Abstract
Cell cycle-dependent, site-specific phosphorylation of the retinoblastoma protein, pRB, is mediated by cyclin-dependent kinases (CDKs) and regulates the binding of pRB to many proteins. We previously showed that the interaction of pRB with E2F on DNA was regulated by the accumulation of phosphate groups on pRB. Here we show that positively charged lysine residues in the B domain of pRB are necessary for the release of pRB from E2F on DNA following phosphorylation by cyclin E-cdk2 kinase. These lysine residues are also important in the binding of the simian virus 40 large T antigen (TAg) to pRB, and mutation of these lysines to arginines alters the dependency of the pRB-TAg interaction on phosphorylation of pRB.
The primary function of the retinoblastoma protein, pRB, is regulation of the activities of transcription factors in a cell cycle and cell-type-specific manner. Best studied is negative regulation in which pRB binds to and represses the activities of transcription factors such as E2F, Elf-1, and PU.1 (21, 25, 26, 30, 31, 59). However, positive regulation of transcription by pRB also occurs through interactions with transcription factors such as MyoD, NF-IL6, and CCAAT enhancer binding protein (C/EBP), involved in terminal differentiation of several cell types (10, 11, 24), and with general transcription factors such as SP1, ATF-2, and hBrm (9, 37, 38, 55, 58).
Cell cycle regulation by pRB is a result of repression of E2F activation of transcription of genes necessary for S-phase entrance (reviewed in references 18 and 29). Early studies showed that pRB binds the transactivation domain of E2F, thereby directly blocking the ability of E2F to activate transcription (21, 30). The ability of pRB to block cells in G1 is dependent on E2F binding and repression of transcription (54). Zhang and coauthors showed that accumulation of hypophosphorylated pRB induced G1 arrest by forming pRB-E2F complexes on the promoters of cell cycle genes (62).
pRB is a nuclear phosphoprotein, the activity of which is regulated by phosphorylation (2, 5, 12, 14, 15, 17, 20, 32-35, 44, 45). In the G0/G1 phase of the cell cycle, hypophosphorylated pRB actively represses transcription of genes required for S phase. At the G1/S transition, pRB is inactivated by phosphorylation on serine and threonine residues through the action of cyclin-dependent kinases (cdks) (reviewed in reference 50). Cyclins D and E form complexes with cdk4/6 and cdk2, respectively, at the G1/S transition and phosphorylate pRB. Progressive hyperphosphorylation of pRB continues through S phase by cyclin A-cdk2 and through mitosis by cyclin B-cdc2 (50). Dephosphorylation and activation of pRB occur after the beginning of mitosis due to the activity of phosphoprotein phosphatase type 1 (PP1) (46).
Studies defining the mechanism of phosphorylation of pRB in the G1 phase of the cell cycle suggest that both cyclin D and cyclin E are required (28, 47). In yeast, cyclins D1 and E can substitute for G1 cyclins Cln2 and Cln3, and both are required to phosphorylate pRB (28). The use of inhibitors of pRB phosphorylation (p16ink4A and cdk2DN inhibit cyclin D-cdk4/6 and cyclin E-cdk2 kinase activity, respectively) in U2OS cells containing wild-type pRB showed that cyclin D-cdk4/6 only partially phosphorylated pRB; complete phosphorylation required cyclin E-cdk2 activity (47). In addition, cyclin D-associated phosphorylation of pRB did not release pRB from its interaction with E2F-1 (47). It is suggested that cyclin D-associated phosphorylation of pRB allows the phosphorylated C terminus of pRB to fold over and interact with residues in the B domain, thereby releasing both histone deacetylase 1 (HDAC1) and active transcriptional repression (27). An intramolecular interaction between the phosphorylated C terminus of pRB and the B domain is necessary for cyclin E-cdk2 to further phosphorylate pRB and release it from E2F.
pRB contains 16 potential phosphorylation sites for specific regulation of pRB binding to different protein complexes. Phosphorylation of pRB at Ser807 and Ser811 released its interaction with c-Abl, whereas phosphorylation at Thr821 and Thr826 was required to release simian virus 40 (SV40) large T antigen (TAg) (39). Regulation of binding of free E2F to pRB occurred by phosphorylation either at a number of C-terminal sites or at two serines in the insert domain of pRB between the A and B pocket regions (40).
Our experiments suggested that regulation of the pRB-E2F interaction on DNA by phosphorylation occurs by accumulation of phosphate groups on the pRB molecule (4). RB proteins containing mutations in six or more phosphorylation sites repressed E2F-mediated transcriptional activation 25-fold better than wild-type pRB in the presence of cyclin E-cdk2 kinase activity. Phosphorylation of six or more sites on pRB may release the interaction of pRB with E2F on DNA.
There are a number of possibilities to explain how phosphorylation releases pRB from E2F, including conformational changes within the E2F-binding domain of pRB, release of hydrophobic contacts with E2F, or a preferential interaction of the E2F-binding domain with another part of pRB. Studies of the structure of pRB bound to an LXCXE peptide (43) suggested that release of the LXCXE peptide from pRB upon phosphorylation might occur by competition of the phosphorylated pRB polypeptide segment for the LXCXE peptide binding site. A six-lysine basic patch on the rim of the LXCXE binding site may bind the phosphorylated peptide segment of pRB (43). A mechanism such as this could explain our data, which showed that six or more phosphorylation sites were necessary for release of pRB from E2F by phosphorylation. Although the binding site for E2F is different from the LXCXE binding site (the A-B interface rather than the B box), it is possible that this competitive mechanism also releases pRB from E2F bound to DNA.
To test the role of the six-lysine basic patch on pRB function, we measured the ability of the cyclin E complex to relieve E2F-mediated repression by phosphorylation of pRB molecules containing mutations in five of the six B-domain lysine residues. We conclude that the phosphorylated polypeptide of pRB interacts with the basic lysine patch in the B domain when pRB releases E2F bound to DNA. We also show that the basic lysine patch is involved in the interaction of pRB with TAg. Interestingly, mutation of the lysines to arginines alters the dependency of the pRB-TAg interaction on phosphorylation of pRB.
MATERIALS AND METHODS
Cell culture.
The C33A cervical carcinoma cell line (from the American Type Culture Collection) and COS-1 monkey kidney cells were grown in alpha modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. Cells were grown in 5% CO2 at 37°C. Metabolic labeling was carried out with [32P]phosphoric acid at 800 μCi/ml in phosphate-free medium supplemented with 10% dialyzed fetal bovine serum for 4.5 h.
Plasmids.
ΔBXHA (wild-type murine pRB) and Δp34HA constructs in the SV40 promoter-containing pECE vector were described previously (3). 5lysRB was made by using ΔBXHA as a template and three PCR primers containing single-base-pair mismatches in the codons of interest. RB3K mutated lysine 713 and lysine 715 to alanines, RB2K mutated lysine 722 and lysine 733 to alanines, and RB1K mutated lysine 758 to alanine. RB1K overlapped a unique restriction site (StuI) 3" of all of the lysines mutated. An additional primer spanning another unique restriction site (SacI) 5" of the mutated lysines was used, and the PCR fragment was cloned into ΔBXHA digested with SacI and StuI to give 5lysRB. 5ARGRB was similarly constructed, except that the three primers contained lysine-to-arginine mutations. pECE constructs expressing cyclin E and cdk2 and the reporter construct E2(−80/−70)-chloramphenicol acetyltransferase (CAT) were described previously (3). The E2(−80/−70)-CAT construct is driven by 160 bp of the adenovirus EIIaE promoter and contains two E2F consensus sites. The RBCAT construct contains the CAT gene driven by 2.0 kb of the genomic RB1 promoter and contains one E2F consensus site (22).
Transfection and CAT assay.
