Abstract
In transcriptional regulation, RNA polymerase II (pol II) interacts and forms complexes with a number of protein factors. To isolate and identify the pol II-associated proteins, we constructed a Schizosaccharomyces pombe strain carrying a FLAG tag sequence fused to the rpb3 gene encoding the pol II subunit Rpb3. By immunoaffinity purification with anti-FLAG antibody-resin, a pol II complex containing the Rpb1 subunit with a nonphosphorylated carboxyl-terminal domain (CTD) was isolated. In addition to the pol II subunits, the complex was found to contain three subunits of a transcription factor TFIIF (TFIIFα, TFIIFβ, and Tfg3) and TFIIF-interacting CTD-phosphatase Fcp1. The same type of pol II complex could also be purified from an Fcp1-tagged strain. The isolated Fcp1 showed CTD-phosphatase activity in vitro. The fcp1 gene is essential for cell viability. Fcp1 and pol II interacted directly in vitro. Furthermore, by chemical cross-linking, glutathione S-transferase pulldown, and affinity chromatography, the Fcp1-interacting subunit of pol II was identified as Rpb4, which plays regulatory roles in transcription. We also constructed an S. pombe thiamine-dependent rpb4 shut-off system. On repression of rpb4 expression, the cell produced more of the nonphosphorylated form of Rpb1, but the pol II complex isolated with the anti-FLAG antibody contained less Fcp1 and more of the phosphorylated form of Rpb1 with a concomitant reduction in Rpb4. This result indicates the importance of Fcp1-Rpb4 interaction for formation of the Fcp1/TFIIF/pol II complex in vivo.
RNA polymerase II (pol II), which is involved in the synthesis of all mRNAs, is a highly structured complex consisting of as many as 12 subunits, Rpb1 to Rpb12 (30, 37, 60, 67, 75), but for accurate transcription, pol II is controlled by a number of factors through protein-protein interactions (56). In preinitiation complex (PIC) formation, a general transcription factor (GTF), TFIIF, associates with pol II to recruit it to the complex on a promoter, which is formed of GTFs, including TFIIA, TFIIB, and TFIID (19). TFIIB (22, 39, 68) and one of the TATA binding protein (TBP)-associating factor (TAF) subunits of TFIID (7) interacts with pol II, and the TBP subunit of TFIID also binds to the nonphosphorylated carboxy-terminal domain (CTD) of Rpb1 (69). TFIIE assembles into the complex through direct interaction with pol II (39, 46) and then promotes association of TFIIH, which phosphorylates the CTD (17, 43, 51). The kinase subunit of TFIIH binds to pol II (18).
The alternative pathway of PIC formation is the prior assembly of pol II and factors to form pol II holoenzyme (38, 50). This large complex consists of pol II, a subset of GTFs, and a mediator complex, and it is recruited to a promoter through the interaction of mediators with DNA-binding activators. In the holoenzyme, the mediator complex, which is composed of SRBs (for suppressor of RNA pol B), mediators, and other subunits, is attached to the CTD (49) and possibly other parts of pol II (3). Srb10 in the mediator complex has CTD-kinase activity (23). Another holoenzyme-like complex, which contains Paf1, Cdc73, Hpr1, Ccr4, and other factors, was also isolated from Saccharomyces cerevisiae (11, 12).
The pol II elongation process is controlled by a number of factors. Interactions of pol II with SII (or TFIIS) (61, 65), ELL (62), elongator (53), and DSIF (for DRB sensitivity-inducing factor) (70, 76) have been reported, and elongin interacts with the pol II holoenzyme (54). P-TEFb stimulates elongation by phosphorylating the CTD (45). After transcription termination, pol IIO, containing the IIo form of Rpb1 with a phosphorylated CTD, is thought to be dephosphorylated into pol IIA, containing the nonphosphorylated IIa form of Rpb1 and to be used for reinitiation, because only pol IIA can be recruited to the PIC (42).
Recently, the CTD-specific phosphatase Fcp1 from S. cerevisiae (2, 34) and humans (1, 13) was identified. TFIIF and TFIIB bind to Fcp1 competitively (10, 35), and TFIIF stimulates CTD-phosphatase activity (1, 10). CTD-phosphatase has a docking site on pol II that is distinct from the CTD (10), but the site has not yet been specified. Moreover, direct binding between Fcp1 and pol II has not been clearly proved, although Fcp1 has been identified as a component of the pol II holoenzyme (1), and the eluate from a pol II affinity column showed CTD-phosphatase activity (10).
These pol II-factor interactions were identified by various methods. The pol II interaction of GTFs, TFIIB (22, 68), TFIID (7, 69), TFIIE (46), TFIIH (18), and most of the elongation factors (45, 62, 76) was established by in vitro binding assays using the purified factors. The direct interaction between mediators and the CTD was also confirmed in vitro (49). The pol II binding of TFIIF and SII was established by the purification method of pol II affinity chromatography (61, 65). Besides these binding methods, copurification of native complexes provides strong evidence for protein-protein interaction. The elongator was copurified with pol II through columns (53). The holoenzyme, a complex of mediators and pol II, was also purified through several steps of conventional column chromatography (38, 50, 66), followed by SII and elongin affinity methods (54). The immunoaffinity method with anti-CTD antibody was also successfully employed for the isolation of an alternative pol II complex, although the complex was dissociated in the elution process (71).
In this study, we carried out the isolation of a pol II complex from S. pombe using the FLAG-tagging method. For this purpose, a DNA sequence encoding the FLAG epitope was inserted into the chromosomal rpb3 gene, and the cell extract was directly applied to anti-FLAG M2 monoclonal antibody (MAb)-agarose beads.
MATERIALS AND METHODS
Plasmids.
pUC-f-rpb3-ura4 was constructed by replacing a His tag sequence between the NheI and ApaI sites in plasmid pUC-rpb3H-ura4 (31) with oligonucleotides encoding the FLAG epitope. For construction of pBS-f-fcp1-ura4, an fcp1 genomic DNA fragment covering nucleotides (nt) +2263 to +2913 (from the translation initiation) was amplified by PCR and inserted into pBluescript II SK+ (Clontech) between the KpnI and XbaI sites. Tandem NheI and ApaI sites were generated by in vitro manipulation between the carboxyl terminus and the termination codons of fcp1, and oligonucleotides for the FLAG epitope were inserted between them. Finally, the ura4+ marker fragment (21) was ligated into a HindIII site within the 3" noncoding region of the fcp1 gene.
To create pBS-fcp1::ura4, two fcp1 genomic DNA fragments, one covering nt −435 to −39 with EcoRI and HindIII sites on the 5" and 3" termini, respectively, and the other covering nt +2356 to +2623 with HindIII and XhoI sites on the 5" and 3" termini, respectively, were PCR amplified and cloned tandemly into pBluescript II SK+ between the EcoRI and XhoI sites. The ura4+ fragment was inserted at the HindIII site. pREP41-f-fcp1 was created by ligating a PCR-amplified fcp1 cDNA fragment between the ApaI and BamHI sites of pREP41XF. pREP41XF was prepared from pREP41X (20) by inserting oligonucleotides containing the XhoI, Met codon, FLAG, NdeI, ApaI, and BamHI sequences between the XhoI and BamHI sites. pREP81-rpb4 was made by inserting a PCR-amplified rpb4 cDNA fragment into pREP81 (6) between the NdeI and BamHI sites.
pET-fcp1-H was constructed by inserting a PCR-amplified fcp1 cDNA fragment into pET21d (Novagen) between the NcoI and XhoI sites. pET-fcp1c-H was constructed by inserting a PCR fragment encoding amino acids 369 to 723 of Fcp1 into pET21b between the NdeI and XhoI sites. pET-tfg3-H was made by inserting a PCR fragment of tfg3 cDNA into pET21b between the NdeI and XhoI sites. To construct pET-GST-fcp1, a PCR fragment encoding glutathione S-transferase (GST) with NdeI and ApaI sites at the 5" and 3" termini, respectively, and an ApaI-BamHI fcp1 fragment from the pREP-f-fcp1 were ligated into pET-21b between the NdeI and BamHI sites. pET-GST-rpb4/7 was created from pET-rpb7 and pET-GST-rpb4. To prepare pET-rpb7, the rpb7 cDNA was cloned into pET21b between the NdeI and BamHI sites. To prepare pET-GST-rpb4, the GST-rpb4 fusion gene was created on a vector and transferred into pET21b between the XbaI and BamHI sites. The shorter SphI-BamHI fragment of pET-GST-rpb4 was inserted between the SphI and BglII sites of pET-Rpb7 to make pET-GST-rpb4/7. pET-rpb4/7-H was described previously as pET-Sp4/Sp7CH (60).
Recombinant proteins.
Plasmids pET-fcp1-H, pET-fcp1c-H, pET-tfg3-H, and pET-rpb4/7-H were used for expression of His-tagged proteins Fcp1-H, Fcp1c-H, Tfg3-H, and Rpb4/7-H, respectively, whereas pET-GST-fcp1 and pET-GST-rpb4/7 were used for GST fusion proteins GST-Fcp1 and GST-Rpb4/7, respectively. Escherichia coli BL21(DE3) was transformed with each plasmid, and protein expression was induced as described previously (60). The harvested cells were lysed in five times the cell volume of 0.5 M NaCl-buffer A (50 mM Tris-HCl [pH 8.0], 10% glycerol, 0.5 mM EDTA, 1 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride [PMSF]) containing 0.3 mg of lysozyme per ml for 20 min and centrifuged at 150,000 × g for 2 h. The supernatants were loaded onto Ni2+-agarose (Qiagen) or glutathione (GSH)-Sepharose 4B (Pharmacia) columns equilibrated with the 0.5 M NaCl-buffer A, and the columns were washed with the same buffer. The Ni2+ columns were washed with 0.1 M NaCl-buffer A containing 20 mM imidazole, and the proteins were eluted with the same buffer containing 200 mM imidazole. The GSH columns were washed with 0.1 M NaCl-buffer A, and the proteins were eluted with the same buffer containing 20 mM GSH. The eluted proteins were purified by high-pressure liquid chromatography on a DEAE-5PW column (Tosoh) with 0.1 to 0.3 M NaCl gradient elution in buffer A. GST protein for control experiments was expressed in E. coli JM109 transformed with pGEX-5X-1 (Pharmacia) and purified on a GSH-Sepharose column.
