Abstract
Polyglutamine expansion causes Huntington disease (HD) and at least seven other neurodegenerative diseases. In HD, N-terminal fragments of huntingtin with an expanded glutamine tract are able to aggregate and accumulate in the nucleus. Although intranuclear huntingtin affects the expression of numerous genes, the mechanism of this nuclear effect is unknown. Here we report that huntingtin interacts with Sp1, a transcription factor that binds to GC-rich elements in certain promoters and activates transcription of the corresponding genes. In vitro binding and immunoprecipitation assays show that polyglutamine expansion enhances the interaction of N-terminal huntingtin with Sp1. In HD transgenic mice (R6/2) that express N-terminal-mutant huntingtin, Sp1 binds to the soluble form of mutant huntingtin but not to aggregated huntingtin. Mutant huntingtin inhibits the binding of nuclear Sp1 to the promoter of nerve growth factor receptor and suppresses its transcriptional activity in cultured cells. Overexpression of Sp1 reduces the cellular toxicity and neuritic extension defects caused by intranuclear mutant huntingtin. These findings suggest that the soluble form of mutant huntingtin in the nucleus may cause cellular dysfunction by binding to Sp1 and thus reducing the expression of Sp1-regulated genes.
Huntington disease (HD) is an autosomally dominant degenerative disorder resulting from expansion (>37 units) of a polyglutamine repeat in huntingtin, a 350-kDa protein of unknown function (15). The polyglutamine repeat is localized in the N-terminal region of huntingtin and is encoded by exon1 of the HD gene. Full-length huntingtin is predominantly distributed in the cytoplasm, whereas N-terminal fragments of huntingtin with expanded polyglutamine tracts accumulate in the nucleus (7, 8, 10). N-terminal huntingtin fragments containing expanded polyglutamine tracts are also toxic to cells. For example, transgenic mice expressing N-terminal fragments of mutant huntingtin develop rapidly progressing neurological symptoms (7, 37) that are more severe than those of mice expressing full-length mutant huntingtin (13, 34). Smaller N-terminal huntingtin fragments, when transfected into cultured cells, kill more cells than do larger huntingtin fragments (11, 27).
The mechanisms for the cellular pathology associated with N-terminal-mutant huntingtin are unknown. N-terminal fragments of mutant huntingtin also form intranuclear aggregates (7, 8, 10, 37), a pathological hallmark that is found in many other polyglutamine diseases (43). Recent studies suggest that nuclear polyglutamine inclusions recruit transcription factors and that this recruitment affects gene expression (29, 39). Indeed, intranuclear huntingtin alters the expression of a number of genes, both in HD cells (22) and in transgenic animals (3, 26). The nuclear effect of mutant huntingtin may stem from its interactions with a number of transcription factors, such as the nuclear receptor corepressor (N-CoR) (2), cyclic AMP-responsive element-binding protein (CREB)-binding protein (CBP) (18, 29, 38, 39), and TATA-binding proteins (TBP) (14, 30). It has been found that, of these transcription factors, TBP and CBP are recruited by polyglutamine inclusions (14, 29, 30, 39). However, several studies have suggested that nuclear polyglutamine inclusions are not associated with neurodegeneration (19, 36). Thus, the role of recruitment of transcription factors by nuclear inclusions remains to be defined. In addition, how the interaction of huntingtin with transcription factors affects gene expression is unclear. The down-regulation of numerous genes in HD animals (3, 26) also suggests that intranuclear huntingtin binds to other transcription factors that are vitally important for the expression of a large number of genes. Based on these ideas, we sought to uncover transcription factors whose function is essential for the expression of many genes and whose binding to huntingtin is influenced by polyglutamine expansion.
Our previous studies and others have shown that intranuclear huntingtin can inhibit the expression of a number of genes (3, 22, 26). We noticed that the expression of many of these genes is mediated by Sp1, a transcription activator that is essential for the transcription of numerous genes (5, 6, 9). In this study, we show that the soluble form of N-terminal-mutant huntingtin binds more tightly to Sp1 than does aggregated huntingtin. Using the nerve growth factor (NGF) receptor (NGFR) promoter as a probe, we found that mutant huntingtin inhibits the binding of Sp1 to the NGFR promoter and suppresses NGFR transcriptional activity. Overexpression of Sp1 reduces cellular dysfunction and toxicity associated with intranuclear mutant huntingtin. These results suggest that the interaction between Sp1 and mutant huntingtin also contributes to HD pathology.
MATERIALS AND METHODS
Antibodies and reagents.
Rabbit antihuntingtin antibody EM48 was generated using a glutathione S-transferase (GST) fusion protein containing the first 256 amino acids of human huntingtin with a deletion of polyglutamine and polyproline stretches (10, 21). The same antigen was also used for the production of mouse monoclonal antibodies by the Auburn University Hybridoma Facility. Of seven hybridoma cell lines extensively characterized, one produced a mouse monoclonal antibody, mEM48, which had immunoreactivity similar to that of rabbit antibody EM48 and which was used in the present study. Other reagents used in the study included antibodies against tubulin (Sigma, St. Louis, Mo.), mouse monoclonal antibody 12CA5 (Boehringer Mannheim, Indianapolis, Ind.), a rabbit antibody against Sp1 (SC-59; Santa Cruz Biotechnology, Santa Cruz, Calif.), a rabbit antibody against green fluorescent protein (GFP), and Hoechst 33258 (Molecular Probes Inc., Eugene, Oreg.).
Production of fusion proteins.
cDNAs encoding N-terminal human huntingtin (amino acids 1 to 171) were inserted in-frame into the pET-28a vector (Novagen Inc., Madison, Wis.) to generate huntingtin fusion proteins that were tagged with six histidines (His) at the N terminus. These fusion proteins contained either 23 (His-23Q) or 120 (His-120Q) glutamines in the repeat region. BL21 bacteria were transformed with these constructs and grown at room temperature to optical density at 600 nm of 0.6. After induction with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 2 h, the cells were spun down and lysed in binding buffer (5 mM imidazole, 500 mM NaCl, 20 mM Tris-HCl [pH 8.0], 25 mM beta-mercaptoethanol, 0.1% NP-40, 1 mM phenylmethylsulfonyl fluoride [PMSF]). The lysates were clarified by centrifugation (14,000 × g for 30 min) and passed through a nickel column. The column was washed three times with binding buffer containing 50 mM imidazole, and the proteins bound to the column were eluted with elution buffer (400 mM imidazole, 500 mM NaCl, 20 mM Tris-HCl [pH 8.0], 20 mM beta-mercaptoethanol, 0.1% NP-40, 1 mM PMSF). Eluate fractions were collected, dialyzed against phosphate-buffered saline (PBS), and analyzed by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE). Coomassie staining and Western blotting were used to examine the purified huntingtin.
Rat cDNA encoding full-length Sp1 was isolated using reverse transcription-PCR with primers rSp1-S1 5"-GTGAATTCATGAGCGACCAAGAT-3") and rSp1-A2 (5"-TGTCTCGAGTCAGAAACCATTGC-3"). Full-length Sp1 cDNA encoding Sp1 amino acids 1 to 788 (Sp1-F) was inserted in-frame into GST fusion protein vector pGEX-4T-1. This construct was cut at SacI or AccI sites to generate two constructs encoding GST fusion proteins containing N-terminal Sp1 (amino acids 1 to 166 [Sp1-N] and 1 to 565 [Sp1-P]). A GST C-terminal construct encoding Sp1 amino acids 569 to 788 (Sp1-C) was generated by PCR using primers 5"-rSp1-CT (5"-GTGGATCCCTTGGCCTTCATGGA-3") and 3"-rSp1-CT (5"-TAGAATTCGAAACCATTGCCACTGATA-3"). These GST fusion proteins were produced in BL21 bacteria and purified using methods described previously (23).
Cell lines.
GFP was fused in-frame to the N terminus of the HD exon1 protein containing 20 (20Q) or 120 (120Q) glutamine repeats using GFP vector pEGFP-C3 (Clontech, Palo Alto, Calif.). The cDNAs were transfected into human embryonic kidney (HEK) 293 cells using Lipofectamine (Life Technologies, Inc.). Cells stably expressing 20Q or 120Q were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, 100 μg of penicillin/ml, 100 μg of streptomycin/ml, and selective antibiotic G-418 (500 μg/ml). To identify stably transfected cells, antibodies against EM48 or GFP were used for fluorescence microscopy and Western blotting. During the selection, we obtained four independent lines of GFP-120Q cells and six lines of GFP-20Q cells. A representative cell line of each group was used for in vitro binding assays.
GST pull down assay.