C33A cells were transfected by the calcium phosphate-mediated transfection procedure (53). COS-1 cells were transfected with Superfect (Qiagen). CAT assays were performed 48 to 72 h posttransfection by the method of Sleigh (56). Three micrograms of the reporter construct E2(−80/−70)CAT was transfected along with the pRB constructs and 5 μg each of the cyclin E and cdk2 constructs. To prevent competition effects from the promoter carrying the effector genes on the reporter construct, an SV40 promoter-driven luciferase gene (SVLuc) was used as filler DNA. One microgram of plasmid pCMVβgal was included in the transfection to normalize CAT activity for transfection efficiency. The CAT activity from the reporter construct in the absence or presence of cyclin E and cdk2 was arbitrarily set to 100%. Experiments with the RBCAT promoter contained 0.5 μg of the reporter construct. Trichostatin A (TSA) was added at a concentration of 100 nM.
Immunoprecipitation and Western blotting.
Transfected cells were lysed in a mixture of 50 mM sodium flouride, 50 mM Tris-HCl (pH 8), 120 mM NaCl, and 0.5% NP40 with the proteinase inhibitors leupeptin, aprotinin, and phenylmethylsulfonyl fluoride (PMSF) added immediately before use. Immunoprecipitation of pRB was performed with 2.5 μg of antihemagglutinin (HA) antibody (12CA5; Roche) or anti-HA probe (F-7; Santa Cruz) followed by Western blotting with either anti-HA antibody (12CA5) or anti-pRB (14001A; Pharmingen). Coimmunoprecipitation of pRB with E2F-1 was performed with 2.5 μg of anti-pRB antibody (14001A) followed by Western blotting with anti-E2F-1 antibody (C-20; Santa Cruz) or immunoprecipitation with 2 μg of anti-E2F-1 (C-20) followed by Western blotting with anti-pRB antibody (14001A). Coimmunoprecipitation of pRB with TAg was performed with 2.5 μg of anti-HA probe (F-7) followed by Western blotting with anti-TAg (14111A; Pharmingen) or immunoprecipitation with 2.5 μg of anti-TAg (14111A) followed by Western blotting with anti-HA (12CA5). Coimmunoprecipitation of pRB with HDAC1 was performed by immunoprecipitation of HDAC1 with anti-Flag antibody (M2; Sigma) followed by Western blotting with anti-pRB (14001A; Pharmingen). Acetylation of pRB was shown by immunoprecipitation with polyclonal antiacetylated lysine antibody (9441S; New England Biolabs) followed by Western blotting with anti-pRB antibody (14001A). Lambda phosphatase (λPP) experiments were performed following immunoprecipitation of pRB with anti-HA antibody (12CA5). Four hundred units of λPP was added to the protein on the beads, which were incubated for 30 min at 30°C and then loaded on sodium dodecyl sulfate (SDS)-polyacrylamide gels.
RESULTS
E2(−80/−70)-CAT is not regulated by HDAC1 activity.
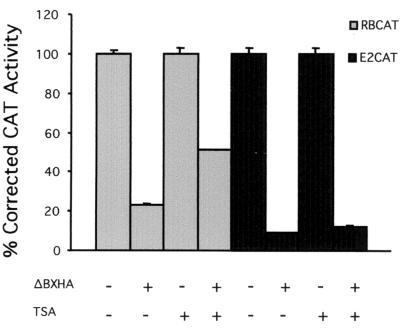
Others have suggested that the basic lysines in the B domain are required for the release of HDAC1 from pRB following phosphorylation by cyclin D kinase activity; phosphorylation by cyclin E could then release E2F (27). However, not all promoters are regulated by HDAC activity (48). Therefore, we tested if the E2(−80/−70)-CAT reporter construct (hereby labeled E2CAT) was regulated by HDAC. Repression of an E2F-regulated promoter, RBCAT, by wild-type murine pRB (ΔBXHA) was partially relieved by treatment with the HDAC inhibitor TSA (Fig. 1), suggesting that both HDAC-dependent repression and HDAC-independent repression occur on this promoter. However, repression by ΔBXHA of the E2CAT promoter was unaffected by TSA treatment, suggesting that HDAC does not regulate the E2CAT promoter.
FIG. 1.
E2CAT promoter is not regulated by HDAC activity. Three micrograms of E2CAT or 0.5 μg of RBCAT promoter constructs was transfected into C33A cells along with 3 μg of ΔBXHA. One microgram of pCMVβgal was included in the transfection to normalize CAT activity for transfection efficiency. TSA was added where indicated at a concentration of 100 nM. The activity obtained with the reporter construct in the absence of RB plasmid was arbitrarily set to 100%.
Characterization of pRB B-domain lysine mutants.
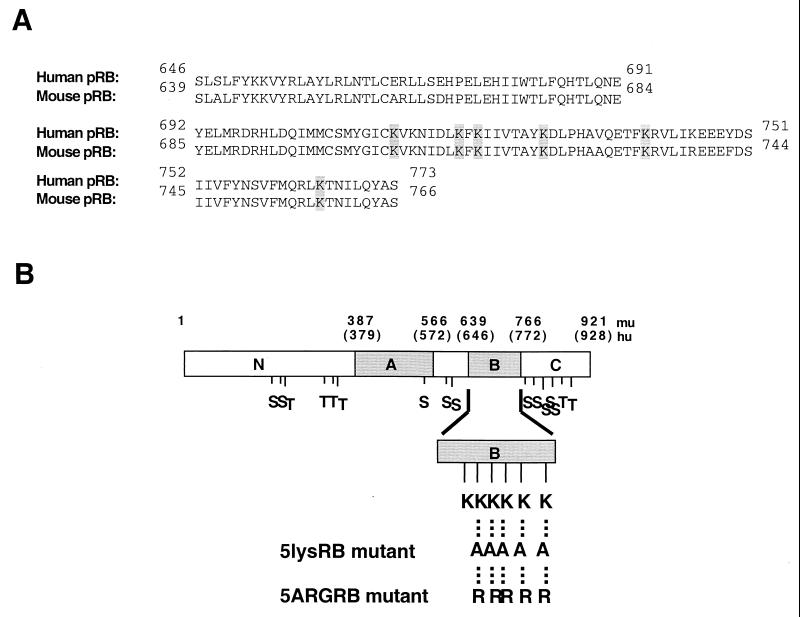
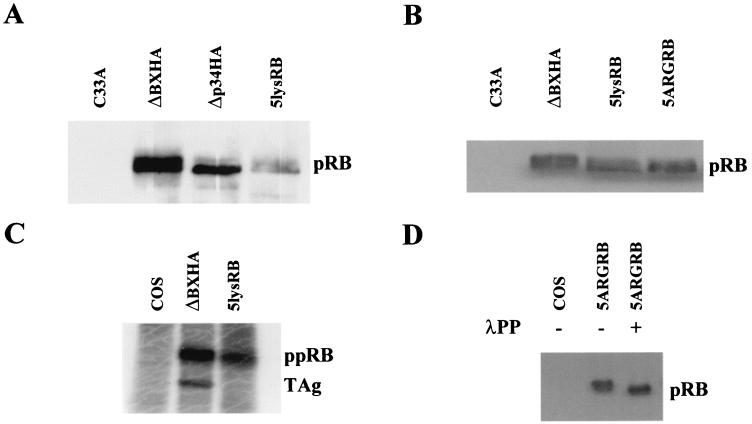
To test the possibility that phosphorylated pRB interacts with positive charges of lysine residues within its B domain, we mutated to alanine or arginine five of the six lysines of the basic patch on the rim of the LXCXE peptide binding site. Site-directed PCR mutagenesis was used to mutate lys713, lys715, lys722, lys733, and lys758 of the murine RB cDNA (Fig. 2A), and the resulting PCR fragment was subcloned into ΔBXHA (murine pRB) under control of the SV40 promoter. The resulting proteins were designated 5lysRB (lysines mutated to alanines) and 5ARGRB (lysines mutated to arginines) (Fig. 2B). Protein expression was confirmed by transfection of the plasmid into C33A cells, immunoprecipitation with anti-HA antibody, and visualization by Western blotting with anti-pRB antibody. Both 5lysRB and 5ARGRB proteins were expressed approximately three times lower than wild-type pRB (ΔBXHA) (shown for 5lysRB in Fig. 3A), but equal expression could be obtained by transfecting threefold less wild-type pRB plasmid (Fig. 3B).
FIG. 2.
