Abstract
Many nuclear and nucleolar small RNAs are accumulated as nonpolyadenylated species and require 3"-end processing for maturation. Here, we show that several genes coding for box C/D and H/ACA snoRNAs and for the U5 and U2 snRNAs contain sequences in their 3" portions which direct cleavage of primary transcripts without being polyadenylated. Genetic analysis of yeasts with mutations in different components of the pre-mRNA cleavage and polyadenylation machinery suggests that this mechanism of 3"-end formation requires cleavage factor IA (CF IA) but not cleavage and polyadenylation factor activity. However, in vitro results indicate that other factors participate in the reaction besides CF IA. Sequence analysis of snoRNA genes indicated that they contain conserved motifs in their 3" noncoding regions, and mutational studies demonstrated their essential role in 3"-end formation. We propose a model in which CF IA functions in cleavage and polyadenylation of pre-mRNAs and, in combination with a different set of factors, in 3"-end formation of nonpolyadenylated polymerase II transcripts.
snoRNAs belong to a complex family of RNA molecules localized in the nucleolus, where they participate in rRNA processing (35) and in the modification of several classes of RNA substrates: rRNAs (5, 7, 33, 40, 48, 54), snRNAs (24, 30, 55), and possibly mRNAs (13, 20). They work in combination with specific sets of proteins, forming ribonucleoprotein complexes; both the structure of the particles and their activity are highly conserved in evolution, as they are present and perform the same activity in archaebacteria and in eukaryotes (42). snoRNA coding units have quite a peculiar gene organization: the majority of metazoan and a few yeast snoRNAs are encoded in introns of protein-coding genes, while most yeast snoRNAs and a few vertebrate ones derive from independent transcription units, either monocistronic or polycistronic (59). Despite this heterogeneous organization, snoRNA biosynthesis relies on a common mechanism: entry sites for 5"-3" and 3"-5" exonucleases are produced from precursor molecules and allow the release of mature snoRNAs (2, 43, 44, 56). In independently transcribed snoRNAs, such entry sites are often generated by the Rnt1p endonuclease (15, 16). In many cases, however, cleavage sites are absent in the 3" portion of the pre-snoRNAs, suggesting that processing starts from the 3" end of the primary transcript.
Like genes coding for mRNAs, snoRNA genes are transcribed by RNA polymerase II. 3"-end formation of pre-mRNAs is accomplished by a two-step reaction, which involves endonucleolytic cleavage followed by addition of a poly(A) tail to the upstream cleavage product (60). The yeast cleavage and polyadenylation machinery consists of several complexes: for specific cleavage, cleavage factor IA (CF IA), CF IB, and CF II are required, while polyadenylation occurs when CF IA, CF IB, Pap1p, and polyadenylation factor I (PF I) are present (3, 18, 31, 32, 36, 37). A complex containing PF I and CF II activity (named cleavage and polyadenylation factor [CPF]) has been isolated from yeast extracts by affinity purification (41). Since PF I and CF II subunits were found in a stable and stoichiometric association, it has been suggested that this complex forms a functional unit in vivo.
Previously it was shown that the presence of a pre-mRNA-derived cleavage and polyadenylation signal, such as the one found in the CYC1 gene, is not compatible with the production of a functional box C/D snoRNA. In this case, only polyadenylated unprocessed RNA molecules which did not associate with snoRNP-specific factors and which were unable to methylate their physiological rRNA substrates accumulated (22). Instead, sequences downstream of the snR13 snoRNA coding unit were able to direct efficient 3"-end formation of the upstream snoRNA, and only the cleavage activity of the pre-mRNA 3"-processing machinery was required.
In the present paper we show that this is a more general phenomenon, since box C/D and H/ACA snoRNAs and snRNAs require this type of 3"-end maturation. Thus, uncoupling of cleavage and polyadenylation is necessary for the biosynthesis of a large class of cellular RNAs, in particular for species that do not require the poly(A) tail but instead require 3"-end processing.
Analysis of a large number of mutant alleles in CF IA and CPF components in vivo suggests that CF IA but not CPF is essential for 3"-end formation of small RNAs. Furthermore, sequence comparison and mutational analysis have pointed out the importance of two conserved sequence elements which are presumptive binding sites for processing factors. These findings suggest that, for snoRNA and snRNA 3"-end formation, CF IA can work in cooperation with factors different from those involved in the cleavage and polyadenylation of pre-mRNAs.
MATERIALS AND METHODS
Strains and media.
The Saccharomyces cerevisiae strains used in this study are listed in Table 1. Standard techniques were used to grow and handle yeast cells. Epitope TAP tagging of Nop56p (YAF2 strain) was performed as described by Rigaut et al. (46). Yeast cells were transformed as described by Villa et al. (57) and grown in the appropriate selective media.
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| W303-1B | MATaura3-1 trp1-1 ade2-1 leu2-3,112 his3-11,15 | 3 |
| rna15-2 strain | MATaura3-1 trp1-1 ade2-1 leu2-3,112 his3-11,15 rna15-2 | 3 |
| LM88 | MATaura3-1 trp1-1 ade2-1 leu2-3,112 his3-11,15 rna14-1 | 36 |
| Ypap1-5 | MATaura3-1 trp1-1 ade2-1 leu2-3,112 his3-11,15 PAP1::LEU2 [pA pap1-5] | 36 |
| SB17 | MATaade2 leu2 ura3 trp1 his3 yth1::TRP1 pNOPPATA-yth1-1 | 8 |
| SB18 | MATaade2 leu2 ura3 trp1 his3 yth1::TRP1 pNOPPATA-yth1-4 | 8 |
| SB13 | MATaade2 leu2 ura3 trp1 his3 yth1::TRP1 pNOPPATA-yth1-7 | 8 |
| YSH1-1 strain | MATaura3-1 trp1Δ ade2-1 leu2-3,112his3-11,15 TRP1::ysh1 [ysh1-1-HIS3-CEN] | Unpublished |
| YHH1-3 strain | MAT? ura3-1 trp1Δ ade2-1 leu2-3,112 his3-11,15 TRP1::yhh1 [yhh1-3-HIS3-CEN] | Unpublished |
| YAF2 | MATα trp1Δ his3Δ ura3-52 lys2-801 ade2-101 URA3::U24 NOP56::TAP::TRP1 | 22 |
| FWY1 | MATα ura3-52 leu2-3,112 sen1-1 pep4-3 | 45 |
| D174 | MATaade2-1 xrn1::URA3 rat1-1 | 28 |
The induction experiments were performed as described by Fatica et al. (22) for both the wild-type strains and temperature-sensitive strains. The temperature- and formamide-sensitive strains were grown in the appropriate selective media supplemented with 3% formamide.
Oligonucleotides.