S. pombe strains.
JY741 (h− ade6-M216 ura4-D18 leu1) and JY746 (h+ ade6-M210 ura4-D18 leu1) are the parental strains used in this work. JY741/f-rpb3 was constructed by two-step gene replacement using pUC-f-rpb3-ura4 as described (31). JY741/f-fcp1 was constructed by one-step gene replacement in which the sequence corresponding to nt +2263 to +2913 of fcp1 in pBS-f-fcp1-ura4 was PCR amplified and used for transformation. To generate the fcp1+/fcp1::ura4+ diploid strain, a fragment corresponding to nt −435 to +2623 of the fcp1 sequence in pBS-fcp1::ura4 was PCR amplified and used for transformation of the diploid strain generated by mating JY741 and JY746. To construct the thiamine-dependent rpb4 shut-off strain JY741/f-rpb3±rpb4, strain JY741/f-rpb3 was transformed with pREP81-rpb4, and the rpb4 gene on the chromosome was disrupted as described (60).
Antibodies and Western blotting.
Anti-Fcp1 and anti-Tfg3 antibodies were raised in rabbits immunized with purified Fcp1c-H and Tfg3-H, respectively. Antibodies against pol II subunits have been described previously (26, 60). Anti-phosphorylated CTD MAb H5 (8) was from Babco. Quantitative Western blotting was carried out as described (33). For Fcp1 quantitation, the GST-Fcp1 protein was used as a standard.
Purification of FLAG-tagged complex.
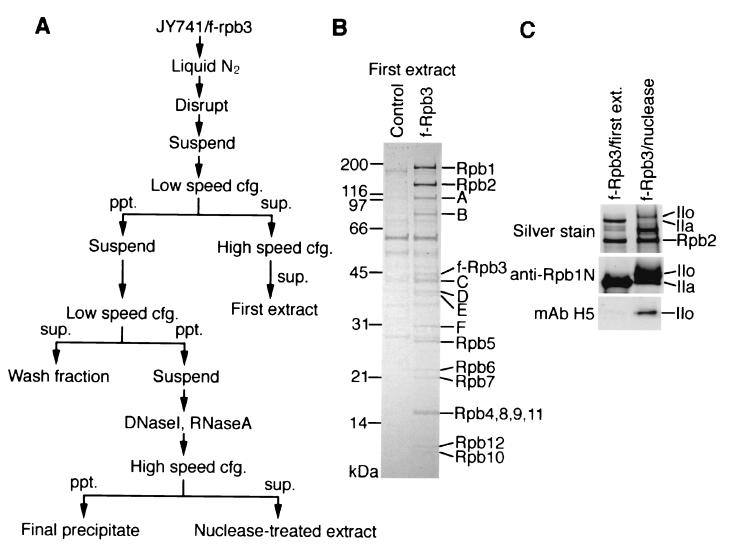
JY741/f-rpb3 or the JY741/f-fcp1 strain was cultured in YE medium (48) containing 50 μg each (of adenine and uracil) per ml at 30°C. Cells were harvested at the exponential phase, frozen in liquid N2, and disrupted with a Cryopress (Microtech Nichion). The cell powder was suspended in one times the cell weight of buffer B (100 mM potassium acetate, 100 mM Tris-acetate [pH 7.8], 20% glycerol, 1 mM EDTA, 1 mM EGTA, 2 mM DTT, 0.4% Nonidet P-40, 1 mM PMSF) containing a proteinase inhibitor mixture (PIM) (31) and centrifuged at 15,000 × g for 10 min. The soluble fraction was centrifuged at 100,000 × g for 2 h, and the resulting supernatant was designated the first extract.
The pellet of the first centrifugation was suspended in one times the cell weight of buffer C (50 mM potassium acetate, 50 mM Tris-acetate [pH 7.8], 10% glycerol, 0.5 mM EDTA, 0.5 mM EGTA, 1 mM DTT, 0.2% Nonidet P-40, 0.5 mM PMSF) containing the PIM and centrifuged at 15,000 × g for 10 min. The supernatant was designated the wash fraction, while the pellet was suspended in buffer D (50 mM potassium acetate, 50 mM Tris-acetate [pH 7.8], 10% glycerol, 10 mM MnCl2, 1 mM DTT, 0.2% Nonidet P-40, 0.5 mM PMSF) containing the PIM and 10 μg each (of DNase I and RNase A) per ml. After incubation at 30°C for 20 min, the digested pellet was centrifuged at 100,000 × g for 2 h, and the supernatant and the pellet were designated the nuclease-treated extract and the final precipitate, respectively. For 1 ml of the first extract and the nuclease-treated extract, 20 μl of M2-agarose (Sigma) equilibrated with buffer C was added and gently mixed for 2 h. The M2-agarose was washed four times with 1 ml of buffer C, and the bound proteins were eluted with one times the resin volume of buffer C containing 100 μg of FLAG-peptide (Sigma) per ml.
Strain JY741/f-rpb3±rpb4 was cultured in Edinburgh minimal medium (EMM) (48) containing 150 μg of adenine per ml. When the culture reached 106 cells/ml, 30 μg of thiamine per ml was added, and at 0, 6, 12, and 18 h after this, cell aliquots were harvested. FLAG-tagged complex was isolated from the first extracts as above.
Purification of f-pol II.
JY741/f-rpb3 was cultured, harvested, and disrupted as above. The cell powder was suspended in two times the cell weight of buffer E [0.15 M (NH4)2SO4, 75 mM Tris-HCl (pH 8.0), 15% glycerol, 0.15 mM EDTA, 1.5 mM DTT, 0.75 mM PMSF] containing the PIM, sonicated, and centrifuged at 18,000 × g for 20 min. The supernatant was diluted fourfold with buffer F [0.1 M (NH4)2SO4, 50 mM Tris-HCl (pH 8.0), 10% glycerol, 0.1 mM EDTA, 1 mM DTT, 0.5 mM PMSF], and 0.1% polyethyleneimine was added. After incubation for 1 h, the precipitate was collected by centrifugation at 18,000 × g for 20 min and extracted by two times the cell weight of buffer G [0.2 M (NH4)2SO4, 50 mM Tris-HCl (pH 8.0), 10% glycerol, 0.1 mM EDTA, 1 mM DTT, 0.5 mM PMSF]. After centrifugation at 18,000 × g for 20 min, FLAG-pol II (f-pol II) was isolated from the supernatant with M2-agarose in buffer G as above.
Mass spectrometry.
Proteins in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels were alkylated with iodoacetamide, digested with trypsin or endoproteinase Glu-C, and extracted as described (27). Peptide masses were measured with a matrix-assisted laser desorption/ionization time of flight mass spectrometer Voyager (PerSeptive Biosystems) using α-cyano-4-hydroxycinnamic acid as the matrix and provided calibration mixture 2. The mass spectra were fitted to databases by the program MS-Fit (University of California, San Francisco).
Gel filtration chromatography.
Proteins were loaded onto a Superose 6 PC3.2/30 column in buffer C without PMSF using Smart System (Pharmacia) at a flow rate of 40 μl/min. Fractions of 50 μl were collected, beginning 15 min after the injection.
CTD-phosphatase assay.
Various amounts of recombinant Fcp1-H or Fcp1c-H protein were incubated with approximately 50 ng of the purified pol II preparation containing both IIO and IIA forms in buffer C with 10 mM MgCl2 for 60 min at 30°C. Dephosphorylation was analyzed by Western blotting with anti-Rpb1 N-terminal domain (NTD) antibody and anti-phosphorylated CTD MAb H5.
Effect of Fcp1 overproduction.
JY741(pREP41) and JY741(pREP-f-fcp1) were cultured at 30°C in EMM (48) containing 150 μg (each) of adenine and uracil per ml, and cell lysates for Western blotting were prepared as described (33).
Tetrad analysis.
The diploid strain with the genotype ura4-D18/ura4-D18 fcp1+/fcp1::ura4+ was grown on a malt extract plate (48) for 2 days at 27°C and then streaked on a YE plate (48) containing 150 μg (each) of adenine and uracil per ml. The asci formed were isolated with a manipulator, and then the plate was incubated for several hours at 30°C. The spores that appeared were dissected, and the plate was incubated for 3 days at 30°C.
Chemical cross-linking.
Protein-protein cross-linking with 2-iminothiolane (ITL) was carried out as described (26). The cross-linked and non-cross-linked complexes were loaded on the same SDS-PAGE gel with two wide wells. The proteins were electrotransferred onto a membrane. For separate immunostaining, the membrane was set in an apparatus Miniblot (Immunetics), which divides the membrane into a number of vertical slots, and each slot was immunostained with one of the antibodies.
GST pulldown assay.
For GST-Fcp1 pulldown assay of pol II, 10 pmol of nondegraded GST-Fcp1 or GST protein was mixed with 250 ng of pol II in buffer C and incubated at 30°C for 4 h. Then, 5 μl of GSH-Sepharose equilibrated with buffer C was added, and the mixture was incubated on ice for 1 h. The resin was washed four times with 1 ml of buffer C, and the proteins were eluted by SDS-PAGE loading buffer. For GST-Rpb4/7 pulldown assay of Fcp1-H, 10 μg of GST-Rpb4/7 or GST was mixed with 2 μg of Fcp1-H in buffer C and processed as above. For GST-Fcp1 pulldown assay of Rpb4/7-H, 10 pmol of nondegraded GST-Fcp1 or GST protein was mixed with 10 pmol of Rpb4/7-H and processed similarly. The contents of nondegraded protein were measured by Sypro Orange (Molecular Probes) staining intensities of SDS-PAGE gels with bovine serum albumin as a standard.