(A) NCBI alignment of the B domains of human versus mouse pRB. The six lysine residues involved in the basic patch of the rim of the LXCXE binding site are highlighted. (B) Diagram of the pRB B-domain mutants. Numbers above the schematic designate the amino acids (human in parentheses). Hu corresponds to human pRB, and mu corresponds to murine pRB. The A and B regions required for binding TAg are shown. Positions of potential phosphorylation sites (S [serine] and T [threonine]) in the protein are shown. Positions of the lysine residues in the B domain are indicated. The Lys713, Lys715, Lys722, Lys733, and Lys758 residues that were mutated to alanines or arginines are indicated.
FIG. 3.
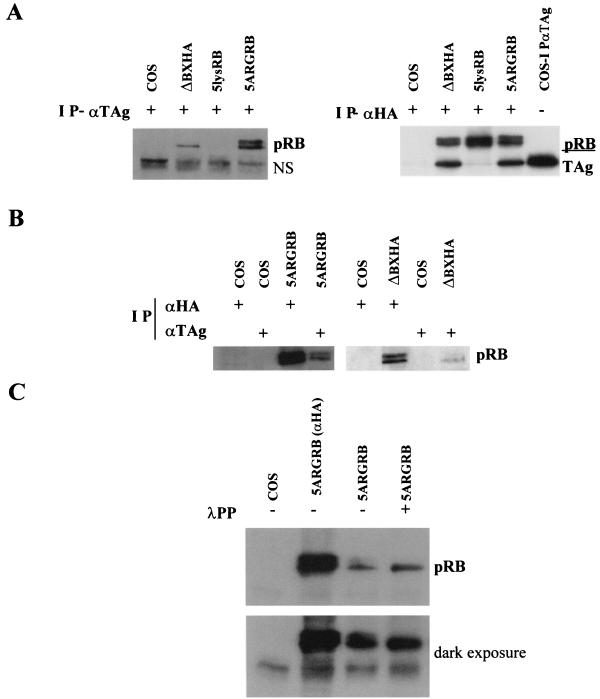
(A) Expression of 5lysRB mutant. Three micrograms of RB plasmid was transfected into C33A cells. Quantities of cell lysates corrected for transfection efficiency with cotransfected pCMVβgal were immunoprecipitated with 2.5 μg of anti-HA probe F-7 (Santa Cruz) followed by Western blotting with anti-pRB antibody 14001A (Pharmingen). (B) Expression of 5lysRB and 5ARGRB. One microgram of wild-type pRB (ΔBXHA) and 3 μg of 5lysRB or 5ARGRB were transfected into C33A cells. Quantities of cell lysates corrected as in panel A were immunoprecipitated with 2.5 μg of anti-HA antibody 12CA5 (Roche) followed by Western blotting with anti-pRB antibody 14001A. (C) Orthophosphate labeling of 5lysRB. At 36 h after transfection of pRB mutants, COS-1 cells were incubated in 1.2 ml of phosphate-deficient medium, containing 800 μCi of 32PO4 per ml at 37°C for 4.5 h. Phosphorylated pRB was immunoprecipitated with anti-HA antibody 12CA5, loaded on SDS-7.5% polyacrylamide gel, and visualized by autoradiography. Coprecipitated large TAg is indicated. (D) 5ARGRB is phosphorylated. Three micrograms of 5ARGRB was transfected into COS-1 cells. Quantities of cell lysates corrected as in panel A were immunoprecipitated with 2.5 μg of anti-HA antibody 12CA5 followed by Western blotting with anti-HA antibody 12CA5. Four hundred units of λPP was added where indicated.
The ability of 5lysRB to become phosphorylated was confirmed by immunoprecipitation of orthophosphate-labeled 5lysRB protein from COS-1-transfected cells (Fig. 3C). In vivo phosphorylation of 5ARGRB was shown by λPP treatment following immunoprecipitation of 5ARGRB with anti-HA antibody from COS-1-transfected cells (Fig. 3D). pRB runs as multiple bands on SDS-polyacrylamide gel electrophoresis, with the lower band representing the hypophosphorylated form of pRB and the upper band(s) representing the hyperphosphorylated form of pRB. Phosphatase treatment resulted in a shift of the upper 5ARGRB band to a lower band, suggesting that this mutant was phosphorylated in vivo.
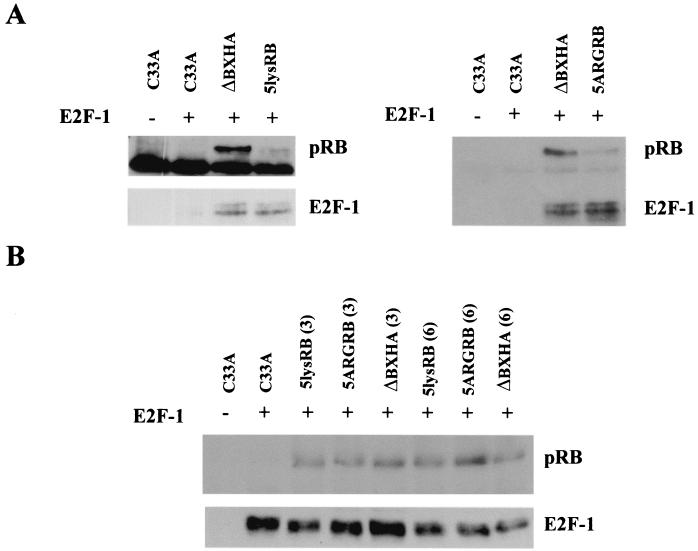
Since the B domain of pRB is required for binding to E2F, we examined the effect of mutation of the lysines in the B domain of pRB on the binding of E2F to pRB. Ten micrograms of each mutant was cotransfected into C33A cells with an E2F-1 expression plasmid, and pRB was immunoprecipitated with anti-pRB antibody. Coprecipitation of E2F-1 was assessed by Western blot analysis with anti-E2F-1 antibody. The top of each nitrocellulose was cut and separately probed with anti-HA antibody to show pRB expression. Both 5lysRB and 5ARGRB coprecipitated E2F-1, suggesting that the mutations in the B domain did not significantly alter the protein's conformation in a way that might decrease binding to E2F-1 (Fig. 4A). Upon initial inspection, it appeared that although each B-domain mutant was expressed much less than wild-type pRB, each mutant precipitated the same amount of E2F-1 protein as wild-type pRB. To test whether the mutant proteins bound more E2F-1 than the wild type, we transfected various amounts of mutant pRB plasmid with a constant amount of E2F-1 into C33A cells. If a mutant bound more E2F-1 than wild-type pRB, then we should see more of that mutant protein coprecipitating with E2F-1, compared to the wild type, when equal amounts of E2F-1 are immunoprecipitated. Western blot analysis of pRB following immunoprecipitation of E2F-1 suggests that similar amounts of mutant and wild-type pRB coprecipitated with equal amounts of E2F-1, whether 3 or 6 μg of RB plasmid was expressed (Fig. 4B), suggesting that the mutant RB proteins binds E2F-1 similar to wild-type pRB.
FIG. 4.
Coimmunoprecipitation of 5lysRB and 5ARGRB with E2F-1. (A) Ten micrograms of RB plasmid was transfected with 0.5 μg of pCMV-E2F-1 (HA tagged) in C33A cells. Quantities of cell lysates corrected, as in Fig. 3A, were immunoprecipitated with 2.5 μg of anti-pRB antibody 14001A. pRB and coprecipitating proteins were separated on an SDS-7.5% polyacrylamide gel. The top half of the nitrocellulose was Western blotted with anti-HA antibody 12CA5, and the bottom half was Western blotted with anti-E2F-1 antibody C-20 (Santa Cruz). (B) Mutant RB proteins do not bind more E2F-1. pCMV-E2F-1 (0.25 μg) (HA tagged) was transfected with 3 or 6 μg of 5lysRB, 5ARGRB, or ΔBXHA in C33A cells. Quantities of cell lysates corrected as in Fig. 3A were immunoprecipitated with 2 μg of anti-E2F-1 antibody C-20, followed by Western blotting with anti-HA antibody 12CA5 to visualize E2F-1. The blot was reprobed with anti-pRB 14001A to visualize pRB.