Sequences of the oligonucleotides used for the different cloning steps are as follows (5"-3"): R13ter-a, CCGCTCGAGCTTTTAACTTCCTCGTAGA; R13ter100-b, GGGGTACCCCCAACGTACTAACATCTTT; R13ter30-b, GGGGTACCGCAGCGCTACGATACAAT; R13ter25-b, GGGGTACCAAGATTTTCTACGAGGAAGTTA; R13ter100-30-a, CCGCTCGAGCGCTGCATATATAATGCGT; R50ter-a, CCGCTCGAGACTAATGTAAGAAACATTTCC; R50ter-b, GGGGTACCGTTGAAATAATTAGTGGCAAC; R47ter-a, CCGCTCGAGATATATTTTCGCGTCATTCTTGC; R47ter-b, GGG GTACCTTTGTACTGCAGATACACAGATTC; R189ter-a, CCGCTCGAGCTATTCTTGTTACTCACTGAT; R189ter-b, GGGGTACCCCAATTCATTTCAACTGTCTAC; RU5ter-a, CCGCTCGAGCTCATTTTCTTGTTTCGGT; RU5ter-b, GGGGTACCCGCGAAACTTTAAAAGCGC; RU2ter-a, CCGCTCGAGTACCACGCAACCAAATATGTA; RU2ter-b, GGGGTACCATTATGAGATGCTGGAGGTAG; RTLC1ter-a, CCGCTCGAGATAATAAAGCCCACCAAATGG; RTLC1ter-b, GGGGTACCCAGTACGGACAACATACTTTA; 1-2a, TCGAGGCGGTAACCTTCTTACATGTAAATCAGAGTAGGTAC; 1-2b, CTACTCTGATTTACATGTAAGAAGGTTACCGCC; Δ1a, TCGAGGCGGTAACCACATGTAAATCAGAGTAGGTAC; Δ1b, CTACTCTGATTTACATGTGGTTACCGCC; Δ2a, TCGAGGCGGTAACCTTCTTACACAGAGTAGG TA; Δ2b, CTACTCTGTGTAAGAAGGTTACCGCC; Δ1-2a, TCGAGGCGGTAACCACACAGAGTAGGTAC; Δ1-2b, and CTACTCTGTGTGGTTACCGCC. Oligonucleotides used for Northern hybridization or primer extension were as follows (5"-3"): αtag, TGCGGACTGCCTGGATCGCG; αsnR13, TTCCACACCGTTACTGATTT; αU5, CCTGTTTCTATGGAGACAACACCCGGATGGTTCTG; αsnR50, GGGGTACCACGTACTTGTGAAAGTAAATAC; αU2, CCAGTTATGGTGTGTGGCGA; αsnR47, GGGGTACCTTTGTACTGCAGATACACAGATTC; and αTRS31, CGCTTGCTTAGGCCCAACAG.
Plasmid construction.
The starting construct GAL/U24* (here renamed CYC1) and T100 construct were described by Fatica et al. (22). The constructs used in this study were obtained by removing the CYC1 terminator from the CYC1 construct by XhoI-KpnI digestion and replacing it with PCR products obtained with the following oligonucleotides: R13ter-a and R13ter100-b (construct C/D13 [named T100 + 20 in Fig. 9C]); R13ter-a and R13ter30-b (construct T30 + 20); R13ter-a and R13ter25-b (construct T25); R13ter100-30-a and R13ter100-b (construct T100-30); R50ter-a and R50ter-b (construct C/D50); R47ter-a and R47ter-b (construct C/D47); R189ter-a and R189ter-b (construct ACA189); RU5ter-a and RU5ter-b (construct U5); RU2ter-a and RU2ter-b (construct U2); RTLC1ter-a and RTLC1ter-b (construct TLC1). To obtain1-2, Δ1, Δ2, and Δ1-2 constructs, the appropriate oligonucleotides (1-2a and1-2b, Δ1a and Δ1b, Δ2a and Δ2b, and Δ1-2a and Δ1-2b) were annealed, phosphorylated, and subsequently cloned in the XhoI-KpnI-digested CYC1 construct.
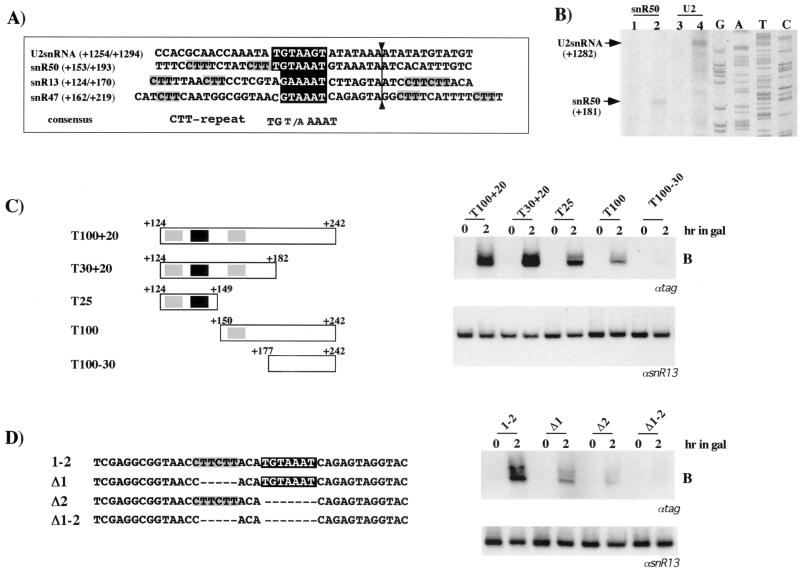
FIG. 9.
Analysis of the sequences required for 3"-end processing. (A) Alignment of the sequences around the cleavage sites mapped in vivo in the rat1-1 xrn1Δ strain. The conserved elements are boxed, and their consensus sequences are shown underneath. Grey and black boxes represent the CTT repeat and the TGTAAAT element, respectively. Numbers indicate distance from the 5" end of the corresponding transcript. (B) Oligonucleotides downstream from the U2 and snR50 coding regions were used as primers for a reverse transcription reaction on RNA extracted from W303 (lanes 1 and 3) and the rat1-1 xrn1Δ strain (lanes 2 and 4). A sequence reaction was run in parallel to map the extension of the 3" cutoff molecules (arrows). The numbers indicate distance from the 5" end of the corresponding transcript. (C and D) Deletion mutants of the natural snR13 3" downstream region (C) and artificial sequences designed on the basis of the two conserved elements found in the snoRNA 3" downstream regions (D). Numbers indicate the extension of the cloned regions according to panel A. The different constructs were transformed into W303; cells were grown in 2% raffinose-0.08% glucose (0 h) and then shifted to 2% galactose for 2 h. Northern blot analysis of 5 μg of total RNA was performed with a 32P-labeled tag-specific oligonucleotide (αtag). The lower panels show control hybridizations with an snR13 snoRNA-specific probe (αsnR13).
The C/D13 was digested with BamHI and KpnI, and the inserts obtained were cloned in the BamHI and KpnI sites of the Bluescript plasmid to obtain a substrate for in vitro transcription. The recombinant plasmid were linearized with KpnI. The CYC1 substrate was described by Minvielle-Sebastia et al. (37).
RNA analysis.
Total RNA was extracted from exponentially growing yeast cultures by the hot-phenol method as previously described (47). RNA concentrations were calibrated by absorbance at 260 nm and normalized by hybridization with snR13- and U5-specific oligonucleotides. For Northern blot analysis, typically 5 μg of total RNA was resolved on 6% polyacrylamide-7 M urea gels and electrotransferred at 4°C to Amersham Hybond-N+ filters in 0.5× Tris-borate-EDTA buffer for 16 h at 10 V. All hybridizations and primer extensions were carried out as previously described (57). Oligonucleotides (10 pmol) were routinely 5"-end labeled with 30 μCi of [γ-32P]ATP.
Yeast extract and immunoprecipitation experiments were performed as previously described (23, 34).
Preparation of extracts and CPF and CF IA.
Extracts competent for in vitro 3"-end processing (processing extracts) were prepared as described previously (12) by cell homogenization in liquid nitrogen (4). For production of whole-cell extracts, the fractionation step using 40% ammonium sulfate was omitted.
CPF was obtained by affinity purification from yeast extracts as previously described (41). Briefly, extracts from cells expressing protein A-tagged Pfs2p fusion protein (strain MO20) were first incubated with immunoglobulin G (IgG)-agarose, the matrix was extensively washed, and bound proteins were released by cleavage with TEV protease (46). The purification of CF IA will be described elsewhere (B. Dichtl and W. Keller, unpublished results).
In vitro transcription and in vitro cleavage assays.