Rpb4/7 affinity chromatography of whole-cell extract.
The purified Rpb4/7-H was coupled to 1 ml of a HiTrap N-hydroxysuccinimide-activated column (Pharmacia) per the manufacturer's instruction. JY741 whole-cell extract was prepared in 0.2 M potassium acetate buffer H (50 mM HEPES-KOH [pH 7.9], 20% glycerol, 0.5 mM EDTA, 0.5 mM EGTA, 2.5 mM DTT, 0.5 M PMSF) containing the PIM by a similar method as employed for the first extract preparation. After dialysis to reduce the potassium acetate concentration to 0.1 M, the extract was loaded onto the Rpb4/7 column equilibrated with the same buffer. The column was washed with 1 M NaCl-buffer H, and the proteins were eluted with 4 M urea-buffer H.
RESULTS
Isolation and identification of pol II-associating proteins from the FLAG-tagged rpb3 strain.
In order to set up a simple but effective isolation system of protein complexes containing pol II, S. pombe strain JY741/f-rpb3, which carries the modified chromosomal rpb3 gene encoding Flag-Rpb3 (f-Rpb3), was constructed. This strain showed no particular phenotype other than relatively slow growth (data not shown). Low-salt buffer was used for extraction of pol II complexes to preserve protein-protein interactions, although the yield of protein extraction was decreased. After anti-FLAG M2-agarose affinity chromatography of the first extract (Fig. 1A and see Materials and Methods), a pol II complex containing f-Rpb3 was isolated. SDS-PAGE in parallel with the control fraction revealed that this complex contained all 12 pol II subunits and six additional proteins (A to F in Fig. 1B). The non-pol II proteins in the complex were identified by mass fingerprinting, and the results are summarized in Table 1. Protein A was identified as an S. pombe homologue of the TFIIF-interacting CTD-phosphatase Fcp1, which has been identified in S. cerevisiae (2, 34) and humans (1, 13).
FIG. 1.
Isolation of the Fcp1/TFIIF/pol IIA complex from strain JY741/f-rpb3. (A) Scheme for extract preparation. The first extract and the nuclease-treated extract were subjected to anti-FLAG M2-agarose affinity chromatography. (B) Proteins isolated with M2-agarose from the first extract (see Materials and Methods) of JY741/f-rpb3 (f-Rpb3 lane) or nontagged strain JY741 (control lane) were separated by SDS-10 to 20% PAGE. The gel was stained with Coomassie brilliant blue (CBB). Non-pol II protein bands that appeared in the f-Rpb3 lane but not in the control lane are designated A to F. Positions of molecular weight markers are indicated on the left. (C) Proteins isolated with M2-agarose from the first extract or the nuclease-treated extract (see Materials and Methods) of JY741/f-rpb3 were separated in SDS-6% PAGE and silver-stained or processed for Western blotting with anti-Rpb1 NTD antibody or anti-phosphorylated CTD MAb H5. Positions of the IIo and IIa forms of Rpb1 and Rpb2 are indicated on the right.
TABLE 1.
Identification of pol II-associated proteinsa
| Band | Size (kDa)
|
Accession no.b | Protein species | MOWSE scorec | |
|---|---|---|---|---|---|
| SDS-PAGE | Calculated | ||||
| A | 105 | 82.0 | 9588462 | Fcp1 | 3.63 × 106 |
| B | 75 | 55.9 | 7493059 | TFIIFα | 2.62 × 105 |
| C | 41 | 34.6 | 7493495 | TFIIFβ | 1.01 × 105 |
| D | 36 | 35.9 | 2494642 | Glyceraldehyde 3-phosphate dehydrogenase 1 | 4.89 × 103 |
| E | 35 | 35.9 | 2494642 | Glyceraldehyde 3-phosphate dehydrogenase 1 | 4.04 × 106 |
| F | 30 | 27.6 | 7493063 | TFIIF small subunit (Tfg3) | 2.95 × 105 |
The protein bands are marked in Fig. 1.
NCBI GI accession number for protein identification.
MOWSE score in MS-fit peptide mass fingerprinting.
To characterize the S. pombe Fcp1 in detail, we cloned the cDNA from S. pombe. The cDNA sequence agreed completely with the sequence of the predicted Fcp1 coding region in the database. The amino acid identity of the S. pombe Fcp1 with the S. cerevisiae and the human homologues is 36 and 27%, respectively (Fig. 2). The His-tagged recombinant Fcp1-H protein (Fig. 3C and also 7B) expressed from the cDNA showed almost the same mobility in SDS-PAGE as protein A (data not shown), indicating that the cloned sequence was full length and that Fcp1 has a lower mobility in SDS-PAGE than that expected from the molecular weight (Table 1). The antibody raised against the recombinant Fcp1 reacted with protein A (see Fig. 3B), but did not react with any protein shown in the control lane in Fig. 1B (data not shown). Because of the high MOWSE scores in MS-fit, proteins B and C were identified as the homologues of α and β subunits of TFIIF, respectively, which are known as pol II-associating proteins RAP74 and RAP30 (64). Again, their mobilities in SDS-PAGE were slightly different from those estimated from the molecular masses (Table 1).
FIG. 2.
Sequence comparison of Fcp1. Protein sequences of S. pombe (accession no. CAC00553), S. cerevisiae (NP_014004), and H. sapiens (AAD42088) Fcp1 were aligned using ClustalW. Black boxes, identity; grey boxes, similarity. Domain structure is indicated according to Kobor et al. (34, 35). Black line, catalytic Fcp1 homology domain (FCPH); grey line, BRCA1 C-terminal domain (BRCT); dashed line, TFIIF- and TFIIB-interacting region; asterisks, motif characteristic of a family of small-molecule phosphotransferases (15).
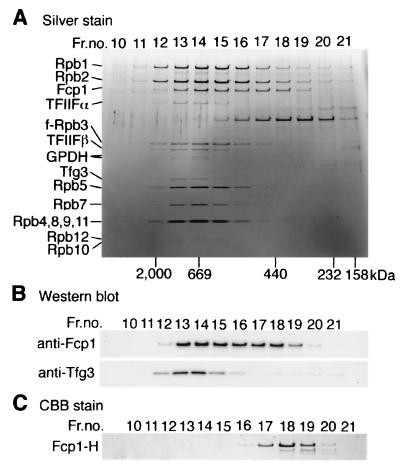
FIG. 3.
Gel filtration chromatography of the Fcp1/TFIIF/pol IIA complex. (A) The proteins isolated with M2-agarose from the first extract of JY741/f-rpb3 were loaded onto a Superose-6 PC3.2/30 column. Aliquots of the eluted fractions were analyzed by SDS-10 to 20% PAGE, and the gel was silver stained. Components of the complex are indicated on the left. GPDH, glyceraldehyde 3-phosphate dehydrogenase 1. Elution positions of molecular mass standards are indicated on the bottom. (B) The gel was processed for Western blotting with anti-Fcp1 and anti-Tfg3 antibodies. (C) The purified recombinant Fcp1-H was fractionated by gel filtration chromatography. The fractions were analyzed by SDS-PAGE, and the gel was stained with CBB.
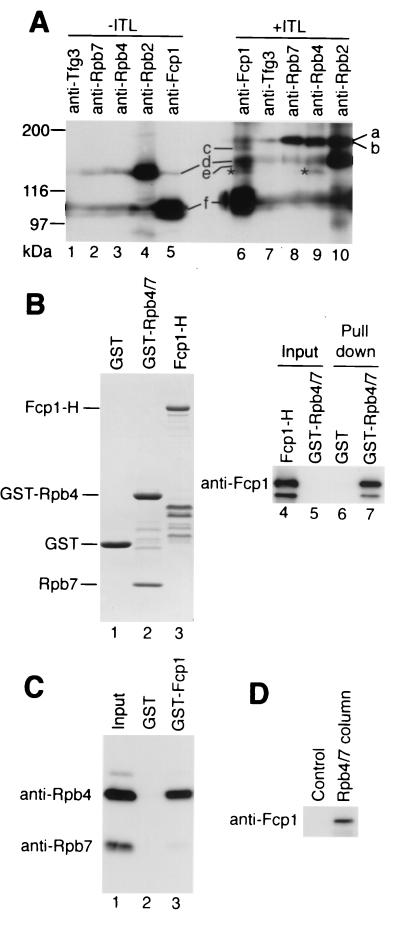
FIG. 7.
