Abstract
Homologous recombination is a versatile DNA damage repair pathway requiring Rad51 and Rad54. Here we show that a mammalian Rad54 paralog, Rad54B, displays physical and functional interactions with Rad51 and DNA that are similar to those of Rad54. While ablation of Rad54 in mouse embryonic stem (ES) cells leads to a mild reduction in homologous recombination efficiency, the absence of Rad54B has little effect. However, the absence of both Rad54 and Rad54B dramatically reduces homologous recombination efficiency. Furthermore, we show that Rad54B protects ES cells from ionizing radiation and the interstrand DNA cross-linking agent mitomycin C. Interestingly, at the ES cell level the paralogs do not display an additive or synergic interaction with respect to mitomycin C sensitivity, yet animals lacking both Rad54 and Rad54B are dramatically sensitized to mitomycin C compared to either single mutant. This suggests that the paralogs possibly function in a tissue-specific manner. Finally, we show that Rad54, but not Rad54B, is needed for a normal distribution of Rad51 on meiotic chromosomes. Thus, even though the paralogs have similar biochemical properties, genetic analysis in mice uncovered their nonoverlapping roles.
DNA double-strand breaks (DSBs) are among a plethora of lesions that threaten the integrity of the genome. If not properly processed, DSBs can lead to cell cycle arrest or illegitimate DNA rearrangements such as translocations, inversions, or deletions. These rearrangements can contribute to cell dysfunction, cell death, or carcinogenesis (22). DSBs can arise through the action of exogenous DNA-damaging agents, but they also arise from endogenous sources, such as oxidative DNA damage and as a consequence of DNA replication (10, 22). Homologous recombination is a major DNA repair pathway by which DSBs are repaired. Homologous recombination is generally a precise way of resolving DSBs, because it uses homologous sequence, usually provided on the sister chromatid, as a repair template (54).
Homologous recombination is a complex process requiring a number of proteins of the RAD52 epistasis group, including Rad51 and Rad54. Rad51 is the key player in this process because it is critical for homology recognition and performs strand exchange between recombining DNA molecules. A pivotal intermediate in these reactions is the Rad51 nucleoprotein filament. This forms when Rad51 polymerizes on single-stranded DNA that results from DNA damage processing (54). Rad54 is an important accessory factor for Rad51 (56). A number of biochemical characteristics of Rad54 have been well defined for different species ranging from yeasts to humans (8, 18, 24, 31, 37, 38, 42, 47, 48, 53, 55, 59). Rad54 is a double-stranded-DNA-dependent ATPase that can translocate on DNA, thereby affecting DNA topology. Biochemically, Rad54 has been implicated in participation in multiple steps of homologous recombination. It can stabilize the Rad51 nucleoprotein filament in an early stage of recombination (30). At a subsequent stage it can promote chromatin remodeling (1, 2, 23) and stimulate Rad51-mediated formation of a joint molecule between the broken DNA and the repair template, referred to as a D loop (37). In later stages of the reaction it can displace Rad51 from DNA (49).
Cell biological experiments have revealed that Rad54 accumulates to form dynamic foci at sites of DNA damage (29, 55) that display rapid turnover of Rad54 (16). In those foci Rad54 colocalizes with and stabilizes Rad51 (55, 60). Chromatin immunoprecipitation experiments using Saccharomyces cerevisiae cells underscore the cooperation between Rad51 and Rad54 (50, 63). In the absence of Rad54, Rad51 is still able to pair homologous sequences, but the joint molecules are qualitatively different from those formed in the presence of Rad54 (50, 62).
Genetic analysis of RAD54 has been performed in a number of species including yeast and mice. S. cerevisiae cells with mutated RAD54 are DNA damage sensitive, including to ionizing radiation; are severely defective in gene conversion; and exhibit increased chromosome loss (21, 26, 43, 46). Mouse Rad54 knockout embryonic stem (ES) cells are ionizing radiation and mitomycin C sensitive, show reduced homologous recombination efficiency as measured by gene targeting, and display aberrant DSB repair (14, 15). Interestingly, while Rad54 knockout mice are sensitive to the interstrand DNA cross-linking agent mitomycin C, they are not ionizing radiation sensitive (17). The contribution of Rad54-mediated homologous recombination to repair of ionizing radiation-induced damage in adult animal is revealed when nonhomologous DNA end joining, an alternative and mechanistically distinct DSB repair pathway, is also impaired (9, 17, 32). A possible explanation for this observation is the existence of redundancy in Rad54 function in mammalian cells.
In S. cerevisiae, a RAD54 homolog, RDH54 (also know as TID1), has been identified (13, 27, 45). Its biochemical properties are similar to those of Rad54; for example, Rdh54 is an ATPase, and it stimulates D-loop formation by Rad51 (39). The phenotypes of rad54 and rdh54 mutants are distinct but do appear to be related to defects in homologous recombination. While rad54 mutants are sensitive to the alkylating agent methyl methanesulfonate, rdh54 mutants are not or are less so (27, 45). However, in the absence of RAD54, the contribution of RDH54 to cell survival is uncovered, because rad54 rdh54 double mutants are more sensitive to methyl methanesulfonate than either single mutant. While RAD54 affects both intra- and interchromosomal recombination, RDH54 seems to be more important for interchromosomal recombination than for intrachromosomal recombination (3, 27, 45). An interaction between the two genes has been found in meiosis. While sporulation efficiency and spore viability are reduced in the rad54 and rdh54 single mutants, these parameters are synergistically reduced in rad54 rdh54 double mutants, likely reflecting partial overlapping functions of RAD54 and RDH54 during meiotic recombination (44).
A RAD54 homolog, named RAD54B, has also been identified in human cells (33, 57). This gene has been labeled the mammalian homolog of yeast RDH54. However, this classification is based on amino acid sequence similarity and not on extensive functional analysis. Here we report the biochemical and genetic characterization of mammalian Rad54B. We show that mammalian Rad54B has biochemical properties similar to those of Rad54. However, the results of genetic experiments using Rad54 knockout, Rad54B knockout, and Rad54 Rad54B double knockout cells and mice suggests that mammalian Rad54B is unlikely to be the true S. cerevisiae RDH54 homolog because the genes are not functionally equivalent.
MATERIALS AND METHODS
Isolation of human and mouse RAD54B DNAs.
The hRAD54B cDNA was initially cloned as two fragments from a human testis cDNA library by PCR using the following primers: fragment 1, 5′-GCAGGGCCAGTGGTTTCTGTC and 5′-GTGGTCCTGATCAACAGTAAAT; fragment 2, 5′-ATTTACTGTTGATCAGGACCAC and 5′-GAAGAGCAATGGAATGTCAGAA. The two PCR fragments were digested with BclI, ligated together, and used as a template for amplification of the full-length hRAD54B cDNA with the primers 5′-CGGGATCCCATATGAGACGATCTGCAGCACC and 5′-CGGGATCCCCTATGTGCCAGTAGCTTGAG (BamHI sites are underlined, a NdeI site is italicized, and start and stop codons are in boldface). The PCR product was cloned into the BamHI site of pUC18 and sequenced. The 2,658-bp mouse Rad54B cDNA was isolated using a combination of reverse transcription-PCR with degenerate primers (25) and cDNA library screening.
Expression and purification of hRad54B.
The hRAD54B cDNA was subcloned from pUC18 into the BamHI site of pFastBac (Invitrogen). The hRad54B-pFastBac was transformed into DH10Bac cells, and the resulting bacmid was isolated and used to generate a recombinant hRad54B baculovirus in Sf9 insect cells. The virus was amplified by infecting 100 ml of Sf9 suspension culture at a multiplicity of infection of 0.1 for 48 h. To express hRad54B, Hi-Five insect cells were infected with the hRad54B recombinant virus at a multiplicity of infection of 10 and harvested after 72 h. Cells collected from 150 ml of insect cell culture were suspended in 20 ml cell breakage buffer (50 mM Tris-HCl, pH 7.5, 10% sucrose, 10 mM EDTA, 600 mM KCl, 1 mM dithiothreitol [DTT], and the protease inhibitors aprotinin, chymostatin, leupeptin, and pepstatin A [each at 5 μg/ml] and 1 mM phenylmethylsulfonyl fluoride) and lysed by passage through a French press. The lysate was clarified by centrifugation (100,000 × g), and the resulting supernatant was treated with ammonium sulfate (0.21 g/ml). The protein precipitate was harvested by centrifugation and redissolved in 20 ml K buffer (20 mM KH2PO4, pH 7.4, 10% glycerol, 0.5 mM EDTA, 0.01% Igepal, and 1 mM DTT). The protein solution, with a conductivity equivalent to 150 mM KCl, was passed in tandem over Q-Sepharose and SP-Sepharose columns (7 ml each), with hRad54B protein passing through the Q column but being retained on the SP column. To elute hRad54B, the SP column was developed with a 60-ml, 150 to 700 mM KCl gradient in K buffer. The hRad54B-containing fractions (350 mM KCl) were pooled, diluted with an equal volume of K buffer, and loaded onto a 5-ml macro-hydroxyapatite column, which was developed with a 60-ml, 0 to 300 mM KH2PO4 gradient in K buffer, with hRad54B eluting at 150 mM KH2PO4. The peak fractions were diluted with an equal volume of T buffer (25 mM Tris-HCl, pH 7.5, 10% glycerol, 0.5 mM EDTA, 0.01% Igepal, and 1 mM DTT) and applied to a 1-ml Mono S column, which was eluted with a 30-ml, 100 to 700 mM KCl gradient in K buffer, with hRad54B eluting at 350 mM KCl. Following the addition of 1/10 volume of 3 M KCl, the peak fractions were pooled and concentrated to 500 μl in a Centricon-30 microconcentrator (Millipore). The concentrated protein was loaded onto a 20-ml Sephacryl S-300 column preequilibrated with K buffer containing 500 mM KCl and eluted with the same buffer. The peak fractions were pooled, concentrated to 3 to 3.5 mg/ml, and stored in 3-μl portions at −80°C.
Generation of hRad54B antibodies.