Runoff transcription to obtain internally 32P-labeled RNAs was performed as described in reference 37. Conditions for in vitro cleavage of pre-mRNAs were as previously described (36). For processing with factors, 1 μl of CPF and 1 μl of CF IA were incubated in either the absence or presence of 100 ng of glutathione S-transferase (GST)-Nab4p (CF IB) (38) with 20 fmol of labeled RNA for 1 h at 30°C (36). When extracts were assayed, approximately 10 μg of total protein in processing extract or whole-cell extract, respectively, replaced purified factors. To allow cleavage only, magnesium acetate was replaced by EDTA and ATP was replaced by CTP.
RESULTS
Downstream regions of different small RNA genes direct efficient 3"-end processing.
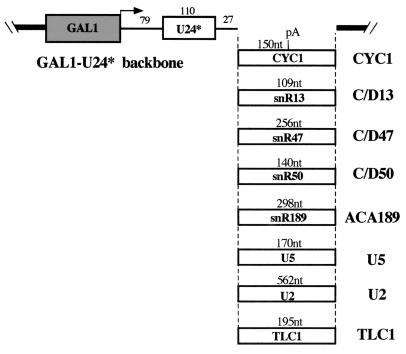
A model system which allowed the analysis of the activity of different 3" downstream regions on the biosynthesis and processing of a specific snoRNA was recently described (22). Plasmid GAL/U24* (22) (here renamed CYC1) contains the yeast U24 snoRNA (6) placed under the control of the inducible GAL1 promoter and the CYC1 cleavage and polyadenylation signal of vector p416GAL1 (39). Because no processing sites are present in its flanking regions, only trimming from the ends of the primary transcript can produce the U24 snoRNA from this construct. In order to distinguish the snoRNA derived from the transforming plasmid from the endogenous species, a tagged U24 snoRNA (U24*) was utilized. The tag was previously shown to affect neither snoRNP formation (58) nor U24 processing and stability (22).The cleavage and polyadenylation cassette was removed from the CYC1 construct and replaced with different 3" downstream regions. Figure 1 shows the GAL1-U24* backbone as well as the lengths and names of all the regions tested in this work.
FIG. 1.
Schematic representation of the constructs used in this study. The GAL1 promoter and the U24* coding region are boxed. The lengths of the regions downstream of the mature 3" end of the corresponding transcript are indicated (in nucleotides [nt]); in the case of U2 and U5, the regions 3" to the Rnt1p site were utilized. All these fragments were amplified by PCR and were cloned in the GAL/U24* backbone.
The lengths of the regions analyzed were chosen on the basis of their distance from the 3" downstream open reading frame. In S. cerevisiae, intergenic distances, including the promoter sequences, are quite short: on average, only a few hundred nucleotides. In the case of the RNAs analyzed here, these values ranged from 280 (snR13) to 820 (snR50) nucleotides. Because of this short distance between the 3"-flanking region of the upstream coding sequence and the downstream open reading frame, mechanisms preventing readthrough must operate quite efficiently.
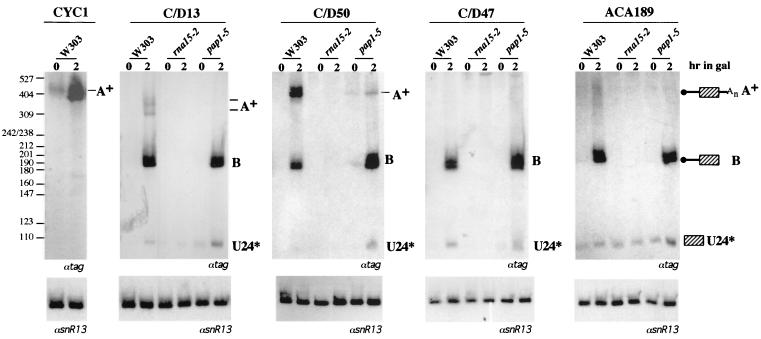
Figure 2 shows the behavior of the sequences present in the 3" downstream regions of several snoRNA genes: snR13 (C/D13), snR50 (C/D50), and snR47 (C/D47) belong to the box C/D family, whereas snR189 (ACA189) is a member of the H/ACA class. The constructs were expressed in strains carrying temperature-sensitive mutant alleles for the RNA15 (rna15-2) and PAP1 (pap1-5) genes and in the isogenic W303 strain. After a 1-h shift to the nonpermissive temperature, transcription was induced by addition of galactose (time zero), and the cells were grown for 2 h more. 3"-end processing can be monitored by the accumulation of product B, whose 3" end corresponds to the 3" end of the mature U24* snoRNA. This species still contains the 5" trailer sequences present in the construct utilized. Since the 5" end of this RNA is capped and no endonucleolytic cleavage sites are present, 5"-3" exo-trimming can occur only after decapping. For this reason the accumulation of a U24 snoRNA with a matured 5" end (band U24*) is quite inefficient.
FIG. 2.
Analysis of different 3" downstream regions on snoRNA processing. The rna15-2 and pap1-5 mutant strains and the isogenic strain W303 were transformed with the constructs indicated above each panel. After a 1-h shift to the nonpermissive temperature (0 h), cultures were induced with galactose for the indicated times. Northern blot analysis of 5 μg of total RNA was performed with a 32P-labeled tag-specific oligonucleotide (αtag). A schematic representation of the A+, B, and U24* molecules is given on the far right. A+ bands correspond to polyadenylatedproducts, while B and U24* molecules have mature 3" ends. The lower panels show control hybridizations with a snR13 snoRNA-specific probe (αsnR13). The migration of the size marker (MspI-digested pBR322) is indicated on the far left; sizes are in nucleotides.
In contrast to CYC1, the other test constructs show efficient accumulation of correctly 3"-processed U24 (band B) and only minor amounts of polyadenylated transcripts (bands A+); furthermore, no accumulation of 3"-end-processed U24* RNA was observed in rna15-2 mutant strains, while a pap1-5 strain had no adverse effect. These data indicate that 3"-end formation of snoRNAs requires CF I activity and not Pap1, consistent with previous results (22).
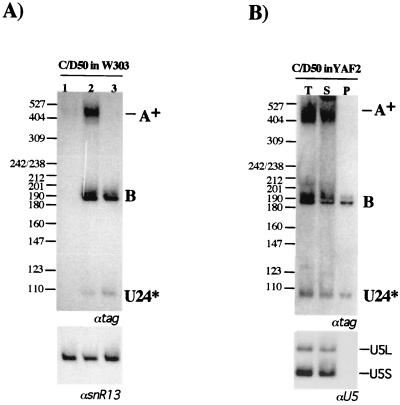
The presence of polyadenylated species originating from the same coding unit prompted us to check whether these molecules have a different fate than the 3"-processed transcripts. To test whether these species represent stable and functional RNAs, we performed a transcription pulse-chase experiment and an immunoprecipitation analysis. C/D50 was chosen because it showed the highest abundance of A+ products. After 2 h of growth in galactose-based medium, cultures were shifted to glucose-containing medium, and the accumulation of the A+ and B molecules was tested after one additional hour of incubation. As shown in Fig. 3A, the A+ band completely disappeared after 1 h, whereas the B band remained stable. Construct C/D50 was also expressed in strain YAF2 (22), which contains the Nop56p protein, a box C/D-specific factor, fused to the TAP tag (46). The incorporation of the different RNA species into snoRNPs was tested by IgG immunoprecipitation. Figure 3B shows that the RNA present in band B is coprecipitated with the Nop56p, whereas the polyadenylated species is not. The observation that the A+ species is not found in snoRNP complexes argues against the possibility that it is rapidly converted into B molecules. Further support for the hypothesis that A+ molecules do not chase into B molecules comes from the observation that the A+ transcripts produced by the CYC1 construct are not converted to 3"-processed U24 RNA (Fig. 2) (22).