Interaction of Fcp1 with Rpb4. (A) ITL cross-linking of the Fcp1/TFIIF/pol IIA complex. The complex without (lanes 1 to 5) and with (lanes 6 to 10) ITL cross-linking was separated by SDS-6% PAGE, and the gel was subjected to Western blotting with anti-Fcp1, anti-Tfg3, anti-Rpb2, anti-Rpb4, and anti-Rpb7 antibodies. Each lane was immunostained with one of these antibodies. A portion of the membrane, including Fcp1-Rpb4 cross-link, is shown. No other Fcp1 cross-link was detected with nine other species of anti-pol II subunit antibodies. Positions of molecular size standards are indicated on the left. The asterisks indicate the Fcp1-Rpb4 cross-link. The assignments of other bands are: a, Rpb2-Rpb7 cross-link; b, Rpb2-Rpb4 cross-link; c, presumed Fcp1-TFIIFα cross-link; d, Rpb2; e, presumed Fcp1-TFIIFβ cross-link; f, Fcp1. The mobilities of Fcp1 and Rpb2 correspond to 105 and 150 kDa, respectively, but they shifted upward after ITL treatment because of intramolecular cross-linking and/or binding of ITL. Because of high protein dosages, all the antibodies and/or the second antibody showed nonspecific cross-reaction against Fcp1 and Rpb2. (B) Interaction between GST-Rpb4/7 and Fcp1-H. (Lanes 1 to 3) SDS-PAGE of purified GST, GST-Rpb4/7, and Fcp1-H proteins used in the GST pulldown assay. The gel was stained with CBB. Positions of the full-length proteins are indicated on the left, and other bands are degradation products. (Lanes 4 to 7) Fcp1-H was incubated with GST or GST-Rpb4/7. The complexes formed were isolated with GSH-Sepharose. The resin-bound fraction (lanes 6 and 7) and 10% of the proteins used (lanes 4 and 5) were subjected to Western blotting with anti-Fcp1 antibody. (C) Interaction between GST-Fcp1 and Rpb4/7-H. Rpb4/7-H was incubated with GST or GST-Fcp1 and processed as above. The anti-Rpb4 and anti-Rpb7 antibodies were used for the Western blotting. Lane 1 shows 10% of the input Rpb4/7-H. (D) Rpb4/7 affinity chromatography of the whole-cell extract. The whole-cell extract prepared from JY741 was loaded onto an Rpb4/7-H-coupled or a control column. After washing the columns, the proteins bound were eluted with urea-buffer, and the eluted proteins were subjected to Western blotting with anti-Fcp1 antibody.
Protein F was identified as S. pombe TFIIF small subunit Tfg3. In S. cerevisiae, Tfg3 (25) is also known as Anc1 (73), TAFII30 (24), SWI/SNF29 (9), and one of the subunits of NuA3 (28). The human homologues, ENL and AF-9, known as leukemogenic proteins (73), are, however, not TFIIF subunits. The S. pombe tfg3 cDNA was cloned, and the sequence agreed with the tfg3 coding sequence in the database. The antibody raised against the recombinant Tfg3 reacted with protein F (see Fig. 3B), but did not react with any protein shown in the control lane in Fig. 1B (data not shown). Mass spectrometry indicated that proteins D and E have sequences of glyceraldehyde 3-phosphate dehydrogenase 1. At present we assume that its pol II binding was fortuitous.
The pol II complex isolated as above should not have been engaged in transcription, because it was extracted easily with the low-salt buffer. To isolate the engaged pol II, the pellet of first extraction was washed and then extracted after digestion of nucleic acids with DNase I and RNase A. The pol II purified with M2-agarose from the nuclease-treated extract contained an Rpb1 subunit that migrated in SDS-PAGE slower than that in the nonengaged complex (Fig. 1C). We assigned this subunit as the phosphorylated IIo form, because the anti-phosphorylated CTD MAb H5 (8) reacted with it but not with that isolated from the first extract (Fig. 1C). The nuclease-treated pol II contained stoichiometric amounts of all other pol II subunits (data not shown). Thus, we concluded that the nonengaged Fcp1/TFIIF/pol II complex contained the CTD-nonphosphorylated IIa form of Rpb1.
To examine the association of these proteins with pol II, the eluted fraction from M2-agarose was subjected to Superose-6 gel filtration (Fig. 3A). All the components identified were coeluted with the pol II subunits in fractions 13 to 15, indicating that these components indeed formed a large complex. However, the Fcp1 peak formed a shoulder at fractions 17 to 19. To examine whether this represents the dissociated form of Fcp1, the purified Fcp1-H protein was fractionated by the same procedure (Fig. 3C). It was recovered in fractions 17 to 19, presumably as dimers (13). Thus, we concluded that a fraction of Fcp1 was dissociated from the Fcp1/TFIIF/pol II complex after elution from M2-agarose. Likewise, some dissociated components of TFIIF such as Tfg3 were also detected in fractions 18 to 21.
We also constructed a strain, JY741/f-fcp1, containing the FLAG-tagged fcp1. The strain showed no particular phenotype. We then tried to isolate Fcp1 complexes by M2 affinity purification as above. As expected, the f-Fcp1/TFIIF/pol II complex was isolated from the first extract (Fig. 4A). The recovery of Rpb1 and Fcp1 in each step is summarized in Table 2. The pol II content in the f-Fcp1 complex was estimated to be about one pol II molecule per Fcp1 dimer. The stoichiometry of Fcp1 in this f-Fcp1 complex was more than that in the f-Rpb3 complex. Thus, it appears that only a fraction of pol II is associated with Fcp1. Here we assumed that Fcp1 assembles into the complex as a dimer based on the gel filtration profile of Fcp1-H (see Fig. 3C). In confirmation is the finding that a heterodimer can be reconstituted from the recombinant Fcp1-H and GST-Fcp1 (M. Kimura, unpublished result). However, the possibility that Fcp1 is present as a monomer in the complex and a fraction of f-Fcp1 isolated was not bound to pol II cannot be ruled out.
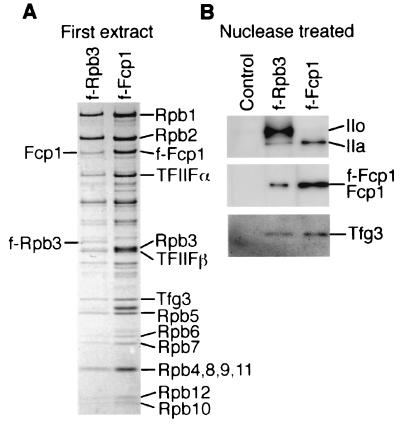
FIG. 4.
Isolation of f-Fcp1 complexes. (A) The Fcp1/TFIIF/pol IIA complex isolated from the first extracts of JY741/f-rpb3 and JY741/f-fcp1 with M2-agarose were separated in SDS-10 to 20% PAGE, and the gel was stained with CBB. (B) The complexes isolated with M2-agarose from the nuclease-treated extract of JY741, JY741/f-rpb3, and JY741/f-fcp1 were separated on SDS-PAGE, and the gel was processed for Western blotting with anti-Rpb1 NTD, anti-Fcp1, and anti-Tfg3 antibodies.
TABLE 2.
Protein yielda
| Fraction | Yield (pmol/g [wet wt] of cells)
|
||||
|---|---|---|---|---|---|
| JY741/f-Rpb3
|
JY741/f-Fcp1
|
||||
| Rpb1 | Fcp1 | Rpb1 | f-Fcp1 | ||
| Supernatant of low-speed centrifugation | 37 | 10 | 33 | 7.6 | |
| First extract | 16 | 9.9 | 16 | 8.0 | |
| M2-agarose-bound fraction | 5.0 | 2.7 | 1.7 | 2.4 | |
| Wash fraction | 5.6 | 1.0 | 3.1 | 0.65 | |
| Nuclease-treated extract | 0.61 | 0.15 | 0.57 | 0.11 | |
| M2-agarose-bound fraction | 0.12 | 0.04 | 0.04 | 0.03 | |
| Final precipitateb | >22 | >7.9 | >11 | >4.6 | |
One gram of cells corresponds to 6 × 109 cells. The purification was repeated using different amounts of cells, and a typical result is presented.
Rpb1 detected in the final precipitate was mostly in the IIo form (data not shown).
The nuclease-treated extract of JY741/f-Fcp1 was also subjected to M2-agarose affinity chromatography, and the protein composition of isolated complexes was compared with that from JY741/f-Rpb3 (Fig. 4B). The species and content of proteins were essentially identical between the f-Rpb3 and the f-Fcp1 complexes. However, the f-Rpb3 complex contained a high level of the IIo form of Rpb1, whereas the Rpb1 in the f-Fcp1 complex was mostly in the IIa form. This indicates that the majority of pol II in the nuclease-treated extract contained the phosphorylated IIo form of Rpb1, but Fcp1 is associated specifically with a minor population of pol II containing the unphosphorylated IIa form.
Functions of S. pombe Fcp1.
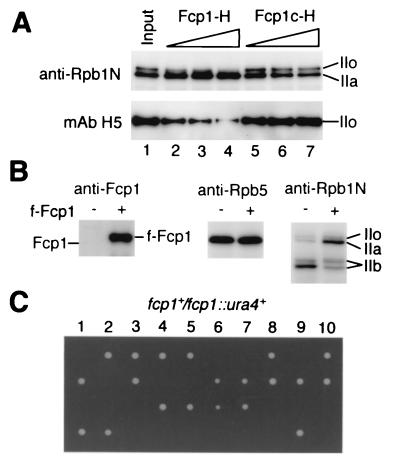
In order to examine whether the identified S. pombe Fcp1 actually has CTD-phosphatase activity, both the recombinant Fcp1-H and the C-terminal fragment, Fcp1c-H, which consists of amino acids 369 to 723 and lacks the phosphotransferase motif (see Fig. 2 for location), were purified by the same procedure and tested for phosphatase activity. One microgram of Fcp1-H hydrolyzed 13 ± 3 nmol of the artificial substrate p-nitrophenyl phosphate (41) into p-nitrophenol per h at 30°C in buffer C containing MgCl2, whereas Fcp1c-H did not hydrolyse it at all, indicating that Fcp1-H (but not any contaminants) had the phosphatase activity. Next we checked the CTD-phosphatase activity. When Fcp1-H and Fcp1c-H were incubated under the same conditions with purified pol II which contained both IIO and IIA forms, the IIo form of Rpb1 was converted into the IIa form by Fcp1-H in a dose-dependent manner (Fig. 5A), whereas Fcp1c-H did not dephosphorylate IIo. Hence, we conclude that S. pombe Fcp1 has CTD-phosphatase activity.
FIG. 5.