GFP-huntingtin-transfected HEK 293 cells were collected in buffer (PBS, 0.2% Triton X-100, 1 mM PMSF, 1:1,000-diluted protease inhibitor cocktail [P8340; Sigma]) and sonicated for 30 s. After centrifugation at 6,500 × g for 5 min at 4°C, the clarified supernatant was incubated with 20 μg of purified GST-Sp1 fusion proteins linked to glutathione beads in binding buffer (PBS, 0.5% Triton X-100) for 2 h. For the interaction of GST-Sp1 with purified His-huntingtin, 5 μg of GST or GST-Sp1 linked to glutathione beads was incubated with 100 ng of His-huntingtin in 400 μl of PBS for 2 h at 4°C. The beads were precipitated and washed twice with 1 ml of PBS. The washed beads were then subjected to Western blotting with mEM48 (1:500 dilution). To assess the relative amount of huntingtin bound to Sp1, the ratio of the bound protein to the input (bound/input ratio) was determined using personal densitometer S1 (Molecular Dynamics).
Coimmunoprecipitation assay.
Full-length Sp1 cDNA was inserted into a PRK vector that provides the hemagglutinin (HA) epitope (YPYDVPDYA) at the C terminus of the transfected protein (23). HEK 293 cells in a 10-cm-diameter plate were cotransfected with 7 μg of huntingtin (NLS-20Q or NLS-150Q) and Sp1-HA for 48 h using Lipofectamine. The transfected cells were lysed in 1 ml of NP-40 buffer (10 mM Tris [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40) at 4°C for 5 min and were spun down at 800 × g for 3 min. The supernatant and the pellet (nuclear fraction) were collected for immunoprecipitation. The pellet was resuspended in 600 μl of NP-40 lysis buffer and sonicated for 10 s on ice. Both soluble and nuclear fractions were incubated with 5 μl of 12CA5 at 4°C for 2 h. Protein A-Sepharose (30 μl, 1:1 dilution) was then added to the incubation mixture for 1 h at 4°C to precipitate the immunocomplex. After centrifugation at 325 × g for 20 s, the precipitated Sepharose was washed twice in NP-40 lysis buffer and subjected to Western blot analysis with EM48 and 12CA5 antibodies. HEK 293 cells transfected with huntingtin alone served as a control to verify the specific coimmunoprecipitation of transfected Sp1-HA and huntingtin by antibody 12CA5.
To precipitate Sp1 protein complexes from brain tissue, brain cortices from R6/2 mice at 4 weeks of age were used. These mice (B6CBA-TgN [HDexon1] strain 62; Jackson Laboratory, Bar Harbor, Maine) express both soluble and aggregated forms of mutant huntingtin in the cortex. The brain tissue (0.1 g/ml) was lysed in buffer (10 mM HEPES [pH. 7.5], 0.2% Triton X-100, 10 mM EDTA, 1 mM PMSF, protease inhibitor cocktail [1,000×; P8340; Sigma]). The lysates were clarified by centrifugation at 8,200 × g for 5 min. The supernatant (300 μl) was subjected to immunoprecipitation with 2.5 μl of rabbit anti-Sp1 for 2 h at 4°C. Protein A-Sepharose (15 μl) was added to the mixture, which was incubated for an additional 1 h. The same brain sample incubated with rabbit immunoglobulin G (IgG) was included as a control. The protein A beads were precipitated by centrifugation, washed twice with lysis buffer, resolved by SDS-8 to 10% PAGE, and analyzed with mEM48 (1:500 dilution).
Immunostaining.
For immunostaining of HD mouse brain, R6/2 mice at 12 weeks of age were anesthetized and perfused intracardially with PBS for 30 s, followed by 4% paraformaldehyde in 0.1 M phosphate buffer at pH 7.2. Brains were removed, cryoprotected in 30% sucrose at 4°C, and sectioned at 40 μm using a freezing microtome. Free-floating sections were preblocked in 4% normal goat serum in PBS-0.1% Triton X-100-avidin (10 μg/ml). The sections were incubated with EM48 or the anti-Sp1 antibody at room temperature for 24 h. The immunoreactive product was visualized with the avidin-biotin complex kit (ABC Elite; Vector, Burlingame, Calif.).
For immunostaining of cultured HEK 293 cells, the cells were transfected using Lipofectamine with the HD exon1 protein containing 150 glutamines in the repeat. The nuclear localization sequence (NLS; PKKKRKV) of the simian virus large T antigen was placed at the N terminus of huntingtin (NLS-150Q) in the PRK vector to facilitate the nuclear accumulation of the transfected huntingtin. Sp1-F and Sp1-N cDNAs were transferred from GST fusion protein constructs to the PRK-HA vector for their transfection into HEK 293 cells. Cells were plated in six-well plates, transfected, fixed with 4% paraformaldehyde in PBS for 15 min, permeabilized with 0.2% Triton X-100 in PBS for 30 min, blocked with 3% bovine serum albumin (BSA) in PBS for 1 h, and incubated with EM48 and an anti-HA antibody (12CA5) in 3% BSA in PBS overnight at 4°C. After several washes, the cells were incubated with secondary antibodies conjugated with either fluorescein isothiocyanate or rhodamine (Jackson Immunoresearch Lab, West Grove, Pa.) and Hoechst dye (1 μg/ml), which labels the nucleus. A Zeiss fluorescence inverted microscope (Axioskop 2) and a 3CCD camera video system (Dage-MTI Inc., Michigan City, Ind.) were used to capture fluorescent images with different optical filters. The captured images were stored and processed using Photoshop software (Adobe Systems, San Jose, Calif.).
EMSA.
Rat adrenal pheochromocytoma (PC12) cells from two 10-cm-diameter dishes were resuspended in NP-40 lysis buffer (10 mM Tris [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40), vortexed briefly, and incubated on ice for 5 min. The nuclear fraction was pelleted at 1,100 × g for 5 min. Nuclear extracts were prepared by incubating the nuclear pellet in buffer (10 mM HEPES [pH 8.0], 25% glycerol, 0.4 M NaCl, 0.1 mM EDTA with protease inhibitors) for 30 min at 4°C. After centrifugation at 13,000 × g for 15 min at 4°C to remove nuclear debris, the nuclear extracts were saved for electromobility supershift assays (EMSA).
To perform EMSA, the sense and antisense oligonucleotides corresponding to bases −80 to −41 of the rat NGFRp75 promoter (33) (5"-GGTAGTTGGGCGGGCTGGGCGGGGAGGAGGCGGGGCTGC-3") were synthesized (DNA Core Facility, Emory University). This region contains three GC-rich boxes, which are Sp1 binding sites (underlined). Mutant sense and antisense oligonucleotides (5"-GGTAGTTGGaatGGCTGGaatGGGAGGAGGaatGGCTGC-3"), in which the Sp1 binding sites were mutated (lowercase letters), were also synthesized as a control. Double-stranded oligonucleotides were obtained by annealing the sense and antisense oligonucleotides at 70°C for 10 min in 100 μl of buffer (25 mM Tris-HCl [pH 8.0], 0.5 mM MgCl2, 25 mM NaCl) and cooling to room temperature. An aliquot (1 pmol) of the double-stranded oligonucleotides was 5" end labeled with [32P]ATP (13.72 μCi/μl; Amersham Pharmacia Biotech) and T4 polynucleotide kinase for 1 h at 37°C, followed by incubation at 70°C for 10 min, and then cooled to room temperature. The probe was purified on a Sephadex G-25 column (Amersham Pharmacia Biotech). A reaction for binding DNA to proteins was performed as described previously (4). Briefly, 5 μg of nuclear extracts of PC12 cells was incubated with 10 fmol of the 32P-radiolabeled probe (20,000 cpm) in 20 μl of binding buffer (20 mM HEPES [pH 7.9], 80 mM KCl, 5 mM MgCl2, 0.2 mM dithiothreitol, 0.1 mM EDTA, 2% Ficoll, 5% glycerol, 0.1% NP-40, 0.5 μg of tRNA/μl, 0.04 μg of polydeoxyinosinic-deoxycytidylic acids Σ/μl). The reaction mixture was incubated for 25 min at room temperature. For competition assays, 100- or 200-fold unlabeled oligonucleotides or mutant oligonucleotides (1 or 2 pmol) were included in the incubation mixture. Anti-Sp1 antibody (1:20 dilution) was also added to the reaction mixture to examine the supershift of the DNA probe. Purified GST-huntingtin fusion proteins were included at a concentration of 0.05 to 0.1 μg/μl to examine their effects on the DNA probe. The DNA-protein complexes were resolved on 4% nondenaturing polyacrylamide gels in 0.5% Ficoll-0.5× Tris-borate-EDTA buffer. Autoradiography was carried out by exposure of the gel to Kodak X-Omat XAR2 film with an intensifying screen at −70°C for 48 to 72 h. Quantification of the intensities of the bands was performed using personal densitometer S1 (Molecular Dynamics).
Transcription assay.
