Abstract
Basal transcription factor TFIIH phosphorylates the RNA polymerase II (RNApII) carboxy-terminal domain (CTD) within the transcription initiation complex. The catalytic kinase subunit of TFIIH is a member of the cyclin-dependent kinase (Cdk) family, designated Kin28 in Saccharomyces cerevisiae and Cdk7 in higher eukaryotes. Together with TFIIH subunits cyclin H and Mat1, Cdk7 kinase is also found in a trimer complex known as Cdk activating kinase (CAK). A yeast trimer complex has not previously been identified, although a Kin28-Ccl1 dimer called TFIIK has been isolated as a breakdown product of TFIIH. Here we show that a trimeric complex of Kin28-Ccl1-Tfb3 exists in yeast extracts. Several Kin28 point mutants that are defective in CTD phosphorylation were created. Consistent with earlier studies, these mutants have no transcriptional defect in vitro. Like other Cdks, Kin28 is activated by phosphorylation on T162 of the T loop. Kin28 T162 mutants have no growth defects alone but do demonstrate synthetic phenotypes when combined with mutant versions of the cyclin partner, Ccl1. Surprisingly, these phosphorylation site mutants appear to destabilize the association of the cyclin subunit within the context of TFIIH but not within the trimer complex.
The carboxy-terminal domain (CTD) of RNA polymerase II (polII) is phosphorylated in a manner correlated with ongoing transcription. Phosphorylation is important for the coupling of transcription to mRNA processing and for release of polymerase into the productive elongation phase. Four complexes with in vitro CTD kinase activity and transcription-related phenotypes have been identified in Saccharomyces cerevisiae. Interestingly, all four are in the cyclin-dependent kinase (Cdk) family, members of which minimally consist of a catalytic subunit and a cyclin-like regulatory subunit. The nonessential Srb10-Srb11 complex (known as Cdk8-cyclin C in higher eukaryotes) is associated with the mediator complex and appears to have a negative regulatory role (32, 74). The CTDKI kinase complex (CTK1-CTK2-CTK3) is nonessential for viability, but loss results in reduction of phosphorylated CTD levels in vivo (43, 46). The closest higher eukaryotic relative is Cdk9 and its associated cyclins. A second potential Cdk9 homologue is Bur1/Sgv1, which is associated with a divergent cyclin-like protein named Bur2 (79). This complex appears to be essential for viability. Mutant Bur1 or Bur2 alleles lead to increased transcription from a promoter lacking any activator binding sites as well as other transcription-related phenotypes (54). Finally, TFIIH contains the Kin28-Ccl1 complex (known as Cdk7-cyclin H in higher eukaryotes) (19). Kin28 activity is essential for viability in S. cerevisiae (65). Mutations in Kin28 result in a loss of phosphorylated RNA polII CTD and severely reduced mRNA levels (10, 55, 74).
In higher eukaryotes, Cdk7 has been isolated as an RNA polII CTD kinase associated with basal transcription factor TFIIH (56, 62, 63) and also as a Cdk-activating kinase or CAK. CAK phosphorylates and thereby activates cell cycle regulatory Cdks such as Cdc2 (67), Cdk2 (24), Cdk4 (50), and Cdk6 (35). Most of the evidence for Cdk7 acting as a CAK comes from in vitro experiments, although some genetic experiments support this role (31, 37, 45). In S. cerevisiae, the physiological CAK is a monomeric 44-kDa protein kinase distantly related to Cdks, known as Cak1 or Civ1 (38, 71). Yeast Cak1 has been shown to phosphorylate multiple Cdk family members, activating both Kin28 and Cdc28 but not Pho85 or Srb10 (11, 17, 38, 41). No homologue of Cak1 has yet been discovered in higher eukaryotes, suggesting that Cdk7's role as a CAK may not be conserved throughout eukaryotes.
Cdk activity is modulated by at least two mechanisms (52, 53). The first is by association with regulatory proteins. Cyclins and other proteins bind the kinase subunit to increase activity and help determine substrate specificity. Many Cdks are also regulated by inhibitory proteins. A second mechanism of Cdk regulation involves a series of regulatory phosphorylations. Three phosphorylation sites have been identified on the prototypical Cdk, Schizosaccharomyces pombe cell cycle regulator Cdc2. Thr14 and Tyr15 can be phosphorylated by the Myt1 and Wee1 kinases to inactivate Cdc2; dephosphorylation by the Cdc25 phosphatase reactivates the kinase (52, 66). A third site (Thr169) in the “T loop” is phosphorylated by CAKs to greatly increase kinase activity. A number of phosphatases that dephosphorylate the T-loop threonine of Cdks have been identified (8, 29, 30). Phosphorylation of the T loop appears to be widely conserved in the Cdk family. Mutation of the activating threonine residue in Cdc28 (38, 71), Cdk4 (39), or Cdk6 (35) abolishes kinase activity and function in vivo. T-loop phosphorylation alters the Cdk substrate interface (57) and stabilizes the Cdk-cyclin interaction (16).
Yeast TFIIH subunit Kin28 contains residues corresponding to the three Cdc2 phosphorylation sites: T162, T17, and Y18 (12, 13, 26). Higher eukaryotic homologues of Kin28 (Cdk7) retain the T-loop phosphorylation site, but the two inhibitory sites are occupied by residues that cannot be phosphorylated. Thus Kin28 kinase activity is potentially regulated by (i) assembly with its cyclin partner Ccl1, (ii) T-loop phosphorylation by Cak1 (17, 41), (iii) other regulatory phosphorylations, and (iv) further association with other proteins such as Tfb3 and other subunits of TFIIH.
Electrophoretic analysis of Kin28 from wild-type cells shows that the protein migrates as a doublet and that the faster-migrating band disappears after phosphatase treatment (10, 17, 23, 41). The phosphorylation site responsible for this shift is T162 in the T loop (17, 41). Kin28 phosphorylation by Cak1 occurs at higher efficiency when Kin28 is in a complex with Ccl1 and Tfb3 (17). Surprisingly, Kin28 proteins with mutations at T162 support normal growth (41). A deleterious effect of T-loop phosphorylation is seen only when the T162 mutation is combined with other mutations in Kin28, Ccl1, or Tfb3 (this report; 17, 41). These results suggest that phosphorylation of Kin28 may be functionally connected to its interactions with other proteins.
Kin28 has been isolated within the nine-subunit holo-TFIIH complex as well as within a TFIIH-derived Kin28-Ccl1 dimer complex designated TFIIK (68). Yeast two-hybrid assays suggest that Tfb3 forms the link between the core and kinase modules of TFIIH by bridging Rad3 and Kin28 (19, 21). In higher eukaryotes, Cdk7 is found both in holo-TFIIH and in a trimer complex consisting of Cdk7, cyclin H, and Mat1 (the homologue of yeast Tfb3). This trimer complex can also associate with XPD/ERCC2, the homologue of Rad3 (15). These findings have led to the proposal that the Tfb3/Mat1 protein distribution differs in the yeast and mammalian systems: it is in both holo-TFIIH and the kinase subcomplex in higher eukaryotes but only in holo-TFIIH in yeast. Here we demonstrate the existence of a yeast Kin28-Ccl1-Tfb3 complex, thereby reconciling the yeast system with that of other eukaryotes. Interestingly, the T-loop phosphorylation of Kin28 is necessary for stable association with Ccl1 but only within the context of the TFIIH holocomplex.
MATERIALS AND METHODS
Plasmids.
The pRS series plasmids have been described previously (64). The hemagglutinin (HA)-tagged KIN28 plasmid was a gift from Mark Solomon (10). An XhoI/EcoRI fragment from this plasmid was transferred into pRS313 for use as a mutagenesis template. Single Kin28 point mutants were generated using PCR-mediated site-directed mutagenesis (33). Oligonucleotide T3 (5"-AATTAACCCTCACTAAAGG-3") and the appropriate mutagenic primer were used to amplify the 5" end of the HA-coding-sequence-tagged KIN28 gene in pRS313. The resulting PCR product was gel purified and utilized as a 5" megaprimer in a second PCR with the 3" primer, oligonucleotide T7 (5"-TAATACGACTCACTATAGGGAGA-3"), for amplification of full-length Kin28 from the same template. The resulting 1.3-kb amplified fragments were cloned into PCR-Script (Stratagene), and mutations were confirmed by sequencing. A 1.3-kb XhoI/EcoRI fragment containing the mutated KIN28 gene was subcloned into pRS313 for further analyses in vivo. T17D-T162A/D and K36A-T162A/D double mutants were created by subcloning the appropriate fragments from the single-mutant plasmids.
The Gal-inducible 2μm plasmids utilized for the examination of whether Ptc2 or Ptc3 is a Kin28 phosphatase were obtained from Mark Solomon [YepLac112Gal, YepLac112Gal*Ptc2, YepLac112 Gal*Ptc3, YepLac112Gal*Ptc2(D234A), YepLac112Gal*Ptc3(D234A), YepLac195Gal*Ptc2, YepLac195Gal*Ptc3] (8). The gene expressing His-tagged Tfb1 was transferred as an SpeI fragment from YCp50/Tfb1-6his (70) (a gift from Roger Kornberg) to pRS313.
Yeast strains, genetic manipulations, and media.
