Abstract
MDC9 (ADAM9/meltrin γ) is a widely expressed and catalytically active metalloprotease-disintegrin protein that has been implicated in the ectodomain cleavage of heparin-binding epidermal growth factor-like growth factor (HB-EGF) and as an α secretase for the amyloid precursor protein. In this study, we evaluated the expression of MDC9 during development and generated mice lacking MDC9 (mdc9−/− mice) to learn more about the function of this protein during development and in adults. During mouse development, MDC9 mRNA is ubiquitously expressed, with particularly high expression levels in the developing mesenchyme, heart and brain. Despite the ubiquitous expression of MDC9, mdc9−/− mice appear to develop normally, are viable and fertile, and do not have any major pathological phenotypes compared to wild-type mice. Constitutive and stimulated ectodomain shedding of HB-EGF is comparable in embryonic fibroblasts isolated from mdc9−/− and wild-type mice, arguing against an essential role of MDC9 in HB-EGF shedding in these cells. Furthermore, there were no differences in the production of the APP α and γ secretase cleavage product (p3) and of β- and γ-secretase cleavage product (Aβ) in cultured hippocampal neurons from mdc9−/− or wild-type mice, arguing against an essential major role of MDC9 as an α-secretase in mice. Further studies, including functional challenges and an evaluation of potential compensation by, or redundancy with, other members of the ADAM family or perhaps even with other molecules will be necessary to uncover physiologically relevant functions for MDC9 in mice.
Metalloprotease-disintegrins (ADAMs) are a family of membrane-anchored glycoproteins that have been implicated in fertilization, myogenesis, neurogenesis, and protein ectodomain shedding (for recent reviews, see references 3, 36, and 42). The first recognized ADAMs were the α and β subunits of the sperm protein fertilin, a protein with an essential role in fertilization (5, 10, 35). Since the discovery of fertilin, a total of 33 ADAMs have been identified in a variety of organisms, 24 of which are found in the mouse (see the following URLs for details: www.people.Virginia.EDU/∼jag6n/adams.html and www.uta.fi/∼loiika/HADAMs.htm). About half of these proteins have a catalytic-site consensus sequence (HEXXH) in their metalloprotease domain and are predicted to be catalytically active. The remaining ADAMs do not have a catalytic site in their metalloprotease domain and are therefore not predicted to be catalytically active, even though the domain is otherwise quite highly conserved. ADAMs are also thought to have roles in cell-cell adhesion, for example through interactions with integrins (9, 12, 31) or syndecans (17).
Despite the substantial number of ADAMs that have been identified to date, only a few have known biological functions. Mice lacking functional ADAM17/TACE die shortly after birth, apparently as a result of a defect in protein ectodomain shedding of epidermal growth factor (EGF) receptor ligands such as transforming growth factor α (TGF-α) (34). ADAM10/KUZ has been implicated in Notch signaling in Drosophila (13, 40, 48) and mice (D. J. Pan, personal communication), and the related SUP-17 has been linked to Notch signaling in Caenorhabditis elegans (51). The catalytically active ADAM13 has a role in neural crest migration in Xenopus laevis (1). Furthermore, the sperm proteins ADAM2 (fertilin β) and ADAM3 (cyritestin), both of which lack a catalytic site (HEXXH), have essential roles in fertilization (10, 11, 32, 43). Finally, ADAM 23, which also lacks a catalytic site, has an essential role in mouse brain development (23).
Targeted deletions and genetic and cell biological studies of ADAMs have thus uncovered a variety of diverse and interesting functions for these molecules. These studies raise questions about the roles of other ADAMs. Based on our current knowledge, one would predict that catalytically active ADAMs would have roles in protein ectodomain shedding or in the degradation, for example, of matrix proteins or in both processes. ADAMs lacking a catalytic site most probably mediate cell-cell or cell-matrix interactions. Catalytically active ADAMs may also combine proteolytic and adhesive functions. We have previously described the identification and biochemical characterization of MDC9/ADAM9, a widely expressed and catalytically active ADAM (39, 52). MDC9 is highly conserved between mouse, human, and Xenopus, suggesting a conserved role in most or all cells and tissues (8). MDC9 is highly expressed at the blastocyst implantation site in the uterus in rabbits, suggesting a possible role in implantation (33). Biochemical studies have shown that recombinant MDC9 has a different peptide cleavage specificity and inhibitor profile from those of TACE, suggesting that MDC9 has different substrates in cells (39). Furthermore, MDC9 has been implicated in heparin-binding EGF-like growth factor (HB-EGF) shedding in assays using overexpressed wild-type and mutant forms of this protein (18), in the processing of the amyloid precursor protein (19), and as a ligand for the integrin α6β1 (31). To learn more about the function of MDC9 during mouse development and in the adult animal, we have generated mice lacking functional MDC9. These animals develop normally, are viable and fertile, and do not suffer from evident pathological abnormalities in a controlled laboratory environment.
MATERIALS AND METHODS
Materials.
Chemicals and reagents were purchased from Sigma (St. Louis, Mo.), and restriction enzymes and reagents for molecular biology were purchased from Roche Diagnostics (Mannheim, Germany), except in cases where a different supplier is indicated.
Construct design.
Genomic clones of the MDC9 gene were isolated from a 129/SvJ genomic library using the mouse MDC9 cDNA (nucleotides 1 to 396 of GenBank sequence U41765) as a probe and further analyzed by restriction mapping and sequence analysis using gene-specific primers. To generate a mutation in the mouse MDC9 gene, we inserted the neomycin selection marker cassette in an unique ApaI site located in the exon encoding amino acids 32 to 66 of MDC9. The final targeting construct consisted of a 14.5-kb genomic DNA fragment, which was interrupted by the insertion of pMC1neoPolyA (49) and contained a diphtheria toxin A cassette as a negative selection marker against random integration (53).
Generation of targeted ES cells.
129/Sv embryonic stem (ES) cells were grown on mitotically inactivated mouse primary fibroblasts (38) and electroporated with 30 μg of NotI-linearized targeting vector. After 7 to 10 days in selection medium (350 μg of G418 per ml), colonies were picked, expanded, and screened by Southern blot analysis (25). Correctly targeted ES clones were identified using 5"- and 3"-specific probes and a neo probe. The targeting efficiency of this construct was about 10%.
Generation of mice lacking MDC9.
Correctly targeted ES cells were injected into C57BL/6J blastocysts to generate chimeras. To obtain germ line transmission, chimeric male mice with a high degree of chimerism (as judged by the agouti coat color) were mated to C57BL/6J females. Chimeric founder mice were also mated with 129/SvJ mice to produce inbred animals of the 129/SvJ background lacking MDC9. Furthermore, mixed-background 129/SvJ/C57BL/6J mice carrying a targeted allele of MDC9 were backcrossed six times with C57BL/6J mice.
Monoclonal antibody production.