Repression of E2F transactivation by 5lysRB and 5ARGRB.
We hypothesized that if the phosphorylated polypeptide of pRB interacts with the positive charges of the lysines in the B domain, thereby releasing E2F, then the 5lysRB mutant from which the positive charges have been removed would remain bound to E2F and continue to repress E2F-mediated transactivation despite phosphorylation. Repression of E2F-mediated transcription was determined with the E2CAT reporter construct. Three micrograms of 5lysRB and 5ARGRB plasmids (to correct for reduced protein expression) and 1 μg of ΔBXHA and Δp34HA plasmids were cotransfected with the reporter construct into C33A cells, and the ability to repress transcription was determined. In the absence of kinase activity, 5lysRB, 5ARGRB, ΔBXHA, and Δp34HA repressed E2F-mediated transcriptional activity similarly (Fig. 5).
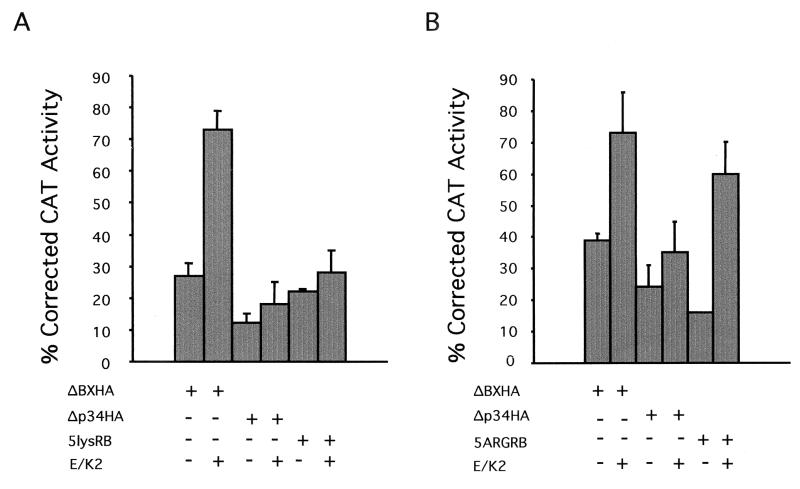
FIG. 5.
Repression of E2CAT reporter construct. Three micrograms of the E2CAT was transfected into C33A cells with 1 μg of ΔBXHA or Δp34HA or 3 μg of 5lysRB (A) or 5ARGRB (B). Five micrograms each of cyclin E (E) and cdk2 (K2) plasmids was also transfected where indicated. One microgram of pCMVβgal was included in the transfection to normalize CAT activity for transfection efficiency. SVLuc was used as filler DNA to prevent promoter competition. The activity obtained with the reporter construct in the absence of RB plasmid or in the presence of cyclin E and cdk2 was arbitrarily set to 100%. The ability of each mutant to suppress transcription was determined by the reduction in CAT activity. Error bars represent standard errors of three experiments performed in duplicate.
In the presence of cyclin E-cdk2 kinase activity, repression of reporter activity by ΔBXHA was released to 75 to 80%. Despite kinase activity, repression by 5lysRB was maintained at approximately 28%, similar to the 20% repression by Δp34HA, which is relatively refractory to phosphorylation due to mutations in 8 of the 16 potential phosphorylation sites (Fig. 5A). The inability of cyclin E-cdk2 to relieve repression by 5lysRB was not due to lack of protein expression (data not shown). Therefore, mutation of five lysines in the B domain of 5lysRB to neutral alanine residues resulted in a protein that remains bound to E2F and continues to repress E2F-mediated transcription, despite phosphorylation. However, cyclin E-cdk2 kinase activity was able to relieve repression by 5ARGRB, in which the positively charged lysines were replaced by positively charged arginines, similar to ΔBXHA (Fig. 5B). These data confirm the importance of the positive charges in the B domain of pRB for inactivation of transcriptional repression by phosphorylation.
In summary, our data (Fig. 1 and 5) indicate that the lysines in the B domain of pRB are important in the relief of repression of E2F activity by phosphorylation of pRB, unrelated to HDAC activity.
TAg binding by the B-domain mutants 5lysRB and 5ARGRB.
Oncoproteins have evolved with acidic residues surrounding the LXCXE residues that are important in binding to pRB. These acidic residues may interact with the positive charges on the six-lysine basic patch on the rim of the LXCXE binding site of pRB (19, 43, 51). To test the importance of the B-domain lysine patch of pRB in binding to TAg, 5lysRB was transfected into COS-1 cells, immunoprecipitated with antibody against TAg, and Western blotted with anti-HA antibody. Unlike wild-type pRB, 5lysRB did not coimmunoprecipitate with TAg (Fig. 6A, left panel) (compare lanes 2 and 3). To confirm that 5lysRB was expressed, but did not bind TAg, 5lysRB was immunoprecipitated with anti-HA antibody, the nitrocellulose was cut in half, and the top half was Western blotted with anti-HA antibody, while the bottom half was Western blotted with anti-TAg antibody (Fig. 6A, right panel). Although 5lysRB was expressed, no TAg was coprecipitated, suggesting that these residues are important in regulation of binding of TAg to pRB. TAg was immunoprecipitated with anti-TAg antibody from COS-1 cells as a positive control (Fig. 6A, right panel, lane 5).
FIG. 6.
5lysRB and 5ARGRB binding to TAg. (A, left panel) One microgram of ΔBXHA or 3 μg of 5lysRB or 5ARGRB was transfected into COS-1 cells. Quantities of cell lysates corrected as in Fig. 3A were immunoprecipitated (IP) with 2.5 μg of anti-TAg antibody 14111A and Western blotted with anti-HA antibody 12CA5. NS, nonspecific band. (Right panel) COS-1 cells were transfected as described above and immunoprecipitated with 2.5 μg of anti-HA antibody 12CA5. Lane 5 contains untransfected COS-1 cells immunoprecipitated with anti-TAg 14111A. The nitrocellulose was cut in half, and the top half was Western blotted with anti-pRB 14001A, while the bottom half was blotted with anti-TAg 14111A. (B) Three micrograms of 5ARGRB (left panel) or 1 μg of ΔBXHA (right panel) was transfected into COS-1 cells. Quantities of cell lysates corrected as in panel A were immunoprecipitated with either 2.5 μg of anti-HA antibody (12CA5 in left panel or probe F-7 in right panel) or 2.5 μg of anti-TAg antibody 14111A as indicated. Western blotting was performed with anti-HA antibody 12CA5. (C) Three micrograms of 5ARGRB was transfected into COS-1 cells. Quantities of cell lysates corrected as in panel A were immunoprecipitated with 2.5 μg of anti-TAg 14111A or 2.5 μg of anti-HA antibody 12CA5 where indicated. Four hundred units of λPP was added to the beads where indicated, following immunoprecipitation and prior to SDS-polyacrylamide electrophoresis.
To confirm the importance of these positive charges in the interaction of pRB with TAg, 5ARGRB, in which positively charged arginines replace the five positively charged lysines, was tested for its ability to bind TAg. Figure 6A shows that 5ARGRB can bind TAg, unlike 5lysRB (left panel compare lanes 3 and 4), confirming the importance of the positive charges in the B domain of pRB in binding to TAg. Surprisingly, both hypophosphorylated and hyperphosphorylated 5ARGRB bound to large TAg. Even though both hyper- and hypophosphorylated pRB were expressed in COS-1 cells, only the hypophosphorylated form of wild-type pRB binds to TAg (Fig. 6B). To confirm that the slower-migrating band that is precipitated with TAg is actually the phosphorylated form of pRB, λPP was added to the beads following immunoprecipitation with anti-TAg antibody, and the coprecipitating proteins were Western blotted with anti-HA antibody (Fig. 6C). In the presence of λPP, there was a shift of 5ARGRB to its faster-migrating form (compare lanes 3 and 4), suggesting that the upper band of 5ARGRB that binds TAg is phosphorylated.
5lysRB and 5ARGRB and HDAC1 binding.