Yeast strains used in this study are listed in Table 1. Strain YSB591, containing an episomal copy of the wild-type KIN28 gene on a URA3-marked plasmid and a deletion of the chromosomal KIN28 gene, was used for plasmid shuffling of KIN28. Yeast strains were transformed by the lithium acetate procedure (27). Standard methods for medium preparation, sporulation, and tetrad analysis were used (4, 28). Double-knockout Kin28Δ/Tfb1Δ yeast strain YSB722 was created by first shuffling pTS313-TFB1-6his into YSB207 and then mating the resulting strain to YSB591. The resulting diploid was sporulated, and tetrads were screened for the appropriate markers. Strains YSB760, YSB761, and YSB762 were created by plasmid shuffling of plasmids expressing HA-tagged wild-type Kin28 and the T162A and T162D mutants, respectively, into YSB722.
TABLE 1.
Yeast strains used in this study
| Strain | Genotypea |
|---|---|
| YSB207 | MATatfb1Δ::LEU2 ura3-52 leu2-3,112 his3Δ200 [pRS316-TFB1] |
| YSB251 | MATatfb1Δ::LEU2 ura3-52 leu2-3,112 his3Δ200 [pRS313-tfb1-6] |
| YSB260 | MATatfb1Δ::LEU2 ura3-52 leu2-3,112 his3Δ200 [pRS313-tfb1-101] |
| YSB347 | MATα kin28Δ::LEU2 ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 ade3-22 can1-100 [YCplac22-KIN28] |
| YSB591 | MATα kin28Δ::LEU2 ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 ade3-22 can1-100 [pRS426-KIN28] |
| YSB722 | MATakin28Δ::LEU2 tfb1Δ::LEU2 ura3 leu2-3,112 trp1-1 his3 ade2-1 [pRS426-KIN28; pRS313-TFB1-6his] |
| YSB759 | MATakin28Δ::LEU2 tfb1Δ::LEU2 ura3 leu2-3,112 trp1-1 his3 ade2-1 [pRS314-haKIN28; pRS313-TFB1-6his] |
| YSB760 | MATakin28Δ::LEU2 tfb1Δ::LEU2 ura3 leu2-3,112 trp1-1 his3 ade2-1 [pRS314-hakin28(T162A); pRS313-TFB1-6his] |
| YSB761 | MATakin28Δ::LEU2 tfb1Δ::LEU2 ura3 leu2-3,112 trp1-1 his3 ade2-1 [pRS314-hakin28(T162D); pRS313-TFB1-6his] |
| YF82 | MATa/MATα KIN28/kin28Δ::LEU2 ura3-1/ura3-1 leu2-3,112/leu2-3,112 trp1-1 his3-11,15/his3-11,15 ade2-1/ade2-1 ade3-22/ade3-22 can1-100/can1-100 |
Brackets indicate episomal plasmids.
Protein analysis and immunoprecipitations.
For the preparation of whole-cell extracts, cells were grown to an optical density (600 nm) of 1.0 and pelleted. Glass beads were used to disrupt cells in lysis buffer (20 mM HEPES [pH 7.6], 200 mM potassium acetate [KOAc], 10% glycerol, 1 mM EDTA) supplemented with phosphatase inhibitors (1 mM NaF and 0.5 mM Na3VO4) and protease inhibitors (1 mM phenylmethylsulfonyl fluoride and 1 μg of aprotinin, leupeptin, and pepstatin-A/ml). Equivalent amounts of protein were then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis. Immunoblotting was done using standard methods. Polyclonal rabbit anti-Kin28 was from Covance. FLAG-M2 agarose beads and horseradish peroxidase-conjugated polyclonal goat anti-rabbit and goat anti-mouse antibodies were from Sigma. Rabbit polyclonal anti-Tfb1 and anti-Tfb3 against the appropriate His-tagged recombinant proteins were made by Covance. The B3 monoclonal antibody was a gift from Ben Blencowe, and polyclonal anti-Ccl1 was from Roger Kornberg.
HA-tagged Kin28 mutant proteins were immunoprecipitated from whole-cell extracts or chromatographic fractions using 12CA5 bound to protein A beads. Complexes were prepared by mixing 10 μl of protein A resin and 2 μl of 12CA5 ascites fluid per sample in Tris-EDTA (TE; pH 8.0) followed by incubation with gentle rolling for 30 min at 4°C. Beads were washed twice with 1 ml of TE (pH 8.0) and diluted 1:1 with TE (pH 8.0). Ten microliters of this mixture was then added to each yeast protein sample. Reaction mixtures were incubated with gentle rolling overnight at 4°C. Finally, beads were pelleted by centrifugation and washed three times in 1 ml of lysis buffer (20 mM HEPES [pH 7.6], 200 mM KOAc, 10% glycerol, 1 mM EDTA) prior to analysis by SDS-PAGE.
For λ-phosphatase treatment, immunoprecipitations were performed as described above except that lysis buffer did not contain phosphatase inhibitors NaF and Na3VO4. Following two washes in lysis buffer, the beads were resuspended to 50 μl in lysis buffer supplemented with 50 mM Tris-HCl, 0.1 mM Na2-EDTA, 5 mM dithiothreitol (DTT), 0.01% Brij 35 (pH 7.5 at 25°C), 2 mM MnCl2, with or without λ-phosphatase (200 U; New England BioLabs). Reaction mixtures were incubated at 30°C for 30 min and further analyzed.
Biochemical separation of Kin28 complexes.
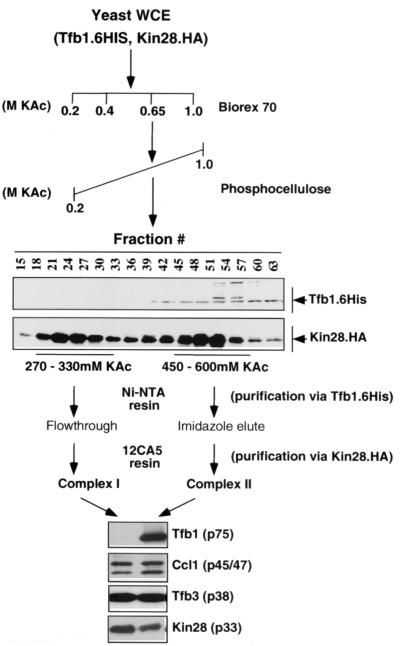
Extracts were made from yeast strains containing both six-His-tagged Tfb1 and HA-tagged Kin28 (wild type and T162A and T162D mutants). The procedure for purification of TFIIH with BioRex-70 and a Ni-nitrilotriacetic acid (NTA) column was modified from the procedure of Svejstrup et al. (69). Twelve liters of each strain was grown overnight in a fermentor and disrupted as described previously (59). All manipulations from this point on were performed at 4°C, and all buffers contained phosphatase and protease inhibitors. One-seventh volume of 4 M KOAc (pH 7.6) was added dropwise with stirring, and after 30 min the mixture was centrifuged in a Beckman 45Ti rotor at 40,000 rpm for 90 min. The supernatant (∼120 ml) was removed and diluted with buffer A-0 (20 mM HEPES-KOH [pH 7.6], 20% glycerol, 1 mM EDTA, 1 mM DTT) until the conductivity matched that of buffer A-0.2 (buffer A plus 0.2 M KOAc) and then chromatographed on a 150-ml BioRex-70 (Bio-Rad) column equilibrated in buffer A-0.2. The column was washed with 200 ml of buffer A-0.2 and 400 ml of buffer A-0.4 before elution with 300 ml of buffer A-0.65. The peak protein fraction (∼120 ml) was pooled and dialyzed against two 2-liter volumes of buffer A-0.2. The conductivity of the eluate was adjusted to match that of buffer A-0.2 before chromatography on a 50-ml phosphocellulose (Whatman) column equilibrated in buffer A-0.2. The column was washed with 50 ml of buffer A-0.2 and eluted with a 300-ml gradient of 0.2 to 1.0 M KOAc in buffer I (20 mM HEPES-KOH [pH 7.6], 20% glycerol, 0.01% NP-40, 0.2% Tween 20, 10 mM imidazole, 5 mM β-mercaptoethanol). Fractions (3.5 ml) were collected from this gradient and subjected to immunoblotting analysis with antibodies against components of TFIIH (Kin28, Tfb3, Ccl1, and Tfb1). Two peaks were identified (see Fig. 5).
FIG. 5.
S. cerevisiae contains two Kin28-containing complexes. S. cerevisiae strains containing His-tagged Tfb1 (Tfb1.6His) and HA-tagged Kin28 (Kin28.HA) were created. Whole-cell extract (WCE) was prepared and passed over BioRex-70 and phosphocellulose columns. Two peaks of Kin28 eluted from the phosphocellulose column, although only one peak also contained Tfb1 (center blots). The two peaks were individually subjected to further purification over Ni-NTA agarose and immunoprecipitation via the HA tag on Kin28. Immunoblotting analysis was carried out to determine the composition of resulting purified complexes I and II. Both contained Kin28, Ccl1, and Tfb3. Complex II also contained Tfb1, identifying this peak as TFIIH.
Each peak was pooled and bound overnight to 2 ml of Ni-NTA beads (Qiagen) equilibrated in buffer I plus 0.5 M KOAc, and the beads were collected by centrifugation. The supernatant was collected from the first peak (trimer) and stored in aliquots at −80°C, while the beads from the TFIIH peak were washed in a column (0.7 cm in diameter) with 20 ml of buffer J-1.0 and 20 ml of buffer J-0.4 (20 mM Tris-acetate [pH 7.6], 20% glycerol, 5 mM β-mercaptoethanol, 0.4 M KOAc) containing 20 mM imidazole. TFIIH was then eluted with 10 ml of buffer J-0.4 containing 100 mM imidazole). One milliliter fractions were collected and assayed by Western analysis for Tfb1. Positive fractions were pooled and stored in aliquots at −80°C. Immunoprecipitations were performed from the Kin28-Ccl1-Tfb3 trimer and TFIIH fractions as described above.