The extracellular domain of mMDC9 (bp 28 to 2118) was cloned in frame to the human Fc-pcDNA expression vector (26). The resulting fusion protein was expressed in CHO cells and purified on protein A-Sepharose as described previously (26). mdc9−/− mice were immunized by intraperitoneal injection with approximately 25μg of mMDC9-EC-Fc emulsified 1:1 in Titermax at 3-week intervals. Once a good immune response was detected in serum from immunized mice by Western blot analysis and enzyme-linked immunosorbent assay, splenocytes were isolated and fused to exponentially growing Sp2/0-Ag14 cells using polyethylene glycol 1500. Fused hybridoma cells were selected by growth in hybridoma serum-free medium (Gibco/BRL) containing 1× hypoxanthine-aminopterin-thymidine (HAT), 15% fetal bovine serum, 1× hybridoma cloning factor (IGEN Inc. International, Gaithersburg, Md.), and 10 mg of gentamicin per ml. Supernatants of immunoglobulin (Ig)-producing cells were first screened by enzyme-linked immunosorbent assay on mMDC9-EC-Fc immobilized on 96-well plates, using Fas-Fc as a negative control. Positive clones were then screened by Western blot analysis on nonreduced antigen and on extracts of COS-7 cells transfected with full-length mouse MDC9 (52). Hybridoma cells secreting monoclonal antibodies against MDC9 were cloned by limited dilution. Hybridomas producing monoclonal antibodies against MDC9 were subcloned, adapted to serum-free conditions, and produced in a MiniPerm bioreactor system (Sartorious, Edgewood, N.Y.). For this study, clone 1A10 (Isotype IgG1 kappa) was used for Western blot analysis of mouse embryonic fibroblasts (see below).
Western blot analysis.
The absence of the MDC9 protein in various tissues of mdc9−/− mice was confirmed by Western blot analysis as described previously (52). Briefly, brain and lung tissues were isolated from a 3-month-old male mdc9−/− or wild-type mouse and homogenized with a polytron homogenizer (Kinematica) in cell lysis buffer supplemented with a protease inhibitor cocktail. For cultured fibroblast cells, lysis buffer was added directly to tissues culture plates. After removal of nuclei and cell debris by centrifugation at 20,000 × g for 10 min, glycoproteins were enriched using concanavalin A-Sepharose beads (Amersham Pharmacia Biotech, Piscataway, N.J.). Bound glycoproteins were eluted in sample loading buffer, separated on a sodium dodecyl sulfate (SDS)-10% polyacrylamide gel, and transferred to a nitrocellulose membrane. The blot was then incubated with either polyclonal serum against the cytoplasmic domain of MDC9 or MDC15 (at 1:1,000) or monoclonal antibody against MDC9-EC-Fc (at 1 μg/ml). Bound antibodies were visualized with the appropriate horseradish peroxidase-labeled second antibody and a chemiluminescence detection system (ECL; Kodak) to expose Bio-MAX film (Kodak, Rochester, N.Y.).
In situ hybridization.
To analyze the expression of MDC9 during embryogenesis by RNA in situ hybridization, timed matings were set up to generate embryos at different stages of gestation (E7.5, E9.5, E11.5, and E15.6). Embryos were fixed in 4% paraformaldehyde overnight at 4°C. For adult mouse brain preparation, mice were anesthetized and perfused transcardially with phosphate-buffered saline (PBS) followed by 4% paraformaldehyde. The brains were removed and postfixed in 4% paraformaldehyde overnight at 4°C. The fixed embryos and brains were dehydrated with a graded series of ethanol, cleared with Histoclear, and then embedded in paraffin. Paraffin sections 8 μm thick were cut and mounted on Fisher Superfrost Plus slides. 33P-labeled single-stranded RNA probes for MDC9 were prepared from linearized plasmids using T7/T3 RNA polymerases and ribonucleoside triphosphate mixture containing 12 μM cold and 4 μM hot UTP. The antisense probe was prepared from a 646-bp MDC9 cDNA C-terminal fragment from the ClaI to XhoI sites. The sense RNA strand was used as a negative control for each probe. The specificities of the labeled cRNA probes were confirmed by Northern blot analysis of the RNAs isolated from mouse embryos.
The RNA in situ hybridization procedure was performed as previously described (27). Briefly, after rehydration, the sections were postfixed in 4% paraformaldehyde and treated with proteinase K followed by deacetylation. Prehybridization and hybridization treatments were performed at 65°C. Slides were preincubated for 2.5 h in the hybridization buffer containing 50% formamide and 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Subsequently, overnight hybridization with 3 × 106 cpm/slide for each probe was carried out. The washes were with 2× SSC at 50°C for 30 min, followed by 50% formamide-2× SSC at 65°C for 30 min and a solution of 0.3 M NaCl, 10 mM Tris (pH 8.0), and 5 mM EDTA, containing 50 μg of RNase A per ml at 37°C for 30 min. The formamide wash was repeated and was followed by two 15-min washes in 2× SSC and two 15-min washes in 0.1× SSC at room temperature. After ethanol dehydration, the slides were dipped in autoradiographic emulsion (NTB-2; Kodak). The slides were exposed for 2 to 3 weeks and developed in Kodak developer D-19, fixed in Kodak fixer, and counterstained with hematoxylin and eosin.
Histopathology and clinical pathology.
For a histopathological analysis of mdc9−/− mice, 1-year-old mice (five males and five females) of mixed genetic background were sacrificed. The spleen, liver, kidneys, testes, heart, and brain were harvested from each mouse, weighed, and fixed in 10% buffered formalin. Eyes and testes were collected in Bouin's fixative. Terminal blood samples were collected for clinical chemistry analysis (Roche Diagnostics; Hitachi analyzer model 911/917), and the following parameters were measured: total protein, albumin, alkaline phosphatase, alanine aminotransferase, aspartate aminotransferase total bilirubin, blood urea nitrogen, cholesterol, glucose, Na+, K+, and Cl−. The following tissues were stained with hematoxylin and eosin after fixation and examined microscopically: adrenals, aorta, brain, cecum, colon, duodenum, epididymides, eyes (including optic nerve), gallbladder, harderian gland, heart, ileum, jejunum, kidneys, liver, lungs, lymph nodes (mandibular and mesenteric), esophagus, ovaries, pancreas, peripheral nerve, pituitary, prostate, rectum, salivary glands, seminal vesicles, skeletal muscle, skin (and mammary glands), spinal cord (cervical, thoracic, and lumbar), spleen, sternum (and bone marrow), stomach, testes, thymus, thyroids (and parathyroids), tongue, trachea, urinary bladder, uterus, and vagina. Brains were also stained with Congo Red and Bielschowsky's Silver stain. Furthermore, 19-month-old male and female mdc9−/− and wild-type mice of mixed genetic background (five each), as well as 14- to 18-month-old male and female mdc9−/− and wild-type mice of the 129/SvJ background (five each) were subjected to a more limited analysis of liver, heart, mammary gland, spleen, and uterus because of minor pathological findings in these tissues in some of the 1-year-old mdc9−/− mice. No major histopathological defects were observed in mdc9−/− mice that were not also seen in wild-type mice.
For hematological analysis, male and female wild-type mice and age matched mice lacking MDC9 were anesthesized using metofluorane. Blood was collected in heparinized syringes and transferred to EDTA-containing collection tubes. Then 1μl was applied to a microscope slide for differential blood counts. No significant differences in cell counts or differential blood counts were seen in samples from mdc9−/− and wild-type mice.
Transfections and cell culture of mouse primary embryonic fibroblasts.