The rat NGFR promoter was isolated with primers rNGFRP-5"-1411 (5"-GGGGTACCTCATCAGTAGGAGAGC-3") and rNGFRP-3"-1720 (5"-CTACTCGAGGGAGCTGGCGGAGGC-3") based on a published sequence (28). The resulting fragment covers promoter region −1411 to −1720 of NGFR. This promoter was linked to the firefly luciferase reporter gene in vector pGL-2 (Promega, Madison, Wis.). The Sp1 binding region between BglI (−1545) and PvuII (−1455) restriction sites was deleted from this reporter construct, resulting in a mutated construct that served as a control to examine the effect of Sp1 sites. The thromboxane synthase (TS) promoter (−90 bp to +30 bp), which has no Sp1 binding site (16, 40), was inserted in the Renilla luciferase vector (pRL; Promega) for comparison. HEK 293 cells cultured in six-well dishes were cotransfected with these promoters (1 μg/well) and NLS-150Q or the PRK vector (1 μg/well) to examine the effect of mutant huntingtin on the activity of these promoters. To examine dose-dependent transcriptional repression by intranuclear huntingtin, NLS-20Q or NLS-150Q (0.1 or 0.5 μg/well) and the NGFR promoter construct (1 μg/well) were cotransfected into HEK 293 cells. Since transfected huntingtin did not significantly affect the activity of pRL-TS, this construct (0.02 μg) was also included in transfection to normalize transfection efficiency. After transfection for 24 h, the luciferase activity of HEK 293 cells was measured using the Dual-Luciferase reporter assay system (Promega) with a TD20/20 luminometer (Turner Designs, Sunnyvale, Calif.). The ratio of firefly luciferase (pGL-2-NFGR promoter) to Renilla luciferase (pRL-TS) reflects the relative activity of the NGFR promoter. All results are expressed as means ± standard errors (SE) of four independent transfection experiments. Western blot analysis of transfected HEK 293 cells with EM48 was also performed to verify the expression of the transfected huntingtin.
Cell viability and neurite outgrowth assays.
HEK 293 cells were plated in six-well dishes and transfected with 1 μg of NLS-150Q and 1 μg of either the PRK vector, full-length Sp1, or N-terminal Sp1 (amino acids 1 to 166). HEK 293 cells transfected with 2 μg of the PRK vector alone served as a control. After transfection for 48 h using Lipofectamine, >60% of HEK 293 cells expressed transfected proteins. The viability of the transfected cells was then determined by a modified 3-(4,5-dimethyl thiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTS) assay using a microplate reader (SPECTRAmax Plus) (22). To examine dead cells, trypan blue exclusion was performed to count ∼3,000 HEK 293 cells from four independent transfections.
For measuring neurites of huntingtin-transfected PC12 cells, wild-type PC12 cells in six-well dishes were transfected using Lipofectamine with the same amount (2 μg/well) of plasmid cDNAs of the PRK vector alone, NLS-150Q with the PRK vector, or NLS-150Q with Sp1-HA. After transfection for 8 h, fresh medium containing 100 ng of nerve growth factor (NGF) was added to the transfected cells for another 36 h. The transfected PC12 cells were then fixed for immunofluorescence staining. Since the transfection efficiency for PC12 cells is low (5 to 10%), cells expressing huntingtin and Sp1 were identified by immunofluorescence labeling with rabbit antibody EM48 for NLS-huntingtin and 12CA5 for Sp1-HA. Phase-contrast imaging was used to examine the neurites of transfected cells. Hoechst was used to show nuclear staining. All these images were captured with the 3CCD camera video system (Dage-MTI Inc.). The percentage of transfected PC12 cells containing neurites longer than two cell body diameters was obtained by measuring 63 to 104 transfected cells in each transfection. Four transfections were performed.
Statistical analysis.
All values were expressed as means ± SE. Statistical significance was assessed by the use of Student's t test, and a P value of <0.05 was considered significant.
RESULTS
Interaction of Sp1 with huntingtin in vitro.
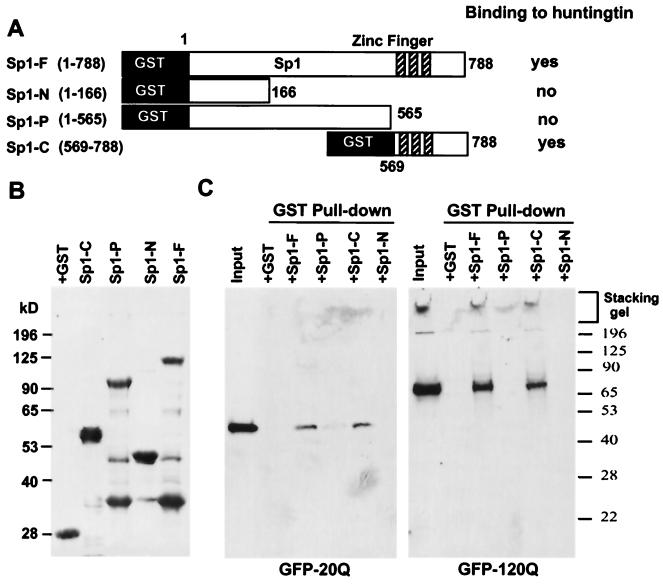
To study whether Sp1-mediated gene expression is altered by mutant huntingtin, we first investigated whether Sp1 interacts with mutant huntingtin. Rat Sp1 cDNA was isolated by reverse transcription-PCR to make GST fusion proteins containing full-length (Sp1-F), partial (Sp1-P), N-terminal (Sp1-N), and C-terminal (Sp1-C) Sp1 (Fig. 1A and B). We also established HEK 293 cells that stably expressed GFP fusion proteins containing huntingtin exon1 with 20 (GFP-20Q) or 120 (GFP-120Q) glutamines. These cells provided us with normal and mutant-N-terminal huntingtin for in vitro binding assays. A GST pull down assay showed that Sp1-F and Sp1-C bound to transfected huntingtin, whereas Sp1-P or Sp1-N did not (Fig. 1C). It appears that the binding site(s) resides in the C-terminal region of Sp1, which contains three zinc finger motifs. The GST pull down assay result also suggested that more GFP-120Q than GFP-20Q was bound by Sp1.
FIG. 1.
Production of huntingtin and Sp1 proteins. (A) Schematic structures of GST-Sp1 fusion proteins containing full-length rat Sp1 (Sp1-F, amino acids 1 to 788), partial Sp1 (Sp1-P, amino acids 1 to 565), N-terminal Sp1 (Sp1-N, amino acids 1 to 166), or C-terminal Sp1 (Sp1-C, amino acids 569 to 788). (B) Coomassie staining of SDS gel containing purified GST and GST-Sp1 fusion proteins. (C) Western blot analysis of GST pull down samples in which HEK 293 cell lysates containing GFP-tagged HD exon1 protein with 20 (GFP-20Q) or 120 (GFP-120Q) glutamines were incubated with GST-Sp1 fusion proteins. EM48 immunoblotting revealed that aggregated huntingtin remained in the stacking gel and that Sp1-F and Sp1-C precipitated GFP-huntingtin.
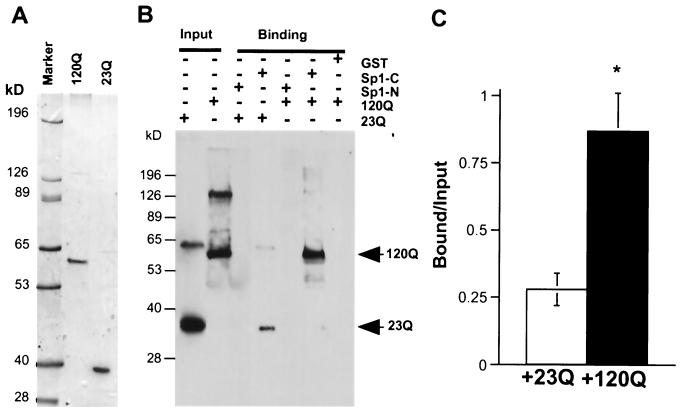
To examine whether Sp1 directly binds to huntingtin and to quantitatively compare its binding to huntingtin proteins containing normal and expanded glutamine repeats, we generated His-tagged huntingtin containing the first 171 amino acids with 23 (His-23Q) or 120 (His-120Q) glutamines in the repeat. Purification of these proteins with a nickel column yielded soluble huntingtin, which was seen as a single major band on a Coomassie-stained gel (Fig. 2A). A weaker band, possibly representing dimerized huntingtin, was also seen. Equal amounts of purified His-23Q and His-120Q proteins were then incubated with GST fusion proteins containing the N- or C-terminal region of Sp1. Only C-terminal Sp1 (Sp1-C), not N-terminal Sp1 or GST alone, bound to the purified His-huntingtin (Fig. 2B). C-terminal Sp1 bound more tightly to the monomeric form of His-huntingtin, as only a weak band of dimerized His-huntingtin was seen in the Sp1-C precipitates. Sp1-C also bound to the 23Q protein. However, comparison of the ratio of the bound to the input clearly revealed that more His-120Q than His-23Q bound to Sp1-C (Fig. 2C). This result clearly shows that polyglutamine expansion enhances the interaction of Sp1 with N-terminal huntingtin in vitro.