In vitro CTD kinase assay.
Immunoprecipitated Kin28 complexes were used to phosphorylate recombinant glutathione S-transferase (GST)-CTD with [γ-32P]ATP essentially as described previously (10, 40). Final concentrations per 25 μl of reaction mixture were 20 mM HEPES-NaOH (pH 7.6), 7.5 mM magnesium acetate, 2 mM DTT, 100 mM KOAc, 2% glycerol, 25 μM ATP, 0.5 μl [γ-32P]ATP, and 100 ng of GST-CTD. Reaction mixtures were incubated at room temperature for 30 min, resolved on an SDS-12% PAGE gel, transferred to nitrocellulose, and exposed to X-ray film or a phosphorimager plate.
In vitro transcription assays.
Assays were performed essentially as described previously (9, 76, 77) with minor modifications. One liter of each strain was grown overnight to mid-log phase and centrifuged, and pellets were washed twice with 500 ml ice-cold double-distilled water. All manipulations from this point on were performed on ice at 4°C, and all buffers contained protease inhibitors. The yeast pellet was resuspended in 2 ml of transcription buffer A (200 mM Tris [pH 7.9], 390 mM NH4SO4, 10 mM MgSO4, 1 mM EDTA, 20% glycerol, 2 mM DTT; note that this buffer A is different from that used in the TFIIH purification) per g wet weight and subjected to glass bead disruption. The lysate was collected and centrifuged in a Sorvall SS-34 rotor at 9,000 rpm for 20 min. The supernatant was collected, 1/7 volume of 4 M KOAc (pH 7.6) was added dropwise, and the resulting mixture was stirred on ice for 30 min. The mixture was centrifuged in a Beckman 45Ti rotor at 40,000 rpm for 90 min, and the supernatant was carefully collected, avoiding the pellet. An equal volume of saturated NH4SO4 (pH 7) was added dropwise with mixing, and the mixture was stirred for 30 min and centrifuged in a Beckman 70Ti rotor at 20,000 rpm for 30 min. The supernatant was discarded, and the pellet was resuspended gently in 50 μl of transcription buffer B (20 mM HEPES-KOH [pH 7.5], 10 mM magnesium acetate, 150 mM KOAc, 10 mM EGTA, 20% glycerol, 5 mM DTT)/g original wet weight. The resulting solution was dialyzed against two changes of 100 volumes of transcription buffer B for 4 h until the conductivity of the lysate was similar to that of the dialysis buffer. Lysates (30 to 60 mg/ml) were stored in aliquots at −80°C.
Transcription reaction mixtures (30 μl each) were assembled on ice and contained 12 μl of mixture A (25 mM HEPES-KOH {pH 7.8}, 10% glycerol, 100 mM KOAc {pH 7.8}, 10 mM Mg acetate {pH 7.8}, 5 mM EGTA, 2.5 mM DTT, 10 mM phosphocreatine {Sigma}, 1 U of creatine kinase {Sigma}, 10 U of RNasin {Promega}, 3 μl of deoxynucleoside triphosphate mixture {500 μM ATP and CTP, 10 μM UTP, 0.5 μl [α-32P]UTP [NEN-Life Science Products], 10 μM 3"-o-methyl-GTP [Amersham Pharmacia Biotech]}), whole-cell extract (200 μg), 100 ng of plasmid template (pCYC-GAL4 CG−), and 100 ng of activator (GAL4-VP16). Reactions were allowed to proceed at 25°C for 45 to 60 min and were terminated by the addition of 120 μl of RNase T1 buffer (10 mM Tris-HCl [pH 7.5], 300 mM NaCl, 5 mM EDTA, 50 U of RNase T1), followed by incubation at room temperature for 15 min. SDS was then added to 0.5%, and proteinase K was added to 100 μg/ml, followed by incubation for 20 min at 37°C. Ten micrograms of tRNA was then added, and the reaction mixtures were extracted with phenol-chloroform and precipitated with ethanol. RNA was resuspended in 10 μl of 0.1% SDS in 50% formamide and resolved on an acrylamide gel. Gels were dried onto gel blot paper (Whatman) and exposed to X-ray film for autoradiography.
RESULTS
Site-directed mutagenesis of Kin28.
To test the importance of potential phosphorylation sites, site-directed mutagenesis of Kin28 residues T17, Y18, and T162 was performed. Also targeted were several residues within the predicted catalytic site. Based on the human Cdk2 structure (36, 57), K36 and D147 should be involved in coordinating magnesium and ATP phosphate while E54 is part of the PSTAIRE helix, which contributes to the active-site and cyclin interactions (53). When possible, conservative amino acid substitutions were made. HA-tagged Kin28 mutants were used to replace the product of the wild-type gene in vivo by plasmid shuffling, and phenotypes were tested under various growth conditions (Table 2).
TABLE 2.
Growth phenotypes of Kin28 mutants
| Mutant class | Amino acid change(s) | Growth phenotypea |
|---|---|---|
| NAb (wild type) | None | +++ |
| Catalytic site | K36A | + |
| E54Q | − | |
| D147N | − | |
| Potential inhibitory phosphorylation site | T17V, -E, or -Q | +++ |
| T17D | + | |
| Y18F | +++ | |
| T17V, Y18F | +++ | |
| Potential activating phosphorylation site | T162A, -D, -V, -E, or -Q | +++ |
| S163A | +++ | |
| T162A, S163A | +++ | |
| Double | T162A, T17D | − |
| T162A, K36A | − |
+++, wild-type growth; +, slow growth; −, inviable. The same results were obtained at 16, 23, 30, and 37°C, as well as when galactose was substituted for glucose in the media.
NA, not applicable.
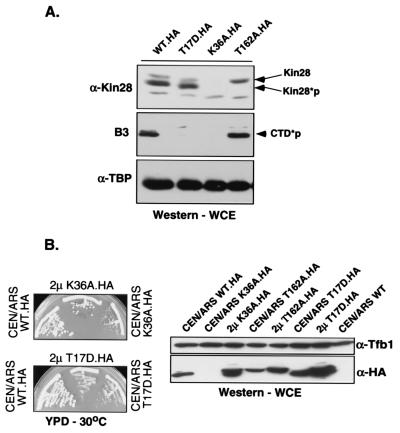
As expected, two of the catalytic-site mutants (D147N and E54Q mutants) were unable to support viability. Both proteins were made at normal levels but were inactive for CTD phosphorylation in immunoprecipitates (data not shown). Surprisingly, a strain carrying the third catalytic-site mutant (K36A mutant) was viable albeit very slow growing. Immunoblotting showed that the K36A protein expressed from a centromeric plasmid is present at greatly reduced levels (Fig. 1A). Kin28 mRNA levels of the mutant were normal (data not shown), suggesting that the protein is unstable. To determine whether slow growth in this mutant is due only to low Kin28 levels or is also due to a defect in catalytic activity, the K36A mutant gene was introduced on a high-copy-number plasmid. Increased levels of the mutant K36A protein are detectable under these conditions, and the growth defect is partially suppressed (Fig. 1B). However, growth was still slower than that of the wild type and in vivo levels of phosphorylated (pol II) were still not at wild-type levels (data not shown). The overexpressed protein also exhibited reduced Rpb1 CTD kinase activity in vitro (Fig. 2A). Therefore, we conclude that the K36 residue is important for both catalytic activity and structural integrity of the Kin28 protein.
FIG. 1.
In vivo analysis of Kin28 mutants. (A) Protein levels. The indicated HA-tagged Kin28 mutants were used to replace the product of the wild-type (WT) gene in yeast by plasmid shuffling. Total proteins extracted from exponentially growing cultures at 30°C in yeast extract-peptone-dextrose (YPD) medium were resolved by SDS-PAGE, and levels of Kin28 (anti-Kin28 [α-Kin28], rabbit polyclonal antibody), phosphorylated Rpb1 CTD (CTD*p; B3 monoclonal antibody), and TATA-binding protein (TBP) (anti-TBP [α-TBP], rabbit polyclonal antibody) were assayed by immunoblotting. Low levels of the K36A protein are detectable following prolonged exposure (not shown). WCE, whole-cell extract. (B) Overexpression of Kin28(K36A) partially suppresses the slow-growth phenotype the mutant produces. Plasmid shuffling was carried out using low-copy-number (CEN/ARS) or high-copy-number (2μm) plasmids carrying the indicated mutant kin28 genes. Strains were streaked on YPD plates and incubated at 30°C for 3 days (left). The mutant strains were also assayed by immunoblotting (right) for Kin28 protein levels using the 12CA5 monoclonal antibody, which recognizes the HA tag (α-HA). A polyclonal antibody against Tfb1 (α-Tfb1) was used as a control for core TFIIH levels.