E13.5 embryos from MDC9 knockout and wild-type mice were used to generate mouse embryonic fibroblasts (38). Briefly, the head and viscera were removed from the embryo. The remaining tissue was minced and then trypsinized in 0.25% trypsin for 15 min at 37°C. Cells were released through mechanical trituration and grown in Dulbecco’s modified Eagle medium, 10% fetal calf serum, and 100 μg of penicillin-streptomycin per ml. To evaluate potential shedding defects in mdc9−/− cells compared to wild-type cells, a cDNA construct coding for human HB-EGF (pSS-AIPh [50]) was used in the following manner. A total of 105 primary fibroblasts were plated on gelatin-coated 10-cm2 wells and were transfected with the alkaline phosphatase (AP)-tagged HB-EGF (HB-EGF-AP) plasmid using LipofectAMINE (Invitrogen Life Technologies, Carlsbad, Calif.) as specified by the manufacturers. Phorbol myristate acetate (PMA) (20 ng/ml) and/or BB-94 (batimastat) (1 μM) (6) was added the next day for 1 h. Supernatants were collected, and cells were lysed in PBS-1% Triton X-100 with protease inhibitors (4). Both supernatants and cell lysates were centrifuged at 25,000 × g for 30 min at 4°C. Cleared supernatants were then precipitated for 1 h with 20 μl of heparin-Sepharose (Amersham Pharmacia Biotech, Piscataway, N.J.), and bound proteins were eluted for 10 min at 37°C with SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer containing 1% SDS and 1 mM dithiothreitol and subjected to gel electrophoresis. SDS-polyacrylamide gels were run at 100 V and then incubated for two 30-min stretches in 2.5% Triton X-100 in double-distilled water and once for 10 min in 100 mM Tris (pH 9.5)-100 mM NaCl-20 mM MgCl2. The AP activity was then visualized by adding nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate NBT/BCIP. After incubation at 37°C, enzyme reactions were stopped in 50% methanol-10% glacial acetic acid. The gels were scanned using a UMAX Astra 2100 scanner and Adobe Photoshop software. For quantification, images were imported into MacBAS software (Fujifilm).
APP shedding in mouse hippocampal cells.
Dissociated primary hippocampal cells were isolated from neonatal mice on embryonic day E17.5. Pooled hippocampi were incubated in 0.1% trypsin for 5 min at 37°C and then triturated with a fire-polished Pasteur pipette. Hippocampal cells were pelleted and resuspended in neurobasal medium with B27 supplement (Invitrogen Life Technologies) and 100 μg of penicillin-streptomycin per ml. Neurons from two hippocampi were plated on 2-cm2 poly-d-lysine-coated plates. For the first 4 days, 25 μM glutamate was added to the medium. The neurons were grown for 2 weeks and then metabolically labeled overnight in Dulbecco’s modified Eagle medium-cysteine-methionine-100 μCi of Promix ([35S]cysteine and [35S]methionine; Amersham Pharmacia Biotech). Supernatants were collected, and β-amyloid precursor protein (β-APP) and the Aβ and p3 degradation products were bound with antibody 3129 (kindly provided by Joseph Buxbaum) overnight. Immune complexes were precipitated with 30 μl of a protein A-Sepharose slurry. Beads were washed with PBS-1% Triton X-100 and run on 10% Nupage Tris-Tricine gels in morpholineethanesulfonic acid (MES) buffer (Invitrogen Life Technologies).
Flow cytometric analysis.
Peripheral lymph nodes (axillary and brachial) were removed from four age-matched wild-type and mdc9−/− mice and used to make single-cell suspensions; they were stained with fluorescein isothiocyanate-conjugated anti-T-cell receptor (clone H57-597) and phycoarythrin-conjugated anti-CD24 (clone M1/69). All antibodies were produced, purified, and conjugated at Memorial Sloan-Kettering Cancer Center. Analysis was performed on an LSR analytical cytometer (BD Immunocytometry Systems, San Jose, Calif).
RESULTS
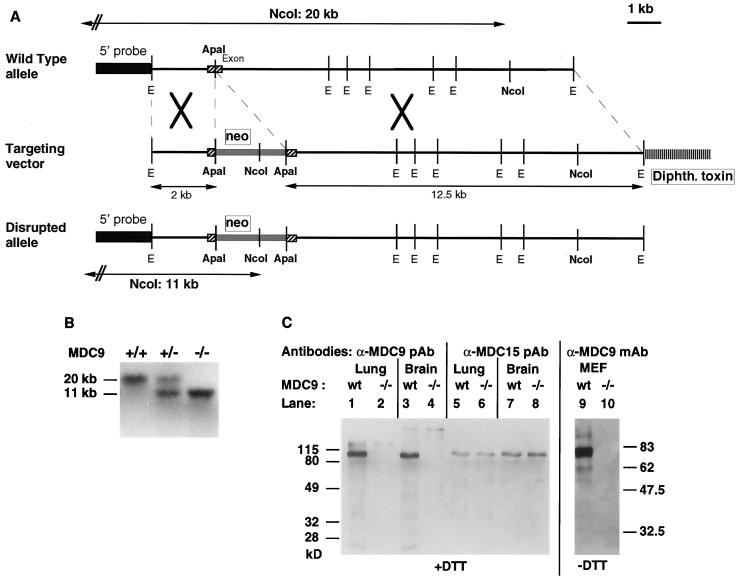
Mice carrying a targeted mutation in mdc9 were generated by homologous recombination (Fig. 1A) (see Materials and Methods for details). Matings of heterozygous mice of inbred backgrounds (C57BL/6J or 129/SvJ [Table 1]) or mixed genetic background (C57BL/6J and 129/SvJ [data not shown]) produced offspring with a Mendelian distribution of wild-type, heterozygous, and homozygous mutant genotypes. To confirm that mice homozygous for the targeted mutation of mdc9 (referred to as mdc9−/− mice) do not express MDC9, Western blots of concanavalin A-enriched lung or brain glycoproteins from mdc9−/− mice were probed with a polyclonal antibody against the cytoplasmic domain of MDC9. In both cases, MDC9 could not be detected in tissue extracts of mdc9−/− mice under conditions where it was clearly detected in tissue extracts of wild-type mice (Fig. 1C, lanes 1 to 4). Identical protein samples were probed with antibodies against MDC15 (lanes 5 to 8). The expression of MDC15 in lung or brains from wild-type or mdc9−/− mice was comparable, arguing against a compensatory upregulation of this widely expressed and catalytic site-containing ADAM (21, 26).
FIG. 1.
Targeted mutation of mdc9. (A) The genomic structure of the wild-type mdc9 allele is shown at the top. The position of the 5" probe used for Southern blot analysis is indicated, as well as the targeted exon, EcoRI (E) sites, and ApaI and NcoI sites. The targeting vector is shown in the middle. The targeted exon was disrupted by insertion of a pMC1neoPolyA cassette, which introduces an additional NcoI site. A diphtheria toxin (Diphth. toxin) gene cassette was included for negative selection. The bottom panel shows the genomic structure of the targeted allele. The additional NcoI site introduced into the targeted allele by homologous recombination reduces the size of the NcoI-digested genomic fragment that is recognized by the 5" probe from ∼20 to ∼11 kb. (B) Southern blot analysis of NcoI-digested genomic DNA from wild-type, heterozygous mdc9+/−, and homozygous mdc9−/− mice. (C) Western blot analysis of MDC9 expression in mouse lung (lanes 1 and 2), brain (lanes 3 and 4) or embryonic fibroblasts (MEF; lanes 9 and 10) from wild-type or mdc9−/− mice confirms the lack of MDC9 expression in mdc9−/− mice. Lanes 1 to 4 were probed with a polyclonal antiserum (pAb) against the MDC9 cytoplasmic tail. Lanes 9 and 10 were probed with a mouse monoclonal antibody (mAb) against mouse MDC9 which was generated from mdc9−/− mice immunized with an IgG-Fc fusion protein with the ectodomain of MDC9 (see Materials and Methods). This monoclonal antibody recognizes only nonreduced MDC9. Identical samples to those in lanes 1 to 4 were probed with a polyclonal antiserum against the cytoplasmic domain of MDC15 (lanes 5 to 8).
