mRNA level of nucleus-encoded chloroplast σ factor oscillates in a circadian manner in several photosynthetic species (10, 19, 30). In Chlamydomonas, the mRNA levels of a large number of nucleus-encoded chloroplast ribosomal proteins are under circadian control (23), and cytoplasmic protein synthesis is required for transcription of several chloroplast genes during circadian peaks (22). However, it is unclear to what extent the nucleus contributes to circadian rhythmicity of the chloroplast in these species. On the other hand, it has been known that in the green macroalga Acetabularia, the chloroplast photosynthetic rhythm persists even in an enucleated cell (46), and the nucleus contributes only to phase determination of the rhythm (41).
To facilitate study of chloroplast gene expression rhythms, here we developed a real-time monitoring system for chloroplast gene expression in Chlamydomonas. Although a bacterial luciferase gene for the Chlamydomonas chloroplast has been developed (a fusion gene of luxA and luxB [25]), we focused on the firefly luciferase gene because (i) it is shorter in nucleotide length than the luxAB fusion gene and (ii) it has been successfully used for circadian rhythm analysis in a wide range of organisms (2, 4, 29, 40, 48, 50). We synthesized a codon-optimized firefly luciferase gene for the chloroplast of C. reinhardtii (lucCP) and succeeded in real-time monitoring of chloroplast gene expression rhythms. Using this luminescence reporting system, we provide direct evidence that the circadian period of chloroplast gene expression rhythms is determined by the nucleus-encoded circadian oscillator.
MATERIALS AND METHODS
Plasmid construction.
De novo synthesis of the coding region of the lucCP gene was carried out according to the method of Stemmer et al. (42). A NdeI site was attached at the initiation codon and an XbaI site was attached just downstream of the stop codon. The resulting product was cloned into pCR2.1-TOPO (Invitrogen) to generate the plasmid pCR2.1-TOPO/lucCP.
The 5′-flanking regions of psbD (nucleotides 175630 to 176055; GenBank/DDBJ/EMBL accession no. BK000554) and tufA (11951 to 12783; BK000554) were amplified from genomic DNA by PCR with the primer sets psbD-F1 (5′-TATGAAATTAAATGGATATT-3′)/psbD-R1 (5′-CATATGGTGTATCTCCAAAA-3′) (underlining denotes the restriction site created for subsequent cloning) and psbK-F1 (5′-TTGTTTGGCAAGCAGCTGTTAGTTTCCGTT-3′)/tufA-R1 (5′-TTTACCGTGGTCAACGTGACCAATAGTACC-3′), respectively. The 3′-terminator region of atpB (159952 to 160185; BK000554) was amplified with the primer set atpB-F1M (5′-GCTTCTAGAAAAGCTGCTTCATTAAAATAA-3′)/atpB-R1M (5′-TCCCGGGACGTTTCCTTTAGTTTTTTGCTG-3′). These three fragments were cloned into pT7Blue-T (Novagen) to obtain pT7BT/PpsbD, pT7BT/PtufA, and pT7BT/TatpB.
The 0.46-kb SmaI-XbaI fragment of pT7BT/PpsbD was subcloned into the HincII/XbaI-digested pT7BT/TatpB, generating pT7BT/PpsbD::TatpB. The 0.75-kb HindIII-EcoRI fragment of pT7BT/PpsbD::TatpB was subcloned into the HindIII/EcoRI-digested pHSG398 vector (Takara, Kusatsu, Japan), generating pHSG/PpsbD::TatpB. The 1.66-kb NdeI-XbaI fragment of pCR2.1-TOPO/lucCP was ligated to NdeI/XbaI-digested pHSG/PpsbD::TatpB, generating pHSG/PpsbD::lucCP::TatpB. Similarly, the 0.87-kb SmaI-XbaI fragment of pT7BT/PtufA was subcloned into pT7BT/TatpB to obtain pT7BT/PtufA::TatpB, and then the 1.2-kb HindIII-EcoRI fragment of pT7BT/PtufA::TatpB was subcloned into the pHSG398, generating pHSG/PtufA::TatpB. The 1.66-kb NdeI-XbaI fragment of pCR2.1-TOPO/lucCP was inserted into NdeI/XbaI-digested pHSG/PtufA::TatpB, generating pHSG/PtufA::lucCP::TatpB.
We constructed reporter vectors pCL208 and pCL218 as follows. A polylinker (top strand, 5′-CCCGGGATCCACTAGTCGACGCATGCAGATCTAGGCCTGCA-3′; bottom strand, 5′-GGCCTAGATCTGCATGCGTCGACTAGTGGATCCCGGGTGCA-3′) was inserted into the unique PstI site (nucleotide 79351; accession no. BK000554) of pBB21 (J. Minagawa, unpublished data), which contains the 6.3-kb StuI fragment of chloroplast genome (75838 to 82122; BK000554) in the opposite direction to the psbT gene, generating pCTS2. An aadA expression cassette that consists of the psbA 5′-flanking region, the aadA coding sequence, and the atpB 3′-terminator region (K. Onai and M. Ishiura, unpublished data) was inserted into the polylinker region (SpeI/SmaI) in the same direction as the psbT gene, generating pCTS2A. Another aadA cassette derived from pBD110 (45), containing the aadA cassette of pUC-atpX-AAD (13), was inserted into the polylinker region (SalI/SmaI) in the same direction as the psbT gene, generating pCTS2B. The 2.3-kb BamHI fragment of pHSG/PpsbD::lucCP::TatpB was inserted into the BglII site of the polylinker region of pCTS2A and pCTS2B in the opposite direction to the aadA cassette, generating pCL208 and pCL218, respectively.