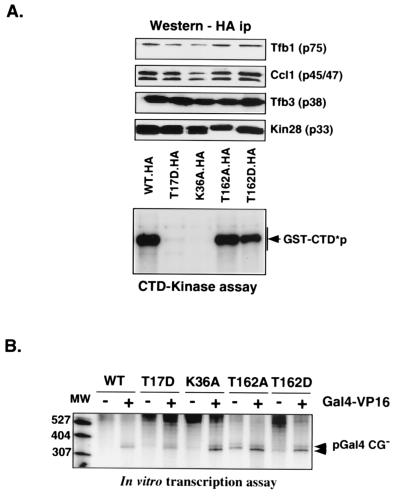
FIG. 2.
Functional analysis of Kin28 mutants. (A) CTD kinase activity. Kin28-containing complexes were immunoprecipitated (ip) via the HA tag with 12CA5 protein A beads. Immunoblotting analyses for the indicated proteins (top) and in vitro CTD kinase assays (bottom) were performed as described in Materials and Methods. Note that the K36A mutant allele was carried on a high-copy-number plasmid while the other alleles were on low-copy-number plasmids so that Kin28 protein levels would be comparable. WT, wild type; CTD*p, phosphorylated Rpb1 CTD. (B) Transcription activity. Whole-cell extracts were prepared from the indicated strains, and in vitro transcription was assayed using a linear plasmid template. Both basal and activated transcription was tested (without [−] and with [+] Gal4-VP16, respectively). Lane MW, radiolabeled molecular size markers (nucleotides).
Mutation of the two potential inhibitory phosphorylation sites failed to provide any evidence for phosphorylation at these sites. Kin28 proteins with substitutions at T17 and/or Y18 supported growth as well as wild-type Kin28 at all temperatures and growth conditions tested (with the exception of the T17D mutant [see below]). Furthermore, immunoblots of extracts from these strains did not show any Kin28 mobility shifts, which would be expected upon changes in phosphorylation pattern. Immunoprecipitated wild-type Kin28 is not recognized by an antiphosphotyrosine antibody (data not shown). Therefore, it appears unlikely that Kin28 is subject to inhibitory phosphorylations, a conclusion also reached by Kimmelman et al. (41).
The Kin28(T17D) mutant grew slowly under all conditions tested. This phenotype is unlikely to be due to loss of a regulatory phosphorylation because the T17E, T17V, and T17Q mutants grow normally (Table 2). Immunoblotting of the T17D strain shows that the mutant protein is present at levels equivalent to that for the wild type. Unlike what was found for the K36A mutant, overexpression does not partially suppress the growth defect conferred by the T17D mutant (Fig. 1B). In the Cdk2 kinase, phosphorylation of the site analogous to Kin28 T17 interferes with the ATP-binding site (52, 66). Therefore, we suggest that the aspartate residue in the T17D change mimics this inhibitory phosphorylation by interfering with the catalytic site of Kin28. In agreement with this hypothesis, phosphorylated-CTD levels are dramatically reduced in the T17D strain (55) (Fig. 1A) and Kin28 immunoprecipitates from T17D mutant cells are deficient in CTD phosphorylation (Fig. 2A).
T-loop phosphorylation of Kin28p.
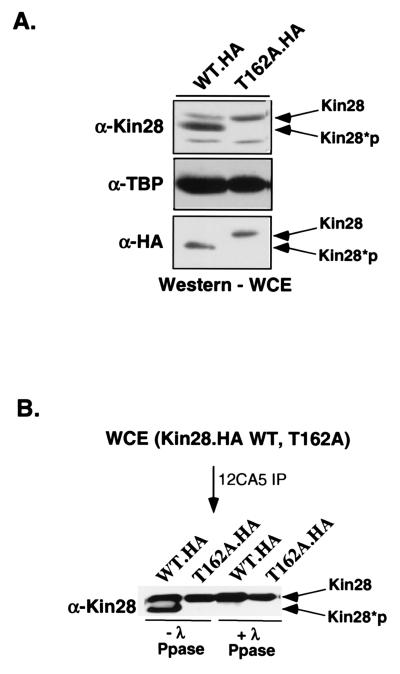
In wild-type cells, two species of the Kin28 protein are resolvable by gel electrophoresis (Fig. 3A). These represent phosphorylated and unphosphorylated proteins since treatment of the protein with λ-phosphatase leads to loss of the lower band (17, 41) (Fig. 3B). The ratio of the proteins suggests that the majority of Kin28 in the cell is phosphorylated. Mutation of T162 in the Kin28 T loop leads to loss of the phosphorylated Kin28 band, providing evidence that this residue is the site of phosphorylation in vivo.
FIG. 3.
Kin28 is phosphorylated on T162 within the T loop. (A) Immunoblot analysis of wild-type (WT) Kin28 whole-cell extracts (WCE) shows that the protein migrates as a doublet corresponding to the phosphorylated and unphosphorylated species. The T162A mutant protein migrates as a single band. Kin28*p, phosphorylated Kin28; TBP, TATA-binding protein. (B) HA-tagged Kin28 was immunoprecipitated (IP) via the HA tag using the 12CA5 monoclonal antibody and treated with λ-phosphatase (Ppase). The faster-migrating band disappears, indicating that the mobility shift is phosphorylation dependent. The HA-tagged Kin28(T162A) [Kin28.HA(T162A)] mutant was used as a gel mobility standard for the unphosphorylated form.
Previous work has been in disagreement concerning the requirement for T162 phosphorylation. Kimmelman et al. (41) found that replacement of T162 by alanine did not produce any observable phenotypes despite the fact that the mutated Kin28 had significantly reduced activity in vitro. Espinoza et al. originally reported that T162 substitutions were lethal but later found that their T162 mutant also contained two other substitutions that contributed to the phenotype (17). In agreement with Kimmelman et al., we found that none of the T162 substitutions tested (A, V, E, D, and Q) conferred an obvious growth phenotype (Table 1).
This result was somewhat surprising, since T-loop phosphorylation has been found essential for the in vivo activity of several other Cdks. Although the T162 substitutions led to the apparent loss of the phosphorylated form of Kin28, we considered the possibility that the adjacent residue (serine 163) might also serve as a site of an activating phosphorylation. This seemed particularly plausible given that higher-eukaryote Cdk7 proteins contain two phosphorylation sites within the T loop (3). However, an S163A substitution produced no mutant phenotypes, either alone or in combination with the T162A substitution. Therefore, there is no indication that S163 can function as an alternate phosphorylation site.
It was previously reported that Kin28 immunoprecipitates from T162A cells had partially reduced in vitro CTD kinase activity (17, 41). Using a different extract procedure, we found that Kin28 immunoprecipitates did not show any marked defect in an in vitro CTD kinase assay (Fig. 2A). Furthermore, immunoblotting of extracts from T162 mutant cells revealed that in vivo levels of phosphorylated RNA polII CTD were roughly equivalent to that of the wild type (Fig. 1A). Therefore, under these conditions we did not find a requirement for T-loop phosphorylation in activating Kin28 kinase activity. A potential explanation for the apparent discrepancy between our results and those of Kimmelman et al. (41) is presented below.
It has been previously shown that the T162A substitution can be synthetically lethal when combined with a second substitution in Kin28 (41). To test whether this interaction was unique to the Kin28 double mutant created by Kimmelman et al., mutants with T162A/D in combination with T17D or K36A were created. All proteins with double mutations in these residues were unable to support viability (data not shown), indicating that T-loop phosphorylation becomes critical when Kin28 activity is limiting in vivo. Kin28(T162A) is also synthetically lethal in combination with a TFB3 mutant (41).
We also combined the Kin28(T162A) allele with alleles of CCL1 encoding amino-terminal truncations leading to slow growth (NΔ48 and NΔ54), resulting in synthetic slow growth and lethality, respectively (data not shown). Interestingly T162A also shows synthetic slow-growth interactions with a Ccl1 mutation (S52A-S53A) that removes potential phosphorylation sites identified in mammalian cyclin H (2). The homologous region of cyclin H is phosphorylated by Cdk8-cyclin C (Srb10/11), an event which negatively regulates the transcriptional activity of mammalian TFIIH (2). We found no evidence for this site being subject to phosphorylation in yeast (data not shown), but these serines may be important for Ccl1 interactions with Kin28.
Transcriptional activity of Kin28 mutants.
Immunoblotting of whole-cell extracts showed that the mutants producing slow-growth phenotypes (T17D and K36A) have reduced levels of CTD phosphorylation in vivo (Fig. 1A). This reduction was shown to be a direct effect by immunoprecipitating Kin28 complexes from extracts and using them to phosphorylate a GST-CTD protein. Although all the mutants coprecipitated roughly equivalent levels of associated TFIIH subunits, the T17D and K36A mutants were severely defective for CTD kinase activity (Fig. 2A). Under these conditions, the T162 substitutions did not result in major defects in kinase activity, mirroring the lack of effect seen in vivo.