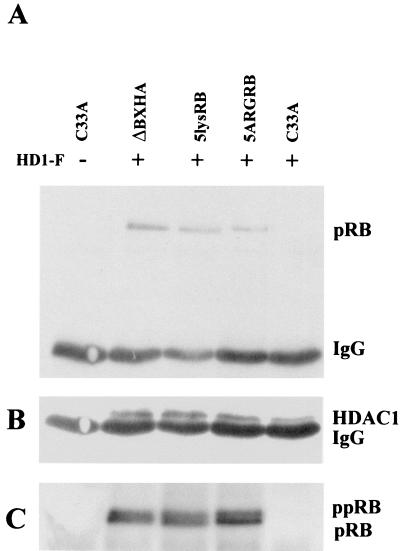
A number of cellular pRB binding proteins rely on an LXCXE motif for binding to pRB. Since we showed that the positively charged lysine residues at the B domain are important for the interaction of pRB with TAg, we tested whether these residues were also necessary for binding of pRB to other LXCXE cellular binding proteins. Wild-type ΔBXHA, 5lysRB, and 5ARGRB were cotransfected into C33A cells with a vector expressing Flag-tagged HDAC1, which contains an LXCXE motif. The wild type and both mutants, 5lysRB and 5ARGRB, coprecipitated with HDAC1 following immunoprecipitation with anti-Flag antibody and Western blotting with anti-pRB antibody (Fig. 7A). Expression of HDAC1 alone did not precipitate pRB from C33A cells that have a nonfunctional pRB (Fig. 7A, lane 5). The presence of HDAC1 in the immunoprecipitate was confirmed in Fig. 7B by reprobing the blot from Fig. 7A with anti-Flag antibody. A duplicate experiment was performed to show the presence of each pRB mutant in the immunoprecipitate (Fig. 7C).
FIG. 7.
Binding of 5lysRB and 5ARGRB with HDAC1. (A) Five micrograms of ΔBXHA, 5lysRB, and 5ARGRB was cotransfected into C33A cells along with the HD1-F plasmid expressing the Flag-tagged HDAC1 protein. Lysates were immunoprecipitated with anti-Flag antibody (M2; Sigma) and Western blotted with anti-pRB antibody 14001A. (B) The above blot was reprobed with anti-Flag antibody to show HDAC1 expression. (C) A duplicate experiment transfected as in panel A was immunoprecipitated with anti-HA antibody 12CA5 followed by Western blot analysis with anti-pRB antibody 14001A.
Acetylation of wild-type pRB and 5lysRB, 5ARGRB mutants.
Mutation of the lysine residues in the B domain resulted in proteins that were expressed approximately three times less than wild-type pRB. Lysine residues are known to be the target for acetylation. Acetylases modify a number of proteins, including transcription factors, and regulate several functions, including protein stability (42). Recent data showed that acetylation stabilizes E2F-1 and increases its DNA binding and activation potential (49). Modification of p53 by acetylation resulted in increased sequence-specific DNA binding (23). Recent experiments also demonstrated that pRB can be acetylated following binding to E1a (8). Therefore, it is possible that mutating the lysines in the B domain of pRB may result in reduced protein because of a reduced ability to be acetylated. Alternatively, removing the positive charges of the lysines may affect the structure in a manner that results in smaller amounts of protein, irrespective of acetylation. If the only requirement is the presence of positive charges, the 5ARGRB mutant protein level should be wild type. However, 5ARGRB was expressed approximately three times less than wild-type pRB, similar to 5lysRB.
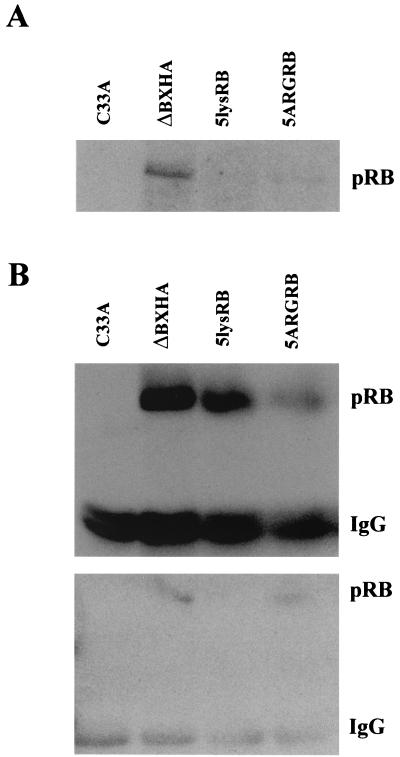
To test for acetylation of pRB, wild-type and mutant RB proteins were expressed in C33A cells, immunoprecipitated with antiacetylated lysine antibody, and Western blotted with anti-pRB antibody. While wild-type pRB was acetylated, acetylation of 5lysRB was absent and acetylation of 5ARGRB was reduced (Fig. 8A). To confirm that the lack of acetylation was not the result of the lack of protein expression, each mutant was expressed in C33A cells, one-quarter of the lysate was immunoprecipitated with anti-HA antibody, and the remainder was immunoprecipitated with antiacetylated lysine antibody. The top panel of Fig. 8B shows that each mutant is expressed, although 5ARGRB was expressed to a much lower level. The bottom panel shows that while pRB is acetylated, 5lysRB is not acetylated even when it is expressed at a level similar to that of wild-type pRB. Replacement of the positive charges in the B domain in the 5ARGRB mutant restored the ability of this protein to become acetylated, although it did not restore the expression levels. This suggests that these positively charged lysine residues in the B domain of pRB might be important in providing structural features that regulate different protein interactions. The inability of 5lysRB to bind TAg is consistent with this hypothesis (Fig. 6). Replacement of the positive charges in 5ARGRB may allow binding of an acetyltransferase to restore acetylation, which is lost in the 5lysRB mutant, but may not affect the binding of protein “X” that regulates pRB protein stability.
FIG. 8.
Acetylation of ΔBXHA, 5lysRB, and 5ARGRB. (A) Ten micrograms of RB plasmid was transfected into C33A cells in triplicate. The lysate from each triplicate was combined and immunoprecipitated with a 1:100 dilution of antiacetylated lysine antibody (9441S; New England Biolabs) followed by Western blotting with anti-pRB antibody 14001A. (B) One-quarter of lysates transfected as in panel A were immunoprecipitated with 2.5 μg of anti-HA antibody 12CA5, and the remaining three-quarters were immunoprecipitated with 1:100 dilution of antiacetylated lysine antibody. The top panel shows a Western blot analysis with anti-HA antibody 12CA5 of the anti-HA immunoprecipitation, while the bottom panel is a Western blot analysis with anti-pRB antibody 14001A of the antiacetylated lysine immunoprecipitation.
DISCUSSION
In recent years, the precise mechanism by which pRB represses transcription has been studied extensively. Besides blocking the transactivation domain of transcription factors or preventing the interaction of transcription factors with the basal transcriptional machinery, pRB recruits chromatin remodeling proteins and corepressors to help silence promoters. Formation of complexes between pRB and HDAC was shown to regulate some but not all E2F-regulated genes (48, 61). Recently Zhang and coworkers showed that complexes between pRB and another chromatin remodeling protein, BRG1, also repress E2F-regulated genes, thereby controlling exit from the G1 and S phases (61). pRB-BRG1 complexes regulated genes such as cyclin A and cdc2, whereas pRB-HDAC complexes regulated cyclin E (61). Repression by pRB of other E2F-regulated genes, such as those coding for E2F-1, B-myb, thymidine kinase, and dihydrofolate reductase, was TSA insensitive, suggesting that these genes are regulated in an HDAC-independent manner (16). However, the interpretation of which genes are regulated by these chromatin remodeling proteins is complicated, since the thymidine kinase and dihydrofolate reductase genes have been shown to be HDAC dependent or HDAC independent, depending on the assay used (48, 61).
We show by TSA treatment that regulation of the E2CAT reporter in C33A cells is not dependent on HDAC activity. In addition, the E2F sites in this promoter act as activators in the presence of E2F and addition of pRB represses the promoter (31). When the E2F sites are removed, pRB no longer represses the promoter (31). Therefore, we examined the role of pRB phosphorylation on inhibition of E2F transactivation.