FIG. 2.
Direct interaction of Sp1 with huntingtin. (A) Purified His-huntingtin proteins that contain the first 171 amino acids with 23 (23Q) and 120 (120Q) glutamines are revealed by Coomassie staining of an SDS gel. (B) Western blot analysis of the interaction of His-huntingtin (23Q and 120Q) and GST-Sp1 fusion proteins. Huntingtin was detected with mEM48. Note that Sp1-C bound more to His-120Q than to His-23Q, whereas GST and Sp1-N did not. Input, 38% of the purified His-huntingtin that was used in the incubations. (C) Densitometry of the ratio of the bound huntingtin to the input huntingtin (bound/input) following the interaction of Sp1-C with His-23Q (+23Q) or His-120Q (+120Q). The data (means ± SE) were obtained from three experiments. ∗, P < 0.05 compared to the binding of His-23Q.
Coimmunoprecipitation of mutant huntingtin with Sp1.
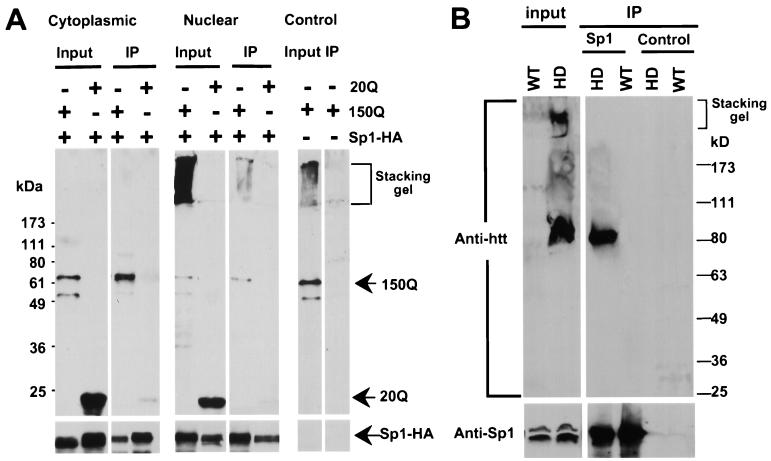
Since N-terminal-mutant huntingtin is able to accumulate in the nucleus, we wanted to examine whether Sp1 interacts with N-terminal-mutant huntingtin in cells. We cotransfected HEK 293 cells with cDNAs for full-length Sp1 and the HD exon1 huntingtin containing 20 (20Q-E) or 150 (150Q-E) glutamines. Sp1 was tagged with the HA epitope (Sp1-HA) to allow precipitation by mouse monoclonal antibody 12CA5. The precipitates were then examined by Western blotting with EM48 (Fig. 3A). Transfection of 150Q-E gave rise to both soluble huntingtin, which was mainly in the cytoplasmic fraction, and aggregated huntingtin, which was predominantly in the nuclear fraction. More soluble mutant huntingtin than aggregated huntingtin was precipitated with Sp1. Less 20Q-E protein was precipitated with Sp1 in both the cytoplasmic and nuclear fractions, also suggesting that polyglutamine expansion increased the binding to Sp1. More importantly, transfection of HEK 293 cells with 150Q-E alone produced no precipitation of transfected huntingtin by 12CA5, indicating that the presence of transfected Sp1 was necessary for the coprecipitation of huntingtin (Fig. 3A).
FIG. 3.
Coimmunoprecipitation of huntingtin and Sp1. (A) Coimmunoprecipitation of transfected huntingtin and Sp1-HA expressed in 293 cells. Transfected proteins were precipitated with anti-HA antibody 12CA5. The blot was probed with EM48 (anti-htt; top) and reprobed with 12CA5 (bottom). The control is the transfection of huntingtin without Sp1-HA. Input, 10% of cell lysate that was used in the incubation. IP, immunoprecipitation. (B) Immunoprecipitation of huntingtin and Sp1 from the brain cortex of an R6/2 mouse at 4 weeks of age. Anti-Sp1 (Sp1) or rabbit IgG (control) was used for immunoprecipitation, and mEM48 was used for Western blotting. Bottom, same blot reprobed with an anti-Sp1 antibody. Input, 10% of cell lysate that was used for coimmunoprecipitation. WT, wild type.
We then examined the interaction of Sp1 and huntingtin in vivo in mouse brain using coimmunoprecipitation. It was difficult to see the coprecipitation of Sp1 and normal mouse huntingtin (data not shown), perhaps because mouse huntingtin contains only seven glutamines in the repeat so its binding to Sp1 is weak. Also, the cytoplasmic localization of full-length huntingtin may hinder its interaction with nuclear Sp1. We then used R6/2 mouse brain in which N-terminal-mutant huntingtin containing a repeat of 115 to 150 glutamines is enriched in the nucleus (7). Although EM48 reacts very weakly with the endogenous full-length huntingtin in mouse brain, it reacts strongly with mutant huntingtin (10, 21). As a result, strong MEM48 immunoreactive products were found specifically in transgenic mouse brain (Fig. 3B). Two forms of transgenic protein were seen: aggregated huntingtin, which remained on the top of the gel, and soluble mutant huntingtin, seen as a band of ∼85 kDa. However, only soluble mutant huntingtin was precipitated with Sp1. Normal mouse huntingtin was not seen in the precipitates from wild-type or transgenic mouse brain. No precipitation of huntingtin was seen in the control in which rabbit IgG was used for immunoprecipitation. Thus, the in vivo interaction data also support the idea that soluble mutant huntingtin binds more tightly to Sp1 than does aggregated huntingtin.
Immunostaining of HD mouse brain and transfected cells.
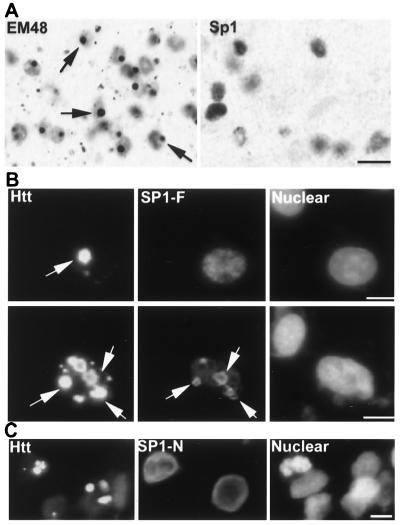
To examine whether huntingtin aggregates recruit Sp1, we stained R6/2 mouse brain using antibodies to Sp1 and huntingtin. We did not see any obvious Sp1 staining of nuclear inclusions, which, on the other hand, could be strongly labeled by EM48 (Fig. 4A). There was no obviously altered subcellular distribution of Sp1 in HD mouse brain compared to control brain (data not shown). It is possible that aggregated huntingtin may have reduced its binding to Sp1 during the course of forming nuclear inclusions. Since transfected mutant huntingtin forms inclusions rapidly in vitro, we examined whether nuclear inclusions formed by transfected huntingtin could recruit Sp1. Transfected huntingtin was tagged with NLS to facilitate its nuclear accumulation following transient transfection. In HEK 293 cells that were doubly transfected with full-length Sp1 and huntingtin, the majority of transfected cells did not show Sp1 staining of nuclear inclusions (Fig. 4B, top). Only about 3% of transfected cells showed some nuclear inclusions that were labeled by both huntingtin and Sp1 antibodies (Fig. 4B, bottom). This result is consistent with mouse brain staining in which we saw no nuclear inclusion staining of Sp1. The colocalization of Sp1 and huntingtin in some transfected cells reflects the interaction of Sp1 with some huntingtin proteins during the formation of nuclear inclusions. However, this interaction is also specific to the C-terminal region of Sp1, as cells cotransfected with Sp1-N and huntingtin did not show any Sp1 staining of nuclear inclusions (Fig. 4C).
FIG. 4.
Subcellular localization of Sp1 and nuclear mutant huntingtin. (A) Immunostaining of brain cortices of R6/2 mice at 12 weeks of age with EM48 and Sp1 antibodies. Diffuse nuclear labeling is seen with both antibodies. Neuropil aggregates (outside of the nuclei) and nuclear inclusions (arrows) were labeled by EM48. Scale bar: 10 μm. (B) Cotransfection of HEK 293 cells with NLS-150Q and full-length Sp1. In the majority of transfected cells, nuclear huntingtin (Htt) inclusions were not Sp1 immunoreactive (top). Only 3% of transfected cells displayed some nuclear inclusions (arrows) labeled by antibodies to both huntingtin and Sp1 (bottom). Scale bars: 5 μm. (C) Cotransfection of Sp1-N and NLS-150Q showing no colocalization of Sp1-N with the inclusions. Immunofluorescence double labeling was performed with rabbit antihuntingtin (EM48, green) and mouse anti-HA (12CA5, red), which labeled the His-tagged Sp1. Hoechst dye (blue) was used to show the nuclei. Scale bar: 5 μm.