The portion of hRad54B encompassing amino acid residues 13 to 395 was fused to glutathione S-transferase (GST) in the vector pGEX-3X. The GST-hRad54B fusion protein was expressed in Escherichia coli BL21(DE3) cells and purified from inclusion bodies by preparative sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to use as antigen for raising polyclonal antibodies in rabbits (Strategic Biosolutions). The same antigen was covalently coupled to cyanogen bromide-activated Sepharose 4B to use as an affinity matrix for purifying antibodies from the rabbit antisera, as described previously (52).
DNA substrates.
Topologically relaxed φX174 DNA was prepared by treatment with calf thymus topoisomerase I, as described previously (59), and pBluescript SK DNA was made in E. coli DH5α as described previously (38). The oligonucleotide used in the D-loop reaction is complementary to positions 1932 to 2022 of the pBluescript SK DNA and had the sequence 5′-AAATCAATCTAAAGTATATATGAGTAAACTTGGTCTGACAGTTACCAATGCTTAATCAGTGAGGCACCTATCTCAGCGATCTGTCTATTT-3′. This oligonucleotide was 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase to use in D-loop reactions.
Protein activity assays.
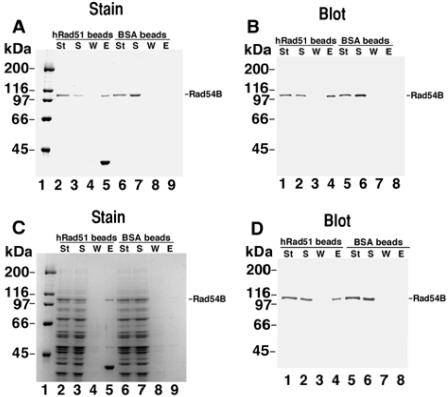
Binding of hRad54B to hRad51 was assessed with the use of immobilized hRad51 protein. hRad51 protein and bovine serum albumin (BSA) were coupled to Affi-Gel 15 beads according to the instructions of the manufacturer (Bio-Rad). The matrices contained 4 and 12 mg/ml of hRad51 and BSA, respectively. hRad54B (1.3 μg) was incubated with 5 μl of Affi-BSA or Affi-hRad51 at 4°C for 30 min in 30 μl of K buffer containing 50 mM KCl and 0.1% Trition X-100 with constant gentle tapping. This was followed by washing the beads twice with the same buffer (50 μl each time) before treating them with 30 μl of 2% SDS at 37°C for 5 min to elute bound hRad54B. For Fig. 1C, hRad54B (1.3 μg) was mixed with yeast extract containing 100 μg total protein in 30 μl of buffer and mixed with the hRad51 and BSA beads as described above. The various fractions (10 μl each) were analyzed by SDS-PAGE and Coomassie blue staining or immunoblotting for their hRad54B content. Note that in Fig. 1C, lane 5, some hRad51 protein is detected, which is probably due to the multimeric nature of hRad51. Since not all of the subunits in this multimeric structure become covalently conjugated to the Affi-Gel beads, the noncovalently linked subunits will be eluted by the SDS treatment. The amount of BSA covalently conjugated to the Affi-Gel beads was quantified by determining the protein coupling efficiency by SDS-PAGE.
FIG. 1.
Direct protein-protein interactions between human Rad54B and human Rad51. hRad51 protein or BSA was coupled to Affi-Gel beads, and binding of hRad54B was investigated. (A) The starting material (St), supernatant (S), wash (W), and eluate (E) from hRad51- or BSA-containing beads were separated in a denaturing gel and stained with Coomassie blue. Interaction of hRad54B and hRad51 is evident from the presence of hRad54B in lane 5 compared to its absence in lane 9. (B) Immunoblot of an experiment similar to that shown in panel A, probed with anti-hRad54B antibodies. (C) To assess the specificity of the interaction between hRad54B and hRad51, purified hRad54B was mixed with a protein extract from S. cerevisiae cells and the mixture was incubated with the hRad51- or BSA-containing beads. Human Rad54B was specifically retained on the hRad51-containing beads (lane 5). (D) An experiment similar to that shown in panel C, except the gel was probed with anti-hRad54B antibodies. Sizes of protein molecular mass markers are indicated.
DNA supercoiling and helix-opening activities of hRad54B were assessed as follows. The indicated amount of hRad54B was incubated with topologically relaxed φX174 double-stranded DNA (5 μM base pairs) for 2 min at 23°C in 11.8 μl of reaction buffer (50 mM Tris-HCl, pH 7.8, 3 mM MgCl2, 1 mM dithiothreitol, 2 mM ATP, and an ATP-regenerating system consisting of 10 mM creatine phosphate and 28 μg/ml creatine kinase). Following the addition of 200 ng of E. coli topoisomerase I in 0.7 μl, the reaction mixtures were incubated at 37°C for 10 min and then deproteinized by treatment with 0.5% SDS and proteinase K (0.5 mg/ml) for 10 min at 37°C. Reaction products were separated in a 1% agarose gel in TAE buffer (40 mM Tris-acetate, pH 7.4, containing 0.5 mM EDTA) and stained with ethidium bromide. The sensitivity of DNA to P1 nuclease was assayed in the same manner, except that E. coli topoisomerase I was replaced by 0.4 unit of P1 nuclease (Roche) and the DNA species were resolved in a 1% agarose gel containing 10 μM ethidium bromide. In some instances, the indicated amount of hRad51 was also included in the initial incubation period.
ATP hydrolysis activity of hRad54B was determined by incubating hRad54B protein (120 nM) with φX174 replicative-form I (RFI) DNA (23 μM base pairs) and 1.5 mM ATP with or without 300 nM yRad51 or hRad51 K133R in 10 μl of reaction buffer (20 mM Tris-HCl, pH 7.4, 25 mM KCl, 1 mM DTT, 4 mM MgCl2, 100 μg/ml BSA) at 30°C for the indicated times. The level of ATP hydrolysis was determined by thin-layer chromatography, as described previously (37).
Reactions to measure the effect of hRad54B on hRad51-mediated D-loop formation were performed as follows. hRad51 (800 nM) was incubated with the 5′-labeled 90-mer oligonucleotide (2.5 μM nucleotides) for 4 min at 37°C in 40 μl of reaction buffer (20 mM Tris-HCl, pH 7.4, 100 μg/ml BSA, 1 mM MgCl2, 2 mM ATP, and the ATP-generating system described above). This was followed by the addition of hRad54B (400 nM) in 4 μl and a 2-min incubation at 23°C. The reaction was completed by adding pBluescript SK replicative-form I DNA (190 μM base pairs) in 6 μl. The reaction mixtures were incubated at 30°C, and 6.3-μl aliquots were withdrawn at the indicated times, deproteinized, and run in 1% agarose gels in TAE buffer. The gels were dried, and the level of D loop was quantified by phosphorimaging analysis. The reactions containing hRad51K133R were assembled in buffer that contained 5 mM MgCl2. The percent D loop refers to the proportion of the radiolabeled oligonucleotide that was incorporated into the pBluescript RFI DNA.
Generation of a mRad54B disruption construct and mRad54B knockout mice.
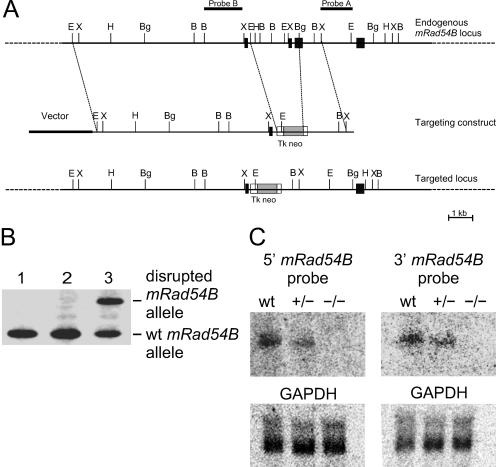
A 129 mouse genomic library (Stratagene catalog no. 946308) was screened with a DNA fragment from the mRad54B cDNA sequence. A positive clone was picked, and the DNA was subsequently subcloned into pBluescript KS. The restriction map of the genomic DNA was determined using EcoRI, BglII, BamHI, and XbaI; it was also noted that this particular stretch of genomic DNA contained seven exons. A 1,851-bp fragment between EcoRI and BglII sites in the genomic DNA was replaced with 1,108-bp XhoI/HindIII fragment from pMC1neo (Stratagene). This pMC1neo fragment contained the neomycin gene under the control of the thymidine kinase promoter. The neomycin marker gene was flanked by an approximately 9-kb EcoRI fragment and a 1,097-bp BglII-XbaI fragment of the mRad54B locus. When used in gene targeting, this construct was expected to eliminate 28 highly conserved amino acids, in effect knocking out functional mRad54B. The targeting construct was electroporated into E14 ES cells, which were then put under G418 selection. Positive clones were screened by DNA blot analysis using probe A (see Fig. 5). One out of 238 clones showed a fragment of a size expected for targeted integration of the construct. This was further confirmed using several restriction enzymes and both upstream (A) and downstream (B) probes (see Fig. 5B). ES cells that contained this targeted event were injected into blastocysts. This gave rise to 17 chimeric males, which were then backcrossed to BDF1 females in order to get pure knockout mice.
FIG. 5.
Characterization of part of the mouse Rad54B genomic locus and generation of mouse ES cells carrying a disrupted mRad54B allele. (A) Part of the mRad54B genomic locus and structure of the targeting construct. Exons 12 to 15 are indicated by black boxes. Shown are the locations of selected restriction sites, EcoRI (E), BamHI (B), BglII (Bg), HindIII (H), and XbaI (X). The positions of two different probes, named A and B, are indicated. (B) DNA blot analysis of G418-resistant ES clones with probe A and EcoRI digested DNA. The wild-type (wt) allele yields a 3.0-kb band, while the disrupted allele results in a 3.6-kb band. Lane 1, wild-type ES cell; lane 2, clone with a randomly integrated targeting construct; lane 3, clone with a homologously integrated targeting construct. (C) RNA blot analysis of mRad54B transcripts in mice carrying the disrupted allele. Total RNA (15 μg) isolated from testes of wild-type, mRad54B+/−, and mRad54B−/− males was probed with 5′ and 3′ mRad54B cDNA probes. A GAPDH (glyceraldehyde-3-phosphate dehydrogenase) cDNA probe served as a loading control.
Homologous targeting assays.