We constructed reporter vector pCL302 as follows. The 5.8-kb HindIII-SacI fragment of pBD101 (J. Minagawa, unpublished data), containing the HindIII-PstI fragment of the chloroplast genome (nucleotides 172207 to 178021; accession no. BK000554), was subcloned into the HindIII/SacI-digested pUC118 vector, and then the BglII linker was inserted into the NdeI site (176052; BK000554) to obtain pCTS3. The 2.7-kb BamHI fragment of pHSG/PtufA::lucCP::TatpB was inserted into the BglII site in the same direction as the psbD gene, generating pCL302.
Strains and chloroplast transformation.
We used the following C. reinhardtii strains in this study: wild-type 2137 mt+ (CC-1021) and 137c mt+ (CC-125), the nonphotosynthetic mutant ΔD2-2 (a psbD deletion strain on the 2137 mt+ genetic background) (J. Minagawa, unpublished data), and the per-1 mt+ (CC-1117) and per-4 mt+ (CC-1119) rhythm mutant strains. The per mutants were obtained from the Chlamydomonas Genetic Center (Duke University, Durham, N.C.). These cells were maintained on Tris-acetate-phosphate (TAP) (14) or high-salt medium (HSM) (44) plates containing 1.5% agar (INA Food Industry, Nagano, Japan) at 24°C under constant light conditions (LL) (10 μmol m−2 s−1 from white fluorescence lamps). Chloroplast transformation with a particle gun was performed as described previously (27). For transformation with pCL208 or pCL218, we selected transformants for resistance to spectinomycin (100 μg/ml). For transformation with pCL302, we used the ΔD2-2 strain as the recipient and selected transformants for restored photosynthetic growth on HSM.
Automated continuous culture of C. reinhardtii.
Cells were grown in TAP medium with aeration at 24°C under LL (30 μmol m−2 s−1) until the optical density at 750 nm reached 0.35 (1.7 × 106 cells/ml), and then luciferin potassium salt (Biosynth, Staad, Switzerland) was added to a final concentration of 100 μM. To maintain a constant cell density, we developed a computer-controlled system that continuously monitored the optical density of cultures with an optical sensor (E3SA-DS50C43A; Omron, Kyoto, Japan) and automatically diluted the cultures with fresh medium containing luciferin to the preset concentration (K. Okamoto and M. Ishiura, unpublished data). Using this system, we maintained the optical density at 750 nm of cultures at 0.33 to 0.35 throughout the experimental period. Cultures were automatically sampled every hour with a peristaltic pump, and bioluminescence of the samples was measured with a photomultiplier tube detector (H7360-01; Hamamatu Photonics, Hamamatu, Japan).
Southern and Northern blot analyses.
For Southern blot analysis, total genomic DNA samples were digested with restriction endonucleases, electrophoresed, and then blotted onto a nylon membrane (GeneScreen Plus; NEN). Using a random primer labeling kit (Prime-It II; Stratagene), we generated 32P-radiolabeled probes from a DNA fragment of an intergenic region between psbT and psbN (nucleotides 78658 to 79007; BK000554) for pCL208 transformants and from that of the 5′ region of ORF2971 (177559 to 178017; BK000554) for pCL302 transformants.
For Northern blot analysis, total RNA was extracted with TRIzol reagent (Invitrogen). Cell pellets were lysed in TRIzol by vortexing for 1 min. After phenol-chloroform extraction and ethanol precipitation, 10-μg samples of total RNA were electrophoresed in a denaturing agarose gel and blotted onto a nylon membrane (Biodyne A; Pall). We generated 32P-radiolabeled probes from DNA fragments of psbD (nucleotides 174701 to 175486; BK000554) and lucCP (160 to 1657; AB190814) as described above. The radioactivity of each hybridization signal was quantified with the BAS2000 system (Fujifilm, Tokyo, Japan).
In vitro luciferase assay.
Cell pellets were lysed in Glo lysis buffer (Promega) by pipetting and vortexing for 5 min. Insoluble debris was pelleted by centrifugation, and supernatant samples (25 ng total protein) were subjected to the Steady-Glo luciferase assay system (Promega). Luminescence was measured with automated bioluminescence-monitoring apparatuses (33, 35).
Bioluminescence monitoring using 96-well microtiter plates.
Cells were grown under LL (10 μmol m−2 s−1) at 24°C for 5 days on TAP plates or for 2 to 3 weeks on HSM plates. Agar blocks with C. reinhardtii colonies were punched out from the plates with a glass tube (6-mm inner diameter; 10 to 20 colonies on the block), and one block was transferred into each well in 96-well microtiter plates. Luciferin was added to the wells at a final concentration of 200 μM. Before bioluminescence monitoring, the circadian clocks of the cells were synchronized by exposure to a 12-hour light (30 μmol m−2 s−1)/12-hour dark (LD) cycle. Bioluminescence was monitored under the LD cycle, LL (30 μmol m−2 s−1), and constant darkness (DD) with our bioluminescence-monitoring apparatuses (33, 35). Bioluminescence was measured for 5 seconds every hour in darkness. For light phase measurements, cells were first subjected to darkness for 210 seconds at each time point to decrease the delayed light emission of chlorophyll. Period lengths and phases of bioluminescence rhythms were estimated by the cosinor method or the visual inspection-peak method using the RAP program (34).
RESULTS
Construction of a chloroplast luciferase gene and reporter strains.