Mutant huntingtin inhibits the binding of Sp1 to DNA.
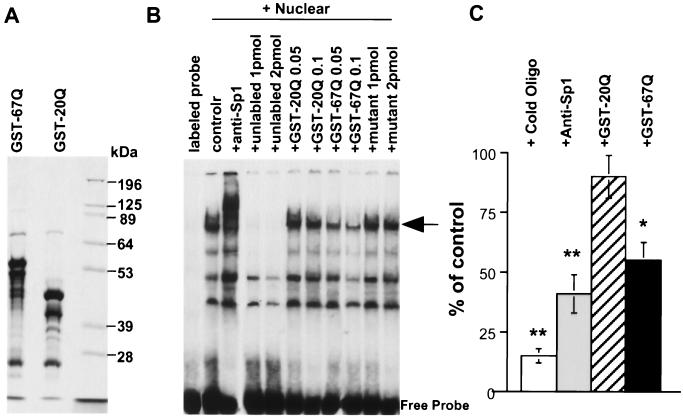
We have previously shown that intranuclear huntingtin decreases the expression of NGFR p75 in PC12 cells (22). The promoter of NGFR, which contains 5 or 6 GC boxes and which binds to Sp1 (33), served as a target to study the effect of mutant huntingtin on the binding of Sp1 to DNA. We synthesized oligonucleotides encoding the promoter region of NGFR and radiolabeled them with [32P] for EMSA. To produce purified huntingtin for these assays, we generated GST fusion proteins containing the HD exon1 with 67 (GST-67Q) or 20 (GST-20Q) glutamines (Fig. 5A). These proteins were tested for their effects on the NGFR promoter, and we observed several nucleoprotein complexes containing the radiolabeled probe (Fig. 5B). The addition of unlabeled oligonucleotides specifically eliminated two bands. The major one of these two bands (Fig. 5B) was also diminished and shifted upwards by the anti-Sp1 antibody, suggesting that this band represents the Sp1-DNA probe complex. To confirm that the formation of this protein-DNA complex is dependent on the Sp1 binding sites in the DNA probe, we also used unlabeled mutant oligonucleotides which lack an Sp1 binding site for competition. As expected, these mutant oligonucleotides did not reduce the intensity of the specific protein-DNA complex (Fig. 5B, last two lanes).
FIG. 5.
Effect of mutant huntingtin on DNA-Sp1 interaction. (A) Purified GST fusion proteins containing the HD exon1 protein with 20 (GST-20Q) or 67 (GST-67Q) glutamines were examined by SDS-PAGE and Coomassie blue staining. (B) EMSA of PC12 cell nuclear proteins that were incubated with 32P-labeled double-stranded oligonucleotides containing Sp1 binding sites of the NGFR promoter. Competitors used were an anti-Sp1 antibody, unlabeled oligonucleotides (unlabeled, 1 to 2 pmol), and purified GST-20Q or GST-67Q (0.05 to 0.1 μg). Unlabeled oligonucleotides with mutated Sp1 binding sites (mutant, 1 to 2 pmol) were also included as a control. The intensity of one specific nucleoprotein complex (arrow) was reduced by unlabeled oligonucleotides, the anti-Sp1 antibody, and GST-67Q. Note that the anti-Sp1 antibody also supershifted this nucleoprotein complex. controlr, 32P-labeled DNA-protein complex without competitors. (C) Densitometric analysis of the intensity of the DNA-Sp1 complex (arrow in panel B) in the presence of unlabeled oligonucleotides (1 pmol), anti-Sp1, and GST-20Q or GST-67Q (0.1 μg). The percentage of the control is the intensity of the band without any competitors. The results are means ± SE of four independent assays. ∗, P < 0.05; ∗∗, P < 0.01 (both compared to the value for GST-20Q treatment).
We then examined whether purified GST-huntingtin proteins could compete with or inhibit the binding of the DNA probe to Sp1. GST-20Q did not significantly decrease the intensity of the Sp1-DNA probe complex, whereas GST-67Q obviously reduced the intensity of this complex (Fig. 5B). No obvious higher-molecular-weight bands were observed, suggesting that GST-67Q might inhibit the binding of Sp1 to the DNA probe rather than forming a larger DNA-protein complex. Similar to the unlabeled oligonucleotides, GST-67Q inhibited the DNA binding in a dose-dependent manner. Quantifying the intensities of the bands on the blots using densitometry also showed that GFP-67Q (0.1 μg/ml), like unlabeled oligonucleotides and the antibody, inhibits the binding of Sp1 to the NGFR promoter sequences (Fig. 5C).
Mutant huntingtin inhibits the transcriptional activity of the NGFR promoter.
Since the transcriptional activity of NGFR is dependent on Sp1 (1, 33), the NGFR promoter also provided us with a tool to examine the effect of mutant huntingtin on Sp1-dependent gene transcription. To do so, we used PCR to isolate the promoter region of rat NGFR. We also used the TS promoter (−90 to +30 bp), which does not contain any Sp1 binding site. TS has been previously used as an internal control for luciferase assays (16, 40).
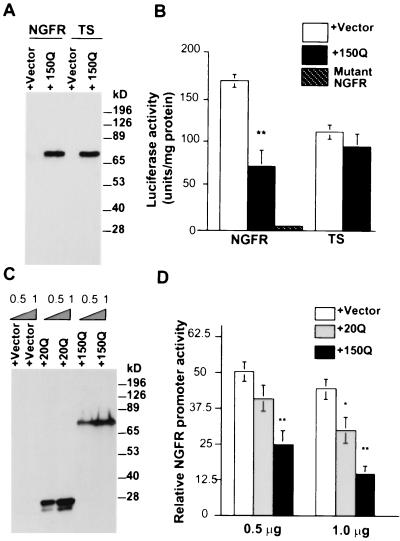
We first transfected these promoters with NLS-150Q into HEK 293 cells to examine whether mutant huntingtin could inhibit their activity. Western blots showed that the expression level of NLS-150Q did not change when it was cotransfected with the NGFR or TS promoter construct (Fig. 6A). Both promoters showed a high activity in transfected cells when cotransfected with the PRK vector (Fig. 6B). However, coexpression of NLS-150Q inhibited the activity of the NGFR promoter but not the TS promoter, suggesting that the inhibition was Sp1 dependent.
FIG. 6.
Suppression of the activity of the NGFR promoter by mutant huntingtin. (A) EM48 Western blot analysis of HEK 293 cells expressing the NGFR or TS promoter and cotransfected with the PRK vector or NLS-150Q. (B) Luciferase assays of the cotransfected cells. The luciferase units were normalized for the amount of proteins used. ∗∗, P < 0.01 compared to the vector. The mutated NGFR promoter construct with deletion of the Sp1 binding sites (mutant NGFR) was used as a control. (C) Western blot analysis of the expression of transfected huntingtin in HEK 293 cells in which the pGL-NGFR promoter (1 μg) was cotransfected with 0.5 or 1 μg of the PRK vector, NLS-20Q, or NLS-150Q. The pRL-TS Renilla luciferase construct (0.02 μg) was also cotransfected in each sample to normalize transfection efficiency. (D) Dual-luciferase reporter assay of the ratio of firefly to Renilla luciferase was performed to measure the relative activity of the NGFR promoter. The data are reported as means ± SE from four sets of transfections. ∗, P < 0.05; ∗∗, P < 0.01 (both compared to the results for the PRK vector transfection).
We then used the pRL-TS vector as an internal control to further assess the effect of polyglutamine expansion on the activity of the NGFR promoter. A dual-luciferase reporter assay was performed to quantitatively measure the relative activity of the NGFR promoter. In this way, we could normalize the results to account for the transfection efficiencies for NLS-20Q and NLS-150Q. We also used a Western blot to confirm that the expression levels of NLS-20Q and NLS-150Q in the transfected cells were similar (Fig. 6C). In the luciferase assay, both NLS-20Q and NLS-150Q inhibited the activity of the NGFR promoter (Fig. 6D). However, NLS-20Q produced a significant inhibition only at the higher dose. It is possible that overexpressed NLS-20Q might affect NGFR promoter activity through its interaction with Sp1, though this interaction is weak in vitro (Fig. 2 and 3). Importantly, the inhibitory effect of NLS-150Q was almost twofold greater than that of NLS-20Q (Fig. 6D), suggesting a glutamine repeat-dependent inhibition. This repeat length-dependent effect is consistent with the greater binding of mutant huntingtin to Sp1.