FIG. 3.
Polyadenylated snoRNAs are unstable and do not associate with snoRNP proteins. (A) W303 cells were transformed with the C/D50 construct and grown in 2% raffinose-0.08% glucose (lane 1). Cultures were shifted to 2% galactose for 2 h (lane 2) and then transferred to 4% glucose-containing medium and incubated for 1 h more (lane 3). Northern blot analysis of 5 μg of total RNA was performed with a 32P-labeled tag-specific oligonucleotide (αtag). The lower panel shows a control hybridization with a snR13 snoRNA-specific probe (αsnR13). (B) Strain YAF2, containing the NOP56-TAP fusion, was transformed with the C/D50 construct. After 2 h of growth in galactose, cells were lysed and immunoprecipitated with IgG beads. RNA was extracted from pellets (P) and supernatants (S) and run on a 6% polyacrylamide-urea gel. Lane T, RNA extracted from nonimmunoprecipitated extracts. Northern blot analysis was performed with αtag. The lower panel shows a control hybridization with a U5 snRNA-specific probe (αU5) which monitors the specificity of the immunoprecipitation. The migration of the size marker (MspI-digested pBR322) is indicated on the left; sizes are in nucleotides.
We therefore conclude that snoRNA transcripts which are polyadenylated are directed to a degradative pathway. The disappearance of A+ molecules in glucose-containing medium is in agreement with the finding that the RNA-degradative pathways are very active under these growth conditions (11).
The 3" downstream regions of snoRNA genes are sensitive to CF I but not to CPF.
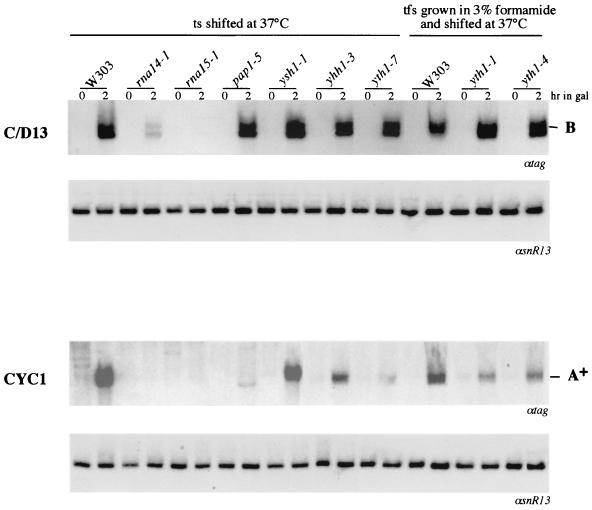
The pre-mRNA cleavage and polyadenylation machinery in yeast is made up of many different factors associated in complexes which have been separated biochemically and classified on the basis of their in vitro activity (60). To test whether CPF activity was required for 3"-end formation of snoRNAs, we expressed the C/D13 construct in strains carrying temperature-sensitive mutant alleles for individual CPF components. As controls, the same strains were transformed with construct CYC1. Figure 4 shows that several mutant alleles of the YTH1 gene, which codes for a CPF component (8), show a strong reduction of the A+ species transcribed from the CYC1 construct (lanes yth1-7, yth1-1, and yth1-4). Similarly, alleles with mutations of the CPF component Yhh1p have a lower level of transcript A+. As expected, rna14-1, rna15-2, and pap1-5 display strong inhibition of A+-RNA accumulation (22). The levels of the A+ CYC1 transcripts were the same in all strains following growth at the permissive temperature (not shown). In comparison to CYC1, the construct containing the 3" downstream region of the snR13 gene is affected only by mutations in proteins belonging to the CF IA complex (rna14-1 and rna15-1), and it is totally unaffected by mutations within CPF subunits.
FIG. 4.
Analysis of 3"-end processing in mutants with mutations in different components of the pre-mRNA 3"-processing machinery. The C/D13 and CYC1 constructs were transformed into the mutant strains and into the isogenic strain W303. After a 1-h shift to the nonpermissive temperature (0 h), cultures were induced with galactose for 2 h. Temperature- and formamide-sensitive mutants (tfs) were grown in 3% formamide. Northern blot analysis of 5 μg of total RNA was performed with a 32P-labeled tag-specific oligonucleotide (αtag). The lower panels show control hybridizations with a snR13 snoRNA-specific probe (αsnR13).
Rna15p is required for 3"-end formation of chromosome-encoded snoRNAs.
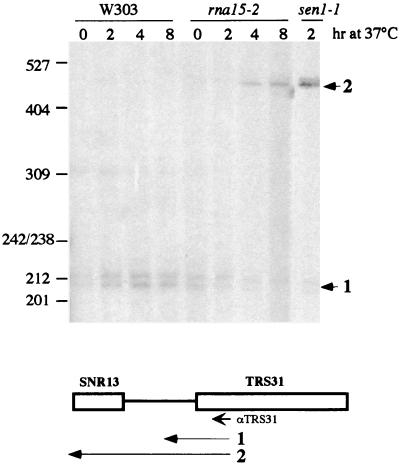
In order to verify whether CFIA is involved in 3"-end formation of endogenous snoRNAs, we tested the production of snoRNA read-through products in the strain carrying the rna15-2 mutant allele when shifted to the nonpermissive temperature. Primer extension analysis was carried out with a primer complementary to the TRS31 mRNA which is transcribed from the gene downstream to the snR13 coding region (Fig. 5) (52). The data for rna15-2 in Fig. 5 show that at the permissive temperature (0 h), a signal corresponding to the 5" end of the TRS31 mRNA is visualized; after a shift to the nonpermissive temperature, this signal decreases while a proportional increase of a read-through product extending to the 5" end of the snR13 snoRNA is observed (lanes 4 and 8). As a control, reverse transcription analysis was also performed on RNA from a sen1-1 strain which was previously shown to affect snoRNA 3" processing and to have a read-through phenotype (45, 52): the sen1-1 lane shows that the read-through product observed in the rna15-1 mutant is produced. These results confirm that Rna15p is essential for 3"-end formation of snoRNAs transcribed from the chromosomal genes.
FIG. 5.
Rna15p is required for 3"-end formation of the chromosomal snR13 transcripts. An oligonucleotide complementary to the coding strand of the TRS31 gene was utilized for primer extension analysis on RNA extracted from the rna15-2 and sen1-1 mutant strains and from the isogenic strain W303. Cultures were grown at 25°C up to an optical density of 0.2 and then shifted to 37°C for the indicated times. As shown schematically below the gel, extended product 1 identifies the 5" end of the TRS31 mRNA, while product 2 corresponds to an elongated RNA starting at the 5" end of the snR13 transcript. Migration of the size marker (MspI-digested pBR322) is indicated on the left; sizes are in nucleotides.
The 3" downstream regions of the U5 and U2 snRNA genes behave like those of snoRNAs.