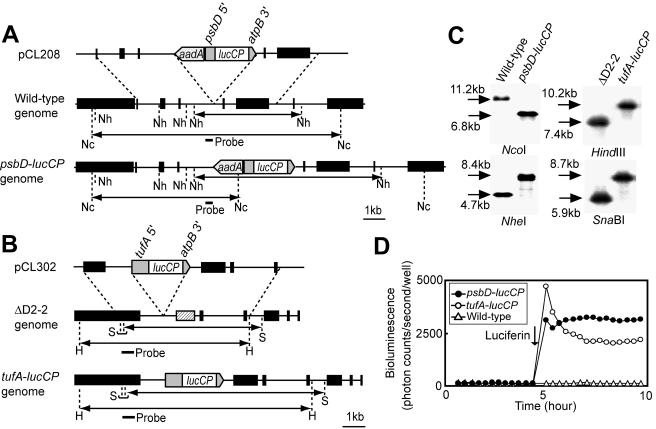
Since codon usage in the C. reinhardtii chloroplast is highly biased toward preferred codons (32), we synthesized a codon-optimized firefly luciferase gene for the C. reinhardtii chloroplast (lucCP). The amino acid sequence for lucCP is identical to that for the luc+ gene (Promega), but all codons are the most frequently used codons in the C. reinhardtii chloroplast (32) (Fig. 1). To express lucCP in the chloroplast, we generated two reporter strains by transformation of the chloroplast genome. The psbD-lucCP strain, a transformant of the wild-type 2137 mt+ strain with the reporter vector pCL208, has a lucCP expression cassette that consists of the promoter and 5′ untranslated region (5′-UTR) of psbD, the lucCP coding region, and the 3′-UTR of atpB. The cassette was located between the psbN and psbT genes in the chloroplast genome (Fig. 2A). The tufA-lucCP strain, a transformant of the ΔD2-2 strain with pCL302, has a lucCP cassette that consists of the promoter and 5′-UTR of tufA, the lucCP coding region, and the 3′-UTR of atpB, located between the ORF2971 and psbD genes (Fig. 2B). Southern blot analysis of these strains detected fragments of the expected sizes (Fig. 2C). No fragments of the wild-type or ΔD2-2 size were detected in the lanes of reporter strains. In the absence of luciferin, no significant luminescence was detected in the psbD-lucCP, tufA-lucCP, and wild-type strains (Fig. 2D). Once luciferin was added to the medium, we observed a dramatic increase in luminescence only in the reporter strains (Fig. 2D). These results demonstrate that the psbD-lucCP and tufA-lucCP strains were homoplasmic for the correctly integrated lucCP expression cassette and that a functional luciferase protein was being expressed.
FIG. 1.
Nucleotide sequence of the lucCP coding region. Nucleotides that were changed from those of the original firefly luciferase gene are denoted by black boxes.
FIG. 2.
Construction of bioluminescence reporter strains. (A) Schematic representation of the reporter construct and the chloroplast genomes of psbD-lucCP and wild-type strains. psbD 5′ represents the promoter and 5′-UTR of the psbD gene; atpB 3′ represents the 3′-UTR of the atpB gene. The black boxes of the chloroplast genomes denote genes: from left to right, wendy, trnE2, psbH, psbN, psbT, psbB, trnD, and rpoA. The small bar indicates the location of the probe used for Southern blot analysis. The double-headed arrows indicate fragments expected to be detected by Southern blot analysis. Restriction sites: Nc, NcoI; Nh, NheI. (B) Schematic representation of the reporter construct and the chloroplast genomes of tufA-lucCP and ΔD2-2 strains. tufA 5′ represents the promoter and 5′-UTR of the tufA gene. The black boxes denote genes: from left to right, ORF2971, psbD (replaced with the 483-bp repeats [12] in the ΔD2-2 genome [hatched box]), psaA exon 2, psbJ, atpI, psaJ, and rps12. Restriction sites: H, HindIII; S, SnaBI. (C) Southern blot analysis of genomic DNAs. Genomic DNAs digested with the restriction enzyme were hybridized with the probes indicated in panels A and B. Sizes of detected bands are indicated. (D) Representative traces of bioluminescence from the wild-type and reporter strains. One hundred microliters of mid-log-phase cultures grown in TAP medium was transferred into individual wells of a 96-well microtiter plate, and bioluminescence was measured every 20 min with the automated bioluminescence-monitoring apparatus. The arrow indicates when luciferin was added (final concentration, 100 μM).
Circadian rhythms in lucCP gene expression and bioluminescence.
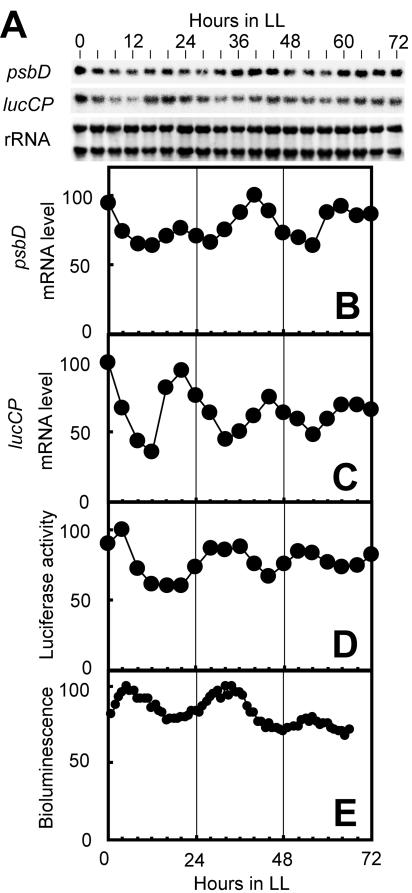
We examined circadian fluctuation in both lucCP gene expression and bioluminescence under LL. Using a continuous culture of the psbD-lucCP strain, we measured the levels of psbD transcript, lucCP transcript, luciferase activity, and bioluminescence in identical cultures. The level of native psbD gene transcript oscillated in a circadian manner (Fig. 3A and B). The level of the ∼1.8-kb lucCP transcript also exhibited a circadian oscillation (Fig. 3A and C). To assess the intracellular level of the lucCP gene product, we extracted total protein from the cells and measured luciferase activity in vitro. The activity exhibited a circadian oscillation, but the phase was delayed with respect to that of the lucCP transcript by 8 to 12 h (Fig. 3C and D). This implies the existence of a regulation mechanism at the posttranscriptional level. In vivo bioluminescence oscillated in a circadian manner that correlated well with the luciferase activity determined by in vitro assay (Fig. 3D and E). These results demonstrate that the psbD-lucCP strain exhibited an endogenous oscillation of bioluminescence that correlated well with the circadian fluctuation of the intracellular level of lucCP gene product.