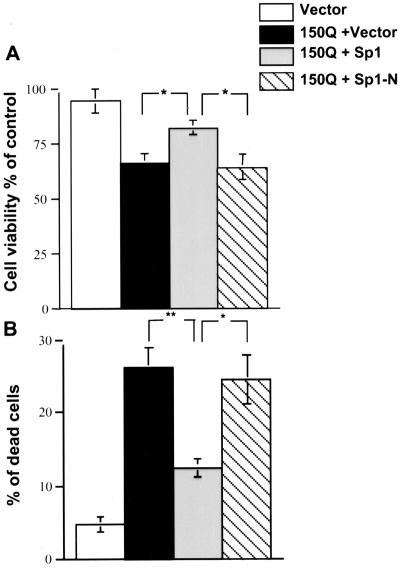
To investigate whether overexpression of Sp1 prevents or reduces the cellular toxicity associated with intranuclear huntingtin, we measured the viability of HEK 293 cells transfected with mutant huntingtin and Sp1. After a 48-h transfection, more than 60% of HEK 293 cells expressed transfected proteins, thus allowing us to measure the viability of the transfected cells by an MTS assay. Transfection of NLS-150Q with the PRK vector significantly reduced cellular viability to 65.9% of that for untransfected cells (Fig. 7A). This result is consistent with many other previous studies showing that intranuclear huntingtin increases cellular toxicity (22, 29, 32, 36). When full-length Sp1 was cotransfected with NLS-150Q, the cellular viability improved significantly to 82.8% (Fig. 7). Importantly, cotransfection of NLS-150Q with N-terminal Sp1, which does not bind to mutant huntingtin in vitro (Fig. 1), failed to improve cell viability (64.1%), suggesting that the interaction of transfected Sp1 with mutant huntingtin is important for the inhibition of cytotoxicity.
FIG. 7.
Effect of Sp1 overexpression on cellular toxicity induced by intranuclear mutant huntingtin. (A) MTS assay of the viability of HEK 293 cells transfected with the PRK vector alone, NLS-150Q with the PRK vector, or NLS-150Q with full-length Sp1 or Sp1-N. The viability is expressed as the percentage of control (untransfected cells). The data are means ± SE from four assays. ∗, P < 0.05 compared to transfection with NLS-150Q plus Sp1. (B) Cell death was evaluated with trypan blue exclusion staining of HEK 293 cells transfected with the PRK vector or cotransfected with NLS-150Q and the PRK vector, full-length Sp1, or Sp1-N. The percentages of dead cells were the means ± SE from four transfections. ∗, P < 0.05; ∗∗, P < 0.01 (both compared to transfection with NLS-150Q plus Sp1).
We also measured the percentage of dead cells using trypan blue exclusion assays. There was significantly more cell death in NLS-150Q-transfected cells (26.1%) than in those transfected with the PRK vector alone (4.7%) or Sp1 and NLS-150Q together (12.5%). Cotransfection of N-terminal Sp1 with NLS-150Q did not significantly improve cell viability (24.5% dead cells).
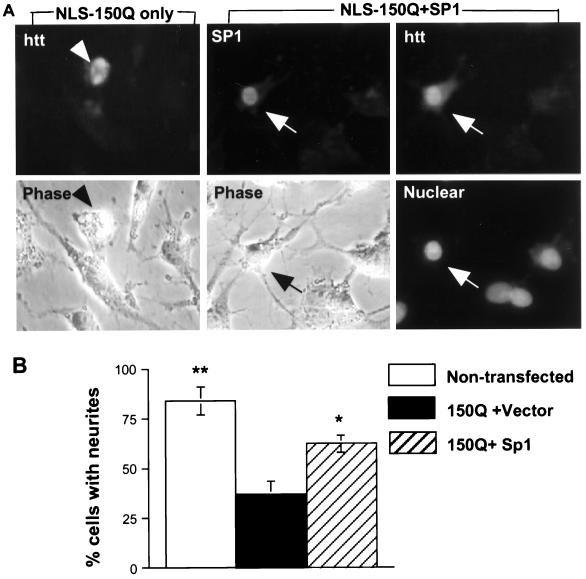
Neurite extension is dependent on the expression of NGFR p75 and the TrkA receptor; both require Sp1 for their transcriptional activation (1, 33, 35,). We have shown that PC12 cells stably expressing intranuclear mutant huntingtin express less of these receptors and have defective neurite extension (22). We wanted to examine whether Sp1 overexpression could also reverse the neurite extension defect. Since PC12 cells stably transfected with the huntingtin gene have a very low transfection efficiency for other genes, we used wild-type PC12 cells to cotransfect NLS-150Q and Sp1. Immunofluorescence and phase-contrast imaging allowed us to identify huntingtin-transfected neurons and their neurites. Cells transfected with NLS-150Q alone showed huntingtin in their nuclei and were unable to grow neurites in the presence of NGF. Coexpression of Sp1 resulted in PC12 cells that had significantly longer neurites, even when mutant huntingtin accumulated in the nucleus (Fig. 8A). To confirm this observation, we counted the transfected PC12 cells and found that 36.7% of NLS-150Q-transfected cells had neurites longer than two cell body diameters, whereas 83.7% of wild-type PC12 cells and 62.1% of cells cotransfected with NLS-150Q and Sp1 displayed such neurites (Fig. 8B). Cotransfection of N-terminal Sp1 with mutant huntingtin did not improve neurite extension. The protection of Sp1 against huntingtin-induced reduction of neurite extension also supports the idea that overexpressed Sp1 could, at least in part, prevent the cellular dysfunction induced by intranuclear mutant huntingtin.
FIG. 8.
Effect of Sp1 overexpression on the inhibition of neurite extension by intranuclear mutant huntingtin. Wild-type PC12 cells were transfected with the PRK vector alone or cotransfected with NLS-150Q and Sp1-F or Sp1-N for 8 h. The transfected PC12 cells were then incubated with NGF (100 ng/ml) for 36 h and stained with EM48 (htt) or anti-HA (Sp1). Nuclei were stained with Hoechst. Phase-contrast images show the neurites. (A) A representative cell expressing NLS-150Q alone (arrowheads) and a double-transfected cell (arrows). Scale bar: 10 μm. (B) Quantitative assessment of transfected cells with neurites that exceed two cell body diameters. The percentages of transfected cells were the means ±SE obtained by counting transfected cells in four transfection experiments. ∗, P < 0.05; ∗∗, P < 0.01 (both compared to transfection with NLS-150Q plus the vector).
DISCUSSION
The present study demonstrates that expansion of the polyglutamine tract can cause huntingtin to bind more tightly to transcriptional activator Sp1, and this binding may induce cellular dysfunction and toxicity. Sp1 acts through its binding to GC boxes and activates the expression of a variety of genes. Its C-terminal region contains a DNA binding domain composed of three zinc finger motifs (17). Several glutamine-rich domains in its N-terminal region regulate transcriptional activity (5, 6, 9, 12). If mutant huntingtin, when abnormally localized in the nucleus, binds to either glutamine-rich domains or to the C-terminal region of Sp1, it could affect transcriptional regulation or prevent Sp1 from binding to DNA, resulting in a decrease in the expression of certain genes.
Our findings suggest that mutant huntingtin binds to the C-terminal region of Sp1 and inhibits the interaction of Sp1 with the NGFR promoter. The gel shift assay also suggests that mutant huntingtin did not form a larger DNA-Sp1-huntingtin complex, as no supershifted bands were observed when mutant huntingtin was incubated with nuclear extracts and the NGFR promoter probe. It appears that mutant huntingtin, through its interaction with the DNA binding region of Sp1, prevents the binding of Sp1 to promoters. The prevention of the usual binding of Sp1 to promoters may constitute the molecular basis for the effect of mutant huntingtin on Sp1-dependent gene expression.
Our findings also suggest that the soluble form of mutant huntingtin binds to Sp1. This is because very little Sp1 binds to aggregated huntingtin or is recruited into polyglutamine inclusions. Other studies observed that Sp1 was in nuclear inclusions in the brains of dentatorubral-pallidoluysian atrophy and spinocerebellar ataxia type 3 patients (38). It is likely that the protein context of polyglutamine proteins may influence the recruitment of Sp1 into nuclear inclusions or affect the conformation of recruited Sp1 such that it becomes unrecognizable by its antibody. However, the coimmunoprecipitation and in vitro binding results support the idea that the soluble form of mutant huntingtin binds more tightly to Sp1 than does aggregated huntingtin. It is possible that aggregated huntingtin binds to Sp1 but that precipitation with Sp1 may be difficult. However, the failure by the anti-Sp1 antibody to stain brain huntingtin inclusions favors the idea that aggregated huntingtin may not bind to Sp1 well. It is likely that, in HD brain, mutant huntingtin in the inclusions loses its binding to Sp1 because of an increase in its self-aggregation during the formation of nuclear inclusions. Since nuclear inclusion formation does not correlate with neuronal degeneration (10, 19, 20, 36), the interaction between Sp1 and soluble mutant huntingtin is more likely to be involved in HD pathology. The repeat length-dependent binding of huntingtin to Sp1 also implies that their interaction has a role in HD pathology.