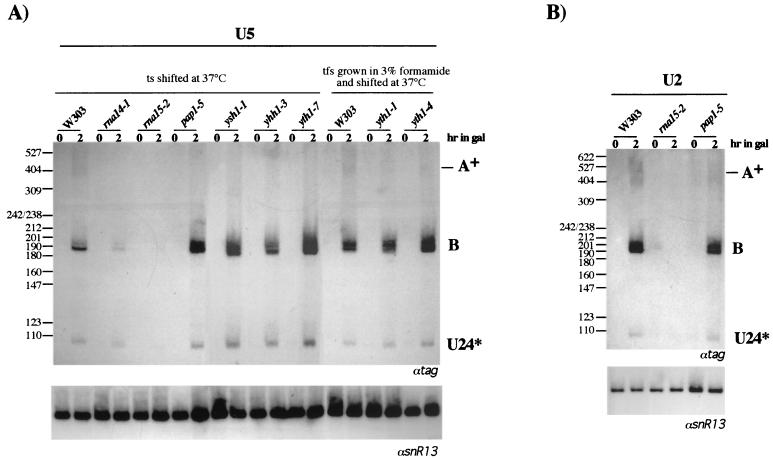
U5 snRNA is found in two forms that differ in length at their 3" ends (U5L and U5S). A temperature-sensitive mutation in the endonuclease Rnt1p was shown to block accumulation of the U5L form in vivo, whereas the production of U5S was unaffected (14, 57). To analyze whether the U5 snRNA gene contains sequences able to direct 3"-end formation similarly to those for snoRNA genes, 170 nucleotides downstream of the Rnt1p recognition hairpin were inserted into the reporter construct (Fig. 1) and tested in vivo. The U5 construct directs efficient 3"-end formation of U24* snoRNA, and only minimal accumulation of polyadenylated species occurs (Fig. 6A, W303 lanes). Expression of this construct in the different strains carrying mutant alleles in CF IA and CPF components and analysis of the RNA products indicated a behavior similar to that of C/D13, in that only mutations in CF IA components resulted in inhibition of accumulation of 3"-processed U24 molecules.
FIG. 6.
The U5 and U2 genes contain sequences that respond only to the cleavage activity of the pre-mRNA 3"-processing machinery. The U5 and U2 constructs were transformed into different mutant strains and into the isogenic strain W303. After a 1-h shift to the nonpermissive temperature (0 h), cultures were induced with galactose for 2 h. Temperature- and formamide-sensitive (tfs) mutants were grown in 3% formamide. Northern blot analysis of 5 μg of total RNA was performed with a 32P-labeled tag-specific oligonucleotide (αtag). Migration of the size marker (MspI-digested pBR322) is indicated on the left; sizes are in nucleotides. The lower panels show control hybridizations with an snR13 snoRNA-specific probe (αsnR13).
The U2 snRNA also needs 3"-5" trimming for 3"-end formation. As is the case for U5 snRNA, the U2 primary transcript contains an Rnt1 cleavage site. In the absence of Rnt1p-mediated cleavage, both mature and extended polyadenylated forms accumulate (1). A total of 562 nucleotides downstream of the U2 Rnt1p site were cloned into the test construct and were tested for U24* 3"-end formation. Figure 6B shows that the majority of the transcripts accumulate as correctly 3"-processed molecules, whereas only a minority are found as poly(A)+ species. Furthermore, the U2 3" downstream region is also sensitive to mutations in CF I (rna15-2 lanes) and not in Pap1 (pap1-5 lanes).
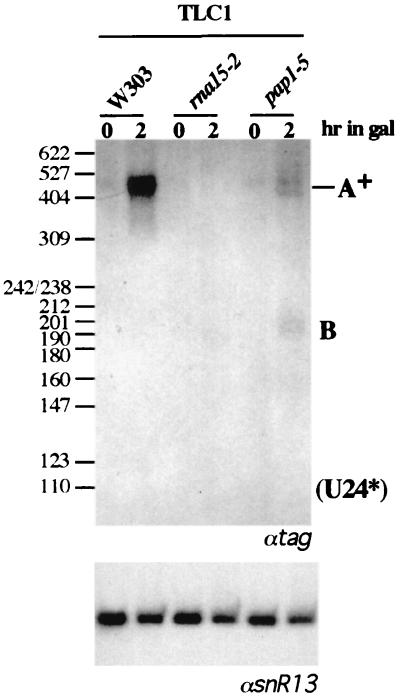
So far, only one small nuclear RNA has been described as being transcribed as a poly(A)+ species; this is the RNA component (encoded by the TLC1 gene) of telomerase (17). It was shown that the synthesis of TLC1 RNA under the control of a GAL-inducible promoter produced at the steady-state level 60% of polyadenylated RNA that was then converted into poly(A)− molecules (17). A 195-nucleotide sequence, derived from the 3" downstream region of the TLC1 gene, was cloned in the U24* reporter plasmid and tested for the ability to produce correctly 3"-processed U24* molecules. Figure 7 shows that all observed transcripts accumulate as poly(A)+ species. In agreement with this, the accumulation of the transcripts is affected not only by the rna15-2 mutation but also by pap1-5. This suggests that 3"-end formation of the TCL1 RNA is directed by sequences that respond to the pre-mRNA cleavage and polyadenylation machinery.
FIG. 7.
The telomerase RNA downstream region produces only polyadenylated transcripts. The TCL1 construct was transformed into the rna15-2 and pap1-5 mutant strains and the isogenic strain W303. After a 1-h shift to the nonpermissive temperature (0 h), cultures were induced with galactose for 2 h. Northern blot analysis of 5 μg of total RNA was performed with a 32P-labeled tag-specific oligonucleotide (αtag). Migration of the size marker (MspI-digested pBR322) is indicated on the left; sizes are in nucleotides. A control hybridization with a snR13 snoRNA-specific probe (αsnR13) is shown at the bottom.
In vitro analysis of snoRNA 3"-end formation.
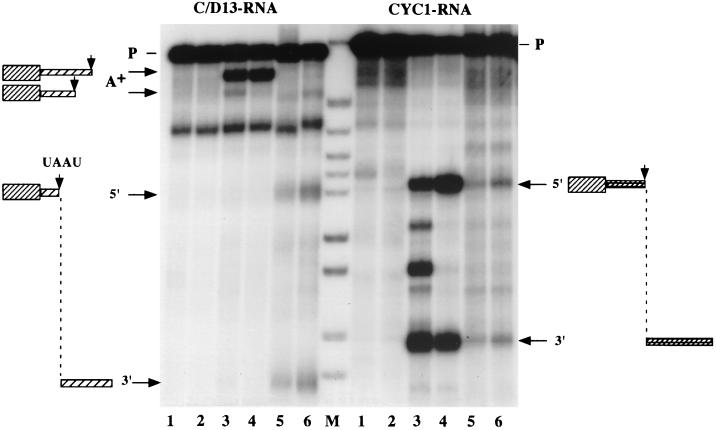
Two in vitro-transcribed 32P-labeled RNAs were used to analyze the 3"-end processing with purified factors or yeast extracts. CYC1 RNA is a well-studied pre-mRNA substrate (37), and C/D13 RNA contains the U24* coding region and 100 nucleotides of snR13 gene 3" downstream sequences. These RNAs were incubated with CF IA alone or in combination with CPF in the absence and presence of recombinant CF IB (GST-Nab4p). Reaction conditions were chosen to allow cleavage only (see Materials and Methods). The left panel of Fig. 8 shows that C/D13 RNA is not cleaved by CF IA alone (lane 2); the addition of CF IA and CPF (lane 3) produced two bands (A+), one of which is intensified in the presence of CF IB (lane 4). These cleavages correspond to the sites that are polyadenylated when construct C/D13 is tested in vivo (Fig. 2). When processing extracts or whole-cell extracts are used in the reaction, cleavage is observed at the site which promotes correct 3"-end processing and which was mapped in vivo on the endogenous snR13 primary transcript (Fig. 8) (22). The right panel of Fig. 8 shows the results of such an analysis on CYC1 RNA. This substrate behaves as expected (21) in that specific cleavage is obtained when CF IA, CPF, and CF IB are incubated together (lane 4). No cleavage is obtained with CF IA alone (lane 2), and additional cleavage products were seen in the absence of CF IB (lane 3); CF IB was shown to act in the selection of the correct cleavage site in a concentration-dependent manner (38). In contrast to the snoRNA substrate, CYC1 RNA was processed with much lower efficiency when unfractionated whole-cell extracts were used (lanes 5 and 6). These results show that factors present in unfractionated extracts that are different from CPF and CF IB are necessary for cleavage at the site utilized for 3" processing of pre-snoRNAs.