In vitro and in vivo functions of Fcp1. (A) Detection of CTD-phosphatase activity in vitro. Approximately 50 ng of the pol II preparation containing both pol IIO and IIA was incubated at 30°C for 1 h with 0 (lane1), 0.25 (lanes 2 and 5), 0.5 (lanes 3 and 6), or 1 μg (lanes 4 and 7) of recombinant Fcp1-H (lanes 2 to 4) or Fcp1c-H (lanes 5 to 7), which is a C-terminal fragment of Fcp1 lacking the catalytic domain, and subjected to SDS-PAGE followed by Western blotting with anti-Rpb1 NTD antibody and anti-phosphorylated CTD MAb H5. The positions of the IIo and IIa forms of Rpb1 are indicated. (B) Effect of Fcp1 overexpression on CTD phosphorylation in vivo. Whole-cell lysates of JY741(pREP41), indicated as f-Fcp1 minus (−), and JY741(pREP-f-fcp1), indicated as plus (+), were subjected to SDS-PAGE followed by Western blotting with anti-Fcp1, anti-Rpb5, and anti-Rpb1 NTD antibodies. (C) Tetrad analysis of the ura4-D18/ura4-D18 fcp1+/fcp1::ura4+ diploid strain of S. pombe. Ten asci were dissected, and spores from the same tetrad are arrayed vertically.
Next we examined possible effects in vivo of overexpression of Fcp1. f-Fcp1 was overexpressed in JY741 with plasmid pREP41-f-fcp1, and the whole-cell lysate was analyzed by Western blotting (Fig. 5B). The expression level of f-Fcp1 was very high (left panel), and it caused slow growth (data not shown). Although the concentration of pol II subunits in the overexpressed strain was not affected (center panel), the level of the IIa form of Rpb1 was higher than in the control strain (right panel). When the cells are grown in minimal medium and disrupted by the glass beads method, the IIb form of Rpb1, which lacks the CTD, is generated due to proteolytic cleavage (33). The generation of the IIb form was much less in the Fcp1-overexpressing cell lysate than in the control lysate (right panel), suggesting that Fcp1 protects Rpb1 from protease accession. These observations also support the formation of the Fcp1/pol II complex.
Tetrad analysis of an S. pombe diploid strain, which carries ura4-D18/ura4-D18 fcp1+/fcp1::ura4+, produced two viable and two nonviable spores from each of the 10 asci dissected (Fig. 5C). The viable progeny showed the ura− phenotype (data not shown). These indicated that the fcp1 gene is essential for cell viability in S. pombe, as was reported for S. cerevisiae (2).
Fcp1-pol II direct interaction.
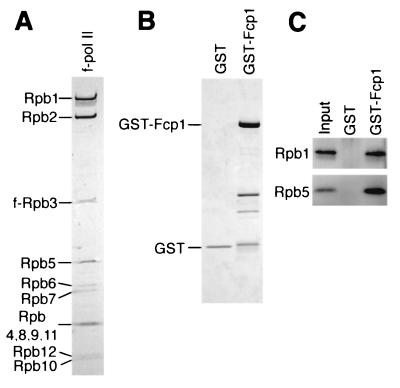
Next we examined the direct interaction between Fcp1 and pol II from S. pombe. f-pol II purified through polyethyleneimine precipitation and the M2-agarose affinity method in the presence of a high concentration of (NH4)2SO4 did not contain any detectable TFIIF subunits (Fig. 6A). The absence of Tfg3 was further confirmed by immunoblotting with the anti-Tfg3 antibody (data not shown). The recombinant GST-Fcp1 or GST (Fig. 6B) was incubated with the f-pol II, and the complexes formed were isolated with GSH-Sepharose (Fig. 6C). The result clearly indicated the direct interaction between Fcp1 and pol II.
FIG. 6.
Direct interaction between Fcp1 and pol II. (A) The f-pol II isolated from strain JY741/f-rpb3 by (NH4)2SO4 extraction, polyethyleneimine precipitation, and M2 affinity chromatography was subjected to SDS-10 to 20% PAGE. The gel was stained with CBB. The pol II subunits are indicated on the left. (B) Purified GST and GST-Fcp1 proteins were separated by SDS-PAGE. The gel was stained with CBB. Migration positions of the full-length proteins are indicated. Fast-migrating bands in the GST-Fcp1 lane are degraded proteins. (C) GST pulldown assay. f-pol II (250 ng) was incubated with 10 pmol of GST or GST-Fcp1, and the complexes formed were isolated with GSH-Sepharose. The resin-bound fractions were subjected to SDS-PAGE followed by Western blotting with anti-Rpb1 NTD and anti-Rpb5 antibodies. The input shows 10% of the f-pol II used.
Fcp1 interacts with Rpb4.
To identify the Fcp1-interacting subunit of pol II, the isolated Fcp1/TFIIF/pol II complex was subjected to a chemical cross-linking experiment using ITL (26). Both untreated complex (Fig. 7A, lanes 1 to 5) and cross-linked complex (lanes 6 to 10) were fractionated by SDS-PAGE, and each lane was immunostained with one of the five species of antibody against Fcp1, Rpb2, Rpb4, Rpb7, and Tfg3. The bands d and f of the untreated complex with mobilities of 150 and 105 kDa (lanes 4 and 5), respectively, correspond to Rpb2 and Fcp1. The weak signals at the same positions in other lanes represent nonspecific cross-reaction owing to high protein dosage.
In the analysis of the cross-linked complex (lanes 6 to 10), the non-cross-linked Rpb2 (band d) and Fcp1 (band f) migrated more slowly than the corresponding proteins in the non-cross-linked sample (lanes 1 to 5) owing to intramolecular cross-linking and/or binding of the reagent ITL. As the molecular sizes of pol II small subunits range between 7.2 and 34 kDa, if Fcp1 is cross-linked to one of the small subunits, the cross-linked product should migrate between Rpb2 and Fcp1, and it should react with both anti-Fcp1 and antisubunit antibodies. The band between Rpb2 and Fcp1, marked by asterisks in lanes 6 and 8, was immunostained with both anti-Fcp1 and anti-Rpb4 antibodies. We concluded that this band represents a cross-linked product between Fcp1 and Rpb4. No other Fcp1 cross-link was detected within that area, as checked with nine species of other antisubunit antibodies (data not shown). Cross-linking, if any, between the two large pol II subunits and Fcp1 could hardly be detected within the resolution limit employed (data not shown). Likewise, formation of cross-link products including more than three components cannot be expected due to low cross-linking efficiency. Assignments of other bands are described in the figure legend.
Since ITL has a length of 14 Å between the reactive sites for cross-linking, cross-linking does not always represent a protein-protein direct interaction. In order to detect direct binding between Fcp1 and Rpb4, we carried out a GST pulldown assay using purified recombinant proteins. The Rpb4 protein was prepared as heterodimers with Rpb7 because of the insolubility of overexpressed Rpb4. GST-Rpb4/7 and Rpb4/7-H (60) could be purified from the soluble fractions. The GST-Rpb4/7 preparation contained no protein immunoreactive with anti-Fcp1 antibody (Fig. 7B, lane 5).
Fcp1-H was incubated with GST-Rpb4/7 or GST, and the complexes formed were isolated with GSH-Sepharose. Fcp1-H bound only to GST-Rpb4/7 (lanes 6 and 7), as revealed by Western blotting of the isolated fractions. When Rpb4/7-H was incubated with GST-Fcp1 or GST (see Fig. 6B) and processed similarly, Rpb4/7-H bound only to GST-Fcp1 (Fig. 7C). The recovery of Rpb7-H in the GSH affinity fraction was less than that of Rpb4. This indicates partial dissociation of Rpb7-H and specific binding of GST-Fcp1 to Rpb4. Fcp1 may induce the dissociation of the Rpb4-Rpb7 complex. A stoichiometric amount of Rpb7 was, however, contained in the f-Fcp1/TFIIF/pol II complex, in agreement with the finding that Rpb7 binds to pol II independently of Rpb4 (Fig. 8C). Another possibility is that the dissociation of Rpb4/7-H was spontaneous and independent of Fcp1.
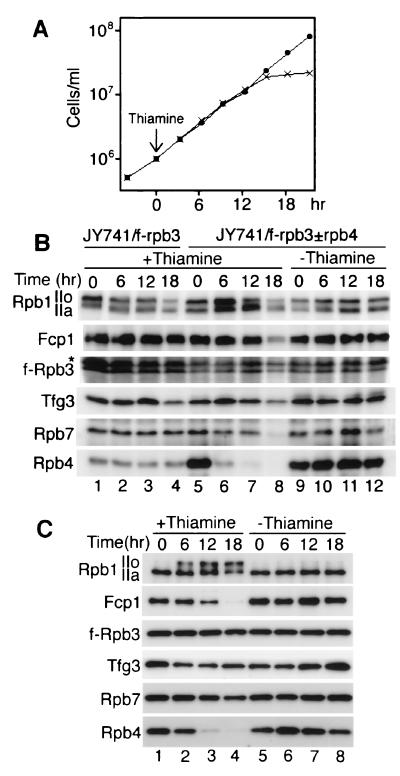
FIG. 8.
Fcp1/TFIIF/pol II complex isolated from the rpb4 shut-off strain. (A) Growth curve of the thiamine-dependent rpb4 shut-off strain JY741/f-rpb3±rpb4. The strain contains the f-rpb3 gene and a plasmid carrying the rpb4 cDNA under a thiamine-repressible promoter, and the chromosomal rpb4 gene was disrupted. The cells were cultured in EMM medium containing 150 μg of adenine per ml at 30°C. When the cell density was 106 cells/ml, 30 μg of thiamine per ml was added. The horizontal axis indicates the time after thiamine addition. ×, plus thiamine; •, without thiamine. (B) Intracellular contents of Fcp1, Tfg3, and pol II subunits. The cells were harvested at the indicated time from cultures of JY741/f-rpb3 with thiamine, JY741/f-rpb3±rpb4 with thiamine, and JY741/f-rpb3±rpb4 without thiamine. Whole-cell lysates were processed for Western blotting with anti-Rpb1 NTD, anti-Fcp1, anti-Rpb3, anti-Tfg3, anti-Rpb7, and anti-Rpb4 antibodies. The same amount of total protein was loaded. An asterisk indicates a nonspecific cross-reaction. (C) Fcp1/TFIIF/pol II complex isolated from the rpb4 shut-off strain. The first extracts were prepared from the same JY741/f-rpb3±rpb4 cells as in panel B. The f-Rpb3 complexes isolated from them were processed for Western blotting as in panel B. Samples containing the same amount of Rpb3 were loaded.