The efficiency of homologous recombination was assessed by homologous gene targeting assays to different loci, mRad54 and CTCF. The mRad54-puro targeting construct and experiments have been described previously (15). DNA blot analysis was carried out to distinguish between targeted events versus random integration of the mRad54 construct. In the case of CTCF targeting, PCR was carried out on genomic DNA of transfected clones. The appearance of a 5-kb PCR product indicated a homologous targeting event. Mouse Rad54B-specific PCR served as an internal control. χ2 tests were performed to evaluate the significance of differences in homologous recombination frequencies.
DNA damage sensitivity assays.
Cellular clonogenic survival assays have been described previously (15). Every measurement was performed in triplicate. For ionizing radiation survival assays, cells were exposed to the specified dose of gamma rays. For mitomycin C survival assays, cells were incubated in medium containing the specified concentration of mitomycin C for an hour. The cells were then washed with phosphate-buffered saline and replenished with fresh medium. The cells were allowed to grow for 10 days, after which the colonies were stained and counted. The mitomycin C survival experiments were performed four times. Cloning efficiencies of untreated cells varied between 10 to 30%.
Sensitivities to ionizing radiation and mitomycin C were assessed with the use of 2- to 4-month-old littermates from the various genotypes (wild type, mRad54−/−, mRad54B−/−, and mRad54−/− mRad54B−/−). They were irradiated with a 7-Gy dose (137Cs source) and monitored for 21 days. Surviving animals were euthanatized. Male and female mice were injected with 15, 10, 7.5, 5, 2.5, and 1 mg of mitomycin C per kg body weight and monitored for 14 days.
DNA damage processing was indirectly assessed by the micronucleus assay. One hundred microliters of peripheral blood was collected by orbital puncture, and the micronucleus assay was performed as described previously (19). Five hundred polychromatic erythrocytes were scored for the presence of micronuclei with an Axioplan fluorescence microscope.
Immunocytochemistry of meiotic chromosomes.
Testes were isolated from 1-, 2-, and 5-month-old 129/BL6 mice and processed as previously described (36). Immunofluorescence was performed as previously described (4). Sycp3 mouse monoclonal antibody was a kind gift from C. Heyting, Wageningen, The Netherlands. The anti-hRad51 antibody was as previously described (16).
Measuring of recombination by single-sperm typing.
Isolation of individual sperm, sperm lysis, single-sperm multiplex PCR, and statistical analysis of the frequency of crossing over were carried out as described previously (12, 28, 41). Recombination in two regions was analyzed. The 13.1-cM chromosome 2 interval was between (CA)n microsatellite markers D2Mit213 and D2Mit412, while the 24.1-cM chromosome 7 interval was between D7Mit268 and D7Mit353 (http://www.broad.mit.edu/cgi-bin/mouse/index). The primer sequences are given in Table S1 in the supplemental material.
Nucleotide sequence accession number.
The 2,658-bp mouse Rad54B cDNA has the GenBank identification number NM009015.
RESULTS
Isolation of a cDNA encoding a mammalian Rad54 paralog.
DNA oligonucleotides of degenerate sequence based on conserved amino acid motifs in the SNF2/SWI2 family of proteins were used to isolate a Rad54 paralog, which we named mouse Rad54B (mRad54B), from mouse cDNA libraries. The mRad54B cDNA consisted of a 2,658-bp open reading frame with the potential to code for an 886-amino-acid protein with a predicted molecular mass of 103 kDa. Amino acid sequence comparison of mRad54 and mRad54B revealed 33% sequence identity that extends over the entire length of the proteins. The predicted amino acid sequence of mRad54B showed 80% sequence identity with human Rad54B (hRad54B) (20). Further sequence analysis indicated that human and mouse Rad54B displayed 34% sequence identity to both S. cerevisiae Rad54 and Rdh54. To obtain insight into the function of mammalian Rad54B, we characterized it biochemically and genetically.
Human Rad54B forms a complex with human Rad51.
The hRAD54B cDNA was cloned, expressed in High-five insect cells, and purified (see Fig. S1 in the supplemental material). In contrast to mammalian Rad54, which can directly interact with mammalian Rad51 (18, 55), it has been suggested that hRad54B does not interact directly with hRad51. This conclusion was reached because no association between hRad51 and hRad54B was detected either in a yeast-two hybrid assay or in in vitro pull-down assays with purified proteins (58). Since genetic evidence implicates Rad54B in homologous recombination (33), we reexamined whether hRad51 and hRad54B form a complex. The hRad51 protein was immobilized on Affi-Gel beads, and interaction of hRad54B with these beads was assessed. As shown in Fig. 1A and B, hRad54B was retained on the Affi-hRad51 beads but did not associate with beads conjugated to BSA. To further establish the specificity of the hRad51/hRad54B interaction, a small amount of purified hRad54B was mixed with an excess of yeast extract proteins and this mixture was incubated with the Affi-hRad51 beads. As shown in Fig. 1C and D, among the many proteins that were present, only hRad54B was retained on the hRad51 affinity beads. As expected, hRad54B did not bind to the control beads that contained BSA. Taken together, the results demonstrate a direct and highly specific interaction between hRad54B and hRad51.
Human Rad54B has DNA translocase activity and promotes DNA helix opening.
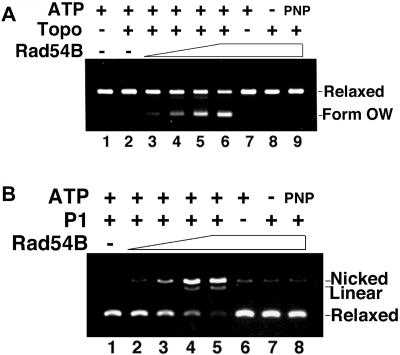
The Rad54 and Rdh54 proteins use the energy from ATP hydrolysis to translocate along duplex DNA, inducing positive supercoils ahead of protein movement and compensatory negative supercoils behind (39, 42, 47, 59). Since, like Rad54 and Rdh54, hRad54B possesses the seven conserved helicase motifs common to SWI2/SNF2 family members (51) and double-stranded-DNA-dependent ATPase activity (58), it was of interest to examine hRad54B protein for translocase activity. To this end we employed an assay that is based on the principle that E. coli topoisomerase I removes negative supercoils from DNA but is unable to act upon positive supercoils. For the reaction, a topologically relaxed plasmid substrate was incubated with hRad54B, and then E. coli topoisomerase I was added to remove any negative supercoils generated in the substrate. An ability of hRad54B to translocate on DNA will result in the formation an overwound (form OW) DNA species (38, 42, 47, 59). Utilizing this assay system, we found that hRad54B generates form OW DNA (Fig. 2). This supercoiling reaction was dependent on ATP hydrolysis by hRad54B, as evidenced by the lack of form OW when ATP was omitted from the reaction or replaced with the nonhydrolyzable analog AMP-PNP (Fig. 2A, lanes 8 and 9).
FIG. 2.
Human Rad54B has ATP-dependent DNA translocase activity that promotes opening of the DNA double helix. (A) A topologically relaxed plasmid substrate was incubated with hRad54B, after which E. coli topoisomerase I (Topo) was added to remove any negative supercoils generated in the substrate. The ability to translocate along DNA will result in the formation of an overwound (form OW) DNA species due to remaining positive supercoils in the substrate. Human Rad54B generated form OW DNA, which increased with increasing amount of protein (lanes 3 to 6). The assay depended on E. coli topoisomerase I (lane 7), and hRad54B translocase activity required ATP (lane 8) and its hydrolysis (lane 9, AMP-PNP). The concentrations of hRad54B used were 300, 400, and 500 nM in lanes 3 to 5, respectively, and 650 nM in lanes 6 to 9. (B) The presence of single-stranded DNA that results from an opening of double-stranded DNA by hRad54B translocation was probed with the use of single-strand nuclease P1. With an increasing amount of hRad54B, there was more single-stranded DNA present, resulting in P1 digestion of the single-stranded DNA and formation of the nicked form of the DNA substrate (lanes 2 to 5). Nicking activity was greatly diminished upon the omission of P1 nuclease (lane 6) or ATP (lane 7) or when ATP was replaced with the nonhydrolyzable ATP analog AMP-PNP (lane 8). DNA incubated with P1 (lane 1) was included as a control. The concentrations of hRad54B used were 100, 200, and 300 nM in lanes 2 to 4, respectively, and 400 nM in lanes 5 to 8.
The ability of Rad54 to generate negative supercoils in duplex DNA leads to the transient opening of the DNA strands in the duplex (47, 59), as indicated by the sensitivity of a topologically relaxed DNA template to the single-strand-specific nuclease P1. We used the same strategy devised for Rad54 to examine whether hRad54B renders topologically relaxed DNA sensitive to P1. As seen in Fig. 2B, the DNA substrate alone was not digested by P1 nuclease because of the lack of single-stranded character in the DNA, but with increasing amounts of hRad54B, progressively more of the DNA substrate was converted into the nicked form. These results show that the negative supercoiling induced by hRad54B translocation causes transient opening of the DNA double helix. As expected, the DNA strand-opening activity of hRad54B was dependent upon ATP hydrolysis, as sensitivity to P1 nuclease was greatly attenuated when ATP was absent or replaced with AMP-PNP (Fig. 2B, lanes 7 and 8).
Stimulation of human Rad54B activities by human Rad51.
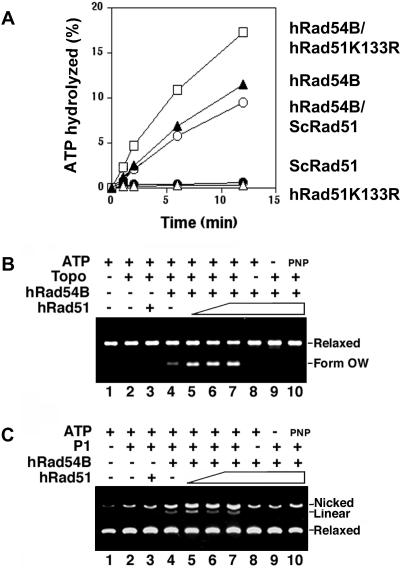
Since hRad51 and hRad54B physically interact (Fig. 1), we tested whether the proteins functionally cooperate in biochemical reactions they catalyze. We examined the effect of hRad51 on the hRad54B activities described above. In contrast to a previous report indicating that hRad51 had no effect on the hRad54B ATPase activity (58), we consistently detected a significant stimulation of this activity by hRad51 K133R, a mutant variant of hRad51 that is capable of ATP binding but not hydrolysis (Fig. 3A). The species specificity of hRad54B ATPase stimulation was established by showing that S. cerevisiae Rad51 was unable to enhance the ATP hydrolytic reaction (Fig. 3A). We next tested whether hRad51 could stimulate the ability of hRad54B to promote DNA supercoiling and DNA strand opening. Using the same systems described above, hRad51 indeed stimulated the supercoiling activity of hRad54B (Fig. 3B) as well as its ability to open the DNA double helix (Fig. 3C), while Rad51 alone did not possess either of these activities (Fig. 3B and C, lanes 3).