FIG. 3.
Circadian rhythms of lucCP gene expression and bioluminescence. A continuous culture of the psbD-lucCP strain was synchronized by exposure to 12 h of darkness, and the culture was sampled under LL every 4 h for RNA and protein analysis and every hour for bioluminescence measurement. (A) Northern blot analyses of psbD and lucCP. RNA was stained with methylene blue, and the stained rRNA bands are shown as loading references. (B to E) The graphs show the temporal patterns of the psbD transcript (B), the lucCP transcript (C), luciferase activity determined by an in vitro assay (D), and in vivo bioluminescence (E). The maximum values were adjusted to 100. Similar results were obtained in two independent experiments.
Real-time monitoring of bioluminescence rhythms in a 96-well plate format.
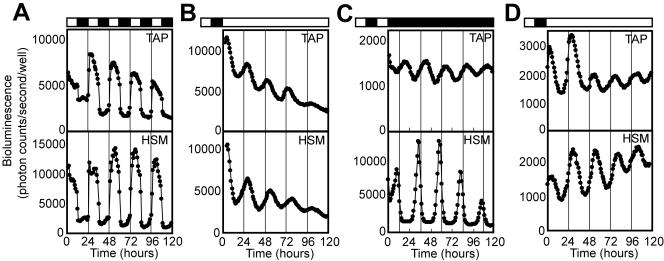
To analyze a large number of samples in a single assay, we examined whether the bioluminescence rhythm assay was adaptable to a 96-well plate format. Colonies of the psbD-lucCP strain grown on TAP or HSM agar plates were transferred into each well of 96-well microtiter plates, and the bioluminescence was monitored. The colonies exhibited a robust diurnal bioluminescence rhythm under LD (high during the light phase and low during the dark phase) on both media (Fig. 4A). Under LL condition, they exhibited clear bioluminescence oscillations that peaked around the middle of the subjective day, with period lengths of 24.0 ± 0.9 h (mean ± standard deviation; n = 129) on TAP agar (Fig. 4B, top), and 24.8 ± 0.6 h (n = 44) on HSM agar (Fig. 4B, bottom). The rhythm on TAP agar was similar in phase and amplitude to the bioluminescence rhythms observed under continuous culture conditions in TAP medium (Fig. 3E). Under DD condition on TAP agar, bioluminescence was relatively low, but the strain exhibited bioluminescence rhythms with a period length of 23.5 ± 0.3 h (n = 34) (Fig. 4C, top). On HSM agar under DD, the strain exhibited high-amplitude bioluminescence rhythms with sharp peaks at the transition of subjective night to subjective day, with a period of 24.1 ± 0.2 h (n = 33) (Fig. 4C, bottom). Interestingly, the phase and amplitude of the rhythm under this condition were distinctly different from those under LL and under DD on TAP (Fig. 4B and C top), suggesting a different circadian regulation of chloroplast gene expression. The tufA-lucCP strain also exhibited bioluminescence rhythms under LL conditions, with periods of 24.2 ± 0.4 h (n = 21) on TAP agar and 24.7 ± 0.7 h (n = 55) on HSM agar (Fig. 4D). These results demonstrate that the bioluminescence rhythm assay of these reporter strains could be adapted to a 96-well plate format under various culture conditions.
FIG. 4.
Real-time monitoring of circadian rhythms in a 96-well plate format. (A to C) Representative bioluminescence rhythms of the psbD-lucCP strain monitored under LD cycles (A), LL (B), and DD (C) at 24°C. (D) Representative bioluminescence rhythms of the tufA-lucCP strain monitored under LL at 24°C. Lighting conditions are indicated above the graphs: White bar, light period; black bar, dark period. The thin vertical lines mark the time of light onset (A) or that of the LD cycle preceding measurement (B to D).
Phase resetting and temperature compensation of bioluminescence rhythms.