TABLE 1.
Results of mdc9+/− × mdc9+/− matings in 129/SvJ and C57BL/6J genetic backgrounds
| Background | No. (%) of offspring that were:
|
||
|---|---|---|---|
| mdc9+/+ | mdc9+/− | mdc9−/− | |
| 129/SvJ | 49 (23.4%) | 106 (50.7%) | 54 (25.8%) |
| C57BL/6J | 40 (26.8%) | 75 (50.6%) | 34 (22.8%) |
| Total | 89 (24.9%) | 181 (50.6%) | 88 (24.6%) |
Mice lacking mdc9 and age-matched wild-type littermates were indistinguishable in appearance and average weight from birth until 2 years of age (data not shown). No behavioral changes were seen when handling mdc9−/− mice compared to wild-type mice. Furthermore, male and female mdc9−/− mice were fertile and gave birth to viable and apparently normal and healthy offspring. The average litter size of matings of mdc9−/− mice in a mixed genetic background (129/SvJ/C57BL/6J) was 5.1 (standard deviation, 2.1), while the average litter size of wild-type mice of the same mixed background was 7.5 (standard deviation, 3.4). In the 129/SvJ genetic background, the average litter size of mdc9−/− mice was 3.7 (standard deviation, 1.6) compared to 4.6 (standard deviation, 2.0) for wild-type mice. Thus the litter size of mdc9−/− mice was slightly, but not significantly, reduced compared to that of wild-type mice. Furthermore, there was no significant difference in mortality between groups of 35 male or female mdc9−/− or age-matched wild-type mice of mixed genetic background over the course of 2 years (data not shown). A histopathological analysis of age-matched adult mdc9−/− and wild-type mice did not uncover morphological aberrations (see Materials and Methods for a listing of the tissues and organs analyzed). Likewise, we found no abnormalities in a differential blood count and analysis of blood chemistry or in the ratio of CD4+ to CD8+ cells in lymph nodes and thymus in mdc9−/− mice compared to wild-type mice (data not shown). Finally, levels of angiotensin-converting enzyme, which is shed from the plasma membrane by a yet to be identified metalloprotease that is distinct from TACE/ADAM17 (41), in serum were slightly elevated in mdc9−/− mice versus wild-type mice (data not shown). This argues against an essential role for MDC9 in the shedding of angiotensin-converting enzyme.
Generation of monoclonal antibodies against mouse MDC9 in mdc9−/− mice.
Attempts to raise monoclonal antibodies against mouse proteins in mice are usually considered futile because a mouse antigen should not be recognized as foreign. Since mdc9−/− mice are healthy and viable, this presented a unique opportunity to generate mouse monoclonal antibodies against mouse MDC9. mdc9−/− mice were immunized with a purified Ig-Fc fusion protein with the ectodomain of mouse MDC9. B cells from immunized mice were used to generate hybridomas producing antibodies against MDC9 (see Materials and Methods). A Western blot of mdc9−/− or wild-type mouse embryonic fibroblasts with one of the resulting monoclonal antibodies is shown in Fig. 1C (lanes 9 and 10).
Analysis of MDC9 expression during mouse development.
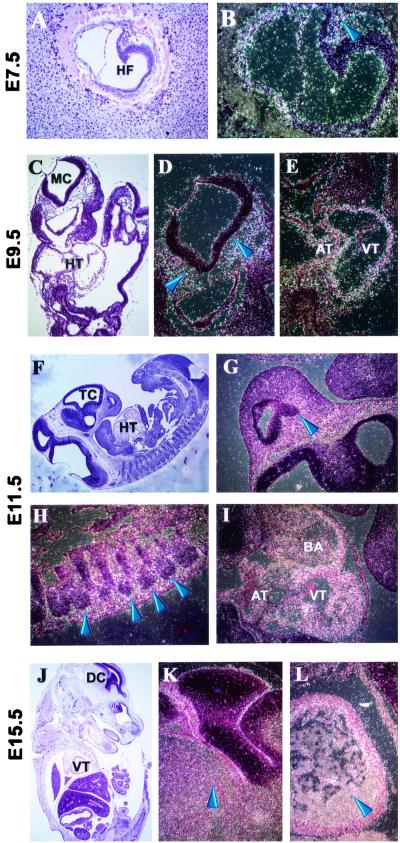
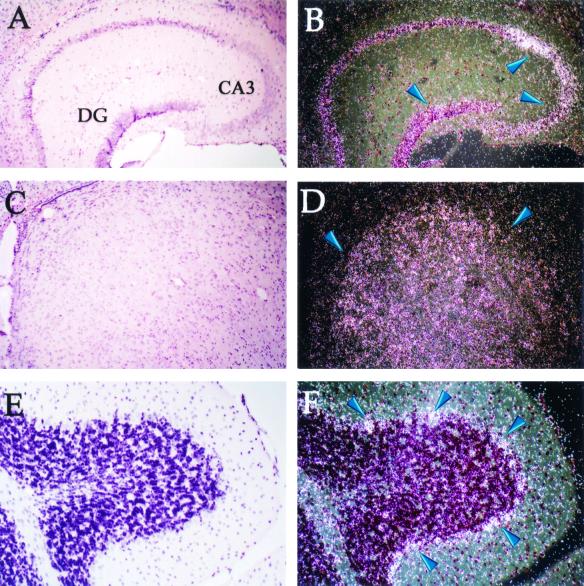
To gather clues about potential functions of MDC9 during development, we analyzed its mRNA expression pattern at different stages of embryogenesis by in situ hybridization. At E 7.5, MDC9 mRNA was ubiquitously expressed in all three germ layers, the ectoderm, mesoderm, and endoderm, with the most prominent expression occurring in heart mesoderm (Fig. 2B). The strong expression of MDC9 mRNA in the developing heart was also clearly visible at later stages of development, in particular in the wall of the ventricle and to a lesser extent in the bulbus arteriosus and atrium (Fig. 2E, I, and L). Furthermore, elevated levels of MDC9 mRNA expression were observed in mesenchymal cells surrounding the brain (Fig. 2D, G, and K). In 11.5-day-old embryos, MDC9 mRNA expression was detected in the mesenchyme condensing around the spinal cord (Fig. 2H). In addition to the regions of relatively high expression of MDC9 mRNA described above, lower levels of expression were observed ubiquitously. In the adult, MDC9 mRNA expression is also ubiquitous, confirming previous results from Western blots of MDC9 protein in different tissues (52). However, certain cells and tissues, in particular the pyramidal cells of the hippocampus (Fig. 3B), the hypothalamus (Fig. 3D), and Purkinje cells in the cerebellum (Fig. 3F), express markedly higher levels of MDC9 mRNA.
FIG. 2.