FIG. 3.
Human Rad54B and human Rad51 functionally interact. (A) Human Rad51 stimulates the ATPase activity of hRad54B. Human Rad54B (120 nM) was incubated with [γ-32P]ATP and various forms of Rad51, and the percentage of ATP hydrolyzed was determined. A significant stimulation of hRad54B ATPase activity was detected with hRad51 K133R (300 nM), a mutant variant of hRad51 that can bind but not hydrolyze ATP. The effect is species specific, because Rad51 from S. cerevisiae (ScRad51; 300 nM) did not affect the ATPase activity of hRad54B. (B) Human Rad51 stimulates hRad54B in the translocase assay. The formation of form OW DNA was determined in the presence of topoisomerase I (Topo), hRad54B (300 nM), and hRad51 (0, 200, 300, and 400 nM, respectively, in lanes 4 to 7). In control reactions, hRad51 (400 nM) and DNA were incubated with topoisomerase and ATP but no hRad54B (lane 3), with hRad54B and ATP but no topoisomerase (lane 8), with hRad54B and topoisomerase but no ATP (lane 9), or with hRad54B, topoisomerase, and AMP-PNP (lane 10). DNA alone (lane 1) and DNA incubated with topoisomerase (lane 2), both in the presence of ATP, were also analyzed. Form OW is the positively supercoiled DNA species generated. (C) Human Rad51 stimulates hRad54B in the P1 nuclease assay. The ability of hRad54B to open up the DNA double helix is enhanced by the presence of hRad51. hRad54B (100 nM) was incubated with topologically relaxed DNA, ATP, P1 nuclease, and hRad51 (100, 200, and 300 nM in lanes 5 to 7, respectively) or without hRad51 (lane 4). In control reactions, hRad51 (300 nM) and DNA were incubated with P1 and ATP but no hRad54B (lane 3), with hRad54B and ATP but no P1 (lane 8), with hRad54B and P1 but no ATP (lane 9), or with hRad54B, P1, and AMP-PNP (lane 10). DNA alone (lane 1) and DNA incubated with P1 (lane 2), both in the presence of ATP, were also analyzed.
D-loop formation by human Rad54B and human Rad51.
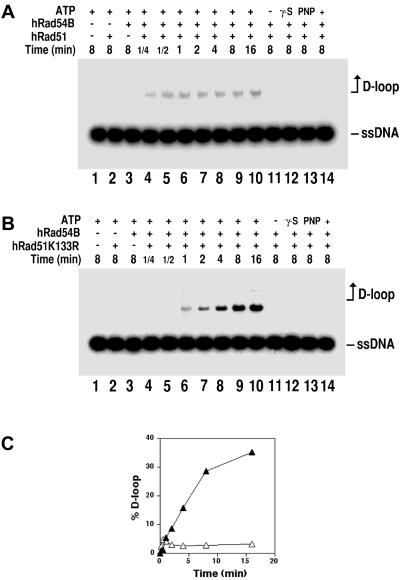
During homologous recombination, homologous pairing between the two recombining DNA molecules results in the formation of a D loop (51). We showed previously that the hRad51 and hRad54 proteins functionally cooperate in D-loop formation (47). Here, we examined the influence of hRad54B on the ability of hRad51 to mediate the D-loop reaction. As reported before (7, 47), in buffer that contains Mg2+, hRad51 by itself had little ability to form D loops (Fig. 4A and B, lanes 2). Importantly, in the presence of hRad54B protein, hRad51-dependent D-loop formation could be readily detected even after only 15 s of incubation (Fig. 4A). Similar to our published results on the hRad51/hRad54-mediated D-loop reaction (47), the D loop made by hRad51 and hRad54B appeared to be unstable. The instability of the D loop made by hRad51/hRad54 has been attributed to ATP hydrolysis by hRad51, which is expected to cause its turnover from the newly made DNA joint to initiate a second round of homologous pairing that dissociates the D loop. In support of this premise, when hRad51 was replaced by the ATPase-defective hRad51 K133R protein, the D loop formed with hRad54B accumulated to a much higher level (Fig. 4B and C). Regardless of whether hRad51 or hRad51 K133R was used, D-loop formation required ATP, which could not be replaced by ATPγS or AMP-PNP (Fig. 4A and B, lanes 11 to 13, respectively). As expected, the D-loop reaction requires homology between the single-stranded and double-stranded DNA substrates (Fig. 4A and B, lanes 14).
FIG. 4.
Human Rad54B stimulates D-loop formation by human Rad51. The formation and the subsequent stability of D loops formed in the presence of hRad51 and hRad54B were studied. (A) The radiolabeled 90-mer oligonucleotide was incubated with hRad51 and ATP to allow filament formation, after which hRad54B and the homologous duplex target (pBluescript RFI DNA) were incorporated, and a portion of the reaction mixture was removed at the indicated times for analysis (lanes 4 to 10). The controls include incubation of DNA substrates with ATP but no recombination protein (lane 1), incubation of DNA substrates with ATP and either Rad51 (lane 2) or hRad54B (lane 3), and incubation of DNA substrates with hRad51 and hRad54B but no ATP (lane 11) or with ATP-γ-S (lane 12) or AMP-PNP (lane 13). As expected, replacement of pBluescript DNA with heterologous duplex DNA (φX174) abolished D-loop formation (lane 14). ssDNA, single-stranded DNA. (B) D-loop reactions were carried out as for panel A using hRad51K133R. (C) The results from lanes 4 to 10 of panel A (open triangles) and panel B (solid triangles) are plotted.
Generation of Rad54B-disrupted mouse ES cells and mice.
The biochemical activities of the mammalian Rad54B protein determined above are consistent with a role for the protein in homologous recombination. To determine its role in vivo and to assess the biological relevance of mammalian Rad54B, we generated Rad54B knockout mouse ES cells and mice. A clone spanning the 3′ region of mRad54B was isolated from a mouse genomic library and subsequently characterized by restriction analysis and intro-exon border mapping (Fig. 5A) and chromosomal localization (see Fig. S2 in the supplemental material) (40, 61). From this clone, a targeting vector was derived that upon homologous integration into the endogenous mRad54B locus would eliminate 28 highly conserved amino acids spanning the last conserved SNF2/SWI2 family member motif. The targeting vector was electroporated into E14 ES cells, and after selection, correctly targeted clones were identified by DNA blotting using a unique probe outside the targeting construct (Fig. 5B). A targeted clone was propagated and injected into blastocysts to generate mice carrying the disrupted mRad54B allele. Inactivation of the mRad54B gene in mRad54B−/− mice was confirmed using RNA blot analysis (Fig. 5C). In RNA samples prepared from testes from both wild-type and mRad54B+/− mice, a 2.3-kb mRad54B transcript was observed. No transcript was detected with either a 5′ or 3′ probe for mRad54B in testes from mRad54B−/− mice.
Interbreeding of mRad54B+/− mice resulted in a Mendelian segregation of the disrupted mRad54B allele. Thus, mRad54B disruption did not result in embryonic or neonatal lethality. No statistically significant difference in weight was observed among mRad54B−/−, mRad54B+/−, and mRad54B+/+ littermates. Importantly, the mRad54B−/− mice exhibited no macroscopic abnormalities up to at least 6 months of age. mRad54B+/− animals were crossed to obtained blastocyts, which were then used to isolate mRad54B−/− ES cells. Two independent mRad54B−/− ES cell lines were obtained, one in a 129 background and the other in a 129/BL6 background. To obtain cells and animals in which both mRad54 paralogs were disrupted, mRad54B+/− mice were crossed with mRad54+/− mice. Similar to mRad54−/− and mRad54B−/− mice, mRad54−/− mRad54B−/− mice displayed no overt phenotypes and appeared normal. mRad54−/−, mRad54B−/−, and mRad54−/− mRad54B−/− ES cells were isolated from blastocysts obtained from intercrossing mice carrying different combinations of the mRad54 and mRad54B knockout alleles.
Frequencies of targeted integration in mouse Rad54B-deficient cells.
Previously, we have shown the involvement of mammalian Rad54 in homologous recombination by demonstrating that the efficiency of homologous gene targeting is reduced by 3- to10-fold in mRad54−/− ES cells (15). To test whether mRad54B is involved in homologous recombination as well, we examined the capacities of wild-type, mRad54B−/−, and mRad54−/− mRad54B−/− cells for gene targeting. Cells were transfected with linearized targeting constructs for either the mRad54 or the CTCF locus. Both constructs carried a puromycin resistance selectable marker flanked by regions of homology. Homologous integrations into mRad54 and CTCF loci were detected by DNA blotting and PCR, respectively (Table 1 and data not shown). Interestingly, the efficiency of homologous recombination as measured by gene targeting was not reduced in mRad54B−/− cells compared to wild-type cells (Table 1). However, the involvement of mRad54B in homologous recombination was revealed in absence of mRad54. Hardly any homologous integration events were detected in mRad54−/− mRad54B−/− ES cells.
TABLE 1.