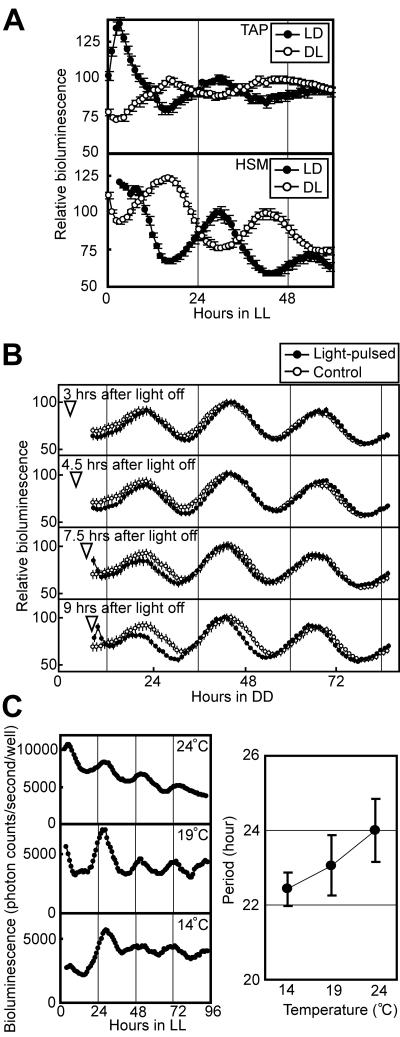
We examined whether the bioluminescence rhythms of the Chlamydomonas chloroplast satisfied the fundamental criteria for circadian rhythms (i.e., persistence under constant conditions, phase resetting by external stimuli, and temperature compensation). Persistence under constant conditions was confirmed as described above (Fig. 3E, 4B, 4C, and 4D). To evaluate phase resetting by light/dark cues, we exposed colonies of the psbD-lucCP strain to LD or a 12-hour dark/12-hour light cycle (DL) and monitored bioluminescence under LL. The bioluminescence from both cultures oscillated with opposite phases that depended upon the prior light/dark phase (Fig. 5A). Furthermore, we examined whether a brief light pulse can shift the phase of the bioluminescence rhythm. Light pulses given at the early subjective night (3 or 4.5 h after light off) delayed the phases of bioluminescence rhythms (by 1.3 or 1.1 h) (Fig. 5B), and light pulses given at the late subjective night (7.5 or 9 h after light off) advanced them (by 1.0 or 2.3 h) (Fig. 5B). These results indicate that the phase of bioluminescence rhythm can be reset by light/dark cues. Next, to evaluate temperature compensation, we measured the period lengths of bioluminescence rhythms at 14°C (n = 41), 19°C (n = 34), and 24°C (n = 129). The period lengths were 22.4 ± 0.4 h, 23.1 ± 0.8 h, and 24.0 ± 0.9 h, respectively (Fig. 5C). The Q10 value for rhythm frequency (1/period) was 0.93, indicating that these rhythms were temperature compensated. Thus, these results suggest that the oscillation of chloroplast gene expression is a bona fide circadian rhythm. In addition, the fact that the Q10 value was less than 1 means that the rhythm runs slightly faster at lower temperatures, as do other rhythms observed in this alga (e.g., phototaxis, stickiness to glass, cell division cycle, and nuclear gene expression) (6, 15, 21, 43).
FIG. 5.
Phase resetting and temperature compensation of the period length of bioluminescence rhythms. (A) Phase resetting of the bioluminescence rhythm. The bioluminescence rhythm of the psbD- lucCP strain exposed to LD or DL was monitored under LL at 24°C. Data points and bars represent means ± standard deviations of the bioluminescence levels of 12 to 20 replicate samples. For precise phase comparisons, the maximum values of the bioluminescence on the second day were adjusted to 100, and the lower portions of the vertical axes were omitted. (B) Phase shifting of the bioluminescence rhythm. The psbD-lucCP strain on TAP agar was exposed to LD and transferred to DD at 24°C, and then a 15-minute light pulse (30 μmol m−2 s−1) was given at 3, 4.5, 7.5, or 9 h after light off (arrow heads). For phase comparisons, the maximum values on the second day were adjusted to 100, and the traces of the control without a light pulse are shown. (C) Temperature compensation of the bioluminescence rhythm. The bioluminescence rhythm of the psbD-lucCP strain on TAP agar was monitored under LL at three different temperatures. The left panel shows representative rhythms. In the right panel, data points and bars represent means ± standard deviations of the period lengths of the rhythms.
Chloroplast gene expression rhythms in per mutants.
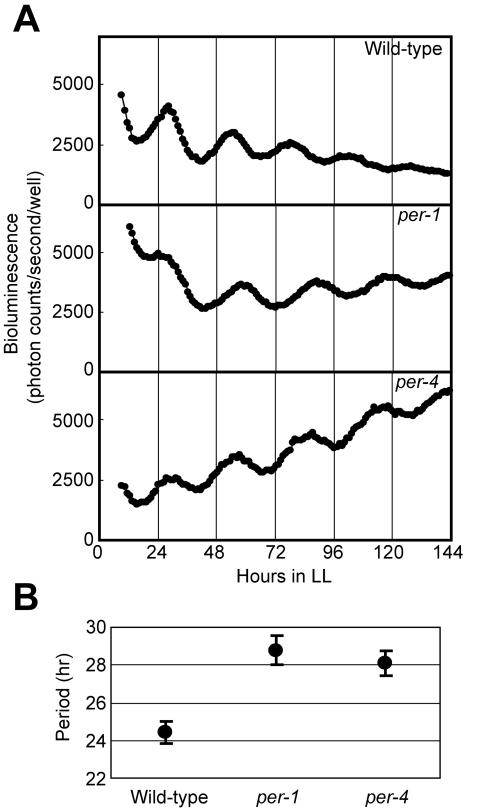
The per-1 and per-4 mutants of Chlamydomonas exhibit a lengthened period (27 to 28 h) in phototactic circadian rhythm, and each of them has a long-period gene in its nuclear genome but at different loci (7). We examined whether per mutations affect chloroplast gene expression rhythms by transforming the per-1 and per-4 mutants with the reporter vector pCL218. The per-1 mutant exhibited bioluminescence rhythms with significantly longer periods than the wild-type 137c strain (per-1, 28.8 ± 0.8 h [n = 127]; 137c, 24.4 ± 0.6 h [n = 36] [P < 0.001 by Student's t test]) (Fig. 6A and B). The per-4 mutant also exhibited bioluminescence rhythms with longer periods (28.1 ± 0.7 h; n = 20 [P < 0.001]) (Fig. 6A and B). These results demonstrate that the period length of bioluminescence rhythms depended upon the genotype of the nucleus, thus suggesting that the circadian rhythmicity of chloroplast gene expression rhythm is under the control of the nucleus-encoded circadian oscillator.