Expression of MDC9 mRNA in mouse embryos. All sections were hybridized with an antisense MDC9 mRNA probe. Digital images were acquired using bright-field (A, C, F, and J) or dark-field (B, D, E, G, H, I, K, and L) microscopy. (A and B) A section of E7.5 at magnifications of 10× (A) and 20× (B). MCD9 mRNA is expressed in all cell layers. Within the embryonic portion, a stronger signal was detected in the heart mesoderm (blue arrow in panel B) than in other regions of the embryo, including the head fold region (HF in panel A), which has a weaker signal despite having a higher cell density. (C to E) A section of E9.5. (C) Image at magnification of 5×, showing the mesencephalon (MC) and the heart (HT). (D and E) Images of the MC and HT regions in panel C at magnifications of 10×. Blue arrows in panel D point to autoradiographic grains in the head mesenchyme. Strong expression signals in panel E outline the heart atrium (AT) and ventricle (VT). (F to I) A sagittal section of E11.5. (F) Image at magnification of 2.5×. TC, telencephalon; HT, heart. (G, H, and J) images of regions in panel F at a magnification of 10×. The blue arrow in panel G points to a strong signal in the facial mesenchyme, anterior to the TC. Panel H represents part of the dorsal region of the embryo, where the loose mesenchyme surrounding the somites (indicated by blue arrows) shows strong gene expression. The white autoradiographic grains of the image in panel I show strong signal in the atrium (AT), ventricle (VT), and bulbus arteriosus (BA) of the heart. (J to L) Images of E15.5. (J) Image of a lateral section at a magnification of 1.25×. DC, diencephalon; VT, heart ventricle. (K and L) portions of the image in panel J, at a magnification of 10×. The blue arrow in panel K shows an area of strong expression in the head mesenchyme close to DC, and the blue arrow in panel L shows strong expression in VT.
FIG. 3.
Expression of MCD9 mRNA in adult mouse brain. Brain sections were hybridized with an antisense MDC9 mRNA probe and analyzed using bright-field (A, C, and E) and corresponding dark-field (B, D, and F) images. Coronal sections through the hippocampus (A and B) showed high expression (blue arrows) in the pyramidal layer, particularly in the CA3 region, as well as in the dentate gyrus (DG). Abundant expression (blue arrows) was detected in the central part of the hypothalamus (C and D) and in Purkinje cerebellar neurons (E and F).
Despite the strong expression of MDC9 in a number of tissues during development, notably the heart and mesenchymal tissues, examination of fixed and stained sections of mdc9−/− embryos of different developmental stages revealed no evident histopathological defects compared to wild-type embryos (data not shown). Likewise, in a histopathological analysis, sections of adult brains from mdc9−/− mice were indistinguishable from age-matched wild-type brains (data not shown).
HB-EGF processing in embryonic fibroblasts lacking MDC9.
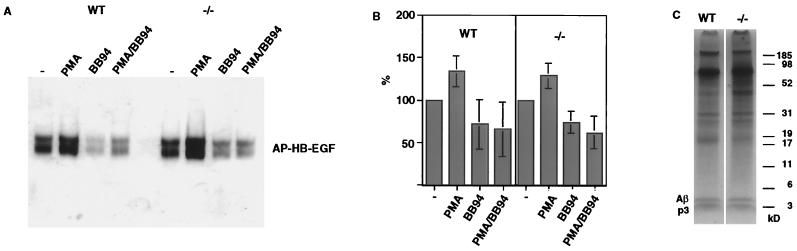
Overexpression studies using mutant forms of MDC9 have implicated it in shedding of the EGF receptor (EGFR) ligand HB-EGF (18). Therefore, we evaluated whether MDC9 is required for HB-EGF shedding in primary mouse embryonic fibroblasts isolated from E13.5 mdc9−/− or wild-type embryos. Expression of MDC9 in wild-type mouse embryo fibroblasts and the lack of MDC9 in mdc9−/− mouse embryo fibroblasts was confirmed in experiments using a monoclonal antibody against MDC9 (see above) (Fig. 1C, lanes 9 and 10). Wild-type and mdc9−/− mouse embryo fibroblasts were transfected with HB-EGF-AP (50). HB-EGF-AP was precipitated from the culture supernatant with heparin-Sepharose, subjected to SDS-PAGE, renatured, and then visualized by incubation of the gel in an AP substrate (see Materials and Methods). HB-EGF-AP shedding was evaluated in untreated cells and in cells stimulated with the phorbol ester PMA, in the presence or absence of the hydroxamic acid-type metalloprotease inhibitor BB-94. No significant difference in the constitutive and stimulated metalloprotease-dependent processing of HB-EGF-AP was observed between wild-type and mdc9−/− mouse embryo fibroblasts (Fig. 4A and B).
FIG. 4.
Shedding of HB-EGF and of APP in mdc9−/− cells. (A) Mouse embryonic fibroblasts were isolated from E13.5 wild-type or mdc9−/− embryos and transfected with HB-EGF-AP cDNA (see Materials and Methods). Cells were incubated with or without the phorbol ester PMA in the presence or absence of the hydroxamic acid-type metalloprotease inhibitor BB-94 for 1 h. The soluble ectodomain of HB-EGF-AP was precipitated with heparin-Sepharose, separated by SDS-PAGE, and renatured, and AP activity was visualized after incubating the gel in AP substrates (see Materials and Methods). (B) Bar graph of the results from eight separate experiments similar to the one shown in panel A, quantified using MacBas image analysis software. (C) Hippocampal neurons were isolated from E18.5 wild-type or mdc9−/− embryos. 35S-labeled APP and its degradation products p3 and Aβ were immunoprecipitated from cell supernatants and separated by SDS-PAGE. No difference in the level of p3 and Aβ released from wild-type and mdc9−/− hippocampal neurons was seen in three separate experiments.
APP α-secretase activity in hippocampal neurons lacking MDC9.
When MDC9 is overexpressed in COS cells, an increase in α-secretase cleavage of APP, a protein which has been linked to the pathogenesis of Alzheimer's disease, has been reported (19). Furthermore, recombinant MDC9 can cleave an APP peptide containing the human α-secretase cleavage site in a position at which APP is processed in hippocampal neurons in the rat (39, 45). The high expression of MDC9 in the hippocampus observed here (Fig. 3B) would thus be consistent with a potential role for MDC9 as a hippocampal α-secretase. To test for a role of MDC9 in processing mouse APP, we isolated hippocampal neurons from mdc9−/− and wild-type mice to evaluate production of the APP cleavage products Aβ and p3. The Aβ peptide, which results from cleavage of APP by β- and γ-secretase, and p3, which results from cleavage by α- and γ-secretase, and APP were immunoprecipitated from supernatants of 35S-labeled dissociated hippocampal neurons (Fig. 4C). The relative amounts of Aβ and P3 released into the culture supernatant in three different experiments were quantitated using a Fuji phosphorimager system and found to be comparable in wild-type and mdc9−/− mice (data not shown).