We show that relief of transcriptional repression by phosphorylation of pRB is dependent on the presence of positively charged residues at the lysine residues in the B domain of pRB. Our previously published data showed that mutation of six or more phosphorylation sites resulted in pRB, which repressed transcription 25-fold better than the wild type in the presence of cyclin E-cdk2 kinase (4). The discovery of a six-lysine basic patch in the B domain of pRB at the rim of the LXCXE binding site offered an explanation of our data. We hypothesized that these basic residues may interact with the minus charges from the phosphates attached to phosphorylation sites on pRB and that this interaction allowed pRB to be removed from its interaction with E2F. 5lysRB contains mutations to alanine of five of the six lysine sites and resulted in a protein that repressed transcription of an E2F promoter even when phosphorylated by cyclin E-cdk2 kinase. However, the 5ARGRB mutant, with positively charged arginines replacing the lysines, lost the ability to repress transcription when phosphorylated by cyclin E-cdk2 kinase activity, similar to wild-type pRB.
Recent studies have shown that phosphorylation of the C terminus of pRB by cyclin D-cdk4 kinase is necessary to release LXCXE-containing proteins such as HDAC, which allows access of cyclin E-cdk2 kinase to phosphorylate Ser567 of pRB and remove pRB from E2F. We showed by treatment with TSA that E2CAT is not regulated by HDAC. Therefore, the repression that we see by pRB on this promoter in C33A cells is HDAC independent. In these cells, either phosphorylation of pRB by cyclin D-cdk4 is not necessary, or there is sufficient cyclin D-cdk4 to phosphorylate pRB prior to cyclin E-cdk2 phosphorylation. We do not see relief of pRB repression of the E2CAT promoter by cyclin D-cdk4 kinase (data not shown).
Our data suggest that phosphorylation of Ser567 by cyclin E-cdk2 kinase activity may not be important in the release of HDAC-independent repression by pRB. All of our previous phosphorylation site mutants contained wild-type Ser567, and some, but not all, were removed from E2F by phosphorylation by cyclin E-cdk2 kinase (4). Additionally, phosphorylation of Ser567 has yet to be shown in vivo (1). Therefore, different mechanisms may release pRB inhibition of E2F activity. Consistent with this conclusion, Zhang and coworkers found that BRG1 interacts with a different site on pRB than HDAC, and phosphorylation of pRB by cyclin D-cdk4 complexes released HDAC, but not BRG1 from pRB (61). Phosphorylation by cyclin E-cdk2 did release pRB from the BRG1 complex, although it was unclear whether cyclin E-cdk2 was acting on pRB or BRG1. C33A cells have little or no BRG1 (57). We showed release of repression of the E2CAT promoter following phosphorylation of pRB by cyclin E-cdk2 in C33A cells, suggesting that the kinase acts on pRB and not BRG1. Therefore, which mechanism is used to release pRB from E2F may depend on which proteins form complexes on a particular promoter.
Harbour et al. (27) also showed that mutation of the B-domain lysines resulted in progressive loss in the ability of CDK to block pRB repressor activity, consistent with our data. They suggest that phosphorylation of the C terminus by cyclin D-cdk4 causes the phosphorylated C terminus to interact with the lysines to relieve pRB repressor activity. However, their experiments were performed with the A, B, and C regions of pRB fused to the GAL4 DNA binding domain. The structure of pRB may be different when it is brought to the promoter through E2F or when bound to the promoter directly via the GAL4 DNA binding domain. We and others showed that the phosphorylation sites required to relieve pRB from E2F were different in the contexts of full-length versus partial protein and binding to free E2F versus E2F bound to a promoter (4, 40). Here we show that when full-length pRB is brought to the promoter through E2F, cyclin E can phosphorylate pRB in the absence of exogenous cyclin D activity and that interaction of the phosphorylated polypeptide with the B-domain lysines is important in releasing pRB from E2F in an HDAC-independent fashion.
The six-lysine basic patch in the B domain of pRB was also hypothesized to be important for binding to oncoproteins such as TAg (43). Oncoproteins have evolved with acidic residues surrounding the LXCXE residues necessary for binding to pRB. The positive charges on the six-lysine basic patch on the rim of the LXCXE binding site of pRB may interact with the acidic charges on the oncoproteins. Mutation to alanine of five of these lysine residues in 5lysRB disrupted binding to TAg, implying that this hypothesis may be correct. However, the recent X-ray crystal structure of the A and B domains of pRB bound to the N terminus of TAg showed that the acidic residues of TAg are very flexible and therefore do not make tight contact with pRB (36). However, three of the lysine residues in the B domain of pRB form hydrogen bonds with the LXCXE motif and other residues surrounding the LXCXE motif of TAg. Mutation of two of these three lysines to alanines in 5lysRB removes three hydrogen bonds and disrupts the stability of the interaction. The importance of these positive charges is confirmed by the binding of TAg to 5ARGRB, in which five of the lysines are replaced by positively charged arginines. Therefore, while the positive charges on the rim of the LXCXE binding site of pRB may not be necessary for interaction with the acidic residues surrounding the LXCXE binding site of TAg, they are necessary for the proper formation of hydrogen bonds with other residues surrounding the LXCXE site, which stabilize the pRB-TAg interaction.
Our protein binding data with the B-domain lysine mutants are consistent with those of another study examining conserved amino acids of pRB that are predicted to make close contacts with the LXCXE peptide (16). Mutation to alanine of tyrosine 709, lysine 713, isoleucine 753, tyrosine 756, asparagine 757, and methionine 761 in human pRB resulted in proteins that were defective in binding to adenovirus E1a and human papillomavirus type 16 E7 oncoproteins, but retained the ability to bind to E2F (16). They also retained the ability to bind to the cellular LXCXE binding proteins, CtIP and HDAC, suggesting that these sites are more important for the binding of viral oncoproteins to pRB than for the binding of cellular LXCXE binding proteins (16). 5lysRB was capable of binding E2F-1 and HDAC1, but was unable to bind SV40 TAg, consistent with the study mentioned above.
Of interest is the finding that mutation of the B-domain lysines to arginines produced an RB protein whose hypo- and hyperphosphorylated forms both bind TAg. Only the hypophosphorylated form of wild-type pRB binds TAg. It is possible that although mutation of the lysines to arginines preserved binding of pRB to TAg, the addition of the larger amino acid may prevent the conformational change necessary for release of TAg when pRB is phosphorylated. Interestingly, both the hypo- and hyperphosphorylated forms of p107 are able to bind TAg as well as E1a (41, 52). It is possible that the replacement of lysines with arginines in the B domain of pRB resulted in a protein with a structure similar to that of p107. Alignment of the B-domain residues from pRB, p107, and p130 is shown in Fig. 9; the residues that correspond to the B-domain lysines are shown in Table 1. While Lys706 and Lys758 of pRB are conserved in p107, only Lys706 is conserved in p130. Lys706 actually makes contact with the LXCXE peptide, whereas the other lysines form the basic rim around the LXCXE binding site (43). The other four lysines are not conserved between pRB and p107 or pRB and p130. Interestingly, two of these residues in p107 and p130 are arginines (Arg868 and Arg879 in p107 and Arg903 and Arg914 in p130). It is possible that by mutating the B-domain lysines of pRB to arginines, we have changed the structure so that pRB functions like p107 in binding to TAg. The binding of p107 and p130 to TAg was necessary for TAg to provide a growth advantage to RB−/− mouse embryo fibroblasts (13, 60).
FIG. 9.
NCBI alignment of the B domains of pRB and p107 (A) and pRB and p130 (B). The six lysine residues involved in the basic patch of the rim of the LXCXE binding site are highlighted. pRB amino acids 639 to 766 were used in the alignment. p107 amino acids 786 to 946 were used in the alignment. p130 amino acids 821 to 1010 were used in the alignment.
TABLE 1.