FIG. 6.
Bioluminescence rhythms of per mutants. (A) Representative bioluminescence rhythms of the wild-type, per-1, and per-4 strains. The bioluminescence rhythms were monitored under LL at 17°C on HSM agar. (B) Period length of bioluminescence rhythm in per mutants. Data points and bars represent means ± standard deviations.
DISCUSSION
We developed reporter strains of C. reinhardtii for real-time monitoring of circadian gene expression in the chloroplast. We demonstrated that the bioluminescence rhythm of the reporter strain correlated well with the circadian expression pattern of the chloroplast lucCP gene and satisfied the three criteria for circadian rhythms. This is the first demonstration that the period length of the chloroplast gene expression rhythm is temperature compensated. Furthermore, we demonstrated that the period length of the bioluminescence rhythm depended upon the genotype of the nucleus.
We thus provide direct evidence that the circadian period of chloroplast gene expression rhythm is determined by the nucleus-encoded circadian oscillator. Our results emphasize the necessity of mediators linking the nuclear clock with the chloroplast gene expression system. However, we cannot exclude the possibility of the existence of a chloroplast-specific clock. If such a clock exists, it should be a “slave oscillator,” that is, under the control of the nuclear “master oscillator,” at least under our experimental conditions. The intracellular desynchronization of circadian oscillators has been demonstrated for the dinoflagellate Gonyaulux (38, 49). It will be of interest to examine whether the chloroplast bioluminescence rhythm and other rhythms observed in Chlamydomonas (e.g., phototaxis, cell division cycle, stickiness to glass, and chemotaxis) (5, 9, 15, 43) can be desynchronized under specific conditions.
Surprisingly, the psbD-lucCP reporter strain exhibited a robust bioluminescence rhythm with sharp peaks and very low baseline even under DD on HSM agar (Fig. 4C, bottom). Under this condition, there are no available external energy sources, and thus cells must use their starch in stock to maintain their cellular functions. At present, we cannot exclude the possibility that the rhythm does not reflect lucCP gene expression level, because we have not examined the LucCP protein level under this condition. However, if the rhythm reflects the gene expression, a possible explanation for the bioluminescence pattern is that in order to minimize cellular energy consumption for cell survival under this condition, the cell may limit chloroplast gene expression to the most efficient time zone for preparation of the photosynthetic apparatus (i.e., immediately before dawn).
For C. reinhardtii, a wide range of molecular genetic approaches, including gene tagging by insertional mutagenesis and complementation cloning by using an indexed genomic library, are now available (47, 51). Our bioluminescence reporter strains will enable the same forward genetic approach to the clock components of the alga as was used for Arabidopsis (26, 36) and cyanobacteria (20). Until now, the molecular mechanisms of the circadian oscillators have been studied with Drosophila, mice, Neurospora, cyanobacteria, and Arabidopsis. Although oscillator mechanisms are conserved through evolution, actual clock components do not seem to be (11, 17). Database searches of the draft sequence of the entire C. reinhardtii genome did not identify any genes homologous to the key components of the circadian oscillator in other species (e.g., period, timeless, frequency, and kaiC) (28). Thus, C. reinhardtii seems to have its own clock components. Understanding the algal circadian oscillator will provide new insights into the evolution of the circadian clock.
Acknowledgments
We thank Kosuke Shimogawara (Teikyo University) for providing the wild-type 137c strain, Hiroshi Hashimoto and Genzo Ito (Hokkaido System Science Co., Ltd.) for synthesis of the lucCP gene, Shingen Yamamoto (Nagoya University) for assistance in the early phase of this project, Syuzo Ishikawa and Chiyomi Miwa (Nagoya University) for advice on automated continuous culture, and Miriam Bloom (SciWrite Biomedical Writing and Editing Services) for professional editing.
This work was supported by grants to M.I. from the Japanese Ministry of Education, Culture, Sports, Science and Technology (MEXT); the Program for Promotion of Basic Research Activities for Innovative Biosciences (BRAIN); Research for the Future Novel Gene Function Involved in Higher-Order Regulation of Nutrition-Storage in Plants (Japan Society for the Promotion of Science); Ground-Based Research for Space Utilization (Japan Space Forum); and the Promoting Cooperative Research Project (Aichi Science and Technology Foundation) and by grants to T.M. from Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists. The Division of Biological Science, Graduate School of Science, Nagoya University, was supported by a 21st Century COE grant from MEXT.
REFERENCES
- 1.Alberts, B., A. Johnson, J. Lewis, M. Raff, K. Roberts, and P. Walter. 2002. Molecular biology of the cell, 4th ed. Garland Science, New York, N.Y.