DISCUSSION
MDC9 (ADAM9) is a widely expressed and catalytically active metalloprotease-disintegrin that is highly conserved among humans, mice, and Xenopus laevis (8, 52). Here we have evaluated the role of MDC9 during mouse development and in the adult animal by generating mice that lack this protein. No major morphological changes or histopathological defects during development or in the adult were evident in mdc9−/− mice compared to wild-type control animals. mdc9−/− mice are fertile and do not have a significant difference in litter size compared to wild-type controls, arguing against an essential role for MDC9 in blastocyst implantation (33). The lack of any apparent developmental defect or severe pathological phenotype in mdc9−/− mice, despite the ubiquitous expression of this protein during development and in the adult, could result from functional compensation by or redundancy with other members of the ADAM family or perhaps even other molecules. Alternatively, MDC9 may have advantageous but nonessential functions during development or in adults. In this context it is interesting that MDC9 does not appear to have an orthologue in D. melanogaster or in C. elegans (data not shown). It has been suggested that vertebrate proteins without orthologues in D. melanogaster or in C. elegans may have functions in cells or organs that are more highly evolved in vertebrates, such as the immune system, cardiovascular system, or the brain (16). These possibilities remain to be addressed through specific challenges of mdc9−/− mice.
ADAMs with a catalytic site consensus sequence, such as MDC9, have been predicted to play a role in protein ectodomain shedding (3, 42). This metalloprotease-dependent process leads to the release of a variety of structurally and functionally diverse proteins from the plasma membrane (15, 29). ADAMs are considered good candidate sheddases because many shedding events are inhibited by hydroxamic acid-type metalloprotease inhibitors but not by tissue inhibitors of metalloproteases (TIMPs) 1 or 2, which in combination are expected to inhibit all currently known matrix metalloproteinases (15). Furthermore, the first identified sheddase, the tumor necrosis factor alpha (TNF-α) convertase is an ADAM (2, 30). While ADAM17/TNF-α convertase has an essential role in ectodomain shedding of TGF-α and possibly also of other EGFR ligands during development (34), metalloprotease-dependent ectodomain shedding of some membrane proteins, such as angiotensin-converting enzyme, occurs in the absence of functional TACE (41). This suggests that other metalloproteases besides TACE are also involved in ectodomain shedding. Indeed, a dominant negative form of ADAM19 interferes with processing of the EGFR ligand neuregulin (44). Furthermore, ADAM10 has also been implicated in the shedding of APP (20, 22) as well as ephrin2A (14), and in cleavage of Delta, a ligand for Notch (37). Finally, overexpression of a mutant form of MDC9 reportedly interferes with shedding of the EGFR ligand HB-EGF (18). However, constitutive and PMA-stimulated shedding of HB-EGF was unaffected in mdc9−/− cells compared to wild-type cells, arguing against an essential role of MDC9 in HB-EGF shedding in fibroblasts. Further studies are necessary to understand why overexpressing a mutant form of MDC9 blocks HB-EGF shedding (18) whereas loss of MDC9 protein does not have this effect.
MDC9 has also been implicated as an α-secretase for APP (19). Furthermore, it is highly expressed in the hippocampus, an area of the brain where Aβ production and deposition are thought to contribute to the pathogenesis of Alzheimer's disease. In dissociated hippocampal neurons, no difference in the generation of Aβ and p3 or in the ratio of these two cleavage products was observed. This argues against an essential and major role for MDC9 as a hippocampal α-secretase in mice. We note that the α-secretase peptide used for in vitro cleavage studies with mouse MDC9 (39) was derived from the human APP sequence, which differs from the mouse APP sequence at the MDC9 cleavage site (HH-QK in human APP versus RH-QK in mouse APP). Our results therefore do not rule out the possibility that MDC9 may act as a hippocampal α-secretase in humans. Currently the best candidate α secretases are TACE/ADAM17 (7, 46, 47) and ADAM10 (20, 22, 24, 28).
In summary, the targeted deletion of the widely expressed and catalytically active MDC9 has shown that this protein is not essential for normal development of mice or for fertility or adult homeostasis. Further studies will address the possibility of functional compensation or redundancy between MDC9 and other widely expressed and catalytically active ADAMs. Physiologically relevant functions of MDC9 may also be uncovered through an assessment of potential shedding defects in mdc9−/− cells and specific challenges of mdc9−/− mice.
Acknowledgments
This work was supported by National Institutes of Health grant RO1 GM58668, the Memorial Sloan-Kettering Cancer Center support grant NCI-P30-CA-08748, the Samuel and May Rudin Foundation, and the DeWitt Wallace Fund.
We thank Vera Suarez, Willie H. Mark, Jia-Hui Dong, Joanne Ingenito, and Liz Lacy for assistance and advice in generating mdc9−/− mice; Lawrence Lum, Byung Lee, Eleanor Spumberg, Thadeous Kacmarczyk, Kenya Parks, and Anthony Zayas for assistance in mouse breeding and genotyping; Howard Petrie for the analysis of B- and T-cell-ratio spleen and peripheral lymphocytes; Joe Buxbaum for antibodies against the amyloid precursor protein; Indira Sen for analysis of serum angiotensin-converting enzyme levels; Taha Merghoub and Pier-Paolo Pandolfi for differential blood counts and hematological analysis; and the MSKCC Transgenic Mouse Facility, Molecular Cytology Core Facility, and Research Animal Resources Facility.
REFERENCES
- 1.Alfandari, D., H. Cousin, A. Gaultier, K. Smith, J. M. White, T. Darribere, and D. W. DeSimone. 2001. Xenopus ADAM 13 is a metalloprotease required for cranial neural crest-cell migration. Curr. Biol. 11:918-930. [DOI] [PubMed]
- 2.Black, R., C. T. Rauch, C. J. Kozlosky, J. J. Peschon, J. L. Slack, M. F. Wolfson, B. J. Castner, K. L. Stocking, P. Reddy, S. Srinivasan, N. Nelson, N. Boiani, K. A. Schooley, M. Gerhart, R. Davis, J. N. Fitzner, R. S. Johnson, R. J. Paxton, C. J. March, and D. P. Cerretti. 1997. A metalloprotease disintegrin that releases tumour-necrosis factor-α from cells. Nature 385:729-733. [DOI] [PubMed] [Google Scholar]
- 3.Black, R. A., and J. M. White. 1998. ADAMs: focus on the protease domain. Curr. Opin. Cell Biol. 10:654-659. [DOI] [PubMed] [Google Scholar]