Whole-cell extracts from strains containing the Kin28 mutants were tested in an in vitro transcription assay (Fig. 2B). Basal and activated transcription was normal in all the mutant whole-cell extracts. Thus, although the T17D and K36A mutants are dramatically reduced in CTD kinase activity, this does not affect in vitro transcription. This result is in agreement with multiple studies that have failed to find a requirement for CTD phosphorylation in transcription in vitro (7, 61). However, several studies of yeast have shown that shifting some Kin28 temperature-sensitive mutants to the nonpermissive temperature results in a rapid drop in mRNA levels (10, 74). We propose two possible explanations for this apparent discrepancy. First, since Kin28 phosphorylation of the polII CTD is required for recruitment of RNA-processing factors (42, 55, 60), RNAs produced in vivo after inactivating Kin28 will not be capped, spliced, and polyadenylated properly. Such RNAs are rapidly degraded by several pathways (5, 34), and it is likely that this degradation contributes to the reduction in steady-state mRNA levels. This effect would not be seen in vitro. Second, the nature of the Kin28 allele tested is likely to be important. The temperature-sensitive alleles tested in earlier studies (10, 74) produce unstable mutant Kin28 proteins. In contrast, the Kin28(T17D) allele makes a catalytically defective but stable protein. Despite strong reductions in CTD phosphorylation at the promoter, normal levels of elongating polymerase are observed in the T17D mutant strain (42). Therefore, the presence of the Kin28 protein within TFIIH may be necessary for proper function of the complex in transcription initiation, although the kinase activity per se is not. Tirode et al. have previously suggested that it is the physical presence of CAK within TFIIH rather than its kinase activity that is required for optimal transcription (72). A similar situation has been found for Rad3; here the protein is essential for transcription although helicase activity is not (22).
Effect of Kin28 mutants on DNA repair.
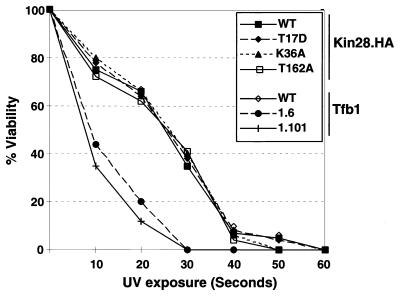
The TFIIH core subunits are utilized in both transcription and nucleotide excision repair (NER). It has been proposed that there are two TFIIH-like complexes: transcription is dependent on a holo-TFIIH containing Kin28-Ccl1, while NER is carried out by a “repairosome” containing the core TFIIH (but not Kin28-Ccl1) and several other proteins (58). Several studies suggest that core TFIIH may be reversibly redistributed between transcription and NER complexes following UV damage (1, 80). To test whether the T162 phosphorylation might serve as a signal for Kin28 dissociation following UV irradiation, wild-type and Kin28 mutant cells were subjected to increasing doses of UV irradiation and tested for viability. In marked contrast to TFIIH core subunit Tfb1 mutants with carboxy-terminal truncations (49), the Kin28(T162A) mutant exhibited no increased sensitivity to DNA damage (Fig. 4). The T17D and K36A mutants were also tested to determine whether the CTD kinase activity of Kin28 was important for DNA repair. Again, no differences compared to wild-type cells were observed. These results support the hypothesis that Kin28, like Ccl1 (73), is not involved in NER. This is in marked contrast to the UV sensitivity exhibited by subunits of core TFIIH (19, 20, 49, 70).
FIG. 4.
Kin28 mutants are not sensitive to UV light-induced DNA damage. Plates containing the indicated Kin28 and Tfb1 mutants were exposed to increasing doses of short-wave UV (200 μW per s per cm2), and surviving colonies were counted. The resulting viability at each time point is expressed as a percentage of the untreated control for each strain. UV-sensitive Tfb1 carboxy-terminal truncation mutants (products of tfb1-6 and tfb1-101 [49]) were used as controls. WT, wild type.
A trimer complex containing Kin28, Ccl1, and Tfb3.
Kin28 purifies as a component of the nine-protein holo-TFIIH complex. Holo-TFIIH can be separated by high salt into “core” TFIIH and a Kin28-Ccl1 dimer complex designated TFIIK (68, 69). Tfb3 remains associated with core TFIIH under these conditions (19, 21). Higher eukaryotes contain two Cdk7-containing complexes: TFIIH and Cdk-activating kinase (or CAK). CAK is a trimer containing Cdk7, cyclin H, and MAT1 (the homologs of Kin28, Ccl1, and Tfb3, respectively). In higher eukaryotes, CAK and TFIIH appear to have different functional roles. Cdk7 within CAK preferentially phosphorylates Cdk2 relative to the polII CTD, while TFIIH exhibits the opposite preference (75, 78). Because TFIIK is an in vitro derivative of TFIIH, it is unlikely that it corresponds to the higher-eukaryote CAK complex.
To determine whether yeast contained a Kin28-Ccl1-Tfb3 trimer complex, Kin28-containing complexes were separated by ion-exchange chromatography over BioRex-70 and phosphocellulose. Two components of TFIIH were tagged: Tfb1 was fused to six histidines (69), and Kin28 was tagged with the HA epitope (10). The majority of the Kin28 protein eluted in the 0.65 M KOAc eluate on the BioRex-70 column, and this fraction was then resolved by gradient elution on phosphocellulose. Kin28 eluted in two peaks (270 to 330 and 450 to 600 mM KOAc), the second of which contained Tfb1 and other members of core TFIIH (Fig. 5; data not shown). Fractions from each peak (complexes I and II) were pooled and subjected to sequential affinity purification on Ni-NTA agarose and an anti-HA antibody column. The resulting eluates were subjected to immunoblotting for components of the TFIIH complex.
The Kin28 found in the fraction eluting at the higher salt concentration (complex II; 450 to 600 mM KOAc) from phosphocellulose was retained on the Ni-NTA column, indicating that it is in a complex with the histidine-tagged Tfb1. This was confirmed by immunoaffinity purification with anti-HA antibodies. Immunoprecipitates contained Kin28, Ccl1, Tfb3, and the Tfb1 proteins. Based on this result and other analyses, we assign this peak of Kin28 to the TFIIH complex.
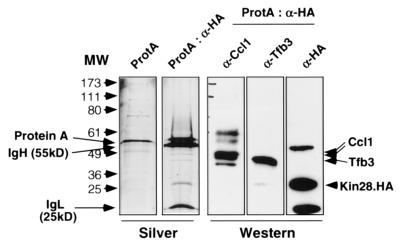
The Kin28 fractions eluting from phosphocellulose at lower salt concentrations (complex I; 270 to 330 mM KOAc) did not contain core subunit Tfb1. This was in agreement with the finding that Kin28, Ccl1, and Tfb3 in this fraction were not retained on a Ni-NTA column. Silver staining of the anti-HA immunoprecipitate from the lower-salt fraction revealed three specific bands, which were found to correspond to Ccl1, Tfb3, and HA-tagged Kin28 by immunoblotting analysis (Fig. 6). The fact that all three proteins coprecipitated indicates that they are in a complex. Based on the copurification of these three proteins away from other core TFIIH subunits, we propose that this species may be analogous to the CAK complex isolated from higher eukaryotes.
FIG. 6.
Complex I is a trimer of Kin28, Ccl1, and Tfb3. Immunopurified complex I (ProtA:α-HA) was subjected to SDS-PAGE followed by silver staining and immunoblotting analysis. As a negative control, the Ni-NTA flowthrough fraction for complex I was precipitated without the anti-HA antibody (ProtA). The positions of protein A and the immunoglobulin heavy and light chains (IgH and IgL, respectively, are indicated. Immunoblotting with antibodies against Ccl1, Tfb3, or Kin28 shows that these proteins align with silver-stained species of the appropriate size. Note that the immunoglobulin proteins are also detected in the anti-HA immunoblot.
This trimer complex is distinct from the TFIIK identified by Svejstrup and coworkers (69), since TFIIK copurified extensively with holo-TFIIH and did not contain Tfb3. Indeed, TFIIK was only partially separated from TFIIH by late-stage gradient chromatography and apparently represents a dissociated subcomplex (68, 69). Since they were purifying TFIIH based on transcriptional activity and/or Tfb1 content rather than Kin28 content, it is not surprising that they did not previously identify the Kin28-Ccl1-Tfb3 trimer complex. The discovery of a trimer complex in yeast suggests that the distribution of Kin28-Cdk7 complexes is more conserved than previously suspected. Whether this conservation extends to a functional role remains to be seen.
Effect of T162 phosphorylation within TFIIH and the trimer complex.
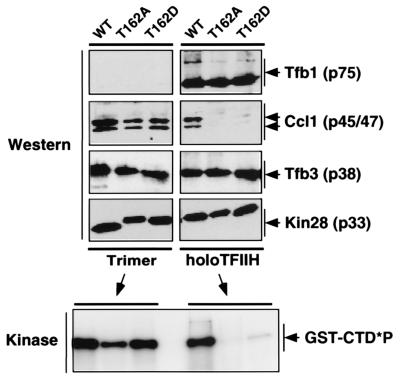
Since Kin28 could be found in two distinct complexes, the role of T162 phosphorylation on each complex was tested after chromatographic separation. Complexes containing wild-type, T162A, or T162D Kin28 were purified as described above. The resulting TFIIH and trimer complexes were analyzed by immunoblotting and an in vitro CTD kinase assay. The distributions of Kin28, Ccl1, and Tfb3 between the two complexes were not affected by the T162 mutations during the phosphocellulose gradient chromatography (data not shown). However, during the anti-HA immunoprecipitation step it was noted that upon stringent washing the Ccl1 subunit was less stably associated with the TFIIH complex (Fig. 7). Both the alanine and aspartate substitutions at T162 produced a similar effect, indicating that aspartate, with a negative charge, cannot substitute for a phosphorylated threonine residue. Surprisingly, Ccl1 within the trimer complex did not appear to be similarly affected.
FIG. 7.