- 2.Aoki, S., S. Kato, K. Ichikawa, and M. Shimizu. 2004. Circadian expression of the PpLhcb2 gene encoding a major light-harvesting chlorophyll a/b-binding protein in the moss Physcomitrella patens. Plant Cell Physiol. 45:68-76. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharya, D., and L. Medlin. 1998. Algal phylogeny and the origin of land plants. Plant Physiol. 116:9-15. [Google Scholar]
- 4.Brandes, C., J. D. Plautz, R. Stanewsky, C. F. Jamison, M. Straume, K. V. Wood, S. A. Kay, and J. C. Hall. 1996. Novel features of Drosophila period transcription revealed by real-time luciferase reporting. Neuron 16:687-692. [DOI] [PubMed] [Google Scholar]
- 5.Bruce, V. G. 1970. The biological clock in Chlamydomonas reinhardi. J. Protozool. 17:328-334. [Google Scholar]
- 6.Bruce, V. G. 1972. Mutants of the biological clock in Chlamydomonas reinhardi. Genetics 70:537-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruce, V. G. 1974. Recombinants between clock mutants of Chlamydomonas reinhardi. Genetics 77:221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bünning. 1973. The physiological clock: circadian rhythms and biological chronometry, 3rd ed. Springer-Verlag, New York, N.Y.
- 9.Byrne, T. E., M. R. Wells, and C. H. Johnson. 1992. Circadian rhythms of chemotaxis to ammonium and of methyammonium uptake in Chlamydomonas. Plant Physiol. 98:876-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carter, M. L., A. C. Smith, H. Kobayashi, S. Purton, and D. L. Herrin. 2004. Structure, circadian regulation and bioinformatic analysis of the unique sigma factor gene in Chlamydomonas reinhardtii. Photosynth. Res. 82:339-349. [DOI] [PubMed] [Google Scholar]
- 11.Dunlap, J. C. 1999. Molecular bases for circadian clocks. Cell 96:271-290. [DOI] [PubMed] [Google Scholar]
- 12.Fischer, N., O. Stampacchia, K. Redding, and J. D. Rochaix. 1996. Selectable marker recycling in the chloroplast. Mol. Gen. Genet. 251:373-380. [DOI] [PubMed] [Google Scholar]
- 13.Goldschmidt-Clermont, M. 1991. Transgenic expression of aminoglycoside adenine transferase in the chloroplast: a selectable marker of site-directed transformation of chlamydomonas. Nucleic Acids Res. 19:4083-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorman, D. S., and R. P. Levine. 1965. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA 54:1665-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goto, K., and C. H. Johnson. 1995. Is the cell division cycle gated by a circadian clock? The case of Chlamydomonas reinhardtii. J. Cell Biol. 129:1061-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harmer, S. L., J. B. Hogenesch, M. Straume, H. S. Chang, B. Han, T. Zhu, X. Wang, J. A. Kreps, and S. A. Kay. 2000. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290:2110-2113. [DOI] [PubMed] [Google Scholar]
- 17.Harmer, S. L., S. Panda, and S. A. Kay. 2001. Molecular bases of circadian rhythms. Annu. Rev. Cell Dev. Biol. 17:215-253. [DOI] [PubMed] [Google Scholar]
- 18.Hwang, S., R. Kawazoe, and D. L. Herrin. 1996. Transcription of tufA and other chloroplast-encoded genes is controlled by a circadian clock in Chlamydomonas. Proc. Natl. Acad. Sci. USA 93:996-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ichikawa, K., M. Sugita, T. Imaizumi, M. Wada, and S. Aoki. 2004. Differential expression on a daily basis of plastid sigma factor genes from the moss Physcomitrella patens. Regulatory interactions among PpSig5, the circadian clock, and blue light signaling mediated by cryptochromes. Plant Physiol. 136:4285-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishiura, M., S. Kutsuna, S. Aoki, H. Iwasaki, C. R. Andersson, A. Tanabe, S. S. Golden, C. H. Johnson, and T. Kondo. 1998. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 281:1519-1523. [DOI] [PubMed] [Google Scholar]
- 21.Jacobshagen, S., and C. H. Johnson. 1994. Circadian rhythms of gene expression in Chlamydomonas reinhardtii: circadian cycling of mRNA abundances of cab II, and possibly of β-tubulin and cytochrome c. Eur. J. Cell Biol. 64:142-152. [PubMed] [Google Scholar]
- 22.Kawazoe, R., S. Hwang, and D. L. Herrin. 2000. Requirement for cytoplasmic protein synthesis during circadian peaks of transcription of chloroplast-encoded genes in Chlamydomonas. Plant Mol. Biol. 44:699-709. [DOI] [PubMed] [Google Scholar]
- 23.Kucho, K., K. Okamoto, S. Tabata, H. Fukuzawa, and M. Ishiura. 2005. Identification of novel clock-controlled genes by cDNA macroarray analysis in Chlamydomonas reinhardtii. Plant Mol. Biol. 57:889-906. [DOI] [PubMed] [Google Scholar]
- 24.Leu, S., D. White, and A. Michaels. 1990. Cell cycle-dependent transcriptional and post-transcriptional regulation of chloroplast gene expression in Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1049:311-317. [DOI] [PubMed] [Google Scholar]
- 25.Mayfield, S. P., and J. Schultz. 2004. Development of a luciferase reporter gene, luxCt, for Chlamydomonas reinhardtii chloroplast. Plant J. 37:449-458. [DOI] [PubMed] [Google Scholar]
- 26.Millar, A. J., I. A. Carre, C. A. Strayer, N. H. Chua, and S. A. Kay. 1995. Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267:1091-1092. [DOI] [PubMed] [Google Scholar]