Efficiency of homologous recombination in wild-type (mRad54+/+ mRad54B+/+), mRad54−/−, mRad54B−/−, and mRad54−/− mRad54B−/− ES cells as measured by homologous gene targetinga
| ES cell genotype | % of clones containing targeted locusb:
|
|
|---|---|---|
| mRad54 | CTCF | |
| mRad54+/+ mRad54B+/+ | 69.0 (87/126) | 60.0 (54/90) |
| mRad54+/+ mRad54B−/− | 64.7 (178/275) | 54.0 (61/113) |
| mRad54−/− mRad54B+/+ | 2.1c (6/284) | 21.3 (36/169) |
| mRad54−/− mRad54B−/− | <0.17 (0/560) | 2.1 (7/332) |
ES cells of the indicated genotype were electroporated with the indicated gene targeting constructs. After selection under the appropriate conditions individual clones were isolated and expanded. Genomic DNA from the clones was isolated. For clones electroporated with the mRad54 targeting construct, genomic DNA was digested with the appropriate restriction enzyme, electrophoresed through an agarose gel, and transfered to a nylon membrane. Membranes were hybridized with radiolabeled probes that discriminated between homologously and randomly integrated targeting construct. For clones electroporated with the CTCF targeting construct, genomic DNA was used for PCRs that discriminated between random and homologous integration events.
The percentage of clones containing the homologously integrated targeting construct relative to the total number of analyzed clones is indicated. Absolute numbers are indicated in parentheses. The differences in targeting efficiency between wild-type and mRad54−/− cells, between wild-type and mRad54−/− mRad54B−/− cells, between mRad54−/− and mRad54−/−mRad54B−/− cells, between mRad54B−/− and mRad54−/− cells, and between mRad54B−/− and mRad54−/− mRad54B−/− cells are statistically significant for both loci (P < 0.001 by χ2 analysis).
Previously reported (34).
Mouse Rad54B deficiency confers hypersensitivity to ionizing radiation and mitomycin C.
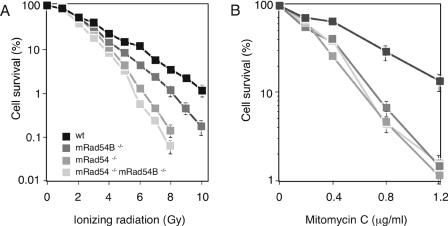
To determine whether the contribution of mRad54B to homologous recombination impinges on the ability of the cell to repair DNA damage, we examined the effect of ionizing radiation and mitomycin C on the survival of wild-type, mRad54−/−, mRad54B−/−, and mRad54−/− mRad54B−/− ES cells. While mRad54−/− ES cells are two- to threefold more sensitive to ionizing irradiation than wild-type ES cells (15), mRad54B−/− cells were only 1.5-fold more sensitive than wild-type ES cells (Fig. 6A). The ionizing radiation sensitivity of the double mutant mRad54−/− mRad54B−/− ES cells was similar to that of mRad54−/− ES cells. For mitomycin C, mRad54−/− and mRad54B−/− single mutant ES cells showed a hypersensitivity similar to that of the double mutant mRad54−/− mRad54B−/− ES cells (Fig. 6B). We conclude that, in addition to mRad54, mRad54B also contributes to repair of ionizing radiation- and mitomycin C-induced DNA damage.
FIG. 6.
Effect of ionizing radiation and mitomycin C on wild-type, mRad54−/−, mRad54B−/−, and mRad54−/− mRad54B−/− ES cells. (A) Clonogenic survival of wild-type (wt) and mutant ES cells after irradiation with increasing doses of gamma rays. The percentage of surviving cells, measured by their colony-forming ability, is plotted as function of the gamma-ray dose. (B) Clonogenic survival of wild-type, mRad54−/−, mRad54B−/−, and mRad54−/− mRad54B−/− ES cells after treatment with mitomycin C. Error bars (some obscured by the symbols) represent standard errors of the means.
Mice lacking both Rad54 paralogs are extremely sensitive to mitomycin C.
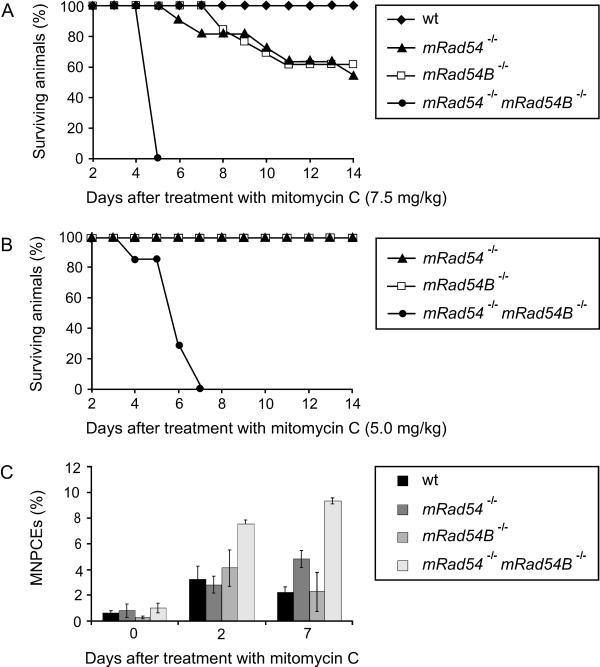
To establish the impact of mRad54B on protection from the adverse effects of induced DNA damage in the adult animal, wild-type, mRad54−/−, mRad54B−/−, and mRad54−/− mRad54B−/− mice were exposed to ionizing radiation and mitomycin C. As has been found for mRad54−/− mice, neither mRad54B−/− nor mRad54−/− mRad54B−/− mice were sensitive to ionizing radiation. All the 2- to 4-month-old littermates survived exposure to 7 Gy of ionizing radiation (data not shown).
Previously, we showed that mRad54−/− mice are hypersensitive to mitomycin C (17). To reveal whether mRad54B also contributes to protection from the mitomycin C-induced DNA damage, wild-type, mRad54−/−, mRad54B−/−, and mRad54−/− mRad54B−/− mice were injected peritoneally with different doses of mitomycin C and monitored for 14 days. As is the case for mRad54−/− mice, mRad54B−/− animals were hypersensitive to mitomycin C (Fig. 7). At a dose of 7.5 mg mitomycin C per kg of body weight, approximately 60% of the mRad54−/− and mRad54B−/− mice survived, compared to 100% of the wild-type mice. By contrast, none of the mRad54−/− mRad54B−/− mice survived the treatment. The latency period of death for mRad54B−/− mice was comparable to that for mRad54−/− mice. At the lower dose of 5 mg/kg mitomycin C, all mRad54−/− and mRad54B−/− mice survived. In stark contrast, all of the mRad54−/− mRad54B−/− mice died within 7 days. We conclude that the mammalian Rad54 paralogs function synergistically to protect mice from the deleterious effects of mitomycin C.
FIG. 7.
Impact of mRad54B on the mitomycin C sensitivity of mice. (A) Survival curve of wild-type (wt) (n = 6), mRad54−/− (n = 11), mRad54B−/− (n = 13), and mRad54−/− mRad54B−/− (n = 4) mice after a single intraperitoneal injection of 7.5 mg of mitomycin C per kg body weight. (B) Survival curve of mRad54−/− (n = 2), mRad54B−/− (n = 2), and mRad54−/− mRad54B−/− (n = 7) mice after treatment with 5 mg/kg mitomycin C. (C) Induction of micronuclei by mitomycin C in polychromatic erythrocytes. Mice of the indicated genotypes were intraperitoneally injected with 2.5 mg/kg body weight mitomycin C. Plotted are percentages of micronucleus-containing polychromatic erythrocytes (MNPCEs) per 500 polychromatic erythrocytes at days 0, 2, and 7 after treatment. Data points represent averages from three independently treated animals. The standard errors of the mean are indicated.
The bone marrow is a major target for mitomycin C-inflicted damage in vivo. Therefore, we tested whether mitomycin C treatment affected cells in the blood to different extents in mRad54−/− mRad54B−/− and mRad54−/− and mRad54B−/− animals by using the peripheral blood micronucleus assay. The presence of micronuclei in polychromatic erythrocytes provides a measure of chromosomal aberrations. A single dose of 2.5 mg mitomycin C per kg body weight was administered to 6- to 8-week-old animals. This treatment resulted in increases in the frequency of micronucleus-containing polychromatic erythrocytes (Fig. 7C). Before the mitomycin C treatment, the percentages of micronucleus-containing polychromatic erythrocytes were similar in wild-type, mRad54−/−, mRad54B−/−, and mRad54−/− mRad54B−/− animals. Consistent with the mitomycin C hypersensitivity of mRad54−/− mRad54B−/− mice, the cross-linking agent induced significantly higher levels of micronucleus-containing polychromatic erythrocytes in mRad54−/− mRad54B−/− mice than in mRad54−/− and mRad54B−/− mice (Fig. 7C).
Abnormal chromosomal distribution of mouse Rad51 during meiosis in the absence of Rad54.