Conservation of B-domain lysines in the pRB familya
| pRB (5ARGRB) residue | Corresponding residue in:
|
|
|---|---|---|
| p107 | p130 | |
| K706 | K852 | K887 |
| K713-R | T859 | S894 |
| K715-R | Q861 | Q896 |
| K722-R | R868 | R903 |
| K733-R | R879 | R914 |
| K758-R | K938 | |
The residues that correspond to the B-domain lysines of pRB are shown for p107 and p130 following alignment with the NCBI BLAST program. The 5ARGRB mutant has lysine-to-arginine mutations as indicated by −R. Arginine residues common to 5ARGRB, p107, and p130 are shown in boldface.
It is interesting to speculate that 5ARGRB could have some therapeutic value. SV40 viral DNA and/or TAg have been reported to be present in certain types of human cancers (6, 7). Since both the hypo- and hyperphosphorylated forms of 5ARGRB can bind to TAg, 5ARGRB could potentially soak up the TAg in a cell so that it cannot interrupt the cell cycle by binding endogenous pRB.
The discovery that pRB is acetylated is relatively novel. Acetylation has been shown to play an important role in both activation of transcription as well as repression of transcription (42). An important target of pRB, E2F-1, has been shown to be acetylated, and acetylation stimulates the function of the non-pRB-bound free form of E2F-1 (49). E2F-1 was also shown to be deacetylated by pRB-associated HDAC, suggesting that pRB's ability to repress E2F activity involves deacetylating E2F, thereby decreasing its DNA binding ability and activation potential (49). Generally, acetylation appears to reduce protein-protein interactions as in the binding of the Drosophila TCF transcription factor to its coactivator, armadillo, and the association of nuclear receptors with their coactivator, ACTR (see reference 42 and references cited therein). Additionally, acetylation may change the conformation of a protein in such a manner to generate a new binding site, such as in the generation of a recognition site for the bromodomain following acetylation of histones (42). The role of acetylation of pRB remains to be determined. However, like phosphorylation of pRB, regulation of pRB by acetylation will probably be complicated.
Acknowledgments
This work was supported in part by the National Cancer Institute of Canada with funds from the Terry Fox Run, the Medical Research Council of Canada, and Apotex, Inc., through the University-Industry Program.
We thank Nick Dyson for the gift of the HDAC1 plasmid and Sanja Pajovic for helpful discussions and critical reading of the manuscript.
REFERENCES
- 1.Adams, P. D. 2001. Regulation of the retinoblastoma tumor suppressor protein by cyclin/cdks. Biochim. Biophys. Acta 1471:M123-M133. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama, T., T. Ohuchi, S. Sumida, K. Matsumoto, and K. Toyoshima. 1992. Phosphorylation of the retinoblastoma protein by cdk2. Proc. Natl. Acad. Sci. USA 89:7900-7904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bremner, R., B. L. Cohen, M. Sopta, P. A. Hamel, C. J. Ingles, B. L. Gallie, and R. A. Phillips. 1995. Direct transcriptional repression by pRB and its reversal by specific cyclins. Mol. Cell. Biol. 15:3256-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, V. D., R. A. Phillips, and B. L. Gallie. 1999. Cumulative effect of phosphorylation of pRB on regulation of E2F activity. Mol. Cell. Biol. 19:3246-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchkovich, K., L. A. Duffy, and E. Harlow. 1989. The retinoblastoma protein is phosphorylated during specific phases of the cell cycle. Cell 58:1097-1105. [DOI] [PubMed] [Google Scholar]
- 6.Butel, J. S., and J. A. Lednicky. 1999. Cell and molecular biology of simian virus 40: implications for human infections and disease. J. Natl. Cancer Inst. 91:119-134. [DOI] [PubMed] [Google Scholar]
- 7.Carbone, M., P. Rizzo, and H. I. Pass. 1997. Simian virus 40, poliovaccines and human tumors: a review of recent developments. Oncogene 15:1877-1888. [DOI] [PubMed] [Google Scholar]
- 8.Chan, H. M., M. Krstic-Demonacos, L. Smith, C. Demonacos, and N. B. La Thangue. 2001. Acetylation control of the retinoblastoma tumor-suppressor protein. Nat. Cell Biol. 3:667-674. [DOI] [PubMed] [Google Scholar]
- 9.Chen, L. I., T. Nishinaka, K. Kwan, I. Kitabayashi, K. Yokoyama, Y.-H. F. Fu, S. Grunwald, and R. Chiu. 1994. The retinoblastoma gene product RB stimulates Sp1 mediated transcription by liberating Sp1 from a negative regulator. Mol. Biol. Cell 14:4380-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, P.-L., D. J. Riley, Y. Chen, and W.-H. Lee. 1996. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 10:2794-2804. [DOI] [PubMed] [Google Scholar]
- 11.Chen, P.-L., D. J. Riley, S. Chen-Kiang, and W.-H. Lee. 1996. Retinoblastoma protein directly interacts with and activates the transcription factor NF-IL6. Proc. Natl. Acad. Sci. USA 93:465-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, P.-L., P. Scully, J.-Y. Shew, J. Y. J. Wang, and W.-H. Lee. 1989. Phosphorylation of the retinoblastoma gene product is modulated during the cell cycle and cellular differentiation. Cell 58:1193-1198. [DOI] [PubMed] [Google Scholar]
- 13.DeCaprio, J. A. 1999. The role of the J domain of SV40 large T in cellular transformation. Biologicals 27:23-28. [DOI] [PubMed] [Google Scholar]
- 14.DeCaprio, J. A., Y. Fusuke, F. Ajchenbaum, J. D. Griffin, and D. M. Livingston. 1992. The retinoblastoma-susceptibility gene product becomes phosphorylated in multiple stages during cell cycle entry and progression. Proc. Natl. Acad. Sci. USA 89:1795-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeCaprio, J. A., J. W. Ludlow, D. Lynch, Y. Furukawa, J. Griffin, H. Piwnica-Worms, C.-M. Huang, and D. M. Livingston. 1989. The product of the retinoblastoma susceptibility gene has properties of a cell cycle regulatory element. Cell 58:1085-1095. [DOI] [PubMed] [Google Scholar]
- 16.Dick, F. A., E. Sailhamer, and N. J. Dyson. 2000. Mutagenesis of the pRB pocket reveals that cell cycle arrest functions are separable from binding to viral oncoproteins. Mol. Cell. Biol. 20:3715-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dowdy, S. F., P. W. Hinds, K. Louie, S. I. Reed, A. Arnold, and R. A. Weinberg. 1993. Physical interaction of the retinoblastoma protein with human D cyclins. Cell 73:499-511. [DOI] [PubMed] [Google Scholar]
- 18.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 19.Dyson, N., P. M. Howley, K. Munger, and E. Harlow. 1989. The human papilloma virus-16 E7 oncoprotein is able to bind the retinoblastoma gene product. Science 243:934-937. [DOI] [PubMed] [Google Scholar]
- 20.Ewen, M. E., H. K. Sluss, C. J. Sherr, H. Matsushime, J.-Y. Kato, and D. M. Livingston. 1993. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell 73:487-497. [DOI] [PubMed] [Google Scholar]
- 21.Flemington, E. K., S. H. Speck, and W. G. Kaelin. 1993. E2F-1-mediated transactivation is inhibited by complex formation with the retinoblastoma susceptibility gene product. Proc. Natl. Acad. Sci. USA 90:6914-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gill, R. M., P. A. Hamel, Z. Jiang, E. Zacksenhaus, B. L. Gallie, and R. A. Phillips. 1994. Characterization of the human RB1 promoter and of elements involved in transcriptional regulation. Cell Growth Differ. 5:467-474. [PubMed] [Google Scholar]
- 23.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595-606. [DOI] [PubMed] [Google Scholar]
- 24.Gu, W., J. W. Schneider, G. Condorelli, S. Kaushal, V. Mahdavi, and B. Nadal-Ginard. 1993. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell 72:309-324. [DOI] [PubMed] [Google Scholar]