Despite the unclear role of nuclear inclusions, the intranuclear accumulation of huntingtin is well correlated with disease progression in HD animal models (7, 24, 41, 42) and is associated with altered gene expression (3, 22, 26). Using the NGFR promoter, whose transcription requires Sp1, we also provided evidence that expression of mutant huntingtin in the nucleus can inhibit the activity of this promoter. This is consistent with our previous findings that intranuclear mutant huntingtin decreases the expression of NGFR in stably transfected PC12 cells (22) and also implies an effect of mutant huntingtin on the expression of other Sp1-mediated genes. Interestingly, overexpressed NLS-20Q also inhibits NGFR promoter activity, though this effect is significantly less than that of NLS-150Q. Such an effect may only occur in vitro when huntingtin is overexpressed. In HD brain, N-terminal fragments of mutant huntingtin are able to accumulate in the nucleus (8, 10, 21). Thus, it is possible that in HD brain only N-terminal-mutant huntingtin fragments are available to bind to Sp1 and to affect gene expression.
Given the finding that Sp1 is important for the expression of many genes involved in the regulation of vital cellular functions (5), an interesting question is why mutant huntingtin affects the expression of only a subset of genes. We hypothesize that the effect of mutant huntingtin on Sp1 may depend on how tightly Sp1 binds to a particular promoter region. The Sp1 binding sites in the promoters of different genes vary to a significant degree (5). Thus, the promoters whose binding to Sp1 is readily inhibited by mutant huntingtin are more affected. Accordingly, mutant huntingtin may affect the function of those genes whose expression level is more dependent on Sp1. This hypothesis also explains why another study found that the transcription of a GC box (6xGC-luc) was not inhibited by mutant huntingtin (29).
Our findings also show that overexpression of Sp1 partially reduces cellular toxicity induced by mutant huntingtin. However, Nucifora et al. (29) examined cell death in GFP-cotransfected N2a cells and found that Sp1 overexpression did not significantly reduce the cellular toxicity of mutant HD exon1. In the present study, we used the MTS assay and trypan blue staining, which may be more sensitive than cell morphology in detecting early cellular pathology. The partial protection of huntingtin toxicity by Sp1 overexpression is consistent with the findings that huntingtin also binds to other nuclear proteins to induce multiple cellular toxicity (2, 18, 29, 38, 39). Considering that Sp1 is involved in the expression of many genes, the interaction between mutant huntingtin and Sp1 may play an important role in the pathology of HD.
Another intriguing question is how the abnormal interaction of Sp1 and mutant huntingtin contributes to the selective neuropathology of HD. Cell-type-specific processing and accumulation of N-terminal huntingtin fragments may be a critical feature of this selectivity. We and others have demonstrated that striatal neurons, which are preferentially affected in HD, show nuclear localization and aggregation of mutant huntingtin much earlier than other types of neurons (13, 21, 24, 41). Thus, mutant huntingtin, once it is degraded into toxic fragments and accumulates in the nucleus in HD-affected neurons, may interact with Sp1 and other transcription factors. The resulting alteration in gene expression could be an early pathological event that ultimately leads to neuronal degeneration (25). Identification of the interaction between Sp1 and mutant huntingtin will help develop an effective therapeutic approach for protecting or reducing HD-induced cellular toxicity.
Acknowledgments
We thank Russ Price at Emory University for providing the pRL-TS vector, Po-Yung Cheng for his advice on gel shift assays, Dimitri Krainc for discussing unpublished data, and Jennifer Macke for critical reading of the manuscript.
This work was supported by grants from the NIH (NS41669 and AG19206) and Huntington's Disease Society of America.
S.-H. Li and A. L. Cheng contributed equally to this work.
REFERENCES
- 1.Bai, G., and J. W. Kusiak. 1997. Nerve growth factor up-regulates the N-methyl-D-aspartate receptor subunit 1 promoter in PC12 cells. J. Biol. Chem. 272:5936-5942. [DOI] [PubMed] [Google Scholar]
- 2.Boutell, J. M., P. Thomas, J. W. Neal, V. J. Weston, J. Duce, P. S. Harper, and A. L. Jones. 1999. Aberrant interactions of transcriptional repressor proteins with the Huntington's disease gene product, huntingtin. Hum. Mol. Genet. 8:1647-1655. [DOI] [PubMed] [Google Scholar]
- 3.Cha, J. H., C. M. Kosinski, J. A. Kerner, S. A. Alsdorf, L. Mangiarini, S. W. Davies, J. B. Penney, G. P. Bates, and A. B. Young. 1998. Altered brain neurotransmitter receptors in transgenic mice expressing a portion of an abnormal human huntington disease gene. Proc. Natl. Acad. Sci. USA 95:6480-6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng, P. Y., N. Kagawa, Y. Takahashi, and M. R. Waterman. 2000. Three zinc finger nuclear proteins, Sp1, Sp3, and a ZBP-89 homologue, bind to the cyclic adenosine monophosphate-responsive sequence of the bovine adrenodoxin gene and regulate transcription. Biochemistry 39:4347-4357. [DOI] [PubMed] [Google Scholar]
- 5.Cook, T., B. Gebelein, and R. Urrutia. 1999. Sp1 and its likes: biochemical and functional predictions for a growing family of zinc finger transcription factors. Ann. N. Y. Acad. Sci. 880:94-102. [DOI] [PubMed] [Google Scholar]
- 6.Courey, A. J., and R. Tjian. 1988. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell 55:887-898. [DOI] [PubMed] [Google Scholar]
- 7.Davies, S. W., M. Turmaine, B. A. Cozens, M. DiFiglia, A. H. Sharp, C. A. Ross, E. Scherzinger, E. E. Wanker, L. Mangiarini, and G. P. Bates. 1997. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell 90:537-548. [DOI] [PubMed] [Google Scholar]
- 8.DiFiglia, M., E. Sapp, K. O. Chase, S. W. Davies, G. P. Bates, J. P. Vonsattel, and N. Aronin. 1997. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277:1990-1993. [DOI] [PubMed] [Google Scholar]
- 9.Gerber, H. P., K. Seipel, O. Georgiev, M. Hofferer, M. Hug, S. Rusconi, and W. Schaffner. 1994. Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science 263:808-811. [DOI] [PubMed] [Google Scholar]
- 10.Gutekunst, C. A., S. H. Li, H. Yi, J. S. Mulroy, S. Kuemmerle, R. Jones, D. Rye, R. J. Ferrante, S. M. Hersch, and X. J. Li. 1999. Nuclear and neuropil aggregates in Huntington's disease: relationship to neuropathology. J. Neurosci. 19:2522-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hackam, A. S., R. Singaraja, C. L. Wellington, M. Metzler, K. McCutcheon, T. Zhang, M. Kalchman, and M. R. Hayden. 1998. The influence of huntingtin protein size on nuclear localization and cellular toxicity. J. Cell Biol. 141:1097-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagen, G., S. Muller, M. Beato, and G. Suske. 1994. Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J. 13:3843-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hodgson, J. G., N. Agopyan, C. A. Gutekunst, B. R. Leavitt, F. LePiane, R. Singaraja, D. J. Smith, N. Bissada, K. McCutcheon, J. Nasir, L. Jamot, X. J. Li, M. E. Stevens, E. Rosemond, J. C. Roder, A. G. Phillips, E. M. Rubin, S. M. Hersch, and M. R. Hayden. 1999. A YAC mouse model for Huntington's disease with full-length mutant huntingtin, cytoplasmic toxicity, and selective striatal neurodegeneration. Neuron 23:181-192. [DOI] [PubMed] [Google Scholar]
- 14.Huang, C. C., P. W. Faber, F. Persichetti, V. Mittal, J. P. Vonsattel, M. E. MacDonald, and J. F. Gusella. 1998. Amyloid formation by mutant huntingtin: threshold, progressivity and recruitment of normal polyglutamine proteins. Somat. Cell Mol. Genet. 24:217-233. [DOI] [PubMed] [Google Scholar]