- 4.Blobel, C. P., D. G. Myles, P. Primakoff, and J. W. White. 1990. Proteolytic processing of a protein involved in sperm-egg fusion correlates with acquisition of fertilization competence. J. Cell Biol. 111:69-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blobel, C. P., T. G. Wolfsberg, C. W. Turck, D. G. Myles, P. Primakoff, and J. M. White. 1992. A potential fusion peptide and an integrin ligand domain in a protein active in sperm-egg fusion. Nature 356:248-252. [DOI] [PubMed] [Google Scholar]
- 6.Botos, I., L. Scapozza, D. Zhang, L. A. Liotta, and E. F. Meyer. 1996. Batimastat, a potent matrix metalloprotease inhibitor, exhibits an unexpected mode of binding. Proc. Natl. Acad. Sci. USA 93:2749-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buxbaum, J. D., K. N. Liu, Y. Luo, J. L. Slack, K. L. Stocking, J. J. Peschon, R. S. Johnson, B. J. Castner, D. P. Cerretti, and R. A. Black. 1998. Evidence that tumor necrosis factor alpha converting enzyme is involved in regulated alpha-secretase cleavage of the alzheimer amyloid protein precursor. J. Biol. Chem. 273:27765-27767. [DOI] [PubMed] [Google Scholar]
- 8.Cai, H., J. Krätzschmar, D. Alfandari, G. Hunnicutt, and C. P. Blobel. 1998. Neural crest-specific and general expression of distinct metalloprotease-disintegrins in early Xenopus laevis development. Dev. Biol. 204:508-524. [DOI] [PubMed] [Google Scholar]
- 9.Cal, S., J. M. Freije, J. M. Lopez, Y. Takada, and C. Lopez-Otin. 2000. ADAM 23/MDC3, a human disintegrin that promotes cell adhesion via interaction with the alphavbeta3 integrin through an RGD-independent mechanism. Mol. Biol. Cell 11:1457-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho, C., D. O. Bunch, J. E. Faure, E. H. Goulding, E. M. Eddy, P. Primakoff, and D. G. Myles. 1998. Fertilization defects in sperm from mice lacking fertilin beta. Science 281:1857-1859. [DOI] [PubMed] [Google Scholar]
- 11.Cho, C., H. Ge, D. Branciforte, P. Primakoff, and D. G. Myles. 2000. Analysis of mouse fertilin in wild-type and fertilin beta (−/−) sperm: evidence for C-terminal modification, alpha/beta dimerization, and lack of essential role of fertilin alpha in sperm-egg fusion. Dev. Biol. 222:289-295. [DOI] [PubMed] [Google Scholar]
- 12.Eto, K., W. Puzon-McLaughlin, D. Sheppard, A. Sehara-Fujisawa, X. P. Zhang, and Y. Takada. 2000. RGD-independent binding of integrin α9β1 to the ADAM-12 and -15 disintegrin domains mediates cell-cell interaction. J. Biol. Chem. 275:34922-34930. [DOI] [PubMed] [Google Scholar]
- 13.Fambrough, D., D. Pan, G. M. Rubin, and C. S. Goodman. 1996. The cell surface metalloprotease/disintegrin kuzbanian is required for axonal extension in Drosophila. Proc. Natl. Acad. Sci. USA 93:13233-13238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hattori, M., M. Osterfield, and J. G. Flanagan. 2000. Regulated cleavage of a contact-mediated axon repellent. Science 289:1360-1365. [DOI] [PubMed] [Google Scholar]
- 15.Hooper, N. M., E. H. Karran, and A. J. Turner. 1997. Membrane protein secretases. Biochem. J. 321:265-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hynes, R. O., and Q. Zhao. 2000. The evolution of cell adhesion. J. Cell Biol. 150:F89-F96. [DOI] [PubMed] [Google Scholar]
- 17.Iba, K., R. Albrechtsen, B. Gilpin, C. Frohlich, F. Loechel, A. Zolkiewska, K. Ishiguro, T. Kojima, W. Liu, J. K. Langford, R. D. Sanderson, C. Brakebusch, R. Fassler, and U. M. Wewer. 2000. The cysteine-rich domain of human ADAM 12 supports cell adhesion through syndecans and triggers signaling events that lead to β1 integrin-dependent cell spreading. J. Cell Biol. 149:1143-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Izumi, Y., M. Hirata, H. Hasuwa, R. Iwamoto, T. Umata, K. Miyado, Y. Tamai, T. Kurisaki, A. Sehara-Fujisawa, S. Ohno, and E. Mekada. 1998. A metalloprotease-disintegrin, MDC9/meltrin-gamma/ADAM9 and PKCdelta are involved in TPA-induced ectodomain shedding of membrane-anchored heparin-binding EGF-like growth factor. EMBO J. 17:7260-7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koike, H., S. Tomioka, H. Sorimachi, T. C. Saido, K. Maruyama, A. Okuyama, A. Fujisawa-Sehara, S. Ohno, K. Suzuki, and S. Ishiura. 1999. Membrane-anchored metalloprotease MDC9 has an alpha-secretase activity responsible for processing the amyloid precursor protein. Biochem J. 343:371-375. [PMC free article] [PubMed] [Google Scholar]
- 20.Kojro, E., G. Gimpl, S. Lammich, W. Marz, and F. Fahrenholz. 2001. Low cholesterol stimulates the nonamyloidogenic pathway by its effect on the alpha-secretase ADAM 10. Proc. Natl. Acad. Sci. USA 98:5815-5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krätzschmar, J., L. Lum, and C. P. Blobel. 1996. Metargidin, a membrane-anchored metalloprotease-disintegrin protein with an RGD integrin binding sequence. J. Biol. Chem. 271:4593-4596. [DOI] [PubMed] [Google Scholar]
- 22.Lammich, S., E. Kojro, R. Postina, S. Gilbert, R. Pfeiffer, M. Jasionowski, C. Haass, and F. Fahrenholz. 1999. Constitutive and regulated alpha-secretase cleavage of Alzheimer's amyloid precursor protein by a disintegrin metalloprotease. Proc. Natl. Acad. Sci. USA 96:3922-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leighton, P. A., K. J. Mitchell, L. V. Goodrich, X. Lu, K. Pinson, P. Scherz, W. C. Skarnes, and M. Tessier-Lavigne. 2001. Defining brain wiring patterns and mechanisms through gene trapping in mice. Nature 410:174-179. [DOI] [PubMed] [Google Scholar]
- 24.Lopez-Perez, E., Y. Zhang, S. J. Frank, J. Creemers, N. Seidah, and F. Checler. 2001. Constitutive alpha-secretase cleavage of the beta-amyloid precursor protein in the furin-deficient LoVo cell line: involvement of the pro-hormone convertase 7 and the disintegrin metalloprotease ADAM10. J. Neurochem. 76:1532-1539. [DOI] [PubMed] [Google Scholar]
- 25.Ludwig, T., C. E. Ovitt, U. Bauer, M. Hollinshead, J. Remmler, P. Lobel, U. Ruther, and B. Hoflack. 1993. Targeted disruption of the mouse cation independant mannose 6-phosphate receptor results in partial missorting of multiple lysosomal enzymes. EMBO J. 12:5225-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lum, L., M. S. Reid, and C. P. Blobel. 1998. Intracellular maturation of the mouse metalloprotease disintegrin MDC15. J. Biol. Chem. 273:26236-26247. [DOI] [PubMed] [Google Scholar]
- 27.Manova, K., K. Nocka, P. Besmer, and R. Bachvarova. 1990. Gonadal expression of c-kit encoded at the W locus of the mouse. Development 110:1057-1069. [DOI] [PubMed] [Google Scholar]