Functional analysis of complex I trimer and TFIIH from the T162 mutants. Trimer and TFIIH complexes were biochemically purified from strains containing wild-type (WT) Kin28 or the T162A or T162D mutants as described for Fig. 5 and in Materials and Methods. Following the final purification via the HA epitope tag, the resulting complexes were subjected to SDS-PAGE and immunoblot analysis (Western; top). The immunoprecipitates were also used for an in vitro CTD phosphorylation assay (bottom). GST-CTD*P, phosphorylated GST-CTD.
The CTD phosphorylation assay showed that kinase activity closely paralleled the presence of the Ccl1 subunit (Fig. 7). TFIIH complexes containing the Kin28 T162 mutants and lacking Ccl1 had very little CTD kinase activity. In contrast, trimer complexes containing the T162 substitutions retained Ccl1 and had essentially wild-type levels of kinase activity. It has been previously reported that the T162A mutation causes a partial reduction (75 to 80%) in CTD kinase activity of Kin28-containing immunoprecipitates (41). This is consistent with a mixture of inactive TFIIH and active trimer complex. We note that there may be variability in Ccl1 association dependent on the particular purification or immunoprecipitation conditions used in various studies. To allow clear conclusions, future studies should monitor Ccl1 association.
We conclude that T162 phosphorylation regulates the affinity of the interaction between Kin28 and Ccl1. T-loop phosphorylation has been found to promote the association of other Cdk-cyclin pairs (16, 44, 48, 52). Interestingly, it has been previously noted that T-loop phosphorylation of Xenopus and mouse Cdk7 promotes in vitro interaction with cyclin H to activate the kinase within the dimer pair but becomes much less important within a Cdk7-cyclin H-MAT1 trimer (14, 25). Our results suggest that a differential sensitivity to T-loop phosphorylation between the trimer and TFIIH complexes also exists in vivo.
DISCUSSION
It is clear that the Kin28 kinase (known as Cdk7 or mo15 in metazoans) performs an essential function in eukaryotic cells. However, many questions remain about whether this kinase has multiple roles and whether these roles have been conserved over evolution. In all organisms studied so far, the Kin28 kinase and its associated cyclin (Ccl1 or cyclin H) have been identified as subunits of basal transcription factor TFIIH. Within this context, Kin28/Cdk7 phosphorylates the CTD of the largest subunit of RNA polII. Although this modification is not necessary for transcription initiation (7, 61), it is required for proper cotranscriptional mRNA processing (9, 42, 51, 55, 60) and perhaps regulation of elongation. In mammalian cells, Cdk7 is found in a second complex known as CAK (CDK-activating kinase). This trimer complex also contains cyclin H and another TFIIH subunit known as Mat1. The role of this complex appears to be to phosphorylate other Cdks on the T loop, thereby activating their kinase activity and regulating the Cdk-cyclin association. However, the function of Cdk7 as a CAK is mostly suggested by in vitro experiments, with some in vivo evidence only recently coming to light (31, 43). In S. cerevisiae, it is clear that Kin28 is not required for CAK activity, because another essential kinase (Cak1/Civ1) performs this function (11, 17, 18).
Until now, it was not known whether S. cerevisiae contains a CAK-like trimer complex. A dimer complex of Kin28 and Ccl1 can be dissociated from TFIIH, and this has been designated TFIIK (69). It is not clear if this is a physiologically relevant complex in yeast, although a dimer complex can also be isolated from Xenopus extracts (48). We now report that yeast extracts contain a complex containing Kin28, Ccl1, and Tfb3 (the homologue of Mat1). We presume that this complex is a trimer, but we cannot rule out higher-order assemblies (e.g., hexamers) at this point. It appears that at least one-half of these proteins are found in the trimer complex. Since it apparently is not used for activation of Cdks, what is the role of this complex? One possibility is that the trimer complex is an evolutionary vestige that has been functionally replaced by Cak1/Civ1 in S. cerevisiae. In support of this idea, there is some evidence from S. pombe that both the Kin28 and Cak1 homologues can function as CAKs (47). It has also been shown that the mammalian trimer complex can inhibit in vitro transcription, although it is not clear whether this is a physiological function (6). Another possibility is that the trimer complex (in both yeast and higher eukaryotes) functions to phosphorylate other unknown substrates. Finally, the trimer complex may be an intermediate in the assembly of TFIIH, joining the core complex that is required for both transcription and DNA repair (70). To explore these possibilities, it will be necessary to find conditions or mutants that specifically affect the trimer complex but not TFIIH.
The discovery of the yeast trimer complex also provides some insight into the role of Kin28 T-loop phosphorylation. In agreement with other groups, we find that Kin28 is phosphorylated at threonine 162. We also tested whether serine 163 could serve as an alternative phosphoacceptor and found that it could not (data not shown). T162 phosphorylation is dependent on Cak1/Civ1 (11, 17). Interestingly, a recombinant trimer complex was preferentially phosphorylated relative to a Kin18-Ccl1 dimer (17). Substitution mutations at threonine 162 produce no apparent phenotype unless they are combined with a second mutation in Kin28 (this report; 17), Tfb3 (41), or Ccl1 (this report). Levels of phosphorylated RNA polII in Kin28(T162) yeast extracts are similar to those in wild-type extracts. In contrast to the mild in vivo effect of the T162A mutation, it has been reported that immunoprecipitates of Kin28(T162) are 75 to 80% defective for CTD phosphorylation. In apparent contradiction, we did not see a strong effect on in vitro phosphorylation by T162 mutant immunoprecipitates (Fig. 2). Several of our findings help resolve these apparent discrepancies. First, we find that TFIIH and the trimer complex are differentially sensitive to the T162A mutation. The mutant TFIIH complex is prone to loss of the Ccl1 subunit as well as CTD kinase activity (Fig. 7). Depending on the ratio of the two complexes within a particular extract, quantitatively varying results for total-extract immunoprecipitates would be expected. Furthermore, we found that the stringency of immunoprecipitate washing can affect Ccl1 association because under milder conditions the Ccl1 subunit can remain associated with the Kin28(T162A) mutant TFIIH (data not shown). When Ccl1 remains associated, CTD kinase activity is also maintained. Taken together, the available evidence indicates that T-loop phosphorylation stabilizes, but is not absolutely required for, the interaction between Kin28 and Ccl1. When other mutations in Kin28, Ccl1, or Tfb3 occur, this stabilization becomes critical.
Does the phosphorylation of Kin28 play a regulatory role or is it simply constitutive? Kimmelman et al. showed that there was no variation in Kin28 phosphorylation during the cell cycle. Also, the Kin28 T162 mutants are not hypersensitive to UV light (Fig. 4) (41), arguing that T-loop phosphorylation is not used to regulate the switch between the TFIIH transcription and DNA repair functions. The Ptc2 and Ptc3 phosphatases have been shown to dephosphorylate the T loop of Cdc28 (8), so they might also act on Kin28. Since T162A-T17D double mutants are inviable, we tested whether overexpression of phosphatases Ptc2 or Ptc3 would exacerbate the slow-growth phenotype of a T17D mutant. We saw no further decrease in growth rate and no change in the ratio of phosphorylated to unphosphorylated Kin28 (data not shown), arguing against a role for these phosphatases in regulating Kin28 activity. So far, there is no evidence to support a regulatory role for phosphorylation of the Kin28 T loop.
We also found no evidence for an inhibitory phosphorylation of Kin28 on threonine 17 or tyrosine 18. The analogous sites in S. cerevisiae Cdc28, S. pombe Cdk2, and some other Cdks are phosphorylated in vivo to down-regulate their kinase activities. In the process of constructing mutants with point mutations at these sites within Kin28, we discovered that a T17D mutation mimicked such a down-regulation. The mutant produced a stable protein with significantly reduced CTD kinase activity, resulting in slow growth. We have used this mutant Kin28 to show that CTD phosphorylation by Kin28 is necessary for recruitment of a capping enzyme to RNA polymerase (42, 55).
Several other mutations of Kin28 that change residues predicted to be key parts of the catalytic site were made. As expected, E54Q and D147N mutants did not support viability and did not have kinase activity. These two residues are thought to be part of the binding pocket for ATP-Mg2+ (36, 53). Lysine 36 is also predicted to be involved in coordinating the alpha phosphate of ATP. Surprisingly, we find that mutation of this residue to alanine results in destabilization of the Kin28 protein, arguing that it must in part play a structural role in the protein. A high-copy-number plasmid encoding the K36A mutant restores proteins levels, but the kinase activity of the mutant protein is still reduced relative to that of the wild type. Therefore, K36 probably functions in both protein stability and catalytic activity.
In conclusion, our data support the general model that T-loop phosphorylation of Cdks serves to stabilize cyclin association. Our identification of a Kin28-Ccl1-Tfb1 trimer extends the conservation of yeast and higher eukaryotes, yet raises new questions about the CAK/TFIIH difference among species. It remains to be determined whether the yeast trimer has a function in vivo. The yeast trimer efficiently phosphorylates the Rpb1 CTD and retains specificity for serine 5 within the heptapeptide repeat (not shown). Further studies will determine whether this complex plays a role in transcription independently of TFIIH.
Acknowledgments
We thank Roger Kornberg, John Feaver, Errol Friedberg, Mark Solomon, David Morgan, and Ben Blencowe for the gift of plasmids, yeast strains, and antibodies. We are grateful to David Hess for help in the early stages of this project.
This work is supported by a grant from NIH to S.B. S.B. is a Scholar of the Leukemia and Lymphoma Society.