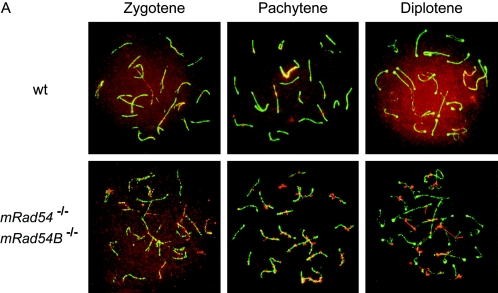
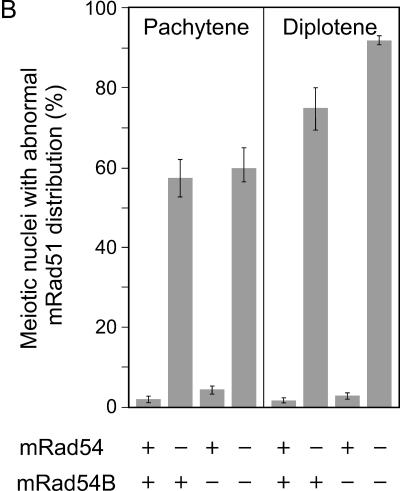
In S. cerevisiae, the function of the Rad54 paralogs appears to overlap to some extent as indicated by their effects on sporulation efficiency and spore viability (27, 45). To assess the effect of the mammalian Rad54 homologs on meiosis, we analyzed meiotic prophase chromosomes in spread nuclei of primary spermatocytes isolated from control, mRad54−/−, mRad54B−/−, and mRad54−/− mRad54B−/− mice. The chromosomes were immunostained for Sycp3 to identify the meiosis-specific synaptonemal complex that organizes the paired homologous chromosomes (Fig. 8A). Given the functional interaction between mammalian Rad54 and Rad54B in mitotic cells, we costained the chromosomes for mRad51. During the early stages of meiotic prophase, leptotene and zygotene, the distributions of mRad51 were similar in spread nuclei from all genotypes. However, in later stages, pachytene and diplotene, abnormal mRad51 localization was observed in spread nuclei from mRad54−/− and mRad54−/− mRad54B−/− mice. In control and mRad54B−/− nuclei, the mRad51 signal was present on the pachytene chromosomes as well as in an overall staining of the nucleus. However, in spread nuclei derived from mRad54−/− and mRad54−/− mRad54B−/− animals, the mRad51 signal was concentrated in abnormal focus-like structures on the chromosomes only (Fig. 8A and B). The defective mRad51 distribution is even more pronounced in diplotene, where abnormally long stretches of mRad51 were observed, instead of the normal homogenous nuclear staining. Interestingly, despite the cytological abnormalities in mRad51 localization in the absence of mRad54, meiotic homologous recombination appeared to be affected only mildly. Using single-sperm typing, crossing over was compared between mRad54−/− and wild-type animals, each heterozygous for 129/J and C57/BL6 markers flanking the chosen regions. The recombination fraction was 0.14 (95% confidence interval [CI] = 0.09 to 0.18) in an mRad54−/− animal and 0.20 (95% CI = 0.15 to 0.25) in the wild-type control for the chromosome 2 region. Using two other mice, the chromosome 7 region recombination fraction was 0.25 (95% CI = 0.21 to 0.30) in the mRad54−/− animal and 0.32 (95% CI = 0.27 to 0.37) in the wild type. The 95% confidence intervals for the mRad54−/− and wild-type individuals overlapped in both regions, indicating there was no statistically significant difference between the two genotypes. Note, however, that the recombination fraction for the mRad54−/− animals was reduced to about the same degree compared to that for wild-type animals for the genetic intervals on both chromosomes 2 and 7 (30% and 22%, respectively). There were no overt defects in fertility of mRad54−/−, mRad54B−/−, and mRad54−/− mRad54B−/− mice (data not shown).
FIG. 8.
Analysis of mouse Rad51 localization on meiotic chromosomes from mRad54- and mRad54B-proficient and -deficient mice. (A) Localization of mRad51 (red) and Sycp3 (green) on spread nuclei of primary spermatocytes as detected by immunofluorescence. Shown are chromosome spreads from zygotene, pachytene, and diplotene stages of meiosis from wild-type (wt) (upper panels) and mRad54−/− mRad54B−/− (lower panels) mice. In leptotene (not shown) and zygotene, no aberrant mRad51 and/or Sycp3 staining was observed, irrespective of genotype. In pachytene and diplotene, an abnormal mRad51 distribution was detected in a high percentage of meiotic chromosome spreads derived from mRad54−/− and mRad54−/− mRad54B−/− mice. (B) Quantification of meiotic nuclei displaying aberrant mRad51 staining. Proficiency and deficiency of the mRad54 and mRad54B proteins in the animals from which the spermatocytes were taken are indicated by + and −, respectively. Spermatocytes were isolated from mRad54- and mRad54B-proficient mice (n = 8), mRad54-deficient and mRad54B-proficient mice (n = 9), mRad54-proficient and mRad54B-deficient mice (n = 8), and mRad54- and mRad54B-deficient mice (n = 4). The percentage of meiotic nuclei with aberrant mRad51 distributions is indicated. One hundred meiotic nuclei per animal were analyzed. Error bars indicated standard errors of the means.
DISCUSSION
Homologous recombination is a versatile DNA damage repair pathway that is essential for preservation of genome integrity. Among lesions that initiate homologous recombination are single-stranded DNA gaps and DSBs. This feature makes homologous recombination ideally suited to underpin DNA replication, because single-stranded DNA gaps and DSBs occur at corrupted replication forks that arise due to either spontaneous or induced DNA damage. Homologous recombination between sister chromatids can rebuild these disabled replication forks in situ in the absence of a replication origin (10). Due to this important function, homologous recombination is essential for mammalian cell viability. The requirement for homologous recombination is not limited to mitotically dividing cells; the process is also central in meiosis to generate genetic diversity by repairing meiosis-specific DSBs with the homologous chromosome as a recombination partner.
A key player in homologous recombination is Rad51, the homology recognition and DNA strand exchange protein. The essential role of homologous recombination for mammalian cell viability is underscored by the lethality of Rad51 knockout cells and mice (54). However, not all proteins involved in homologous recombination are essential, implying either redundancy in function or the existence of subpathways of recombination that by themselves are not essential for cell viability. One example of a homologous recombination protein that is not essential for cell viability is the Rad51 accessory factor Rad54 (6, 15). To address whether there is potential redundancy in Rad54 function or recombination subpathways requiring a specialized Rad54 protein, we biochemically and genetically characterized the mammalian Rad54 paralog Rad54B.
The mammalian Rad54 paralogs have similar biochemical activities.
We performed biochemical analyses of hRad54B to compare and contrast its activities with those of hRad54. We find that, like hRad54, hRad54B is a double-stranded DNA-dependent ATPase (Fig. 3) (57). However, its ATP turnover rate is about sixfold lower than that of hRad54 (53, 58). In addition, we show that hRad54B has DNA translocase and DNA double-helix-opening activities similar to those of hRad54 (Fig. 2; (37, 39, 42, 55). Another well-explored activity of hRad54 is its interaction with hRad51 (18, 31, 55, 59). Interestingly, it has been reported that hRad54B is different from hRad54 in this respect (58). However, using more direct assays, we demonstrate that hRad54B does interact with hRad51 and that this interaction is highly specific (Fig. 1). Furthermore, we demonstrate that this interaction is functional. Human Rad51 stimulates the ATPase activity, DNA translocase activity, and DNA double-helix-opening activity of hRad54B (Fig. 3). Conversely, hRad54B stimulates the formation of D loops, a critical intermediate in homologous recombination, generated by hRad51 (Fig. 4). We conclude that hRad54B has biochemical activities similar to those of its paralog, hRad54. Therefore, hRad54 and hRad54B could provide redundant functions with respect to homologous recombination-mediated DNA damage repair or could provide similar functions but in a tissue-specific manner.
A contribution of mRad54B to homologous recombination in vivo is revealed in the absence of mRad54.
The biochemical activities of the mammalian Rad54B suggest a role for the protein in homologous recombination in vivo. Previously, it has been reported that human Rad54B contributes to homologous recombination, because homologous gene targeting is reduced in the human colon cancer cell line HCT116 in which Rad54B had been inactivated (33). Curiously, this cell line is not sensitive to any DNA-damaging agents tested, including ionizing radiation and interstrand DNA cross-linkers. The disadvantage of this cell line is its genetic instability; it is mismatch repair deficient, displays microsatellite instability, and harbors numerous chromosomal aberrations. To assess the biological impact on homologous recombination and DNA damage repair of mammalian Rad54B, we generated Rad54B knockout ES cells. In contrast to the HCT116 cells, these cells are nontransformed, display a stable normal karyotype, and carry no additional adverse mutations that complicate the interpretation of the results. As a further advantage, the ES cells allowed us to generate double mutants that in addition to inactivated mRad54B alleles, also carried disrupted mRad54 alleles.
As a reporter on homologous recombination, we measured the efficiency of homologous gene targeting in wild-type ES cells and derivatives in which either one or both Rad54 paralogs are inactivated. In accordance with previous results, the absence of mRad54 results in a reduced homologous recombination efficiency (15). Depending on the locus, the reduction varies between 3- and 30-fold (Table 1). In the absence of Rad54B, we consistently observe an approximately 10% reduction in recombination efficiency, but even with more than 400 events analyzed, this reduction is not statistically significant. However, cells lacking both Rad54 paralogs have a 30- to 400-fold-reduced homologous recombination efficiency compared to wild-type cells (Table 1). This is a reduction of a further 10-fold compared to the absence of mRad54 alone. Thus, the involvement of mRad54B in the homologous recombination subpathway that mediates gene targeting is uncovered in the absence of mRad54.
Both mammalian Rad54 paralogs contribute to cell survival in response to DNA damage.
Because homologous recombination is such a versatile mechanism to repair DNA damage compared to repair mechanisms that rely on a specific DNA damage recognition protein to initiate the reaction, it is remarkable that an hRad54B-deficient cancer cell line is not hypersensitive to DNA-damaging agents (33). Given the caveats mentioned above, we examined DNA damage sensitivity in mouse ES cells instead. The lack of Rad54 in ES cells results in cellular hypersensitivity to ionizing radiation and mitomycin C (15). In contrast to the hRad54B-deficient cancer line, ES cells lacking mRad54B are hypersensitive to ionizing radiation and mitomycin C (Fig. 6). At the cellular level, our results reveal no strong indication for an additive or synergic interaction between the two Rad54 paralogs with respect to the repair of ionizing radiation- and mitomycin C-induced DNA damage repair.
The Rad54 paralogs synergistically contribute to mitomycin C resistance in mice.
The contribution of Rad54 to survival of mice in response to DNA damage differs from that in ES cells. While mRad54−/− ES cells are ionizing radiation and mitomycin C hypersensitive, mRad54−/− mice are mitomycin C but not ionizing radiation hypersensitive (17). We tested whether the lack of ionizing radiation hypersensitivity in mice is due to redundancy in mRad54 function provided by mRad54B. However, this is not the case, because mRad54−/− mRad54B−/− mice are also not overtly ionizing radiation hypersensitive (data not shown). The contribution of homologous recombination to repair of ionizing radiation-induced DNA damage is revealed in the absence of nonhomologous DNA end-joining, an alternative, mechanistically distinct DSB repair pathway (17). Possibly, while homologous recombination is an important DNA repair pathway for two-ended breaks, such as those induced by ionizing radiation, in rapidly dividing ES cells, nonhomologous DNA end-joining is much better suited for repair of these lesions in the many nondividing cells of the adult mice. In contrast, mitomycin C-induced DNA interstrand cross-links are processed into single-ended DSBs by DNA replication (35). Single-ended DSBs cannot be acted upon efficiently by nonhomologous DNA end-joining and require homologous recombination for repair instead (11). As is the case for mRad54−/− mice, mRad54B−/− mice are mitomycin C hypersensitive (Fig. 7). Furthermore, mRad54−/− mRad54B−/− double mutant mice are extremely mitomycin C hypersensitive. This synergistic effect of the Rad54 paralogs could be due to their functions in distinct subpathways of interstrand DNA cross-link repair. However, given their biochemical similarities and the lack of a significant difference in mitomycin C hypersensitivity of mRad54B−/− versus mRad54−/− mRad54B−/− ES cells, this is unlikely. Alternatively, the Rad54 paralogs might have a tissue-specific function by differentially contributing to interstrand DNA cross-link repair in different cell types of mice. This premise predicts that there should be differences in expression of the Rad54 paralogs among different tissues. Although no direct comparison is currently available, the existing data do suggest that this could be the case. For example, while expression of hRAD54B is extremely low in the spleen (see Fig. S2 in the supplemental material), expression of mRad54 in the spleen is robust (25).