- 15.Huntington Disease Collaborative Research Group. 1993. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 72:971-983. [DOI] [PubMed] [Google Scholar]
- 16.Ibrahim, N. M., A. C. Marinovic, S. R. Price, L. G. Young, and O. Frohlich. 2000. Pitfall of an internal control plasmid: response of Renilla luciferase (pRL-TK) plasmid to dihydrotestosterone and dexamethasone. BioTechniques 29:782-784. [DOI] [PubMed] [Google Scholar]
- 17.Kadonaga, J. T., K. R. Carner, F. R. Masiarz, and R. Tjian. 1987. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell 51:1079-1090. [DOI] [PubMed] [Google Scholar]
- 18.Kazantsev, A., E. Preisinger, A. Dranovsky, D. Goldgaber, and D. Housman. 1999. Insoluble detergent-resistant aggregates form between pathological and nonpathological lengths of polyglutamine in mammalian cells. Proc. Natl. Acad. Sci. USA 96:11404-11409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klement, I. A., P. J. Skinner, M. D. Kaytor, H. Yi, S. M. Hersch, H. B. Clark, H. Y. Zoghbi, and H. T. Orr. 1998. Ataxin-1 Nuclear localization and aggregation: role in polyglutamine-induced disease in SCA1 transgenic mice. Cell 95:41-53. [DOI] [PubMed] [Google Scholar]
- 20.Kuemmerle, S., C. A. Gutekunst, A. M. Klein, X. J. Li, S. H. Li, M. F. Beal, S. M. Hersch, and R. J. Ferrante. 1999. Huntington aggregates may not predict neuronal death in Huntington's disease. Ann. Neurol. 46:842-849. [PubMed] [Google Scholar]
- 21.Li, H., S. H. Li, H. Johnston, P. F. Shelbourne, and X. J. Li. 2000. Amino-terminal fragments of mutant huntingtin show selective accumulation in striatal neurons and synaptic toxicity. Nat. Genet. 25:385-389. [DOI] [PubMed] [Google Scholar]
- 22.Li, S. H., A. L. Cheng, H. Li, and X. J. Li. 1999. Cellular defects and altered gene expression in PC12 cells stably expressing mutant huntingtin. J. Neurosci. 19:5159-5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, S. H., C. A. Gutekunst, S. M. Hersch, and X. J. Li. 1998. Interaction of huntingtin-associated protein with dynactin P150Glued. J. Neurosci. 18:1261-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin, C. H., S. Tallaksen-Greene, W. M. Chien, J. A. Cearley, W. S. Jackson, A. B. Crouse, S. Ren, X. J. Li, R. L. Albin, and P. J. Detloff. 2001. Neurological abnormalities in a knock-in mouse model of Huntington's disease. Hum. Mol. Genet. 10:137-144. [DOI] [PubMed] [Google Scholar]
- 25.Lin, X., B. Antalffy, D. Kang, H. T. Orr, and H. Y. Zoghbi. 2000. Polyglutamine expansion down-regulates specific neuronal genes before pathologic changes in SCA1. Nat. Neurosci. 3:157-163. [DOI] [PubMed] [Google Scholar]
- 26.Luthi-Carter, R., A. Strand, N. L. Peters, S. M. Solano, Z. R. Hollingsworth, A. S. Menon, A. S. Frey, B. S. Spektor, E. B. Penney, G. Schilling, C. A. Ross, D. R. Borchelt, S. J. Tapscott, A. B. Young, J. H. Cha, and J. M. Olson. 2000. Decreased expression of striatal signaling genes in a mouse model of Huntington's disease. Hum. Mol. Genet. 9:1259-1271. [DOI] [PubMed] [Google Scholar]
- 27.Martindale, D., A. Hackam, A. Wieczorek, L. Ellerby, C. Wellington, K. McCutcheon, R. Singaraja, P. Kazemi-Esfarjani, R. Devon, S. U. Kim, D. E. Bredesen, F. Tufaro, and M. R. Hayden. 1998. Length of huntingtin and its polyglutamine tract influences localization and frequency of intracellular aggregates. Nat. Genet. 18:150-154. [DOI] [PubMed] [Google Scholar]
- 28.Metsis, M., T. Timmusk, R. Allikmets, M. Saarma, and H. Persson. 1992. Regulatory elements and transcriptional regulation by testosterone and retinoic acid of the rat nerve growth factor receptor promoter. Gene 121:247-254. [DOI] [PubMed] [Google Scholar]
- 29.Nucifora, F. C., Jr., M. Sasaki, M. F. Peters, H. Huang, J. K. Cooper, M. Yamada, H. Takahashi, S. Tsuji, J. Troncoso, V. L. Dawson, T. M. Dawson, and C. A. Ross. 2001. Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science 291:2423-2428. [DOI] [PubMed] [Google Scholar]
- 30.Perez, M. K., H. L. Paulson, S. J. Pendse, S. J. Saionz, N. M. Bonini, and R. N. Pittman. 1998. Recruitment and the role of nuclear localization in polyglutamine-mediated aggregation. J. Cell Biol. 143:1457-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perutz, M. F., T. Johnson, M. Suzuki, and J. T. Finch. 1994. Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc. Natl. Acad. Sci. USA 91:5355-5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peters, M. F., F. C. Nucifora, Jr., J. Kushi, H. C. Seaman, J. K. Cooper, W. J. Herring, V. L. Dawson, T. M. Dawson, and C. A. Ross. 1999. Nuclear targeting of mutant Huntingtin increases toxicity. Mol. Cell. Neurosci. 14:121-128. [DOI] [PubMed] [Google Scholar]
- 33.Poukka, H., P. J. Kallio, O. A. Janne, and J. J. Palvimo. 1996. Regulation of the rat p75 neurotrophin receptor promoter by GC element binding proteins. Biochem. Biophys. Res. Commun. 229:565-570. [DOI] [PubMed] [Google Scholar]
- 34.Reddy, P. H., M. Williams, V. Charles, L. Garrett, L. Pike-Buchanan, W. O. Whetsell, Jr., G. Miller, and D. A. Tagle. 1998. Behavioural abnormalities and selective neuronal loss in HD transgenic mice expressing mutated full-length HD cDNA. Nat. Genet. 20:198-202. [DOI] [PubMed] [Google Scholar]
- 35.Sacristan, M. P., J. G. de Diego, M. Bonilla, and D. Martin-Zanca. 1999. Molecular cloning and characterization of the 5" region of the mouse trkA proto-oncogene. Oncogene 18:5836-5842. [DOI] [PubMed] [Google Scholar]
- 36.Saudou, F., S. Finkbeiner, D. Devys, and M. E. Greenberg. 1998. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell 95:55-66. [DOI] [PubMed] [Google Scholar]
- 37.Schilling, G., M. W. Becher, A. H. Sharp, H. A. Jinnah, K. Duan, J. A. Kotzuk, H. H. Slunt, T. Ratovitski, J. K. Cooper, N. A. Jenkins, N. G. Copeland, D. L. Price, C. A. Ross, and D. R. Borchelt. 1999. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum. Mol. Genet. 8:397-407. [DOI] [PubMed] [Google Scholar]
- 38.Shimohata, T., T. Nakajima, M. Yamada, C. Uchida, O. Onodera, S. Naruse, T. Kimura, R. Koide, K. Nozaki, Y. Sano, H. Ishiguro, K. Sakoe, T. Ooshima, A. Sato, T. Ikeuchi, M. Oyake, T. Sato, Y. Aoyagi, I. Hozumi, T. Nagatsu, Y. Takiyama, M. Nishizawa, J. Goto, I. Kanazawa, I. Davidson, and N. Tanese. 2000. Expanded polyglutamine stretches interact with TAFII130, interfering with CREB-dependent transcription. Nat. Genet. 26:29-36. [DOI] [PubMed] [Google Scholar]
- 39.Steffan, J. S., A. Kazantsev, O. Spasic-Boskovic, M. Greenwald, Y. Z. Zhu, H. Gohler, E. E. Wanker, G. P. Bates, D. E. Housman, and L. M. Thompson. 2000. The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc. Natl. Acad. Sci. USA 97:6763-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tazawa, R., E. D. Green, K. Ohashi, K. K. Wu, and L. H. Wang. 1996. Characterization of the complete genomic structure of human thromboxane synthase gene and functional analysis of its promoter. Arch. Biochem. Biophys. 334:349-356. [DOI] [PubMed] [Google Scholar]
- 41.Wheeler, V. C., J. K. White, C. A. Gutekunst, V. Vrbanac, M. Weaver, X. J. Li, S. H. Li, H. Yi, J. P. Vonsattel, J. F. Gusella, S. Hersch, W. Auerbach, A. L. Joyner, and M. E. MacDonald. 2000. Long glutamine tracts cause nuclear localization of a novel form of huntingtin in medium spiny striatal neurons in HdhQ92 and HdhQ111 knock-in mice. Hum. Mol. Genet. 9:503-513. [DOI] [PubMed] [Google Scholar]
- 42.Yamamoto, A., J. J. Lucas, and R. Hen. 2000. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington's disease. Cell 101:57-66. [DOI] [PubMed] [Google Scholar]
- 43.Zoghbi, H. Y., and H. T. Orr. 2000. Glutamine repeats and neurodegeneration. Annu. Rev. Neurosci. 23:217-247. [DOI] [PubMed] [Google Scholar]