REFERENCES
- 1.Adamczewski, J. P., M. Rossignol, J. P. Tassan, E. A. Nigg, V. Moncollin, and J. M. Egly. 1996. MAT1, cdk7 and cyclin H form a kinase complex which is UV light-sensitive upon association with TFIIH. EMBO J. 15:1877-1884. [PMC free article] [PubMed] [Google Scholar]
- 2.Akoulitchev, S., S. Chuikov, and D. Reinberg. 2000. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature 407:102-106. [DOI] [PubMed] [Google Scholar]
- 3.Akoulitchev, S., and D. Reinberg. 1998. The molecular mechanism of mitotic inhibition of TFIIH is mediated by phosphorylation of CDK7. Genes Dev. 12:3541-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed). 1991. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 5.Beelman, C. A., A. Stevens, G. Caponigro, T. E. LaGrandeur, L. Hatfield, D. M. Fortner, and R. Parker. 1996. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382:642-646. [DOI] [PubMed] [Google Scholar]
- 6.Bochar, D. A., Z.-Q. Pan, R. Knights, R. P. Fisher, A. Shilatifard, and R. Shiekhattar. 1999. Inhibition of transcription by the trimeric cyclin-dependent kinase 7 complex. J. Biol. Chem. 274:13162-13166. [DOI] [PubMed] [Google Scholar]
- 7.Buratowski, S., and P. A. Sharp. 1990. Transcription initiation complexes and upstream activation with RNA polymerase II lacking the C-terminal domain of the largest subunit. Mol. Cell. Biol. 10:5562-5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, A., K. E. Ross, P. Kaldis, and M. J. Solomon. 1999. Dephosphorylation of cyclin-dependent kinases by type 2C protein phosphatases. Genes Dev. 13:2946-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho, E. J., and S. Buratowski. 1999. Evidence that transcription factor IIB is required for a post-assembly step in transcription initiation. J. Biol. Chem. 274:25807-25813. [DOI] [PubMed] [Google Scholar]
- 10.Cismowski, M. J., G. M. Laff, M. J. Solomon, and S. I. Reed. 1995. KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase activity. Mol. Cell. Biol. 15:2983-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cross, F. R., and K. Levine. 1998. Molecular evolution allows bypass of the requirement for activation loop phosphorylation of the Cdc28 cyclin-dependent kinase. Mol. Cell. Biol. 18:2923-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desai, D., Y. Gu, and D. O. Morgan. 1992. Activation of human cyclin-dependent kinases in vitro. Mol. Biol. Cell 3:571-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desai, D., H. C. Wessling, R. P. Fisher, and D. O. Morgan. 1995. Effects of phosphorylation by CAK on cyclin binding by CDC2 and CDK2. Mol. Cell. Biol. 13:345-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devault, A., A. M. Martinez, D. Fesquet, J. C. Labbe, N. Morin, J. P. Tassan, E. A. Nigg, J. C. Cavadore, and M. Doree. 1995. MAT1 (‘menage a trois') a new RING finger protein subunit stabilizing cyclin H-cdk7 complexes in starfish and Xenopus CAK. EMBO J. 14:5027-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drapkin, R., R. G. Le, H. Cho, S. Akoulitchev, and D. Reinberg. 1996. Human cyclin-dependent kinase-activating kinase exists in three distinct complexes. Proc. Natl. Acad. Sci. USA 93:6488-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ducommun, B., P. Brambilla, M. A. Felix, B. J. Franza, E. Karsenti, and G. Draetta. 1991. cdc2 phosphorylation is required for its interaction with cyclin. EMBO J. 10:3311-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Espinoza, F., A. Farrell, J. Nourse, H. Chamberlin, O. Gileadi, and D. Morgan. 1998. Cak1 is required for Kin28 phosphorylation and activation in vivo. Mol. Cell. Biol. 18:6365-6373. (Erratum, 20:1898, 2000.) [DOI] [PMC free article] [PubMed]
- 18.Espinoza, F. H., A. Farrell, H. Erdjument-Bromage, P. Tempst, and D. O. Morgan. 1996. A cyclin-dependent kinase-activating kinase (CAK) in budding yeast unrelated to vertebrate CAK. Science 273:1714-1717. [DOI] [PubMed] [Google Scholar]
- 19.Feaver, W. J., N. L. Henry, Z. Wang, X. Wu, J. Q. Svejstrup, D. A. Bushnell, E. C. Friedberg, and R. D. Kornberg. 1997. Genes for Tfb2, Tfb3, and Tfb4 subunits of yeast transcription/repair factor IIH. Homology to human cyclin-dependent kinase activating kinase and IIH subunits. J. Biol. Chem. 272:19319-19327. [DOI] [PubMed] [Google Scholar]
- 20.Feaver, W. J., W. Huang, and E. C. Friedberg. 1999. The TFB4 subunit of yeast TFIIH is required for both nucleotide excision repair and RNA polymerase II transcription. J. Biol. Chem. 274:29564-29567. [DOI] [PubMed] [Google Scholar]
- 21.Feaver, W. J., W. Huang, O. Gileadi, L. Myers, C. M. Gustafsson, R. D. Kornberg, and E. C. Friedberg. 2000. Subunit interactions in yeast transcription/repair factor TFIIH. Requirement for Tfb3 subunit in nucleotide excision repair. J. Biol. Chem. 275:5941-5946. [DOI] [PubMed] [Google Scholar]
- 22.Feaver, W. J., J. Q. Svejstrup, L. Bardwell, A. J. Bardwell, S. Buratowski, K. D. Gulyas, T. F. Donahue, E. C. Friedberg, and R. D. Kornberg. 1993. Dual roles of a multiprotein complex from S. cerevisiae in transcription and DNA repair. Cell 75:1379-1387. [DOI] [PubMed] [Google Scholar]
- 23.Feaver, W. J., J. Q. Svejstrup, N. L. Henry, and R. D. Kornberg. 1994. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell 79:1103-1109. [DOI] [PubMed] [Google Scholar]
- 24.Fesquet, D., J. C. Labbe, J. Derancourt, J. P. Capony, S. Galas, F. Girard, T. Lorca, J. Shuttleworth, M. Doree, and J. C. Cavadore. 1993. The MO15 gene encodes the catalytic subunit of a protein kinase that activates cdc2 and other cyclin-dependent kinases (CDKs) through phosphorylation of Thr161 and its homologues. EMBO J. 12:3111-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fisher, R. P., P. Jin, H. M. Chamberlain, and D. O. Morgan. 1995. Alternative mechanisms of CAK assembly require an accessory factor or an activating kinase. Cell 83:47-57. [DOI] [PubMed] [Google Scholar]
- 26.Fisher, R. P., and D. O. Morgan. 1994. A novel cyclin associates with MO15/CDK7 to form the CDK-activating kinase. Cell 78:713-724. [DOI] [PubMed] [Google Scholar]
- 27.Gietz, D., A. St. Jean, R. A. Woods, and R. Schiestl. 1992. Improved method for high-efficiency transformation of intact yeast cells.Nucleic Acids Res. 20:1425.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guthrie, C., and G. R. Fink (ed.). 1991. Methods in enzymology, vol. 194. Guide to yeast genetics and molecular biology. Academic Press, Inc., Boston, Mass. [PubMed]
- 29.Gyuris, J., E. Golemis, H. Chertkov, and R. Brent. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75:791-803. [DOI] [PubMed] [Google Scholar]
- 30.Hannon, G. J., D. Casso, and D. Beach. 1994. KAP: a dual specificity phosphatase that interacts with cyclin-dependent kinases. Proc. Natl. Acad. Sci. USA 91:1731-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harper, J. W., and S. J. Elledge. 1998. The role of Cdk7 in CAK function, a retro-retrospective. Genes Dev. 12:285-289. [DOI] [PubMed] [Google Scholar]
- 32.Hengartner, C. J., V. E. Myer, S.-M. Liao, C. J. Wilson, S. S. Koh, and R. A. Young. 1998. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell 2:43-53. [DOI] [PubMed] [Google Scholar]
- 33.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 34.Hsu, C. L., and A. Stevens. 1993. Yeast cells lacking 5"3" exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5" cap structure. Mol. Cell. Biol. 13:4826-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iavarone, A., and J. Massague. 1997. Repression of the CDK activator Cdc25A and cell-cycle arrest by cytokine TGF-β in cells lacking the CDK inhibitor p15. Nature 387:417-422. [DOI] [PubMed] [Google Scholar]
- 36.Jeffrey, P. D., A. A. Russo, K. Polyak, E. Gibbs, J. Hurwitz, J. Massague, and N. P. Pavletich. 1995. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature 376:313-320. [DOI] [PubMed] [Google Scholar]
- 37.Kaldis, P. 1999. The cdk-activating kinase (CAK): from yeast to mammals. Cell Mol. Life Sci. 55:284-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaldis, P., A. Sutton, and M. J. Solomon. 1996. The Cdk-activating kinase (CAK) from budding yeast. Cell 86:553-564. [DOI] [PubMed] [Google Scholar]
- 39.Kato, J.-Y., M. Matsuoka, D. K. Storm, and C. J. Sherr. 1994. Regulation of cyclin D-dependent kinase 4 (cdk4) by cdk4-activating kinase. Mol. Cell. Biol. 14:2713-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim, Y. J., S. Bjorklund, Y. Li, M. H. Sayre, and R. D. Kornberg. 1994. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77:599-608. [DOI] [PubMed] [Google Scholar]