Mammalian Rad54 affects the distribution of Rad51 on meiotic chromosomes.
Both Rad54 paralogs in S. cerevisiae contribute to meiosis, and therefore we analyzed the effect of the mammalian Rad54 paralogs on mouse meiotic prophase chromosomes. Because both mammalian Rad54 paralogs functionally interact with Rad51, we investigated the distribution of mRad51 on meiotic prophase chromosomes. During meiosis, mRad51 is first observed in the leptotene and zygotene stages as foci distributed throughout the nucleus (5). Subsequently, mRad51 foci arrange into linear arrays that colocalize with the axial elements of the synaptonemal complex. mRad51 staining disappears during late pachytene and diplotene. Interestingly, the absence of mRad54 results in an aberrant mRad51 distribution on meiotic chromosomes in spread nuclei from primary spermatocytes, consisting of focus- and thread-like mRad51 structures localized on the chromosomal loops emanating from the synaptonemal complex that persist into the diplotene stage (Fig. 8A and B). In contrast, mRad54B deficiency does not have a dramatic impact on mRad51 localization during meiosis. Possibly, mRad54B has a minor role, because when mRad54 is also absent, the percentage of meiotic spread nuclei with abnormal mRad51 distributions increases only slightly in diplotene.
Even though the absence of the Rad54 paralogs causes an abnormal mRad51 distribution, meiosis appears not to be affected with regard to other features. mRad54−/−, mRad54B−/−, and mRad54−/− mRad54B−/− mice are fertile, no increase in apoptosis during spermatogenesis is detected, and there is no difference in the number of MLH1 foci on meiotic chromosomes in spread nuclei (data not shown). Furthermore, the absence of Rad54 only slightly affects the frequency of meiotic crossing-over. Meiotic homologous recombination in mammals might depend for most of the functions provided by Rad54 paralogs on a yet-unidentified, potentially meiosis-specific, Rad54 paralog. The aberrant mRad51 distribution is consistent with a role suggested for Rad54 in S. cerevisiae, namely, its ability to remove Rad51 filaments from double-stranded DNA (49). This would be a late function of mRad54 during homologous recombination, at a stage when actual repair of DSBs has already taken place or at a stage at which this can be accomplished by mechanisms that no longer require mRad54.
Comparison of the yeast and mammalian Rad54 paralogs.
Taken together, our data lead us to conclude that the premise that mammalian Rad54B is the functional homolog of S. cerevisiae Rdh54 is unlikely. While S. cerevisiae rdh54 mutant cells display no overt DNA damage sensitivities, mRad54B−/− ES cells do. Furthermore, both S. cerevisiae Rad54 paralogs make significant contributions to meiotic homologous recombination. In particular, Rdh54 plays an important role in interhomolog recombination (3). In contrast, we have not detected an essential role of the mammalian Rad54 paralogs in meiosis. It is possible that a yet-undiscovered meiosis-specific Rad54 paralog exists in mammals. Alternatively, rather than being strictly assigned to meiotic DSB repair, this hypothetical Rad54 paralog might overlap in DSB repair function with Rad54 and Rad54B and take over part of the function of both Rad54 and Rad54B in their absence, which would be consistent with the viability of mRad54−/− mRad54B−/− mice and the essential role of homologous recombination for mammalian cell viability. However, when challenged with exogenous DNA-damaging agents such as mitomycin C, the DNA damage load might exceed the threshold of its ability to repair on its own, and this is reflected by the mitomycin C hypersensitivity of Rad54- and Rad54B-deficient mice. The DNA damage threshold may also explain the apparent normal viability of the Rad54- and Rad54B-deficient mice, because the level of endogenous DNA damage might be low enough to be effectively handled by a yet-unidentified Rad54 paralog.
Supplementary Material
Acknowledgments
We thank Ellen van Drunen and Miranda Boeve for technical assistance.
This work was supported by grants from the Dutch Cancer Society (KWF), The Netherlands Organization for Scientific Research (NWO), and the European Commission, and by the NIH (grants GM57814 and CA110415 to P.S. and GM36745 to N.A.).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alexeev, A., A. Mazin, and S. C. Kowalczykowski. 2003. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat. Struct. Biol. 10:182-186. [DOI] [PubMed] [Google Scholar]
- 2.Alexiadis, V., A. Lusser, and J. T. Kadonaga. 2004. A conserved N-terminal motif in Rad54 is important for chromatin remodeling and homologous strand pairing. J. Biol. Chem. 279:27824-27829. [DOI] [PubMed] [Google Scholar]
- 3.Arbel, A., D. Zenvirth, and G. Simchen. 1999. Sister chromatid-based DNA repair is mediated by RAD54, not by DMC1 or TID1. EMBO J. 18:2648-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baarends, W. M., E. Wassenaar, J. W. Hoogerbrugge, G. van Cappellen, H. P. Roest, J. Vreeburg, M. Ooms, J. H. Hoeijmakers, and J. A. Grootegoed. 2003. Loss of HR6B ubiquitin-conjugating activity results in damaged synaptonemal complex structure and increased crossing-over frequency during the male meiotic prophase. Mol. Cell. Biol. 23:1151-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barlow, A. L., F. E. Benson, S. C. West, and M. A. Hulten. 1997. Distribution of the Rad51 recombinase in human and mouse spermatocytes. EMBO J. 16:5207-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bezzubova, O., A. Silbergleit, Y. Yamaguchi-Iwai, S. Takeda, and J. M. Buerstedde. 1997. Reduced X-ray resistance and homologous recombination frequencies in a RAD54−/− mutant of the chicken DT40 cell line. Cell 89:185-193. [DOI] [PubMed] [Google Scholar]
- 7.Bugreev, D. V., and A. V. Mazin. 2004. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl. Acad. Sci. USA 101:9988-9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clever, B., H. Interthal, J. Schmuckli-Maurer, J. King, M. Sigrist, and W. D. Heyer. 1997. Recombinational repair in yeast: functional interactions between Rad51 and Rad54 proteins. EMBO J. 16:2535-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Couedel, C., K. D. Mills, M. Barchi, L. Shen, A. Olshen, R. D. Johnson, A. Nussenzweig, J. Essers, R. Kanaar, G. C. Li, F. W. Alt, and M. Jasin. 2004. Collaboration of homologous recombination and nonhomologous end-joining factors for the survival and integrity of mice and cells. Genes Dev. 18:1293-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox, M. M., M. F. Goodman, K. N. Kreuzer, D. J. Sherratt, S. J. Sandler, and K. J. Marians. 2000. The importance of repairing stalled replication forks. Nature 404:37-41. [DOI] [PubMed] [Google Scholar]
- 11.Cromie, G. A., J. C. Connelly, and D. R. Leach. 2001. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol. Cell 8:1163-1174. [DOI] [PubMed] [Google Scholar]
- 12.Cui, X. F., H. H. Li, T. M. Goradia, K. Lange, H. H. Kazazian, Jr., D. Galas, and N. Arnheim. 1989. Single-sperm typing: determination of genetic distance between the G gamma-globin and parathyroid hormone loci by using the polymerase chain reaction and allele-specific oligomers. Proc. Natl. Acad. Sci. USA 86:9389-9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dresser, M. E., D. J. Ewing, M. N. Conrad, A. M. Dominguez, R. Barstead, H. Jiang, and T. Kodadek. 1997. DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics 147:533-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dronkert, M. L., H. B. Beverloo, R. D. Johnson, J. H. Hoeijmakers, M. Jasin, and R. Kanaar. 2000. Mouse RAD54 affects DNA double-strand break repair and sister chromatid exchange. Mol. Cell. Biol. 20:3147-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Essers, J., R. W. Hendriks, S. M. Swagemakers, C. Troelstra, J. de Wit, D. Bootsma, J. H. Hoeijmakers, and R. Kanaar. 1997. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell 89:195-204. [DOI] [PubMed] [Google Scholar]
- 16.Essers, J., R. W. Hendriks, J. Wesoly, C. E. Beerens, B. Smit, J. H. Hoeijmakers, C. Wyman, M. L. Dronkert, and R. Kanaar. 2002. Analysis of mouse Rad54 expression and its implications for homologous recombination. DNA Repair (Amsterdam) 1:779-793. [DOI] [PubMed]
- 17.Essers, J., H. van Steeg, J. de Wit, S. M. Swagemakers, M. Vermeij, J. H. Hoeijmakers, and R. Kanaar. 2000. Homologous and non-homologous recombination differentially affect DNA damage repair in mice. EMBO J. 19:1703-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golub, E. I., O. V. Kovalenko, R. C. Gupta, D. C. Ward, and C. M. Radding. 1997. Interaction of human recombination proteins Rad51 and Rad54. Nucleic Acids Res. 25:4106-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayashi, M., T. Morita, Y. Kodama, T. Sofuni, and M. Ishidate, Jr. 1990. The micronucleus assay with mouse peripheral blood reticulocytes using acridine orange-coated slides. Mutat. Res. 245:245-249. [DOI] [PubMed] [Google Scholar]
- 20.Hiramoto, T., T. Nakanishi, T. Sumiyoshi, T. Fukuda, S. Matsuura, H. Tauchi, K. Komatsu, Y. Shibasaki, H. Inui, M. Watatani, M. Yasutomi, K. Sumii, G. Kajiyama, N. Kamada, K. Miyagawa, and K. Kamiya. 1999. Mutations of a novel human RAD54 homologue, RAD54B, in primary cancer. Oncogene 18:3422-3426. [DOI] [PubMed] [Google Scholar]