To examine binding of native Fcp1 to the recombinant Rpb4/7-H, whole-cell extract of JY741 was chromatographed on an Rpb4/7-H-coupled column. Fcp1 was detected in the column-bound fraction (Fig. 7D), indicating that the extract contained the unassembled form of Fcp1 that can bind to Rpb4/7-H.
Involvement of Rpb4 in Fcp1/TFIIF/pol II complex formation in vivo.
To examine the role of the Fcp1-Rpb4 interaction in the formation in vivo of the Fcp1/TFIIF/pol II complex, we constructed an S. pombe strain in which rpb4 expression can be shut off by the addition of thiamine. Plasmid pREP81-rpb4, which carries the rpb4 cDNA under a thiamine-repressible promoter, was introduced into strain JY741/f-rpb3, and then the chromosomal rpb4 gene was disrupted. The resultant strain was designated JY741/f-rpb3±rpb4. Growth curves of the strain in EMM medium with and without thiamine are shown in Fig. 8A. Thiamine was added when the culture reached 106 cells/ml, and cell growth stopped after three to four cycles of cell division. This agrees with the observation that the rpb4 is essential for cell growth in S. pombe (60).
After the shut-off of Rpb4 synthesis, the intracellular contents of Fcp1, Tfg3, and pol II subunits were determined by Western blotting (Fig. 8B). The cells were harvested at various times after thiamine addition and disrupted by the freezing method to prevent protein degradation (see Fig. 5B). Under the thiamine-minus condition, strain JY741/f-rpb3±rpb4 expressed more Rpb4 (lanes 5 and 9 to 12) than the parental strain (lanes 1 to 4). After the addition of thiamine, however, Rpb4 was reduced gradually (lanes 6 to 8), and at 6 h, when the cells were still growing, the Rpb4 content was reduced to almost the same level as that of the parental strain (compare lanes 6 and 1 to 4). In parallel with the reduction of Rpb4, both the IIo and IIa forms of Rpb1 were increased by an unknown mechanism. At 12 h, when the cells stopped growing, less Rpb4 was detected, and remarkably, the IIo form began to decrease but IIa remained at a high level (lane 7). The decrease in the IIo form might be due to inhibition of transcription initiation because of the lack of Rpb4, or it could be due to a decreased interaction between pol II and a CTD-kinase. The levels of other pol II subunits and Fcp1 remained constant up to 12 h. At 18 h, when cell growth stopped, Rpb4 was hardly detected and other proteins were also degraded (lane 8).
The Fcp1/TFIIF/pol II complex containing f-Rpb3 was isolated from the first extracts of JY741/f-rpb3±rpb4, and the protein components were analyzed by Western blotting (Fig. 8C). The Rpb4 in the complex was reduced gradually after the addition of thiamine, with concomitant reduction of Fcp1 (lanes 1 to 4). In the absence of thiamine, the Rpb1 in the complex was the IIa form (lanes 1 and 5 to 8). However, 6 h after thiamine addition, the IIo form increased, and at 18 h, the level of IIo was higher than that of IIa (lanes 1 to 4). The contents of other components such as Rpb7 and Tfg3 remained almost constant.
In contrast to the case with S. cerevisiae (16), the S. pombe Rpb7 seems to be associated with pol II in the absence of Rpb4. The complex isolated from the cells cultured without thiamine did not show any change in the components during this period (lanes 5 to 8). All these observations, especially the one that both Fcp1 binding to pol II and CTD-dephosphorylation were defective within the period when the cells were growing normally (6 to 12 h after thiamine addition), indicate that Rpb4 is important for the assembly in vivo of Fcp1 into the pol II complex and the dephosphorylation of CTD.
DISCUSSION
We have isolated the Fcp1/TFIIF/pol IIA complex from S. pombe for the first time. This type of complex has not been reported from other organisms. The successful isolation resulted from the use of low-salt buffer throughout the purification and the simple method for complex isolation. Fcp1, however, was not contained in the S. cerevisiae pol II complex isolated with the anti-CTD antibody (71). This discrepancy might have originated from the use of different antibodies. Otherwise, the S. cerevisiae Fcp1 does not form a complex with pol II.
We used the fission yeast S. pombe because the transcriptional mechanism is more similar to that of higher eukaryotes (40, 55, 60), all the pol II subunit genes have been cloned (4, 5, 29, 47, 58, 59, 60, 63, 64), and the subunit-subunit interactions have been well characterized (31, 32, 33). A large complex of pol II holoenzyme has been isolated from S. pombe (66). A similar holoenzyme could not be purified, at least by the method employed. It might not be extracted, or the FLAG tag on the N terminus of Rpb3 or C terminus of Fcp1 might be masked by other components in the holoenzyme. In fact, a large amount of pol II remained unextracted in the final precipitates (see Table 2). Large amounts of Fcp1 were also left unsolved in the final precipitates, suggesting the existence of other Fcp1-containing complexes.
The stoichiometry of Fcp1 in the Rpb3-tagged complex was less than those of pol II subunits, indicating that Fcp1 is associated with a portion of pol II molecules. The total number of Fcp1 molecules in strain JY741, which contains 10,000 molecules/cell of pol II at exponential phase in a rich medium (33), was measured as 3,500 ± 300 molecules/cell by quantitative Western blotting (data not shown). The amount of Fcp1-asociated pol II complex herein determined is consistent with the intracellular level of Fcp1. This ratio supports the binding of Fcp1 to pol II in a certain stage of the transcription cycle for the processive reaction of CTD dephosphorylation.
The association of Fcp1 with pol II that is not engaged in transcription may prevent phosphorylation of the CTD by CTD-kinases. The pol II isolated from the nuclease-treated extract was also associated with a small amount of Fcp1. Most of the nuclease-treated pol II was the pol IIO form, but Fcp1 was found to be associated only with pol IIA. This agrees with the notion that elongation control includes phosphorylation and dephosphorylation of the CTD (45, 70, 76).
Although the phosphatase activity of the recombinant Fcp1-H was reasonably high with the artificial substrate p-nitrophenyl phosphate (see Results), the CTD-phosphatase activity in vitro was rather low as measured using purified pol II substrate. Efficient dephosphorylation of CTD may require an additional factor or condition. In the case of S. cerevisiae and humans, TFIIFα interacts directly with Fcp1 and stimulates its CTD-phosphatase activity (1, 10, 35). As expected from the function of Fcp1, the IIa form of Rpb1 increased in the Fcp1-overexpressing cells. When the cells were disrupted by the glass beads method, Rpb1 degraded into the IIb form. However, the degradation was less in the Fcp1-overexpressing cells, suggesting that formation of the Fcp1/pol II complex protects the CTD from degradation by proteinases. Since fcp1 disruptants are inviable, there seems to be no other phosphatase that can substitute for Fcp1.
Several lines of evidence indicated direct binding between pol II and Fcp1. From the previous results (1, 10), however, the possibility cannot be excluded that a factor such as TFIIF bridges pol II and Fcp1 in the absence of Fcp1-pol II direct binding. We concluded that Rpb4, which is a pol II-specific subunit, is one of the Fcp1-interacting subunits of pol II. This is consistent with the observation that neither recombinant CTD nor purified Rpb1 is the substrate of CTD-phosphatase (10). The C-terminal region of S. cerevisiae Fcp1 was shown to interact with TFIIFα (35), and human TFIIFβ was reported to interact with Rpb5 (72). Taken together, the whole protein-protein interactions among Fcp1, TFIIF, and pol II have been elucidated.
In S. pombe, Rpb4 is a stably associated subunit of pol II, and the rpb4 gene is essential for cell growth (60). In contrast, it is one of the dissociable subunits in S. cerevisiae (14, 36), and the rpb4 gene is nonessential for cell viability (74). Taking advantage of its nonessential nature, the functions of Rpb4 were characterized in detail using S. cerevisiae. Altogether, the results indicate the importance of Rpb4 in transcription initiation (16, 52), especially at high temperature (44, 57). Here we propose another function of Rpb4: it plays a role in assembly of the Fcp1-pol II complex and thereby promotes CTD dephosphorylation for the reutilization of pol II in a new cycle of transcription. This conclusion was strongly supported by the result of the rpb4 shut-off experiment. After the shut-off of Rpb4 synthesis, apparent defects were clearly observed in both Fcp1 binding to pol II and CTD dephosphorylation, long before the cells showed any notable growth defect.
Another notable point in this work is the identification of S. pombe Tfg3 in the same complex with TFIIFα and -β. This suggested that S. pombe TFIIF, like S. cerevisiae (25) but different from human TFIIF, is composed of three species of subunit, although a functional assay is needed to define the essentiality of the Tfg3 subunit in TFIIF function. This finding is rather unexpected, because the transcription mechanism of S. pombe has been reported to be more similar in several aspects to that of higher eukaryotes than to that of S. cerevisiae; for example, the start site position (40), the functionality of different classes of activator (55), and the structure and stoichiometry of Rpb4 (60). The S. pombe transcription mechanism might have in-between characteristics useful to connect the knowledge of S. cerevisiae and higher eukaryotes.