- 41.Kimmelman, J., P. Kaldis, C. J. Hengartner, G. M. Laff, S. S. Koh, R. A. Young, and M. J. Solomon. 1999. Activating phosphorylation of the Kin28p subunit of yeast TFIIH by Cak1p. Mol. Cell. Biol. 19:4774-4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Komarnitsky, P., E.-J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuchin, S., and M. Carlson. 1998. Functional relationships of Srb10-Srb11 kinase, carboxy-terminal domain kinase CTDK-I, and transcriptional corepressor Ssn6-Tup1. Mol. Cell. Biol. 18:1163-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Larochelle, S., J. Chen, R. Knights, J. Pandur, P. Morcillo, H. Erdjument-Bromage, P. Tempst, B. Suter, and R. P. Fisher. 2001. T-loop phosphorylation stabilizes the CDK7-cyclin H-MAT1 complex in vivo and regulates its CTD kinase activity. EMBO J. 20:3749-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larochelle, S., J. Pandur, R. P. Fisher, H. K. Salz, and B. Suter. 1998. Cdk7 is essential for mitosis and for in vivo Cdk-activating kinase activity. Genes Dev. 12:370-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee, J. M., and A. L. Greenleaf. 1991. CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Expr. 1:149-167. [PMC free article] [PubMed] [Google Scholar]
- 47.Lee, K. M., J. E. Saiz, W. A. Barton, and R. P. Fisher. 1999. Cdc2 activation in fission yeast depends on Msc6 and Csk1, two partially redundant Cdk-activating kinases (CAKs). Curr. Biol. 9:441-444. [DOI] [PubMed] [Google Scholar]
- 48.Martinez, A.-M., M. Afshar, F. Martin, J.-C. Cavadore, J.-C. Labbé, and M. Dorée. 1997. Dual phosphorylation of the T-loop in cdk7: its role in controlling cyclin H binding and CAK activity. EMBO J. 16:343-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsui, P., J. DePaulo, and S. Buratowski. 1995. An interaction between the Tfb1 and Ssl1 subunits of yeast TFIIH correlates with DNA repair activity. Nucleic Acids Res. 23:767-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsuoka, M., J.-Y. Kato, R. P. Fisher, D. O. Morgan, and C. J. Sherr. 1994. Activation of cyclin-dependent kinase 4 (Cdk4) by mouse MO15-associated kinase. Mol. Cell. Biol. 14:7265-7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCracken, S., E. Rosonina, N. Fong, M. Sikes, A. Beyer, K. O'Hare, S. Shuman, and D. Bentley. 1998. Role of RNA polymerase II carboxy-terminal domain in coordinating transcription with RNA processing. Cold Spring Harbor Symp. Quant. Biol. 63:301-309. [DOI] [PubMed] [Google Scholar]
- 52.Morgan, D. O. 1997. Cyclin-dependent kinases: Engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13:261-291. [DOI] [PubMed] [Google Scholar]
- 53.Pines, J. 1995. Conformational change. Nature 376:294-295. [DOI] [PubMed] [Google Scholar]
- 54.Prelich, G., and F. Winston. 1993. Mutations that suppress the deletion of an upstream activating sequence in yeast: involvement of a protein kinase and histone H3 in repressing transcription in vivo. Genetics 135:665-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodriguez, C. R., E.-J. Cho, M.-C. Keogh, C. L. Moore, A. L. Greenleaf, and S. Buratowski. 2000. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol. Cell. Biol. 20:104-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roy, R., J. P. Adamczewski, T. Seroz, W. Vermeulen, J. P. Tassan, L. Schaeffer, E. A. Nigg, J. H. Hoeijmakers, and J. M. Egly. 1994. The MO15 cell cycle kinase is associated with the TFIIH transcription-DNA repair factor. Cell 79:1093-1101. [DOI] [PubMed] [Google Scholar]
- 57.Russo, A. A., P. D. Jeffrey, and N. P. Pavletich. 1996. Structural basis of cyclin-dependent kinase activation by phosphorylation. Nat. Struct. Biol. 3:696-700. [DOI] [PubMed] [Google Scholar]
- 58.Sancar, A. 1996. DNA excision repair. Annu. Rev. Biochem. 65:43-81. [DOI] [PubMed] [Google Scholar]
- 59.Sayre, M. H., H. Tschochner, and R. D. Kornberg. 1992. Reconstitution of transcription with five purified initiation factors and RNA polymerase II from Saccharomyces cerevisiae. J. Biol. Chem. 267:23376-23382. [PubMed] [Google Scholar]
- 60.Schroeder, S. C., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Serizawa, H., J. W. Conaway, and R. C. Conaway. 1993. Phosphorylation of CTD of RNA polymerase II is not required in basal transcription. Nature 363:371-374. [DOI] [PubMed] [Google Scholar]
- 62.Serizawa, H., T. P. Makela, J. W. Conaway, R. C. Conaway, R. A. Weinberg, and R. A. Young. 1995. Association of Cdk-activating kinase subunits with transcription factor TFIIH. Nature 374:280-282. [DOI] [PubMed] [Google Scholar]
- 63.Shiekhattar, R., F. Mermelstein, R. P. Fisher, R. Drapkin, B. Dynlacht, H. C. Wessling, D. O. Morgan, and D. Reinberg. 1995. Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature 374:283-287. [DOI] [PubMed] [Google Scholar]
- 64.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Simon, M., B. Seraphin, and G. Faye. 1986. KIN28, a yeast split gene coding for a putative protein kinase homologous to cdc28. EMBO J. 5:2697-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Solomon, M. J., and P. Kaldis. 1998. Regulation of Cdks by phosphorylation. Results Probl. Cell Differ. 22:79-109. [DOI] [PubMed] [Google Scholar]
- 67.Solomon, M. J., T. Lee, and M. W. Kirschner. 1992. Role of phosphorylation in p34cdc2 activation: identification of an activating kinase. Mol. Biol. Cell 3:13-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Svejstrup, J. Q., W. J. Feaver, and R. D. Kornberg. 1996. Subunits of yeast RNA polymerase II transcription factor TFIIH encoded by the CCL1 gene. J. Biol. Chem. 271:643-645. [DOI] [PubMed] [Google Scholar]
- 69.Svejstrup, J. Q., W. J. Feaver, J. LaPointe, and R. D. Kornberg. 1994. RNA polymerase transcription factor IIH holoenzyme from yeast. J. Biol. Chem. 269:28044-28048. [PubMed] [Google Scholar]
- 70.Svejstrup, J. Q., Z. Wang, W. J. Feaver, X. Wu, D. A. Bushnell, T. F. Donahue, E. C. Friedberg, and R. D. Kornberg. 1995. Different forms of TFIIH for transcription and DNA repair: holo-TFIIH and a nucleotide excision repairosome. Cell 80:21-28. [DOI] [PubMed] [Google Scholar]
- 71.Thuret, J. Y., J. G. Valet, G. Faye, and C. Mann. 1996. Civ1 (CAK in vivo), a novel Cdk-activating kinase. Cell 86:565-576. [DOI] [PubMed] [Google Scholar]
- 72.Tirode, F., D. Busso, F. Coin, and J. M. Egly. 1999. Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol. Cell 3:87-95. [DOI] [PubMed] [Google Scholar]
- 73.Valay, J. G., M. F. Dubois, O. Bensaude, and G. Faye. 1996. Ccl1, a cyclin associated with protein kinase Kin28, controls the phosphorylation of RNA polymerase II largest subunit and mRNA transcription. C. R. Acad. Sci. Ser. III 319:183-189. [PubMed] [Google Scholar]
- 74.Valay, J. G., M. Simon, M. F. Dubois, O. Bensaude, C. Facca, and G. Faye. 1995. The KIN28 gene is required for both RNA polymerase II mediated transcription and phosphorylation of the Rbb1p CTD. J. Mol. Biol. 249:535-544. [DOI] [PubMed] [Google Scholar]
- 75.Watanabe, Y., H. Fujimoto, T. Watanabe, M. Maekawa, C. Masutani, F. Hanaoka, and Y. Ohkuma. 2000. Modulation of TFIIH-associated kinase activity by complex formation and its relationship with CTD phosphorylation of RNA polymerase II. Genes Cells 5:407-423. [DOI] [PubMed] [Google Scholar]
- 76.Woontner, M., and J. A. Jaehning. 1990. Accurate initiation by RNA polymerase II in a whole cell extract from Saccharomyces cerevisiae. J. Biol. Chem. 265:8979-8982. [PubMed] [Google Scholar]
- 77.Woontner, M., P. A. Wade, J. Bonner, and J. A. Jaehning. 1991. Transcriptional activation in an improved whole-cell extract from Saccharomyces cerevisiae. Mol. Cell. Biol. 11:4555-4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yankulov, K. Y., and D. L. Bentley. 1997. Regulation of CDK7 substrate specificity by MAT1 and TFIIH. EMBO J. 16:1638-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yao, S., A. Neiman, and G. Prelich. 2000. BUR1 and BUR2 encode a divergent cyclin-dependent kinase-cyclin complex important for transcription in vivo. Mol. Cell. Biol. 20:7080-7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.You, Z., W. J. Feaver, and E. C. Friedberg. 1998. Yeast RNA polymerase II transcription in vitro is inhibited in the presence of nucleotide excision repair: complementation of inhibition by Holo-TFIIH and requirement for RAD26. Mol. Cell Biol 18:2668-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]