- 21.Ho, K. S., and R. K. Mortimer. 1975. X-ray-induced lethality and chromosome breakage and repair in a radiosensitive strain of yeast. Basic Life Sci. 5B:545-547. [DOI] [PubMed] [Google Scholar]
- 22.Hoeijmakers, J. H. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411:366-374. [DOI] [PubMed] [Google Scholar]
- 23.Jaskelioff, M., S. Van Komen, J. E. Krebs, P. Sung, and C. L. Peterson. 2003. Rad54p is a chromatin remodeling enzyme required for heteroduplex DNA joint formation with chromatin. J. Biol. Chem. 278:9212-9218. [DOI] [PubMed] [Google Scholar]
- 24.Jiang, H., Y. Xie, P. Houston, K. Stemke-Hale, U. H. Mortensen, R. Rothstein, and T. Kodadek. 1996. Direct association between the yeast Rad51 and Rad54 recombination proteins. J. Biol. Chem. 271:33181-33186. [DOI] [PubMed] [Google Scholar]
- 25.Kanaar, R., C. Troelstra, S. M. Swagemakers, J. Essers, B. Smit, J. H. Franssen, A. Pastink, O. Y. Bezzubova, J. M. Buerstedde, B. Clever, W. D. Heyer, and J. H. Hoeijmakers. 1996. Human and mouse homologs of the Saccharomyces cerevisiae RAD54 DNA repair gene: evidence for functional conservation. Curr. Biol. 6:828-838. [DOI] [PubMed] [Google Scholar]
- 26.Kim, P. M., K. S. Paffett, J. A. Solinger, W. D. Heyer, and J. A. Nickoloff. 2002. Spontaneous and double-strand break-induced recombination, and gene conversion tract lengths, are differentially affected by overexpression of wild-type or ATPase-defective yeast Rad54. Nucleic Acids Res. 30:2727-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein, H. L. 1997. RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics 147:1533-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, H. H., U. B. Gyllensten, X. F. Cui, R. K. Saiki, H. A. Erlich, and N. Arnheim. 1988. Amplification and analysis of DNA sequences in single human sperm and diploid cells. Nature 335:414-417. [DOI] [PubMed] [Google Scholar]
- 29.Lisby, M., J. H. Barlow, R. C. Burgess, and R. Rothstein. 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118:699-713. [DOI] [PubMed] [Google Scholar]
- 30.Mazin, A. V., A. A. Alexeev, and S. C. Kowalczykowski. 2003. A novel function of Rad54 protein. Stabilization of the Rad51 nucleoprotein filament. J. Biol. Chem. 278:14029-14036. [DOI] [PubMed] [Google Scholar]
- 31.Mazin, A. V., C. J. Bornarth, J. A. Solinger, W. D. Heyer, and S. C. Kowalczykowski. 2000. Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol. Cell 6:583-592. [DOI] [PubMed] [Google Scholar]
- 32.Mills, K. D., D. O. Ferguson, J. Essers, M. Eckersdorff, R. Kanaar, and F. W. Alt. 2004. Rad54 and DNA ligase IV cooperate to maintain mammalian chromatid stability. Genes Dev. 18:1283-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyagawa, K., T. Tsuruga, A. Kinomura, K. Usui, M. Katsura, S. Tashiro, H. Mishima, and K. Tanaka. 2002. A role for RAD54B in homologous recombination in human cells. EMBO J. 21:175-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niedernhofer, L. J., J. Essers, G. Weeda, B. Beverloo, J. de Wit, M. Muijtjens, H. Odijk, J. H. Hoeijmakers, and R. Kanaar. 2001. The structure-specific endonuclease Ercc1-Xpf is required for targeted gene replacement in embryonic stem cells. EMBO J. 20:6540-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niedernhofer, L. J., H. Odijk, M. Budzowska, E. van Drunen, A. Maas, A. F. Theil, J. de Wit, N. G. Jaspers, H. B. Beverloo, J. H. Hoeijmakers, and R. Kanaar. 2004. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol. Cell. Biol. 24:5776-5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peters, A. H., A. W. Plug, M. J. van Vugt, and P. de Boer. 1997. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 5:66-68. [DOI] [PubMed] [Google Scholar]
- 37.Petukhova, G., S. Stratton, and P. Sung. 1998. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature 393:91-94. [DOI] [PubMed] [Google Scholar]
- 38.Petukhova, G., S. A. Stratton, and P. Sung. 1999. Single strand DNA binding and annealing activities in the yeast recombination factor Rad59. J. Biol. Chem. 274:33839-33842. [DOI] [PubMed] [Google Scholar]
- 39.Petukhova, G., P. Sung, and H. Klein. 2000. Promotion of Rad51-dependent D-loop formation by yeast recombination factor Rdh54/Tid1. Genes Dev. 14:2206-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinkel, D., T. Straume, and J. W. Gray. 1986. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc. Natl. Acad. Sci. USA 83:2934-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qin, J., S. Baker, H. Te Riele, R. M. Liskay, and N. Arnheim. 2002. Evidence for the lack of mismatch-repair directed antirecombination during mouse meiosis. J. Hered. 93:201-205. [DOI] [PubMed] [Google Scholar]
- 42.Ristic, D., C. Wyman, C. Paulusma, and R. Kanaar. 2001. The architecture of the human Rad54-DNA complex provides evidence for protein translocation along DNA. Proc. Natl. Acad. Sci. USA 98:8454-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmuckli-Maurer, J., M. Rolfsmeier, H. Nguyen, and W. D. Heyer. 2003. Genome instability in rad54 mutants of Saccharomyces cerevisiae. Nucleic Acids Res. 31:1013-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shinohara, M., S. L. Gasior, D. K. Bishop, and A. Shinohara. 2000. Tid1/Rdh54 promotes colocalization of rad51 and dmc1 during meiotic recombination. Proc. Natl. Acad. Sci. USA 97:10814-10819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shinohara, M., E. Shita-Yamaguchi, J. M. Buerstedde, H. Shinagawa, H. Ogawa, and A. Shinohara. 1997. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics 147:1545-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Signon, L., A. Malkova, M. L. Naylor, H. Klein, and J. E. Haber. 2001. Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol. Cell. Biol. 21:2048-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sigurdsson, S., S. Van Komen, G. Petukhova, and P. Sung. 2002. Homologous DNA pairing by human recombination factors Rad51 and Rad54. J. Biol. Chem. 277:42790-42794. [DOI] [PubMed] [Google Scholar]
- 48.Solinger, J. A., and W. D. Heyer. 2001. Rad54 protein stimulates the postsynaptic phase of Rad51 protein-mediated DNA strand exchange. Proc. Natl. Acad. Sci. USA 98:8447-8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Solinger, J. A., K. Kiianitsa, and W. D. Heyer. 2002. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol. Cell 10:1175-1188. [DOI] [PubMed] [Google Scholar]
- 50.Sugawara, N., X. Wang, and J. E. Haber. 2003. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell 12:209-219. [DOI] [PubMed] [Google Scholar]
- 51.Sung, P., L. Krejci, S. Van Komen, and M. G. Sehorn. 2003. Rad51 recombinase and recombination mediators. J. Biol. Chem. 278:42729-42732. [DOI] [PubMed] [Google Scholar]
- 52.Sung, P., L. Prakash, S. Weber, and S. Prakash. 1987. The RAD3 gene of Saccharomyces cerevisiae encodes a DNA-dependent ATPase. Proc. Natl. Acad. Sci. USA 84:6045-6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swagemakers, S. M., J. Essers, J. de Wit, J. H. Hoeijmakers, and R. Kanaar. 1998. The human RAD54 recombinational DNA repair protein is a double-stranded DNA-dependent ATPase. J. Biol. Chem. 273:28292-28297. [DOI] [PubMed] [Google Scholar]
- 54.Symington, L. S. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol Rev. 66:630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan, T. L., J. Essers, E. Citterio, S. M. Swagemakers, J. de Wit, F. E. Benson, J. H. Hoeijmakers, and R. Kanaar. 1999. Mouse Rad54 affects DNA conformation and DNA-damage-induced Rad51 foci formation. Curr. Biol. 9:325-328. [DOI] [PubMed] [Google Scholar]
- 56.Tan, T. L., R. Kanaar, and C. Wyman. 2003. Rad54, a Jack of all trades in homologous recombination. DNA Repair (Amsterdam) 2:787-794. [DOI] [PubMed]
- 57.Tanaka, K., T. Hiramoto, T. Fukuda, and K. Miyagawa. 2000. A novel human rad54 homologue, Rad54B, associates with Rad51. J. Biol. Chem. 275:26316-26321. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka, K., W. Kagawa, T. Kinebuchi, H. Kurumizaka, and K. Miyagawa. 2002. Human Rad54B is a double-stranded DNA-dependent ATPase and has biochemical properties different from its structural homolog in yeast, Tid1/Rdh54. Nucleic Acids Res. 30:1346-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Komen, S., G. Petukhova, S. Sigurdsson, S. Stratton, and P. Sung. 2000. Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol. Cell 6:563-572. [DOI] [PubMed] [Google Scholar]
- 60.van Veelen, L. R., T. Cervelli, M. W. van de Rakt, A. F. Theil, J. Essers, and R. Kanaar. 2005. Analysis of ionizing radiation-induced foci of DNA damage repair proteins. Mutat. Res. 574:22-33. [DOI] [PubMed] [Google Scholar]
- 61.Weeda, G., J. Wiegant, M. van der Ploeg, A. H. Geurts van Kessel, A. J. van der Eb, and J. H. Hoeijmakers. 1991. Localization of the xeroderma pigmentosum group B-correcting gene ERCC3 to human chromosome 2q21. Genomics 10:1035-1040. [DOI] [PubMed] [Google Scholar]
- 62.Wolner, B., and C. L. Peterson. 2005. ATP-dependent and ATP-independent roles for the Rad54 chromatin remodeling enzyme during recombinational repair of a DNA double strand break. J. Biol. Chem. 280:10855-10860. [DOI] [PubMed] [Google Scholar]
- 63.Wolner, B., S. van Komen, P. Sung, and C. L. Peterson. 2003. Recruitment of the recombinational repair machinery to a DNA double-strand break in yeast. Mol. Cell 12:221-232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.