- 28.Marcinkiewicz, M., and N. G. Seidah. 2000. Coordinated expression of beta-amyloid precursor protein and the putative beta-secretase BACE and alpha-secretase ADAM10 in mouse and human brain. J. Neurochem. 75:2133-2143. [DOI] [PubMed] [Google Scholar]
- 29.Massague, J., and A. Pandiella. 1993. Membrane-anchored growth factors. Annu. Rev. Biochem. 62:515-541. [DOI] [PubMed] [Google Scholar]
- 30.Moss, M. L., S.-L. C. Jin, M. E. Milla, W. Burkhart, H. L. Cartner, W.-J. Chen, W. C. Clay, J. R. Didsbury, D. Hassler, C. R. Hoffman, T. A. Kost, M. H. Lambert, M. A. Lessnitzer, P. McCauley, G. McGeehan, J. Mitchell, M. Moyer, G. Pahel, W. Rocque, L. K. Overton, F. Schoenen, T. Seaton, J.-L. Su, J. Warner, D. Willard, and J. D. Becherer. 1997. Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-α. Nature 385:733-736. [DOI] [PubMed] [Google Scholar]
- 31.Nath, D., P. M. Slocombe, A. Webster, P. E. Stephens, A. J. Docherty, and G. Murphy. 2000. Meltrin gamma(ADAM-9) mediates cellular adhesion through alpha(6)beta(1)integrin, leading to a marked induction of fibroblast cell motility. J. Cell Sci. 113:2319-2328. [DOI] [PubMed] [Google Scholar]
- 32.Nishimura, H., C. Cho, D. R. Branciforte, D. G. Myles, and P. Primakoff. 2001. Analysis of loss of adhesive function in sperm lacking cyritestin or fertilin beta. Dev. Biol. 233:204-213. [DOI] [PubMed] [Google Scholar]
- 33.Olson, G. E., V. P. Winfrey, P. E. Matrisian, S. K. NagDas, and L. H. Hoffman. 1998. Blastocyst-dependent upregulation of metalloproteinase/disintegrin MDC9 expression in rabbit endometrium. Cell Tissue Res. 293:489-498. [DOI] [PubMed]
- 34.Peschon, J. J., J. L. Slack, P. Reddy, K. L. Stocking, S. W. Sunnarborg, D. C. Lee, W. E. Russel, B. J. Castner, R. S. Johnson, J. N. Fitzner, R. W. Boyce, N. Nelson, C. J. Kozlosky, M. F. Wolfson, C. T. Rauch, D. P. Cerretti, R. J. Paxton, C. J. March, and R. A. Black. 1998. An essential role for ectodomain shedding in mammalian development. Science 282:1281-1284. [DOI] [PubMed] [Google Scholar]
- 35.Primakoff, P., H. Hyatt, and J. Tredick-Kline. 1987. Identification and purification of a sperm surface protein with a potential role in sperm-egg membrane fusion. J. Cell Biol. 104:141-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Primakoff, P., and D. G. Myles. 2000. The ADAM gene family: surface proteins with an adhesion and protease activity packed into a single molecule. Trends Genet. 16:83-87. [DOI] [PubMed] [Google Scholar]
- 37.Qi, H., M. D. Rand, X. Wu, N. Sestan, W. Wang, P. Rakic, T. Xu, and S. Artavanis-Tsakonas. 1999. Processing of the notch ligand delta by the metalloprotease Kuzbanian. Science 283:91-94. [DOI] [PubMed] [Google Scholar]
- 38.Robertson, E. J. 1987. Embryo derived stem cell lines, p. 71-112. In E. J. Robertson (ed.), Teratocarcinomas and embryonic stem cells: a practical approach. IRL Press, Oxford, United Kingdom.
- 39.Roghani, M., J. D. Becherer, M. L. Moss, R. E. Atherton, H. Erdjument-Bromage, J. Arribas, R. K. Blackburn, G. Weskamp, P. Tempst, and C. P. Blobel. 1999. Metalloprotease-disintegrin MDC9: intracellular maturation and catalytic activity. J. Biol. Chem. 274:3531-3540. [DOI] [PubMed] [Google Scholar]
- 40.Rooke, J., D. Pan, T. Xu, and G. M. Rubin. 1996. KUZ, a conserved metalloprotease-disintegrin protein with two roles in Drosophila neurogenesis. Science 273:1227-1230. [DOI] [PubMed] [Google Scholar]
- 41.Sadhukhan, R., K. R. Santhamma, P. Reddy, J. J. Peschon, R. A. Black, and I. Sen. 1999. Unaltered cleavage and secretion of angiotensin-converting enzyme in tumor necrosis factor-alpha-converting enzyme-deficient mice. J. Biol. Chem. 274:10511-10516. [DOI] [PubMed] [Google Scholar]
- 42.Schlöndorff, J., and C. Blobel. 1999. Metalloprotease-disintegrins: modular proteins capable of promoting cell-cell interactions and triggering signals by protein ectodomain shedding. J. Cell Sci. 112:3603-3617. [DOI] [PubMed] [Google Scholar]
- 43.Shamsadin, R., I. M. Adham, K. Nayernia, U. A. Heinlein, H. Oberwinkler, and W. Engel. 1999. Male mice deficient for germ-cell cyritestin are infertile. Biol. Reprod. 61:1445-1451. [DOI] [PubMed] [Google Scholar]
- 44.Shirakabe, K., S. Wakatsuki, T. Kurisaki, and A. Fujisawa-Sehara. 2001. Roles of Meltrin beta/ADAM19 in the processing of neuregulin. J. Biol. Chem. 276:9352-9358. [DOI] [PubMed] [Google Scholar]
- 45.Simons, M., B. de Strooper, G. Multhaup, P. J. Tienari, C. G. Dotti, and K. Beyreuther. 1996. Amyloidogenic processing of the human amyloid precursor protein in primary cultures of rat hippocampal neurons. J. Neurosci. 16:899-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skovronsky, D. M., D. B. Moore, M. E. Milla, R. W. Doms, and V. M. Lee. 2000. Protein kinase C-dependent alpha-secretase competes with beta-secretase for cleavage of amyloid-beta precursor protein in the trans-Golgi network. J. Biol. Chem. 275:2568-2575. [DOI] [PubMed] [Google Scholar]
- 47.Slack, B. E., L. K. Ma, and C. C. Seah. 2001. Constitutive shedding of the amyloid precursor protein ectodomain is up-regulated by tumour necrosis factor-alpha converting enzyme. Biochem. J. 357:787-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sotillos, S., F. Roch, and S. Campuzano. 1997. The metalloprotease-disintegrin Kuzbanian participates in Notch activation during growth and patterning of Drosophila imaginal discs. Development 124:4769-4779. [DOI] [PubMed] [Google Scholar]
- 49.Thomas, K. R., and M. R. Capecchi. 1987. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 51:503-512. [DOI] [PubMed] [Google Scholar]
- 50.Tokumaru, S., S. Higashiyama, T. Endo, T. Nakagawa, J. Miyagawa, K. Yamamori, Y. Hanakawa, H. Ohmoto, K. Yoshino, Y. Shirakata, Y. Matsuzawa, K. Hashimoto, and N. Taniguchi. 2000. Ectodomain shedding of epidermal growth factor receptor ligands is required for keratinocyte migration in cutaneous wound healing. J. Cell Biol. 151:209-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wen, C., M. M. Metzstein, and I. Greenwald. 1997. SUP-17, a Caenorhabditis elegans ADAM protein related to Drosophila KUZBANIAN, and its role in LIN-12/NOTCH signaling. Development 124:4759-4767. [DOI] [PubMed] [Google Scholar]
- 52.Weskamp, G., J. R. Krätzschmar, M. Reid, and C. P. Blobel. 1996. MDC9, a widely expressed cellular disintegrin containing cytoplasmic SH3 ligand domains. J. Cell Biol. 132:717-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yagi, T., Y. Ikawa, K. Yoshida, Y. Shigetani, N. Takeda, I. Mabuchi, T. Yamamoto, and S. Aizawa. 1990. Homologous recombination at c-fyn locus of mouse embryonic stem cells with use of diphtheria toxin A-fragment gene in negative selection. Proc. Natl. Acad. Sci. USA 87:9918-9922. [DOI] [PMC free article] [PubMed] [Google Scholar]