FIG. 8.
CPF, CF IA, and CF IB are not sufficient for correct cleavage of a box C/D snoRNA substrate at the in vivo processing site. The RNA precursors and processing products are indicated on the sides of the panels. Internally 32P-labeled RNAs were incubated with CF IA (lanes 2), CF IA and CPF (lane 3), CF IA, CPF, and 100 ng of GST-Nab4p (lanes 4), processing extract (lanes 5), or whole-cell extract (lanes 6). Input RNA alone was loaded in lanes 1. RNAs were resolved on a 6% polyacrylamide-8.3 M urea gel. HpaII-digested pBR322 fragments were 5"-end labeled and served as markers (lane M). A+ bands identify the sites that become polyadenylated, while the 5" and 3" bands are the 5" and 3" cutoff products when cleavage occurs at the UA"AU sequence, which is utilized as the entry site for 3" processing in vivo.
Mutant analysis of the snR13 3" downstream region.
In order to study the sequences involved in this peculiar mechanism of 3"-end formation, we initially performed an alignment of the cleavage sites of the primary transcripts of different RNA coding units. This was possible thanks to the identification of the 3" cutoff products in a strain depleted of the 5"-3" Rat1p and Xrn1p exonucleases. Under normal conditions, such molecules are rapidly degraded in vivo since they are uncapped; in contrast, their half-life increases in a rat1-1 xrn1Δ strain. Total RNA extracted from this strain was subjected to reverse transcription analysis with primers specific for regions approximately 200 nucleotides downstream from the 3" end of the mature transcript. This analysis was carried out on box C/D snoRNAs (snR13, snR47, and snR50) and on snRNAs (U2 and U5). It was not possible to map such cleavage sites on U5 snRNA, since most of the signals detected corresponded to the products of U5L snoRNA, which is produced by Rnt1p cleavage (data not shown). Figure 9A shows the alignment of mapped snoRNA and snRNA downstream regions with respect to the cleavage sites, while Fig. 9B shows an example of such mappings on the U2 and snR50 genes. Conserved sequences can be identified: a TGT/AAAAT element is found in all four genes at a fixed distance (seven nucleotides) from the cleavage site (A"Pu). In addition, several CTT repeats were found upstream and downstream of the cleavage site; however, this sequence was exclusively found in box C/D snoRNA genes. To analyze the significance of these elements, we generated several deletion mutants of the snR13 downstream region and tested their ability to direct 3"-end formation of the reporter U24* snoRNA. Figure 9C shows that a 58-nucleotide region containing all the conserved elements (construct T30 + 20) has the highest activity in terms of the amount of 3"-processed molecules. If only the elements upstream of the cleavage site are maintained (construct T25), cleavage still occurs efficiently, while if they are deleted (construct T100), processing is less efficient. The removal of all three conserved elements (construct T100-30) leads to the almost complete absence of processed RNA.
In order to dissect more precisely the contribution of the two conserved sequences, we made additional constructs in which the two elements (CTTCTT and TGTAAAT) are cloned together (clone 1-2) or separately (clones Δ1 and Δ2) in the 3" downstream region of our model construct. The in vivo analysis shows that the highest 3"-end formation activity is obtained when the two elements are present together, while lower activity is revealed when they were cloned separately. Quantitative analysis of the bands indicated that the TGTAAAT element is at least twice as efficient as the CTT repeat. The deletion of both elements completely abolishes 3"-end formation. These results indicate that each single element is able to drive 3"-end formation even though a synergistic effect is obtained when the two sequences are linked.
DISCUSSION
mRNA precursors are processed at their 3" ends by a two-step reaction that involves endonucleolytic cleavage followed by the addition of a poly(A) tail to the upstream cleavage product. The poly(A) tail is required for several functions: it stabilizes the 3" end of the mRNA (10), it facilitates transport to the cytoplasm (29), and it increases the efficiency of translation (53). Small nuclear and nucleolar RNAs are also transcribed by RNA polymerase II; however, they accumulate as nonpolyadenylated species which are confined to the nucleus and which require 3"-end processing in order to reach the mature form.
We showed previously that the presence of a pre-mRNA-derived cleavage and polyadenylation signal downstream of a box C/D snoRNA-coding unit was incompatible with snoRNA maturation and function. Instead, a specific sequence which directed efficient 3"-end processing of the test snoRNA and which required only the cleavage components of the CF I complex and not the Pap1 activity was found (22). This finding led us to propose that for a specific set of polymerase II transcripts, the cleavage activity of the pre-mRNA 3"-end formation machinery could be uncoupled from polyadenylation.
In the present paper we extend this mechanism of 3"-end formation to a larger group of nuclear and nucleolar RNAs. Several box C/D snoRNA genes were analyzed, as well as box H/ACA and the U2 and U5 snRNA genes. In all cases, the 3" downstream regions of these genes directed efficient 3"-end processing of a reporter RNA and only minimal amounts of transcripts accumulated as polyadenylated species. Both U2 and U5 snRNA genes contain a downstream Rnt1p cleavage site that was suggested to be the entry site for 3" processing. Nevertheless, it was found that in strains depleted of the Rnt1p factor, mature U2 and U5 snRNA continued to accumulate, although to a lower extent (1, 14, 57). This indicated that processing should have occurred from the 3" end of the primary transcript. Regions downstream of the Rnt1p site were cloned in the test construct and were shown to direct 3"-end processing in a polyadenylation-independent manner.
The only exception among the genes tested is the one encoding the RNA component of telomerase. It was previously shown that when TLC1 was expressed under the control of the GAL1 promoter, poly(A)+ transcripts accumulated and were then chased into poly(A)− species. This suggested a model in which the telomerase RNA is first polyadenylated and then rapidly processed to give the stable poly(A)− form (17). When the 3" downstream region of the TLC1 gene was analyzed in our test system, we found that only polyadenylated RNA species were produced. This case represents an exception to a general model of 3"-end formation by uncoupled cleavage and polyadenylation, but it demonstrates that the model system we used is able to respond specifically to the different 3"-processing signals. Analysis of the endogenous transcripts produced in a rna15-2 strain at the nonpermissive temperature showed the accumulation of a read-through RNA extending inside the gene downstream to the snR13 coding region, showing that CF IA is indeed involved in the 3"-end formation of chromosomal snoRNA transcripts.
The pre-mRNA cleavage and polyadenylation reaction is a very well characterized process mediated by the CF I (A and B) and CPF complexes (38, 41). CPF is essential for both steps of pre-mRNA 3"-end formation, i.e., cleavage and polyadenylation (41). Recently we showed that one function of CPF in pre-mRNA cleavage lies in recognition of the site of polyadenylation (21). Clearly, this activity of CPF would not be required for the recognition of sequences which direct snoRNA and snRNA 3"-end formation, since those are different from cis-acting elements which mediate cleavage and polyadenylation of pre-mRNAs (see below). But it remains unclear whether the requirement for CPF in cleavage of pre-mRNAs is limited to substrate recognition or whether CPF is more directly involved in catalysis of cleavage. Our analysis of mutant CPF subunits indicates that CPF activity is dispensable for 3"-end processing of snoRNAs and snRNAs. We cannot entirely exclude, however, the possibility that mutations in other CPF subunits might have an effect. In contrast to CPF, CF IA is essential for 3"-end formation of snoRNAs and snRNAs. In vitro analysis showed that CF IA alone is not able to cleave pre-snoRNA molecules and that CF IA and CPF together directed cleavage only at those sites that in vivo become polyadenylated and produce nonfunctional molecules. Cleavage at the specific site that in vivo directs 3"-end processing (22) was obtained only when the pre-snoRNA was incubated with whole-cell extracts. Combining these results with the in vivo data on the requirement of CF IA indicates that factors other than CPF and CF IB must cooperate with CF IA for the production of correct 3"-end processing of pre-snoRNA molecules.