- 27.Minagawa, J., and A. R. Crofts. 1994. A robust protocol for site-directed mutagenesis of the D1 protein in Chlamydomonas reinhardtii: a PCR-spliced psbA gene in a plasmid conferring spectinomycin resistance was introduced into a psbA deletion strain. Photosynth. Res. 42:121-131. [DOI] [PubMed] [Google Scholar]
- 28.Mittag, M., S. Kiaulehn, and C. H. Johnson. 2005. The circadian clock in Chlamydomonas reinhardtii. What is it for? What is it similar to? Plant Physiol. 137:399-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgan, L. W., A. V. Greene, and D. Bell-Pedersen. 2003. Circadian and light-induced expression of luciferase in Neurospora crassa. Fungal Genet. Biol. 38:327-332. [DOI] [PubMed] [Google Scholar]
- 30.Morikawa, K., S. Ito, Y. Tsunoyama, Y. Nakahira, T. Shiina, and Y. Toyoshima. 1999. Circadian-regulated expression of a nuclear-encoded plastid sigma factor gene (sigA) in wheat seedlings. FEBS Lett. 451:275-278. [DOI] [PubMed] [Google Scholar]
- 31.Nakahira, Y., K. Baba, A. Yoneda, T. Shiina, and Y. Toyoshima. 1998. Circadian-regulated transcription of the psbD light-responsive promoter in wheat chloroplasts. Plant Physiol. 118:1079-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura, Y., T. Gojobori, and T. Ikemura. 2000. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okamoto, K., K. Onai, T. Furusawa, and M. Ishiura. 2005. A portable integrated automatic apparatus for the real-time monitoring of bioluminescence in plants. Plant Cell Environ. 28:1305-1315. [Google Scholar]
- 34.Okamoto, K., K. Onai, and M. Ishiura. 2005. RAP, an integrated program for monitoring bioluminescence and analyzing circadian rhythms in real time. Anal. Biochem. 240:193-200. [DOI] [PubMed] [Google Scholar]
- 35.Okamoto, K., K. Onai, N. Ezaki, T. Ofuchi, and M. Ishiura. 2005. An automated apparatus for the real-time monitoring of bioluminescence in plants. Anal. Biochem. 340:187-192. [DOI] [PubMed] [Google Scholar]
- 36.Onai, K., K. Okamoto, H. Nishimoto, C. Morioka, M. Hirano, N. Kami-Ike, and M. Ishiura. 2004. Large-scale screening of Arabidopsis circadian clock mutants by a high-throughput real-time bioluminescence monitoring system. Plant J. 40:1-11. [DOI] [PubMed] [Google Scholar]
- 37.Panda, S., M. P. Antoch, B. H. Miller, A. I. Su, A. B. Schook, M. Straume, P. G. Schultz, S. A. Kay, J. S. Takahashi, and J. B. Hogenesch. 2002. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109:307-3020. [DOI] [PubMed] [Google Scholar]
- 38.Roenneberg, T., and D. Morse. 1993. Two circadian oscillators in one cell. Nature 362:362-364. [DOI] [PubMed] [Google Scholar]
- 39.Salvador, M. L., U. Klein, and L. Bogorad. 1993. Light-regulated and endogenous fluctuations of chloroplast transcript levels in Chlamydomonas. Regulation by transcription and RNA degradation. Plant J. 3:213-219. [DOI] [PubMed] [Google Scholar]
- 40.Schmitz, O., M. Katayama, S. B. Williams, T. Kondo, and S. S. Golden. 2000. CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science 289:765-768. [DOI] [PubMed] [Google Scholar]
- 41.Schweiger, E., H. G. Wallraff, and H. G. Schweiger. 1964. Endogenous circadian rhythm in cytoplasm of Acetabularia: influence of the nucleus. Science 146:658-659. [DOI] [PubMed] [Google Scholar]
- 42.Stemmer, W. P., A. Crameri, K. D. Ha, T. M. Brennan, and H. L. Heyneker. 1995. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene 164:49-53. [DOI] [PubMed] [Google Scholar]
- 43.Straley, S. C., and V. G. Bruce. 1979. Stickiness to glass. Circadian changes in the cell surface of Chlamydomonas reinhardtii. Plant Physiol. 63:1175-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sueoka, N. 1960. Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 46:83-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sugiura, M., Y. Inoue, and J. Minagawa. 1998. Rapid and discrete isolation of oxygen-evolving His-tagged photosystem II core complex from Chlamydomonas reinhardtii by Ni2+ affinity column chromatography. FEBS Lett. 426:140-144. [DOI] [PubMed] [Google Scholar]
- 46.Sweeney, B. M., and F. T. Haxo. 1961. Persistence of a photosynthetic rhythm in enucleated Acetabularia. Science 134:1361-1363. [DOI] [PubMed] [Google Scholar]
- 47.Tam, L. W., and P. A. Lefebvre. 1993. Cloning of flagellar genes in Chlamydomonas reinhardtii by DNA insertional mutagenesis. Genetics 135:375-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vallone, D., S. B. Gondi, D. Whitmore, and N. S. Foulkes. 2004. E-box function in a period gene repressed by light. Proc. Natl. Acad. Sci. USA 101:4106-4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von der Heyde, F., A. Wilkens, and L. Rensing. 1992. The effects of temperature on the circadian rhythms of flashing and glow in Gonyaulax polyedra: are the two rhythms controlled by two oscillators? J. Biol. Rhythms 7:115-123. [DOI] [PubMed] [Google Scholar]
- 50.Yamazaki, S., R. Numano, M. Abe, A. Hida, R. Takahashi, M. Ueda, G. D. Block, Y. Sakaki, M. Menaker, and H. Tei. 2000. Resetting central and peripheral circadian oscillators in transgenic rats. Science 288:682-685. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, H., P. L. Herman, and D. P. Weeks. 1994. Gene isolation through genomic complementation using an indexed library of Chlamydomonas reinhardtii DNA. Plant Mol. Biol. 24:663-672. [DOI] [PubMed] [Google Scholar]