Acknowledgments
We are grateful to Akira Iwata and Susumu Ueda (Nippon Institute for Biological Science) for preparing antibodies. We thank Nobuyuki Fujita and Hiroshi Mitsuzawa for instruction of mass spectrometry and tetrad analysis.
This work was supported by the Core Research for Evolutional Science and Technology and grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- 1.Archambault, J., G. Pan, G. K. Dahmus, M. Cartier, N. Marshall, S. Zhang, M. E. Dahmus, and J. Greenblatt. 1998. FCP1, the RAP74-interacting subunit of a human protein phosphatase that dephosphorylates the carboxyl-terminal domain of RNA polymerase IIO. J. Biol. Chem. 273:27593-27601. [DOI] [PubMed] [Google Scholar]
- 2.Archambault, J., R. S. Chambers, M. S. Kobor, Y. Ho, M. Cartier, D. Bolotin, B. Andrews, C. M. Kane, and J. Greenblatt. 1997. An essential component of a C-terminal domain phosphatase that interacts with transcription factor IIF in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:14300-14305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asturias, F. J., Y. W. Jiang, L. C. Myers, C. M. Gustafsson, and R. D. Kornberg. 1999. Conserved structure of mediator and RNA polymerase II holoenzyme. Science 283:985-987. [DOI] [PubMed] [Google Scholar]
- 4.Azuma, Y., M. Yamagishi, and A. Ishihama. 1993. Subunits of the Schizosaccharomyces pombe RNA polymerase II: enzyme purification and structure of the subunit 3 gene. Nucleic Acids Res. 21:3749-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azuma, Y., M. Yamagishi, R. Ueshima, and A. Ishihama. 1991. Cloning and sequence determination of the Schizosaccharomyces pombe rpb1 gene encoding the largest subunit of RNA polymerase II. Nucleic Acids Res. 19:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basi, G., E. Schmid, and K. Maundrell. 1993. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123:131-136. [DOI] [PubMed] [Google Scholar]
- 7.Bertolotti, A., T. Melot, J. Acker, M. Vigneron, O. Delattre, and L. Tora. 1998. EWS, but not EWS-FLI-1 is associated with both TFIID and RNA polymerase II: interactions between two members of the TET family, EWS and hTAFII68, and subunits of TFIID and RNA polymerase II complexes. Mol. Cell. Biol. 18:1489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bregman, D. B., L. Du, S. van der Zee, and S. L. Warren. 1995. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J. Cell Biol. 129:287-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cairns, B. R., N. L. Henry, and R. D. Kornberg. 1996. TFG3/TAF30/ANC1, a component of the yeast SWI/SNF complex that is similar to the leukemogenic proteins ENL and AF-9. Mol. Cell. Biol. 16:3308-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chambers, R. S., B. Q. Wang, Z. F. Burton, and M. E. Dahmus. 1995. The activity of COOH-terminal domain phosphatase is regulated by a docking site on RNA polymerase II and by the general transcription factors IIF and IIB. J. Biol. Chem. 270:14962-14969. [DOI] [PubMed] [Google Scholar]
- 11.Chang, M., D. French-Cornay, H-Y Fan, H. Klein, C. L. Denis, and J. A. Jaehning. 1999. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol. Cell. Biol. 19:1056-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, M., and J. A. Jaehning. 1997. A multiplicity of mediators: alternative forms of transcription complexes communicate with transcriptional regulators. Nucleic Acids Res. 25:4861-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho, H., T.-K. Kim, H. Mancebo, W. S. Lane, O. Flores, and D. Reinberg. 1999. A protein phosphatase functions to recycle RNA polymerase II. Genes Dev. 13:1540-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choder, M., and R. A. Young. 1993. A portion of RNA polymerase II molecules has a component essential for stress responses and stress survival. Mol. Cell. Biol. 13:6984-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collet, J. F., V. Stoobant, M. Pirard, G. Delpierra, and E. Van Schaftingen. 1998. A new class of phosphotransferases phosphorylated on an aspartate residue in an amino-terminal DX(T/V) motif. J. Biol. Chem. 273:14107-14112. [DOI] [PubMed] [Google Scholar]
- 16.Edwards, A. M., C. M. Kane, R. A. Young, and R. D. Kornberg. 1990. Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro. J. Biol. Chem. 266:71-75. [PubMed] [Google Scholar]
- 17.Feaver, W. J., O. Gileadi, Y. Li, and R. D. Kornberg. 1991. CTD kinase associated with yeast RNA polymerase II initiation factor b. Cell 67:1223-1230. [DOI] [PubMed] [Google Scholar]
- 18.Feaver, W. J., J. Q. Svejstrup, N. L. Henry, and R. D. Kornberg. 1994. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell 79:1103-1109. [DOI] [PubMed] [Google Scholar]
- 19.Flores, O., H. Lu, M. Killeen, J. Greenblatt, Z. F. Burton, and D. Reinberg. 1991. The small subunit of transcription factor IIF recruits RNA polymerase II into the preinitiation complex. Proc. Natl. Acad. Sci. USA 88:9999-10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forsburg, S. L. 1993. Comparison of Schizosaccharomyces pombe expression system. Nucleic Acids Res. 21:2955-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grimm, C., J. Kohli, J. Murray, and K. Maundrell. 1988. Genetic engineering of Schizosaccharomyces pombe: a system for gene disruption and replacement using the ura4 gene as a selectable marker. Mol. Gen. Genet. 215:81-86. [DOI] [PubMed] [Google Scholar]
- 22.Ha, I., S. Roberts, E. Maldonado, X. Sun, L.-U. Kim, M. Green, and D. Reinberg. 1993. Multiple functional domains of human transcription factor IIB: distinct interactions with two general transcription factors and RNA polymerase II. Genes Dev. 7:1021-1032. [DOI] [PubMed] [Google Scholar]
- 23.Hengartner, C. J., V. E. Myer, S.-M. Liao, C. J. Wilson, S. S. Koh, and R. A. Young. 1998. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell 2:43-53. [DOI] [PubMed] [Google Scholar]
- 24.Henry, N. L., A. M. Campbell, W. J. Feaver, D. Poon, P. A. Weil, and R. D. Kornberg. 1994. TFIIF-TAF-RNA polymerase II connection. Genes Dev. 8:2868-2878. [DOI] [PubMed] [Google Scholar]
- 25.Henry, N. L., M. H. Sayre, and R. D. Kornberg. 1992. Purification and characterisation of yeast RNA polymerase II general initiation factor g. J. Biol. Chem. 267:23388-23392. [PubMed] [Google Scholar]
- 26.Ishiguro, A., M. Kimura, K. Yasui, A. Iwata, S. Ueda, and A. Ishihama. 1998. Two large subunits of the fission yeast RNA polymerase II provide platforms for the assembly of small subunits. J. Mol. Biol. 279:703-712. [DOI] [PubMed] [Google Scholar]
- 27.Jiménez, C. R., L. Huang, Y. Que, and A. L. Burlingame. 1998. In-gel digestion of proteins for MALDI-MS fingerprint mapping, p. 16.4.1-16.4.5. In J. E. Coligan, B. M. Dunn, H. L. Ploegh, D. W. Speicher, and P. T. Wingfield (ed.), Current protocols in protein science. John Wiley & Sons, Inc., New York, N.Y. [DOI] [PubMed]
- 28.John, S., L. Howe, S. T. Tafrov, P. A. Grant, R. Sternglanz, and J. L. Workman. 2000. The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAFII30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc67/Pob3)-FACT complex. Genes Dev. 14:1196-1208. [PMC free article] [PubMed] [Google Scholar]
- 29.Kawagishi, M., M. Yamagishi, and A. Ishihama. 1993. Cloning and sequence of the Schizosaccharomyces pombe rpb2 gene encoding the subunit 2 of RNA polymerase II. Nucleic Acids Res. 21:469-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khazak, V., J. Estojak, H. Cho, J. Majors, G. Sonoda, J. R. Testa, and E. A. Golemis. 1998. Analysis of the interaction of the novel RNA polymerase II (pol II) subunit hsRPB4 with its partner hsRPB7 and with pol II. Mol. Cell. Biol. 18:1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimura, M., A. Ishiguro, and A. Ishihama. 1997. RNA polymerase II subunits 2, 3, and 11 form a core subassembly with DNA binding activity. J. Biol. Chem. 272:25851-25855. [DOI] [PubMed]
- 32.Kimura, M., and A. Ishihama. 2000. Involvement of multiple subunit-subunit contacts in the assembly of RNA polymerase II. Nucleic Acids Res. 28:952-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kimura, M., H. Sakurai, and A. Ishihama. 2001. Intracellular contents and assembly states of all 12 subunits of the RNA polymerase II in the fission yeast Schizosaccharomyces pombe. Eur. J. Biochem. 268:612-619. [DOI] [PubMed] [Google Scholar]
- 34.Kobor, M. S., J. Archambault, W. Lester, F. C. P. Holstege, O. Gileadi, D. B. Jansma, E. G. Jennings, F. Kouyoumdjian, A. R. Davidson, R. A. Young, and J. Greenblatt. 1999. An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S. cerevisiae. Mol. Cell 4:55-62. [DOI] [PubMed] [Google Scholar]
- 35.Kobor, M. S., L. D. Simon, J. Omichinski, G. Zhong, J. Archambault, and J. Greenblatt. 2000. A motif shared by TFIIF and TFIIB mediates their interaction with the RNA polymerase II carboxy-terminal domain phosphatase Fcp1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:7438-7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolodziej, P. A., N. Woychik, S-M. Liao, and R. A. Young. 1990. RNA polymerase II subunit composition, stoichiometry, and phosphorylation. Mol. Cell. Biol. 10:1915-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larkin, R. M., G. Hagen, and T. J. Guifoil. 1999. Arabidopsis thaliana RNA polymerase II subunits related to yeast and human RPB5. Gene 231:41-47. [DOI] [PubMed] [Google Scholar]