Sequence comparison and mutational analysis revealed the presence of two conserved motifs (CTT repeat and TGTAAAT) which are essential for directing 3"-end processing of pre-snoRNAs. These elements are clearly distinct from cis-acting signals which direct 3"-end formation of pre-mRNAs (21, 26, 27). Consequently, recognition of pre-snoRNA cleavage sites is expected to involve a different set of trans-acting factors. Similar to the elements that are involved in pre-mRNA processing, the snoRNA elements are redundant and only one of them is sufficient for 3"-end formation. Our experiments indicate that the CTT repeat and the TGTAAAT element can function separately, even though the second is twice as efficient as the first. A synergistic effect is obtained when the two sequences are linked. The CTT repeat was previously described to be involved in controlling 3"-end maturation of snoRNAs and to respond to the Sen1p factor (45). The TGTAAA sequence was shown to produce the appearance of truncated transcripts when present in an intron downstream of a U6 snRNA coding region. The Nrd1 protein was shown to interact directly with the UGUAAA element and to mediate 3"-end formation in concert with the Sen1 factor (50, 51); Nrd1p was also shown to physically interact with the C-terminal domain (CTD) of polymerase II, with the RNA-binding protein Nab3p, and with the CTD kinase CTDK-1 (19). For this reason, this factor was previously proposed to be involved in transcription termination or transcription elongation, leading to a model in which it would act at the interface of transcription and processing (19, 49). Recent experiments have proved that the Nrd1 protein, together with Nab3, Sen1, and the CTD of polymerase II, is an essential factor for poly(A)-independent 3"-end formation of snRNAs and snoRNAs (52).
Taking all these data together, we propose a model in which CF IA not only functions in association with factors involved in cleavage and polyadenylation of pre-mRNAs but also acts in the 3"-end formation of nonpolyadenylated transcripts in cooperation with a different set of proteins, possibly Nrd1 and its associated factors. It remains to be shown whether these distinct complexes occur independently from each other in the cell or whether they are associated in a highly regulated supramolecular complex. Further work is also necessary for clarifying the role of these factors in processing or termination and for understanding the relationship between these two events. It is interesting that many of the components participating in this reaction also interact with the CTD of the largest subunit of RNA polymerase II (9, 19). This could indicate that specific commitments, with respect to 3"-end formation, could occur very early, possibly during the first steps of transcription. An important point to be addressed in future experiments is whether the type of 3"-processing complex, which forms on the nascent RNA, depends on the type of transcript and on the protein interactions that occur on it. In the case of snoRNAs it would be interesting to test whether snoRNP-specific factors, which are known to bind their substrates at the level of precursor molecules (22, 25), influence the type of 3"-end commitment complex. It seems reasonable to assume that efficient 3"-end formation should occur only when snoRNPs are formed; in contrast, when snoRNP particles are not assembled, the newly transcribed snoRNA would be polyadenylated and directed to the discard pathway.
Acknowledgments
We thank M. Arceci and G. Ricci for skillful technical help.
This work was partially supported by grants to I.B. from MURST-CNR Biotechnology Program L.95/95, from MURST Biotecnologie and PRIN 40%, and from CNR Target Project on Biotechnology and Tecnologie di base della post-genomica. B.D. was the recipient of an EMBO Long-Term Fellowship and was supported by the University of Basel, the Swiss National Science Foundation, and the Louis-Jeanlet Foundation of Medicine.
REFERENCES
- 1.Abou Elela, S., and M. Ares, Jr. 1998. Depletion of yeast RNase III blocks correct U2 3" end formation and results in polyadenylated but functional U2 snRNA. EMBO J. 17:3738-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allmang, C., J. Kufel, G. Chanfreau, P. Mitchell, E. Petfalski, and D. Tollervey. 1999. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 19:5399-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amrani, N., M. Minet, M. Le Gouar, F. Lacroute, and F. Wyers. 1997. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol. Cell. Biol. 17:3694-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ansari, A., and B. Schwer. 1995. SLU7 and a novel activity, SSF1, act during the PRP16-dependent step of yeast pre-mRNA splicing. EMBO J. 14:4001-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bachellerie, J. P., and J. Cavaillé. 1998. Small nucleolar RNAs guide the ribose methylations of eukaryotic rRNAs, p. 255-272. In H. Grosjean and R. Benne (ed.), Modification and editing of RNA. ASM Press, Washington, D.C.
- 6.Bachellerie, J. P., M. Nicoloso, L. H. Qu, B. Michot, M. Caizergues-Ferrer, J. Cavaille, and M. H. Renalier. 1995. Novel intron-encoded small nucleolar RNAs with long sequence complementarities to maturer RNAs involved in ribosome biogenesis. Biochem. Cell Biol. 73:835-843. [DOI] [PubMed] [Google Scholar]
- 7.Balakin, A. G., L. Smith, and M. J. Fournier. 1996. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell 86:823-834. [DOI] [PubMed] [Google Scholar]
- 8.Barabino, S. M., M. Ohnacker, and W. Keller. 2000. Distinct roles of two Yth1p domains in 3"-end cleavage and polyadenylation of yeast pre-mRNAs. EMBO J. 19:3778-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barilla, D., B. A. Lee, and N. J. Proudfoot. 2001. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98:445-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beelman, C. A., and R. Parker. 1995. Degradation of mRNA in eukaryotes. Cell 81:179-183. [DOI] [PubMed] [Google Scholar]
- 11.Bousquet-Antonelli, C., C. Presutti, and D. Tollervey. 2000. Identification of a regulated turnover pathway for nuclear pre-mRNA. Cell 102:765-775. [DOI] [PubMed] [Google Scholar]
- 12.Butler, J. S., P. P. Sadhale, and T. Platt. 1990. RNA processing in vitro produces mature 3" ends of a variety of Saccharomyces cerevisiae mRNAs. Mol. Cell. Biol. 10:2599-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavaillé, J., K. Buiting, M. Kiefmann, M. Lalande, C. I. Brannan, B. Horsthemke, J. P. Bachellerie, J. Brosius, and A. Huttenhofer. 2000. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. USA 97:14311-14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chanfreau, G., S. Abou Elela, M. Ares, Jr., and C. Guthrie. 1997. Alternative 3"-end processing of U5 snRNA by RNaseIII. Genes Dev. 11:2741-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chanfreau, G., P. Legrain, and A. Jacquier. 1998. Yeast Rnase III as a key processing enzyme in small nucleolar RNA metabolism. J. Mol. Biol. 284:975-988. [DOI] [PubMed] [Google Scholar]
- 16.Chanfreau, G., G. Rotondo, P. Legrain, and A. Jacquier. 1998b. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 17:3726-3737. [DOI] [PMC free article] [PubMed]
- 17.Chapon, C., T. R. Cech, and A. J. Zaug. 1997. Polyadenylation of telomerase RNA in budding yeast. RNA 3:1337-1351. [PMC free article] [PubMed] [Google Scholar]