- 25.Hagemeier, C., A. J. Bannister, A. Cook, and T. Kouzarides. 1993. The activation domain of transcription factor PU.1 binds the retinoblastoma (RB) protein and the transcription factor TFIID in vitro: RB shows sequence similarity to TFIID and TFIIB. Proc. Natl. Acad. Sci. USA 90:1580-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamel, P. A., R. M. Gill, R. A. Phillips, and B. L. Gallie. 1992. Transcriptional repression of the E2-containing promoters EIIaE, c-myc, and RB1 by the product of the RB1 gene. Mol. Cell. Biol. 12:3431-3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harbour, J. W., R. X. Luo, A. D. Santi, A. A. Postigo, and D. C. Dean. 1999. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell 98:859-869. [DOI] [PubMed] [Google Scholar]
- 28.Hatakeyama, M., J. A. Brill, G. R. Fink, and R. A. Weinberg. 1994. Collaboration of G1 cyclins in the functional inactivation of the retinoblastoma protein. Genes Dev. 8:1759-1771. [DOI] [PubMed] [Google Scholar]
- 29.Helin, K. 1998. Regulation of cell proliferation by the E2F transcription factors. Curr. Opin. Genet. Dev. 8:28-35. [DOI] [PubMed] [Google Scholar]
- 30.Helin, K., E. Harlow, and A. Fattaey. 1993. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol. Cell. Biol. 13:6501-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hiebert, S. W., S. P. Chellappan, J. M. Horowitz, and J. R. Nevins. 1992. The interaction of RB with E2F coincides with an inhibition of the transcriptional activity of E2F. Genes Dev. 6:177-185. [DOI] [PubMed] [Google Scholar]
- 32.Hinds, P. W., S. Mittnacht, V. Dulic, A. Arnold, S. I. Reed, and R. A. Weinberg. 1992. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell 70:993-1006. [DOI] [PubMed] [Google Scholar]
- 33.Horton, L. E., Y. Qian, and D. J. Templeton. 1995. G1 cyclins control of the retinoblastoma gene product growth regulation activity via upstream mechanisms. Cell Growth Differ. 6:395-407. [PubMed] [Google Scholar]
- 34.Hu, Q., J. A. Lees, K. J. Buchkovich, and E. Harlow. 1992. The retinoblastoma protein physically associates with the human cdc2 kinase. Mol. Cell. Biol. 12:971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato, J.-Y., H. Matsushime, S. W. Hiebert, M. E. Ewen, and C. J. Sherr. 1993. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 7:331-342. [DOI] [PubMed] [Google Scholar]
- 36.Kim, H.-Y., B.-Y. Ahn, and Y. Cho. 2001. Structural basis for the inactivation of retinoblastoma tumor suppressor by SV40 large T antigen. EMBO J. 20:295-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, S.-J., U. S. Onwuta, Y. I. Lee, R. Li, M. R. Botchan, and P. D. Robbins. 1992. The retinoblastoma gene product regulates Sp1-mediated transcription. Mol. Cell. Biol. 12:2455-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim, S.-J., S. Wagner, F. Liu, M. A. Reilly, P. D. Robbins, and M. R. Green. 1992. Retinoblastoma gene product activates expression of the human TGF-beta 2 gene through transcription factor ATF-2. Nature 358:331-334. [DOI] [PubMed] [Google Scholar]
- 39.Knudsen, E. S., and J. Y. J. Wang. 1996. Differential regulation of retinoblastoma protein function by specific cdk phosphorylation sites. J. Biol. Chem. 271:8313-8320. [DOI] [PubMed] [Google Scholar]
- 40.Knudsen, E. S., and J. Y. J. Wang. 1997. Dual mechanisms for the inhibition of E2F binding to RB by cyclin-dependent kinase-mediated RB phosphorylation. Mol. Cell. Biol. 17:5771-5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knudsen, E. S., and J. Y. J. Wang. 1998. Hyperphosphorylated p107 and p130 bind to T-antigen: identification of a critical regulatory sequence present in Rb but not in p107/p130. Oncogene 16:1655-1663. [DOI] [PubMed] [Google Scholar]
- 42.Kouzarides, T. 2000. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 19:1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee, J.-O., A. A. Russo, and N. P. Pavletich. 1998. Structure of the retinoblastoma tumor-suppressor pocket domain bound to a peptide from HPV E7. Nature 391:859-865. [DOI] [PubMed] [Google Scholar]
- 44.Lees, J. A., K. J. Buchkovich, D. R. Marshak, C. W. Anderson, and E. Harlow. 1991. The retinoblastoma protein is phosphorylated on multiple sites by human cdc2. EMBO J. 10:4279-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin, B. T.-Y., S. Gruenwald, A. O. Morla, W.-H. Lee, and J. Y. J. Wang. 1991. Retinoblastoma cancer suppressor gene product is a substrate of the cell cycle regulator cdc2 kinase. EMBO J. 10:857-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ludlow, J. W., C. L. Glendening, D. M. Livingston, and J. A. DeCaprio. 1993. Specific enzymatic dephosphorylation of the retinoblastoma protein. Mol. Cell. Biol. 13:367-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lundberg, A. S., and R. A. Weinberg. 1998. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol. Cell. Biol. 18:753-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo, R. X., A. A. Postigo, and D. C. Dean. 1998. Rb interacts with histone deacetylase to repress transcription. Cell 92:463-473. [DOI] [PubMed] [Google Scholar]
- 49.Martinez-Balbas, M. A., U.-M. Bauer, S. J. Nielsen, A. Brehm, and T. Kouzarides. 2000. Regulation of E2F1 activity by acetylation. EMBO J. 19:662-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mittnacht, S. 1998. Control of pRB phosphorylation. Curr. Opin. Genet. Dev. 8:21-27. [DOI] [PubMed] [Google Scholar]
- 51.Munger, K., B. A. Werness, N. Dyson, W. C. Phelps, E. Harlow, and P. M. Howley. 1989. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 8:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parreno, M., J. Garriga, A. Limón, X. Mayol, G. R. Beck, Jr., E. Moran, and X. Graña. 2000. E1A blocks hyperphosphorylation of p130 and p107 without affecting the phosphorylation status of the retinoblastoma protein. J. Virol. 74:3166-3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 54.Sellers, W. R., B. G. Novitch, S. Miyake, A. Heith, G. A. Otterson, F. J. Kaye, A. B. Lassar, and W. G. Kaelin. 1998. Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 12:95-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh, P., J. Coe, and W. Hong. 1995. A role for retinoblastoma protein in potentiating transcriptional activation by the glucocorticoid receptor. Nature 374:562-565. [DOI] [PubMed] [Google Scholar]
- 56.Sleigh, M. J. 1986. A nonchromatographic assay for expression of the chloramphenicol acetyltransferase gene in eukaryotic cells. Anal. Biochem. 156:251-256. [DOI] [PubMed] [Google Scholar]
- 57.Strobeck, M. W., K. E. Knudsen, A. F. Fribourg, M. F. DeCristofaro, B. E. Weissman, A. N. Imbalzano, and E. S. Knudsen. 2000. BRG-1 is required for RB-mediated cell cycle arrest. Proc. Natl. Acad. Sci. USA 97:7748-7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Udvadia, A. J., K. T. Rogers, P. D. R. Higgins, Y. Murata, K. H. Martin, P. A. Humphrey, and J. M. Horowitz. 1993. Sp-1 binds promoter elements regulated by the RB protein and Sp-1 mediated transcription is stimulated by RB coexpression. Proc. Natl. Acad. Sci. USA 90:3265-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, C.-Y., B. Petryniak, C. B. Thompson, W. G. Kaelin, and J. M. Leiden. 1993. Regulation of the Ets-related transcription factor Elf-1 by binding to the retinoblastoma protein. Science 260:1330-1335. [DOI] [PubMed] [Google Scholar]
- 60.Zalvide, J., and J. A. DeCaprio. 1995. Role of pRb-related proteins in simian virus 40 large-T-antigen-mediated transformation. Mol. Cell. Biol. 15:5800-5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang, H. S., M. Gavin, A. Dahiya, A. A. Postigo, D. Ma, R. X. Luo, J. W. Harbour, and D. C. Dean. 2000. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell 101:79-89. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, H. S., A. A. Postigo, and D. C. Dean. 1999. Active transcriptional repression by the Rb-E2F complex mediates G1 arrest triggered by p16INK4a, TGFβ, and contact inhibition. Cell 97:53-61. [DOI] [PubMed] [Google Scholar]