- 38.Lee, T. I., and R. A. Young. 2000. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34:77-137. [DOI] [PubMed] [Google Scholar]
- 39.Leuther, K. K., D. A. Bushnell, and R. G. Kornberg. 1996. Two-dimensional crystallography of TFIIB- and TFIIE-RNA polymerase II complex: implications for start site selection and initiation complex formation. Cell 85:773-779. [DOI] [PubMed] [Google Scholar]
- 40.Li, Y., P. M. Flanagan, H. Tschochner, and R. D. Kornberg. 1994. RNA polymerase II initiation factor interactions and transcription start site selection. Science 263:805-807. [DOI] [PubMed] [Google Scholar]
- 41.Lowry, O. H. 1957. Micromethods for the assay of enzymes. Methods Enzymol. 4:366-381. [Google Scholar]
- 42.Lu, H., O. Flores, R. Weinmann, and D. Reinberg. 1991. The nonphosphorylated form of RNA polymerase II preferentially associates with the preinitiation complex. Proc. Natl. Acad. Sci. USA 88:10004-10008. [DOI] [PMC free article] [PubMed]
- 43.Lu, H., L. Zawel, L. Fisher, J.-M. Egly, and D. Reinberg. 1992. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature 358:641-645. [DOI] [PubMed] [Google Scholar]
- 44.Maillet, I., J. M. Buhler, A. Sentenac, and J. Labarre. 1999. Rpb4p is necessary for RNA polymerase II activity at high temperature. J. Biol. Chem. 274:22586-22590. [DOI] [PubMed] [Google Scholar]
- 45.Marshall, N. F., J. Peng, Z. Xie, and D. H. Price. 1996. Control of RNA polymerase II elongation potential by a novel carboxy-terminal domain kinase. J. Biol. Chem. 271:27176-27183. [DOI] [PubMed] [Google Scholar]
- 46.Maxon, M. E., J. A. Goodrich, and R. Tjian. 1994. Transcription factor IIE binds preferentially to RNA polymerase IIa and recruits TFIIH: a model for promoter clearance. Genes Dev. 8:515-524. [DOI] [PubMed] [Google Scholar]
- 47.Miyao, T., K. Yasui, H. Sakurai, M. Yamagishi, and A. Ishihama. 1996. Molecular assembly of RNA polymerase II from the fission yeast Schizosaccharomyces pombe: subunit-subunit contact network involving Rpb5. Genes Cells 1:843-854. [DOI] [PubMed] [Google Scholar]
- 48.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 49.Myers, L. C., C. M. Gustafsson, D. A. Bushnell, M. Lui, H. Erdjument-Bromage, P. Tempst, and R. D. Kornberg. 1998. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 12:45-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Myers, L. C., and R. D. Kornberg. 2000. Mediator of transcriptional regulation. Annu. Rev. Biochem. 69:729-749. [DOI] [PubMed] [Google Scholar]
- 51.Ohkuma, Y., and R. G. Roeder. 1994. Regulation of TFIIH ATPase and kinase activities by TFIIE during active initiation complex formation. Nature 368:160-163. [DOI] [PubMed] [Google Scholar]
- 52.Orlicky, S. M., P. T. Tran, M. H. Sayer, and A. M. Edwards. 2001. Dissociable Rpb4-Rpb7 subassembly of RNA polymerase II binds to single-stranded nucleic acid and mediates a post-recruitment step in transcription initiation. J. Biol. Chem. 276:10097-10102. [DOI] [PubMed] [Google Scholar]
- 53.Otero, G., J. Fellows, Y. Li, T. de Bizemont, A. M. G. Dirac, C. M. Gustafsson, H. Erdjument-Bromage, P. Tempst, and J. Q. Svejstrup. 1999. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 3:109-118. [DOI] [PubMed] [Google Scholar]
- 54.Pan, G., T. Aso, and J. Greenblatt. 1997. Interaction of elongation factors TFIIS and elongin A with a human RNA polymerase II holoenzyme capable of promoter-specific initiation and responsive to transcriptional activators. J. Biol. Chem. 272:24563-24571. [DOI] [PubMed] [Google Scholar]
- 55.Remacle, J. E., G. Albrecht, R. Brys, G. H. Braus, and D. Huylebroeck. 1997. Three classes of mammalian transcription activation domain stimulate transcription in Schizosaccharomyces pombe. EMBO J. 16:5722-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roeder, R. G. 1996. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21:327-334. [PubMed] [Google Scholar]
- 57.Rosenhech, S., and M. Choder. 1998. Rpb4, a subunit of RNA polymerase II, enables the enzyme to transcribe at temperature extremes in vitro. J. Bacteriol. 180:6187-6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sakurai, H., and A. Ishihama. 1997. Gene organization and protein sequence of the small subunits of Schizosaccharomyces pombe RNA polymerase II. Gene 196:165-174. [DOI] [PubMed] [Google Scholar]
- 59.Sakurai, H., M. Kimura, and A. Ishihama. 1998. Identification of the gene and the protein of RNA polymerase II subunit 9 (Rpb9) from the fission yeast Schizosaccharomyces pombe. Gene 221:11-16. [DOI] [PubMed] [Google Scholar]
- 60.Sakurai, H., H. Mitsuzawa, M. Kimura, and A. Ishihama. 1999. The Rpb4 subunit of fission yeast Schizosaccharomyces pombe RNA polymerase II is essential for cell viability and similar in structure to the corresponding subunits of higher eukaryotes. Mol. Cell. Biol. 19:7511-7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sawadogo, M., A. Sentenac, and P. Fromageot. 1980. Interaction of a new polypeptide with yeast RNA polymerase B. J. Biol. Chem. 255:12-15. [PubMed] [Google Scholar]
- 62.Shilatifard, A., D. Haque, R. C. Conaway, and J. W. Conaway. 1997. Structure and function of RNA polymerase II elongation factor ELL. J. Biol. Chem. 272:22355-22363. [DOI] [PubMed] [Google Scholar]
- 63.Shpakovski, G. V. 1994. The fission yeast Schizosaccharomyces pombe rpb6 gene encodes the common phosphorylated subunit of RNA polymerase and complements a mutation in the corresponding gene of Saccharomyces cerevisiae. Gene 147:63-69. [DOI] [PubMed] [Google Scholar]
- 64.Shpakovski, G. V., J. Acker, M. Wintzerith, J. F. Lacroix, P. Thriaux, and M. Vigneron. 1995. Four subunits that are shared by the three classes of RNA polymerase are functionally interchangeable between Homo sapiens and Saccharomyces cerevisiae. Mol. Cell. Biol. 15:4702-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sopta, M., R. W. Carthew, and J. Greenblatt. 1985. Isolation of three proteins that bind to mammalian RNA polymerase II. J. Biol. Chem. 260:10353-10360. [PubMed] [Google Scholar]
- 66.Spåhr, H., J. Bève, T. Larsson, J. Bergström, K.-A. Karlsson, and C. M. Gustafsson. 2000. Purification and characterization of RNA polymerase II holoenzyme from Schizosaccharomyces pombe. J. Biol. Chem. 275:1351-1356. [DOI] [PubMed] [Google Scholar]
- 67.Thuriaux, P., and A. Sentenac. 1992. Yeast nuclear RNA polymerases, p. 1-48. In J. R. Broach, J. R. Pingle, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces: gene expression, vol. II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 68.Tschochner, H., M. H. Sayre, P. M. Flanagan, W. J. Feaver, and R. D. Kornberg. 1992. Yeast RNA polymerase II initiation factor e: isolation and identification as the functional counterpart of human transcription factor IIB. Proc. Natl. Acad. Sci. USA 89:11292-11296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Usheva, A., E. Maldonado, A. Goldring, H. Lu, C. Houbavi, D. Reinberg, and Y. Aloni. 1992. Specific interaction between the nonphosphorylated form of RNA polymerase II and the TATA-binding protein. Cell 69:871-881. [DOI] [PubMed] [Google Scholar]
- 70.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. H. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12:343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wade, P. A., W. Werel, R. C. Fentzke, N. E. Thompson, J. F. Leykam, R. R. Burgess, J. A. Jaehning, and Z. F. Burton. 1996. A novel collection of accessory factors associated with yeast RNA polymerase II. Protein Expr. Purif. 8:85-90. [DOI] [PubMed] [Google Scholar]
- 72.Wei, W., D. Dorjsuren, Y. Lin, W. Qin, T. Nomura, N. Hayashi, and S. Murakami. 2001. Direct interaction between the subunit RAP30 of transcription factor IIF (TFIIF) and RNA polymerase subunit 5, which contribute to the association between TFIIF and RNA polymerase II. J. Biol. Chem. 276:12266-12273. [DOI] [PubMed] [Google Scholar]
- 73.Welch, M. D., and D. G. Drubin. 1994. A nuclear protein with sequence similarity to proteins implicated in human acute leukemias is important for cellular morphogenesis and actin cytoskeletal function in Saccharomyces cerevisiae. Mol. Biol. Cell 5:617-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Woychik, N. A., and R. A. Young. 1989. RNA polymerase II subunit RPB4 is essential for high- and low-temperature yeast cell growth. Mol. Cell. Biol. 9:2854-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Woychik, N. A., and R. A. Young. 1994. Exploring RNA polymerase II structure and function, p. 227-242. In R. C. Conaway and J. W. Conaway (ed.), Transcription: mechanisms and regulation. Raven Press, New York, N.Y.
- 76.Yamaguchi, Y., T. Wada, D. Watanabe, T. Takagi, J. Hasegawa, and H. Handa. 1999. Structure and function of the human transcription elongation factor DSIF. J. Biol. Chem. 274:8085-8092. [DOI] [PubMed] [Google Scholar]