- 18.Chen, J., and C. Moore. 1992. Separation of factors required for cleavage and polyadenylation of yeast pre-mRNA. Mol. Cell. Biol. 12:3470-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conrad, N. K., S. M. Wilson, E. J. Steinmetz, M. Patturajan, D. A. Brow, M. S. Swanson, and J. L. Corden. 2000. A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics 154:557-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de los Santos, T., J. Schweizer, C. A. Rees, and U. Francke. 2000. Small evolutionarily conserved RNA, resembling C/D box small nucleolar RNA, is transcribed from PWCR1, a novel imprinted gene in the Prader-Willi deletion region, which is highly expressed in brain. Am. J. Hum. Genet. 67:1067-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dichtl, B., and W. Keller. 2001. Recognition of polyadenylation sites in yeast pre-mRNAs by cleavage and polyadenylation factor. EMBO J. 20:3197-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fatica, A., M. Morlando, and I. Bozzoni. 2000. Yeast snoRNA accumulation relies on a cleavage-dependent/polyadenylation-independent 3"-processing apparatus. EMBO J. 19:6218-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ganot, J.-P., M. Caizergues-Ferrer, and T. Kiss. 1997. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 11:941-956. [DOI] [PubMed] [Google Scholar]
- 24.Ganot, P., B. E. Jàdy, M. L. Bortolin, X. Darzacq, and T. Kiss. 1999. Nucleolar factors direct the 2"-O-ribose methylation and pseudouridylation of U6 spliceosomal RNA. Mol. Cell. Biol. 19:6906-6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giorgi, C., A. Fatica, R. Nagel, and I. Bozzoni. 2001. Release of U18 snoRNA from its host intron requires interaction of Nop1p with the Rnt1p endonuclease. EMBO J. 20:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graber, J. H., C. R. Cantor, S. C. Mohr, and T. F. Smith. 1999. Genomic detection of new yeast pre-mRNA 3"-end-processing signals. Nucleic Acids Res. 27:888-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo, Z., and F. Sherman. 1996. 3"-end-forming signals of yeast mRNA. Trends Biochem. Sci. 21:477-481. [DOI] [PubMed] [Google Scholar]
- 28.Henry, Y., H. Wood, J. P. Morrissey, E. Petfalski, S. Kearsey, and D. Tollervey. 1994. The 5" end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 13:2452-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang, Y., and G. G. Carmichael. 1996. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell. Biol. 16:1534-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jady, B. E., and T. A. Kiss. 2001. Small nucleolar guide RNA functions both in 2"-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J. 20:541-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kessler, M. M., J. Zhao, and C. L. Moore. 1996. Purification of the Saccharomyces cerevisiae cleavage/polyadenylation factor I. Separation into two components that are required for both cleavage and polyadenylation of mRNA 3" ends. J. Biol. Chem. 271:27167-27175. [DOI] [PubMed] [Google Scholar]
- 32.Kessler, M. M., M. F. Henry, E. Shen, J. Zhao, S. Gross, P. A. Silver, and C. L. Moore. 1997. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3"-end formation in yeast. Genes Dev. 11:2545-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiss, T. 2001. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 20:3617-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lafontaine, D. L., and D. Tollervey. 2000. Synthesis and assembly of the box C+D small nucleolar RNPs. Mol. Cell. Biol. 20:2650-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maxwell, E. S., and M. J. Fournier. 1995. The small nucleolar RNAs. Annu. Rev. Biochem. 64:897-934. [DOI] [PubMed] [Google Scholar]
- 36.Minvielle-Sebastia, L., P. J. Preker, and W. Keller. 1994. RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3"-end processing factor. Science 266:1702-1705. [DOI] [PubMed] [Google Scholar]
- 37.Minvielle-Sebastia, L., P. J. Preker, T. Wiederkehr, Y. Strahm, and W. Keller. 1997. The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in pre-messenger RNA 3"-end formation. Proc. Natl. Acad. Sci. USA 94:7897-7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minvielle-Sebastia, L., K. Beyer, A. M. Krecic, R. E. Hector, M. S. Swanson, and W. Keller. 1998. Control of cleavage site selection during mRNA 3"-end formation by a yeast hnRNP. EMBO J. 17:7454-7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mumberg, D., R. Muller, and M. Funk. 1994. Regulatable promoters of S. cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22:5767-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ni, J., A. L. Tien, and M. J. Fournier. 1997. Small nucleolar RNAs direct site-specific synthesis of pseudouridines in ribosomal RNA. Cell 89:565-573. [DOI] [PubMed] [Google Scholar]
- 41.Ohnacker, M., S. Barabino, P. J. Preker, and W. Keller. 2000. The WD-repeat protein Pfs2p bridges two essential factors within the yeast pre-mRNA 3"-end-processing complex. EMBO J. 19:37-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Omer, A. D., T. M. Lowe, A. G. Russell, H. Ebhardt, S. R. Eddy, and P. P. Dennis. 2000. Homologs of small nucleolar RNAs in Archaea. Science 288:517-522. [DOI] [PubMed] [Google Scholar]
- 43.Ooi, S. L., D. Samarsky, M. J. Fournier, and J. D. Boeke. 1998. Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: intron length effects and activity of a precursor snoRNA. RNA 4:1096-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petfalski, E., T. Dandekar, Y. Henry, and D. Tollervey. 1998. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol. 18:1181-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rasmussen, T. P., and M. R. Culbertson. 1998. The putative nucleic acid helicase Sen1p is required for formation and stability of termini and for maximal rates of synthesis and levels of accumulation of small nucleolar RNAs in Saccharomyces cerevisiae. Mol. Cell. Biol 18:6885-6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 47.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith, C. M., and J. A. Steitz. 1997. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell 89:669-672. [DOI] [PubMed] [Google Scholar]
- 49.Steinmetz, E. J. 1997. Pre-mRNA processing and the CTD of RNA polymerase II: the tail that wags the dog? Cell 89:491-494. [DOI] [PubMed] [Google Scholar]
- 50.Steinmetz, E. J., and D. A. Brow. 1996. Repression of gene expression by an exogenous sequence element acting in concert a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol. Cell. Biol. 16:6993-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steinmetz, E. J., and D. A. Brow. 1998. Control of pre-mRNA accumulation by the essential yeast protein Nrd1 requires high-affinity transcript binding and a domain implicated in RNA polymerase II association. Proc. Natl. Acad. Sci. USA 95:6699-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steinmetz, E. J., N. K. Conrad, D. A. Brow, and J. L. Corden. 2001. RNA-binding protein Nrd1 directs poly(A)-independent 3"-end formation of RNA polymerase II transcripts. Nature 413:327-331. [DOI] [PubMed] [Google Scholar]
- 53.Tarun, S. Z., Jr., and A. B. Sachs. 1996. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 15:7168-7177. [PMC free article] [PubMed] [Google Scholar]
- 54.Tollervey, D., and T. Kiss. 1997. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 9:337-342. [DOI] [PubMed] [Google Scholar]
- 55.Tycowski, K. T., Z. H. You, P. J. Graham, and J. A. Steitz. 1998. Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol. Cell 5:629-638. [DOI] [PubMed] [Google Scholar]
- 56.Van Hoof, A., P. Lennertz, and R. Parker. 2000. Yeast exosome mutants accumulate 3"-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 20:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Villa, T., F. Ceradini, C. Presutti, and I. Bozzoni. 1998. Processing of the intron-encoded U18 small nucleolar RNA in the yeast Saccharomyces cerevisiae relies on both exo- and endonucleolytic activities. Mol. Cell. Biol. 18:3376-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Villa, T., F. Ceradini, and I. Bozzoni. 2000. Identification of a novel element required for processing of intron-encoded box C/D small nucleolar RNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:1311-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weinstein, L. B., and J. A. Steitz. 1999. Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol. 11:378-384. [DOI] [PubMed] [Google Scholar]
- 60.Zhao, J., L. Hyman, and C. Moore. 1999. Formation of mRNA 3" ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63:405-445. [DOI] [PMC free article] [PubMed] [Google Scholar]